Abstract
PCR, using primers PIp1 and PIp2, was evaluated for the detection of DNA from Bordetella pertussis in bacterial strains and in nasopharyngeal samples from patients with a cough lasting at least 7 days. The assay could detect DNA from 6 CFU of B. pertussis/10 μl of sample. Results of the PCR assay were compared with those of cultures, a determination of serum antibodies against pertussis toxin and filamentous hemagglutinin, and a clinical evaluation of 2,442 coughing episodes. The overall sensitivity of PCR was 65% (623 of 956), which was higher than the sensitivity of cultures (58%) (P < 0.001). Factors influencing the sensitivity of PCR were the interval between the onset of symptoms and sampling and the vaccination status of the patient. The specificity of PCR was 98% (1,451 of 1,486). The positive and negative predictive values were 95 and 81%, respectively. Parapertussis PCR, using primers BPPA and BPPZ, was positive in 11 of 18 culture-positive cases and was confirmed by serology in another 4 cases. In conclusion, PCR is a valuable complement to cultures and can probably replace cultures for diagnosis of B. pertussis and Bordetella parapertussis infections.
Pertussis can be diagnosed by cultures and serological methods. Isolation of Bordetella pertussis from nasopharyngeal secretions has a specificity close to 100%, but the sensitivity is only about 60% if the culture is taken early during the disease and even lower the longer the disease persists (11, 14). Demonstrations of significant increases in serum antibodies against pertussis toxin (PT) or filamentous hemagglutinin (FHA) are also well-documented methods for diagnosing pertussis with high specificity and sensitivity (3, 26, 27). Serology requires paired sera taken about 1 month apart. Hence, results are not available until the patient is recovering. Thus, there is a need for a specific and sensitive diagnostic method for pertussis that can be used in the early stage of the disease. Detection of B. pertussis-specific DNA by PCR is a promising new technique for rapid and reliable diagnosis of pertussis. Although several studies have shown that PCR is more sensitive than cultures (1, 4, 7, 16, 24), questions about the specificity of PCR have been raised (4, 19). The aim of the present study was to evaluate the sensitivity and specificity of PCR for diagnosis of B. pertussis and Bordetella parapertussis infections by comparison of PCR with cultures, serology, and clinical diagnosis.
MATERIALS AND METHODS
Study design.
All individuals in the present study were participants or family members of participants in a double-blind, placebo-controlled efficacy trial of a monocomponent pertussis toxoid vaccine performed in the Göteborg area of western Sweden (21, 22). In Sweden, there was no licensed pertussis vaccine between 1979 and 1996. Because a Swedish-made whole-cell vaccine was ineffective, pertussis had already recurred during the early 1970s, with a yearly incidence rate in preschool children of about 10% (9, 10).
A total of 3,450 healthy infants were randomized to vaccination with diphtheria-tetanus toxoids with or without pertussis toxoid at 3, 5, and 12 months of age. There were 10,200 family members: 6,900 parents and 3,300 siblings. Families were enrolled in the study between September 1991 and June 1992. The study was completed in January 1995.
The vaccine trial was approved by the institutional review board of the National Institute of Child Health and Human Development; the Food and Drug Administration; the Medical Products Agency, Uppsala, Sweden; and the Ethics Committee, Göteborg University. All parents gave their written consent after receiving oral and written information.
Follow-up of coughing episodes.
Parents were instructed to contact the study nurse if anyone in the family coughed for ≥7 days. In addition a study nurse called each family once a month. Clinical and laboratory investigations were performed in the same way for study children and family members. A nasopharyngeal sample for culture and PCR and a serum sample for antibody determination were obtained. A convalescent serum sample was taken ≥4 weeks later. Information about exposure to pertussis within and outside the family, duration of the cough, paroxysms, whooping, vomiting, fever, rhinitis, and use of antibiotics was recorded.
Laboratory assays. (i) Nasopharyngeal secretions.
Nasopharyngeal secretions were collected by swabbing with a rayon swab (PurFybr Inc., Munster, Ind.) and transported to the laboratory in a modified Stuart medium. The same samples were used for culture and PCR.
