Abstract
We undertook a prospective study to evaluate the accuracy of PCR of serum (aimed at the pneumococcal pneumolysin gene) at detecting pneumococcal infections in infants and children. The assay was positive for all blood and cerebrospinal fluid culture-positive samples and for 38 and 44% of patients with lobar pneumonia and acute otitis media, respectively. It was positive for 17% of healthy controls. There was a marked effect of age on the rate of positivity among healthy controls, with the highest rate (33%) being in 2-year-old children, the age group with the highest rate of nasopharyngeal (NP) carriage; the lowest rate was found among infants <2 months of age (13%) and adults ages 18 to 50 years (0%), age groups with the lowest NP pneumococcal carriage rates. Carriers of pneumococci in the nasopharynges had a higher rate of positivity than noncarriers of pneumococci in the nasopharynges for all groups. Our results suggest that although PCR of serum is a sensitive test for the detection of Streptococcus pneumoniae in sterile fluids, its high rate of positivity for healthy controls, related to NP pneumococcal carriage, might exclude it from being useful in detecting deep-seated pneumococcal infections.
Streptococcus pneumoniae is an important cause of morbidity and mortality in all societies (24). It is a major cause of pneumonia, meningitis, sepsis, and otitis media worldwide. In addition, antibiotic resistance is an important and increasing problem and has an enormous impact on clinicians, microbiologists, drug manufacturers, and public health authorities (1, 9, 18, 27).
Currently, the definitive diagnosis of pneumococcal infection requires the isolation of S. pneumoniae from the site of infection, demanding invasive procedures such as lung puncture, pleural fluid aspiration, and middle ear fluid (MEF) aspiration. Isolation of S. pneumoniae from a blood culture is acceptable indirect evidence of the presence of pneumococcal pneumonia. However, blood cultures are positive for only 20 to 30% of adults and <10% of children with pneumococcal pneumonia (16, 20, 21). Other tests such as the demonstration of pneumococcal capsular antigens in urine or serum by various methods such as coagglutination and counterimmunoelectrophoresis have failed due to a lack of sensitivity and specificity, even in the presence of bacteremic infections (2, 17, 22, 30). Even when present in blood, S. pneumoniae may be missed by culturing a single blood sample due to a low density of the pathogens, the fastidious nature of the organism, and previous administration of antibiotics (15, 31). The antibody response to pneumococcal antigens is not routinely measured and would require paired serum specimens (17).
The recently developed PCR for gene amplification has made it possible to detect low numbers of infectious agents or even fragments of DNA from the agents. This method has shown great promise in improving the diagnosis of infections due to various fastidious agents that cannot routinely be cultured. The PCR method has been tested for the detection of pneumococcal infections, with promising results (10, 12, 23, 26, 28, 31).
We undertook a prospective study to evaluate the accuracy of PCR of serum for the detection of various pneumococcal infections in infants and children.
MATERIALS AND METHODS
Purification of total DNA from serum.
Serum specimens (100 μl) were diluted with 100 μl of 0.1 M Tris-HCl (pH 8.0). The samples were heated to 80°C for 10 min. DNA was extracted with 200 μl of phenol-chloroform-isoamyl alcohol (25:24:1 ratio) and then with an equal volume of chloroform. DNA was precipitated with ethanol by the addition of 2.5 volumes of cold ethanol and 10% sodium acetate (3 M) and was incubated at −70°C for 30 min. The DNA pellet was washed with ethanol and dissolved in distilled water.
Purification of total DNA from bacteria.
Nine pneumococcal strains were tested. Two strains (serotypes 1 and 18C) from the American Type Culture Collection (Rockville, Md.) were given to us by M. Leinonen (Helsinki, Finland). Seven pneumococcal strains (2 isolates of type 14 and 1 isolate each of types 1, 3, 18C, 19A, and 23F) and 19 nonpneumococcal isolates were obtained from routine clinical specimens submitted to the Soroka Medical Center Microbiology Laboratory and identified by standard laboratory methods (for details, see the legend to Fig. 1A).
