Abstract
MicroRNAs are small RNAs with ability to attach to the large number of RNA that regulate gene expression on post-transcriptional level via inhibition or degradation of specific mRNAs. MiRNAs in cells are the primary regulators of functions such as cell growth, differentiation, and apoptosis and considerably influence cell function. The expression levels of microRNAs change in human diseases, including cancer. These changes highlight their essential role in cancer pathogenesis. Ubiquitous irregular expression profiles of miRNAs have been detected in various human cancers using genome-wide identification techniques, which are emerging as novel diagnostic and prognostic cancer biomarkers of high specificity and sensitivity. The measurable miRNAs with enhanced stability in blood, tissues, and other body fluids provide a comprehensive source of miRNA-dependent biomarkers for human cancers. The leading role of miRNAs as potential biomarkers in human cancers is discussed in this article. In addition, the interests and difficulties of miRNAs as biomarkers have been explored.
Keywords: MicroRNA, Biogenesis of miRNA, Cancer diagnosis, Cancer prognosis
Introduction
Cancer is a group of diseases that involve abnormal cell growth with the possibility of invasion or spread to other parts of the body. Genetic, epigenetic, and environmental factors may play a role.1 There are many changes in the tissue and cell surface of the tumor, so the exact mechanism of the pathogen is unknown.2 The complexity of cancer has increased with the discovery of different classes of genes involved, especially tumor inhibitor genes and molecular signaling pathways.3 Cancer is the second leading cause of death after heart disease.4 Of the causes of death from cancer, it is detected at an advanced stage in more than 50% of cases. A strong correlation has been found between the time of diagnosis and the life expectancy of patients. Therefore, an early cancer diagnosis is the best way to reduce the mortality rate and control it long-term.5,6
MicroRNAs (miRNAs) are small non-coding RNAs (ncRNAs) that control gene expression at the post transcription level by targeting messenger RNAs (mRNAs).7 The new types of “oncomiRs” or “tumor suppressors” are called irregular miRNAs, which play a significant role in cancer development and progression.8 Studies show that more than half of the proteins encoding proteins are controlled by miRNAs. One miRNA can affect multiple genes, and multiple miRNAs can regulate one gene. Thus, a particular miRNA can control various cellular processes such as apoptosis, growth, and cell proliferation.9-11 The impact of microRNAs on human cancers was first identified in chronic lymphocytic leukemia studies.12 This evidence is growing and has led to the introduction of miRNAs as very attractive new biomarkers for many human diseases, including cancer. Irregular expression profiles of miRNAs have been detected in various human cancers, indicating the considerable possibility of brand-new biomarkers and cancer diagnosis with high sensitivity and specificity.13 The expression profile of miRNAs has been shown to differ between normal, and tumor tissues and these changes can be used to distinguish tumor tissues from normal tissues.14 the early detection of neoplasms can ensure the selection of the most effective, modern therapy, extend the time to disease progression, extend the overall survival time, and improve its quality, and early detection may be possible thanks to miRNA molecules. In this study, recent advances in cancer diagnosis and abnormalities in tumor-associated miRNAs as diagnostic biomarkers for cancer were reviewed. The advantages and disadvantages of using miRNA biomarkers as a tool for cancer diagnosis were then discussed.
Current diagnosis strategies for cancer
Cancer is a genetic disease that results from uncontrolled cell division. It can be caused by environmental factors and genetic abnormalities.15 Several critical genes, including oncogenes and tumor-inhibitory genes, play a role in cells becoming cancerous.16 When the disease is diagnosed at an early stage, treatment options are more effective. Cancer cells can be detected in several ways. Methods of cancer diagnosis include imaging, laboratory tests, and tumor biopsy.17 There are several imaging techniques that a physician can use to examine and detect cancerous tissue. Most breast cancers originate in the mammary gland tissue, which is associated with anatomical changes in the duct and the appearance of small and large masses.18
One of the most effective ways to combat this disease is to diagnose it in its early stages. Studies have shown that more effective treatment can reduce the potential mortality rate if the cancer is diagnosed early.19 Although mammography is currently one of the most effective screening tools for breast cancer, it does not detect all breast cancers.20 Mammography is very difficult to interpret because the natural tissue of the breast is different and unique in each patient.21 Although mammography is the standard method of diagnosing breast cancer, complementary ultrasonography (complementary screening) can increase the sensitivity of the diagnosis, especially in the younger age group.22 Ultrasound is a painless, non-invasive procedure to examine internal organs. This device records the image of the internal organs with sound waves, which reveals the condition of the internal organs.23 Therefore, it is helpful to find malignant tumors hidden in the abdominal and pelvic cavities, such as gastric tumors and ovarian cancer. However, cancer diagnosis by this method is relative.24 Computerized tomography (CT) is used to find tumors in the body cavities that are not detected by regular clinical examinations for early detection of lung, head, and neck cancers.25 The amount of radiation used in this method has side effects.25 Magnetic resonance imaging (MRI), which uses radio waves instead of X-rays, images soft (non-bony) tissue. However, MRI is not a routine test for the definitive diagnosis of cancer.26 Samples for cancer diagnosis are taken through a simple outpatient procedure, usually under local anesthesia. Sometimes the sample is taken with a needle from the deep parts of the mass.27 However, the anesthesia, the surgery, the removal of the mass, and the associated removal of large amounts of healthy tissue have clear and unclear consequences for the body. Some believe that the formation of a cancerous mass in the body is a defensive measure by the body to control and limit cancer cells. Removal and elimination of this mass may have unintended consequences on the spread of cancer in the body.28
With the advent of Next Generation Sequence (NGS) technology, gene sequencing and cancer diagnosis have entered a new field in the last decade. NGS technology can be used to diagnose many neurological diseases and cancers.29,30 Cancer is a very heterogeneous disease, and different cells are found in a tumor. The primary mutation that causes malignancy may only be found in a small portion of the cell genome that NGS can detect, although detection requires great care and concentration.31 However, since many genetic alterations are not detected in various cancers, the use of NGS is limited. Since accurate gene profiles for various cancers are not available,31,32 miRNAs play an essential role as biological biomarkers in cancer and can be considered for cancer screening and early detection. However, several studies are required to profile miRNAs in different types of cancer.
