Summary
Neutrophils are the first immune responders to bacterial or viral infection and play key roles in the host immune response; however, handling and investigating fresh neutrophils can be challenging. Here, we present a protocol for isolating neutrophils from the peripheral blood of healthy donors using density gradient separation method. We describe steps for morphology analysis by cytospin and immunophenotyping by flow cytometry analysis. This protocol can be used for the isolation of neutrophils from healthy and diseased individuals.
For complete details on the use and execution of this protocol, please refer to Parthasarathy et al.1
Subject areas: Cell Biology, Cell Isolation, Flow Cytometry/Mass Cytometry, Immunology
Graphical abstract

Highlights
-
•
Gradient density separation method to isolate human neutrophils from whole blood
-
•
Flow cytometry to immunophenotype freshly isolated neutrophils
-
•
Morphology analysis of neutrophils using cytospin
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Neutrophils are the first immune responders to bacterial or viral infection and play key roles in the host immune response; however, handling and investigating fresh neutrophils can be challenging. Here, we present a protocol for isolating neutrophils from the peripheral blood of healthy donors using density gradient separation method. We describe steps for morphology analysis by cytospin and immunophenotyping by flow cytometry analysis. This protocol can be used for the isolation of neutrophils from healthy and diseased individuals.
Before you begin
The protocol below describes steps for isolation of neutrophils from human blood as in Parthasarathy et al.1
Institutional permissions
All human samples specified in this protocol were obtained and used following institutional guidelines and were approved by Merck & Co., Inc., Rahway, NJ, USA, IRB#20121234.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC mouse anti-human CD11b, clone ICRF44, used as 1:50 dilution | BD Biosciences | Cat# 562793; RRID: AB_2737798 |
| PerCP-CY5.5 mouse anti-human CD66b, clone G10F5, used as 1:20 dilution | BD Biosciences | Cat# 562254; RRID: AB_11154419 |
| Alexa Fluor 647 mouse anti-human CD63, clone H5C6, used as 1:20 dilution | BD Biosciences | Cat# 561983; RRID: AB_10897006 |
| Alexa Fluor 700 mouse anti-human CD45, clone HI30, used as 1:100 dilution | BD Biosciences | Cat# 560566; RRID: AB_1645452 |
| APC-CY7 mouse anti-human CD62L, clone DREG-56, used as 1:100 dilution | BioLegend | Cat# 304814; RRID: AB_493582 |
| BV711 mouse anti-human CD184, clone 12G5, used as 1:20 dilution | BD Biosciences | Cat# 740799; RRID: AB_2740462 |
| BV510 mouse anti-human CD11c, clone B-ly6, used as 1:20 dilution | BD Biosciences | Cat# 563026; RRID: AB_2737960 |
| BUV737 mouse anti-human CD10, clone HI10α, used as 1:50 dilution | BD Biosciences | Cat# 612826; RRID: AB_2870150 |
| PE mouse anti-human CD16, clone 3G8, used as 1:100 dilution | BD Biosciences | Cat# 555407; RRID: AB_395807 |
| BUV395 mouse anti-human CD300f, clone UP-D1, used as 1:20 dilution | BD Biosciences | Cat# 745678; RRID: AB_2743166 |
| BV786 mouse anti-human CD80, clone L307.4, used as 1:20 dilution | BD Biosciences | Cat# 564159; RRID: AB_2738631 |
| PE-CF594 mouse anti-human CD86, clone 2331(FUN-1), used as 1:20 dilution | BD Biosciences | Cat# 562390; RRID: AB_11154047 |
| BV421 mouse anti-human CD177, clone MEM-166, used as 1:100 dilution | BD Biosciences | Cat# 564240; RRID: AB_2738694 |
| PE-CY7 mouse anti-human CD274, clone MIH1, used as 1:100 dilution | BD Biosciences | Cat# 558017; RRID: AB_396986 |