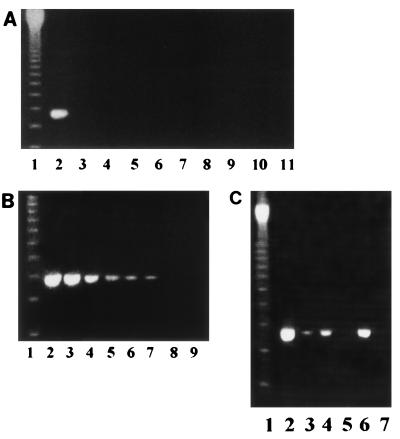
FIG. 1.
Sensitivity and specificity of the PCR assay. (A) Specificity of the PCR-based assay. PCR tests were conducted with 25 pg of purified DNA from the organisms. Purified S. pneumoniae DNA was used for the positive control. Lanes: 1, molecular size marker; 2, S. pneumoniae; 3, Staphylococcus aureus; 4, Streptococcus mitis; 5, group A Streptococcus; 6, Escherichia coli; 7, Enterococcus faecalis; 8, Pseudomonas aeruginosa; 9, Salmonella group D; 10, H. influenzae; 11, Moraxella catarrhalis. (B) Agarose gel electrophoresis of PCR-amplified products from a 10-fold dilution of purified S. pneumoniae DNA. Lane 1, molecular size marker; lanes 2 to 8, dilution series from 10 ng to 10 fg of DNA (corresponding to 106 to 1 CFU/ml); lane 9, negative control. (C) Agarose gel electrophoresis of PCR-amplified products from culture-positive CSF or blood before and after antibiotic treatment. Lane 1, molecular size marker; lanes 2 to 5, samples from culture-positive CSF and blood before (lanes 2 and 4, respectively) and after (lanes 3 and 5, respectively) antibiotic treatment for 48 h; lane 6, positive control (S. pneumoniae DNA); lane 7, negative control (H2O).
Freshly cultured bacteria were washed twice with 40 ml of sterile saline, recovered by centrifugation, and standardized to a concentration of approximately 1.5 × 108 cells/ml. Aliquots of 1 ml of the freshly cultured bacteria were centrifuged at 10,000 × g for 10 min and washed twice with phosphate-buffered saline. The pellet was suspended in a buffer containing 10 mM Tris, 0.14 M NaCl, 0.1 M sodium citrate, and 10 mM EDTA, and the mixture was incubated at 80°C for 10 min. DNA was extracted as described above for the serum specimens.
PCR amplification assay.
The selection of the primers was based on the published sequence of the pneumolysin gene (29). The pair of primers selected (5′-GTGATATTTCTGTAACAGCTACC and 5′-GAGAATTCCCTGTCTTTTCAAAG) amplified a 355-bp region of the published pneumolysin gene sequence. The PCR amplification was performed in a microprocessor-controlled incubation system (Crocodile II; Appligene Inc., Pleasanton, Calif.). The reaction mixture (volume, 50 μl) contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 5 mM deoxynucleotides, 50 pmol of primers, 1.5 U of Taq DNA polymerase (Bet-Haemek, Israel), and 2 μl (about 250 ng) of DNA extracted from the serum specimens. Amplification was done with 30 cycles of denaturation at 94°C, annealing at 60°C for 1 min, and synthesis at 72°C for 1 min. The pneumococcal DNA preparation was used as a positive control. Sterile distilled water or serum samples from healthy adult volunteers were used as negative controls. An 8-μl sample of the completed reaction mixture was run in a 2% agarose gel stained with ethidium bromide. Amplified products were visualized and photographed under UV light.
Samples tested.
The following clinical samples were tested by the PCR assay: (i) S. pneumoniae culture-positive fluid samples from 13 patients (5 serum samples from adults, 4 serum samples from juveniles, and 4 cerebrospinal fluid [CSF] samples from juveniles), (ii) sera from 34 children with putative acute pneumococcal pneumonia, and (iii) sera from 12 infants and children with acute otitis media (AOM) with MEF and nasopharyngeal (NP) swab cultures positive for S. pneumoniae.
