Abstract
This study tested the hypothesis that high frequencies of natural killer (NK) cells are protective against symptomatic SARS-CoV-2 infection. Samples were utilized from the COVID-19 Health Action Response for Marines study, a prospective, observational study of SARS-CoV-2 infection in which participants were enrolled prior to infection and then serially monitored for development of symptomatic or asymptomatic infection. Frequencies and phenotypes of NK cells (CD3−CD14−CD19−CD56+) were assessed by flow cytometry. Individuals that developed asymptomatic infections were found to have higher pre-infection frequencies of total NK cells compared to symptomatic individuals (10.61% [SD 4.5] vs 8.33% [SD 4.6], p = 0.011). Circulating total NK cells decreased over the course of infection, reaching a nadir at 4 weeks, while immature NK cells increased, a finding confirmed by multidimensional reduction analysis. These results indicate that NK cells likely play a key role in controlling the severity of clinical illness in individuals infected with SARS-CoV-2.
Keywords: NK cells, Natural killer cells, SARS-CoV-2, Asymptomatic, Symptomatic
Graphical abstract

Highlights
-
•
High baseline NK cell are frequencies associated with asymptomatic SARS-CoV-2 infection.
-
•
Total NK cell frequencies in blood decline during SARS-CoV-2 infection.
-
•
Nadir of total NK cell frequencies reached at 3–4 weeks post-infection.
-
•
Normalization of total NK cell frequencies by 7–8 weeks post-infection.
-
•
Immature NK cell frequencies increase post-infection, peaking at 4 weeks.
1. Introduction
SARS-CoV-2 has infected over 650 million people worldwide and caused close to 6.6 million deaths as of January 2023. While much has been learned regarding the pathophysiology of infection, little is known regarding the immune factors that provide protection against symptomatic disease. In this study, we tested the hypothesis that high frequencies of NK cells, innate immune cells that are critical for clearing virus-infected cells early in the course of infection, are associated with protection against symptomatic SARS-CoV-2 infection.
In humans, NK cells make up 5–20% of circulating lymphocytes and can target infected, transformed, or stressed cells (Abel et al., 2018; Björkström et al., 2022). There are several subsets of NK cells that are characterized by the receptors they display and their functional activity. Immature NK cells are defined as being CD56bright and CD16− and are primarily cytokine producing cells with limited cytotoxicity (Björkström et al., 2022). Mature CD16+ NK cells are defined as being CD56dim and CD16+ and they are functionally more cytotoxic than immature NK cells. Cells that are CD56dimCD16− are believed to be mature NK cells that have lost CD16 expression, which can occur after NK cell activation (Romee et al., 2013). Another subset are adaptive NK cells, which have been identified in people with chronic viral infections like cytomegalovirus, or human immunodeficiency virus (Freud et al., 2017; Gumá et al., 2004; Béziat et al., 2013; Lopez-Vergès et al., 2011; Gondois-Rey et al., 2017). An increase in adaptive NK cells has also been identified in patients with severe SARS-CoV-2 infection (Maucourant et al., 2020). Adaptive NK cells are memory-like cells that can survive longer than other NK cell subsets, can rapidly proliferate, and have effector functions when re-exposed to a virus (Maucourant et al., 2020). NK cell activation is controlled by a balance between activating and inhibitory signals. Healthy somatic cells typically express surface ligands that are inhibitory for NK cell receptors. However, a reduction in inhibitory ligands and/or an increase in activating ligands induces NK cells to become activated and kill the target cell by lysis or apoptosis (Ralainirina et al., 2007).