(ii) Culture.
The swab was inoculated on Regan-Lowe medium (15) and then inserted into a tube containing enrichment medium. The enrichment medium had the same composition as the culture medium but with only half the concentration of charcoal agar (5, 6). The plates and the enrichment tubes were inoculated at 35 to 37°C. A subculture from the enrichment medium was made after 72 h of incubation. The primary plates were inspected from the third through the seventh days after inoculation. The subcultures from the enrichment medium were incubated at 35 to 37°C for at least 4 days. Colonies of B. pertussis and B. parapertussis were verified by Gram staining, agglutination with specific antisera (Difco, Detroit, Mich.), and biochemical tests (6).
(iii) Pretreatment of the clinical samples for PCR.
While the original swab was put into the enrichment tube for culture, a new sterile swab (OTE Sjukvårdsprodukter, Billdal, Sweden) was put into the transport tube to obtain material for PCR. The swab was tested to ensure that it did not inhibit the PCR (data not shown). The swab was kept in the transport medium for about 10 min before being inserted into an Eppendorf tube containing 100 μl of sterile, double-distilled water. After 5 min of incubation at room temperature, the tube was shaken for 60 s and the swab was removed. The tubes were incubated in a heat block at 95 to 100°C for 10 min. The samples were stored at 4 to 8°C until they were used for PCR. If the samples had to be stored for more than 48 h they were kept frozen at −20°C.
(iv) B. pertussis PCR.
A modified PCR method was used as described by Houard et al. (8). Briefly, the B. pertussis PCR gave rise to a 121-bp fragment from the insertion sequence IS481, with forward primer PIp1 (5′ CCC ATA AGC ATG CCC GAT TGA C 3′) and reverse primer PIp2 (5′ CGC ACA GTC GGC GCG GTG AC 3′), corresponding to bp 110 to 131 and 211 to 230, respectively. The oligonucleotides were purchased from Scandinavian Gene Synthesis (SGS) AB, Köping, Sweden.
The PCR was performed in a total volume of 50 μl containing 20 mM (NH4)2SO4, 75 mM Tris-HCl (pH 9.0), 0.01% (wt/vol) Tween, 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate (dUTP was used instead of dTTP), 30 pmol of PIp1, 15 pmol of PIp2, 2 U of Taq DNA polymerase (Advanced Biotechnologies, Leatherhead, United Kingdom), and 10 μl of the pretreated clinical sample. To be able to use the uracil-DNA-glycosylase enzyme (Roche Molecular Systems, Inc. Branchbury, N.J.), we used dUTP instead of dTTP to destroy any DNA already amplified (13).
The master mixture (containing all of the ingredients mentioned above except the sample) was prepared without MgCl2 in a special area, with pipettes and tips which were used only there, overlaid with 50 μl of mineral oil, and frozen at −20°C until used for PCR, but no longer than 1 month. When used, the tubes were thawed and 6 μl of 12.5 mM MgCl2 was added to each tube under the mineral oil.
The samples were amplified in a thermal reactor (Hybaid, Teddington, United Kingdom). DNA was denatured at 94°C for 30 s. Annealing and DNA chain extension were performed at 66°C for 2.5 min. After 40 cycles, a temperature delay step of 5 min at 66°C was done to complete the elongation. The thermal reactor was programmed to hold at 72°C until stopped, in case the UNG enzyme had to be used. Preparation of the master mixture, pretreatment of clinical samples, and amplification were performed in three different areas to avoid contamination.
Fifteen microliters of each amplified PCR product stained with ethidium bromide, was analyzed on a 2.0% agarose gel. The PCR products were visualized under UV light, and the gel was photographed.
(v) Specificity tests.
To test the specificity of the PCR method for B. pertussis, suspensions of species related to B. pertussis and other potential pathogens and commensals of the respiratory tract were examined (Fig. 1 and 2). One colony of each species was put in 100 μl of sterile double-distilled water, vortexed, and boiled for 10 min. Pertussis PCR was performed as described above.