| BV650 mouse anti-human HLA-DR, clone 46-6, used as 1:20 dilution | BD Biosciences | Cat# 564231; RRID: AB_2738685 |
| Biological samples | ||
| Healthy donor whole blood | MRL volunteer blood donor program | IRB #20121234 |
| Chemicals, peptides, and recombinant proteins | ||
| 10× Phosphate-buffered saline (PBS) | Thermo Fisher | 70011–044 |
| Percoll | Cytiva | 17089101 |
| Ultra-pure H2O | Invitrogen | 10977–015 |
| Fc block | BD Biosciences | 564219 |
| Ammonium chloride solution | STEMCELL Technologies | 07800 |
| 1× Phosphate-buffered saline (PBS) | Thermo Fisher | 14190-136 |
| 1× Roswell Park Memorial Institute (RPMI) 1640 medium | Thermo Fisher | A1049-01 |
| May Grünwald stain | Sigma-Aldrich | MG-500-500ML |
| Giemsa stain | Sigma-Aldrich | GS500-500ML |
| VectaMount mounting medium | Vector Laboratories | H-5000 |
| BD FACS stain buffer | BD Biosciences | 554656 |
| 1M HEPES (N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid)) | Thermo Fisher | 15630080 |
| BSA (bovine serum albumin) | Sigma-Aldrich | A3059-100G |
| 1× Hank’s balanced salt solution (HBSS), no calcium, no magnesium, no phenol red | Thermo Fisher | 14175095 |
| Ultracomp compensation beads | Invitrogen | 01-2222-42 |
| Software and algorithms | ||
| FlowJo V10.8.1 | BD Biosciences | http://www.flowjo.com |
| Prism V9.0.0 | GraphPad | http://graphpad.com |
| BD FACSDiva V9.3 | BD Biosciences | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| Other | ||
| 50 mL Conical tube | Thermo Fisher | 339653 |
| 10 mL Sodium heparin Vacutainer tube | Thermo Fisher | BD 367874 |
| Corning Falcon 96-well U-bottom plate | Fisher Scientific | 08-772-17 |
| Falcon round-bottom polystyrene test tubes | Fisher Scientific | 14-959-1A |
| Glass staining jar | Fisher Scientific | 22-309-247 |
| RA Lamb plastic slide box | Thermo Fisher | E405 |
| Fisherbrand double frosted microscope glass slide | Fisher Scientific | 12-552-5 |
| Fisherbrand cover glass | Thermo Fisher | 12-540A |
| C-Chip disposable hemacytometer | InCyto | DHCS022 |
| Cytospin centrifuge | Thermo Fisher | Cytospin4 |
| Cytospin filter card | Fisher Scientific | 22–030410 |
| Humidified 37°C and 5% CO2 incubator | Thermo Fisher | HERACell Vios 160i |
| Biosafety cabinets | LabGard | Class II TYPE A2 |
| Cell culture microscope | Life Technologies | EVOS XL Core |
| Flow cytometry | BD Biosciences | BD Symphony A5 (Special Order Research Product) |
| Benchtop centrifuge | Thermo Fisher | Sorvall Legend XTR |
| Benchtop centrifuge | Thermo Fisher | Sorvall ST16R |
| Benchtop centrifuge | Thermo Fisher | Sorvall legend micro 21R |
| Benchtop centrifuge | Beckman Coulter | Allegra 6R |
Materials and equipment
62% Percoll gradient solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Percoll | 62% | 31 mL |
| 10× PBS | 10% | 5 mL |
| Ultra-pure H2O | 28% | 14 mL |
| Total | N/A | 50 mL |
Store at 4°C, use within 6 months.
75% Percoll gradient solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Percoll | 75% | 37.5 mL |
| 10× PBS | 10% | 5 mL |
| Ultra-pure H2O | 15% | 7.5 mL |
| Total | N/A | 50 mL |
Store at 4°C, use within 6 months.
%10 BSA
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA | 10 mg/mL (10%) | 10 g |
| 1× PBS | N/A | 100 mL |
| Total | N/A | 100 mL |
Store at 4°C, use within 6 months
Neutrophil buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1× HBSS | N/A | 500 mL |
| 1M HEPES | 5 mM | 2.5 mL |
| 10% BSA buffer | 0.2% | 10 mL |
| Total | N/A | 512.5 mL |
Store at 4°C, use within 6 months.
