Abstract
Giardia duodenalis is a gastrointestinal protozoan ubiquitous in nature. It is a confirmed zoonotic pathogen, and cattle are considered a source of giardiasis outbreaks in humans. This study aimed to evaluate the prevalence and multilocus genotype (MLG) of G. duodenalis in dairy cattle in Central Inner Mongolia. This study was based on the small subunit ribosomal RNA (SSU rRNA), glutamate dehydrogenase (gdh), triosephosphate isomerase (tpi), and beta-giardin (bg) genes of G. duodenalis. DNA extraction, polymerase chain reaction (PCR), and sequence analysis were performed on 505 dairy cattle fecal samples collected in 2021 from six sampling sites and four age groups in Central Inner Mongolia to determine the prevalence and MLG distribution of G. duodenalis. The PCR results of SSU rRNA revealed that the overall prevalence of G. duodenalis was 29.5% (149/505) and that the overall prevalence of the diarrhea and nondiarrhea samples was 31.5% (46/146) and 28.5% (103/359), respectively; the difference was not significant (p > 0.05). SSU rRNA sequence analysis revealed that G. duodenalis assemblage E (91.1%, 133/146) was primarily detected and that assemblage A (8.9%, 13/146) was detected in 13 samples. The G. duodenalis—positive samples were PCR amplified and sequenced for gdh, tpi, and bg, from which 38, 47, and 70 amplified sequences were obtained, respectively. A combination of G. duodenalis assemblages A and E were detected in seven samples. Multilocus genotyping yielded 25 different assemblage E MLGs, which formed six subgroups. To the best of our knowledge, this is the first report regarding G. duodenalis infection in dairy cattle in Inner Mongolia, China. This study revealed that Inner Mongolian cattle pose a risk of giardiasis transmission to humans and that the distribution of local cattle G. duodenalis assemblage E MLGs is diverse. The findings of this study can bridge the knowledge gap in the molecular epidemiological investigation of giardiasis in Central Inner Mongolia.
Subject terms: Parasite biology, Parasite evolution, Parasite genetics
Introduction
Giardia duodenalis, also known as Giardia lamblia or Giardia intestinalis, was first described in 16811,2 and is a group of ubiquitous pathogenic gastrointestinal protozoans3. The hosts include humans, companion animals, livestock, and wildlife4. Human waterborne protozoan parasitic outbreaks worldwide has increased from 325 in an almost 100-year period5 to 199 between 2004 and 20106 and at least 381 outbreaks between 2011 and 20167. In the latter two records, Giardia was confirmed as the pathogen in 70 (35.2%)6 and 141 (37%) cases7, respectively. It is estimated that 8 × 108 global cases of giardiasis occur annually2. Giardiasis is a notifiable disease by the Centers for Disease Control and Prevention in the USA2. Cattle are considered the source of waterborne giardiasis outbreaks in humans4. Calves are more susceptible to acute infections caused by G. duodenalis and adult cattle often do not exhibit clinical signs and tend to have mild G. duodenalis infections. However, adult animals help maintain persistent infections in cattle and environmental contamination, thereby leading to the spread of giardiasis8.
Based on genetic studies using molecular typing techniques, G. duodenalis was classified into at least eight genetically distinct but morphologically identical lineages, i.e., assemblages A–H4,9–11. Of these, assemblages A and B have a broad host spectrum, infect most vertebrates, and have a high zoonotic risk12–14. Assemblages C–H have a high host specificity4,15, with assemblages C and D mainly found in canine animals, assemblage E mainly found in hoofed livestock and wildlife, assemblage F found in cats, assemblage G found in rodents, assemblage H found in seals and other marine mammals9,14, and assemblages C, D, E, and F identified in human patients12. Assemblage E is the most common genotype in cattle globally4,13,14,16–20, followed by assemblages A and B4,13,16, in addition assemblages C, D15, and F have also been reported in cattle8,15. In addition, the frequency of infection with zoonotic assemblages A and B from calves was reportedly higher than that with assemblage E, suggesting that calves are associated with a greater risk of transmitting G. duodenalis infection to humans4,21–24.
The pooled prevalence of bovine G. duodenalis detected using molecular methods was ~ 22% globally, and the difference between the highest pooled prevalence (55.4% in Canada) and the lowest pooled prevalence (4.2% in Iran) of giardiasis in different geographic areas is high14. Recently, in China, molecular epidemiological surveys of giardiasis in cattle have been conducted in several provinces in recently with differences in prevalence4,22–48. Inner Mongolia is located on the northern border of China, spanning 28° 52′ in longitude from east to west with a linear distance of > 2400 km and spanning 15° 59′ in latitude from north to south with a linear distance of 1700 km. Presently, only the prevalence and molecular characterizations of G. duodenalis in 108 beef cattle from one farm in the Southwest Alxa Left Banner have been investigated in Inner Mongolia24. Hence, this study aimed to investigate the prevalence of G. duodenalis in dairy cows in Central Inner Mongolia.
Methods
Study areas and sample collection
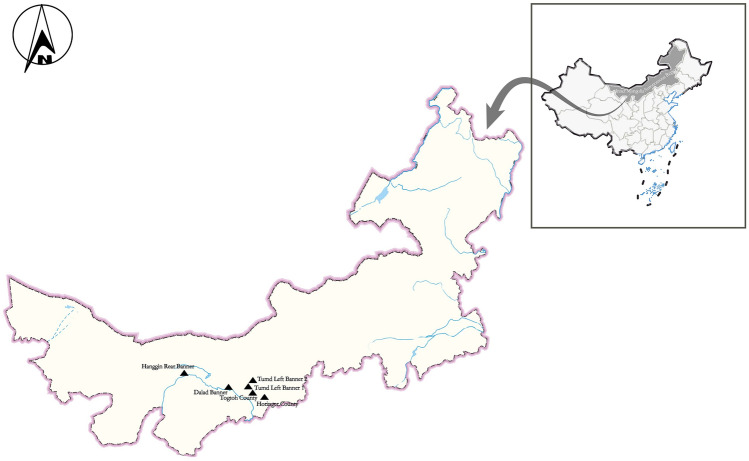
From March to September 2021, 505 fresh fecal samples were randomly collected from six dairy farms in the vicinity of Tumd Left Banner, Horinger County, Togtoh County, Dalad Banner, and Hanggin Rear Banner (107° 28′ E–111° 16′ E, 40° 21′ N–40° 35′ N) in Central Inner Mongolia. We have annotated six dairy farms on the map using phptpshop software (Fig. 1). The fecal samples were collected via rectal sampling from dairy cattle or from the inner top layer of fresh feces. The samples included 103 preweaned calves (0–60 days), 105 postweaned calves (61–180 days), 124 young cattle (181–360 days), and 173 adult cattle (> 361 days). Information regarding whether the animals experienced diseases such as diarrhea was recorded during sampling, and the samples were stored at 4 °C before extracting DNA. In the laboratory, the fresh fecal samples was added to a beaker alongside an appropriate amount of distilled water, and then stirred and filtered. The filtrate was centrifuged at 3500×g for 10 min and the precipitate was used for DNA extraction.
Figure 1.
Specific locations from which specimens were collected for this study. Filled triangle: study locations.
DNA extraction and polymerase chain reaction (PCR) amplification
The genomic DNA of 505 fecal samples was extracted E.Z.N. A® Stool DNA Kit (Omega Biotek, Norcross, GA, USA) according to the manufacturer’s protocol and stored at − 20 °C for subsequent experiments. The small subunit ribosomal RNA (SSU rRNA) gene49 was amplified via nested PCR (the annealing temperatures for two rounds of PCR were 55 °C and 59 °C) using Premix Taq™ (TaKaRa Taq™ Version 2.0 plus dye) (TaKaRa, Beijing, China) and 1 μL extracted DNA as the template. SSU rRNA–positive DNA was subsequently amplified via the nested PCR of bg50 (the annealing temperatures for two rounds of PCR were 62.7 °C and 55 °C), gdh51 (the annealing temperatures for two rounds of PCR were 53.7 °C and 56.2 °C), and tpi52 (the annealing temperatures for two rounds of PCR were 59.5 °C and 56.2 °C). Subsequently, 5 μL PCR products were analyzed via 1.5% agarose gel electrophoresis, and all the PCR products positive for the four genes were sent to a commercial company (Sangon Biotech, Shanghai, China) for sequencing.
Sequence analysis
The sequences were aligned with reference sequences downloaded from GenBank (http://www.ncbi.nlm.nih.gov) using MEGA 7.0 software (http://www.megasoftware.net/) and the obtained results were analyzed using the BLAST online platform. To comprehensively investigate the relationship among the different isolates, phylogenetic analyses were performed using a concatenated dataset of bg, gdh, and tpi sequences. The specimens successfully subtyped at all the three loci were included in the multilocus genotype (MLG) analysis of G. duodenalis, wherein MLG types were identified. Phylogenetic trees were constructed using the neighbor-joining algorithm based on a matrix of evolutionary distances calculated using the Kimura 2-parameter model via MEGA 7.0 software. To assess the robustness of the clusters, 1000 bootstrap replicates were used.
Statistical analyses
Chi-square test was performed and 95% confidence interval (CI) was obtained using SPSS Statistics 21.0 (IBM Corp., New York, NY, USA) to compare G. duodenalis infection rates among the different farms and age groups and diarrhea and nondiarrhea groups. A two-tailed p-value of < 0.05 was considered statistically significant.
Ethics approval and consent to participate
This study was carried out in strict accordance with international standards as published in the “Guide to the feeding, management and use of experimental animals” (8th Edition) and follows the “Regulations on the management of experimental animals” and other relevant laws and regulations. The biomedical research ethics committee of Inner Mongolia Agricultural University specifically approved this study (No. 2020[081]). In addition, permission was obtained from the farm owners before the specimens were collected, and all efforts were made to minimize suffering.