The control sera included the following: (i) sera from 20 infants and children with putative nonpneumococcal otitis media, including 7 patients with Haemophilus influenzae-positive MEF samples and S. pneumoniae-positive NP swabs; 2 patients with culture-negative MEF samples and S. pneumoniae-positive NP swab cultures; and 11 patients with S. pneumoniae-negative NP swab cultures (7 with H. influenzae-positive MEF samples and 4 with culture-negative MEF samples); (ii) sera from 11 infants and toddlers with acute viral upper respiratory infections (URIs) with no clinical evidence of AOM, sinusitis, or pneumonia and no suggestion of bacterial infection according to a peripheral blood count and with an S. pneumoniae-negative NP swab culture; (iii) 174 healthy infants and children enrolled in various vaccine studies; and (iv) 20 adults not exposed at home to children younger than 5 years of age.
The ages of the pediatric patients were as follows: those with bacteremia and meningitis, 10 months to 7 years; those with pneumonia, 12 to 59 months (median, 17 months); and those with AOM, 6 to 33 months (median, 11 months). The ages of the healthy pediatric controls were as follows: 2 months (n = 30), 1 year (n = 42), 2 years (n = 30), 4 to 6 years (n = 40), and 11 to 16 years (n = 32). The ages of the pediatric controls with viral URIs were 7 to 24 months (median, 14 months).
NP swabs were obtained from all patients with lobar pneumonia, AOM, and URIs and from all healthy controls ages 2, 12, and 24 months.
The pediatric patients were seen at the pediatric emergency room of the Soroka University Medical Center, and all the healthy pediatric controls were seen in various Maternal Child Health Centers in the city of Beer-Sheva. All the pediatric healthy controls were part of vaccine studies, and a physician performed a clinical examination to confirm that the subjects were in good health. Blood samples were obtained from patients during the acute phase of the disease and from healthy controls during regular visits for various vaccine studies. The blood samples were obtained with sterile syringes and were processed with sterile tubes and equipment. The blood was kept refrigerated at 4°C until it was processed in the laboratory within 4 h, and the serum was separated and kept at −70°C until it was tested by PCR. Tympanocentesis procedures and bacteriologic diagnosis of otitis media are described elsewhere (3). All blood samples for culture were tested with the BACTEC 660NR system (Beckton Dickinson Diagnostic Instrument Systems, Towson, Md.). Definition and characterization of S. pneumoniae were performed as described previously (4, 5). NP swabs for culture were obtained and processed as described previously (4, 5).
For comparison of the NP carriage rate of healthy controls used in the present study and the NP carriage rate in the community, we used data obtained from our community and published elsewhere (5).
All samples were tested without the technician’s knowledge of the culture results or any other details about the patients.
Statistical analysis.
The statistical package of Epi-Info, version 6, was used to test (i) differences in proportions (by the chi-square test or Fisher’s exact test, as appropriate) and (ii) linear trends (by the chi-square test for linear trend in proportion). A P value of <0.05 was considered significant.
RESULTS
Specificity and sensitivity of PCR.
The nine pneumococcal isolates gave a clear band of the expected molecular size in the PCR. The results for a representative pneumococcal strain are presented in Fig. 1. The specificity of the PCR assay is demonstrated by its negative results for nine other organisms including closely related streptococci (Fig. 1A). To determine the lower limit of detection of pneumococcal DNA, the sensitivity of the PCR assay was evaluated with a 10-fold dilution of the purified S. pneumoniae genomic DNA. The PCR assay was able to detect ≥10 CFU of S. pneumoniae per ml (Fig. 1B). The PCR assay was efficient at detecting pneumococcal DNA in serum or CSF from patients with positive culture results (Fig. 1C). Treatment with intravenous ceftriaxone given over 48 h reduced dramatically the capability of the PCR assay to detect pneumococcal DNA. The intensity of the band in the PCR with CSF, which was very high, was significantly reduced following antibiotic treatment. No pneumococcal DNA could be detected in serum 48 h after the initiation of antibiotic treatment.
PCR of clinical specimens and specimens from controls.