MicroRNAs
MicroRNAs (miRNAs) are a group of gene expression regulators that are synthesized endogenously and can affect the expression of genes. These molecules are single-stranded and non-coding RNAs, so the length of these miRNAs does not exceed about 23 nucleotides in the functional state.33 The critical point is that one miRNA can target multiple mRNAs, thus exerting a regulatory effect on different genes. Moreover, studies have shown that one mRNA is regulated by multiple miRNAs.34 miRNA expression is different in different tissues. This difference in expression is observed in incredibly healthy and cancer cells, indicating the importance of miRNAs in the pathogenesis process.35 Some miRNAs act as tumor suppressors or oncogenes by targeting cancer-related genes. They also play essential roles in apoptosis, proliferation, migration, and cell invasion.36,37 MiRNAs decrease target gene expression by decreasing the stability of mRNA and preventing its translation.38 By decreasing the abundance of specific proteins, miRNAs exert control effects on many physiological processes. Not surprisingly, their dysregulated expression can influence neoplastic diseases.39
Biogenesis and function of miRNA
For the first time, a miRNA gene named lin-4 was identified in Caenorhabditis elegans, which binds to the mRNA of the lin-4 construct and inhibits its translation.40 Later, another miRNA named let-7 was identified in Caenorhabditis Elegans, which regulates cell growth and development.41 MiRNAs are processed and produced in several steps: In the first stage, RNA polymerase II transcribes miRNA genes (in most cases) or RNA polymerase III, which is the product of an extended transcript (hundreds to several thousand nucleotides) called primary miRNA (pri-miRNA).33 Pri-miRNA is seen as a stem-loop hairpin structure with a cap ‘5 and a poly-A tail.42 Subsequently, a ribonuclease enzyme called Drosha cleaves the pri-miRNA with the help of DGCR8. A pre-miRNA molecule of about 60-100 nucleotides is formed with a stem-loop or hairpin structure.42,43 Ran GTP and exportin-5 transfer pre-miRNAs from the nucleus to the cytoplasm. Upon entering the cytoplasm, RanGTP is hydroxylated to RanGDP, leading to the release of pre-miRNA from exportin 5.44 The ribonuclease III, called Dicer, binds with another protein to the pre-miRNA and generates the mature miRNA by cleavage in the cytoplasm. Mature miRNA (miRNA/miRNA*) with about 22 nucleotides is a double-stranded molecule without a stem-loop hairpin structure.45,46 Not all bases are paired in miRNA / miRNA*, and incomplete connections between two strands can be seen. In the next step, one of the duplex miRNA strands, miRNA/miRNA*, is included in the RISC set, and the other miRNA* strand is separated or destroyed. A strand with a “5” end paired with another strand with lower stability enters the RISC complex.9,47 The primary role of the RISC complex is to guide the mature miRNA to the target transcript and prevent the translation process and protein production (Figure 1).
Figure 1.
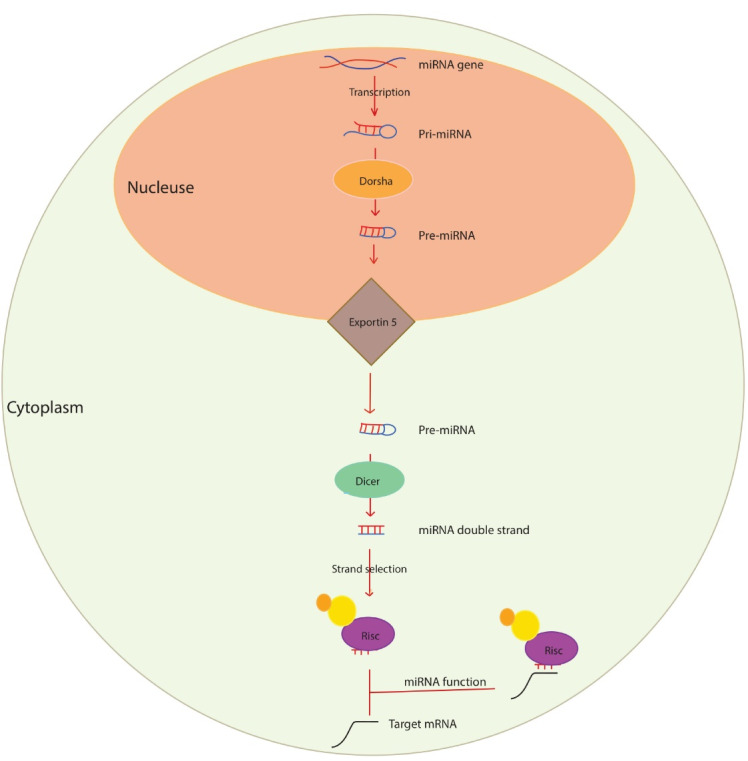
The biogenesis and function of miRNA. miRNA biogenesis takes place in the nucleus; Pri-miRNA is converted by RNase III endonuclease, Drosha, and its Cofactor (Dgcr8) into smaller stem-looped structures called precursor miRNA (pre-miRNA). Pre-miRNAs are transported by the exportin 5 from the nucleus to the cytosol; further processing results in the generation of mature miRNA by a second RNase III enzyme and Dicer
The mature miRNA is associated with the miRNA-induced silencing complex (miRISC), which leads to post-transcriptional gene silencing. Overall, the function of miRNAs is to silence genes.48 There is at least one miRNA binding site in 30% to 80% of coding protein genes, so a miRNA can regulate the expression of numerous mRNAs by partially binding to an mRNA.49,50 MiRNA is also involved in almost all pathological and biological processes.51 MiRNAs specifically identify mRNA and regulate gene expression through several post-transcriptional mechanisms: (1) inhibition of translation and also (2) degradation of mRNA.52 The component of the regulatory mechanism depends primarily on the degree of miRNA-mRNA complementarity. When the degree of complementarity between miRNA and mRNA is high, damage to the target mRNA is possible. However, if the complementarity between miRNA and mRNA is low, the mechanism of translational repression is triggered.53 For this reason, changes in mi expression are observed in various diseases.
The role of miRNAs in cancer
MiRNAs have physiological contributions to various vital processes, both biological and pathological, and to cellular stress. Estimates suggest that conserved miRNA target regions comprise the most damaged DNA checkpoint and repair genes.54 High transcription rates and low translation rates can occur in genes that are primarily regulatory or important for cell functionality.55 Negatively modulated miRNAs allows the cell to produce many mRNA transcripts with a small amount of protein with thorough regulation.56 The miRNA expression profile suggests that the expression of many miRNAs is drastically altered in human cancers; undoubtedly, primary overexpression or loss occurs for some miRNAs in tumors compared to intact tissues. Tumor growth is promoted by oncogenic miRNAs, known as oncomiRs, by negatively affecting tumor-suppressing genes, and tumor-suppressing miRNAs, known as anti-oncomiRs, target oncogenes to inhibit tumor growth.57 MiRNAs related to the DNA damage response (DDR) pathway contribute to tumorigenesis and development. Genes related to cell cycle regulation have been shown to repress three members of the miR-34 family - miR-34a, miR-34c and miR-34b.58 It has been reported that miR-17/20 and miR-221/ 222 clusters target cell cycle regulators, leading to the regulation of cell cycle checkpoints and progression.59,60 Currently, there is evidence for functional enhancement of several miRNA target genes associated with DDR pathway. In cancer, aberrantly expressed miRNAs are detected by mature or precursor miRNA copies that are abnormally expressed compared to the corresponding intact tissues. There is increasing evidence for a typical aberrantly described profile of miRNAs alongside pre-miRNAs in human cancers. Various high-performance sequencing platforms for whole-genome analysis of miRNA gene expression have in recent years uncovered aberrantly expressed profiles of miRNAs that are either down- or up-regulated in a variety of selected human malignancies, including breast,61 colorectal,62 and prostate cancers,63 as well as glioma (Table 1).64
Table 1. MicroRNAs are involved in the cancer-causing process .