Results
Giardia duodenalis infection status
Based on the SSU rRNA gene of G. duodenalis, 149 positive samples were amplified via PCR of 505 samples, with an overall prevalence of 29.5% (149/505). The overall prevalence of diarrhea and nondiarrhea samples was 31.5% (46/146) and 28.7% (103/359), respectively (Table 1), and the difference was not significant (odds ratio [OR] 0.875; 95% CI 0.576–1.328; p = 0.529).
Table 1.
Prevalence of Giardia duodenalis and assemblages determined via sequence analysis of SSU rRNA.
| Farm | Samples size | Age | Total | p-value | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|
| Preweaned calves | Postweaned calves | Young cattle | Adult cattle | |||||
| Tumd left banner 1 | Samples size (diarrhea/nondiarrhea) | 20 (8/12) | 40 (21/19) | 40 (9/31) | 40 (17/23) | 140 (55/85) | 0.006 | 1.785 (1.181–2.697) |
| Positive samples size (Diarrhea/Nondiarrhea) | 3 (0/3) | 22 (9/13) | 13 (2/11) | 16 (7/9) | 54 (18/36) | |||
| Overall prevalence (prevalence of diarrhea/nondiarrhea samples) (%) | 15% (0/25%) | 55% (42.9%/68.4%) | 32.5% (22.2%/35.5%) | 40% (41.2%/39.1%) | 38.6% (32.7%/42.4%) | |||
| SSU rRNA (n) | E (3) | E (17), A (5) | E (13) | E (16) | E (49), A (5) | |||
| Tumd left banner 2 | Samples size (Diarrhea/Nondiarrhea) | 30 (2/28) | 20 (9/11) | 20 (1/19) | 40 (0/40) | 110 (12/98) | 0.197 | 0.728 (0.449–1.181) |
| Positive samples size (Diarrhea/Nondiarrhea) | 8 (0/8) | 8 (3/5) | 5 (1/4) | 6 (0/6) | 27 (4/23) | |||
| Overall prevalence (Prevalence of diarrhea/nondiarrhea samples) (%) | 26.7% (0/28.6%) | 40% (33.3%/45.5%) | 25% (100%/21.1%) | 15% (0/15%) | 24.5% (33.3%/23.5%) | |||
| SSU rRNA (n) | E (8) | E (7) | E (5) | E (5), A (1) | E (25), A (1) | |||
| Horinger county | Samples size (Diarrhea/Nondiarrhea) | 23 (8/15) | 20 (18/2) | 41 (7/34) | 36 (7/29) | 120 (40/80) | 0.031 | 0.589 (0.363–0.956) |
| Positive samples size (Diarrhea/Nondiarrhea) | 0 (0/0) | 9 (7/2) | 9 (2/7) | 8 (2/6) | 26 (11/15) | |||
| Overall prevalence (Prevalence of diarrhea/nondiarrhea samples) (%) | 0 (0/0) | 45% (38.9%/100%) | 22.0% (28.6%/20.6%) | 22.2% (28.6%/20.7%) | 21.7% (27.5%/18.8%) | |||
| SSU rRNA (n) | – | E (8), A (1) | E (8), A (1) | E (7), A (1) | E (23), A (3) | |||
| Togtoh county | Samples size (diarrhea/nondiarrhea) | 30 (12/18) | 20 (20/0) | 20 (3/17) | 40 (4/36) | 110 (39/71) | 0.548 | 1.15 0.729–1.816) |
| Positive samples size (Diarrhea/Nondiarrhea) | 0 (0/0) | 11 (11/0) | 13 (1/12) | 11 (1/10) | 35 (13/22) | |||
| Overall prevalence (prevalence of diarrhea/nondiarrhea samples) (%) | 0 (0/0) | 55% (55%/0) | 65% (33.3%/70.6%) | 27.5% (25%/27.8%) | 31.8% (33.3%/31.0%) | |||
| SSU rRNA (n) | – | E (11) | E (9), A (2) | E (10), A (1) | E (30), A (3) | |||
| Dalad banner | Samples size (diarrhea/nondiarrhea) | 0 (0/0) | 5 (0/5) | 3 (0/3) | 9 (0/9) | 17 (0/17) | 0.594 | 1.316 (0.478–3.626) |
| Positive samples size (Diarrhea/Nondiarrhea) | 0 (0/0) | 0 (0/0) | 3 (0/3) | 3 (0/3) | 6 (0/6) | |||
| Overall prevalence (prevalence of diarrhea/nondiarrhea samples) (%) | 0 (0/0) | 0 (0/0) | 100% (0/100%) | 33.3% (0/33.3%) | 35.3% (0/35.3%) | |||
| SSU rRNA (n) | – | – | E (2), A (1) | E (3) | E (5), A (1) | |||
| Hanggin rear Banner | Samples size (diarrhea/nondiarrhea) | 0 (0/0) | 0 (0/0) | 0 (0/0) | 8 (0/8) | 8 (0/8) | 0.288 | 0.337 (0.041–2.762) |
| Positive samples size (Diarrhea/Nondiarrhea) | 0 (0/0) | 0 (0/0) | 0 (0/0) | 1 (0/1) | 1 (0/1) | |||
| Overall prevalence (prevalence of diarrhea/nondiarrhea samples) (%) | 0 (0/0) | 0 (0/0) | 0 (0/0) | 12.5% (0/12.5%) | 12.5% (0/12.5%) | |||
| SSU rRNA (n) | – | – | – | E (1) | E (1) | |||
| Total | Samples size (diarrhea/nondiarrhea) | 103 (30/73) | 105 (68/37) | 124 (20/104) | 173 (28/145) | 505 (146/359) | – | – |
| Positive samples size (Diarrhea/Nondiarrhea) | 11 (0/11) | 50 (30/20) | 43 (6/37) | 45 (10/35) | 149 (46/103) | |||
| Overall prevalence (prevalence of diarrhea/nondiarrhea samples) (%) | 10.7% (0/15.1%) | 47.6% (44.1%/54.1%) | 34.7% (30%/35.6%) | 26.0% (35.7%/24.1%) | 29.5% (31.5%/28.7%) | |||
| SSU rRNA (n) | E (11) | E (43), A (6) | E (37), A (4) | E (42), A (3) | E (133), A (13) | |||
| p-value | – | < 0.001 | < 0.001 | 0.146 | 0.214 | – | – | – |
| OR (95% CI) | – | 0.229 (0.118–0.442) | 2.764 (1.771–4.314) | 1.377 (0.894–2.122) | 0.771 (0.511–1.163) | |||
The dash (–) indicates that no data were obtained.
The overall prevalence of G. duodenali in the six sampling farms was 38.6% (54/140), 24.5% (27/110), 21.7% (26/120), 31.8% (35/110), 35.3% (6/17), and 12.5% (1/8). The difference in the Tumd Left Banner 1 field was highly significant compared with that in the other fields (OR 1.785; 95% CI 1.181–2.697; p = 0.006); Horinger County field had also showed a significant difference (OR 0.589; 95% CI 0.363–0.956; p = 0.031). The prevalence G. duodenalis in the diarrhea samples from the six sampling sites was 32.7% (18/55), 33.3% (4/12), 27.5% (11/40), 33.3% (13/39), 0 (0/0), and 0 (0/0) (Table 1), and the difference in prevalence among the sampling sites with diarrhea was not significant (p > 0.05).
The overall prevalence of G. duodenalis in all the samples in the four age groups was 10.7% (11/103), 47.6% (50/105), 34.7% (43/124), and 26.0% (45/173). The difference in preweaned calves was highly significant compared with that in the other age groups (OR 0.229; 95% CI 0.118–0.442; p < 0.001); the difference in postweaned calves was highly significant (OR 2.764; 95% CI 1.771–4.314; p < 0.001). The prevalence of G. duodenalis in the diarrhea samples of the four age groups were 0 (0/30), 44.1% (30/68), 30% (6/20), and 35.7% (10/28) (Table 1), with a highly significant difference in postweaned calves compared with the other age groups (OR 3.059; 95% CI 1.476–6.341; p = 0.002).
In total, 149 positive samples were used for the PCR amplification of SSU rRNA; however, sequence analyses revealed that 146 of them were plausible sequences, with 13 (8.9%, 13/146) for G. duodenalis assemblages A and 133 (91.1%, 133/146) for assemblage E. Assemblage E was detected in all the four age groups. Only assemblage E was present in Hanggin Rear Banner field and in some age groups in other fields. Assemblage A was not observed in preweaned calves at any of the sites (Table 1).
Giardia species identification and analysis
In the 149 positive samples, 38, 47, and 70 plausible sequences were obtained via PCR amplification and gene sequencing of gdh, tpi, and bg of G. duodenalis, respectively. Of the 38 isolates of gdh (Table 2), one isolate was identified as 1 assemblage A sequence (A1) and 37 were identified as 12 assemblage E sequences, including E1 (n = 17), E2 (n = 9), E3 (n = 1), E4 (n = 1), E5 (n = 2), and one each for E6–E12. Furthermore, of the 47 isolates of tpi (Table 2), three isolates were identified as 1 assemblage A sequence (A1) and 44 were identified as 23 assemblage E sequences, including E1 (n = 11), E2 (n = 6), E3 (n = 1), E4 (n = 2), E5 (n = 3), E19 (n = 2), E20 (n = 3), E6–E18 (n = 1), and one each for E21–E23. Of the 70 isolates of bg (Table 2), two isolates were identified as 1 assemblage A sequence (A1) and 68 were identified as 24 assemblage E sequences, including E1 (n = 22), E2 (n = 4), E3 (n = 7), E4 (n = 9), E5 (n = 3), E6 (n = 2), E7 (n = 3), E8 (n = 1), E9 (n = 2), and one each for E10–E24. The sequence alignment analysis of the above four genes identified differences in the genotypes of seven samples alongside mixed infections (assemblage A + E) (Table 3).
Table 2.