Step-by-step method details
The following steps describe how to isolate neutrophils from fresh human blood.
Neutrophil isolation
Timing: 1–1.5 h
This step describes the isolation of the neutrophils from human blood using a double-gradient Percoll separation starting from 10 mL of blood.2 The blood was drawn in 10 mL Sodium Heparin vacutainer tubes. Volumes can be adjusted depending on sample availability.
Note: All reagents should be brought at room temperature (20–25°C). All procedures should be performed in a biosafety cabinet.
-
1.
Add 10 mL 75% Percoll into a 50 mL Conical tube using sterile serological pipette.
-
2.
Carefully overlay 10 mL 62% Percoll on top of 75% Percoll using sterile serological pipette.
-
3.
Dilute 10 mL fresh blood 1:1 in 10 mL of serum-free RPMI 1640 without antibiotics.
-
4.
Carefully overlay 20 mL of diluted blood on top of the 62% Percoll using a sterile serological pipette in a dropwise manner (Figure 1), thus bringing to a total volume of 40 mL.
Note: Volumes can be adjusted depending on the amount of blood drawn per donor. We recommend that the amount of diluted blood overlaid is not greater than 1.5 times the volume of the Percoll gradient. For instance, 15 mL of drawn blood (i.e. 30 mL of RPMI 1640 diluted blood) can be overlaid on a gradient of 10 mL of 75% Percoll and 10 mL of 62% Percoll. This protocol can be adapted also to a 15 mL conical tube.
-
5.
Centrifuge the 50 mL conical tubes at 200 × g for 25 min and 400 × g for 15 min.
Note: Perform above step with acceleration and deceleration set to zero on the centrifuge.
-
6.Carefully remove tubes following centrifugation so not to disrupt the gradient. The separation of cells and respective bands are shown in Figure 1.
-
a.Use sterile transfer pipettes or micropipettes to first collect peripheral blood mononuclear cells (PBMC) on top of the 62% Percoll layers.
-
b.Then use sterile transfer pipettes or micropipettes to collect neutrophils positioned between the 75% and 62% Percoll layers and transfer the cells to a new 50 mL Conical tube.
-
a.
Note: PBMCs population is positioned on top of the 62% Percoll layer. If appropriately performed, red blood cells (RBCs) should form a pellet underneath the 75% Percoll layer. Ensure careful collection of neutrophils without disrupting the RBC layer.
-
7.
Add RPMI 1640 to the tube up to a volume of 50 mL. Wash the neutrophils by centrifuging at 300 × g for 5 min.
-
8.
Remove the supernatant completely and resuspend the cell pellet in 1 mL of neutrophil buffer.
-
9.
Add 10 μL of cell suspension to a disposable hemocytometer and count the cell number under the microscope.
Note: This protocol typically yields an average (± SD) of 2.15 × 10+6 (± 0.54 × 10+6) neutrophils/mL blood. Neutrophils can be resuspended at 1.0 × 10+6 cells/mL in neutrophil buffer. In the case that the yield is lesser than above mentioned, adjust volume of resuspension in neutrophil buffer accordingly, to obtain desired cell concentrations. Cells can be further diluted with neutrophil buffer prior to counting.
CRITICAL: In order to obtain good separation, ensure careful overlay of the 62% Percoll on top of the 75% Percoll layer, and subsequent careful overlay of diluted blood on top of the 62% Percoll layer.
Figure 1.
Neutrophils separation from human blood following overlay on Percoll gradient and centrifugation
(A) Drawing and pictures of double layer Percoll gradient before (left) and after (right) centrifugation.
(B) After Percoll gradient, neutrophils are identified in neutrophils layer between the two Percoll layers. (i.) Few low-density granulocytes can be found in the layer between serum and Percoll (ii.) Granulocytes (neutrophils + eosinophils) can be found between the Percoll layers.
Cytospin and staining
Timing: 1–2 h
This part of the protocol describes the steps involved in cytospin. These steps allow us to examine the morphology of the neutrophils after Percoll gradient-based isolation.
-
10.
Place cytospin filter paper on top of the microscope slide (Figure 2A). Place the cytofunnel on top of the filter paper, making sure the holes are aligned.
