Summary
The hypothalamus is a region of the brain that plays an important role in regulating body functions and behaviors. There is a growing interest in human pluripotent stem cells (hPSCs) for modeling diseases that affect the hypothalamus. Here, we established an hPSC-derived hypothalamus organoid differentiation protocol to model the cellular diversity of this brain region. Using an hPSC line with a tyrosine hydroxylase (TH)-TdTomato reporter for dopaminergic neurons (DNs) and other TH-expressing cells, we interrogated DN-specific pathways and functions in electrophysiologically active hypothalamus organoids. Single-cell RNA sequencing (scRNA-seq) revealed diverse neuronal and non-neuronal cell types in mature hypothalamus organoids. We identified several molecularly distinct hypothalamic DN subtypes that demonstrated different developmental maturities. Our in vitro 3D hypothalamus differentiation protocol can be used to study the development of this critical brain structure and can be applied to disease modeling to generate novel therapeutic approaches for disorders centered around the hypothalamus.
Subject areas: Cell biology, Neuroscience, Stem cells research, Transcriptomics
Graphical abstract
Highlights
-
•
We established a novel protocol for generating hypothalamus organoids in vitro
-
•
Mature hypothalamus organoids consisted of 11 broad cell types
-
•
Mature hypothalamus organoids contained 17 neuronal subtypes
-
•
We identified seven dopaminergic neuronal subtypes in hypothalamus organoids
Cell biology; Neuroscience; Stem cells research; Transcriptomics
Introduction
The hypothalamus is a small yet highly conserved structure in the center of the brain that performs a diverse set of functions to maintain homeostasis, including regulating body temperature, circadian rhythm, feeding, and reproduction. This structure works closely with the pituitary gland to regulate hormone secretion and therefore serves as a link between the nervous and endocrine systems.1,2 Due to the multifaceted role of the hypothalamus, its dysfunction can cause a variety of disorders, including disturbances in sleep and temperature regulation, as well as developmental abnormalities.3 The hypothalamus-pituitary-adrenal (HPA) axis regulates the stress response and its dysregulation has been associated with eating and metabolic disorders such as obesity, diabetes mellitus, and diabetes insipidus.2,4,5 The hypothalamus is also affected in several neurodegenerative disorders such as Alzheimer’s disease and is thought to play an important role in a subset of symptoms that are associated with dysregulation of metabolic homeostasis.2,6
It is therefore crucial to understand the structural complexity, cellular composition, and molecular characteristics of the hypothalamus to improve disease modeling approaches. The hypothalamus contains several regions (referred to as nuclei) with a diverse set of neurons and neuroendocrine cells that regulate distinct functions.1,7 Moreover, the hypothalamus contains a variety of neurons such as GABAergic, glutamatergic, and catecholaminergic cell types, including dopaminergic neurons (DNs).8,9 There are several DN subtypes in the hypothalamus, referred to as A11–A15, with various functions such as regulation of sensorimotor integration, nociceptive processing, and prolactin secretion.10,11,12 In the context of the neurodegenerative disorder Parkinson’s disease (PD), the hypothalamic DN subtypes are relatively unaffected compared to the midbrain subtypes A8-A10, although some of the nonmotor symptoms have been attributed to changes in the hypothalamus.13,14 However, much is still unknown about the physiology of hypothalamic DN subtypes. Therefore, modeling these neurons in vitro can enhance our understanding of cell type-specific vulnerability and neuroprotection in PD. Human pluripotent stem cells (hPSCs) are a useful tool for generating cell types of interest in vitro, and hPSC-derived 3D organoid models faithfully recapitulate some aspects of physiology due to the similarity of their cytoarchitecture to that of organs in vivo.15,16
Here, we developed a high-throughput 3D differentiation protocol to derive mature and electrophysiologically active hypothalamus organoids from hPSCs. We used an established tyrosine hydroxylase (TH)-TdTomato reporter to track DNs in hypothalamus organoids over time.17,18 We applied single-cell RNA sequencing (scRNA-seq), a robust method to assay the transcriptome in each cell, to examine hypothalamus cellular diversity in long-term cultures, comparing TH-TdTomato-sorted DNs to other cell types. We identified a diversity of neuronal and non-neuronal cell types, including molecularly distinct hypothalamic DN subtypes representing different developmental stages. The combination of 3D modeling, DN reporter, and long-term cultures is advantageous over other existing hypothalamus differentiation protocols as it allows for examination of hypothalamic DN subtypes, as well as non-neuronal cell types that emerge later during embryonic development.19,20,21,22 Our model can be used to study the spectrum of disorders in which hypothalamic DNs and other cell types are differentially affected, opening the path for novel therapeutic strategies.
Results
Generation and characterization of hPSC-derived hypothalamus organoids
The hypothalamus contains several DN subtypes with various functions.12 To better understand the molecular diversity among these subtypes, we modeled them in vitro using an hPSC line containing a TH-TdTomato reporter (WA01 TH #2; See STAR Methods). We developed and optimized a hypothalamus differentiation protocol with the aim of producing higher percentages of hypothalamic TH+ neurons. We used 3D spinner flasks for high-throughput generation of hypothalamus organoids. During embryonic development, the forebrain gives rise to several brain structures, including the hypothalamus.23 Forebrain patterning in our in vitro protocol began with dual-SMAD inhibition for neural induction, WNT inhibition for rostral patterning, and SHH activation to induce ventralization (Figure 1A). On day 12 (D12) the media was changed to a terminal differentiation medium to promote the maturation of neural progenitor cells (NPCs) into DNs. The organoids increased in size during and after the patterning stage, with an average area of 0.15 mm2 by D30 of differentiation (Figures S1A and S1B).
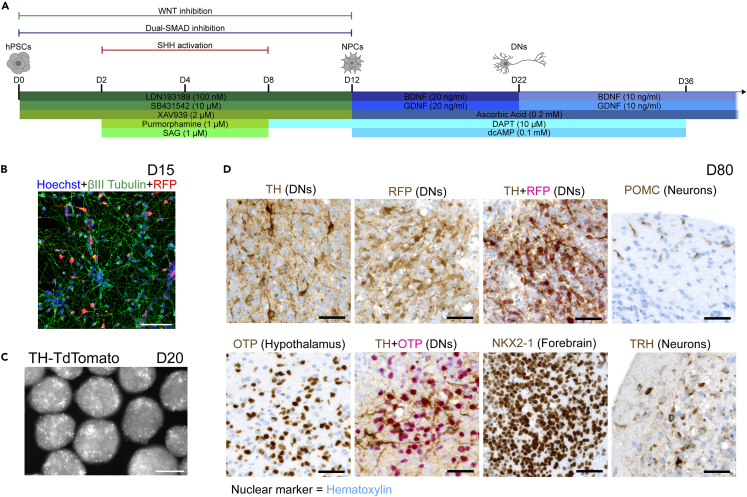
Figure 1.
Characterization of hypothalamus organoids
(A) Schematic overview of the hypothalamus differentiation protocol from hPSCs to NPCs and mature cells including DNs. hPSCs, human pluripotent stem cells; NPCs, neural progenitor cells; DNs, dopaminergic neurons.
(B) IF co-staining in D15 monolayer neurons for βIII Tubulin and RFP. Hoechst was used as a nuclear marker. Scale bar: 100 μm. IF, immunofluorescence; D15, day 15, RFP, red fluorescent protein.
(C) Fluorescent image of D20 organoids showing expression of the TH-TdTomato reporter. Scale bar: 250 μm. TH, tyrosine hydroxylase.
(D) IHC stains for TH and RFP for DNs, hypothalamic and ventral forebrain markers, OTP and NKX2-1, and hypothalamus-specific neuronal markers, POMC and TRH, in D80 hypothalamus organoids. TH + RFP co-staining was used for validation of the TH-TdTomato reporter in DNs, while TH + OTP co-staining showed a diversity of cell types. Hematoxylin was used as a nuclear marker in all images. Scale bars: 50 μm. IHC, immunohistochemistry.
See also Figure S1.
To validate hypothalamic fate, the organoids were dissociated on D12 and plated as a monolayer for immunofluorescence (IF) staining at the NPC stage. By D15, 97% of the total cell population was positive for the ventral forebrain marker, NKX2-1, and approximately 5% expressed the hypothalamus marker, OTP (Figures S1C and S1D). OTP has been shown to have a region-specific expression in the developing hypothalamus.24 The small percentage of OTP+ cells on D15 suggested that these cells began to emerge at the NPC stage as they obtained hypothalamic identity. We previously demonstrated a near 100% overlap between the DN marker, TH, and TdTomato (red fluorescent protein or RFP) to validate our TH-TdTomato reporter lines throughout differentiation.17,18 Almost all cells expressed the neuronal marker, βIII Tubulin, by D15. Furthermore, 7% of all cells and 6% of βIII Tubulin+ cells expressed RFP, suggesting that the hypothalamic DNs emerged at the NPC stage (Figures 1B and S1D). TH expression in full organoids became robust by D20 of differentiation as observed by the TH-TdTomato reporter (Figure 1C).
Immunohistochemistry (IHC) staining in D80 organoids further confirmed DN fate and hypothalamic identity (Figure 1D). A population of hypothalamic DNs was observed using TH and RFP markers. Overlap between these two markers via co-staining validated the TH-TdTomato reporter at this differentiation stage (Figure 1D). We also observed a population of OTP+ cells in D80 organoids. Although these cells did not show region specificity within the organoids, co-staining of TH and OTP revealed a diverse set of cells that expressed TH only, OTP only, or both (Figure 1D). This observation was consistent with prior in vivo work that showed OTP expression is restricted to specific nuclei of the hypothalamus and to specific DN subtypes during development.25,26 NKX2-1 staining confirmed commitment to the forebrain fate by D80 (Figure 1D). Moreover, we detected smaller populations of POMC- and TRH-expressing neurons that have specific neuroendocrine functions in the hypothalamus (Figure 1D).
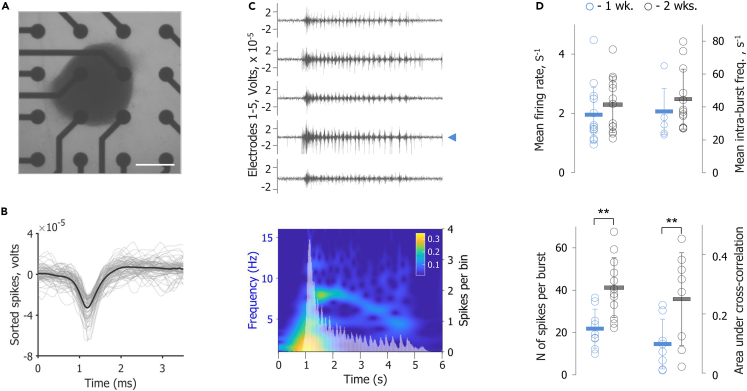
To evaluate hypothalamus organoid functionality, we performed extracellular recordings of spontaneous electrical activity using the multi-electrode array (MEA) approach. Hypothalamus organoids were plated as one organoid per well on D94. Each well contained 16 low-impedance platinum microelectrodes that were 50 μm in diameter and spaced 350 μm apart (Figure 2A). Electrical activity of hypothalamus organoids was recorded one and two weeks after plating on D101 and D108, respectively. Neuronal spikes from individual electrodes covered by the organoid were isolated by sorting for the standard extracellular action potential waveform structure with typical refractory periods (Figure 2B), and the electrodes displayed a robust pattern of activity (Figure 2C; Top). Electrical activity in hypothalamus organoids exhibited periods of relative quiescence and short bursts of spontaneous network-synchronized spiking (network bursts). These network bursts were periodic, occurring roughly every 30 s with the prominent presence of nested faster oscillatory peaks in the theta-frequency range (4–8 Hz) (Figure 2C; Bottom). On average, over the course of two weeks the hypothalamus organoids showed no significant change in the mean firing rate per electrode and the mean intra-burst frequency, or the firing rate during a network burst (Figure 2D; Top). However, the metrics of network synchronicity, such as the number of spikes in the network burst and the area under cross-correlation of spike times in different electrodes, demonstrated a significant increase (Figure 2D; Bottom). Together, our data demonstrated that hypothalamus organoids contained a diverse set of functional neurons with robust electrophysiological properties and maturing network oscillatory activity. In conclusion, the hypothalamus organoids showed successful patterning and contained electrophysiologically active neurons upon maturation.
Figure 2.
Electrophysiological properties of mature hypothalamus organoids
(A) Transmitted light image of a hypothalamus organoid in a well of a 48-well MEA plate. Scale bar: 300 μm. MEA, multi-electrode array.
(B) Representative example of individual action potentials recorded from a single electrode, marked with a blue arrowhead in C (top), of an MEA plate. An overlay of 40 peak-aligned spikes is shown in gray with the average spike waveform in black.
(C) Top: Electrical activity from five individual MEA electrodes recorded during an organoid-wide network activity burst. Bottom: A compound spike-time histogram from all electrodes overlaid with its CWT. Color bar indicates CWT power. CWT, continuous wavelet transform.
(D) Top: Comparison of mean firing rate of hypothalamus organoids (left) and mean intra-burst frequency, or frequency of spiking within the network burst (right), in s −1 for 1 week (blue = D101) and 2 weeks (black = D108) on the MEA plate. Each data point indicates an individual organoid and the error bars represent SD. s−1, inverse second or per second; wk., week; SD, standard deviation. Bottom: Comparison of the mean N of spikes in the network burst (left) and the area under cross-correlation of spike times (right). Statistical significance was derived by unpaired Student’s t test. p values: ∗∗ = p < 0.01. N = number.