| Type of cancer | microRNA | Target gene | Role in cancer | Reference |
| Liver cancer | miR-503 | VEGF-A and FGF2 | Inhibition of angiogenesis | 65 |
| Ovarian cancer | miR-125b | HIF-1a, VEGF, HER2, HER3 | Inhibition of angiogenesis | 66 |
| Bladder cancer | miR-200 family (miR-200a/200b/200c/141/429) | ZEB | EMT/MET process | 67 |
| Colorectal cancer | miR-34a | Snail, ZNF281, IL-6R | EMT process | 68 |
| Breast cancer | miR-335 | SOX4, TNC | migration and invasion | 69 |
| Gastric cancer | MiR-503 | Notch and IGF1R | EMT process | 70 |
| Non-small cell lung cancer | MiR-194 | BMI-1 | migration and invasion | 70 |
| Glioma cancer | MiR-128 | E2F3a | cell growth | 71 |
EMT, Epithelial-mesenchymal transition; MET, mesenchymal-epithelial transition.
MiRNAs and their extracellular stability
The suitability of circulating miRNAs as biomarkers, particularly for cancer, depends on their stability and ability to indicate tumor status and anticipate therapeutic responses. Several systematic studies have shown that circulating miRNAs maintain their stability even when exposed to harsh conditions that typically weaken RNAs, such as high or low pH, boiling, and prolonged storage followed by freezing and thawing cycles.72 The remarkably stable miRNAs have been attributed in part to their relationship with protein complexes, and these miRNAs in circulating microvesicles are referred to as exosomes. In the present study, most circulating miRNAs in plasma were shown to be co-fractionated with Argonaute2 (Ago2), suggesting that the mechanisms of action involved in the stability of miRNAs in plasma are circulating Ago2 complexes.73 Ago2 is a member of an RNA-mediated silencing complex. According to the findings of this recent research, the crucial effector protein of miRNA-induced silencing suggests that the vesicle-related and Ago2 complex-related miRNAs originate in different cell types and show the expression of miRNAs depending on the cell shape.
Nevertheless, circulating vesicle-bound miRNAs accounted for only 10% of plasma. Furthermore, the entire detection was confirmed by ultrafiltration of extracellular miRNAs with Ago2. After ultracentrifugation at 110 000 g, most miRNAs in plasma and cell culture remain in the supernatant, suggesting that extracellular miRNAs emerge as non-vesicular ones.74 Moreover, non-miRNA species (U6 RNA, RNU24, RNU43, RNU44, RNU48, and RNU6B) and mRNAs have no connection with Ago proteins present in the extracellular milieu, present only at low levels. Some other proteins in the supernatant after Ago2 immunoprecipitation suggest a possible link with circulating miRNAs.73 The discovery of another mechanism, likely involving the nSMase2 pathway, shows that high-density lipoproteins can transfer circulating miRNAs and alter gene expression by transporting miRNAs to recipient cells.75
However, miRNAs could additionally be associated with paracrine and autocrine miRNA signaling with exosomes. Existing evidence using only samples from intact pens or culture media suggests that most circulating miRNAs lack exosomes.73,76 Studies did not examine two circulating miRNA populations (i.e., extracellular and exosomal) or compare intact pens with cancer patients. Notable results of such studies show that tumor-derived exosome levels are increased in plasma samples from cancer patients compared to those in healthy donor samples.77,78 In addition, miRNA-containing exosomes were present in the bloodstream and various other body fluids, such as saliva.79,80 A research report suggests that exosomal miRNAs, which remain hidden in the bone marrow stroma, are associated with breast cancer cells and breast cancer recurrence and poor prognosis. In the current study, the contribution of miRNAs to breast cancer cell quiescence was demonstrated by visualizing their transition through intercellular interaction at gap junctions and stroma-derived exosomes between bone marrow stroma and quiescent breast cancer cells.81 The origin of extracellular circulating miRNAs is in dead cells, as Ago2/miRNA complexes with incredible stability are present in the cell cytoplasm.74,82 There is no doubt that miRNAs are produced in body fluids by tumor cells undergoing apoptosis and necrosis and by other sources, including blood cells, the liver, lungs, kidneys, and other body organs with which blood plasma remains in high contact.83 Theoretically, this suggests the need for warning when extracellular circulating miRNAs are used as biomarkers. This results from the fact that cancer-specific miRNAs can be hidden in circulating miRNAs from intact tissues. Studies are needed to determine the true origin, mode of action of formation in circulation, and biological effects of these miRNA molecules in distant regions.
MiRNAs as biomarkers of diagnosis for cancer
As previous observations have shown, cancers of various hematopoietic and epithelial lineages possess complete miRNA profiles that differ in their indications based on their developmental basis. For example, a microarray platform was used to assess the expression levels of 47 miRNAs in 101 formalin-fixed and paraffin-implanted samples from primary or metastatic cancers. Overall accuracies of 100 and 78% were obtained for primary and metastatic cancers, respectively. By applying the signature to a dataset of 170 samples with independent publications, 86% of correctly predicted metastatic cases were confirmed.84 Accurate determination of tumor subtypes in patients with malignancies has a significant impact on treatment decision-making. For example, a group of researchers developed a category system for renal cell carcinoma (RCC) with decision trees capable of distinguishing different RCC subtypes in 94 different subtype samples by analyzing exclusive miRNA signatures.85 The system was 97% sensitive at discriminating between healthy samples and RCC patients. For four RCC types (clear cell RCC (ccRCC), papillary RCC (pRCC), oncocytoma, and chromophobe RCC (chRCC)), 100% accuracy was achieved for the ccRCC subtype against a variety of other subtypes, 97% for the pRCC subtype against oncocytoma and chRCC, and 100% accuracy for distinguishing oncocytoma from chRCC.
In addition, Gilad et al confirmed a miRNA-based assay to distinguish between four primary forms of lung cancer cells in an independent composite of 451 samples. Their results showed an overall accuracy of 94% for over 90% of the samples, and pathological and cytological samples showed the same performance.86 The observations suggest that miRNA signatures accurately categorize different types of cancer in cytological and pathological samples. Biomarkers are urgently needed for the timely diagnosis of cancer, as human survivability and diagnosis depend entirely on clumping at the time of detection. Diagnosis is mainly improved by timely diagnosis. Reports suggest that miRNA signatures are a promising option for timely diagnosis. A report on the primacy of overexpressed miR-21 and miR-205 in ductal adenocarcinomas over phenotypic regulation in ducts shows that aberrantly produced miRNAs are an initial manifestation of cancer progression,87 which also supports their potential application for early detection of cancer (Figure 2).
Figure 2.