Serum PCR was positive for all patients with pneumococcal bacteremia and pneumococcal meningitis. In the four patients with pneumococcal meningitis, CSF was also positive by PCR (Table 1). The rates of positivity for patients with lobar or segmental pneumonia or AOM and healthy controls were 38, 44, and 17%, respectively (P < 0.02 for patients in all categories grouped together versus controls).
TABLE 1.
PCR results for serum specimens from patients with pneumococcal sepsis, meningitis, lobar or segmental pneumonia, or AOM and healthy controls
| Clinical presentation | No. of samples | No. (%) of samples positive by PCR |
|---|---|---|
| Pneumococcal bacteremia | 9 | 9 (100) |
| Pneumococcal meningitisa | 4 | 4 (100) |
| Lobar or segmental pneumonia | 34 | 13 (38) |
| AOM | 32 | 14 (44) |
| Healthy controls | 202 | 35 (17) |
Both CSF and serum samples were positive by culture and PCR.
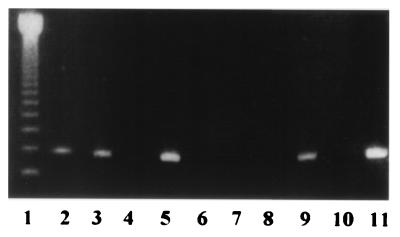
The intensity of the band in the PCR with culture-positive sera was not different from that of the band in the PCR with control sera when the control sera were positive (Fig. 2).
FIG. 2.
Agarose gel electrophoresis of PCR-amplified products from sera. Lane 1, molecular size marker; lanes 2 to 4 and 6 to 8, samples from healthy controls; lanes 5 and 9, samples from blood culture-positive patients; lane 10, negative control; lane 11, positive control.
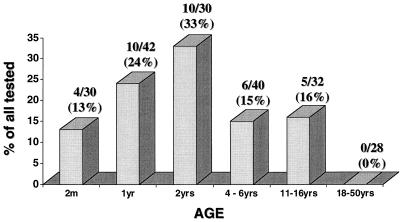
When the rate of positivity among healthy controls was examined in relation to age, a clear pattern could be observed: The PCR positivity rate increased with age from ages 2 to 24 months and decreased thereafter (Fig. 3). None of the 28 serum samples from adults was positive.
FIG. 3.
PCR results for serum samples from 202 healthy controls, by age.
The detailed PCR results for sera from patients with lobar or segmental pneumonia are presented in Table 2. All 3 samples with positive blood culture results were positive by PCR, whereas only 10 of 27 (37%) samples with negative blood culture results but positive NP swab culture results and 1 of the 4 samples with both negative blood culture and NP culture results were positive by PCR (P < 0.03; chi-square for linear trend in proportions). In the case of AOM, this trend was not observed, and a high rate of positivity ranging from 36 to 56% was observed, regardless of the positivity of the MEF or NP swab cultures. Five of the 11 patients (45%) with viral URIs who carried S. pneumoniae in their nasopharynges had a positive PCR result for pneumococci.
TABLE 2.
PCR results for sera from patients with lobar or segmental pneumonia, AOM, and or viral URIs in relation to positive pneumococcal blood culture, positive pneumococcal MEF culture, positive pneumococcal NP swab culture, or negative pneumococcal cultures
| Clinical presentation and culture resultsa | No. of specimens positive/no. tested | % Positive | P for trend in proportions |
|---|---|---|---|
| Lobar or segmental pneumonia (n = 34) | |||
| + blood | 3/3 | 100 | |
| − blood, + NP swab | 10/27 | 37 | <0.03 |
| − blood, − NP swab | 0/4 | 0 | |
| AOM (n = 32) | |||
| + MEF, + NP swab | 5/12 | 42 | |
| − MEF, + NP swab | 5/9 | 56 | Not significant |
| − MEF, − NP swab | 4/11 | 36 | |
| Viral URI (n = 11) | 5/11 | 45 | − |
+, positive for S. pneumoniae by culture; −, negative for S. pneumoniae by culture.
A relatively high proportion (24 of 102 [24%]) of the samples from healthy subjects ages 2, 12, or 24 months were positive by PCR. Therefore, we compared healthy subjects from whom S. pneumoniae was isolated from the nasopharynges to those with negative NP swab culture results (Table 3). PCR of serum was often positive for both NP swab culture-positive and NP swab culture-negative subjects, although a trend toward a higher rate of positivity was observed for NP swab culture-positive subjects (P = 0.08).