Cellular diversity in hypothalamus organoids is revealed by scRNA-seq analysis
We next sought to identify the cell types present in the hypothalamus organoids by applying scRNA-seq (Figure 3A). We used D85 organoids to allow for the emergence of glial cells, such as astrocytes, as previously reported.18,27 First, approximately 60 organoids per replicate were dissociated and cells were sorted into THhi and THlo populations with fluorescence-activated cell sorting (FACS) using the TH-TdTomato (RFP) reporter (Figure S2A). The THhi gate contained the cells with the highest TdTomato intensity which constituted about 3.6% of the population. The THlo gate contained all the remaining cells that either expressed no TH or had relatively lower expression levels. A negative TdTomato (-TdTomato) control consisted of neurons differentiated from the KOLF2.1J hPSC line without the TH-TdTomato reporter. Although approximately 54% of the cells were TH+ on D85 compared to the -TdTomato control (Figure S2B), we selected the highest TH-expressing population by setting a stringent gate for the THhi population. As a result, we enriched for DNs as they are expected to express the highest TH levels.28 Approximately 75,000 THhi and 100,000 THlo cells were sorted for each replicate, thereby enriching the 3.6% THhi population to about 43%. Antibody hashtags were used to delineate triplicates for each sorted population before pooling even proportions of each sample and capturing single cells with 10x Genomics technology.
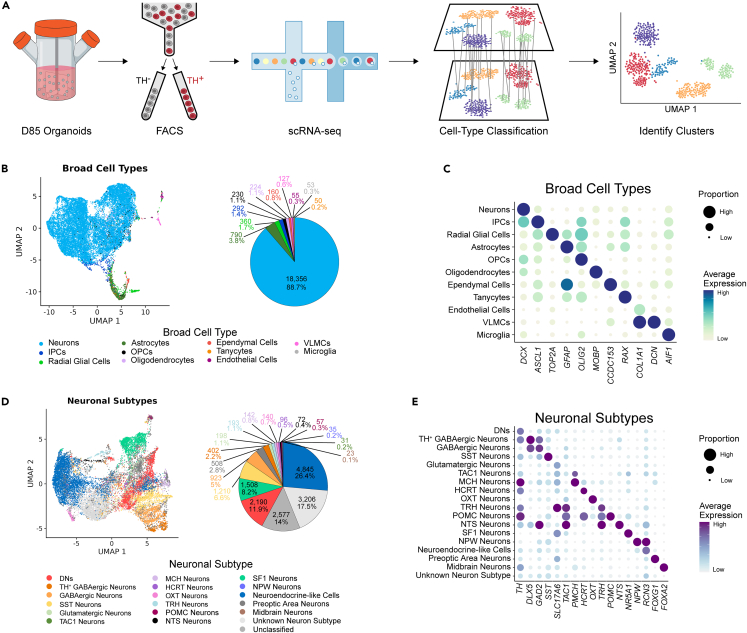
Figure 3.
Identification of cell types in D85 hypothalamus organoids by scRNA-seq
(A) Schematic overview of the scRNA-seq approach from sorting to sequencing and analysis. scRNA-seq, single-cell RNA sequencing.
(B) Left: UMAP plot of the broad cell type categories in the dataset derived by supervised clustering and classification. UMAP, Uniform Manifold Approximation and Projection. Right: Pie chart showing distribution of the different broad cell type populations.
(C) Dot plot showing average gene expression and proportion of cells expressing representative markers for each broad cell type.
(D) Left: UMAP plot of the neuronal subtype categories within the neurons of the broad cell types derived by unsupervised clustering. Right: Pie chart showing distribution of the different neuronal subtype populations.
(E) Dot plot showing average gene expression and proportion of cells expressing representative markers for each neuronal subtype.
See also Figures S2–S8 and Tables S1 and S2.
We captured 27,966 cells (13,902 THhi and 14,064 THlo cells). We used a suite of validated methods to detect and remove empty droplets, doublets, droplets without detectable hashtags, and cells with high mitochondrial percent (See STAR Methods). Each replicate demonstrated similar distributions for common quality metrics before and after filtering for high-quality cells (Figure S2C). After the quality control (QC) steps a total of 21,001 cells (2,647–4,015 cells per THhi replicate for a total of 10,464 THhi cells and 3,262–3,663 cells per THlo replicate for a total of 10,537 THlo cells) were used for analysis. The TdTomato reporter sequence was localized at the 3′ end of the TH gene of one allele in the WA01 TH #2 hPSC line (heterozygous TH-TdTomato knock-in). Due to the 3′ capture of 10x Genomics sequencing, we anticipated that the reads from the TH-TdTomato allele would map to the TdTomato sequence. As a result, TH+ cells could express TH only, TdTomato only, or both (Figures S2D and S2E).
We then annotated the cell populations in our dataset using scPred, a supervised method that annotates cells based on similarity to reference transcriptomes.29 For this purpose, we leveraged three publicly available hypothalamus datasets; two mouse scRNA-seq datasets that were obtained at several developmental timepoints and one human single nucleus RNA-seq (snRNA-seq) dataset, which consisted of samples from multiple neonatal developmental ages.19,30,31 The three datasets were processed and combined before annotating 12 cell types (Figures S3A and S3B; See STAR Methods). A group of 304 cells that could not be annotated were labeled as “unassigned” and were excluded from the analysis (Table 1). The mitochondrial percent demonstrated similar distributions across all cell types (Figure S3C). The combined datasets were used to train a model with high sensitivity and specificity (Figure S3D). Application of the prediction model identified all 12 broad cell types in the hypothalamus organoids with high prediction probabilities (Figures 3B, 3C, and S3E). The organoids primarily comprise neurons, and, as expected, this cell type was especially enriched in the TH+ flow sorted cells (Figures 3B and S4A; Table 1). The organoids contained a larger proportion of neurons compared to the reference datasets, partially due to the neuronal enrichment in the TH+ flow sorted cells as well as technical differences among the studies (Figure S4B; Table S1).
Table 1.
The number of cells classified per category in broad cell types
| # | Name | Number of Cells |
|---|---|---|
| 1 | Neurons | 18,356 |
| 2 | Astrocytes | 790 |
| 3 | Radial Glial Cells | 360 |
| 4 | Unassigned | 304 |
| 5 | IPCs | 292 |
| 6 | OPCs | 230 |
| 7 | Oligodendrocytes | 224 |
| 8 | Ependymal Cells | 160 |
| 9 | VLMCs | 127 |
| 10 | Endothelial Cells | 55 |
| 11 | Microglia | 53 |
| 12 | Tanycytes | 50 |
| Total | 21,001 |
See also Figures S3–S5.
In addition to neurons, we found intermediate progenitor cells (IPCs), radial glial cells, and astrocytes, indicating that the hypothalamus organoids contained progenitor cells that could give rise to young neurons or astrocytes (Figures 3B and 3C). Observation of astrocytes in the hypothalamus organoid cultures was consistent with previous work in which the emergence of this glial cell type was reported in brain organoids starting at around D60 of differentiation.18,27 We also observed unexpected and rare populations of seven other non-neuronal cell types that each constituted less than 1.4% of the entire population. These groups included oligodendrocyte precursor cells (OPCs), oligodendrocytes, ependymal cells, tanycytes, vascular and leptomeningeal cells (VLMCs), and microglia (Figures 3B and 3C; Table 1). Due to the small number of cells and transcriptional similarity of oligodendrocytes (n = 176) and immature oligodendrocytes (n = 48), we considered them a single cell type (oligodendrocytes; Figure S4C). Cell type-specific markers that differentiated the populations in the reference datasets demonstrated similar expression patterns in the hypothalamus organoids (Figure S5A; Table S2). Common markers for the 11 broad cell types further supported the model annotations (Figures 3C and S5B). These included DCX for neurons, ASCL1 for IPCs, TOP2A for radial glial cells, GFAP for astrocytes, OLIG2 for OPCs, MOBP for oligodendrocytes, CCDC153 for ependymal cells, RAX for tanycytes, COL1A1 for endothelial cells, DCN for VLMCs, and AIF1 for microglia. The percentage of each cell type was different in the hypothalamus organoid and reference datasets (Figure S4B; Table S1).
Next, we identified neuronal cell subtypes using an unsupervised clustering approach followed by bootstrapping to identify cells that were misannotated during clustering. Cell labels were updated during bootstrapping if the cell was predicted to be a different neuronal subtype with high probability (Figure S6A; See STAR Methods). We identified a diverse set of 17 neuronal subtypes (Figures 3D, 3E, S6B, and S7A). A group of 3,206 cells did not express any known markers used for clustering and thus was labeled as “unknown neuron subtype.” An additional 2,577 cells across all clusters had high probabilities for two different cell types, as identified by bootstrapping, and were therefore considered “unclassified.” Since the cells were enriched for TH via sorting, we examined TH expression across different cell types. Among the broad cell types, neurons and IPCs had the highest TH expression levels, suggesting that the hypothalamus organoids contained both immature and mature TH+ neurons (Figure S7B). Within the neuronal subtypes the highest TH levels were observed in DNs and TH+ GABAergic neurons, as well as POMC, MCH, HCRT, and TRH neurons (Figure S7C). The largest neuronal subtype was the neuroendocrine-like cells which comprised 26.4% of all neurons (Figures 3D and 3E; Table 2). This population was annotated based on the expression of CREB3, which plays a role in neuroendocrine regulation, as well as PCSK1 and RCN3, which are markers of the anterior pituitary gland.32,33,34 The different expression patterns of CREB3 and RCN3 compared to PCSK1 suggest that further investigation of this cell population may reveal distinct subtypes of neuroendocrine cells (Figures 3D, 3E, and S8A). In addition, SIX3, which is important for both hypothalamic and pituitary development,35 was expressed throughout the neurons in our dataset, including in the neuroendocrine-like cells (Figures 3D and S8A).
Table 2.
The number of cells annotated per category in neuronal subtypes
| # | Name | Number of Cells |
|---|---|---|
| 1 | Neuroendocrine-like Cells | 4,845 |
| 2 | Unknown Neuron Subtype | 3,206 |
| 3 | Unclassified | 2,577 |
| 4 | DNs | 2,190 |
| 5 | SF1 Neurons | 1,508 |
| 6 | SST Neurons | 1,210 |
| 7 | GABAergic Neurons | 923 |
| 8 | Preoptic Area Neurons | 508 |
| 9 | TH+ GABAergic Neurons | 402 |
| 10 | Glutamatergic Neurons | 198 |
| 11 | TRH Neurons | 193 |
| 12 | MCH Neurons | 142 |
| 13 | OXT Neurons | 140 |
| 14 | HCRT Neurons | 96 |
| 15 | NTS Neurons | 72 |
| 16 | POMC Neurons | 57 |
| 17 | NPW Neurons | 35 |
| 18 | TAC1 Neurons | 31 |
| 19 | Midbrain Neurons | 23 |
| Total | 18,356 |
See also Figure S6.
We previously observed high expression of the forebrain marker, NKX2-1, in hypothalamus organoids on D15 and D80 (Figures 1D, S1C, and S1D). To determine whether our organoids had mixed regional identity, we examined markers of non-hypothalamic tissues that are found adjacent to the hypothalamus in vivo in our D85 organoid scRNA-seq dataset. While the majority of the cells expressed NKX2-1 and the hypothalamus marker, OTP, we also observed lower levels of FOXG1 (Figure S8B), which is an important marker for the development and regulation of the ventral telencephalon.36,37 Since FOXG1 expression has been previously shown in preoptic area cells in the developing chicken embryo,38 we annotated these cells as preoptic area neurons which constituted 2.8% of the neurons (Figures 3D and 3E). However, it may be possible that these cells represent a mix of preoptic area and ventral telencephalon neurons.
We then investigated a series of thalamus markers. GBX2, NEUROG2, OLIG2, and OLIG3 have been reported to be expressed in the developing mouse thalamus,39 GATA2 and GATA3 are markers of the developing human thalamus,24 while ESRRG, GBX2, LEF1, POU4F1, PROX1, SIX3, and TCF7L2 have been described as thalamus markers in the adult mouse and human brain.40 Among these markers, we did not detect expression of GATA2, GATA3, GBX2, NEUROG2, OLIG2, OLIG3, or POU4F1 in our dataset. However, we did observe sporadic expression of ESRRG, LEF1, and PROX1 in the neuronal population (Figure S8C). These three markers have been previously reported to be expressed in the developing hypothalamus.41,42,43 Therefore, even though these markers are expressed in the adult thalamus, it is likely that they do not represent thalamic cell types in our dataset since the D85 organoids have a relatively embryonic gene expression profile. Lastly, we observed strong expression of SIX3 throughout the neuronal population (Figure S8A), suggesting that the cells have a hypothalamic/pituitary fate.35 Taken together, we conclude that our organoids do not contain any cells of thalamic identity. We also looked at the midbrain markers, FOXA2 and LMX1A, which showed a similar expression pattern in a small subset of the neurons (Figure S8D). We selected FOXA2 as the representative marker for midbrain neurons which constituted 0.1% of the neuronal population in our dataset (Figures 3D and 3E). In summary, our hypothalamus organoids demonstrated neuronal subtype diversity. Further characterization of the different cell types in our system may help in better understanding their physiology in vivo.