Intra-assemblage substitutions and insertion in gdh, tpi, and bg sequences within Giardia duodenalis assemblage E.
| Subtype (no.) gdh | Nucleotide positions | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 105 | 133 | 141 | 282 | 303 | 318 | 336 | 378 | 435 | 444 | 453 | 455 | 457 | 461 | 468 | 469 | 471 | 472 | 473 | 481 | 483 | 485 | 498 | ||||
| Ref. sequence (GenBank ID: MK645797) | C | A | A | T | C | G | A | G | G | C | T | G | G | T | T | G | A | G | G | C | T | G | G | |||
| E1 (17) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| E2 (9) | T | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| E3 (1) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | A | – | – | – | A | |||
| E4 (1) | T | – | – | G | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| E5 (2) | – | – | G | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| E6 (1) | – | – | – | – | – | – | – | – | – | – | – | A | – | – | – | – | – | – | – | – | – | – | – | |||
| E7 (1) | – | – | – | – | T | A | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| E8 (1) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | A | – | – | – | – | |||
| E9 (1) | – | – | – | – | – | – | – | C | C | G | G | – | C | A | C | C | C | T | – | T | C | T | – | |||
| E10 (1) | T | C | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| E11 (1) | T | – | – | – | – | – | – | – | – | – | – | A | – | – | – | – | – | – | – | – | – | – | – | |||
| E12 (1) | – | – | G | G | – | – | G | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| Tpi | 50 | 71 | 83 | 87 | 88 | 119 | 124 | 138 | 145 | 186 | 243 | 264 | 285 | 302 | 312 | 335 | 338 | 342 | 377 | 381 | 399 | 450 | 455 | 468 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref. sequence (GenBank ID: MH079444) | T | T | T | G | * | A | T | A | A | A | C | G | C | A | C | A | A | T | A | G | C | A | A | A | ||
| E1 (11) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E2 (6) | – | C | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E3 (1) | – | C | – | – | – | – | – | G | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E4 (2) | – | C | – | – | – | – | – | – | – | – | T | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E5 (3) | C | C | G | A | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E6 (1) | – | C | – | – | – | – | – | – | – | – | – | – | – | T | – | G | – | – | – | – | – | – | – | |||
| E7 (1) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | ||
| E8 (1) | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | – | – | – | – | – | – | – | ||
| E9 (1) | – | – | – | – | – | – | – | – | – | G | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E10 (1) | – | – | – | – | – | G | – | – | – | – | – | – | – | – | – | – | – | – | – | A | – | – | – | – | ||
| E11 (1) | – | C | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | – | – | – | – | – | – | ||
| E12 (1) | C | C | – | – | – | – | – | – | – | – | T | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E13 (1) | – | C | – | – | – | – | – | – | G | – | T | – | – | – | – | – | – | – | G | – | – | G | – | G | ||
| E14 (1) | – | C | – | A | – | – | – | – | – | – | T | – | – | – | – | – | – | – | – | – | – | G | C | G | ||
| E15 (1) | – | C | – | A | – | – | – | – | – | – | T | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E16 (1) | – | C | G | A | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | C | – | ||
| E17 (1) | C | C | G | A | G | – | – | – | – | – | T | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E18 (1) | C | C | – | – | – | – | C | – | – | – | – | – | – | – | T | – | G | – | – | – | – | – | – | – | ||
| E19 (1) | – | – | – | – | – | – | – | G | – | – | – | A | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E20 (2) | – | C | – | A | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E21 (3) | C | C | – | A | – | – | – | – | – | – | – | – | – | – | – | G | – | – | – | – | – | – | – | – | ||
| E22 (1) | C | C | – | A | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| E23 (1) | – | C | – | A | – | – | – | – | – | – | – | – | – | – | – | – | – | G | – | – | – | – | – | – |
| Bg | 68 | 69 | 77 | 103 | 104 | 107 | 124 | 129 | 155 | 156 | 175 | 203 | 232 | 259 | 277 | 304 | 316 | 319 | 348 | 368 | 418 | 431 | 436 | 439 | 457 | 463 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref. sequence (GenBank ID: MK610389) | T | * | * | G | A | A | C | A | G | C | A | G | G | A | C | C | G | C | G | G | C | G | C | C | T | G |
| E1 (22) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| E2 (4) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | – |
| E3 (7) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | – | T | – | – | – | – | – |
| E4 (9) | C | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | – |
| E5 (3) | C | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| E6 (2) | – | – | – | – | – | – | – | – | – | – | G | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| E7 (3) | C | – | – | – | – | – | – | – | – | – | G | – | – | G | – | – | – | – | – | – | – | – | – | – | – | – |
| E8 (1) | C | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | – | – | – | – | – | – |
| E9 (2) | C | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | T | – | – | – | – | – |
| E10 (1) | – | – | – | – | – | – | – | – | – | – | G | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – |
| E11 (1) | – | – | – | – | – | – | – | – | A | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| E12 (1) | – | – | – | – | – | – | – | – | – | – | – | A | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| E13 (1) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | – | – | – | – | – | – | – |
| E14 (1) | C | T | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| E15 (1) | – | – | A | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | – |
| E16 (1) | C | – | – | – | T | G | – | G | – | – | – | – | A | – | – | – | – | – | – | – | T | – | – | – | – | – |
| E17 (1) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | – | – | – | – | A |
| E18 (1) | – | – | – | – | – | – | – | – | – | T | G | – | – | – | – | – | – | – | – | – | – | A | – | T | – | – |
| E19 (1) | – | – | – | A | – | – | – | – | – | – | G | – | – | – | – | – | – | – | – | A | – | – | – | – | – | – |
| E20 (1) | – | – | – | – | – | – | – | – | – | – | G | – | – | – | – | – | – | – | A | – | – | – | – | T | – | – |
| E21 (1) | – | – | – | – | – | – | T | – | – | – | G | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| E22 (1) | – | – | – | – | – | – | – | – | – | – | G | – | – | – | T | – | – | – | – | – | T | – | – | – | – | – |
| E23 (1) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | A | T | – | – | T | – | – | – | C | – |
| E24 (1) | – | – | – | – | – | – | – | – | – | – | G | – | – | G | – | – | – | – | – | – | – | – | – | – | – |
The dash (–) indicates that the sequence is the same as the reference sequence. The asterisk (*) indicates the insertion site of the reference sequence.
Table 3.
Multilocus characterization of Giardia duodenalis isolates based on bg, gdh, and tpi sequences.
| Farm | Age | Cattle ID | SSU rRNA (GenBank accession no.) | gdh (GenBank accession no.) | tpi (GenBank accession no.) | bg (GenBank accession no.) | MLG type | Mix |
|---|---|---|---|---|---|---|---|---|
| Tumd left banner 1 | Preweaned calves | T1–2 | OP189375 (E) | OP189521 (E1) | OP189568 (E1) | |||
| T1–4 | OP189376 (E) | |||||||
| T1–15 | OP189377 (E) | |||||||
| Postweaned calves | T1–22 | OP189378 (A) | ||||||
| T1–23 | OP189379 (E) | OP189606 (E2) | ||||||
| T1–26 | OP189380 (E) | OP189550 (A1) | OP189569 (E1) | A + E | ||||
| T1–28 | OP189381 (E) | |||||||
| T1–30 | OP189382 (A) | OP189640 (A1) | OP189570 (E1) | A + E | ||||
| T1–32 | OP189383 (E) | |||||||
| T1–36 | OP189384 (A) | |||||||
| T1–38 | OP189385 (E) | OP189641 (E1) | ||||||
| T1–39 | OP189476 (A) | OP189551 (E1) | A + E | |||||
| T1–44 | OP189386 (E) | |||||||
| T1–46 | OP189387 (E) | OP189611 (E23) | ||||||
| T1–47 | OP189388 (E) | OP189649 (E2) | OP189571 (E4) | |||||
| T1–48 | OP189389 (E) | OP189605 (E4) | ||||||