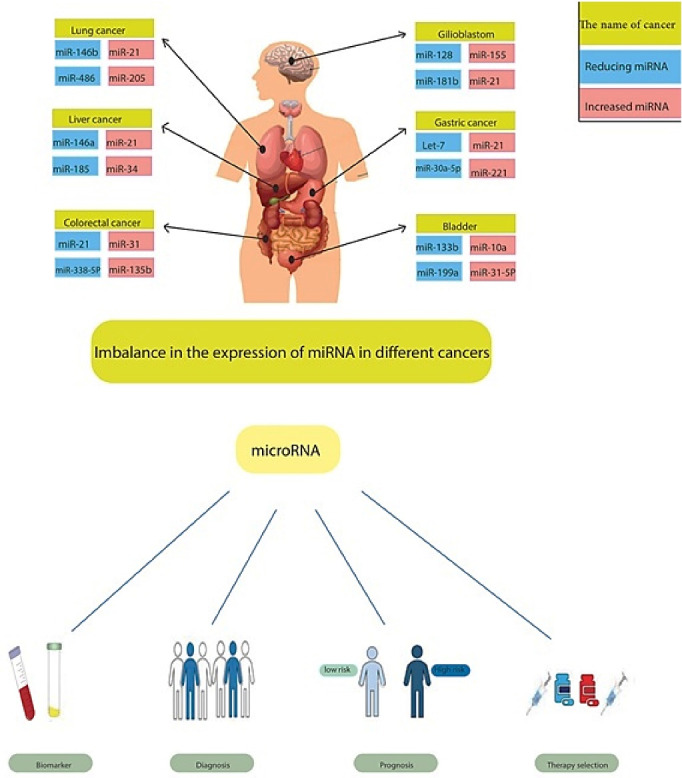
The effect of microRNAs on the clinical treatment of cancer. Since microRNA expression patterns are altered in cancer compared to normal tissues and vary between subtypes, they are likely future biomarkers for stratification and diagnosis prediction in cancer patients. In future cancer treatment, microRNAs could play an essential role in deciding which drugs to select for a patient and whether the patient will respond to the drug
MiRNAs as cancer prognosis biomarkers
In addition to original tissue differentiation, subtypes, and timely cancer diagnosis contributing to cancer detection, miRNA signatures are of additional value for cancer prognostication.88 A signature of five miRNAs in lung cancer has also been established, which can be used to predict therapeutic consequences in non-small cell lung cancer. Highly expressed let-7a leads to many diagnoses, while elevated miR-137, miR-372 and miR-182 are associated with poor prognosis.89,90 In addition, plasma concentrations of miR-10b and miR-373 were investigated in a recent study on the role of metastatic breast cancer. The relationships between these miRNAs were revealed in detecting lymph node metastasis, highlighting the potential of prognostic biomarkers.91 Even a single miRNA can have accurate predictive potential, as studies in breast cancer patients have shown. It was found that overexpressed miR-210 was associated with an increased risk of recurrence and a lower chance of survival without recurrence. A single miR-210 level could predict diagnosis to a similar extent as a 76-gene mRNA signature assay (GENE76).92,93 There was a negative association between miR-410 signature and overall survival in progressive serous ovarian cancer.94 Incredibly, a recently discovered eight-miRNA signature model in gastric cancer can predict both overall survival and recurrence-free survival.95 A six-miRNA-based classifier found in patients with stage II colon cancer was an independent predictor of disease recurrence (Figure 2).96
Limits of miRNAs Biomarkers of cancer diagnosis and prognosis
MiRNAs present in the blood circulation can be quantified from different sources of substances (i.e., plasma, serum, and whole blood),97 demonstrating the influence of blood cell miRNAs (present in erythrocytes, leukocytes, and platelets) on the analysis of circulating miRNAs. According to their data, blood cells contribute significantly to miRNAs in the circulation and, in particular, alter specific miRNA levels. Therefore, it is impossible to consider the whole blood as a specific biological fluid to detect circulating miRNAs. Nevertheless, it is necessary to eliminate cellular components that may interfere with accurate miRNA measurement when assays are performed on plasma and serum. Several research teams have compared different plasma/serum preparation techniques, evaluating the differences between the two biological fluids in the distribution of miRNA levels.
However, indisputable evidence is not available.98,99 EDTA and citrate are anticoagulant chemicals commonly added to blood collection tubes that can detect miRNAs adequately without further treatments.100 Nevertheless, EDTA tubes are preferable to citrate use, as the latter can lead to hemolysis (see the section on hemolysis affecting the analysis of miRNAs).101 Finally, a comparison of fresh and iced liquids has yielded stunning results.
Since miRNAs are very stable in the bloodstream, no or only minor differences were observed between fresh and frozen samples, even after repeated cycles of freezing/thawing.102,103 However, it is recommended to avoid excessive freezing/thawing. In the case of the occurrence of degraded miRNAs (even to a limited extent), it is possible to miss miRNAs with misrepresentation. After obtaining the data, normalization is the next challenge. Techniques can be expected to vary between samples due to variations in the preparing compound, RNA extraction, or reaction performance during labeling or hybridization. Housekeeping transcripts used to analyze miRNAs in tissues (e.g. RNU6 and RNU48) are often unidentifiable in the bloodstream because they are extensively damaged by RNAse mediation.104,105 One proposed housekeeping miRNA is miR-16, which has been most frequently cited in previous reports. It is still strongly influenced by hemolysis and cannot be considered a helpful miRNA reference for normalizing data. Several other housekeeping miRNAs have been proposed in independently conducted studies, but there is still a global lack of agreement.99,106,107 Universal normalization methods are applied throughout the analysis that determines a reasonably high level of miRNAs. A commonly applied method for amplification-based array data uses a universal estimate of miRNA expression profile, e.g., the mean or median, as a calibrator. However, the diversity of miRNAs found in the bloodstream by PCR-based methods is generally around 100, which may not be sufficient to use a universal normalization method.
Furthermore, a minimal variety of potential miRNA markers usually need further confirmation after the detection phase, both in independent case series and technically. Nevertheless, post-detection assays (usually performed by assay alone or a custom map with an insufficient number of miRNAs) indeed do not use the same method to normalize the data. To address this issue, Pizzamiglio et al108 recently introduced an all-inclusive technique that starts with a data-driven normalization procedure based on amplification-based array data and detects a small number of miRNAs to be used as a reference for normalizing data based on subsequent confirmatory assays. Moreover, Kroh et al109 introduced miRNA outright metrology using a standard curve generated by an artificial miRNA (synthetic oligonucleotides) performed by RT-PCR after biological samples. The technique is plausible for quantifying single miRNA markers, but it is not suitable for evaluating complex miRNAs. For microarray-based data, normalization is also a critical step.110 miRNA expression is usually profiled using global normalization procedures that have proven successful in gene expression analysis (e.g., Lowness or quantile normalization).