-
11.
Place the slide, filter paper and cytofunnel into the cytoclip with the cardboard filters facing the center of the cytospin (Figure 2B).
-
12.
Aliquot 1.0–2.0 × 10+5 neutrophils in a volume of 100 μL into the well of the cytofunnel and slide set (Figure 2C).
-
13.
Carefully place the cytoclip into the cytocentrifuge and spin at 800 × g for 5 min at 20–25°C.
Note: Ensure proper balance of the cytofunnel slide set on the cytocentrifuge.
-
14.
Following centrifugation, carefully open the cytoclip set and remove the filters from their slides.
Note: Ensure minimal physical contact with cell smears on the slides. Examine each slide under the microscope to ensure cells have attached well to the slide.
-
15.
Air dry the slide at 20–25°C for 20 min.
-
16.
Stain the slide with May Grünwald stain solution for 5 min at 20–25°C and transfer the slide to 1× PBS for 1.5 min.
-
17.
Stain the slide with diluted Giemsa stain solution (1:20 with Ultrapure H2O) for 20 min at 20–25°C.
-
18.
Wash the slide under running Ultrapure H2O until clean.
-
19.
Air dry the stained slide at 20–25°C. Mount the slide by adding 1 drop of VectaMount mounting medium to the center of the cells and then put the coverslip on top (Figure 2D).
-
20.
Cells can now be imaged using a light microscope with recommended magnifications of 40×/100× magnification, and their morphological characteristics can be evaluated (Figure 2E). Note: Neutrophils stained with May Grünwald stain and Giemsa stain appear with light purple/pink cytoplasmic granules with violet color of multi-lobed nuclei respectively.
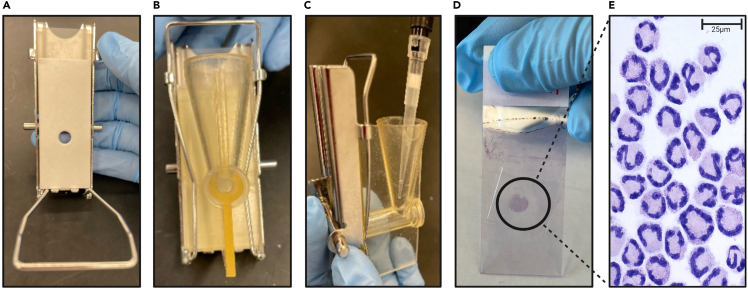
Figure 2.
Step-by-step description of cytospin preparation
(A) Place cytospin filter paper on top of the microscope slide.
(B) Place the slide and filter paper in to the cytoclip.
(C) Add 100 uL of cells into the well of the cytoclip set.
(D, E) Following centrifugation, cells are Giemsa and May Grünwald stained. Representative image (scale bar: 25 μm) is shown.
Antibody titration
Timing: 1–2 h
This step describes the titration of antibodies for flow cytometry. These steps will enable identification of optimal concentrations of antibodies for use in flow cytometry assays.
Note: This section of the protocol describes steps involved in optimizing the neutrophil-related flow cytometry panel and is recommended to be performed beforehand. Titrations and dilutions recommended in this protocol are manufacturer specific and could potentially vary between different lot numbers.
-
21.
Add 0.5–1 × 10+6 neutrophils in 100 μL neutrophil buffer per well in a 96-well U-bottom plate or per tube in round-bottom polystyrene tubes.
Note: These titrations are valid for a total of 0.5–1 × 10+6 neutrophils per sample.
-
22.
Centrifuge the plate (with lid on) or tubes at 300 × g for 5 min at 4°C.
-
23.
Discard supernatant and resuspend cells in 100 μL FACS buffer + Fc Block (diluted 1:100 in FACS buffer) for 20 min on ice or at 4°C.
-
24.
Centrifuge the plate at 300 × g for 5 min at 4°C. Discard supernatant and resuspend cells in 100 μL of FACS buffer per well.
-
25.Perform staining as follows:
-
a.Resuspend unstained control in 100 μL FACS buffer per well.