TABLE 3.
PCR results for sera from 102 healthy control subjects ages 2, 12, and 24 months by NP pneumococcal carriage
| Age group (mo) | No. of subjects tested/no. positive (%)
|
|
|---|---|---|
| NP swab culture negative | NP swab culture positive | |
| 2 (n = 30) | 0/15 (0) | 4/15 (27) |
| 12 (n = 42) | 5/22 (23) | 5/20 (25) |
| 24 (n = 30) | 3/15 (20) | 7/15 (47) |
| All (n = 102) | 8/52 (15)a | 16/50 (32)a |
P = 0.08.
When the results for sera from all children with positive NP swab cultures were compared to those for sera from children with negative NP swab cultures, a significantly higher rate of positivity was found among those with positive pneumococcal NP swab culture results (P = 0.03) (Table 4).
TABLE 4.
PCR results for sera from patients with viral URI, lobar or segmental pneumonia, or nonpneumococcal AOM and healthy controls according to positivity of NP swab culture for pneumococci
| Clinical presentation | No. of samples tested/ no. positive (%)a
|
|
|---|---|---|
| NP − | NP + | |
| Viral URI | ND | 5/11 (45) |
| Lobar or segmental pneumonia | 0/4 (0) | 10/27 (37) |
| Nonpneumococcal AOM | 4/11 (36) | 5/9 (56) |
| Healthy controls | 9/52 (17) | 15/50 (30) |
| Total | 13/67 (19) | 35/97 (36) |
NP −, negative NP swab culture result; NP +, positive nasopharyngeal NP swab culture result; ND, not determined.
P = 0.03.
DISCUSSION
The sensitivity and specificity of the pneumolysin PCR assay for S. pneumoniae were demonstrated by its capability to detect 10 CFU of this organism per ml and its negative results for other organisms including closely related streptococci. Antibiotic treatment for 48 h reduced the rate of detection of pneumolysin in CSF by PCR and abolished its detection in serum. To determine the sensitivity of the test with clinical specimens, we used sera from five adults and four children with culture-proven pneumococcal bacteremia; all of these sera were positive by PCR. However, for 17% of all controls, PCR results were also positive.
A high rate of sensitivity with what could be regarded as a relatively high rate of false-positive results, as found in our study, was also demonstrated in two other studies testing the diagnostic value of PCR of blood or serum. One earlier study (26) detecting pneumolysin in adults by PCR showed that all serum samples from patients with acute pneumococcal pneumonia, confirmed by blood culture, were positive, while 6% of serum samples from healthy elderly controls were positive. Another study testing for the gene for penicillin-binding protein 2B in whole blood had a sensitivity of 80% and a specificity of 84% (31). A possible way of overcoming the high rate of apparently false-positive results might have been the use of a lower PCR assay strength. However, this manipulation could have resulted in false-negative results, since the intensities of the positive PCR bands were similar for both serum with a positive culture result and serum from controls. The PCR method used in our study was at least as sensitive as those used in other studies (10, 25, 31).
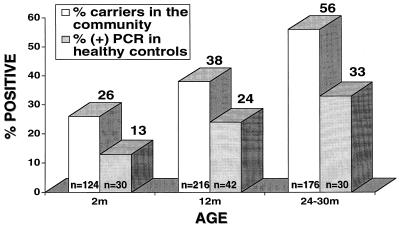
We observed a clear association of PCR-positive sera with age: positivity increased from 13% at 2 months of age to 33% at 2 years of age and then decreased. Since PCR-positive sera were found more frequently among those with positive NP swab cultures than among those with negative cultures, we examined whether the increase in the rate of positivity by PCR with age was associated with a similar increase in the NP pneumococcal carriage rate. This was possible, since during the period of the present study we also studied the epidemiology of NP pneumococcal carriage in healthy children in our community (5), and the serum samples for PCR were obtained from infants belonging to the group from whom the epidemiologic data were obtained. Figure 4 shows clearly that the age-related rate of NP carriage of pneumococci in the community paralleled the rate of positivity of PCR of serum for our subjects. We did not study the carriage rate in older children or adults, but other studies showed that very young infants, older children, and adults are much less likely to carry S. pneumoniae in the nasopharynges than older infants and young children attending day care. Dunlap and Harvey (7) found that the rate of pneumococcal colonization of the nasopharynges at 1 year of age was about 1.2 times higher than at 4 to 5 years of age, about two times higher than that at 7 to 10 years of age, and 17 times higher than that at >25 years of age. Hendley et al. (13) found the carriage rates to be 38, 29, 9, and 19% among subjects ≤5, 6 to 12, 13 to 17, and ≥18 years of age, respectively (13).