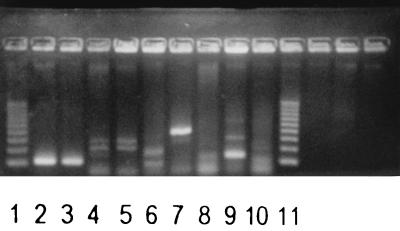
FIG. 1.
Electrophoretic analysis of the PCR products obtained by amplification of DNA belonging to B. pertussis and related species with primers PIp1 and PIp2. Lanes 1 and 11, 100-bp “S′ladder low” (Advanced Biotechnologies); lane 2, B. pertussis; lane 3, B. holmesii; lane 4, Bordetella bronchiseptica; lane 5, B. parapertussis; lane 6, Bordetella avium; lane 7, Bordetella trematum; lane 8, Bordetella hinzii; lane 9, Alcaligenes xylosoxidans; lane 10, P. putida.
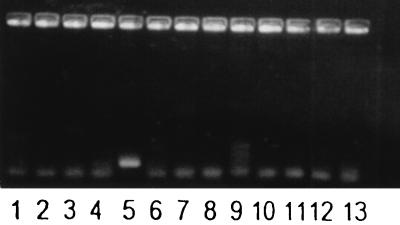
FIG. 2.
Amplification of DNA from B. pertussis and other species appearing in the human respiratory tract with primers PIp1 and PIp2. Lane 1, Moraxella catarrhalis; lane 2, Escherichia coli; lane 3, Haemophilus influenzae; lane 4, Haemophilus parainfluenzae; lane 5, B. pertussis; lane 6, negative control; lane 7, Klebsiella pneumoniae; lane 8, Legionella pneumophila; lane 9, Pseudomonas aeruginosa; lane 10, Staphylococcus aureus; lane 11, coagulase-negative staphylococci; lane 12, Streptococcus pneumoniae; lane 13, Streptococcus pyogenes.
(vi) Controls.
As negative controls, sterile test tubes with sterile, double-distilled water and the master mixture were used. A negative control was processed after every fourth test tube. All specimens which were culture positive and PCR negative were processed once more. No inhibitory phenomenon was found in the examined specimens (data not shown).
(vii) Southern blot analysis of PCR products.
Hybridization was performed by standard procedures (17). Briefly, 15 μl of the PCR product was analyzed on a 2% agarose gel. DNA was transferred from the agarose gel to a nylon membrane (Hybond-N+; Amersham International pl., Little Chalfont, United Kingdom) by diffusion blotting in 0.4 M NaOH overnight. Membranes were preincubated for 20 min at 40°C in a hybridization solution containing 5× SSPE (20× SSPE contains 3 M NaCl, 0.2 M NaH2PO4 · H2O, 20 mM EDTA [pH 7.4]), 0.5% sodium dodecyl sulfate, 5× Denhardt solution (50× Denhardt solution contains 1% Ficoll, 1% polyvinylpyrrolidone, and 1% bovine serum albumin), and 10 μg of sonicated salmon sperm DNA/ml. Hybridization was performed for 20 min at 40°C in 10 ml of hybridization solution with 5 μl of streptavidin peroxidase dehydrogenase conjugate (500 U/ml; Boehringer Mannheim) and 1 pmol of 5′ biotinylated PI probe (5′ GTG CCT GAA GCG GCC CGC GC 3′; SGS AB)/ml (18). Subsequent washings were carried out three times in 20 ml of a solution containing 1× SSPE and 0.1% sodium dodecyl sulfate for 10 min each time at 40°C. Detection procedures were performed according to the supplier’s instructions with an ECL detection system (Amersham International pl.). Autoradiography was conducted for 3 to 5 min with Kodak X-OMAT S film and intensifying screens.
(viii) B. parapertussis PCR.