Validation of cellular diversity in hypothalamus organoids
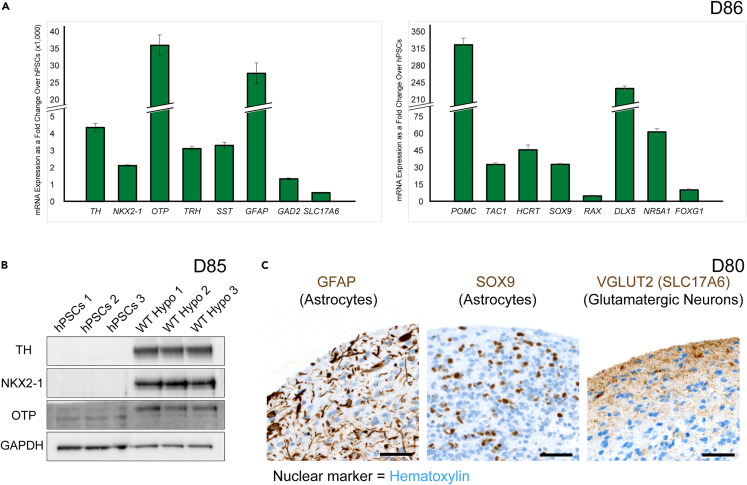
To confirm our scRNA-seq observations, we examined the mRNA expression of several cell type-specific markers for broad categories and neuronal subtypes using validated quantitative reverse transcription polymerase chain reaction (qRT-PCR) primers in D86 hypothalamus organoids (Figures 4A and S9A). These included the DN marker, TH, ventral forebrain/hypothalamus markers NKX2-1 and OTP, neuronal markers, GAD2, SLC17A6, and DLX5, markers for specific neuroendocrine cell types or regions of the hypothalamus, TRH, SST, POMC, TAC1, HCRT, NR5A1, and FOXG1, as well as glial markers, GFAP, SOX9, and RAX. SOX9 is a broad glial cell marker that is found co-expressed with different markers in astrocytes, ependymal cells, and tanycytes.44 We also confirmed protein expression of TH, NKX2-1, and OTP by western blot analysis in D85 hypothalamus organoids (Figure 4B). Moreover, expression of the astrocyte markers, GFAP and SOX9, as well as the glutamatergic neuron marker, VGLUT2 (encoded by the SLC17A6 gene) were confirmed by IHC in D80 hypothalamus organoids (Figure 4C). Several of the markers used in this study were validated in postmortem adult hypothalamus tissue by IHC (Figure S9B). TH, POMC, TRH, GFAP, and VGLUT2 showed robust expression. However, NKX2-1, OTP, and SOX9 had lower expression levels, likely due to the primary roles of these markers in early brain development and decreased levels or specific localization in the adult brain.25,45,46
Figure 4.
Validation of marker expression in hypothalamus organoids
(A) qRT-PCR of hPSCs and D86 organoids for several markers identified by scRNA-seq analysis (n = 3). ACTB was used as a housekeeping gene for normalization. Error bars represent SD. qRT-PCR, quantitative real-time PCR.
(B) Western blot of hPSCs and D85 organoids for TH, NKX2-1, and OTP. GAPDH was used as loading control.
(C) IHC staining of D80 organoids for the astrocyte markers, GFAP and SOX9, as well as the glutamatergic neuron marker, VGlut2 (encoded by the SLC17A6 gene). Hematoxylin was used as a nuclear marker. Scale bars: 50 μm.
See also Figure S9.
Characterization of hypothalamic DN subtypes and developmental stages in hypothalamus organoids
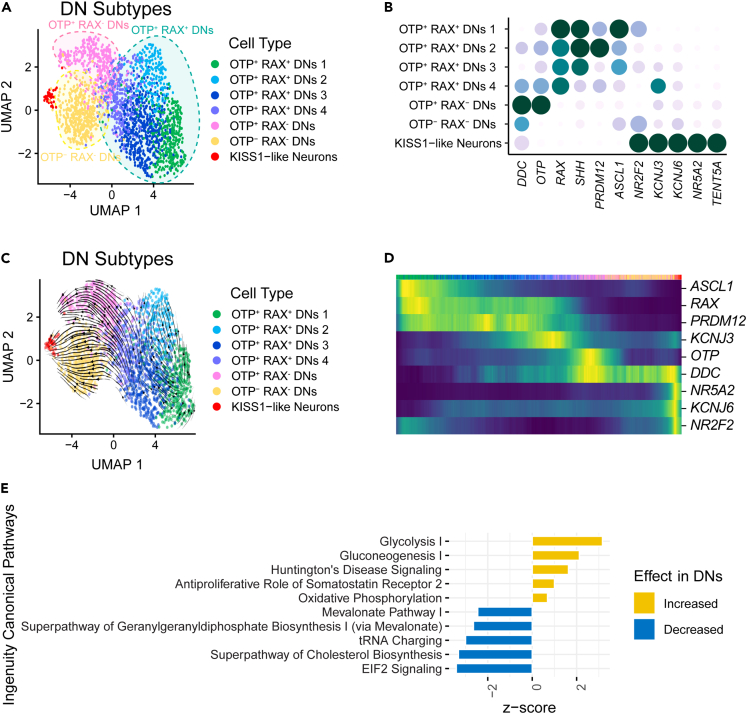
Several hypothalamic DN subtypes have been identified in vivo,12 but their diversity has not yet been explored using single cell approaches. We sought to determine the DN subtypes in the hypothalamus organoids using unsupervised clustering. We identified seven DN subtypes that consisted of four groups of OTP+ RAX+ DNs, as well as OTP+ RAX− DNs, OTP− RAX− DNs, and KISS1-like neurons (Figures 5A, 5B, and S10A; Table 3). These subtypes are consistent with our previous observation that OTP is expressed in a subset of TH+ cells (Figure 1D). OTP is known to be a region-specific hypothalamus marker in the arcuate, paraventricular, and supraoptic nuclei.24 In addition, RAX is known to play an important role in early development of the arcuate and ventromedial nuclei in the hypothalamus.47 KISS1 neurons are hypothalamic neuroendocrine cells that play a role in reproductive processes and have been shown to express TH.24,48,49,50 These DN subtypes were characterized by a collection of markers. Although not highly expressed, expression of the DN marker, DDC, was highest in the two RAX− DN subtypes (Figures 5B and S10A). In addition, KISS1-like neurons (38 cells) expressed NR5A2 and TENT5A which are previously reported markers of KISS1 neurons (Figures 5B and S10A).24,48 The mature DN marker, KCNJ6, was expressed at low levels throughout all DN subtypes, with highest expression in KISS1-like neurons, suggesting that these neurons were relatively mature in comparison to the other DN subtypes (Figures 5B and S10A).
Figure 5.
Molecular diversity within the DN subtypes in hypothalamus organoids
(A) UMAP plots illustrating the distribution of annotated DN subtypes.
(B) Dot plot showing the expression of select markers that were used to annotate the DN subtypes.
(C) RNA velocity arrows overlaid on the DN subtype UMAP, showing pseudotime trajectories.
(D) RNA velocity heatmap showing the distribution of the DN subtype markers across the pseudotime trajectories.
(E) IPA results showing the pathways that were upregulated (yellow = increased) or downregulated (blue = decreased) in DNs compared to non-DN neurons in the organoids based on significance or p value. IPA, Ingenuity Pathway Analysis.
See also Figure S10 and Table S3.
Table 3.
The number of cells annotated and list of markers expressed per category in DN subtypes
| # | Name | Number of Cells | Markers |
|---|---|---|---|
| 1 | OTP+ RAX+ DNs (2) | 491 | OTP, RAX, SHH, PRDM12, ASCL1 |
| 2 | OTP− RAX− DNs | 432 | DDC, NR2F2 |
| 3 | OTP+ RAX+ DNs (1) | 342 | OTP, RAX, SHH, PRDM12, ASCL1, NR2F2 |
| 4 | OTP+ RAX+ DNs (4) | 339 | OTP, RAX, KCNJ3 |
| 5 | OTP+ RAX− DNs | 334 | DDC, OTP |
| 6 | OTP+ RAX+ DNs (3) | 214 | OTP, RAX, SHH, ASCL1 |
| 7 | KISS1-like Neurons | 38 | NR2F2, KCNJ3, KCNJ6, NR5A2, TENT5A |
| Total | 2,190 |
See also Figure S10.
The four OTP+ RAX+ DNs were distinguished based on the expression of several markers (Figures 5A, 5B, and S10A; Table 3). Groups 1–3 had the highest levels of SHH expression, groups 1–2 had relatively high PRDM12 expression, while group 1 had highest levels of ASCL1 and NR2F2 compared to the other OTP+ RAX+ DN groups, with some expression of ASCL1 also in groups 2–3. On the other hand, group 4 had the highest expression levels of KCNJ3. SHH is important in hypothalamus development and has been reported to become more restricted to the neuroepithelium in later developmental stages.51 The transcription factor PRDM12 has been studied in the context of nociceptive neurons, as well as POMC neurons, and plays an important role in neurogenesis and early neuronal development.52,53,54 ASCL1 and NR2F2 are neural progenitor cell markers.55,56 Moreover, KCNJ3 is a relatively mature marker for neurons, playing an important role in neuronal excitability.57 These observations suggest that OTP+ RAX+ DN groups 1-4 may reflect different developmental maturities of these hypothalamic DN subtypes. Outside the OTP+ RAX+ DN groups, OTP− RAX− DNs also expressed NR2F2, while KISS-like neurons had a relatively high expression of both NR2F2 and KCNJ3, suggesting that these DN subtypes are relatively mature (Figures 5B and S10A).
To confirm these findings, we performed RNA velocity analysis and determined the pseudotime trajectories of the DN subtypes. We found that the DNs showed a developmental trajectory from the OTP+ RAX+ to OTP+ RAX− and OTP− RAX− groups, with the KISS1-like neurons showing the most mature subtypes (Figures 5C, 5D, S10B, and S10C). Altogether, these findings suggest that the hypothalamus organoids contain several molecularly distinct DN subtypes representing different developmental stages or maturation levels. We then performed Ingenuity Pathway Analysis (IPA) to examine the signaling pathways that were differentially regulated in DNs (TH+) compared to other neurons (TH−) (Figure 5E; Table S3). We observed an upregulation of pathways such as glycolysis I, gluconeogenesis I, and oxidative phosphorylation in DNs, while EIF2 signaling, superpathway of cholesterol biosynthesis, tRNA charging, and superpathway of geranylgeranyl diphosphate biosynthesis I (via mevalonate) were downregulated. Taken together, we observed that the hypothalamic DNs in our organoids demonstrated molecular diversity and had a transcriptional profile in line with an increase in energy production compared to other neuronal populations.
Discussion
In this study, we introduced a 3D hypothalamus organoid model that contains diverse disease-relevant cell types and applied it to study TH+ DN populations. The hypothalamus plays an important role in hormonal maintenance and its dysregulation can result in various disorders.1,2,4,5 In addition, the hypothalamus contains several DN subtypes, A11-A15, with distinct functions. The A12 subtype is the most well-characterized group and has been shown to inhibit prolactin secretion from the anterior pituitary.10,11 The A11 subtype regulates locomotion, nociception or the detection of pain, and sensory processes.12,58 The A13 subtype also plays a role in nociception, but less is known about the functions of the A14 and A15 subtypes.12,59,60 Unlike the DN subtypes in the midbrain, the hypothalamic DNs are not impacted by neurodegeneration in PD.14 Investigating the mechanisms of neuroprotection in these neurons can help us gain insight into the underlying causes of cell type-specific vulnerability in PD and other neurodegenerative disorders. Here, we adapted a previous 2D monolayer culture approach to develop a 3D hypothalamus differentiation protocol in vitro.20 Unlike other existing hypothalamus protocols, our approach generates organoids with relatively homogeneous structure and size at large-scale using spinner flasks. Moreover, the differentiation strategy enhances DN production and is ideal for studying these neurons in vitro. Starting on D12 of the differentiation protocol, the organoids can continue to be cultured in spinner flasks or transferred to low-attachment plates on a shaker for chemical or genetic perturbations. Alternatively, they can be dissociated and plated as a 2D monolayer or splatted as full organoids on a coated dish to allow for the outgrowth of axons.18 The different culture formats render versatility to our protocol based on experimental needs.
Using our previously established TH-TdTomato reporter, we enriched for DNs and observed diversity of neuronal and non-neuronal cell types in D85 hypothalamus organoids via scRNA-seq analysis (Figure 3). The TH+ flow sorted population contained ∼90% neurons, although we also found smaller populations of IPCs, radial glial cells, and astrocytes that comprised roughly 7% of total cells. In addition, the organoids contained rare populations of OPCs, oligodendrocytes, ependymal cells, tanycytes, endothelial cells, VLMCs, and microglia. Each of these 7 groups constituted only up to about 1.4% of the total population. Due to their low numbers, we could not verify these cell types. However, some of them have been previously reported in brain organoid cultures. For example, studies using forebrain-patterned retinal organoids and whole-brain organoids showed microglial populations in their transcriptomic profiles, while long-term cultures of whole-brain organoids resulted in the emergence of OPCs and oligodendrocytes.61,62,63 We also observed cell type diversity within the neuronal population in D85 hypothalamus organoids, including neuroendocrine-like cells. Although several of the cell types in our dataset are considered neuroendocrine cells due to their roles in hormonal regulation, they were annotated separately from neuroendocrine-like cells based on the expression of specific neuroendocrine cell type markers, such as POMC, TRH, and SST. We examined the expression of TH within the neuronal subtypes and observed that in addition to DNs and TH+ GABAergic neurons, other neuronal subtypes also showed high TH levels, including POMC, MCH, HCRT, and TRH neurons (Figures 3E and S7C). These neurons have been shown to co-localize or interact with TH-expressing cells in different regions of the hypothalamus, although it has not been reported whether they co-express TH.64,65,66,67,68 Examining the transcriptomic profiles of these cell types can reveal their roles in hypothalamus function.