| T1–49 | OP189390 (E) | |||||||
| T1–51 | OP189391 (E) | OP189642 (E12) | OP189631 (E1) | |||||
| T1–52 | OP189476 (E) | |||||||
| T1–53 | OP189392 (E) | OP189670 (E1) | ||||||
| T1–54 | OP189393 (A) | OP189522 (A1) | OP189572 (A1) | |||||
| T1–56 | OP189394 (E) | OP189607 (E4) | ||||||
| T1–57 | OP189395 (E) | |||||||
| T1–58 | OP189396 (E) | OP189552 (E1) | ||||||
| T1–60 | OP189397 (E) | OP189650 (E2) | OP189553 (E23) | OP189573 (E1) | MLG E12 | |||
| Young cattle | T1–105 | OP189480 (E) | ||||||
| T1–110 | OP189410 (E) | OP189643 (E10) | OP189554 (E1) | OP189576 (E8) | MLG E22 | |||
| T1–111 | OP189481 (E) | |||||||
| T1–112 | OP189411 (E) | OP189633 (E3) | ||||||
| T1–113 | OP189482 (E) | OP189651 (E6) | OP189614 (E4) | |||||
| T1–120 | OP189483 (E) | |||||||
| T1–122 | OP189484 (E) | |||||||
| T1–125 | OP189485 (E) | |||||||
| T1–130 | OP189412 (E) | OP189555 (E1) | OP189615 (E1) | |||||
| T1–131 | OP189413 (E) | OP189556 (E2) | ||||||
| T1–133 | OP189414 (E) | |||||||
| T1–139 | OP189486 (E) | OP189566 (E5) | OP189634 (E1) | |||||
| T1–140 | OP189487 (E) | |||||||
| Adult cattle | T1–63 | OP189515 (E) | OP189632 (E1) | |||||
| T1–64 | OP189398 (E) | |||||||
| T1–68 | OP189399 (E) | |||||||
| T1–69 | OP189400 (E) | OP189523 (E2) | ||||||
| T1–70 | OP189401 (E) | |||||||
| T1–76 | OP189402 (E) | |||||||
| T1–78 | OP189478 (E) | |||||||
| T1–79 | OP189403 (E) | OP189612 (E1) | ||||||
| T1–80 | OP189404 (E) | |||||||
| T1–82 | OP189405 (E) | |||||||
| T1–84 | OP189479 (E) | |||||||
| T1–90 | OP189406 (E) | OP189574 (E11) | ||||||
| T1–96 | OP189407 (E) | |||||||
| T1–97 | OP189516 (E) | |||||||
| T1–98 | OP189408 (E) | OP189575 (E12) | ||||||
| T1–99 | OP189409 (E) | OP189524 (E2) | OP189608 (E19) | |||||
| Total | – | – | E (49), A (5) | E (7), A (1) | E (10), A (2) | E (20), A (1) | ||
| Tumd left banner 2 | Preweaned calves | T2–1 | OP189417 (E) | OP189664 (E1) | OP189525 (E5) | OP189577 (E9) | MLG E1 | |
| T2–2 | OP189418 (E) | |||||||
| T2–6 | OP189419 (E) | OP189609 (E3) | ||||||
| T2–12 | OP189489 (E) | |||||||
| T2–21 | OP189420 (E) | |||||||
| T2–22 | OP189421 (E) | OP189665 (E1) | OP189526 (E19) | OP189578 (E1) | MLG E2 | |||
| T2–25 | OP189422 (E) | |||||||
| T2–29 | OP189423 (E) | OP189666 (E2) | OP189527 (E19) | OP189579 (E5) | MLG E13 | |||
| Postweaned calves | T2–31 | OP189424 (E) | OP189644 (E5) | OP189561 (E20) | OP189610 (E1) | MLG E21 | ||
| T2–38 | OP189425 (E) | OP189580 (E1) | ||||||
| T2–42 | OP189426 (E) | OP189667 (E3) | OP189562 (E2) | OP189581 (E4) | MLG E19 | |||
| T2–43 | OP189427 (E) | OP189668 (E2) | OP189563 (E1) | OP189603 (E2) | MLG E14 | |||
| T2–45 | OP189428 (E) | OP189529 (E21) | ||||||
| T2–46 | OP189429 (E) | OP189672 (E5) | OP189528 (E2) | |||||
| T2–49 | OP189430 (E) | OP189582 (E9) | ||||||
| Young cattle | T2–54 | OP189431 (E) | ||||||
| T2–57 | OP189432 (E) | |||||||
| T2–63 | OP189433 (E) | |||||||
| T2–65 | OP189490 (E) | |||||||
| T2–68 | OP189434 (E) | OP189669 (E1) | OP189530 (E16) | OP189583 (E4) | MLG E8 | |||
| Adult cattle | T2–73 | OP189491 (E) | OP189613 (E15) | |||||
| T2–85 | OP189492 (E) | OP189531 (E3) | OP189604 (E5) | |||||
| T2–97 | OP189435 (A) | |||||||
| T2–99 | OP189488 (E) | OP189532 (E13) | OP189584 (E18) | |||||
| T2–103 | OP189415 (E) | OP189674 (E8) | OP189533 (E22) | OP189616 (E20) | MLG E23 | |||
| T2–105 | OP189416 (E) | OP189638 (E1) | OP189635 (E1) | |||||
| Total | – | – | E (25), A (1) | E (10) | E (12) | E (15) | ||
| Horinger county | Preweaned calves | 0 | ||||||
| Postweaned calves | He-101 | OP189461 (A) | ||||||
| He-103 | OP189462 (E) | OP189648 (E11) | OP189567 (E10) | OP189596 (E3) | MLG E25 | |||
| He-104 | OP189463 (E) | OP189547 (E15) | OP189597 (E1) | |||||
| He-107 | OP189464 (E) | |||||||
| He-108 | OP189465 (E) | OP189598 (E1) | ||||||
| He-112 | OP189466 (E) | OP189671 (E4) | OP189548 (E2) | OP189599 (E6) | MLG E20 | |||
| He-117 | OP189467 (E) | |||||||
| He-118 | OP189468 (E) | |||||||
| He-120 | OP189469 (E) | OP189675 (E1) | OP189549 (E1) | OP189630 (E10) | MLG E9 | |||
| Young cattle | He-62 | OP189455 (E) | ||||||
| He-63 | OP189456 (E) | OP189595 (E3) | ||||||
| He-65 | OP189512 (E) | OP189647 (E7) | OP189545 (E18) | OP189626 (E17) | MLG E24 | |||
| He-66 | OP189457 (E) | |||||||
| He-78 | OP189458 (E) | |||||||
| He-89 | OP189459 (E) | OP189546 (E11) | ||||||
| He-92 | OP189513 (E) | OP189629 (E2) | ||||||
| He-93 | OP189460 (A) | |||||||
| He-96 | OP189514 (E) | |||||||
| Adult cattle | He-26 | OP189448 (E) | OP189593 (E1) | |||||
| He-29 | OP189449 (E) | |||||||
| He-32 | OP189450 (E) | |||||||
| He-35 | OP189451 (E) | OP189627 (E22) | ||||||
| He-43 | OP189452 (E) | OP189625 (E6) | ||||||
| He-54 | OP189453 (E) | |||||||
| He-55 | OP189511 (E) | OP189594 (E1) | ||||||
| He-56 | OP189454 (A) | OP189628 (E1) | A + E | |||||
| Total | – | – | E (23), A (3) | E (4) | E (6) | E (13) | ||
| Togtoh county | Preweaned calves | 0 | ||||||
| Postweaned calves | Tuo-31 | OP189436 (E) | OP189652 (E1) | OP189534 (E17) | OP189585 (E7) | MLG E3 | ||
| Tuo-32 | OP189437 (E) | OP189653 (E1) | OP189535 (E4) | OP189586 (E7) | MLG E6 | |||
| Tuo-33 | OP189438 (E) | OP189654 (E2) | OP189536 (E1) | OP189587 (E13) | MLG E16 | |||
| Tuo-34 | OP189439 (E) | OP189655 (E1) | OP189537 (E5) | OP189588 (E14) | MLG E10 | |||
| Tuo-36 | OP189440 (E) | OP189656 (E1) | OP189538 (E12) | OP189589 (E5) | MLG E4 | |||
| Tuo-39 | OP189493 (E) | OP189657 (E2) | OP189539 (E6) | OP189590 (E3) | MLG E17 | |||
| Tuo-40 | OP189494 (E) | OP189639 (E1) | OP189564 (E1) | OP189636 (E3) | MLG E11 | |||
| Tuo-41 | OP189441 (E) | OP189658 (E1) | OP189540 (E20) | OP189591 (E1) | MLG E5 | |||
| Tuo-42 | OP189442 (E) | OP189659 (E1) | OP189541 (E4) | OP189592 (E7) | MLG E6 | |||
| Tuo-44 | OP189443 (E) | OP189660 (E1) | OP189565 (E9) | |||||
| Tuo-45 | OP189444 (E) | OP189661 (E1) | OP189557 (E7) | OP189617 (E24) | MLG E7 | |||
| Young cattle | Tuo-71 | OP189499 (A) | OP189662 (E2) | OP189558 (A1) | A + E | |||
| Tuo-73 | OP189500 (A) | OP189559 (E8) | OP189623 (A1) | A + E | ||||
| Tuo-74 | OP189501 (E) | |||||||
| Tuo-75 | OP189447 (E) | OP189645 (E2) | OP189542 (E14) | OP189600 (E2) | MLG E15 | |||
| Tuo-76 | OP189502 (E) | |||||||
| Tuo-84 | OP189503 (E) | OP189663 (E2) | OP189543 (E1) | OP189601 (E1) | MLG E18 | |||
| Tuo-85 | OP189518 (E) | OP189619 (E4) | ||||||
| Tuo-87 | OP189519 (E) | OP189544 (E1) | OP189602 (E1) | |||||
| Tuo-88 | OP189504 (E) | |||||||
| Tuo-89 | OP189505 (E) | OP189637 (E3) | ||||||
| Tuo-90 | OP189520 (E) | OP189560 (E20) | ||||||
| Adult cattle | Tuo-51 | OP189445 (E) | OP189618 (E21) | |||||
| Tuo-53 | OP189495 (E) | |||||||
| Tuo-54 | OP189496 (E) | |||||||
| Tuo-57 | OP189497 (E) | |||||||
| Tuo-60 | OP189498 (A) | OP189622 (E16) | A + E | |||||
| Tuo-68 | OP189517 (E) | OP189673 (E9) | ||||||
| Tuo-70 | OP189446 (E) | |||||||
| Tuo-92 | OP189506 (E) | OP189646 (E1) | OP189620 (E1) | |||||
| Tuo-93 | OP189507 (E) | |||||||
| Tuo-94 | OP189508 (E) | |||||||
| Tuo-95 | OP189509 (E) | |||||||
| Total | – | – | E (30), A (3) | E (16) | E (16), A (1) | E (18), A (1) | ||
| Dalad banner | Preweaned calves | 0 | ||||||
| Postweaned calves | 0 | |||||||
| Young cattle | Da-6 | OP189470 (A) | ||||||
| Da-7 | OP189471 (E) | OP189621 (E4) | ||||||
| Da-8 | OP189472 (E) | OP189624 (E4) | ||||||
| Adult cattle | Da-11 | OP189473 (E) | ||||||
| Da-15 | OP189474 (E) | |||||||
| Da-17 | OP189475 (E) | |||||||
| Total | – | – | E (5), A (1) | 0 | 0 | E (2) | ||
| Hanggin rear banner | Preweaned calves | 0 | ||||||
| Postweaned calves | 0 | |||||||
| Young cattle | 0 | |||||||
| Adult cattle | Ba-4 | OP189510 (E) | ||||||
| Total | – | – | E (1) | 0 | 0 | 0 | ||
| Total | – | E (133), A (13) | E (37), A (1) | E (44), A (3) | E (68), A (2) | |||
The phylogenetic analysis of gdh (Fig. 2), tpi (Fig. 3), and bg (Fig. 4) sequences based on G. duodenalis revealed that the phylogenetic tree of the three genes was divided into two branches (assemblages A and E).
Figure 2.