-
b.Titrate each antibody by adding 5 μL, 2 μL or 1 μL of the respective antibodies to 100 μL FACS buffer per well.
-
c.Mix each well by pipetting up and down using a multichannel micropipette.
-
a.
-
26.
Wrap the plate in foil to protect from light and incubate for 30 min at 4°C.
-
27.
Following incubation, centrifuge the plate at 300 × g for 5 min at 4°C.
-
28.
Discard supernatant and resuspend cells with 200 μL FACS buffer as a wash step.
-
29.
Centrifuge the plate at 300 × g for 5 min at 4°C. Discard the supernatant and resuspend cells in 200 μL FACS buffer per well. The cells are now ready to be acquired by flow cytometry.
- 30.
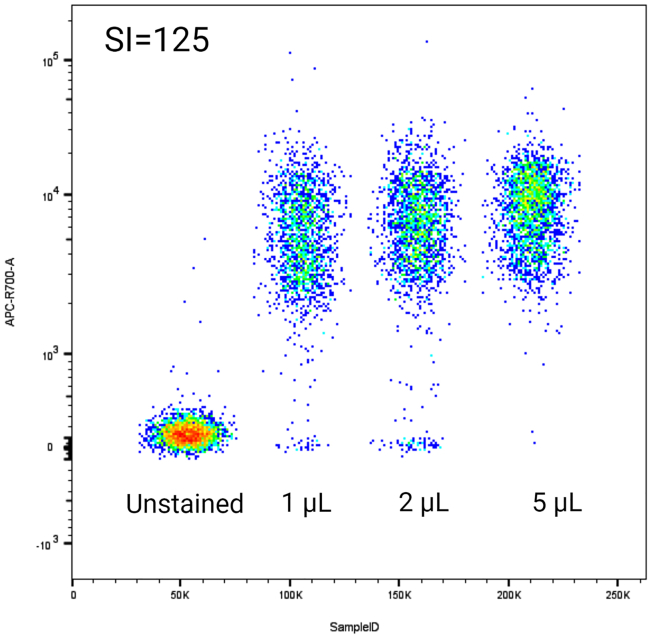
Note: The SI is used as a measure for evaluating the amount of fluorescence for each titration. The higher the SI value, the better the separation between the positive and negative populations.3
Figure 3.
Example of Concatenating antibody titration
Neutrophils were either unstained or stained with 1, 2 or 5 μL anti-CD45 Alexa 700 antibody respectively. The optimal concentration for this specific antibody is 1 μL based on the titration.
Table 1.
Neutrophil antibody master mix
| Marker | Fluorophore | Dilution factor | Antibodies for 1 sample (total volume 100 μL) | Antibodies for 2 samples (total volume 200 μL) |
|---|---|---|---|---|
| CD11b (Mac-1) | FITC | 1:50 | 2 μL | 4 μL |
| CD66b | PerCP-Cy5.5 | 1:20 | 5 μL | 10 μL |
| CD63 | Alexa 647 | 1:20 | 5 μL | 10 μL |
| CD45 | Alexa 700 | 1:100 | 1 μL | 2 μL |
| CD62L | APC-Cy7 | 1:100 | 1 μL | 2 μL |
| CXCR4 (CD184) | BV 711 | 1:20 | 5 μL | 10 μL |
| CD11c | BV510 | 1:20 | 5 μL | 10 μL |
| CD10 | BUV737 | 1:50 | 2 μL | 4 μL |
| CD16 | PE | 1:100 | 1 μL | 2 μL |
| CD300f | BUV395 | 1:20 | 5 μL | 10 μL |
| CD80 | BV785 | 1:20 | 5 μL | 10 μL |
| CD86 | PE-CF594 | 1:20 | 5 μL | 10 μL |
| CD177 | BV421 | 1:100 | 1 μL | 2 μL |
| CD274 | PE Cy7 | 1:100 | 1 μL | 2 μL |
| HLA-DR | BV650 | 1:20 | 5 μL | 10 μL |
| Total volume of antibodies | 49 μL | 98 μL | ||
| Volume of FACS buffer for 2 samples | 51 μL | 102 μL |
Neutrophil staining for flow cytometry
Timing: 1–2 h
This step describes the staining of neutrophils with a panel of antibodies custom designed to identify different neutrophil subsets.1 Fluorescence Minus One (FMO) controls are used to identify the background signal on omitted fluorophores and gate on the positive populations. These steps allow us to confirm the positive staining of the neutrophils.