Nevertheless, the smaller number of functions (compared to the diversity of genes identified in a gene expression microarray) poses some challenges in terms of the relevance of these approaches in the field of miRNA profiling, and newly introduced methods include tailored least-variant set normalization for miRNA microarrays.111,112 Quantification of miRNAs based on sequencing is a good novelty compared to qRT-PCR and microarrays, and the optimal procedure for its use is easily standardized. Methods developed for microarray normalization (e.g., lowness or quantile normalization) of miRNA-seq results have also been used.113 In addition to the potential procedural biases mentioned above, several other critical variables with a potentially intense impact on the accurate interpretation of circular miRNA biomarker studies are associated with inherent interpersonal variation and the influence of factors independent of disease. When considering circulating miRNAs as biomarker molecules for cancer, the tumor itself is the primary source of variation to be considered. The specificity of miRNA expression patterns for individual cancer types, circulating miRNA signatures are expected to be specific for different cancer types or molecular subtypes and exclusive for tumor functionalities.
Merits of miRNA in the diagnosis and prognosis of cancers
Circulating miRNAs can be obtained quickly and without severe injury. Moreover, a plethora of potentially helpful miRNA biomarkers is shown to be stable in normal humans. Although cell-free miRNAs from serum and plasma are among the most common circulating miRNA biomarkers, several other body fluid samples, such as saliva and urine, are also relevant sources of circulating miRNAs.114 Overexpression of miR-186-5p has been observed in tumor tissue, urine, and blood of patients with bladder cancer.115 Several miRNAs, including miR-210-3p, were upregulated in the urine of patients with transient cell carcinoma and could promote cancer detection.116 PCR remains the primary technique for evaluating circulating miRNA. PCR is characterized by amplification as a critical phase that magnifies the main difference between samples, even for relatively insignificant differences. Accordingly, the current detection technique makes circulating miRNAs the most sensitive biomarkers. The miRNAs are formed dynamically and promptly in response to internal or external stimulants, which enhances the ability of miRNAs to be monitored in real-time and dynamically throughout progression changes, from tumor initiation during adhesion to progression.117,118
TNBC patients were found to have considerable downregulation of a miRNA panel consisting of miR-34a-5p, miR-34b-5p and miR-34c-5p. Of the miRNAs mentioned above, the expression of miR-34a-5p and miR-34b-5p were positively correlated with lymph node metastasis and with miR-34c-5p was correlated with tumor grade and distant metastasis.118 These findings demonstrate the potential of circulating miRNAs for assessing tumor stage and progression. The dynamically revealed miRNA pattern could illustrate the progression background of cancer throughout its development.
Conclusion
MiRNAs are stable molecules in the biological fluids of the body. They have great potential to be considered as non-invasive biomarkers for various cancers because they can be obtained rapidly and with minimal risk in biological body fluids such as saliva, serum, and urine. MiRNAs can be valuable in many ways, including screening for early-stage cancer, subclassification, predicting drug susceptibility, selecting treatment strategies, and screening for tumor chemical or radiological resistance to predict outcomes and recurrence. Significantly, more work needs to be done to fully identify and validate increased or decreased miRNAs in any disease.
Competing Interests
The authors declare that there are no conflicts of interest.
Ethical Approval
Not applicable.
References
- 1.Shademan B, Masjedi S, Karamad V, Isazadeh A, Sogutlu F, Saeedi Rad MH, et al. CRISPR technology in cancer diagnosis and treatment: opportunities and challenges. Biochem Genet. 2022;60(5):1446–70. doi: 10.1007/s10528-022-10193-9. [DOI] [PubMed] [Google Scholar]
- 2. Biray Avcı Ç, Sogutlu F, Ozates NP, Shademan B, Gunduz C. Enhanced anti-cancer potency using a combination of oleanolic acid and maslinic acid to control treatment resistance in breast cancer. Res Sq [Preprint]. November 23, 2021. 10.21203/rs.3.rs-1077510/v1. [DOI] [PMC free article] [PubMed]
- 3.Ozates NP, Sogutlu F, Lerminoglu F, Demir B, Gunduz C, Shademan B, et al. Effects of rapamycin and AZD3463 combination on apoptosis, autophagy, and cell cycle for resistance control in breast cancer. Life Sci. 2021;264:118643. doi: 10.1016/j.lfs.2020.118643. [DOI] [PubMed] [Google Scholar]
- 4.Harding MC, Sloan CD, Merrill RM, Harding TM, Thacker BJ, Thacker EL. Transitions from heart disease to cancer as the leading cause of death in US states, 1999-2016. Prev Chronic Dis. 2018;15:E158. doi: 10.5888/pcd15.180151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espina C, Soerjomataram I, Forman D, Martín-Moreno JM. Cancer prevention policy in the EU: Best practices are now well recognised; no reason for countries to lag behind. J Cancer Policy. 2018;18:40–51. doi: 10.1016/j.jcpo.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wardle J, Robb K, Vernon S, Waller J. Screening for prevention and early diagnosis of cancer. Am Psychol. 2015;70(2):119–33. doi: 10.1037/a0037357. [DOI] [PubMed] [Google Scholar]
- 7.Ratti M, Lampis A, Ghidini M, Salati M, Mirchev MB, Valeri N, et al. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Target Oncol. 2020;15(3):261–78. doi: 10.1007/s11523-020-00717-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24(16):R762–76. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soheilyfar S, Velashjerdi Z, Sayed Hajizadeh Y, Fathi Maroufi N, Amini Z, Khorrami A, et al. In vivo and in vitro impact of miR-31 and miR-143 on the suppression of metastasis and invasion in breast cancer. J BUON. 2018;23(5):1290–6. [PubMed] [Google Scholar]
- 10.Xu J, Shao T, Ding N, Li Y, Li X. miRNA-miRNA crosstalk: from genomics to phenomics. Brief Bioinform. 2017;18(6):1002–11. doi: 10.1093/bib/bbw073. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Zhang Z. miRNA regulatory variation in human evolution. Trends Genet. 2013;29(2):116–24. doi: 10.1016/j.tig.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33(6):1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daoud AZ, Mulholland EJ, Cole G, McCarthy HO. MicroRNAs in pancreatic cancer: biomarkers, prognostic, and therapeutic modulators. BMC Cancer. 2019;19(1):1130. doi: 10.1186/s12885-019-6284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian F, Li R, Chen Z, Shen Y, Lu J, Xie X, et al. Differentially expressed miRNAs in tumor, adjacent, and normal tissues of lung adenocarcinoma. Biomed Res Int. 2016;2016:1428271. doi: 10.1155/2016/1428271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shademan B, Karamad V, Nourazarian A, Biray Avcı Ç. CAR T cells: cancer cell surface receptors are the target for cancer therapy. Adv Pharm Bull. 2022;12(3):476–89. doi: 10.34172/apb.2022.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H. MicroRNAs and apoptosis in colorectal cancer. Int J Mol Sci. 2020;21(15):5353. doi: 10.3390/ijms21155353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagpal M, Singh S, Singh P, Chauhan P, Zaidi MA. Tumor markers: a diagnostic tool. Natl J Maxillofac Surg. 2016;7(1):17–20. doi: 10.4103/0975-5950.196135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weledji EP, Tambe J. Breast cancer detection and screening. Med Clin Rev. 2018;4(2):8. doi: 10.21767/2471-299x.1000071. [DOI] [Google Scholar]
- 19.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primers. 2019;5(1):66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 20.Giampietro RR, Cabral MVG, Lima SAM, Weber SAT, Dos Santos Nunes-Nogueira V. Accuracy and effectiveness of mammography versus mammography and tomosynthesis for population-based breast cancer screening: a systematic review and meta-analysis. Sci Rep. 2020;10(1):7991. doi: 10.1038/s41598-020-64802-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itani M, Griffin AT, Whitman GJ. Mammography of breast calcifications. Imaging Med. 2013;5(1):63–74. doi: 10.2217/iim.13.6. [DOI] [Google Scholar]
- 22.Berg WA, Bandos AI, Mendelson EB, Lehrer D, Jong RA, Pisano ED. Ultrasound as the primary screening test for breast cancer: analysis from ACRIN 6666. J Natl Cancer Inst. 2016;108(4):djv367. doi: 10.1093/jnci/djv367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen Thi T. Determination of Thyroid Volume by Ultrasound [thesis]. Jönköping, Sweden: School of Health and Welfare, Jonkoping University; 2016.