A modified PCR method was used as described by van der Zee et al. (23). The specific DNA fragment (498 bp) is from IS1001. The oligonucleotide primers BPPA (5′ CGC CGC TTG ATG ACC TTG ATA 3′), corresponding to bp 1211 to 1232, and BPPZ (5′ CAC CGC CTA CGA GTT GGA GAT 3′), corresponding to bp 734 to 755, purchased from SGS AB, were used. The PCR was performed as described for B. pertussis, except that 12 pmol of primer BPPA and 12 pmol of primer BPPZ were used instead of PIp1 and PIp2. In 67 of 2,442 clinical samples, parapertussis PCR was not performed.
(ix) Serology.
Antibodies against PT and FHA immunoglobulin G (IgG) were measured by enzyme-linked immunosorbent assay in duplicate in eight threefold dilutions starting from 1/10 (20, 22). PT lot 97/98 was obtained from North American Vaccine (Beltsville, Md.), and FHA lot 10,000 was obtained from the Institute Pasteur-Mérieux (Marcy L’Etoile, France). The reference was Food and Drug Administration pertussis antiserum lot 3. Titers were defined as the reciprocal serum dilution corresponding to an absorbance of 0.2 above the background. Based on intra-assay and intraindividual variation, threefold rises were considered significant if the titer in the convalescent serum was ≥200. Titers of ≥6,000 for anti-PT IgG and anti-FHA IgG concurrently in the same convalescent serum were considered a sign of pertussis if an acute serum was not available (20, 22). Anti-PT IgM and IgA antibodies were determined by enzyme-linked immunosorbent assay in patients with significant FHA IgG increase and without significant PT IgG increase in order to differentiate between pertussis and parapertussis (20, 22).
A detailed description of all laboratory methods has been given in a technical report from the vaccine trial (20).
Case definitions.
In the present study, all coughing episodes of the study participants and family members were classified into five categories based on results of cultures, serology, family exposure, and clinical symptoms but without the use of PCR (Table 1).
TABLE 1.
Classification of coughing episodes involving pertussis and parapertussis based on cultures, serology, family exposure, and clinical symptoms
| Classification and criteria |
|---|
| Pertussis |
| 1. B. pertussis isolated from the nasopharynx |
| 2. At least one of the following criteria fulfilled: |
| a. Significant increase in PT IgG |
| b. Significant increase in FHA IgG without other criteria for parapertussis |
| c. Both PT IgG and FHA IgG ≥6,000 in the same convalescent serum |
| d. Family member with pertussis verified by culture or serology |
| 3. Clinical pertussis with at least 3 weeks of paroxysmal cough and known exposure to pertussis outside the family; serum samples lacking or suboptimal timing in relation to onset of symptoms |
| 4. Known exposure to pertussis within or outside the household; clinical symptoms not evaluable because of early erythromycin treatment |
| Parapertussis |
| 1. B. parapertussis isolated from the nasopharynx |
| 2. Significant FHA IgG increase without an increase in PT IgG, IgM, and IgA antibodies |
RESULTS
PCR assay.
Amplifications were carried out with PIp1 and PIp2 as primers and purified DNA from B. pertussis, giving rise to the expected 121-bp fragment. On inspection of ethidium bromide-stained gels, a single fragment was clearly visible when 50 fg of B. pertussis template DNA was present, equivalent to 10 genome copies, assuming a genome size of 3,800 kbp (18). A band of the expected size was also visible when PCR was performed on cultures of B. pertussis serially diluted in double-distilled water and boiled. The same volume of sample that was used for PCR was cultured before the boiling procedure, revealing that 6 CFU per 10 μl of sample, visible by culture, was detected by PCR. This finding is consistent with that of a previous study (8).
Visualization of amplified DNA from B. pertussis and other human respiratory tract pathogens or species genetically related to B. pertussis is shown in Fig. 1 and 2. Amplification of DNA from species closely related to B. pertussis (Fig. 1, lanes 2 to 9) gave rise to DNA fragments of the wrong size in most cases. However, amplification of DNA belonging to Bordetella holmesii (Fig. 1, lane 3) gave rise to a fragment indistinguishable from that of DNA belonging to B. pertussis. This band also hybridized to the PI probe (data not shown). Amplified DNA from Pseudomonas putida is shown in Fig. 1, lane 10. DNA from this species also gave a band of about the same size as that of B. pertussis when amplified with the PIp1 and PIp2 primers. This band, however, did not hybridize to the PI probe (data not shown). Amplification of DNA from 11 other human respiratory tract pathogens or commensals was negative, as they did not give rise to any PCR product (Fig. 2).