FIG. 4.
Age relation of the proportion of positive serum samples by PCR in relation to NP pneumococcal carriage rate among healthy infants and children in the community younger than age 30 months. The data for the NP carriage rate in the community were derived from a previous report (23).
The age effect on the positivity of serum by PCR is clinically important, since most pneumococcal infections occur in those in the age group in which the rate of colonization is highest, namely, ages 6 to 24 months. Furthermore, an increased colonization rate is detected during the seasons that are mainly associated with viral infections that can predispose the individual to systemic and mucosal pneumococcal infections (6, 11). The colonization rates and the concentrations of pneumococci among infants and children with respiratory infections were found to be higher than those among healthy controls (8). Thus, our higher rate of PCR-positive sera during illness might be related, at least in part, to the fact that most sera tested during illness were obtained from children belonging to the age groups in which the heaviest colonization occurs and that the diseases for which we tested (pneumonia, AOM, and URIs) occur often in winter in association with viral illnesses, factors responsible for the higher NP pneumococcal colonization rates. In this regard, a recently published article reporting a high rate of detection of otitis media pathogens, including S. pneumoniae in culture-negative MEF by PCR, should be mentioned (23). The authors speculated that a positive PCR test result indicated the presence in MEF of viable organisms that were undetected by culture. We believe that aspiration of DNA fragments from the nasopharynx into the middle-ear cavity can serve as an alternative speculation.
Although we found a higher rate of PCR-positive sera among subjects who carried S. pneumoniae in their nasopharynges, a nonnegligible rate of PCR-positive sera was also found among subjects in whom NP pneumococcal colonization could not be demonstrated. This was not surprising, since NP swab culture is not the most sensitive method of detecting carriage of pneumococci in the nasopharynges. Other methods, such as the mouse inoculation method, are more sensitive (14), although the latter is not used nowadays for convenience reasons. Since at any given moment a high proportion of healthy subjects and patients have positive NP pneumococcal culture results, it can be assumed that a considerable proportion of those with negative culture results either have low concentration of pneumococci or had recently had a positive culture result and may still have small quantities of circulating pneumococcal DNA which invaded the blood either directly or by phagocytosis of organisms by lymphoid cells that later circulated in the bloodstream. Further support for this speculation is provided by the fact that a systemic immune response to the serotype being carried may be seen following NP pneumococcal carriage (19).
We conclude that the PCR assay used in our study is a sensitive method for detecting pneumococcal bacteremia, but the high rate of detection of pneumococcal DNA in healthy controls associated with NP carriage excluded the test from being useful in determining deep-seated pneumococcal infections, at least in children.