NK cells typically respond vigorously with rapid activation and homing to affected tissues during acute viral infections (Björkström et al., 2022). Additionally, tissue-resident NK cells at the site of infection can become activated and contribute to the response (Björkström et al., 2022). NK cells possess several effector functions that aid in antiviral immunity, including natural cytotoxicity, antibody dependent cellular cytotoxicity (ADCC), and cytokine production (Ralainirina et al., 2007). NK cells play a central role in protection against numerous viral infections (Waggoner et al., 2016). Studies examining individuals with NK cell deficiencies have found increased susceptibility to several viral infections (Björkström et al., 2022; Jordan, 2002). Similarly, mouse studies have found that depletion of NK cells prior to viral challenge results in higher virus titers and more severe disease (Bukowski et al., 1983). In the context of a respiratory viral infection like influenza, mouse studies show that depletion of NK cells increases morbidity and mortality during the early course of medium-dose influenza infection (Stein-Streilein and Guffee, 1986; Stein-Streilein et al., 1988; Nogusa et al., 2008).
Several studies have evaluated NK cell frequency and phenotype during SARS-CoV-2 infection (Maucourant et al., 2020; Varchetta et al., 2021; Osman et al., 2020; Bozzano et al., 2021; Zheng et al., 2020). These studies find that individuals with moderate to severe SARS-CoV-2 infection have lower frequencies of NK cells in the circulation during infection compared to uninfected controls (Maucourant et al., 2020; Varchetta et al., 2021; Osman et al., 2020; Bozzano et al., 2021; Zheng et al., 2020). A limitation of these studies is that the evaluated samples were obtained once symptomatic infection had already been established, limiting the ability to discern if low NK cell frequencies were caused by the infection or were a factor for susceptibility to symptomatic infection. In this study we tested the hypothesis that individuals with high frequencies of NK cells at baseline, prior to infection, would experience less symptomatic infection with SARS-CoV-2 than those with lower NK cell frequencies. Because we wanted to conduct this analysis on individuals who did not have adaptive immunity to SARS-CoV-2, we utilized peripheral blood mononuclear cells (PBMCs) obtained during the COVID-19 Health Action Response for Marines (CHARM) study, a prospective, longitudinal study conducted from May to November of 2020 (prior to widespread vaccination and infection). The CHARM study serially monitored marine recruits for development of symptomatic or asymptomatic SARS-CoV-2 infection with repeated nasal PCR testing and symptom assessments. All participants included in the current analysis were selected on the basis of being SARS-CoV-2 PCR and antibody negative at their first CHARM study visit, becoming PCR positive for SARS-CoV-2 during the CHARM study, and having baseline PBMCs available for analysis. We also evaluated longitudinal specimens to evaluate the kinetics of NK cell subsets during infection.
2. Methods
2.1. Study participants
Details of the CHARM study have been published previously (Letizia et al., 2021). Briefly, from May 11 to November 4, 2020, Marine recruits were placed into a two week supervised-quarantine, prior to reporting for recruit training at Parris Island. Upon entry into the quarantine, recruits were asked if they wanted to participate in the study. Inclusion criteria included being 18 years of age or older and being available for follow-up appointments. Baseline evaluation included medical history, SARS-CoV-2 serology, and polymerase chain reaction (PCR) testing of mid-turbinate swabs for SARS-CoV-2 RNA. Additionally, PBMCs were obtained and cryopreserved at enrollment from every recruit who consented to the study. After enrollment, participants were evaluated once a week for PCR testing of mid-turbinate swabs and for assessment of any viral respiratory symptoms while in quarantine. If negative on the last day of quarantine, they proceeded to training and were assessed every two weeks. If a participant tested PCR positive for SARS-CoV-2, additional PBMCs were collected two times per week for the first two weeks after PCR positivity and every two weeks thereafter until they graduated from boot camp. The subset of participants evaluated for this study were randomly selected from a pool of individuals that A) were SARS-CoV-2 PCR and IgG Spike antibody negative at study entry, B) developed a positive SARS-CoV-2 PCR test during the study, C) had cryopreserved PBMCs available from the baseline study visit, seven days or fewer from time of PCR positivity, and one to two months post initial PCR positivity, and D) completed symptom questionnaires at each clinic visit. The study was approved by the institutional review board of the Naval Medical Research Center and complied with all applicable federal regulations governing the protection of human subjects. All participants provided written informed consent.