We then examined the expression of markers of adjacent non-hypothalamic tissues to determine whether our forebrain-pattern organoids had mixed regional identity. We found small populations of preoptic area/ventral telencephalon and midbrain cells (Figures S8B–S8D). Although we observed sporadic expression of the thalamus markers, ESRRG, LEF1, and PROX1 (Figure S8C), upon further examination of these markers we concluded it was unlikely that our organoids contained cells of thalamic identity. ESRRG and other estrogen receptor genes have been previously reported to be expressed in the POMC neurons of the mouse hypothalamus.42 Similarly, in our organoid dataset the ESRRG-expressing cells were most highly concentrated in the POMC neuron population, and therefore did not represent cells with thalamic identity (Figures 3D, S6B, and S8C). Moreover, both LEF1 and PROX1 have been shown to be expressed in the developing hypothalamus in mice.41,43 Since our hypothalamus organoids have a relatively embryonic gene expression profile, it is likely that these markers of adult thalamus do not represent thalamic cell types in our dataset.
Several markers have been established for DNs, notably TH, DDC, VMAT2, and KCNJ6.69,70 The DNs in our dataset expressed TH and other DN markers, including DDC and KCNJ6 (Figures 5B and S10A). However, they did not express VMAT2. Combinations of the three markers TH, DDC, and VMAT2 have been reported in different DN subtypes in vivo, including DNs that lack VMAT2 expression.28 The midbrain DN subtypes in the substantia nigra and ventral tegmental area express all three of these markers. In contrast, DN subtypes in other brain regions may express TH and DDC only, while DNs in areas such as specific nuclei of the hypothalamus express TH only. We found that the DNs in hypothalamus organoids fell into the two latter categories, as some expressed DDC and some did not. We used two regional markers of early hypothalamus development, OTP and RAX, to cluster the DN subtypes into three main categories.24,47 Using additional markers, we found molecular diversity within the DN population which could be reflective of the DN subtypes found in vivo. Moreover, RNA velocity analysis demonstrated that the different DN groups followed a pseudotime trajectory representing their relative developmental maturity (Figures 5C, 5D, S10B, and S10C). However, defining the DNs subtypes in vitro is not a straightforward process. Other factors, such as the structural organization of the neurons within the hypothalamus in vivo and their interactions with other nearby cells, likely contribute to their subtype specification during development. A more in-depth characterization of the DNs in the hypothalamus organoids as well as comparison of our dataset to additional in vivo references with DN-related clusters will aid in this process. Our observations from IPA suggested that hypothalamic DNs have high energy demands that are met by an increase in glucose and energy production compared to other neuronal subtypes (Figure 5E). At the same time, these neurons downregulate their protein translation and biosynthesis of lipids and other compounds to preserve energy. This view is consistent with previously reported high energy demands of other DN subtypes in the midbrain,71 hinting at a similarity in the physiology of DN subtypes of different origins.
Taken together, our hypothalamus differentiation protocol serves as a useful tool for generating forebrain-patterned DNs and other cell types at large-scale. Long-term culture of these organoids results in their molecular and functional maturity with robust electrophysiological properties. Our model can be applied to studying not only hypothalamic DNs, but also other neuronal and non-neuronal cell types that may be of interest for disease modeling. Future studies using this protocol can help in further understanding the role of the hypothalamus in a wide range of diseases, including metabolic, neurodegenerative, and developmental disorders, with the aim of establishing novel therapeutic strategies.
Limitations of the study
In this study, the hPSC line, H1 (WA01), was used to generate hypothalamus organoids. In future studies, it would be crucial to apply our hypothalamus differentiation protocol to other hPSC lines of both male and female origins to confirm successful patterning across cell lines with different genetic backgrounds. Moreover, this study was performed on a wild-type cell line to examine the cell types that emerge in hypothalamus organoids in the absence of a disease condition. However, future studies using disease models will be important in validating our differentiation protocol as a useful in vitro tool for studying disorders that affect the hypothalamus.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Monoclonal Anti-Beta III Tubulin (Clone 2G10) | Abcam | Cat#ab78078; RRID: AB_2256751 |
| Rabbit Polyclonal Anti-GAPDH | Abcam | Cat#Ab9485; RRID: AB_307275 |
| Mouse Monoclonal Anti-Glial Fibrillary Acidic Protein (Clone GA5) | Millipore | Cat#MAB360; RRID: AB_11212597 |
| Rabbit Polyclonal Anti-OTP | Thermo Fisher Scientific | Cat#PA5-31513; RRID: AB_2548986 |
| Rabbit Monoclonal Anti-POMC (EPR17571) | Abcam | Cat#Ab210605; RRID: N/A |
| Rabbit Polyclonal Anti-RFP | Rockland | Cat#600-401-379; RRID: AB_2209751 |
| Rabbit Monoclonal Anti-SOX9 (EPR14335-78) | Abcam | Cat#Ab185966; RRID: AB_2728660 |
| Mouse Monoclonal Anti-TH (Clone LNC1) | Millipore | Cat#MAB318; RRID: AB_2201528 |
| Rabbit Polyclonal Anti-TRH | Thermo Fisher Scientific | Cat#PA5-97006; RRID: AB_2808808 |
| Rabbit Monoclonal Anti-TTF1 (NKX2-1) (Clone EPR5955(2)) | Abcam | Cat#Ab133737; RRID: AB_2811263 |
| Mouse Monoclonal Anti-VGLUT2 (Clone CL2921) | Atlas Antibodies | Cat#AMAb91081; RRID: AB_2665793 |
| Biological samples | ||
| Postmortem Hypothalamus (PCR) | Neuropathology Brain Bank and Research CoRE, Mount Sinai | Cat#NPBB023; RRID: N/A |
| Postmortem Hypothalamus (IHC) | Neuropathology Brain Bank and Research CoRE, Mount Sinai | Cat#NPBB127; RRID: N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Accutase Cell Detachment Solution | Innovative Cell Technologies | Cat#AT104; RRID: AB_2869384 |
| B-27™ Supplement (50X), Minus Vitamin A | Gibco | Cat#12587010; RRID: N/A |
| Blotto, Non-Fat Dry Milk | ChemCruz | Cat#sc-2324; RRID: N/A |
| Bovine Albumin Fraction V (7.5% solution) | Gibco | Cat#15260037; RRID: N/A |
| DAPT | Cayman Chemical | Cat#13197; RRID: N/A |
| DB-cAMP / Dibutyryl-cAMP | BioLog | Cat#D 009; RRID: N/A |
| DMEM/F-12, GlutaMAX™ Supplement | Gibco | Cat#10565018; RRID: N/A |
| DNase Vial (D2) | Worthington Biochemical Corporation | Cat#LK003170; RRID: N/A |
| Geltrex™ LDEV-Free, hESC-Qualified, Reduced Growth Factor Basement Membrane Matrix | Gibco | Cat#A1413302; RRID: N/A |
| HBSS with 10 mM HEPES, Without Phenol Red | STEMCELL Technologies | Cat#37150; RRID: N/A |
| Hibernate A | BrainBits | Cat#HA; RRID: N/A |
| Hoechst 33342, Trihydrochloride, Trihydrate - 10 mg/mL Solution in Water | Thermo Fisher Scientific | Cat#H3570; RRID: N/A |
| Laminin Mouse Protein, Natural | Gibco | Cat#23017015; RRID: N/A |
| L-Ascorbic Acid (White Crystalline Powder) | Fisher Scientific | Cat#BP351-500; RRID: N/A |
| LDN 193189 Dihydrochloride | Tocris | Cat#6053; RRID: N/A |
| Leibovitz's L-15 Medium | Gibco | Cat#11415064; RRID: N/A |
| N-2 Supplement (100X) | Gibco | Cat#17502048; RRID: N/A |
| Normal Goat Serum | Thermo Fisher Scientific | Cat#31872; RRID: N/A |
| PBS, pH 7.4 (-CaCl2, -MgCl2) | Gibco | Cat#10010023; RRID: N/A |
| PDS Kit, Papain Vial | Worthington Biochemical Corporation | Cat#LK003176; RRID: N/A |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140122; RRID: N/A |
| Pierce™ 16% Formaldehyde (w/v), Methanol-free | Thermo Fisher Scientific | Cat#28908; RRID: N/A |
| Poly-L-Ornithine Hydrobromide | Sigma-Aldrich | Cat#P3655; RRID: N/A |
| Ponceau S Solution | Sigma-Aldrich | Cat#P7170; RRID: N/A |
| Purmorphamine | STEMCELL Technologies | Cat#72204; RRID: N/A |
| Recombinant Human BDNF Protein | R&D Systems | Cat#248-BD; RRID: N/A |
| Recombinant Human GDNF Protein | R&D Systems | Cat#212-GD; RRID: N/A |
| SAG | Cayman Chemical | Cat#11914; RRID: N/A |
| SB431542 | Stemgent | Cat#04-0010-10; RRID: N/A |
| StemFlex™ Medium | Gibco | Cat#A3349401; RRID: N/A |
| Thiazovivin | Selleck Chemicals | Cat#S1459; RRID: N/A |
| Triton™ X-100 Surfact-Amps™ Detergent Solution | Thermo Fisher Scientific | Cat#85111; RRID: N/A |
| XAV939 | Tocris | Cat#3748; RRID: N/A |
| Y-27632 Dihydrochloride | Tocris | Cat#1254; RRID: N/A |
| Critical commercial assays | ||
| 4X Bolt™ LDS Sample Buffer | Thermo Fisher Scientific | Cat#B0007; RRID: N/A |
| Antibody Dilution Buffer | Roche Diagnostics | Cat#05280524001; RRID: N/A |
| cOmplete™, Mini Protease Inhibitor Cocktail | MilliporeSigma | Cat#11836153001; RRID: N/A |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#4368814; RRID: N/A |
| Maestro Multiwell 768 channel MEA system with USB | Axion Biosystems | Cat# AAVMAESTRO768DEA; RRID: N/A |
| Monarch® Total RNA Miniprep Kit | New England BioLabs | Cat#T2010S; RRID: N/A |
| NuPAGE™ MOPS SDS Running Buffer (20X) | Thermo Fisher Scientific | Cat#NP0001; RRID: N/A |
| Phosphatase Inhibitor Cocktail II | Abcam | Cat#ab201113; RRID: N/A |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225; RRID: N/A |
| PowerUp SYBR Green Master Mix | Applied Biosystems | Cat#A25742; RRID: N/A |
| Q5® Hot Start High-Fidelity 2X Master Mix | New England BioLabs | Cat#M0494S; RRID: N/A |
| RIPA Lysis and Extraction Buffer | Thermo Fisher Scientific | Cat#89900; RRID: N/A |
| TBS Buffer, 20X Liquid | VWR | Cat#10791-796; RRID: N/A |
| TWEEN® 20 | Sigma-Aldrich | Cat#P1379; RRID: N/A |
| ULTRA Cell Conditioning (ULTRA CC1) | Roche Diagnostics | Cat#950-224; RRID: N/A |
| Western Lightning Plus, Chemiluminescent Substrate (130 mL) | PerkinElmer | Cat#NEL103001EA; RRID: N/A |
| Deposited data | ||
| scRNA-seq Dataset (GEO) | This paper | GSE237274 |
| scRNA-seq Dataset - processed (Mendeley Data) | This paper | https://doi.org/10.17632/nxtkn2w28v.1 |
| Customs Codes and Script for scRNA-seq Dataset (GitHub) | This paper | https://github.com/powellgenomicslab/hypothalamus_organoid_scRNA |
| Experimental models: Cell lines | ||
| Human: H1 (WA01) hESCs | WiCell Research Institute | Cat#WA01; RRID: CVCL_9771 |
| Human: KOLF2.1J hiPSCs | The Jackson Laboratory | Cat#KOLF2.1J; RRID: N/A |
| Oligonucleotides | ||
| Primer: ACTB Forward: CCTGGCACCCAGCACAAT | Pilegaard et al.72 | RRID: N/A |
| Primer: ACTB Reverse: GCCGATCCACACGGAGTACT | Pilegaard et al.72 | RRID: N/A |
| Primer: DLX5 Forward: GTCTTCAGCTACCGATTCTGAC | PrimerBank | Cat#84043959c3; RRID: N/A |
| Primer: DLX5 Reverse: CTTTGCCATAGGAAGCCGAG | PrimerBank | Cat#84043959c3; RRID: N/A |
| Primer: FOXG1 Forward: TGGCCCATGTCGCCCTTCCT | Nolbrant et al.73 | RRID: N/A |
| Primer: FOXG1 Reverse: GCCGACGTGGTGCCGTTGTA | Nolbrant et al.73 | RRID: N/A |
| Primer: GAD2 Forward: TTTTGGTCTTTCGGGTCGGAA | PrimerBank | Cat#197276619c1; RRID: N/A |
| Primer: GAD2 Reverse: TTCTCGGCGTCTCCGTAGAG | PrimerBank | Cat#197276619c1; RRID: N/A |
| Primer: GFAP Forward: CTGCGGCTCGATCAACTCA | PrimerBank | Cat#334688843c1; RRID: N/A |
| Primer: GFAP Reverse: TCCAGCGACTCAATCTTCCTC | PrimerBank | Cat#334688843c1; RRID: N/A |
| Primer: HCRT Forward: TTCCACAAAGGTCTCCTGGG | This paper | RRID: N/A |
| Primer: HCRT Reverse: TAGAGGCGGCAAGAGCAAG | This paper | RRID: N/A |
| Primer: NKX2-1 Forward: TGAGTCCAAAGCACACGACTCC | This paper | RRID: N/A |
| Primer: NKX2-1 Reverse: CCTCCATGCCCACTTTCTTGTA | This paper | RRID: N/A |
| Primer: NR5A1 Forward: GGGTGGCCGGAACAAGTTT | PrimerBank | Cat#169881245c2; RRID: N/A |
| Primer: NR5A1 Reverse: GCCCGAATCTGTGCCTTCT | PrimerBank | Cat#169881245c2; RRID: N/A |
| Primer: OTP Forward: CGACATCTTTATGCGTGAGGAG | This paper | RRID: N/A |
| Primer: OTP Reverse: GTCTTTTTGCGCTTCTTCCACT | This paper | RRID: N/A |
| Primer: POMC Forward: CTCCCGAGACAGAGCCTCA | Flori et al.74 | RRID: N/A |