REFERENCES
- 1.Appelbaum P C. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin Infect Dis. 1992;15:77–83. doi: 10.1093/clinids/15.1.77. [DOI] [PubMed] [Google Scholar]
- 2.Cerosalettis K M, Roghmann C M, Bentley D W. Comparison of latex particle agglutination and counterimmunoelectrophoresis for the detection of pneumococcal antigen in elderly pneumonia patients. J Clin Microbiol. 1985;22:553–557. doi: 10.1128/jcm.22.4.553-557.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagan R, Abramson O, Leibovitz E, Lang R, Goshen S, Greenberg D, Yagupsky P, Leiberman A, Fliss D. Impaired bacteriologic response to oral cephalosporins in acute otitis media caused by pneumococci with intermediate resistance to penicillin. Pediatr Infect Dis J. 1996;15:980–985. doi: 10.1097/00006454-199611000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Dagan R, Melamed R, Muallem M, Piglansky L, Greenberg D, Abramson O, Mendelman P M, Bohidar N, Yagupsky P. Reduction of nasopharyngeal carriage of pneumococci during the second year of life by a heptavalent conjugate pneumococcal vaccine. J Infect Dis. 1996;174:1271–1278. doi: 10.1093/infdis/174.6.1271. [DOI] [PubMed] [Google Scholar]
- 5.Dagan R, Melamed R, Muallem M, Piglansky L, Yagupsky P. Nasopharyngeal colonization in southern Israel with antibiotic-resistant pneumococci during the first 2 years of life: relation to serotypes likely to be included in pneumococcal conjugate vaccines. J Infect Dis. 1996;174:1352–1355. doi: 10.1093/infdis/174.6.1352. [DOI] [PubMed] [Google Scholar]
- 6.Dowling J N, Sheehe P R, Feldman H A. Pharyngeal pneumococcal acquisition in “normal” families: a longitudinal study. J Infect Dis. 1971;124:9–17. doi: 10.1093/infdis/124.1.9. [DOI] [PubMed] [Google Scholar]
- 7.Dunlap M B, Harvey H S. Host influence on upper respiratory flora. N Engl J Med. 1956;255:640–646. doi: 10.1056/NEJM195610042551403. [DOI] [PubMed] [Google Scholar]
- 8.Faden H, Staniewich J, Brodsky L, Bernstein J, Ogra P L. Changes in nasopharyngeal flora during otitis media of childhood. Pediatr Infect Dis J. 1990;9:623–626. [PubMed] [Google Scholar]
- 9.Friedland T R, McCracken G H., Jr Management of infections caused by antibiotic-resistant Streptococcus pneumoniae. N Engl J Med. 1994;331:377–382. doi: 10.1056/NEJM199408113310607. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie S H, Ullman C, Smith M D, Emery V. Detection of Streptococcus pneumoniae in sputum samples by PCR. J Clin Microbiol. 1994;32:1308–1311. doi: 10.1128/jcm.32.5.1308-1311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray B M, Turner M E, Dillon H C., Jr Epidemiologic studies of Streptococcus pneumoniae in infants. The effect of season and age on pneumococcal acquisition and carriage in the first 24 months of life. Am J Epidemiol. 1982;116:692–703. doi: 10.1093/oxfordjournals.aje.a113452. [DOI] [PubMed] [Google Scholar]
- 12.Hassan-King M, Baldeh I, Secka O, Falade A, Greenwood B. Detection of Streptococcus pneumoniae DNA in blood cultures by PCR. J Clin Microbiol. 1994;32:1721–1724. doi: 10.1128/jcm.32.7.1721-1724.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendley O J, Sande M A, Stewart P M, Gwaltney J M., Jr Spread of Streptococcus pneumoniae in families. I. Carriage rates and distribution of types. J Infect Dis. 1975;132:55–61. doi: 10.1093/infdis/132.1.55. [DOI] [PubMed] [Google Scholar]
- 14.Hodge R G, MacLeod C M, Bernhard W G. Epidemic pneumococcal pneumonia. III. Pneumococcal carrier studies. Am J Hyg. 1946;44:207–230. doi: 10.1093/oxfordjournals.aje.a119090. [DOI] [PubMed] [Google Scholar]
- 15.Isaacman D J, Karasic R B. Utility of collecting blood cultures through newly inserted intravenous catheters. Pediatr Infect Dis J. 1990;9:815–818. doi: 10.1097/00006454-199011000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Korppi M, Koskela M, Jalonen E, Leinonen M. Serologically indicated pneumococcal respiratory infection in children. Scand J Infect Dis. 1992;24:437–443. doi: 10.3109/00365549209052629. [DOI] [PubMed] [Google Scholar]