2.2. Collection of viral respiratory symptoms
At all visits in the CHARM study, participants filled out a detailed questionnaire that asked whether, since the last visit (or in the preceding 14 days for the first visit), they had experienced fever above 100.4 °F, subjective fever (felt feverish), chills, muscle aches, fatigue, runny nose, sore throat, cough, shortness of breath, nausea or vomiting, headache, decreased ability to taste or smell, abdominal pain, diarrhea, or other symptoms to specify with choices to select being yes, no or unknown. Participants were instructed on how to fill out the symptom questionnaire by reviewing each question with a study team member at the first visit. At all subsequent encounters, participants completed the symptoms questionnaire on their own.
2.3. qPCR
For SARS-CoV-2 quantitative PCR testing, all swabs in viral transport media were kept at 4 °C. All assays were performed within 48 h of sample collection at high complexity Clinical Laboratory Improvement Amendments-certified laboratories using the US Food and Drug Administration-authorised Thermo Fisher TaqPath COVID-19 Combo Kit (Thermo Fisher Scientific, Waltham, MA, USA). Lab24Inc (Boca Raton, FL, USA) performed PCR testing from study initiation (May 11, 2020) until Aug 24, 2020, and the Naval Medical Research Center (NMRC, Silver Spring, MD, USA) from Aug 24, 2020, until the conclusion of the study (Nov 2, 2020).
2.4. PBMC isolation and purification
Peripheral blood (4ml/subject) from CHARM participants was collected in one sodium heparin blood collection tube per person (BD Vacutainer Heparin Tubes) and then shipped to NMRC overnight to be processed immediately. Briefly, sterile 15-ml conical tubes (Corning Reference # 430052) were filled with 4 ml of Ficoll-Paque Plus (GE Healthcare #17-1440-03). Vacutainer tubes containing blood were inverted several times to mix the blood. The whole blood was slowly overlaid on top of the Ficoll. The tubes were centrifuged at 800×g for 15 min at room temperature with brake turned off. PBMCs at the interface of Ficoll and plasma were carefully collected into 15-ml tubes (Corning Reference # 430052), and washed with 1x PBS (without Ca++ and Mg++) (Quality Biological, Gaithersburg, MD) twice at 400×g for 10 min at room temperature. PBMCs were gently mixed with 10% Dimethyl Sulphoxide (DMSO) (Fisher Bioreagents, https://www.fishersci.com/)/90%FBS (Hyclone, https://www.gehealthcare.com/) freezing solution and placed in cryo-vials (VMR.com) on ice and then transferred to StrataCooler Cryo Preservation containers (Stratagene, La Jolla, CA) to −80 °C for 24 h prior to transfer into liquid nitrogen for long-term storage.
2.5. Cell preparation and flow cytometry for NK cell phenotyping
Frozen PBMC samples were thawed and then washed with pre-warmed complete RPMI media (RPMI, 10% FBS, and 1% Penicillin/Streptomycin). Cells were then incubated at 37 °C for 1 h with pre-warmed complete RPMI with 50 U/ml DNase (Invitrogen). After incubation, cells were counted and put into tubes at 1 × 106 cells per vial in 100 μl cold PBS. At this time an unstained control tube was made with 1 × 106 cells and a viability control tube made that contained 5 × 105 live cells and 5 × 105 cells killed by heating to 70 °C for 10 min. All tubes except for the unstained control tube were then incubated with 1 μL of LIVE/DEAD™ Fixable Blue (Invitrogen) viability dye for 30 min on ice in the dark. Cells were then washed, resuspended in PBS/0.5% BSA, and incubated for 5 min with BD Horizon™ Brilliant Stain Buffer. Cells were then incubated for 30 min on ice in the dark with fluorochrome-conjugated antibodies to CD16 (BUV496), CD3 (BUV737), NKG2D (Super Bright 436), CD14 (BV510), CD19 (BV510), KIR3DL1 (BV711), CD57 (BV785), CD56 (FITC), CD107a (PE), KIR2DL2/L3/S2 (PE-Cy5.5), NKG2A (PE-Vio770), NKG2C (APC), CD69 (APC-R700), and KIR2DL1 (APC-Vio770) (Supplemental Table 1). The cells were washed, suspended in fixation buffer (BD Biosciences), and then incubated for 15 min at room temperature in the dark. After fixation the cells were washed and resuspended in PBS/0.5% bovine serum albumin (BSA) and stored at 4 °C until flow cytometric analysis. In