| Primer: POMC Reverse: CACTCCAGCAGGTTGCTTTC | This paper | RRID: N/A |
| Primer: RAX Forward: AAGCCCCTCGACCCTACTG | PrimerBank | Cat#126116580c2; RRID: N/A |
| Primer: RAX Reverse: CCGCCGATGCTTTTTCTTGG | PrimerBank | Cat#126116580c2; RRID: N/A |
| Primer: SLC17A6 Forward: TGGACATGGTCAACAACAGCA | PrimerBank | Cat#215820654c2; RRID: N/A |
| Primer: SLC17A6 Reverse: GGAACCGTGGATCATCCCC | PrimerBank | Cat#215820654c2; RRID: N/A |
| Primer: SOX9 Forward: GTACCCGCACTTGCACAAC | Masuda et al.75 | RRID: N/A |
| Primer: SOX9 Reverse: TCTCGCTCTCGTTCAGAAGTC | Masuda et al.75 | RRID: N/A |
| Primer: SST Forward: ACCCAACCAGACGGAGAATGA | PrimerBank | Cat#71979669c1; RRID: N/A |
| Primer: SST Reverse: GCCGGGTTTGAGTTAGCAGA | PrimerBank | Cat#71979669c1; RRID: N/A |
| Primer: TAC1 Forward: CAGCGACCAGATCAAGGAGG | This paper | RRID: N/A |
| Primer: TAC1 Reverse: GGGCGATTCTCTGCAGAAGA | This paper | RRID: N/A |
| Primer: TH Forward: GGAAGGCCGTGCTAAACCT | PrimerBank | Cat#88900504c1; RRID: N/A |
| Primer: TH Reverse: GGATTTTGGCTTCAAACGTCTC | PrimerBank | Cat#88900504c1; RRID: N/A |
| Primer: TRH Forward: TCCTCCGGGAAAACATCCAG | PrimerBank | Cat#306518589c1; RRID: N/A |
| Primer: TRH Reverse: GGAGAGCCAGTCAGATTGAAAGA | PrimerBank | Cat#306518589c1; RRID: N/A |
| Software and algorithms | ||
| Aperio ImageScope | https://www.leicabiosystems.com/digital-pathology/manage/aperio-imagescope/ | RRID: SCR_020993 |
| AxIS | https://www.axionbiosystems.com/products/axis-software | RRID: SCR_016308 |
| CellProfiler Image Analysis Software | https://cellprofiler.org/ | RRID: SCR_007358 |
| Cell Ranger | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger | RRID: SCR_017344 |
| Cyclone | Scialdone et al.76 | RRID: N/A |
| Demuxafy | Neavin et al.77 | RRID: N/A |
| DropletQC | Muskovic and Powell78 | RRID: N/A |
| FSC Express 7 | https://denovosoftware.com/ | RRID: N/A |
| Geneious Prime | http://www.geneious.com/ | RRID: SCR_010519 |
| GraphPad Prism | http://www.graphpad.com/ | RRID: SCR_002798 |
| MAST | Finak et al.79 | RRID: SCR_016340 |
| Microsoft Excel | https://www.microsoft.com/en-gb/ | RRID: SCR_016137 |
| R | https://www.r-project.org/ | RRID: SCR_001905 |
| scDblFinder | Germain et al.80 | RRID: SCR_022700 |
| scds (Single Cell Doublet Scoring) | Bais and Kostka81 | RRID: SCR_021541 |
| scPred | Alquicira-Hernandez et al.29 | RRID: N/A |
| scVelo | https://github.com/theislab/scvelo | RRID: SCR_018168 |
| Seurat | https://satijalab.org/seurat/ | RRID: SCR_007322 |
| Velocyto | http://velocyto.org/ | RRID: SCR_018167 |
| Other | ||
| Aperio VERSA Brightfield, Fluorescence & FISH Digital Pathology Scanner | Leica Biosystems | RRID: N/A |
| BD FACSAria II Cell Sorter | BD Biosciences | RRID: SCR_018934 |
| Benchmark Special Stains Stainer Module | Roche Diagnostics | Cat#06468373001; RRID: N/A |
| CellInsight CX7 HCS Platform | Thermo Fisher Scientific | RRID: N/A |
| Corning® 125 mL Disposable Spinner Flask with 70 mm Top Cap and 2 Angled Sidearms, Sterile | Corning | Cat#3152; RRID: N/A |
| CytoView MEA Plate 48 | Axion Biosystems | Cat#M768-tMEA-48B; RRID: N/A |
| Eclipse Ts2R-fl | Nikon | RRID: N/A |
| Falcon® Round-Bottom Tubes with Cell Strainer Cap, 5 mL | STEMCELL Technologies | Cat#100-0087; RRID: N/A |
| Fisherbrand™ Sterile Cell Strainers (70 μm) | Fisher Scientific | Cat#22-363-548; RRID: N/A |
| iBlot™ 2 Gel Transfer Device | Thermo Fisher Scientific | Cat#IB21001; RRID: N/A |
| iBlot™ Transfer Stack, PVDF, mini | Thermo Fisher Scientific | Cat#IB401002; RRID: N/A |
| NuPAGE™ 4 to 12%, Bis-Tris, 1.0–1.5 mm, Mini Protein Gels | Thermo Fisher Scientific | Cat#NP0321BOX; RRID: N/A |
| NuPAGE™ 4 to 12%, Bis-Tris, 1.0–1.5 mm, Mini Protein Gels | Thermo Fisher Scientific | Cat#NP0323BOX; RRID: N/A |
| pluriStrainer® 300 μm, 25 pcs. – Sterile (Cell Strainer) | pluriSelect | Cat#43-50300-03; RRID: N/A |
| QuantSudio 7 Flex Real-Time PCR System | Applied Biosystems | Cat#4485701; RRID: N/A |
| Stirrers, Magnetic, Nine-position, Dura-Mag | Chemglass Life Sciences | Cat#CLS-4100-09; RRID: N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Joel Blanchard (joel.blanchard@mssm.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
The human cell lines used in this study are detailed in the key resources table. The two cell lines, H1 (WA01) and KOLF2.1J, were cultured in StemFlex medium and maintained in incubators set to 37°C and 5% CO2. Both cell lines are male in origin.
Method details
hPSC culture
Two hPSC lines were used in this study, the human embryonic stem cell (hESC) line, H1 (WA01), and the human induced pluripotent stem cell (hiPSC) line, KOLF2.1J. The WA01 TH #2 line was previously gene edited using CRISPR/Cas9 to contain a TH-TdTomato knock-in reporter.18 hPSCs were maintained in tissue culture incubators set to 37°C and 5% CO2. Cells were cultured on Geltrex-coated (Gibco # A1413302) 10-cm dishes in 8 ml StemFlex medium (Gibco #A3349401) with media changes every other day. When the hPSCs reached a confluency of 70-80%, they were passaged at a split ratio of 1:10 using Accutase (Innovative Cell Technologies #AT104). The media was removed, the cells were washed with 8 ml PBS (Gibco #10010023), and incubated with 3 ml Accutase for 5 minutes (min) at room temperature (RT) with occasional manual shaking. Cells were then gently pipetted 3-5 times, transferred to a 15-ml tube containing PBS supplemented with 2% BSA solution (Gibco #15260037; “Bovine Albumin Fraction V (7.5% solution)” is referred to as “BSA solution” here) and 2 μM Thiazovivin Rho-associated, coiled-coil containing protein kinase (ROCK) inhibitor (Thia; Selleck Chemicals #S1459), and centrifuged at 500 g at RT for 3 min. The supernatant was aspirated and the pellet was resuspended in StemFlex supplemented with 2 μM Thia. The cells were plated in new Geltrex-coated dishes in StemFlex supplemented with 2 μM Thia and the media was changed to fresh StemFlex on the following day to remove Thia.
Hypothalamus organoid differentiation
The spinner flask approach for generation of organoids was adapted from a previous publication.82 hPSCs were collected from four 10-cm dishes and seeded into a 125-ml disposable spinner flask (Corning #3152) in Stemflex supplemented with 10 μM Y-27632 ROCK inhibitor (Tocris #1254) and Penicillin-Streptomycin (Pen/Strep; Gibco #15140122) in a total volume of 120 ml. The flask was placed on a nine-position stir plate (Chemglass Life Sciences #CLS-4100-09) at 65 rpm in the incubator. On day 2 after seeding, half of the culture medium (60 ml) was replaced with fresh Stemflex supplemented with 10 μM Y-27632 and Pen/Strep. On day 4 after seeding, the spheres were collected and filtered through a 300 μm strainer (pluriSelect #43-50300-03) into a 50 ml tube to remove large organoids and clumps of cells. The spheres were settled by gravity and the media was aspirated carefully. The spheres were then transferred back to the flask in 120 ml D0-1 medium to begin hypothalamus patterning. The base medium throughout the differentiation consisted of DMEM/F12+Glutamax (Gibco #10565018) supplemented with 1x B27 Supplement, minus vitamin A (Gibco #12587010) and 1x N-2 Supplement (Gibco #17502048). Half the media (60 ml) was changed daily throughout the patterning stage unless otherwise noted. The D0-1 medium consisted of the BMP inhibitor, LDN193189 (100 nM; Tocris #6053), the TGF-β inhibitor, SB431542 (10 μM; Stemgent #04-0010-10), and XAV939 (2 μM; Tocris #3748) to induce floor plate formation via dual-SMAD inhibition and promote forebrain patterning. The D2-7 medium also contained Purmorphamine (1 μM; STEMCELL Technologies #72202) and SAG (1 μM; Cayman Chemical #11914) for SHH activation and ventralization of the developing NPCs. Purmorphamine and SAG were removed in the D8-11 medium and replaced with DAPT (10 μM; Cayman Chemical #13197). On D8, 80 ml media was changed to reduce the final concentrations of SHH agonists in the remaining media. On D12, 80 ml media was changed as well to switch to the terminal differentiation medium consisting of BDNF (20 ng/ml; R&D Systems #248-BD), GDNF (20 ng/ml; R&D Systems #212-GD), Ascorbic Acid (0.2 mM; Fisher Scientific #BP351-500), DAPT (10 μM), and dcAMP (0.1 mM; BioLog #D 009) to promote DN fate commitment and maturation. Starting on D12 the media was changed every other day. On D22, the concentrations of both BDNF and GDNF were reduced to half (10 ng/ml), as these growth factors have been shown to mask DN-related phenotypes in vitro.83 Beginning on D36, DAPT and dcAMP were excluded from the medium, as DAPT has been reported to inhibit astrocyte proliferation in long-term cultures and both compounds may affect DN-associated phenotypes.84,85,86
Organoid culture options
Starting on D12, hypothalamus organoids could be continued to be cultured in flasks. Alternatively, they could be transferred to low-attachment 6-well plates on a shaker in the incubator for pharmacological or genetic perturbations, and to reduce media usage. Due to the small size of the hypothalamus organoids, it is recommended to culture up to approximately 60 organoids per well of a 6-well plate, although this number may vary depending on the organoid size in different hPSC lines. The organoids could also be cultured in a 2D monolayer format upon dissociation for live imaging and IF staining, or in a 2.5D format (splatting) to examine axon outgrowths and morphology.18 The coating procedure for the culture plates consisted of incubating the wells with 1 mg/ml Poly-L-Ornithine (PLO; Sigma-Aldrich #3655) in borate buffer at 37°C overnight (O/N). On the following day, the PLO solution was collected for re-use and the wells were washed with ddH2O three times for 10 min each. The wells were then coated with 10 μg/ml laminin (Gibco #23017015) in L15 media (Gibco #11415064) supplemented with 2.5% sodium bicarbonate solution (stock concentration: 7.5 g/L) and incubated at 37°C O/N. On the following day, the laminin solution was aspirated, and the cells were plated in culture media.
For organoid dissociation, up to 60 D12 hypothalamus organoids were placed in a well of a 6-well plate and washed with 5 ml PBS. The organoids were then incubated in 5 ml papain (Worthington Biochemical Corporation #LK003178) in HBSS-H (STEMCELL Technologies #37150) supplemented with DNase (Worthington Biochemical Corporation #LK003170) and 10 μM Y-27632 ROCK inhibitor for 20-40 min (depending on organoid size) on a shaker in the incubator. The organoids were gently triturated using fire-polished glass pipets of decreasing tip sizes. The cell suspension was filtered using a 70 μm strainer (Fisher Scientific #22-363-548) into a 50-ml tube containing 10 ml Hibernate-A medium (BrainBits #HA) supplemented with 1x B27 (minus vitamin A), 1% BSA solution, DNase, and 10 μM Y-27632 ROCK inhibitor. The cells were centrifuged at 800 g for 5 min and the pellet was resuspended in media. The suspension was filtered through a 35 μm strainer (STEMCELL Technologies #100-0087) and 40,000 cells were plated per well of a PLO/laminin-coated 96-well plate for imaging or IF staining. Addition of Y-27632 ROCK inhibitor to the media upon plating greatly enhanced cell survival. The media was changed after two days to remove Y-27632 ROCK inhibitor and regular half media changes were performed every 2-3 days depending on changes in the media color.
qRT-PCR
RNA was extracted from full organoids using the Monarch Total RNA Miniprep Kit (New England BioLabs #T2010S). cDNA synthesis was performed on 1-2 μg RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems #4368814). The 20 μl reaction was then diluted at 1:10 and 4 μl cDNA was used per 10 μl qRT-PCR reaction with the PowerUp SYBR Green Master Mix (Applied Biosystems #A25742). The samples were run on a QuantSudio 7 Flex Real-Time PCR System (Applied Biosystems #4485701). The delta-delta Ct (2–ΔΔCt) method was used to analyze the results.87 The key resources table lists the qRT-PCR primers used in this study.