Hybridization.
Sixty-seven samples randomly selected from patients in the study were hybridized with a probe. All 17 of those which were positive according to the expected 121-bp fragment also gave a positive signal with hybridization. The 50 samples that were negative by PCR were also negative by hybridization with the probe.
PCR of nasopharyngeal samples from patients with a cough.
A total of 2,442 nasopharyngeal specimens were available for PCR and culture. The results of PCR in relation to cultures, serology, clinical, and epidemiological data are given in Table 2. Assuming that not only the 942 patients with positive culture, serologic evidence, or known household exposure had pertussis but also the 14 patients with only clinical and epidemiological data, the overall sensitivity of PCR was 65% (623 of 956), the specificity was 98% (1,451 of 1,486), the positive predictive value was 95% (623 of 658), and the negative predictive value was 81% (1,451 of 1,784) (Table 3).
TABLE 2.
Results of cultures, serology, and clinical evaluation in patients with positive and negative pertussis PCR
| Results | Total patients with:
|
|
|---|---|---|
| Positive PCR | Negative PCR | |
| Pertussis confirmed by culture | 522 | 36 |
| Pertussis confirmed by serology and/or household exposure | 87 | 297 |
| Clinical pertussis; sera unavailable or suboptimal timing | 9 | |
| Exposure to pertussis; early erythromycin treatment | 5 | |
| Nonspecific cough; no laboratory data indicating pertussis | 35a | 1,429 |
| Parapertussis | 22 | |
Including one case with known contamination during sampling (11).
TABLE 3.
Results of PCR versus culture, serology, exposure, or clinical pertussis
| PCR | No. of patients with:
|
Total | |
|---|---|---|---|
| Pertussis diagnosed by culture, serology, exposure, or clinical symptoms | Nonspecific cough; no laboratory data indicating pertussis | ||
| Positive | 623 | 35 | 658c |
| Negative | 333 | 1,451 | 1,784d |
| Total | 956a | 1,486b | 2,442 |
Sensitivity, 65% (623 of 956).
Specificity, 98% (1,451 of 1,486).
Positive predictive value, 95% (623 of 658).
Negative predictive value, 81% (1,451 of 1,784).
The sensitivity of PCR was higher than the sensitivity of cultures (65% [623 of 956] versus 58% [558 of 956]; P < 0.001 by the binomial distribution test and McNemar’s test), but it should be noted that PCR was negative in 36 of 558 culture-positive cases (6%).
Of the 35 patients who had a positive PCR but no other laboratory or clinical indications of B. pertussis infection (false positive), one case was the result of contamination with B. pertussis DNA from whole-cell vaccine from equipment used for taking the nasopharyngeal sample at a pediatric outpatient clinic (19).
The capability to diagnose pertussis with PCR and culture decreased with time from the onset of symptoms. The median time from onset to sampling in 522 patients with positive culture and positive PCR was 9 days. In 297 patients with serology-confirmed pertussis who had negative cultures and negative PCR, the corresponding time was 15 days (P < 0.01; unpaired t test).
PCR was less often positive in fully vaccinated than in nonvaccinated children in the study with pertussis confirmed by other methods. In the randomized study the assay was positive for 70 of 148 (47%) participants who had received three doses of pertussis toxoid and in 205 of 324 (63%) nonvaccinated control children (P < 0.001; Fisher’s exact test).
Parapertussis PCR was positive for 11 of 18 culture-positive patients and for another 4 patients with serology-confirmed parapertussis (i.e., increase in FHA IgG without increase in PT IgG, IgM, or IgA). There was no case of positive parapertussis by PCR without other laboratory confirmation, i.e., no false-positive cases. PCR was negative for 7 culture-positive patients and 2,353 culture-negative patients.