addition to the unstained and viability controls using cells, single color reference controls were made using UltraComp eBeads™ Compensation Beads. The antibodies were incubated with compensation beads for 15 min on ice then washed and resuspended in PBS/0.5% BSA. All antibodies were titrated prior to use to determine optimal staining concentrations. Fluorescence minus one (FMO) controls were made by staining control cells with all of the antibodies except one to a tube. FMO cells were used to establish positivity cut-offs for CD69, CD107a, CD56, CD16, CD57, NKG2A, NKG2C, NKG2D, KIR2DL1, KIR2DL2/L3/S2, and KIR3DL1. All samples were run on a Cytek Aurora spectral cytometer (Cytek Biosciences) at the Uniformed Services University Biomedical Instrumentation Center Flow Cytometry facility. The data was analyzed with Flowjo Software v10 (BD Biosciences).
2.6. Dimensionality reduction analysis
For the dimensionality reduction analysis of NK cells, FCS3.0 files were exported from FACSDiva and imported into FlowJo v.10.8.1 for subsequent analysis. The following plug-ins were used: DownSample (3.3) and UMAP (3.1). First, dataset as such was used for the downstream analysis in both manual gating and automated analysis. For the automated analysis, events were first downsampled from the NK gate across all samples using DownSample. Categorical values for each sample were added to downsampled populations as metadata to enable identification of the two groups, and these were then concatenated for analysis. Five thousand cells per sample were exported from the NK gate, making a total of 1.284 x10e6 concatenated events. When assigning categorical groups formed by different clinical parameters and timepoints (T0: baseline, T4-7:days 4 or 7 post-pcr positive, T35: 35 days post-pcr positive), there was an uneven number of patients represented in each group (e.g., asymptomatic donors n-42: T0: 2.14 x10e5 cells, T4-7: 1.92 x10e5 cells, T35: 2.01 x10e5 cells, for symptomatic donors: n-45: T0: 2.24 x10e5 cells, T4-7: 2.22 x10e5 cells, T35: 2.27 x10e5 cells). UMAP was run using all parameters from the panel except Live/Dead, dump, CD3, CD69, CD45 and CD107a.
2.7. Statistical analyses
Mann Whitney U test was performed for comparisons between unpaired groups, Spearman's correlation was performed to evaluate relationship of one factor to another, and Dunnett's multiple comparison's test for comparisons between multiple timepoints. Statistical analyses were performed using GraphPad 9.
3. Results
3.1. Participant demographics and symptomatology
The initial CHARM study enrolled 3472 individuals. Of these, 1205 participants tested PCR positive for SARS-CoV-2 on longitudinal surveillance testing (34.7%). Among those that were PCR positive, 567 (47.1%) were symptomatic and 638 (52.9%) were asymptomatic.
At the time of this analysis, cryopreserved PBMCs from baseline (pre-infection) and post-infection visits were available from 88 participants (46 symptomatic and 42 asymptomatic), once eight samples were excluded due to low recovery of PBMCs. Of the 88 participants analyzed, 88.6% identified as male and the mean age was 19.08 years (SD 1.69) (Table 1). Participants identified their race as White (69.3%), Black (12.5%), American Indian/Alaska Native (2.3%), other (3.4%), multi-racial (3.4%) or non-specified (9.1%). Among the 88 participants, 42 were classified as asymptomatic because they reported no symptoms at the time of first PCR positivity and during subsequent encounters. The other 46 individuals were classified as having symptomatic SARS-CoV-2 infection based upon reported symptoms within the first 14 days of positive SARS-CoV-2 PCR test. Among the symptomatic cohort, the number of symptoms experienced during infection ranged from one to twelve with a mean of 5.3 (SD 3.05) (Supplemental Fig. 1). Individuals experienced a variety of different symptoms, with the most common symptoms being headache (71%), sore throat (54.3%), cough (52.2%), and fatigue (52.2%), as outlined in Supplemental Table 2. All symptomatic participants were treated as outpatients and required, at most, only symptomatic therapy with acetaminophen and/or non-steroid anti-inflammatory medications.