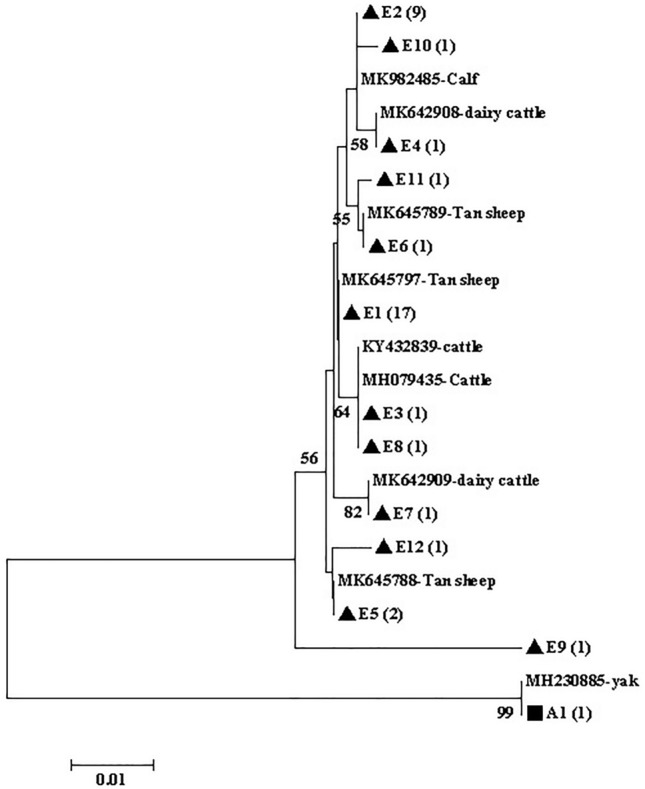
Phylogenetic tree of Giardia duodenalis based on gdh sequences. The phylogenetic tree was inferred via neighbor-joining analysis of genetic distances calculated using the Kimura 2-parameter model. Percent bootstrap values of > 50% from 1000 replicates are shown to the left of nodes. The isolates indicated in black triangles (filled triangle) and black squares (filled square) represent assemblages E and A, respectively, identified in cattle in this study.
Figure 3.

Phylogenetic tree of Giardia duodenalis based on tpi sequences. The phylogenetic tree was inferred via neighbor-joining analysis of genetic distances calculated using the Kimura 2-parameter model. Percent bootstrap values > 50% from 1000 replicates are shown to the left of nodes. Assemblages E and A isolates identified in cattle in this study are indicated in black triangles (filled triangle) and black squares (filled square), respectively.
Figure 4.

Phylogenetic tree of Giardia duodenalis based on bg sequences. The phylogenetic tree was inferred via the neighbor-joining analysis of genetic distances calculated using the Kimura 2-parameter model. Percent bootstrap values > 50% from 1000 replicates are shown to the left of nodes. The black triangles (filled triangle) and black squares (filled square) represent assemblage E and assemblage A, respectively, identified in cattle in this study.
Assemblage E multilocus genotype (MLG) distribution
The sequences of gdh, tpi, and bg were successfully obtained from 26 isolates, and three genes from 26 isolates were combined for genotyping, forming 25 different assemblage E multilocus genotypes (MLGs) (Table 3). Phylogenetic analysis revealed that all assemblage E MLGs formed six subgroups (Fig. 5).
Figure 5.
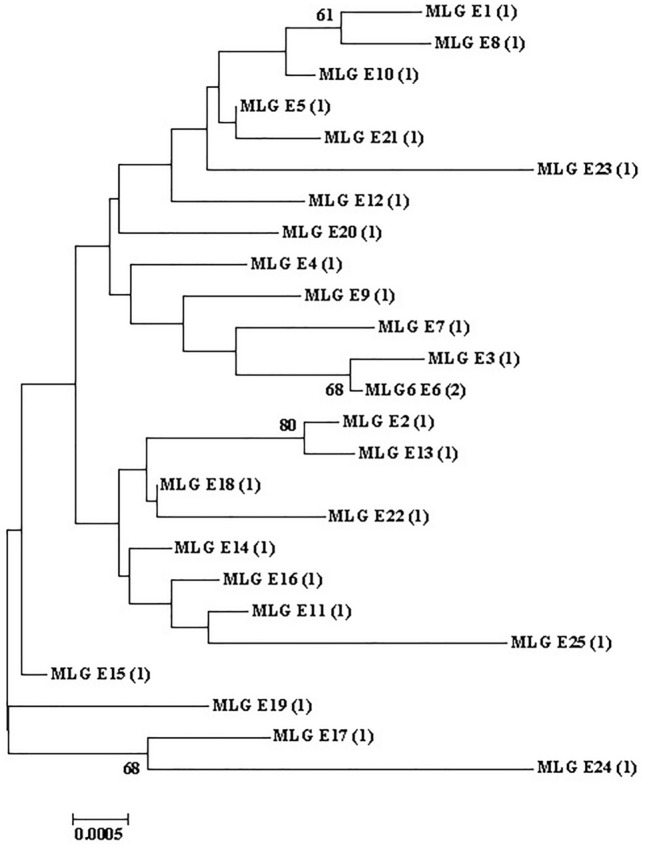
Phylogenetic relationships between Giardia duodenalis assemblage E MLGs. The phylogenetic tree was constructed using a concatenated dataset of bg, tpi, and gdh sequences, and the neighbor-joining analysis revealed identical topologies.
Discussion
Giardia duodenalis is an important intestinal parasite. To date, numerous studies worldwide have reported G. duodenalis infection in cattle14. Bovine giardiasis is also widespread in China4,13,14,24–48. Herein, a molecular epidemiological investigation of G. duodenalis was conducted involving 505 dairy cattle fecal samples from six sites in Central Inner Mongolia. The findings filled the gaps in the data regarding G. duodenalis infection in dairy cattle in Inner Mongolia as well as reconfirmed the existence of G. duodenalis infection in animals in Inner Mongolia44,53–55. The overall prevalence of G. duodenalis was 29.5% (149/505) in this study, which exceeded the global pooled prevalence of 22%14 and was higher than the pooled prevalence of 14.1% in Chinese cattle14. In China, the prevalence of G. duodenalis in this study was only lower than the prevalence of G. duodenalis in cattle in Shanghai (60.1%, 492/818)32, Sichuan (41.2%, 126/306)34, and Guangdong (74.2%, 288/388)38 and higher than the prevalence data reported in Xinjiang (13.4%, 69/514)4; Ningxia (4.38%, 74/1688)37 and (2.12%, 29/1366)24; Heilongjiang (5.2%, 42/814)25,26, (4.98%, 16/321)27 and (15.4%, 8/52)28; Jilin (6.63%, 25/377)28; Liaoning (8.4%, 19/226)28; Hubei ( 22.7%, 77/339)29; Shandong (13.04%, 9/69, PCR) and (18.84%, 13/69, rapid kit)30; Shaanxi (18.87%, 70/371)31; Sichuan (9.4%, 26/278)33; Beijing (1.70%, 14/822)35; Gansu (1.0%, 14/1414)36 and (2.63%, 33/1257)37; Guangdong (2.2%, 31/1440)39; Hebei and Tianjin (4.7%, 49/1040)40; Henan (7.2%, 128/1777)41; Jiangsu (20.6%, 281/1366)42; Jiangxi (9.2%, 52/566)43; Qinghai (10%, 39/389)45; Tibet (3.8%, 17/442)46; Yunnan (10.49%, 41/ 391)47; Taiwan (19.87%, 31/156)48 and that reported in the only survey on G. duodenalis in Inner Mongolia (9.3%, 10/108)44. However, it is difficult to compare prevalence data because they are influenced by a range of factors, including study design, diagnostic method, geographical conditions, total number of samples, age of animals, and sampling season4. The high prevalence of this study may also be related to the high density of local cattle farming. In addition, there were significant differences in the overall prevalence of G. duodenalis among the sampling sites in this study. Furthermore, positive samples were detected in all the age groups, indicating that all the age groups of cows are susceptible to G. duodenalis16,38.
Herein, neither the overall prevalence nor the prevalence in the different sampling sites of G. duodenalis was correlated with the presence of diarrhea in the sampled animals. Additionally, G. duodenalis was not detected in the diarrhea samples of preweaned calves. This observation is inconsistent with the combined global data, as the latter reported a significant correlation between G. duodenalis infection and cattle having diarrhea and preweaned calves14. The causes for diarrhea in animals are complex, with the pathogens including various viruses, bacteria, and parasites56. In addition, in G. duodenalis infection, the appearance of symptoms is also related to the stage of the infection. However, in the diarrhea samples in the present study, the prevalence of G. duodenalis in postweaned calves was significantly higher than that in the other age groups. In addition, the overall prevalence in the preweaned calves was significantly lower than that in the other age groups and that in postweaned calves was significantly higher than that in the other age groups in all the samples, which does not reflect the decrease in G. duodenalis infection rates with increasing age as reported in the literature42. The high prevalence of G. duodenalis in postweaned calves has also been reported in China4,27,29, Norway57, Germany58, USA59,60, and Canada61. However, there are also several studies reporting a relatively high prevalence of G. duodenalis infection in preweaned calves24,28,31,36,37,39–43. Certainly, there are some inconsistencies in the ages of preweaned and postweaned calves, or the age of the sampled cows is not clearly stated in previous studies. If calves aged < 6 months are defined as preweaned calves, the postweaned calves in this study will be classified as preweaned calves, but this is not consistent with the current situation of cattle farming in China. If this was the case, the prevalence of G. duodenalis in preweaned calves in this study was 29.3% (61/208), which is inconsistent with the higher G. duodenalis prevalence in preweaned calves (aged < 6 months) reported in the pooled global data14. As mentioned earlier, several factors influence the prevalence of G. duodenalis infection, age distribution, and diarrhea occurrence, such as the immature and susceptible immune system of younger animals60, asymptomatic immunocompetent individuals14,62, and the increased resistance due to nonspecific immunity acquired via breast milk63.