DISCUSSION
The present study showed that PCR, using the primers PIp1 and PIp2, was specific for B. pertussis with one exception: B. holmesii gave rise to a band indistinguishable from that induced by B. pertussis. This band also hybridized to the PI probe. The risk of B. holmesii giving false-positive results by pertussis PCR is minimal because the organism has not been found in the respiratory tract and does not cause a disease similar to pertussis. The organism has been isolated from the blood of a few septic patients (12, 25). Bacterial suspensions of the other 5 Bordetella species and of another 13 bacterial species that can be found in the respiratory tract gave no false-positive reactions.
The study also showed that PCR for diagnosis of pertussis had a high specificity (98%) and a higher sensitivity than nasopharyngeal culture in a large body of clinical material. A sensitive method such as PCR, which can detect DNA from as few as 6 CFU, is extremely susceptible to contamination that may occur anywhere between the examination room and the laboratory. One of the false-positive assays in this study was due to contamination of the sampling material in a room that was also used for vaccination with a whole-cell pertussis vaccine (19). Though difficult to prove, another source of contamination in the examination room could have been that a child with pertussis was examined in the same room immediately before a participant in this study. Contamination in the laboratory is also a possibility, but this seems less likely in the present study, because there was no clustering of false-positive samples and because PCR and culture for pertussis were performed by different technicians on separate floors in the same building. The negative control used after every fourth sample was negative throughout the study. Transient PCR positivity can also be expected if an immune individual is exposed to pertussis (4, 7).
The sensitivity of cultures for diagnosing pertussis was 58% in this study, which is similar to or slightly higher than that found in previous studies (2, 4, 5, 11). However, some investigators have found a lower sensitivity in adults (4). PCR had a significantly higher sensitivity (65%), even though the study design did not favor PCR, as the original swab was used for culture and a new sterile swab was then put into the transport tube to get material for PCR. There is a possibility that this swab missed DNA-positive material, which would explain why 6% of culture-positive cases were PCR negative.
Several methods for detecting different DNA sequences of B. pertussis by PCR in nasopharyngeal secretions have been described elsewhere. Consistent with other studies, we noted that the sensitivity of PCR is higher than that of cultures (1, 4, 7, 16, 24), but culture-confirmed cases with negative PCR have been reported (16, 24). Previous studies have reported varying sensitivity of pertussis PCR. The overall sensitivity of PCR for diagnosing pertussis was 65% in the present study, where serology was used to confirm the great majority of cases. In two large studies that also used culture and serology for verifying the diagnosis, the sensitivities were 94 (16) and 21% (24), respectively. In these two studies, the target sequences were the PT operon and IS1001, respectively. Such comparisons, however, do not necessarily imply that one method is superior to another, because differences in the clinical material may also influence the results. In our study and in one previous study (24), the intervals between the onset of symptoms and sampling were found to influence the results. The present study also demonstrated that the vaccination status of the patient is of importance, since a higher proportion of samples from nonvaccinated children with pertussis than from vaccinated children with the disease were PCR positive.
The specificity of PCR can be estimated only if a highly sensitive reference method is available. In the present study and some previous studies it was possible to compare PCR and serology. In two of the largest studies, with 2,421 and 833 nasopharyngeal samples, the specificities were considered to be 99 and 98%, respectively (16, 24), i.e., specificities similar to that obtained in the present study.
The present study confirmed that PCR for detection of DNA from B. parapertussis can be a useful addition to cultures, as has been shown elsewhere (24). Four culture-negative patients with serologic evidence of parapertussis had positive parapertussis PCR, and there were no false-positive tests. When comparing numbers of patients with positive cultures and positive PCR for parapertussis, it should be emphasized again that the study design, with priority for cultures, may have treated PCR unfavorably.