Table 1.
Study demographics.
| All n/N (%) | Asymptomatic n/N (%) | Symptomatic n/N (%) | |
|---|---|---|---|
| Sex | |||
| Female | 10/88 (11.4) | 0/42 (0) | 10/46 (21.7) |
| Male | 78/88 (88.6) | 42/42 (100) | 36/46 (78.3) |
| Race | |||
| White | 61/88 (69.3) | 25/42 (59.5) | 36/46 (78.3) |
| Black | 11/88 (12.5) | 7/42 (16.7) | 4/46 (8.7) |
| American Indian\Alaska Native | 2/88 (2.3) | 1/42 (2.4) | 1/46 (2.2) |
| Other | 3/88 (3.4) | 2/42 (4.8) | 1/46 (2.2) |
| Multiracial | 3/88 (3.4) | 1/42 (2.4) | 2/46 (4.3) |
| Non-specified | 8/88 (9.1) | 6/42 (14.2) | 2/46 (4.3) |
| Age | |||
| Mean age (range) | 19 (18–28) | 19 (18–28) | 19 (18–23) |
3.2. Individuals that develop asymptomatic SARS-CoV-2 infection have higher baseline frequencies of total and mature NK cells
Flow cytometry was performed on baseline PBMC samples collected on average 38 (SD 7.9 Range 7–56) days before the date of the first positive PCR test for each participant. NK cells were defined as CD3−CD14−CD19−CD56+ cells (Supplemental Fig. 2). NK cells subsets were defined as mature CD16+ (CD56dimCD16+), mature CD16− (CD56dimCD16-) immature (CD56brightCD16-), and adaptive (CD56dimCD16+NKG2C+CD57+). The percentage of NK cells present in each participant was calculated by dividing the number of cells in the “Total NK Cells” gate out of the number of cells in the “Total Cells” gate (Supplemental Fig. 2). As seen in Fig. 1A, individuals that experienced asymptomatic SARS-CoV-2 infections had significantly greater percentages of total NK cells than individuals that developed symptomatic infections (10.61% [SD 4.5] vs 8.33% [SD 4.6], p = 0.011).
Fig. 1.
Total NK cell frequencies, and mature NK cell frequencies prior to infection are greater in individuals that develop asymptomatic SARS-CoV-2 infection. Percentages of (A) total NK cells (CD3-CD14-CD19-CD56+) (B) immature NK cells (CD56brightCD16-), (C) mature CD16+ NK cells (CD56dimCD16+), (D) mature CD16- NK cells (CD56dimCD16-), and (E) adaptive NK cells (CD56dimCD16+NKG2C+CD57+) were compared between baseline samples from asymptomatic (n = 42) and symptomatic (n = 46) SARS-CoV-2 PCR+ individuals. Data are presented as mean value ± s.d. (*p < 0.05 by Mann Whitney U Test).
Percentages of the four NK cell subsets were determined with respect to the “Total NK Cells” gate (Supplemental Fig. 2). While there were no differences in percentages of immature, mature CD16−, or adaptive NK cells, asymptomatic individuals had significantly greater percentages of mature CD16+ NK cells than symptomatic individuals prior to infection (76.27% [SD 14.32] versus 69.58% [SD 17.04], p = 0.048, Fig. 1C).
Interestingly, we observed that all 10 females who were PCR positive for SARS-CoV-2 exhibited symptomatic infection. Sensitivity analysis excluding females demonstrated that total and mature NK cell frequencies remained higher in asymptomatic males than in symptomatic males (10.61% [SD 4.5] vs 8.96% [SD 4.8] and 76.27% [SD 14.3] vs 73.27% [SD 13.6] respectively). These differences, however, were not statistically different (p = 0.08, p = 0.15), possibly due to the decreased number of symptomatic male participants in the analysis.