Overall, 146 plausible SSU rRNA sequences were identified as 13 assemblage A and 133 as assemblage E, with assemblage E being the dominant genotype. This finding is consistent with the studies reporting that assemblage E is the most common genotype in cattle worldwide4,13,14,16–20. Combining the sequence analysis results of 38 isolates of gdh, 47 isolates of tpi, and 70 isolates of bg, the genetic diversity of these positive G. duodenalis isolates was determined. The results revealed their genetic diversity, and seven isolates were identified to exhibit inconsistent assemblage. This finding is similar to the results reported previously in China and abroad4,64. It is also consistent with the results reported for 89,139 cattle from 48 countries in seven regions, with assemblage E being the most common, followed by assemblages A and A + E14. Transmission via environment (e.g., cyst contaminated water and food) may play a key role in parasite epidemiology65. Assemblages A and B are considered zoonotic4,14,21–24, and assemblage E has also been reported in humans in Egypt66, Brazil67, and Australia68. In China, cows are considered significant reservoirs of human giardiasis69. The results of the present study suggest that dairy cattle in Inner Mongolia pose a risk of causing G. duodenalis infection in humans.
Herein, the comparison of cattle belonging to different age groups for four of G. duodenalis assemblage genes revealed that preweaned calves only contained assemblage E, whereas postweaned calves contained assemblages E and A. Seven assemblage A + E were present in postweaned calves (n = 3), young cattle (n = 2), and adult cattle (n = 2). This finding is consistent with the report of preweaned calves containing only assemblage E in Sichuan, China33, and postweaned calves containing assemblages E, A, and A + E34. Furthermore, it is inconsistent with reports mentioning that assemblages E and A were detected in preweaned calves and postweaned calves in China31,41, USA59,60, and Europe70. A high prevalence of assemblage A in preweaned calves has also been reported4. Assemblage A infection has been reported in dairy cattle of all ages59,60,71,72, and assemblage E is also been reportedly common in adult cattle47. In addition, assemblage E is more common in calves than in adult cattle73. Herein, only assemblage E was found in the Hanggin Rear Banner field; all the other fields were found to have assemblages E and A, and three of them were also found to have assemblage A + E. In China, reports on cattle G. duodenalis assemblage varied with different sites. Assemblage E was only detected in Hubei Province29, Beijing35, Gansu36,37, Inner Mongolia44, and Qinghai45; assemblages E and A were detected in Shaanxi31, Jilin28, Jiangxi43, Tibet46, and Yunnan47; assemblages E and A + E were detected Hebei and Tianjin40; assemblages E, A, and A + E were detected in Xinjiang4, Liaoning28, Sichuan33,34, Guangdong38,39, Henan41, and Jiangsu42; assemblages E, B,24,37, and A were detected37 in Ningxia; assemblages E, A, B, and A + E were detected in Heilongjiang26–28 and Shanghai32; and assemblages E and D were detected in Taiwan48. However, the available data in China do not reflect the geographical distribution pattern of G. duodenalis assemblages.
Reports on genetic variations in G. duodenalis assemblage remain insufficient. The characteristics of individual loci of G. duodenalis often lead to inconsistent genotyping results16. The MLG model was used to better understand the diversity of human and animal G. duodenalis in different geographic regions, which can help reveal the potential and dynamic transmission of zoonosis74. Herein, 26 isolates containing the gdh, tpi, and bg were combined to obtain 25 different assemblage E MLGs with six subgroups. Consistent with the results of previous studies, numerous MLGs were identified in assemblage E. Additionally, G. duodenalis isolates that were classified in the same assemblage may be classified as distinct MLGs4,31,32,34,37–39,41,75.
The results of the present study suggest that dairy cattle in Inner Mongolia pose a risk of causing G. duodenalis infection in humans. In addition, Giardia are commonly found on fruits76, and vegetables77,78, and in various types of water79–89 in other regions of China. Therefore, further studies need to investigate the molecular epidemiology of cattle keepers and neighboring water sources in Inner Mongolia to evaluate the transmission dynamics of G. duodenalis, to adopt effective strategies to prevent and control G. duodenalis transmission among dairy cattle and humans in Inner Mongolia.
Conclusions
To the best of our knowledge, the present study is the first to report G. duodenalis infection in dairy cattle in Inner Mongolia, thereby filling a gap in the molecular epidemiological data regarding giardiasis in Central Inner Mongolia. The results reconfirmed previous findings in other parts of China that G. duodenalis infection is common in dairy cattle. The livestock-specific G. duodenalis assemblage E was the dominant assemblage; however, zoonotic assemblage A was also present in Inner Mongolia. The distribution of bovine G. duodenalis assemblage E MLGs was diverse.
Abbreviations
- MLG
Multilocus genotype
- SSU rRNA
Small subunit ribosomal RNA
- gdh
Glutamate dehydrogenase
- tpi
Triosephosphate isomerase
- bg
Beta-giardin
- PCR
Polymerase chain reaction
- CI
Confidence interval
- OR
Odds ratio
Author contributions
L.Z., Z.S.Z. and Y.H.L. conceived and designed the study and critically revised the manuscript. L.Z., Z.S.Z., H.L.C., W.X.H., B.Y., and Y.H.L. performed sample collection. Z.S.Z. and Y.H.L. prepared Figs. 1, 2, 3, 4 and 5. Z.S.Z., H.L.C., M.Y.W., Y.W., S.Z., W.H.Z., Y.M.M., Y.J.Z., L.F.W., Y.L.D., J.L.W. and L.Z. conducted the laboratory experiments. All the authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (32260887), National Natural Science Foundation of Inner Mongolia (2022MS03023), and Inner Mongolia Agricultural University High-level Talents Research Initiation Fund Project (NDYB2019-3).
Data availability
All the sequences obtained in our laboratory have been uploaded to the GenBank database under the Accession Numbers OP189375 to OP189675.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Li Zhao and Zhan-Sheng Zhang.
References
- 1.Adam RD. Giardia duodenalis: Biology and pathogenesis. Clin. Microbiol. Rev. 2021;34:e0002419. doi: 10.1128/CMR.00024-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortega YR, Raad R. Giardia duodenalis. In: Ortega YR, Raad R, editors. Encyclopedia of Infection and Immunity. Elsevier; 2022. pp. 570–579. [Google Scholar]
- 3.Sadeghi H, Borji H. A survey of intestinal parasites in a population in Qazvin, north of Iran. Asian Pac. J. Trop. Dis. 2015;5:231–233. [Google Scholar]
- 4.Qi M, Wang H, Jing B, Wang R, Jian F, Ning C, Zhang L. Prevalence and multilocus genotyping of Giardia duodenalis in dairy calves in Xinjiang, Northwestern China. Parasit. Vectors. 2016;9:546. doi: 10.1186/s13071-016-1828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karanis P, Kourenti C, Smith H. Waterborne transmission of protozoan parasites: A worldwide review of outbreaks and lessons learnt. J. Water Health. 2007;5:1–38. doi: 10.2166/wh.2006.002. [DOI] [PubMed] [Google Scholar]
- 6.Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2004–2010. Water Res. 2011;45:6603–6614. doi: 10.1016/j.watres.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Efstratiou A, Ongerth JE, Karanis P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2011–2016. Water Res. 2017;114:14–22. doi: 10.1016/j.watres.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 8.Cardona GA, de Lucio A, Bailo B, Cano L, de Fuentes I, Carmena D. Unexpected finding of feline-specific Giardia duodenalis assemblage F and Cryptosporidium felis in asymptomatic adult cattle in Northern Spain. Vet. Parasitol. 2015;209:258–263. doi: 10.1016/j.vetpar.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 9.Xiao LH, Feng YY. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8–9:14–32. doi: 10.1016/j.fawpar.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan U, Hijjawi N, Feng Y, Xiao L. Giardia: An under-reported foodborne parasite. Int. J. Parasitol. 2019;49:1–11. doi: 10.1016/j.ijpara.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Karanis P, Ey PL. Characterization of axenic isolates of Giardia intestinalis established from humans and animals in Germany. Parasitol. Res. 1998;84:442–449. doi: 10.1007/s004360050427. [DOI] [PubMed] [Google Scholar]
- 12.Kiani-Salmi N, Fattahi-Bafghi A, Astani A, Sazmand A, Zahedi A, Firoozi Z, et al. Molecular typing of Giardia duodenalis in cattle, sheep and goats in an arid area of central Iran. Infect. Genet. Evol. 2019;75:104021. doi: 10.1016/j.meegid.2019.104021. [DOI] [PubMed] [Google Scholar]
- 13.Zhang XP, Dan J, Wang L, Liu H, Zhou Z, Ma X, et al. High genetic diversity of Giardia duodenalis assemblage E in Chinese dairy cattle. Infect. Genet. Evol. 2021;92:104912. doi: 10.1016/j.meegid.2021.104912. [DOI] [PubMed] [Google Scholar]
- 14.Taghipour A, Sharbatkhori M, Tohidi F, Ghanbari MR, Karanis P, Olfatifar M, et al. Global prevalence of Giardia duodenalis in cattle: A systematic review and meta-analysis. Prev. Vet. Med. 2022;203:105632. doi: 10.1016/j.prevetmed.2022.105632. [DOI] [PubMed] [Google Scholar]