- 24.Namikawa T, Hanazaki K. Clinicopathological features and treatment outcomes of metastatic tumors in the stomach. Surg Today. 2014;44(8):1392–9. doi: 10.1007/s00595-013-0671-9. [DOI] [PubMed] [Google Scholar]
- 25.Goerres GW, von Schulthess GK, Steinert HC. Why most PET of lung and head-and-neck cancer will be PET/CT. J Nucl Med. 2004;45 Suppl 1:66S–71S. [PubMed] [Google Scholar]
- 26.Engels RRM, Israël B, Padhani AR, Barentsz JO. Multiparametric magnetic resonance imaging for the detection of clinically significant prostate cancer: what urologists need to know Part 1: acquisition. Eur Urol. 2020;77(4):457–68. doi: 10.1016/j.eururo.2019.09.021. [DOI] [PubMed] [Google Scholar]
- 27.Reijnen C, van der Putten LJM, Bulten J, Snijders M, Küsters-Vandevelde HVN, Sweegers S, et al. Mutational analysis of cervical cytology improves diagnosis of endometrial cancer: a prospective multicentre cohort study. Int J Cancer. 2020;146(9):2628–35. doi: 10.1002/ijc.32686. [DOI] [PubMed] [Google Scholar]
- 28.Valastyan S, Weinberg RA. MicroRNAs: crucial multi-tasking components in the complex circuitry of tumor metastasis. Cell Cycle. 2009;8(21):3506–12. doi: 10.4161/cc.8.21.9802. [DOI] [PubMed] [Google Scholar]
- 29. Morganti S, Tarantino P, Ferraro E, D’Amico P, Viale G, Trapani D, et al. Role of next-generation sequencing technologies in personalized medicine. In: Pravettoni G, Triberti S, eds. P5 eHealth: An Agenda for the Health Technologies of the Future. Cham: Springer; 2020. p. 125-54. 10.1007/978-3-030-27994-3_8. [DOI]
- 30.Shademan B, Biray Avcı Ç, Nikanfar M, Nourazarian A. Application of next-generation sequencing in neurodegenerative diseases: opportunities and challenges. Neuromolecular Med. 2021;23(2):225–35. doi: 10.1007/s12017-020-08601-7. [DOI] [PubMed] [Google Scholar]
- 31.Berger MF, Mardis ER. The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol. 2018;15(6):353–65. doi: 10.1038/s41571-018-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh RR. Next-generation sequencing in high-sensitive detection of mutations in tumors: challenges, advances, and applications. J Mol Diagn. 2020;22(8):994–1007. doi: 10.1016/j.jmoldx.2020.04.213. [DOI] [PubMed] [Google Scholar]
- 33.Selvi Gunel N, Birden N, Caliskan Kurt C, Goker Bagca B, Shademan B, Sogutlu F, et al. Effect of valproic acid on miRNAs affecting histone deacetylase in a model of anaplastic thyroid cancer. Mol Biol Rep. 2021;48(8):6085–91. doi: 10.1007/s11033-021-06616-2. [DOI] [PubMed] [Google Scholar]
- 34.Rooda I, Hensen K, Kaselt B, Kasvandik S, Pook M, Kurg A, et al. Target prediction and validation of microRNAs expressed from FSHR and aromatase genes in human ovarian granulosa cells. Sci Rep. 2020;10(1):2300. doi: 10.1038/s41598-020-59186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aksenenko M, Palkina N, Komina A, Tashireva L, Ruksha T. Differences in microRNA expression between melanoma and healthy adjacent skin. BMC Dermatol. 2019;19(1):1. doi: 10.1186/s12895-018-0081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou K, Liu M, Cao Y. New insight into microRNA functions in cancer: oncogene-microRNA-tumor suppressor gene network. Front Mol Biosci. 2017;4:46. doi: 10.3389/fmolb.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frixa T, Donzelli S, Blandino G. Oncogenic microRNAs: key players in malignant transformation. Cancers (Basel) 2015;7(4):2466–85. doi: 10.3390/cancers7040904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M, Marin-Muller C, Bharadwaj U, Chow KH, Yao Q, Chen C. MicroRNAs: control and loss of control in human physiology and disease. World J Surg. 2009;33(4):667–84. doi: 10.1007/s00268-008-9836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bracht JR, Van Wynsberghe PM, Mondol V, Pasquinelli AE. Regulation of lin-4 miRNA expression, organismal growth and development by a conserved RNA binding protein in C. elegans. Dev Biol. 2010;348(2):210–21. doi: 10.1016/j.ydbio.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell. 2016;7(2):100–13. doi: 10.1007/s13238-015-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Y, Yu Z, Han G, Li J, Anh V. Identification of pre-microRNAs by characterizing their sequence order evolution information and secondary structure graphs. BMC Bioinformatics. 2018;19(Suppl 19):521. doi: 10.1186/s12859-018-2518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtsuka M, Ling H, Doki Y, Mori M, Calin GA. MicroRNA processing and human cancer. J Clin Med. 2015;4(8):1651–67. doi: 10.3390/jcm4081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheng P, Fields C, Aadland K, Wei T, Kolaczkowski O, Gu T, et al. Dicer cleaves 5’-extended microRNA precursors originating from RNA polymerase II transcription start sites. Nucleic Acids Res. 2018;46(11):5737–52. doi: 10.1093/nar/gky306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu Y, Chen J, Huang Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA. 2019;1(1):24. doi: 10.1186/s41544-019-0024-y. [DOI] [Google Scholar]
- 47.Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, et al. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem. 2004;279(40):42230–9. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 48.Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17(10):1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu X, Liu P, Dimopoulos G, Zhu J. Dynamic miRNA-mRNA interactions coordinate gene expression in adult Anopheles gambiae. PLoS Genet. 2020;16(4):e1008765. doi: 10.1371/journal.pgen.1008765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017;8(1):45–56. doi: 10.4331/wjbc.v8.i1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei Z, Wong GW. Roles of micro-RNAs in metabolism. In: Encyclopedia of Biological Chemistry. 2nd ed. Elsevier Inc; 2013. p. 191-4. 10.1016/b978-0-12-378630-2.00078-5. [DOI]
- 52.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20(1):21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bellato M, De Marchi D, Gualtieri C, Sauta E, Magni P, Macovei A, et al. A bioinformatics approach to explore microRNAs as tools to bridge pathways between plants and animals. Is DNA damage response (DDR) a potential target process? Front Plant Sci. 2019;10:1535. doi: 10.3389/fpls.2019.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Metzl-Raz E, Kafri M, Yaakov G, Barkai N. Gene transcription as a limiting factor in protein production and cell growth. G3 (Bethesda) 2020;10(9):3229–42. doi: 10.1534/g3.120.401303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaezi Astamal R, Maghoul A, Taefehshokr S, Bagheri T, Mikaeili E, Derakhshani A, et al. Regulatory role of microRNAs in cancer through Hippo signaling pathway. Pathol Res Pract. 2020;216(12):153241. doi: 10.1016/j.prp.2020.153241. [DOI] [PubMed] [Google Scholar]