3.3. No correlation between baseline NK cell frequencies and duration of PCR positivity
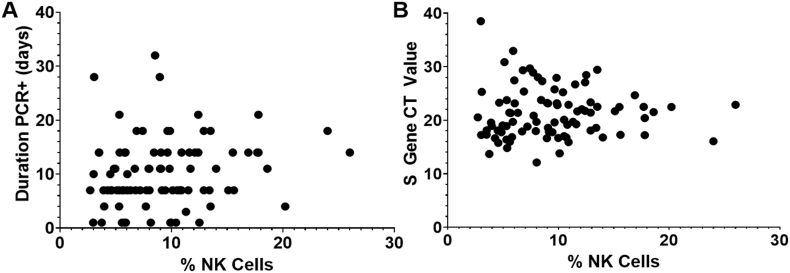
Mean duration of PCR positivity for all participants was 10 days (SD 6.3) and mean CT value was 21.3 (SD 4.7). No correlation was observed between baseline NK cell frequencies and duration of PCR positivity or CT values (Fig. 2).
Fig. 2.
No correlation observed between percentage of NK cells at baseline and duration of PCR positivity or peak S gene CT value for SARS-CoV-2. Correlation between an individual's baseline NK cell percentage and the number of days they were PCR+ for SARS-CoV-2 (A) and peak S gene CT values (B) was analyzed. No significant correlations were found between the percentage of NK cells and duration of PCR+ or S gene CT value by Spearman Correlation analyses (r = 0.177, p = 0.095; r = 0.106, p = 0.322 respectively).
3.4. Baseline expression of activating and inhibitory receptors were not significantly different between asymptomatic and symptomatic groups
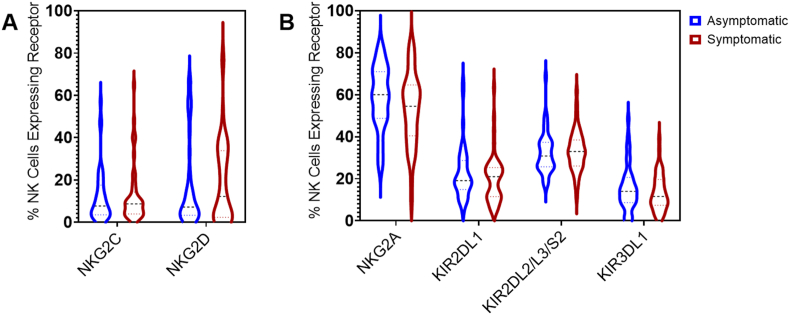
To assess whether baseline predominance of NK cell expression of activating or inhibitory receptors was associated with clinical presentation of SARS-CoV-2 infection, we assessed surface expression of two major activating receptors (NKG2C, NKG2D) and four major inhibitory receptors (NKG2A, KIR2DL1, KIR2DL2/L3/S2, and KIR3DL1) of NK cells using flow cytometry of the baseline PBMC samples. The expression of each receptor was calculated as the number of cells positively expressing a specific receptor out of the number of total NK cells. Expression of each activating and inhibitory receptor was compared between the asymptomatic and symptomatic groups. There were no significant differences between asymptomatic and symptomatic groups in the percentages of cells expressing any of these receptors (Fig. 3).
Fig. 3.
No significant differences in the expression of activating or inhibitory receptors on NK cells at baseline between asymptomatic and symptomatic individuals. Percentages of NK cells expressing (A) the activating receptors NKG2C or NKG2D, or (B), the inhibitory receptors NKG2A, KIR2DL1, KIR2DL2/L3/S2, or KIR3DL1. Percentages were calculated as the number of cells positive for the specified receptor out of the number of total NK cells (CD56+). No significant differences were observed between the asymptomatic (n = 42) vs. symptomatic (n = 46) groups for any of the receptors based on Mann Whitney U test.