PCR validation of qRT-PCR primers
The qRT-PCR primers were confirmed by PCR and Sanger sequencing (Figure S9A). PCR reactions were set up for each primer set using postmortem hypothalamus cDNA and the Q5 Hot Start High-Fidelity 2x Master Mix (New England BioLabs #M0494S) in a total volume of 40 μl. For each reaction mix, 4 μl was run on a 2% agarose gel to screen for an expected band size of between 100 and 200 base pairs. The remaining reaction mix of samples that had one clear band was analyzed by Sanger sequencing with either the forward or reverse primer. The Geneious Prime software was used to align the sequencing results to the cDNA of the genes of interest.
Western Blot
Protein was extracted from organoids using RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific #89900) supplemented with protease and phosphatase inhibitors (MilliporeSigma #11836153001 and Abcam #ab201113, respectively). Protein quantification was performed using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific #23225). Sample mixes were prepared in 1x Bolt LDS Sample Buffer (Thermo Fisher Scientific #B0007) and boiled at 100°C for 5 min. 40 μg protein was loaded per well of a NuPAGE 4-12% Bis-Tris Gel (Thermo Fisher Scientific #NP0321BOX or #NP0323BOX) and the gel was run in 1x NuPAGE MOPS SDS Running Buffer (Thermo Fisher Scientific #NP0001). The samples were transferred to an iBlot PVDF Transfer Stack membrane (Thermo Fisher Scientific #IB401002) using an iBlot 2 Gel Transfer Device (Thermo Fisher Scientific #IB21001). Ponceau S (Sigma-Aldrich #P7170) staining was used to confirm proper protein transfer. The membrane was blocked in 5% milk (ChemCruz #sc-2324) in 1x TBST at RT for 1 hour, followed by incubation with the primary antibody diluted in TBST at 4°C O/N. The 1x TBST solution was prepared using 20x TBS Buffer (VWR #10791-796), ddH2O, and 0.1% TWEEN® 20 (Sigma #P1379). On the following day, the membrane was washed three times with TBST and then incubated in secondary antibody diluted in TBST at RT for 45 min. The membrane was washed again three times with TBST and then imaged using ECL (PerkinElmer #NEL103001EA). Table S4 (top) and the key resources table list the antibodies and dilutions used for Western blot in this study.
IF staining
Monolayer cultures of neurons in a 96-well plate were washed with PBS and fixed in fresh 4% PFA (Thermo Fisher Scientific #28908) diluted in PBS at RT for 3 min. They were then rinsed quickly in PBS followed by two 5 min washes. The cells were blocked in 0.1% Triton-X (Thermo Fisher Scientific #85111) in PBS containing 5% goat serum (Thermo Fisher Scientific #31872) at RT for 1 hour and incubated in primary antibody diluted in blocking buffer at 4°C O/N. On the following day the samples were washed 3 times with PBS and incubated in secondary antibody diluted in blocking buffer at RT for 45 min. The samples were washed 5 times with PBS and in the third wash Hoechst (Thermo Fisher Scientific #H3570) was added at 1:5,000 for nuclear staining. Table S4 (middle) and the key resources table list the antibodies and dilutions used for IF staining.
IHC staining
Organoids were washed with PBS, fixed in fresh 4% PFA at RT for 30-60 min (depending on organoid size) on a shaker and stored in PBS at 4°C until embedding. The samples were embedded in paraffin and 5 μm sections were prepared and mounted on positively charged slides. The sections were baked O/N at 70°C. IHC staining was performed on a Ventana Benchmark XT automatic staining platform (Roche Diagnostics #06468373001) according to the manufacturer's guidelines. Antigen retrieval was performed using the CC1 buffer (Tris/Borate/EDTA buffer, pH 8.0 - 8.5; Roche Diagnostics #950-224) for 1 hour. The samples were incubated in primary antibodies diluted in antibody dilution buffer (ABD24; Roche Diagnostics #05280524001) and the incubation time was optimized for each antibody. The slides were then incubated in either 3,3'-diaminobenzidine (DAB) or alkaline phosphatase for visualization, and both reagents were used for slides that were double-stained. Hematoxylin was used for nuclear counterstaining, and the slides were coverslippped. Table S4 (bottom) and the key resources table list the antibodies and dilutions used for IHC staining in this study.
MEA recording and analysis
A 48-well MEA plate (Axion Biosystems #M768-tMEA-48B) was coated with 100 μg/ml PLO and 10 μg/ml laminin. Hypothalamus organoids were plated at one per well on the MEA plate on D94 with 16 wells containing organoids. The cultures were fed twice a week and measurements were collected 24 hours after the medium was changed for two weeks after plating (on D101 and D108). Recordings were performed using a Maestro MEA system (Axion Biosystems #AAVMAESTRO768DEA) and AxIS software Spontaneous Neural Configuration. Spikes were detected with AxIS software using an adaptive threshold crossing set to 5.5 times the standard deviation (SD) of the estimated noise for each electrode (channel). For the MEA analysis, the electrodes that detected at least 5 spikes/min were classified as active electrodes. Network bursts were defined as events with minimum number of 5 spikes and inter-spike interval <100 ms occurring in at least 25% of active electrodes. The synchrony index was calculated using a cross-correlogram synchrony window of 20 ms. Brightfield images were captured to assess cell density and electrode coverage. Custom MATLAB routine was used for plotting continuous wavelet transform (CWT) scalogram with the absolute value of the CWT plotted as a function of time and frequency of the compound activity signal during the network burst.
Imaging
Brightfield and fluorescent images of live organoids were captured at 4x or 10x magnification using a Nikon Eclipse Ts2R-fl microscope with a DS-Qi2 digital camera head (Nikon). IF staining images were obtained at 20x magnification using the CellInsight CX7 HCS Platform (Thermo Fisher Scientific). IHC images were taken at 20x or 40x magnification using an Aperio VERSA 8 Digital Slide Scanner (Leica Biosystems) and fields of interest were selected using the Aperio ImageScope software.
Image analysis
For IF staining experiments the CellProfiler software was used to calculate the percentage of positive cells. Images of hPSC stains were used as negative controls for each antibody. The threshold for positivity was established based on the hPSC mean intensity distribution. In CellProfiler, the region outside the nucleus was used to calculate mean fluorescence intensity.
FACS
Hypothalamus organoids were dissociated and pelleted as previously described. The cell pellet was resuspended in “FACS buffer” consisting of Hibernate-A medium supplemented with 1x B27 (minus vitamin A), 1% BSA solution, DNase, and 10 μM Y-27632 ROCK inhibitor and was filtered through a 35 μm strainer into a 5-ml Falcon round-bottom tube. The BD FACSAria II Cell Sorter (BD Biosciences) was used to enrich for the TH-TdTomato+ cells for the scRNA-seq experiment. The FACS gates consisted of the P1 gate to exclude cell debris, singlets to exclude doublets, and % live to exclude dead cells (Figure S2A). The TH-TdTomato gate was set based on a negative control using organoids differentiated form the KOLF2.1J hiPSC line (Figure S2B). The FSC Express 7 software was used to analyze the results.
scRNA-seq
Approximately 100,000 cells were sorted for each group (THhi and THlo) in triplicates. Cells were pelleted at a speed of 800 g at 4°C for 5 min, and the pellets were resuspended in PBS supplemented with 0.04% BSA. Viability of single cells was assessed using Trypan Blue staining and debris-free suspensions of >80% viability were deemed suitable for sequencing. scRNA-seq was performed using the Chromium platform (10x Genomics) with the 3’ Gene Expression (3’ GEX) with Feature Barcoding V3 kit, using an input of ∼10,000 cells. First, cells were labeled with barcoded antibodies to differentiate among individual samples. Then, labeled cells were combined and loaded, and Gel Bead-in-Emulsions (GEMs) were generated on the sample chip in the Chromium controller. Barcoded cDNA was extracted from the GEMs by Post-GEM RT-cleanup and amplified for 12 cycles. Amplified cDNA was fragmented and subjected to end-repair, poly A-tailing, adapter ligation, and 10x-specific sample indexing following the manufacturer’s protocol. Libraries were quantified using Bioanalyzer (Agilent) and QuBit (Thermo Fisher Scientific) analysis and were sequenced in paired end mode on a NovaSeq instrument (Illumina) targeting a depth of 50,000 - 100,000 reads per cell. Sequencing data was aligned and quantified using the Cell Ranger Single-Cell software Suite against the provided GRCh38 human reference genome that included TdTomato with default settings (See cellranger.sh code provided on Github). This resulted in 13,949 and 14,093 droplets captured for the TdTomato high and low pools, respectively.
scRNA-seq analysis
We anticipate about 1,568 doublets (droplets that contained two or more cells) since 14,000 droplets captured in a single pool using the 10x platform results in ∼11.2% doublets.77,88 We therefore employed both hashtag and transcriptome-based doublet removal methods. Hashtag demultiplexing was carried out with the Seurat package from the R software using “RC” (Relative Counts) with the MULTIseqDemux function (See Hash_Demultiplexing.R script provided on Github). Droplet-level QC was carried out with DropletQC to detect empty droplets and damaged cells and with scds to detect doublets (See DropletQC_scds.R script provided on Github).78,81 Droplets that were either classified as a doublet, did not have detectable hashtag antibodies from MULTIseqDemux and scds, or were identified as empty droplets by DropletQC, were removed from downstream analysis. Likewise, cells with more than 12.5% mitochondrial genes were also removed. After removal of droplets that did not pass the QC thresholds, 21,001 total cells remained consisting of 10,537 cells from the THhi and 10,464 from the THlo pools. The cell cycle state of each cell was estimated using Cyclone76).
Cell type classification
Broad cell types
We leveraged three publicly available datasets as references for cell type classification in the hypothalamus organoids. Two of those references were mouse scRNA-seq datasets with samples collected at multiple developmental timepoints.30,31 The final reference was a human neonatal snRNA-seq dataset with samples spanning several neonatal developmental ages.19 The three reference datasets were first processed separately. The human neonatal snRNA-seq dataset was filtered for 5% mitochondrial reads, consistent with the original manuscript, and cell types were annotated based on cluster gene expression according to the manuscript.19 Both mouse hypothalamus scRNA-seq datasets were pre-processed and filtered for cells with less than 10% mitochondrial reads (See Mouse_hypothalamus_preprocessing_nature.R and Mouse_hypothalamus_preprocessing_CSC.R scripts provided on Github).30,31 The three datasets were integrated together for Uniform Manifold Approximation and Projection (UMAP) visualization implemented in Seurat v4 (See Integrate.R script provided on Github).89 We used multiple rounds of clustering to annotate broad cell type groups: neurons, IPCs, radial glial cells, astrocytes, OPCs, immature oligodendrocytes, oligodendrocytes, ependymal cells, tanycytes, endothelial cells, VLMCs, and microglia (See Annotate_integrated_ref.R script provided on Github). We then identified and removed doublets using scds and scDblFinder implemented through Demuxafy (v0.0.3; See DoubletDetecting.R script on provided Github).77,80,81 We then applied this reference to predict cell types in the hypothalamus organoids using scPred and a combination of MDA and svmRadial to best train the model (Figure S3D).29 Cells that had high probability assignments for two different cell types (<0.15 difference) were considered low-confidence predictions and were thus labeled as unassigned and excluded from downstream analysis.
Neuronal subtypes
We used clustering to annotate the neurons present in the hypothalamus organoid dataset. Table S5 lists the markers that were used for this step. Following the original cluster annotation, we used bootstrapping to identify cells that were misclassified during clustering. Cells were reclassified to another cell type if their original neuron type annotation probability was less than 0.5 and if they had a probability greater than 0.5 of being another cell type while the other probabilities were at least 0.15 less than the highest probability. Any cells with a second highest neuron cell type probability within 0.15 of the highest neuron cell type probability were removed from downstream analysis since we could not be confident of the correct cell type.
DN subtypes
Cells identified as DNs were separated from the other cells, normalized, and clustered at multiple resolutions to identify a resolution that appropriately resulted in biologically relevant groups. A resolution of 0.7 was found to provide the best separation of different DNs and was used to annotate the subtypes (Figure 5A).
RNA velocity analysis
DNs were processed with velocyto to quantify the spliced and unspliced reads for each gene per cell (See velocyto.sh and velocyto_submit.sh scripts provided on Github). scVelo was used to estimate RNA velocities across DNs (See DN_RNA_velocity.py script provided on Github).
Differential expression and pathway analysis for DNs
We tested for genes that were differentially expressed between the DN population and non-DN neurons. We compared THhi DNs (isolated by FACS and annotated as DNs) that expressed TH or TdTomato mRNA (or both) to non-DN neurons that were THlo (isolated by FACS) and had no expression of either TH or TdTomato mRNA using the MAST algorithm and corrected for ribosomal percentage, the number of unique molecular identifiers (UMIs) per cell, and replicate (See DN_vs_neurons_DE.R script provided on Github).79
Quantification and statistical analysis
All experiments were performed in triplicates. The organoid size and IF staining quantifications were represented as mean ± standard error of the mean (SEM). qRT-PCR and MEA results were shown as mean ± SD. When applicable, the statistical details and information on significance were included in figure legends. The bar charts were generated in Microsoft Excel or GraphPad Prism. Single cell analysis and figures were carried out in R.