- 57.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28(3-4):369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 58.Majidinia M, Yousefi B. DNA damage response regulation by microRNAs as a therapeutic target in cancer. DNA Repair (Amst) 2016;47:1–11. doi: 10.1016/j.dnarep.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 59.Mens MM, Ghanbari M. Cell cycle regulation of stem cells by microRNAs. Stem Cell Rev Rep. 2018;14(3):309–22. doi: 10.1007/s12015-018-9808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, et al. miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules. 2014;19(6):7122–37. doi: 10.3390/molecules19067122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhat SA, Majid S, Hassan T. MicroRNAs and its emerging role as breast cancer diagnostic marker- a review. Adv Biomark Sci Technol. 2019;1:1–8. doi: 10.1016/j.abst.2019.05.001. [DOI] [Google Scholar]
- 62.Xiao Z, Chen S, Feng S, Li Y, Zou J, Ling H, et al. Function and mechanisms of microRNA-20a in colorectal cancer. Exp Ther Med. 2020;19(3):1605–16. doi: 10.3892/etm.2020.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li C, Gao Y, Zhang K, Chen J, Han S, Feng B, et al. Multiple roles of microRNA-100 in human cancer and its therapeutic potential. Cell PhysiolBiochem. 2015;37(6):2143–59. doi: 10.1159/000438572. [DOI] [PubMed] [Google Scholar]
- 64. Tao F, Tian X, Ruan S, Shen M, Zhang Z. miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. FASEB J 2018:fj201800495RR. 10.1096/fj.201800495RR. [DOI] [PubMed]
- 65.Polioudakis D, Abell NS, Iyer VR. miR-503 represses human cell proliferation and directly targets the oncogene DDHD2 by non-canonical target pairing. BMC Genomics. 2015;16(1):40. doi: 10.1186/s12864-015-1279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallach S, Calabuig-Fariñas S, Jantus-Lewintre E, Camps C. MicroRNAs: promising new antiangiogenic targets in cancer. Biomed Res Int. 2014;2014:878450. doi: 10.1155/2014/878450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He SJ, Xiang CQ, Zhang Y, Lu XT, Chen HW, Xiong LX. Recent progress on the effects of microRNAs and natural products on tumor epithelial-mesenchymal transition. Onco Targets Ther. 2017;10:3435–51. doi: 10.2147/ott.s139546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weng YS, Tseng HY, Chen YA, Shen PC, Al Haq AT, Chen LM, et al. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol Cancer. 2019;18(1):42. doi: 10.1186/s12943-019-0988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Almanza G, Rodvold JJ, Tsui B, Jepsen K, Carter H, Zanetti M. Extracellular vesicles produced in B cells deliver tumor suppressor miR-335 to breast cancer cells disrupting oncogenic programming in vitro and in vivo. Sci Rep. 2018;8(1):17581. doi: 10.1038/s41598-018-35968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu X, Li D, Yu F, Jia C, Xie J, Ma Y, et al. miR-194 inhibits the proliferation, invasion, migration, and enhances the chemosensitivity of non-small cell lung cancer cells by targeting forkhead box A1 protein. Oncotarget. 2016;7(11):13139–52. doi: 10.18632/oncotarget.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shan ZN, Tian R, Zhang M, Gui ZH, Wu J, Ding M, et al. miR128-1 inhibits the growth of glioblastoma multiforme and glioma stem-like cells via targeting BMI1 and E2F3. Oncotarget. 2016;7(48):78813–26. doi: 10.18632/oncotarget.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108(9):3713–8. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao C, Sun X, Li L. Biogenesis and function of extracellular miRNAs. ExRNA. 2019;1(1):38. doi: 10.1186/s41544-019-0039-4. [DOI] [Google Scholar]
- 76.Tomasetti M, Lee W, Santarelli L, Neuzil J. Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Exp Mol Med. 2017;49(1):e285. doi: 10.1038/emm.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tian F, Shen Y, Chen Z, Li R, Ge Q. No significant difference between plasma miRNAs and plasma-derived exosomal miRNAs from healthy people. Biomed Res Int. 2017;2017:1304816. doi: 10.1155/2017/1304816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liao J, Liu R, Shi YJ, Yin LH, Pu YP. Exosome-shuttling microRNA-21 promotes cell migration and invasion-targeting PDCD4 in esophageal cancer. Int J Oncol. 2016;48(6):2567–79. doi: 10.3892/ijo.2016.3453. [DOI] [PubMed] [Google Scholar]
- 79.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romano E, Netti PA, Torino E. Exosomes in gliomas: biogenesis, isolation, and preliminary applications in nanomedicine. Pharmaceuticals (Basel) 2020;13(10):319. doi: 10.3390/ph13100319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA, et al. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 2011;71(5):1550–60. doi: 10.1158/0008-5472.can-10-2372. [DOI] [PubMed] [Google Scholar]
- 82.Nassar W, El-Ansary M, Fayyad T, Abdel Aziz M. Extracellular micro-RNAs in health and disease: basic science, biogenesis and release. Am J Mol Biol. 2016;6:1–11. doi: 10.4236/ajmb.2016.61001. [DOI] [Google Scholar]
- 83.Zhou M, Hara H, Dai Y, Mou L, Cooper DK, Wu C, et al. Circulating organ-specific microRNAs serve as biomarkers in organ-specific diseases: implications for organ allo- and xeno-transplantation. Int J Mol Sci. 2016;17(8):1232. doi: 10.3390/ijms17081232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferracin M, Pedriali M, Veronese A, Zagatti B, Gafà R, Magri E, et al. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225(1):43–53. doi: 10.1002/path.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Youssef YM, White NM, Grigull J, Krizova A, Samy C, Mejia-Guerrero S, et al. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur Urol. 2011;59(5):721–30. doi: 10.1016/j.eururo.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 86.Gilad S, Lithwick-Yanai G, Barshack I, Benjamin S, Krivitsky I, Edmonston TB, et al. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J Mol Diagn. 2012;14(5):510–7. doi: 10.1016/j.jmoldx.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 87.du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56(4):603–12. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 88.Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics. 2018;10:59. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Del Vescovo V, Grasso M, Barbareschi M, Denti MA. MicroRNAs as lung cancer biomarkers. World J Clin Oncol. 2014;5(4):604–20. doi: 10.5306/wjco.v5.i4.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao W, Liu L, Lu X, Shu Y. Circulating microRNAs: possible prediction biomarkers for personalized therapy of non-small-cell lung carcinoma. Clin Lung Cancer. 2011;12(1):14–7. doi: 10.3816/CLC.2011.n.001. [DOI] [PubMed] [Google Scholar]