-
31.
Add 0.5–1 × 10+6 neutrophils in 100 μL neutrophil buffer per well into the appropriate wells of a 96-well U-bottom plate.
-
32.
Centrifuge the plate at 300 × g for 5 min at 4°C.
-
33.
Discard supernatant and resuspend cells in 200 μL FACS buffer per well.
-
34.
Centrifuge plate at 300 × g for 5 min at 4°C.
-
35.
Discard supernatant and resuspend cells in 100 μL FACS buffer + Fc Block (diluted 1:100 in FACS buffer) for 20 min on ice or at 4°C.
-
36.
Centrifuge plate at 300 × g for 5 min at 4°C.
-
37.Perform staining as follows:
-
a.Resuspend unstained control in 100 μL FACS buffer per well.
-
b.Stain the cells with appropriate volume of antibodies based on the titration (Table 1).
- c.
-
d.Mix each well by pipetting up and down using a multichannel micropipette
-
a.
-
38.
Wrap plate in foil to protect from light and incubate for 30 min at 4°C.
-
39.
Following incubation, centrifuge plate at 300 × g for 5 min at 4°C. Discard supernatant and wash cells with 200 μL FACS buffer per well.
-
40.
Centrifuge plate at 300 × g for 5 min at 4°C.
-
41.
Discard supernatant and resuspend cells in 200 μL FACS buffer per well.
-
42.
The cells are ready to be acquired by flow cytometer.
Table 2.
Neutrophil FMO controls plate map
| Antigen | FITC | PerCP-Cy55 | Alexa 647 | AF700 | APC-Cy7 | BV711 | BV510 | BUV737 | PE | BUV395 | BV786 | BV421 | PE-Cy7 | BV650 | PE-CF594 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD11b (Mac-1) FMO | ------ | CD66b | CD63 | CD45 | CD62L | CD184 | CD11c | CD10 | CD16 | CD64 | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CD66b FMO | CD11b | ------ | CD63 | CD45 | CD62L | CD184 | CD11c | CD10 | CD16 | CD64 | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CD63 FMO | CD11b | CD66b | ------ | CD45 | CD62L | CD184 | CD11c | CD10 | CD16 | CD64 | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CD45 FMO | CD11b | CD66b | CD63 | ------ | CD62L | CD184 | CD11c | CD10 | CD16 | CD64 | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CD62L FMO | CD11b | CD66b | CD63 | CD45 | ------ | CD184 | CD11c | CD10 | CD16 | CD64 | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CXCR4 (CD184) FMO | CD11b | CD66b | CD63 | CD45 | CD62L | ------ | CD11c | CD10 | CD16 | CD64 | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CD11c FMO | CD11b | CD66b | CD63 | CD45 | CD62L | CD184 | ------ | CD10 | CD16 | CD64 | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CD10 FMO | CD11b | CD66b | CD63 | CD45 | CD62L | CD184 | CD11c | ------ | CD16 | CD64 | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CD16 FMO | CD11b | CD66b | CD63 | CD45 | CD62L | CD184 | CD11c | CD10 | ------ | CD64 | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CD300f FMO | CD11b | CD66b | CD63 | CD45 | CD62L | CD184 | CD11c | CD10 | CD16 | ------ | CD80 | CD177 | CD274 | HLA-DR | CD86 |
| CD80 FMO | CD11b | CD66b | CD63 | CD45 | CD62L | CD184 | CD11c | CD10 | CD16 | CD64 | ------ | CD177 | CD274 | HLA-DR | CD86 |
| CD177 FMO | CD11b | CD66b | CD63 | CD45 | CD62L | CD184 | CD11c | CD10 | CD16 | CD64 | CD80 | ------ | CD274 | HLA-DR | CD86 |
| CD274 FMO | CD11b | CD66b | CD63 | CD45 | CD62L | CD184 | CD11c | CD10 | CD16 | CD64 | CD80 | CD177 | ------ | HLA-DR | CD86 |
| HLA-DR FMO | CD11b | CD66b | CD63 | CD45 | CD62L | CD184 | CD11c | CD10 | CD16 | CD64 | CD80 | CD177 | CD274 | ------ | CD86 |
| CD86 FMO | CD11b | CD66b | CD63 | CD45 | CD62L | CD184 | CD11c | CD10 | CD16 | CD64 | CD80 | CD177 | CD274 | HLA-DR | ------ |
Figure 4.