- 15.Ryan U, Cacciò SM. Zoonotic potential of Giardia. Int. J. Parasitol. 2013;43:943–956. doi: 10.1016/j.ijpara.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Feng YY, Xiao LH. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santin M. Cryptosporidium and Giardia in ruminants. Vet. Clin. N. Am. Food Anim. Pract. 2020;36:223–238. doi: 10.1016/j.cvfa.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Cai WL, Ryan U, Xiao L, Feng Y. Zoonotic giardiasis: An update. Parasitol. Res. 2021;120:4199–4218. doi: 10.1007/s00436-021-07325-2. [DOI] [PubMed] [Google Scholar]
- 19.Minetti C, Taweenan W, Hogg R, Featherstone C, Randle N, Latham SM, Wastling JM. Occurrence and diversity of Giardia duodenalis assemblages in livestock in the UK. Transbound Emerg. Dis. 2014;61:e60–e67. doi: 10.1111/tbed.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang GH, Wang GP, Li XP, Ma LQ, Karanis G, Christodoulou-Vafeiadou E, Karanis P. Detection of Giardia duodenalis assemblage E infections at the Tibetan Plateau area: Yaks are suitable hosts. Acta Trop. 2017;169:157–162. doi: 10.1016/j.actatropica.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 21.Geurden T, Geldhof P, Levecke B, Martens C, Berkvens D, Casaert S, et al. Mixed Giardia duodenalis assemblage A and E infections in calves. Int. J. Parasitol. 2008;38:259–264. doi: 10.1016/j.ijpara.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Dixon B, Parrington L, Cook A, Pintar K, Pollari F, Kelton D, Farber J. The potential for zoonotic transmission of Giardia duodenalis and Cryptosporidium spp. from beef and dairy cattle in Ontario, Canada. Vet. Parasitol. 2011;175:20–26. doi: 10.1016/j.vetpar.2010.09.032. [DOI] [PubMed] [Google Scholar]
- 23.Ng J, Yang R, McCarthy S, Gordon C, Hijjawi N, Ryan U. Molecular characterization of Cryptosporidium and Giardia in pre-weaned calves in Western Australia and New South Wales. Vet. Parasitol. 2011;176:145–150. doi: 10.1016/j.vetpar.2010.10.056. [DOI] [PubMed] [Google Scholar]
- 24.Huang JY, Yue D, Qi M, Wang R, Zhao J, Li J, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet. Res. 2014;10:292. doi: 10.1186/s12917-014-0292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu AQ, Zhang X, Zhang L, Wang R, Li X, Shu J, et al. Occurrence of bovine giardiasis and endemic genetic characterization of Giardia duodenalis isolates in Heilongjiang Province, in the Northeast of China. Parasitol. Res. 2012;111:655–661. doi: 10.1007/s00436-012-2883-0. [DOI] [PubMed] [Google Scholar]
- 26.Liu AQ, Yang F, Shen Y, Zhang W, Wang R, Zhao W, et al. Genetic analysis of the gdh and bg genes of animal-derived Giardia duodenalis isolates in Northeastern China and evaluation of zoonotic transmission potential. PLoS ONE. 2014;9:e95291. doi: 10.1371/journal.pone.0095291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma N, Wang HX, Tao WF, Xue NY, Bai JY, Zhao Q, et al. Detection of point prevalence and assemblages of Giardia spp. in dairy calves and sika deer, Northeast China. Vector Borne Zoonotic Dis. 2021;21:685–691. doi: 10.1089/vbz.2020.2757. [DOI] [PubMed] [Google Scholar]
- 28.Liu G, Su Y, Zhou M, Zhao J, Zhang T, Ahmad W, et al. Prevalence and molecular characterization of Giardia duodenalis isolates from dairy cattle in northeast China. Exp. Parasitol. 2015;154:20–24. doi: 10.1016/j.exppara.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Fan YY, Wang T, Koehler AV, Hu M, Gasser RB. Molecular investigation of Cryptosporidium and Giardia in pre- and post-weaned calves in Hubei Province, China. Parasit. Vectors. 2017;10:519. doi: 10.1186/s13071-017-2463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei XJ, Wang W, Dong Z, Cheng F, Zhou X, Li B, Zhang J. Detection of infectious agents causing neonatal calf diarrhea on two large dairy farms in Yangxin County, Shandong Province, China. Front. Vet. Sci. 2020;7:589126. doi: 10.3389/fvets.2020.589126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang XT, Wang RJ, Ren GJ, Yu ZQ, Zhang LX, Zhang SY, et al. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi Province, northwestern China. Parasitol. Res. 2016;115:1355–1361. doi: 10.1007/s00436-016-4908-6. [DOI] [PubMed] [Google Scholar]
- 32.Wang XL, Cai M, Jiang W, Wang Y, Jin Y, Li N, et al. High genetic diversity of Giardia duodenalis assemblage E in pre-weaned dairy calves in Shanghai, China, revealed by multilocus genotyping. Parasitol. Res. 2017;116:2101–2110. doi: 10.1007/s00436-017-5509-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhong ZJ, Dan J, Yan G, Tu R, Tian Y, Cao S, et al. Occurrence and genotyping of Giardia duodenalis and Cryptosporidium in pre-weaned dairy calves in central Sichuan Province, China. Parasite. 2018;25:45. doi: 10.1051/parasite/2018046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dan JM, Zhang X, Ren Z, Wang L, Cao S, Shen L, et al. Occurrence and multilocus genotyping of Giardia duodenalis from post-weaned dairy calves in Sichuan Province, China. PLoS ONE. 2019;14:e0224627. doi: 10.1371/journal.pone.0224627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li FH, Wang H, Zhang Z, Li J, Wang C, Zhao J, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Vet. Parasitol. 2016;219:61–65. doi: 10.1016/j.vetpar.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 36.Wang YL, Cao J, Chang Y, Yu F, Zhang S, Wang R, Zhang L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Gansu, northwest China. Parasite. 2020;27:62. doi: 10.1051/parasite/2020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang XX, Tan QD, Zhao GH, Ma JG, Zheng WB, Ni XT, et al. Prevalence, risk factors and multilocus genotyping of Giardia intestinalis in dairy cattle, Northwest China. J. Eukaryot. Microbiol. 2016;63:498–504. doi: 10.1111/jeu.12293. [DOI] [PubMed] [Google Scholar]
- 38.Feng YY, Gong X, Zhu K, Li N, Yu Z, Guo Y, et al. Prevalence and genotypic identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in pre-weaned dairy calves in Guangdong, China. Parasit. Vectors. 2019;12:41. doi: 10.1186/s13071-019-3310-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui ZH, Wang L, Cao L, Sun M, Liang N, Wang H, et al. Genetic characteristics and geographic segregation of Giardia duodenalis in dairy cattle from Guangdong Province, southern China. Infect. Genet. Evol. 2018;66:95–100. doi: 10.1016/j.meegid.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 40.Hu SH, Liu Z, Yan F, Zhang Z, Zhang G, Zhang L, et al. Zoonotic and host-adapted genotypes of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in dairy cattle in Hebei and Tianjin, China. Vet. Parasitol. 2017;248:68–73. doi: 10.1016/j.vetpar.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 41.Wang HY, Zhao G, Chen G, Jian F, Zhang S, Feng C, et al. Multilocus genotyping of Giardia duodenalis in dairy cattle in Henan, China. PLoS ONE. 2014;9:e100453. doi: 10.1371/journal.pone.0100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang R, Li N, Jiang W, Guo Y, Wang X, Jin Y, et al. Infection patterns, clinical significance, and genetic characteristics of Enterocytozoon bieneusi and Giardia duodenalis in dairy cattle in Jiangsu, China. Parasitol. Res. 2019;118:3053–3060. doi: 10.1007/s00436-019-06426-3. [DOI] [PubMed] [Google Scholar]
- 43.Li S, Zou Y, Zhang XL, Wang P, Chen XQ, Zhu XQ. Prevalence and multilocus genotyping of Giardia lamblia in cattle in Jiangxi Province, China: Novel assemblage E subtypes identified. Korean J. Parasitol. 2020;58:681–687. doi: 10.3347/kjp.2020.58.6.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu Y, Dong H, Bian X, Qin Z, Han H, Lang J, et al. Molecular characterizations of Giardia duodenalis based on multilocus genotyping in sheep, goats, and beef cattle in Southwest Inner Mongolia, China. Parasite. 2022;29:33. doi: 10.1051/parasite/2022036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jian YN, Zhang X, Li X, Karanis G, Ma L, Karanis P. Prevalence and molecular characterization of Giardia duodenalis in cattle and sheep from the Qinghai-Tibetan Plateau area (QTPA), northwestern China. Vet. Parasitol. 2018;250:40–44. doi: 10.1016/j.vetpar.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Wu YY, Chen Y, Chang Y, Zhang X, Li D, Wang L, et al. Genotyping and identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi from free-range Tibetan yellow cattle and cattle-yak in Tibet, China. Acta Trop. 2020;212:105671. doi: 10.1016/j.actatropica.2020.105671. [DOI] [PubMed] [Google Scholar]
- 47.Liang XX, Zou Y, Li TS, Chen H, Wang SS, Cao FQ, et al. First report of the prevalence and genetic characterization of Giardia duodenalis and Cryptosporidium spp. in Yunling cattle in Yunnan Province, southwestern China. Microb. Pathog. 2021;158:105025. doi: 10.1016/j.micpath.2021.105025. [DOI] [PubMed] [Google Scholar]
- 48.Lam HYP, Chen TT, Tseng YC, Chang KC, Yang TH, Peng SY. Detection and genotyping of Giardia duodenalis from cattle and pigs in Hualien country, Eastern Taiwan. J. Microbiol. Immunol. Infect. 2021;54:718–727. doi: 10.1016/j.jmii.2020.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Appelbee AJ, Frederick LM, Heitman TL, Olson ME. Prevalence and genotyping of Giardia duodenalis from beef calves in Alberta, Canada. Vet. Parasitol. 2003;112:289–294. doi: 10.1016/s0304-4017(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 50.Lalle M, Pozio E, Capelli G, Bruschi F, Crotti D, Cacciò SM. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int. J. Parasitol. 2005;35:207–213. doi: 10.1016/j.ijpara.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 51.Cacciò SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int. J. Parasitol. 2008;38:1523–1531. doi: 10.1016/j.ijpara.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg. Infect. Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye JB, Xiao L, Wang Y, Guo Y, Roellig DM, Feng Y. Dominance of Giardia duodenalis assemblage A and Enterocytozoon bieneusi genotype BEB6 in sheep in Inner Mongolia, China. Vet. Parasitol. 2015;210:235–239. doi: 10.1016/j.vetpar.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 54.Cao LT, Han K, Wang L, Hasi S, Yu F, Cui Z, et al. Genetic characteristics of Giardia duodenalis from sheep in Inner Mongolia, China. Parasite. 2020;27:60. doi: 10.1051/parasite/2020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao SS, Li YH, Zhang Y, Zhou Q, Jing B, Xu CY, et al. Multilocus genotyping of Giardia duodenalis in Bactrian camels (Camelus bactrianus) in China. Parasitol. Res. 2020;119:3873–3880. doi: 10.1007/s00436-020-06905-y. [DOI] [PubMed] [Google Scholar]