In conclusion, PCR detecting a 121-bp fragment from the gene IS481 of B. pertussis had both high sensitivity and high specificity and can be used as a complement to or replacement for cultures. A positive PCR, however, must always be evaluated in relation to the clinical picture because transient colonization or contamination may lead to false-positive results at a low frequency. The decisive advantage of PCR over culture is its rapidity: PCR can be performed in 1 day, while results of culture are available in 4 to 7 days.
ACKNOWLEDGMENTS
We thank Elisabeth Pettersson for excellent technical assistance and Valter Sundh for data management.
The vaccine trial was supported by a contract (NO1-HD-9-2905) from the National Institute of Child Health and Human Development. PCR analyses were financed by Göteborg University and the Göteborg Medical Society (91/178).
REFERENCES
- 1.Bäckman A, Johansson B, Olcén P. Nested PCR optimized for detection of Bordetella pertussis in clinical nasopharyngeal samples. J Clin Microbiol. 1994;32:2544–2548. doi: 10.1128/jcm.32.10.2544-2548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Granström G, Wretlind B, Granström M. Diagnostic value of clinical and bacteriological findings in pertussis. J Infect. 1991;22:17–26. doi: 10.1016/0163-4453(91)90842-g. [DOI] [PubMed] [Google Scholar]
- 3.Granström M, Granström G, Lindfors A, Askelöf P. Serologic diagnosis of whooping cough by an enzyme-linked immunosorbent assay using fimbrial hemagglutinin as an antigen. J Infect Dis. 1982;146:741–745. doi: 10.1093/infdis/146.6.741. [DOI] [PubMed] [Google Scholar]
- 4.Grimprel E, Bégué P, Anjak I, Betsou F, Guiso N. Comparison of polymerase chain reaction, culture, and Western immunoblot serology for diagnosis of Bordetella pertussis infection. J Clin Microbiol. 1993;31:2745–2750. doi: 10.1128/jcm.31.10.2745-2750.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallander H O, Storsaeter J, Möllby R. Evaluation of serology and nasopharyngeal cultures for diagnosis of pertussis in a vaccine efficacy trial. J Infect Dis. 1991;163:1046–1054. doi: 10.1093/infdis/163.5.1046. [DOI] [PubMed] [Google Scholar]
- 6.Hallander H O, Reizenstein E, Renemar B, Rasmuson G, Mardin L, Olin P. Comparison of nasopharyngeal aspirates with swabs for culture of Bordetella pertussis. J Clin Microbiol. 1993;31:50–52. doi: 10.1128/jcm.31.1.50-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Q, Mertsola J, Soini H, Skurnik M, Ruuskanen O, Viljanen M K. Comparison of polymerase chain reaction with culture and enzyme immunoassay for diagnosis of pertussis. J Clin Microbiol. 1993;31:642–645. doi: 10.1128/jcm.31.3.642-645.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houard S, Hackel C, Herzog A, Bollen A. Specific identification of Bordetella pertussis by the polymerase chain reaction. Res Microbiol. 1989;140:477–487. doi: 10.1016/0923-2508(89)90069-7. [DOI] [PubMed] [Google Scholar]
- 9.Isacson J, Trollfors B, Taranger J, Zackrisson G, Lagergård T. How common is whooping cough in a nonvaccinating country? Pediatr Infect Dis J. 1993;12:284–288. doi: 10.1097/00006454-199304000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Krantz I, Taranger J, Trollfors B. Estimating incidence of whooping cough over time: a cross-sectional recall study of four Swedish birth cohorts. Int J Epidemiol. 1989;18:959–963. doi: 10.1093/ije/18.4.959. [DOI] [PubMed] [Google Scholar]
- 11.Kwantes W, Joynson D H M, Williams W O. Bordetella pertussis isolation in general practice: 1977–1979 whooping cough epidemic in West Glamorgan. J Hyg Lond. 1983;90:149–158. doi: 10.1017/s0022172400028825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindquist S W, Weber D J, Mangum M E, Hollis D G, Jordan J. Bordetella holmesii sepsis in an asplenic adolescent. Pediatr Infect Dis J. 1995;14:813–815. [PubMed] [Google Scholar]