Example of flow cytometry data acquisition and gating strategy for the neutrophil panel
(A) Representative PMT voltage for each marker.
(B) Representative pictures of compensation matrix for each marker.
(C) Example of CD16 FMO plots to identify the background signal and the positive populations.
(D) Gating strategy for identification of neutrophils subsets.
Acquisition on the flow cytometer
Timing: 1–2 h
This part details the acquisition of stained neutrophils using a flow cytometer (steps detailed here are specific to BD Symphony, using FACS Diva software). Any flow cytometer with the correct number of channels/colors and analysis software can be used to perform the steps in this protocol.
-
43.Prepare unstained and single-stained controls for every fluorophore that is used in the panel to correct for fluorescence spillover and set appropriate voltages for each channel.
-
a.Add 100 μL FACS buffer per well in a 96-well-U bottom plate or per tube in round-bottom polystyrene tubes.
-
b.Add one drop of compensation beads to each well containing FACS buffer.Note: Vortex the beads for 1 min prior to use.
-
c.Add 1 μL of the antibody to respective wells except the unstained control.
-
a.
-
44.
Wrap plate in foil to protect from light and incubate for 30 min at 4°C.
-
45.
Following incubation, centrifuge plate at 300 × g for 5 min at 4°C.
-
46.
Discard supernatant and resuspend beads in 200 μL FACS buffer per well.
-
47.
Controls and samples (from step 41) are now ready for acquisition.
-
48.
Run the unstained and single-stained beads to set appropriate photomultiplier tubes (PMTs) voltage (Figure 4A) and compensation matrix (Figure 4B), followed by the acquisition of samples.
Note: For best results, we recommended acquiring the samples within an hour of staining to avoid confounding results due to cell activation/damage or death.
Expected outcomes
This protocol details steps for isolation of neutrophils from peripheral human blood. Utilization of a Percoll-gradient to isolate neutrophils provides significant advantages over bead-based methods and yields a pure population with limited contamination and robust repeatability, while avoiding activation of neutrophils. The average yield (± SD) from this protocol is typically 2.15 × 10+6 (± 0.54 × 10+6) neutrophils/mL blood and the average purity (± SD) of the neutrophils (defined as CD45+CD16+ cells) is 97.3 (± 2.4)% (Figure 4D). Further, using the forward scatter (FSC) and side scatter (SSC) parameters in flow cytometry, we observe a pure population of granulocytes between the 62% and 75% Percoll layers (Figure 1B(ii)). Additionally, cytocentrifugation (Figure 2E) confirms the purity and allows for additional classification based on nuclear morphology.
This protocol also provides step-by-step guidance on antibody titration and staining of freshly isolated neutrophils for immunophenotyping by multi-color flow cytometry (Figure 1B and 4D), and the gating strategy to separate neutrophils from red blood cells and other leukocytes (Figure 4D). The debris-removal gate was employed first. This was followed by gating on singlets using the FSC-W and FSC-H parameters. The singlets were then used to separate neutrophils and eosinophils based on expression of CD16 and CD45. Gates on individual markers were drawn based on the FMO controls. For example, the gate for CD16+ population was set based on the FMO control of CD16 (Figure 4C, top), and the associated positive staining control for CD16 (Figure 4C, bottom). CD16+CD45+CD10- immature and CD16+CD45+CD10+ mature neutrophils were then identified based on CD10 expression. Similarly, expression of CD177 and CD184 can be further analyzed in CD16+CD10+/− neutrophil subsets. Detection and in-depth analysis of various neutrophils subsets can be performed using the customized flow cytometry panel published by us previously.1
Limitations
In this study, we focus on isolation of neutrophils from peripheral human blood samples. Although this method yields a pure population of neutrophils in most cases (purity of ≥90%, defined as CD45+CD16+ cells), it necessitates careful handling to avoid contamination. In addition, to capture the adequate biology, this protocol necessitates neutrophils to be analyzed from fresh blood and not from blood stored overnight.