- 56.Rostampour YS, Ghane M, Doudi M, Rezaee A, Naghavi NS. A study of leptospirosis epidemiology in Iran and diagnostic techniques for human, livestock and environment samples. Med. Lab. J. 2020;14:1–9. [Google Scholar]
- 57.Hamnes IS, Gjerde B, Robertson L. Prevalence of Giardia and Cryptosporidium in dairy calves in three areas of Norway. Vet. Parasitol. 2006;140:204–216. doi: 10.1016/j.vetpar.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 58.Gillhuber J, Rügamer D, Pfister K, Scheuerle MC. Giardiosis and other enteropathogenic infections: A study on diarrhoeic calves in Southern Germany. BMC Res. Notes. 2014;7:112. doi: 10.1186/1756-0500-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trout JM, Santín M, Greiner E, Fayer R. Prevalence of Giardia duodenalis genotypes in pre-weaned dairy calves. Vet. Parasitol. 2004;124:179–186. doi: 10.1016/j.vetpar.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 60.Trout JM, Santín M, Greiner E, Fayer R. Prevalence and genotypes of Giardia duodenalis in post-weaned dairy calves. Vet. Parasitol. 2005;130:177–183. doi: 10.1016/j.vetpar.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 61.O’Handley RM, Cockwill C, McAllister TA, Jelinski M, Morck DW, Olson ME. Duration of naturally acquired giardiosis and cryptosporidiosis in dairy calves and their association with diarrhea. J. Am. Vet. Med. Assoc. 1999;214:391–396. [PubMed] [Google Scholar]
- 62.Cacciò SM, Lalle M, Svärd SG. Host specificity in the Giardia duodenalis species complex. Infect. Genet. Evol. 2018;66:335–345. doi: 10.1016/j.meegid.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Mahmud MA, Chappell CL, Hossain MM, Huang DB, Habib M, DuPont HL. Impact of breast-feeding on Giardia lamblia infections in Bilbeis, Egypt. Am. J. Trop. Med. Hyg. 2001;65:257–260. doi: 10.4269/ajtmh.2001.65.257. [DOI] [PubMed] [Google Scholar]
- 64.Gillhuber J, Pallant L, Ash A, Thompson RC, Pfister K, Scheuerle MC. Molecular identification of zoonotic and livestock-specific Giardia-species in faecal samples of calves in Southern Germany. Parasit. Vectors. 2013;6:346. doi: 10.1186/1756-3305-6-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heyworth MF. Giardia duodenalis genetic assemblages and hosts. Parasite. 2016;23:13. doi: 10.1051/parasite/2016013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abdel-Moein KA, Saeed H. The zoonotic potential of Giardia intestinalis assemblage E in rural settings. Parasitol. Res. 2016;115:3197–3202. doi: 10.1007/s00436-016-5081-7. [DOI] [PubMed] [Google Scholar]
- 67.Fantinatti M, Bello AR, Fernandes O, Da-Cruz AM. Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. J. Infect. Dis. 2016;214:1256–1259. doi: 10.1093/infdis/jiw361. [DOI] [PubMed] [Google Scholar]
- 68.Zahedi A, Field D, Ryan U. Molecular typing of Giardia duodenalis in humans in Queensland—First report of assemblage E. Parasitology. 2017;144:1154–1161. doi: 10.1017/S0031182017000439. [DOI] [PubMed] [Google Scholar]
- 69.Feng YY, Xiao LH. Molecular epidemiology of cryptosporidiosis in China. Front. Microbiol. 2017;8:1701. doi: 10.3389/fmicb.2017.01701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geurden T, Vanderstichel R, Pohle H, Ehsan A, von Samson-Himmelstjerna G, Morgan ER, et al. A multicentre prevalence study in Europe on Giardia duodenalis in calves, with molecular identification and risk factor analysis. Vet. Parasitol. 2012;190:383–390. doi: 10.1016/j.vetpar.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 71.Trout JM, Santín M, Fayer R. Prevalence of Giardia duodenalis genotypes in adult dairy cows. Vet. Parasitol. 2007;147:205–209. doi: 10.1016/j.vetpar.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 72.Trout JM, Santín M, Greiner EC, Fayer R. Prevalence and genotypes of Giardia duodenalis in 1–2 year old dairy cattle. Vet. Parasitol. 2006;140:217–222. doi: 10.1016/j.vetpar.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 73.Santín M, Trout JM, Fayer R. A longitudinal study of Giardia duodenalis genotypes in dairy cows from birth to 2 years of age. Vet. Parasitol. 2009;162:40–45. doi: 10.1016/j.vetpar.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 74.Huey CS, Mahdy MA, Al-Mekhlafi HM, Nasr NA, Lim YA, Mahmud R, Surin J. Multilocus genotyping of Giardia duodenalis in Malaysia. Infect. Genet. Evol. 2013;17:269–276. doi: 10.1016/j.meegid.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 75.Zhao GH, Du SZ, Wang HB, Hu XF, Deng MJ, Yu SK, et al. First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi) Infect. Genet. Evol. 2015;34:394–401. doi: 10.1016/j.meegid.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 76.Wei XH, Hou S, Pan X, Xu C, Li J, Yu H, et al. Microbiological contamination of strawberries from U-pick farms in Guangzhou, China. Int. J. Environ. Res. Public Health. 2019;16:4910. doi: 10.3390/ijerph16244910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang Q, Huang S, Kuang W, Yi J, Xiao S, Zhao F, Xiao G. Health risks of Cryptosporidium and Giardia in the application of surface water and septic tank effluent in Chinese agriculture: Impact on cancer patients identified by quantitative microbial risk assessment. Food Microbiol. 2023;111:104213. doi: 10.1016/j.fm.2022.104213. [DOI] [PubMed] [Google Scholar]
- 78.Li XP, Zhang X, Jian Y, Wang G, Ma L, Schou C, Karanis P. Detection of Cryptosporidium oocysts and Giardia cysts in vegetables from street markets from the Qinghai Tibetan Plateau area in China. Parasitol. Res. 2020;119:1847–1855. doi: 10.1007/s00436-020-06661-z. [DOI] [PubMed] [Google Scholar]
- 79.Wei XH, Li J, Hou S, Xu C, Zhang H, Atwill ER, et al. Assessment of microbiological safety of water in public swimming pools in Guangzhou, China. Int. J. Environ. Res. Public Health. 2018;15:1416. doi: 10.3390/ijerph15071416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma LQ, Sotiriadou I, Cai Q, Karanis G, Wang G, Wang G, et al. Detection of Cryptosporidium and Giardia in agricultural and water environments in the Qinghai area of China by IFT and PCR. Parasitol. Res. 2014;113:3177–3184. doi: 10.1007/s00436-014-3979-5. [DOI] [PubMed] [Google Scholar]
- 81.Ma JW, Feng Y, Hu Y, Villegas EN, Xiao L. Human infective potential of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in urban wastewater treatment plant effluents. J. Water Health. 2016;14:411–23. doi: 10.2166/wh.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang CC, Hu Y, Wang L, Wang Y, Li N, Guo Y, et al. Environmental transport of emerging human-pathogenic Cryptosporidium species and subtypes through combined sewer overflow and wastewater. Appl. Environ. Microbiol. 2017;83:e00682. doi: 10.1128/AEM.00682-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang Q, Yang L, Li B, Du H, Zhao F, Han L, et al. Cryptosporidium spp. and Giardia duodenalis emissions from humans and animals in the three gorges reservoir in Chongqing, China. PeerJ. 2020;8:e9985. doi: 10.7717/peerj.9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan YY, Wang X, Yang R, Zhao W, Li N, Guo Y, et al. Molecular characterization of the waterborne pathogens Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. in wastewater and sewage in Guangzhou, China. Parasit. Vectors. 2021;14:66. doi: 10.1186/s13071-020-04566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiao S, Zhang Y, Zhao XY, Sun L, Hu S. Presence and molecular characterization of Cryptosporidium and Giardia in recreational lake water in Tianjin, China: A preliminary study. Sci. Rep. 2018;8:2353. doi: 10.1038/s41598-018-20902-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hsu BM. Evaluation of analyzing methods for Giardia and Cryptosporidium in a Taiwan water treatment plant. J. Parasitol. 2003;89:369–371. doi: 10.1645/0022-3395(2003)089[0369:EOAMFG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 87.Xiao S, Hu S, Zhang Y, Zhao X, Pan W. Influence of sewage treatment plant effluent discharge into multipurpose river on its water quality: A quantitative health risk assessment of Cryptosporidium and Giardia. Environ. Pollut. 2018;233:797–805. doi: 10.1016/j.envpol.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 88.Hsu BM, Huang C, Jiang GY, Hsu CL. The prevalence of giardia and cryptosporidium in Taiwan water supplies. J. Toxicol. Environ. Health A. 1999;57:149–160. doi: 10.1080/009841099157728. [DOI] [PubMed] [Google Scholar]
- 89.Xiao GS, Wang Z, Chen J, Qiu Z, Li Y, Qi J, et al. Occurrence and infection risk of waterborne pathogens in Wanzhou watershed of the three gorges reservoir, China. J. Environ. Sci. 2013;25:1913–1924. doi: 10.1016/s1001-0742(12)60241-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the sequences obtained in our laboratory have been uploaded to the GenBank database under the Accession Numbers OP189375 to OP189675.