- 17.Leinonen M. Serological diagnosis of pneumococcal pneumonia—will it ever become a clinical reality. Semin Respir Infect. 1994;3:189–191. [PubMed] [Google Scholar]
- 18.Munford R S, Murphy T V. Antimicrobial resistance in Streptococcus pneumoniae: can immunization prevent its spread? J Invest Med. 1994;42:613–621. [PubMed] [Google Scholar]
- 19.Musher D M, Groover J E, Reichler M R, Riedo F X, Schwartz B, Watson D A, Baughn R E, Breiman R F. Emergence of antibody to capsular polysaccharides of Streptococcus pneumoniae during outbreaks of pneumonia: association with nasopharyngeal colonization. Clin Infect Dis. 1997;24:441–446. doi: 10.1093/clinids/24.3.441. [DOI] [PubMed] [Google Scholar]
- 20.Nohynek H, Eskola J, Laine E, Halonen P, Ruutu P, Saikku P, Kleemola M, Leinonen M. The causes of hospital-treated acute lower respiratory infections in children. Am J Dis Child. 1991;145:618–622. doi: 10.1001/archpedi.1991.02160060036016. [DOI] [PubMed] [Google Scholar]
- 21.Norrby S R, Pope K A. Pneumococcal pneumonia. J Infect. 1979;1:109–120. [Google Scholar]
- 22.Perlino C A. Laboratory diagnosis of pneumonia due to Streptococcus pneumoniae. J Infect Dis. 1984;150:139–144. doi: 10.1093/infdis/150.1.139. [DOI] [PubMed] [Google Scholar]
- 23.Post J C, Preston R A, Aul J J, Larkins-Pettigrew M, Rydquist-White J, Anderson K W, Wadowsky R M, Reagan D R, Walker E S, Kingsley L A, Magit A E, Erlich G D. Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA. 1995;273:1598–1604. [PubMed] [Google Scholar]
- 24.Robbins J B, Austrian R, Lee C J, Rastogi S C, Schiffman G, Henrichsen J, Makela P H, Broome C V, Facklam R R, Tiesjema R H, Parke J C., Jr Considerations for formulating the second-generation pneumococcal capsular polysaccharide vaccine with emphasis on the cross-reactive types within groups. J Infect Dis. 1983;148:1136–1159. doi: 10.1093/infdis/148.6.1136. [DOI] [PubMed] [Google Scholar]
- 25.Rudolph K M, Parkinson A J, Black C M, Mayer L W. Evaluation of polymerase chain reaction of diagnosis of pneumococcal pneumonia. J Clin Microbiol. 1993;31:2661–2666. doi: 10.1128/jcm.31.10.2661-2666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salo P, Örtqvist A, Leinonen M. Diagnosis of bacteremic pneumococcal pneumonia by amplification of pneumolysin gene fragment in serum. J Infect Dis. 1995;171:479–482. doi: 10.1093/infdis/171.2.479. [DOI] [PubMed] [Google Scholar]
- 27.Tomasz A. The pneumococcus at the gates. N Engl J Med. 1995;333:514–515. doi: 10.1056/NEJM199508243330810. [DOI] [PubMed] [Google Scholar]
- 28.Virolainen A, Salo P, Jero J, Karma P, Eskola J, Leinonen M. Comparison of PCR assay with bacterial culture for detecting Streptococcus pneumoniae in middle ear fluid of children with acute otitis media. J Clin Microbiol. 1994;32:2667–2670. doi: 10.1128/jcm.32.11.2667-2670.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker J A, Allen R L, Falmagne P, Jonson M K, Boulnois G J. Molecular cloning, characterization, and complete nucleotide sequence of the gene for pneumolysin, the sulfhydryl-activated toxin of Streptococcus pneumoniae. Infect Immun. 1987;55:1184–1189. doi: 10.1128/iai.55.5.1184-1189.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitby M, Kristinsson K G, Brown M. Assessment of rapid methods of pneumococcal antigen detection in routine sputum bacteriology. J Clin Pathol. 1985;38:341–344. doi: 10.1136/jcp.38.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Isaacman D J, Wadowsky R M, Rydquist-White J, Post J C, Ehrlich G D. Detection of Streptococcus pneumoniae in whole blood by PCR. J Clin Microbiol. 1995;33:596–601. doi: 10.1128/jcm.33.3.596-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]