Troubleshooting
Problem 1
Variable yield of neutrophils after centrifugation (steps 1–9).
Potential solution
The protocol works best when using blood within 3 h after withdrawing. All the reagents must be at 20°C–25°C before using. This average yield (± SD) from this protocol was 2.15 × 10+6 (± 0.54 × 10+6) neutrophils/mL blood and the average purity (± SD) of the neutrophils (defined as CD45+CD16+ cells) was 97.3 (±2.4)%. Even though this happens extremely rarely, donor-to-donor variability may lead to relatively variable yield of neutrophils. Volumes of Percoll can be adjusted appropriately based on the amount of blood.
Problem 2
Red blood cell (RBC) contamination (steps 1–9).
Potential solution
Following Percoll gradient centrifugation, ensure careful collection of neutrophils without disrupting the RBC pellet. In case of contamination of neutrophils with RBCs, isolated cell populations can be subjected to RBC lysis by treatment with ammonium chloride solution (STEMCELL Technologies, #07800) for 2 min at 20°C–25°C. Treated cells should be centrifuged at 300 × g, for 5–10 min to yield RBC-lysed population.
Problem 3
Too many or too less cells attached to the slide on the cytoclip set post-cytocentrifugation (steps 10–20).
Potential solution
Ensure careful handling of the cytoclip set following cytocentrifugation and avoid movement of the filter paper on the slide. In addition, optimize volume and concentration of the cells for cytocentrifugation to obtain well-attached layer of cells.
Problem 4
Fluorescence for different antibodies is too low or too high (steps 21–30).
Potential solution
Titration of individual antibodies is highly recommended before use to identify the optimal concentration as they are manufacturer specific and could potentially vary between different lot numbers. Additionally, PMT voltages can be further adjusted as required.
Problem 5
Spillover between fluorophores and low resolution of flow cytometry data (steps 37–43).
Potential solution
Ensure set up of appropriate compensation controls to account for spillover and establish appropriate PMT voltages on the instrument to better define neutrophil populations.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Roberta Martinelli roberta.martinelli@merck.com.
Materials availability
This study did not generate new unique reagents.
Author contributions
Conceptualization, R.M. and Y.K.; Methodology and Formal Analysis, Y.K. and R.M.; Validation, Y.K. and U.P.; Resources, Y.K. and R.M.; Writing – Original Draft, Y.K.; Writing – Review and Editing, U.P., Y.K., and R.M.; Visualization, Y.K. and U.P.; Supervision, R.M.
Acknowledgments
This publication was supported by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA. We thank all the subjects for participating and donating blood for this study. Graphical abstract and figures were created using BioRender.com.
Declaration of interests
The authors declare no competing interests.
Data and code availability
This study did not generate any new data or code.
References
- 1.Parthasarathy U., Kuang Y., Thakur G., Hogan J.D., Wyche T.P., Norton J.E., Killough J.R., Sana T.R., Beakes C., Shyong B., et al. Distinct subsets of neutrophils crosstalk with cytokines and metabolites in patients with sepsis. iScience. 2023;26:105948. doi: 10.1016/j.isci.2023.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dri P., Haas E., Cramer R., Menegazzi R., Gasparini C., Martinelli R., Scheurich P., Patriarca P. Role of the 75-kDa TNF Receptor in TNF-Induced Activation of Neutrophil Respiratory Burst. J. Immunol. 1999;162:460–466. doi: 10.4049/jimmunol.162.1.460. [DOI] [PubMed] [Google Scholar]
- 3.Laboratory, U.F.C. 2005. FlowJo for Antibody Titrations: Separation Index and Concatenation.https://cancer.wisc.edu/research/resources/flow/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any new data or code.