- 91.Sheedy P, Medarova Z. The fundamental role of miR-10b in metastatic cancer. Am J Cancer Res. 2018;8(9):1674–88. [PMC free article] [PubMed] [Google Scholar]
- 92.Bavelloni A, Ramazzotti G, Poli A, Piazzi M, Focaccia E, Blalock W, et al. MiRNA-210: a current overview. Anticancer Res. 2017;37(12):6511–21. doi: 10.21873/anticanres.12107. [DOI] [PubMed] [Google Scholar]
- 93.Cheng Q, Li X, Liu J, Ye Q, Chen Y, Tan S, et al. Multiple myeloma-derived exosomes regulate the functions of mesenchymal stem cells partially via modulating miR-21 and miR-146a. Stem Cells Int. 2017;2017:9012152. doi: 10.1155/2017/9012152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Müssnich P, Raverot G, Jaffrain-Rea ML, Fraggetta F, Wierinckx A, Trouillas J, et al. Downregulation of miR-410 targeting the cyclin B1 gene plays a role in pituitary gonadotroph tumors. Cell Cycle. 2015;14(16):2590–7. doi: 10.1080/15384101.2015.1064207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ding B, Gao X, Li H, Liu L, Hao X. A novel microRNA signature predicts survival in stomach adenocarcinoma. Oncotarget. 2017;8(17):28144–53. doi: 10.18632/oncotarget.15961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jacob H, Stanisavljevic L, Storli KE, Hestetun KE, Dahl O, Myklebust MP. A four-microRNA classifier as a novel prognostic marker for tumor recurrence in stage II colon cancer. Sci Rep. 2018;8(1):6157. doi: 10.1038/s41598-018-24519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ramaswamy P, Yadav R, Pal PK, Christopher R. Clinical application of circulating microRNAs in Parkinson’s disease: the challenges and opportunities as diagnostic biomarker. Ann Indian Acad Neurol. 2020;23(1):84–97. doi: 10.4103/aian.AIAN_440_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57(6):833–40. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 99.Felekkis K, Papaneophytou C. Challenges in using circulating micro-RNAs as biomarkers for cardiovascular diseases. Int J Mol Sci. 2020;21(2):561. doi: 10.3390/ijms21020561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glinge C, Clauss S, Boddum K, Jabbari R, Jabbari J, Risgaard B, et al. Stability of circulating blood-based microRNAs - pre-analytic methodological considerations. PLoS One. 2017;12(2):e0167969. doi: 10.1371/journal.pone.0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Markowitz J, Abrams Z, Jacob NK, Zhang X, Hassani JN, Latchana N, et al. MicroRNA profiling of patient plasma for clinical trials using bioinformatics and biostatistical approaches. Onco Targets Ther. 2016;9:5931–41. doi: 10.2147/ott.s106288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD, et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013;8(6):e64795. doi: 10.1371/journal.pone.0064795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matias-Garcia PR, Wilson R, Mussack V, Reischl E, Waldenberger M, Gieger C, et al. Impact of long-term storage and freeze-thawing on eight circulating microRNAs in plasma samples. PLoS One. 2020;15(1):e0227648. doi: 10.1371/journal.pone.0227648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the microRNA spectrum between serum and plasma. PLoS One. 2012;7(7):e41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data normalization strategies for microRNA quantification. Clin Chem. 2015;61(11):1333–42. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Mensah M, Borzi C, Verri C, Suatoni P, Conte D, Pastorino U, et al. MicroRNA based liquid biopsy: the experience of the plasma miRNA signature classifier (MSC) for lung cancer screening. J Vis Exp 2017(128):56326. 10.3791/56326. [DOI] [PMC free article] [PubMed]
- 107.Tay JW, James I, Hughes QW, Tiao JY, Baker RI. Identification of reference miRNAs in plasma useful for the study of oestrogen-responsive miRNAs associated with acquired protein S deficiency in pregnancy. BMC Res Notes. 2017;10(1):312. doi: 10.1186/s13104-017-2636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pizzamiglio S, Bottelli S, Ciniselli CM, Zanutto S, Bertan C, Gariboldi M, et al. A normalization strategy for the analysis of plasma microRNA qPCR data in colorectal cancer. Int J Cancer. 2014;134(8):2016–8. doi: 10.1002/ijc.28530. [DOI] [PubMed] [Google Scholar]
- 109.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheng L, Lo LY, Tang NL, Wang D, Leung KS. CrossNorm: a novel normalization strategy for microarray data in cancers. Sci Rep. 2016;6:18898. doi: 10.1038/srep18898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Suo C, Salim A, Chia KS, Pawitan Y, Calza S. Modified least-variant set normalization for miRNA microarray. RNA. 2010;16(12):2293–303. doi: 10.1261/rna.2345710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sewer A, Gubian S, Kogel U, Veljkovic E, Han W, Hengstermann A, et al. Assessment of a novel multi-array normalization method based on spike-in control probes suitable for microRNA datasets with global decreases in expression. BMC Res Notes. 2014;7:302. doi: 10.1186/1756-0500-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garmire LX, Subramaniam S. Evaluation of normalization methods in mammalian microRNA-Seq data. RNA. 2012;18(6):1279–88. doi: 10.1261/rna.030916.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9(2):276. doi: 10.3390/cells9020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Z, Sha HH, Li HJ. Functions and mechanisms of miR-186 in human cancer. Biomed Pharmacother. 2019;119:109428. doi: 10.1016/j.biopha.2019.109428. [DOI] [PubMed] [Google Scholar]
- 116.Geva GA, Gielchinsky I, Aviv N, Max KEA, Gofrit ON, Gur-Wahnon D, et al. Urine cell-free microRNA as biomarkers for transitional cell carcinoma. BMC Res Notes. 2017;10(1):641. doi: 10.1186/s13104-017-2950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Niu Y, Zhang L, Qiu H, Wu Y, Wang Z, Zai Y, et al. An improved method for detecting circulating microRNAs with S-Poly(T) Plus real-time PCR. Sci Rep. 2015;5:15100. doi: 10.1038/srep15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou L, Lim MYT, Kaur P, Saj A, Bortolamiol-Becet D, Gopal V, et al. Importance of miRNA stability and alternative primary miRNA isoforms in gene regulation during Drosophila development. Elife. 2018;7:e38389. doi: 10.7554/eLife.38389. [DOI] [PMC free article] [PubMed] [Google Scholar]