Acknowledgments
We thank the Genomics Core Facility at Mount Sinai for processing our samples for scRNA-seq. We are also thankful to the Neuropathology Brain Bank and Research CoRE for processing our samples for sectioning and IHC staining, as well as the Microscopy and Advanced Bioimaging Core and Flow Cytometry CoRE for providing training and access to equipment. This work was supported by funding from NIH (R01: 0255E131; T32 [for C.G.]: 2T32AG049688-06), Training Program in Stem Cell Biology fellowship from the New York State Department of Health (for L.S.: NYSTEM-C32561GG), and the Michael J. Fox Foundation (0266-3275). D.R.N. was supported by an Investigator Grant from the National Health and Medical Research Council (NHMRC, 1175781) and the Australian Research Council Discovery Project (190100825).
Author contributions
L.S., G.M.P., and T.A. conceived the project. G.M.P. established and optimized the protocol. L.S. further optimized the protocol, designed, and performed most experiments, and assisted in scRNA-seq analysis. D.R.N. performed the scRNA-seq analysis. I.A.K. performed and analyzed the electrophysiology experiments. K.W. scanned the IHC slides, R.R. quantified the IF images, E.C. and T.K. performed the qRT-PCR primer validation and prepared the samples for sequencing, C.G. generated all graphics, and R.T. maintained the organoid cultures provided by L.S. for electrophysiology. L.S. and D.R.N. wrote the manuscript. All authors provided feedback on the data during manuscript preparation and helped with editing and revisions.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 2, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107525.
Contributor Information
Joel Blanchard, Email: joel.blanchard@mssm.edu.
Tim Ahfeldt, Email: ahfeldt@gmail.com.
Supplemental information
Data and code availability
-
•
All scRNA-seq data have been deposited at GEO and Mendeley Data and are publicly available as of the date of publication. The accession number and DOI are listed in the key resources table.
-
•
All the custom codes and script for the dataset analyzed in this study have been deposited at GitHub and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Pop M.G., Crivii C., Opincariu I. In: Hypothalamus in Health and Diseases. Baloyannis S.J., Gordeladze J.O., editors. IntechOpen; 2018. Anatomy and function of the hypothalamus; pp. 1–14. [Google Scholar]
- 2.Swaab D.F., Hofman M.A., Lucassen P.J., Purba J.S., Raadsheer F.C., Van de Nes J.A. Functional neuroanatomy and neuropathology of the human hypothalamus. Anat. Embryol. 1993;187:317–330. doi: 10.1007/BF00185889. [DOI] [PubMed] [Google Scholar]
- 3.Biran J., Tahor M., Wircer E., Levkowitz G. Role of developmental factors in hypothalamic function. Front. Neuroanat. 2015;9:47. doi: 10.3389/fnana.2015.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bose M., Oliván B., Laferrère B. Stress and obesity: The role of the hypothalamic-pituitary-adrenal axis in metabolic disease. Curr. Opin. Endocrinol. Diabetes Obes. 2009;16:340–346. doi: 10.1097/MED.0b013e32832fa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocco C., Brancia C., Corda G., Ferri G.L. The hypothalamic-pituitary axis and autoantibody related disorders. Int. J. Mol. Sci. 2017;18:2322. doi: 10.3390/ijms18112322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vercruysse P., Vieau D., Blum D., Petersén Å., Dupuis L. Hypothalamic alterations in neurodegenerative diseases and their relation to abnormal energy metabolism. Front. Mol. Neurosci. 2018;11:2. doi: 10.3389/fnmol.2018.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maggi R., Zasso J., Conti L. Neurodevelopmental origin and adult neurogenesis of the neuroendocrine hypothalamus. Front. Cell. Neurosci. 2014;8:440. doi: 10.3389/fncel.2014.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D.W., Washington P.W., Wang Z.Q., Lin S.H., Sun C., Ismail B.T., Wang H., Jiang L., Blackshaw S. The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development. Nat. Commun. 2020;11:4360. doi: 10.1038/s41467-020-18231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palkovits M. Catecholamines in the hypothalamus: An anatomical review. Neuroendocrinology. 1981;33:123–128. doi: 10.1159/000123215. [DOI] [PubMed] [Google Scholar]
- 10.Gudelsky G.A. Tuberoinfundibular dopamine neurons and the regulation of prolactin secretion. Psychoneuroendocrinology. 1981;6:3–16. doi: 10.1016/0306-4530(81)90044-5. [DOI] [PubMed] [Google Scholar]
- 11.Lerant A., Herman M.E., Freeman M.E. Dopaminergic neurons of periventricular and arcuate nuclei of pseudopregnant rats: Semicircadian rhythm in Fos-related antigens immunoreactivities and in dopamine concentration. Endocrinology. 1996;137:3621–3628. doi: 10.1210/endo.137.9.8756525. [DOI] [PubMed] [Google Scholar]
- 12.Lookingland K.J., Moore K.E. In: Handbook of Chemical Neuroanatomy. Dunnett S.B., Bentivoglio M., Björklund A., Hökfelt T., editors. Elsevier; 2005. Functional neuroanatomy of hypothalamic dopaminergic neuroendocrine systems; pp. 435–523. [Google Scholar]
- 13.de Pablo-Fernández E., Warner T.T. In: Handbook of Clinical Neurology. Swaab D.F., Buijs R.M., Kreier F., Lucassen P.J., Salehi A., editors. Elsevier; 2021. Hypothalamic α-synuclein and its relation to autonomic symptoms and neuroendocrine abnormalities in Parkinson disease; pp. 223–233. [DOI] [PubMed] [Google Scholar]
- 14.Matzuk M.M., Saper C.B. Preservation of hypothalamic dopaminergic neurons in Parkinson’s disease. Ann. Neurol. 1985;18:552–555. doi: 10.1002/ana.410180507. [DOI] [PubMed] [Google Scholar]
- 15.Lee K.M., Hawi Z.H., Parkington H.C., Parish C.L., Kumar P.V., Polo J.M., Bellgrove M.A., Tong J. The application of human pluripotent stem cells to model the neuronal and glial components of neurodevelopmental disorders. Mol. Psychiatry. 2020;25:368–378. doi: 10.1038/S41380-019-0495-0. [DOI] [PubMed] [Google Scholar]
- 16.Suarez-Martinez E., Suazo-Sanchez I., Celis-Romero M., Carnero A. 3D and organoid culture in research: Physiology, hereditary genetic diseases and cancer. Cell Biosci. 2022;12:39. doi: 10.1186/s13578-022-00775-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahfeldt T., Ordureau A., Bell C., Sarrafha L., Sun C., Piccinotti S., Grass T., Parfitt G.M., Paulo J.A., Yanagawa F., et al. Pathogenic pathways in early-onset autosomal recessive Parkinson’s disease discovered using isogenic human dopaminergic neurons. Stem Cell Rep. 2020;14:75–90. doi: 10.1016/j.stemcr.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarrafha L., Parfitt G.M., Reyes R., Goldman C., Coccia E., Kareva T., Ahfeldt T. High-throughput generation of midbrain dopaminergic neuron organoids from reporter human pluripotent stem cells. STAR Protoc. 2021;2:100463. doi: 10.1016/j.xpro.2021.100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W.K., Wong S.Z.H., Pather S.R., Nguyen P.T.T., Zhang F., Zhang D.Y., Zhang Z., Lu L., Fang W., Chen L., et al. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell. 2021;28:1657–1670.e10. doi: 10.1016/j.stem.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirwan P., Jura M., Merkle F.T. Generation and characterization of functional human hypothalamic neurons. Curr. Protoc. Neurosci. 2017;81:3.33.1–3.33.24. doi: 10.1002/cpns.40. [DOI] [PubMed] [Google Scholar]
- 21.Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Egli D., Leibel R.L. Efficient generation of hypothalamic neurons from human pluripotent stem cells. Curr. Protoc. Hum. Genet. 2016;90:21.5.1–21.5.14. doi: 10.1002/cphg.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Y., Dorsky R.I. Development of the hypothalamus: Conservation, modification and innovation. Development. 2017;144:1588–1599. doi: 10.1242/dev.139055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X., Lu Y., Zhao F., Dong J., Ma W., Zhong S., Wang M., Wang B., Zhao Y., Shi Y., et al. Deciphering the spatial-temporal transcriptional landscape of human hypothalamus development. Cell Stem Cell. 2022;29:328–343.e5. doi: 10.1016/j.stem.2021.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Lee B., Kim J., An T., Kim S., Patel E.M., Raber J., Lee S.K., Lee S., Lee J.W. Dlx1/2 and Otp coordinate the production of hypothalamic GHRH- and AgRP-neurons. Nat. Commun. 2018;9:2026. doi: 10.1038/s41467-018-04377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryu S., Mahler J., Acampora D., Holzschuh J., Erhardt S., Omodei D., Simeone A., Driever W. Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr. Biol. 2007;17:873–880. doi: 10.1016/j.cub.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Monzel A.S., Smits L.M., Hemmer K., Hachi S., Moreno E.L., van Wuellen T., Jarazo J., Walter J., Brüggemann I., Boussaad I., et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Rep. 2017;8:1144–1154. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weihe E., Depboylu C., Schütz B., Schäfer M.K.H., Eiden L.E. Three types of tyrosine hydroxylase-positive CNS neurons distinguished by dopa decarboxylase and VMAT2 co-expression. Cell. Mol. Neurobiol. 2006;26:659–678. doi: 10.1007/s10571-006-9053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alquicira-Hernandez J., Sathe A., Ji H.P., Nguyen Q., Powell J.E. scPred: Accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 2019;20:264. doi: 10.1186/s13059-019-1862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanov R.A., Tretiakov E.O., Kastriti M.E., Zupancic M., Häring M., Korchynska S., Popadin K., Benevento M., Rebernik P., Lallemend F., et al. Molecular design of hypothalamus development. Nature. 2020;582:246–252. doi: 10.1038/s41586-020-2266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y.H., Xu M., Shi X., Sun X.L., Mu W., Wu H., Wang J., Li S., Su P., Gong L., et al. Cascade diversification directs generation of neuronal diversity in the hypothalamus. Cell Stem Cell. 2021;28:1483–1499.e8. doi: 10.1016/j.stem.2021.03.020. [DOI] [PubMed] [Google Scholar]
- 32.Guo Y., Wu Y., Shi J., Zhuang H., Ci L., Huang Q., Wan Z., Yang H., Zhang M., Tan Y., et al. miR-29a/b1 regulates the Luteinizing hormone secretion and affects mouse ovulation. Front. Endocrinol. 2021;12:636220. doi: 10.3389/fendo.2021.636220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenwood M., Paterson A., Rahman P.A., Gillard B.T., Langley S., Iwasaki Y., Murphy D., Greenwood M.P. Transcription factor Creb3l1 regulates the synthesis of prohormone convertase enzyme PC1/3 in endocrine cells. J. Neuroendocrinol. 2020;32:e12851. doi: 10.1111/jne.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sampieri L., Di Giusto P., Alvarez C. CREB3 transcription factors: ER-Golgi stress transducers as hubs for cellular homeostasis. Front. Cell Dev. Biol. 2019;7:123. doi: 10.3389/fcell.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bando H., Brinkmeier M.L., Castinetti F., Gergics P., Mortensen A.H., Ozel A.B., Ma Q., Li J.Z., Brue T., Camper S.A. SAT-291 SIX3 is essential for hypothalamic and pituitary development. J. Endocr. Soc. 2020;4(Suppl 1):SAT-291. doi: 10.1210/jendso/bvaa046.159. [DOI] [Google Scholar]
- 36.Du A., Wu X., Chen H., Bai Q.R., Han X., Liu B., Zhang X., Ding Z., Shen Q., Zhao C. Foxg1 directly represses Dbx1 to confine the POA and subsequently regulate ventral telencephalic patterning. Cereb. Cortex. 2019;29:4968–4981. doi: 10.1093/cercor/bhz037. [DOI] [PubMed] [Google Scholar]
- 37.Manuel M., Martynoga B., Yu T., West J.D., Mason J.O., Price D.J. The transcription factor Foxg1 regulates the competence of telencephalic cells to adopt subpallial fates in mice. Development. 2010;137:487–497. doi: 10.1242/dev.039800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metwalli A.H., Abellán A., Freixes J., Pross A., Desfilis E., Medina L. Distinct subdivisions in the transition between telencephalon and hypothalamus produce Otp and Sim1 cells for the extended amygdala in sauropsids. Front. Neuroanat. 2022;16:883537. doi: 10.3389/fnana.2022.883537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scholpp S., Lumsden A. Building a bridal chamber: Development of the thalamus. Trends Neurosci. 2010;33:373–380. doi: 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagalski A., Puelles L., Dabrowski M., Wegierski T., Kuznicki J., Wisniewska M.B. Molecular anatomy of the thalamic complex and the underlying transcription factors. Brain Struct. Funct. 2016;221:2493–2510. doi: 10.1007/s00429-015-1052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavado A., Oliver G. Prox1 expression patterns in the developing and adult murine brain. Dev. Dyn. 2007;236:518–524. doi: 10.1002/dvdy.21024. [DOI] [PubMed] [Google Scholar]