- 13.Longo M C, Berninger M S, Hartley J L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 14.Onorato I M, Wassilak G F. Laboratory diagnosis of pertussis: the state of the art. Pediatr Infect Dis J. 1987;6:145–151. doi: 10.1097/00006454-198702000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Regan J, Lowe F. Enrichment medium for the isolation of Bordetella. J Clin Microbiol. 1977;6:303–309. doi: 10.1128/jcm.6.3.303-309.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reizenstein E, Lindberg L, Möllby R, Hallander H O. Validation of nested Bordetella PCR in pertussis vaccine trial. J Clin Microbiol. 1996;34:810–815. doi: 10.1128/jcm.34.4.810-815.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Stibitz S, Garletts T L. Derivation of a physical map of the chromosome of Bordetella pertussis Tohama I. J Bacteriol. 1992;174:7770–7777. doi: 10.1128/jb.174.23.7770-7777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taranger J, Trollfors B, Lind L, Zackrisson G, Beling-Holmquist K. Environmental contamination leading to false-positive polymerase chain reaction for pertussis. Pediatr Infect Dis J. 1994;13:936–937. doi: 10.1097/00006454-199410000-00020. [DOI] [PubMed] [Google Scholar]
- 20.Taranger J, Trollfors B, Lagergård T. Clinical trials of a monocomponent pertussis toxoid vaccine (NICHD Ptxd): a technical report. The Göteborg Pertussis Vaccine Study, Göteborg University. Göteborg, Sweden: Graphic Systems; 1995. [Google Scholar]
- 21.Taranger J, Trollfors B, Lagergård T, Lind L, Sundh V, Zackrisson G, Bryla D A, Robbins J B. Unchanged efficacy of a pertussis toxoid vaccine throughout the 2 years after the third vaccination of infants. Pediatr Infect Dis J. 1997;16:180–184. doi: 10.1097/00006454-199702000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Trollfors B, Taranger J, Lagergård T, Lind L, Sundh V, Zackrisson G, Lowe C U, Blackwelder W, Robbins J B. A placebo-controlled trial of a pertussis-toxoid vaccine. N Engl J Med. 1995;333:1045–1050. doi: 10.1056/NEJM199510193331604. [DOI] [PubMed] [Google Scholar]
- 23.Van der Zee A, Agterberg C, Peeters M, Schellekens J, Mooi F R. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J Clin Microbiol. 1993;31:2134–2140. doi: 10.1128/jcm.31.8.2134-2140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Zee A, Agterberg C, Peeters M, Mooi F, Schellekens J. A clinical validation of Bordetella pertussis and Bordetella parapertussis polymerase chain reaction: comparison with culture and serology using samples from patients with suspected whooping cough from a highly immunized population. J Infect Dis. 1996;174:89–96. doi: 10.1093/infdis/174.1.89. [DOI] [PubMed] [Google Scholar]
- 25.Weyant R S, Hollis D G, Weaver R E, Amin M F M, Steigerwalt A G, O’Connor S P, Whitney A M, Daneshvar M I, Moss C W, Brenner D J. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J Clin Microbiol. 1995;33:1–7. doi: 10.1128/jcm.33.1.1-7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zackrisson G, Krantz I, Lagergård T, Larsson P, Sekura R, Sigurs N, Taranger J, Trollfors B. Antibody response to pertussis toxin in patients with clinical pertussis measured by enzyme-linked immunosorbent assay. Eur J Clin Microbiol Infect Dis. 1988;7:149–154. doi: 10.1007/BF01963068. [DOI] [PubMed] [Google Scholar]
- 27.Zackrisson G, Arminjon F, Krantz I, Lagergård T, Sigurs N, Taranger J, Trollfors B. Serum antibody response to filamentous hemagglutinin in patients with clinical pertussis measured by an enzyme-linked immunosorbent assay. Eur J Clin Microbiol Infect Dis. 1988;7:764–770. doi: 10.1007/BF01975044. [DOI] [PubMed] [Google Scholar]