- 42.Salehi A., Loganathan N., Belsham D.D. Bisphenol A induces Pomc gene expression through neuroinflammatory and PPARγ nuclear receptor-mediated mechanisms in POMC-expressing hypothalamic neuronal models. Mol. Cell. Endocrinol. 2019;479:12–19. doi: 10.1016/j.mce.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Xie Y., Kaufmann D., Moulton M.J., Panahi S., Gaynes J.A., Watters H.N., Zhou D., Xue H.H., Fung C.M., Levine E.M., et al. Lef1-dependent hypothalamic neurogenesis inhibits anxiety. PLoS Biol. 2017;15:e2002257. doi: 10.1371/journal.pbio.2002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen R., Wu X., Jiang L., Zhang Y. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 2017;18:3227–3241. doi: 10.1016/j.celrep.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang P., Lee H.K., Glasgow S.M., Finley M., Donti T., Gaber Z.B., Graham B.H., Foster A.E., Novitch B.G., Gronostajski R.M., Deneen B. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orquera D.P., Tavella M.B., de Souza F.S.J., Nasif S., Low M.J., Rubinstein M. The homeodomain transcription factor NKX2.1 is essential for the early specification of melanocortin neuron identity and activates Pomc expression in the developing hypothalamus. J. Neurosci. 2019;39:4023–4035. doi: 10.1523/JNEUROSCI.2924-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orquera D.P., Nasif S., Low M.J., Rubinstein M., de Souza F.S.J. Essential function of the transcription factor Rax in the early patterning of the mammalian hypothalamus. Dev. Biol. 2016;416:212–224. doi: 10.1016/j.ydbio.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atkin S.D., Owen B.M., Bookout A.L., Cravo R.M., Lee C., Elias C.F., Elmquist J.K., Kliewer S.A., Mangelsdorf D.J. Nuclear receptor LRH-1 induces the reproductive neuropeptide kisspeptin in the hypothalamus. Mol. Endocrinol. 2013;27:598–605. doi: 10.1210/me.2012-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdul Satar N.M., Ogawa S., Parhar I.S. Kisspeptin-1 regulates forebrain dopaminergic neurons in the zebrafish. Sci. Rep. 2020;10:19361. doi: 10.1038/s41598-020-75777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephens S.B.Z., Rouse M.L., Tolson K.P., Liaw R.B., Parra R.A., Chahal N., Kauffman A.S. Effects of selective deletion of tyrosine hydroxylase from kisspeptin cells on puberty and reproduction in male and female mice. eNeuro. 2017;4 doi: 10.1523/ENEURO.0150-17.2017. ENEURO.0150-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blaess S., Szabó N., Haddad-Tóvolli R., Zhou X., Álvarez-Bolado G. Sonic hedgehog signaling in the development of the mouse hypothalamus. Front. Neuroanat. 2014;8:156. doi: 10.3389/fnana.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartesaghi L., Wang Y., Fontanet P., Wanderoy S., Berger F., Wu H., Akkuratova N., Bouçanova F., Médard J.J., Petitpré C., et al. PRDM12 is required for initiation of the nociceptive neuron lineage during neurogenesis. Cell Rep. 2019;26:3484–3492.e4. doi: 10.1016/j.celrep.2019.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hael C.E., Rojo D., Orquera D.P., Low M.J., Rubinstein M. The transcriptional regulator PRDM12 is critical for Pomc expression in the mouse hypothalamus and controlling food intake, adiposity, and body weight. Mol. Metab. 2020;34:43–53. doi: 10.1016/j.molmet.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landy M.A., Goyal M., Casey K.M., Liu C., Lai H.C. Loss of Prdm12 during development, but not in mature nociceptors, causes defects in pain sensation. Cell Rep. 2021;34:108913. doi: 10.1016/j.celrep.2021.108913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castro D.S., Martynoga B., Parras C., Ramesh V., Pacary E., Johnston C., Drechsel D., Lebel-Potter M., Garcia L.G., Hunt C., et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmouth J.F., Castellarin M., Laprise S., Banks K.G., Bonaguro R.J., McInerny S.C., Borretta L., Amirabbasi M., Korecki A.J., Portales-Casamar E., et al. Non-coding-regulatory regions of human brain genes delineated by bacterial artificial chromosome knock-in mice. BMC Biol. 2013;11:106. doi: 10.1186/1741-7007-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada K., Iwayama Y., Toyota T., Ohnishi T., Ohba H., Maekawa M., Yoshikawa T. Association study of the KCNJ3 gene as a susceptibility candidate for schizophrenia in the Chinese population. Hum. Genet. 2012;131:443–451. doi: 10.1007/s00439-011-1089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reinig S., Driever W., Arrenberg A.B. The descending diencephalic dopamine system is tuned to sensory stimuli. Curr. Biol. 2017;27:318–333. doi: 10.1016/j.cub.2016.11.059. [DOI] [PubMed] [Google Scholar]
- 59.Björklund A., Lindvall O., Nobin A. Evidence of an incerto-hypothalamic dopamine neurone system in the rat. Brain Res. 1975;89:29–42. doi: 10.1016/0006-8993(75)90131-6. [DOI] [PubMed] [Google Scholar]
- 60.Moriya S., Yamashita A., Masukawa D., Setoyama H., Hwang Y., Yamanaka A., Kuwaki T. Involvement of A13 dopaminergic neurons located in the zona incerta in nociceptive processing: A fiber photometry study. Mol. Brain. 2020;13:60. doi: 10.1186/s13041-020-00600-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gabriel E., Albanna W., Pasquini G., Ramani A., Josipovic N., Mariappan A., Schinzel F., Karch C.M., Bao G., Gottardo M., et al. Human brain organoids assemble functionally integrated bilateral optic vesicles. Cell Stem Cell. 2021;28:1740–1757.e8. doi: 10.1016/j.stem.2021.07.010. [DOI] [PubMed] [Google Scholar]
- 62.Matsui T.K., Matsubayashi M., Sakaguchi Y.M., Hayashi R.K., Zheng C., Sugie K., Hasegawa M., Nakagawa T., Mori E. Six-month cultured cerebral organoids from human ES cells contain matured neural cells. Neurosci. Lett. 2018;670:75–82. doi: 10.1016/j.neulet.2018.01.040. [DOI] [PubMed] [Google Scholar]
- 63.Ormel P.R., Vieira de Sá R., van Bodegraven E.J., Karst H., Harschnitz O., Sneeboer M.A.M., Johansen L.E., van Dijk R.E., Scheefhals N., Berdenis van Berlekom A., et al. Microglia innately develop within cerebral organoids. Nat. Commun. 2018;9:4167. doi: 10.1038/s41467-018-06684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carter M.E., Brill J., Bonnavion P., Huguenard J.R., Huerta R., de Lecea L. Mechanism for Hypocretin-mediated sleep-to-wake transitions. Proc. Natl. Acad. Sci. USA. 2012;109:E2635–E2644. doi: 10.1073/pnas.1202526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diniz G.B., Bittencourt J.C. The melanin-concentrating hormone (MCH) system: A tale of two peptides. Front. Neurosci. 2019;13:1280. doi: 10.3389/fnins.2019.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Egles C., René F., Varon S., Louis J.C., Félix J.M., Schimchowitsch S. Differentiation of rat hypothalamic dopaminergic neurons is stimulated in vitro by target cells: The melanotrophs. Eur. J. Neurosci. 1998;10:1270–1281. doi: 10.1046/j.1460-9568.1998.00138.x. [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi T., Hirata Y., Nagatsu T., Oda S., Sugimoto T., Matsuura S., Konagaya M., Takayanagi T., Sobue I. Tyrosine hydroxylase and tryptophan hydroxylase activities and its cofactor biopterin level in brain regions of the rolling mouse: The influence of thyrotropin releasing hormone. Neurochem. Int. 1982;4:491–494. doi: 10.1016/0197-0186(82)90037-7. [DOI] [PubMed] [Google Scholar]
- 68.Ye H., Cui X.Y., Ding H., Cui S.Y., Hu X., Liu Y.T., Zhao H.L., Zhang Y.H. Melanin-concentrating hormone (MCH) and MCH-R1 in the locus coeruleus may be involved in the regulation of depressive-like behavior. Int. J. Neuropsychopharmacol. 2018;21:1128–1137. doi: 10.1093/ijnp/pyy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeremic D., Sanchez-Rodriguez I., Jimenez-Diaz L., Navarro-Lopez J.D. Therapeutic potential of targeting G protein-gated inwardly rectifying potassium (GIRK) channels in the central nervous system. Pharmacol. Ther. 2021;223:107808. doi: 10.1016/j.pharmthera.2021.107808. [DOI] [PubMed] [Google Scholar]
- 70.Negishi K., Payant M.A., Schumacker K.S., Wittmann G., Butler R.M., Lechan R.M., Steinbusch H.W.M., Khan A.M., Chee M.J. Distributions of hypothalamic neuron populations coexpressing tyrosine hydroxylase and the vesicular GABA transporter in the mouse. J. Comp. Neurol. 2020;528:1833–1855. doi: 10.1002/cne.24857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pacelli C., Giguère N., Bourque M.J., Lévesque M., Slack R.S., Trudeau L.É. Elevated mitochondrial bioenergetics and axonal arborization size are key contributors to the vulnerability of dopamine neurons. Curr. Biol. 2015;25:2349–2360. doi: 10.1016/j.cub.2015.07.050. [DOI] [PubMed] [Google Scholar]
- 72.Pilegaard H., Saltin B., Neufer P.D. Effect of short-term fasting and refeeding on transcriptional regulation of metabolic genes in human skeletal muscle. Diabetes. 2003;52:657–662. doi: 10.2337/diabetes.52.3.657. [DOI] [PubMed] [Google Scholar]
- 73.Nolbrant S., Heuer A., Parmar M., Kirkeby A. Generation of high-purity human ventral midbrain dopaminergic progenitors for in vitro maturation and intracerebral transplantation. Nat. Protoc. 2017;12:1962–1979. doi: 10.1038/nprot.2017.078. [DOI] [PubMed] [Google Scholar]
- 74.Flori E., Mastrofrancesco A., Mosca S., Ottaviani M., Briganti S., Cardinali G., Filoni A., Cameli N., Zaccarini M., Zouboulis C.C., Picardo M. Sebocytes contribute to melasma onset. iScience. 2022;25:103871. doi: 10.1016/j.isci.2022.103871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masuda T., Wahlin K., Wan J., Hu J., Maruotti J., Yang X., Iacovelli J., Wolkow N., Kist R., Dunaief J.L., et al. Transcription factor sox9 plays a key role in the regulation of visual cycle gene expression in the retinal pigment epithelium. J. Biol. Chem. 2014;289:12908–12921. doi: 10.1074/jbc.M114.556738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scialdone A., Natarajan K.N., Saraiva L.R., Proserpio V., Teichmann S.A., Stegle O., Marioni J.C., Buettner F. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 77.Neavin D., Senabouth A., Hang Lee J.T., Ripoll A., Franke L., Prabhakar S., Ye C.J., McCarthy D.J., Melé M., Hemberg M., Powell J.E. Demuxafy: Improvement in droplet assignment by integrating multiple single-cell demultiplexing and doublet detection methods. bioRxiv. 2022 doi: 10.1101/2022.03.07.483367. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muskovic W., Powell J.E. DropletQC: Improved identification of empty droplets and damaged cells in single-cell RNA-seq data. Genome Biol. 2021;22:329. doi: 10.1186/s13059-021-02547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/S13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Germain P., Lun A., Macnair W., Robinson M.D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Research. 2021 doi: 10.12688/f1000research.73600.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bais A.S., Kostka D. scds: Computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics. 2020;36:1150–1158. doi: 10.1093/bioinformatics/btz698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rigamonti A., Repetti G.G., Sun C., Price F.D., Reny D.C., Rapino F., Weisinger K., Benkler C., Peterson Q.P., Davidow L.S., et al. Large-scale production of mature neurons from human pluripotent stem cells in a three-dimensional suspension culture system. Stem Cell Rep. 2016;6:993–1008. doi: 10.1016/j.stemcr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stahl K., Mylonakou M.N., Skare Ø., Amiry-Moghaddam M., Torp R. Cytoprotective effects of growth factors: BDNF more potent than GDNF in an organotypic culture model of Parkinson’s disease. Brain Res. 2011;1378:105–118. doi: 10.1016/j.brainres.2010.12.090. [DOI] [PubMed] [Google Scholar]
- 84.Momčilović O., Liu Q., Swistowski A., Russo-Tait T., Zhao Y., Rao M.S., Zeng X. Genome wide profiling of dopaminergic neurons derived from human embryonic and induced pluripotent stem cells. Stem Cells Dev. 2014;23:406–420. doi: 10.1089/scd.2013.0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roberts H.L., Schneider B.L., Brown D.R. α-Synuclein increases β-amyloid secretion by promoting β-/γ-secretase processing of APP. PLoS One. 2017;12:e0171925. doi: 10.1371/journal.pone.0171925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y., He K., Wang F., Li X., Liu D. Notch-1 signaling regulates astrocytic proliferation and activation after hypoxia exposure. Neurosci. Lett. 2015;603:12–18. doi: 10.1016/j.neulet.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 87.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 88.10x Genomics Chromium Single Cell 3′ Reagent Kits User Guide (v3.1 Chemistry Dual Index) 2022. https://support.10xgenomics.com/single-cell-gene-expression/library-prep/doc/user-guide-chromium-single-cell-3-reagent-kits-user-guide-v31-chemistry-dual-index
- 89.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All scRNA-seq data have been deposited at GEO and Mendeley Data and are publicly available as of the date of publication. The accession number and DOI are listed in the key resources table.
-
•
All the custom codes and script for the dataset analyzed in this study have been deposited at GitHub and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.