A previous study observed that NKG2A+ NK cells are highly activated during SARS-CoV-2 infection and may play a key role in controlling SARS-CoV-2 activity (Hammer et al., 2022). While no differences were observed in NKG2A expression on total NK cells, as secondary analyses frequencies of immature and mature CD16+ NK cells expressing NKG2A were compared between the asymptomatic and symptomatic groups. We found that the asymptomatic group had significantly greater frequencies of NKG2A+ immature NK cells compared to the symptomatic group (96.32% SD 2.81, vs. 94.27% SD 5.12; p = 0.034) (Supplemental Fig. 3A). No differences were found in NKG2A+ mature CD16+ NK cells between asymptomatic and symptomatic groups (Supplemental Fig. 3B).
3.5. Frequencies of total NK cells decreased during SARS-CoV-2 infection while immature NK cells increased
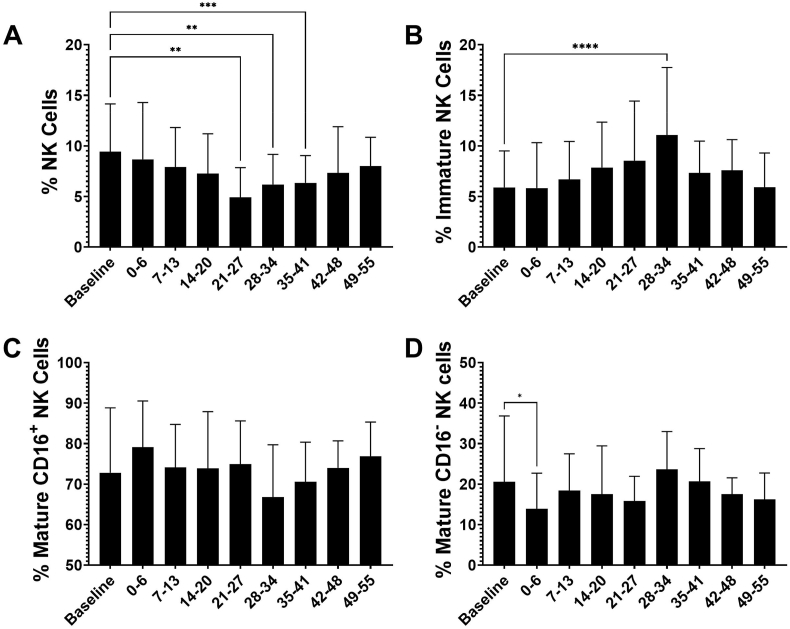
Flow cytometric analyses of total NK cells and NK cell subsets was conducted on longitudinal samples collected from 88 individuals for up to 9 weeks post-infection. As seen in Fig. 4A, a gradual decrease was observed in the frequency of total peripheral NK cells during the first four weeks following infection, dropping from a baseline of 9.4% (SD 4.7) to a nadir of 4.9% (SD 2.9) 21–27 days post first positive PCR test (p = 0.0031) and then gradually returning to baseline levels around 49–55 days (Fig. 4A).
Fig. 4.
Peripheral NK cell kinetics post-infection. Frequencies of (A) total NK cells (CD56+), (B) immature NK cells (CD56brightCD16-), (C) mature CD16+ NK cells (CD56dimCD16+), and (D) mature CD16- NK cells (CD56dimCD16-) were calculated for samples obtained from individuals at baseline and at timepoints after initial PCR+ test for SARS-CoV-2. All available samples were analyzed. Number of samples at baseline n = 87, post-PCR samples analyzed at days 0–6 (n = 27), 7–13 (n = 86), 14–20 (n = 14), 21–27 (n = 12), 28–34 (n = 24), 35–41 (n = 47), 42–48 (n = 6), 49–55 (n = 35). Analyses between timepoints were conducted using Dunnett's multiple comparisons test (**p < 0.01, ***p < 0.001, ****p < 0.0001).
Interestingly, as total NK cell frequencies decreased post-infection, the percentages of NK cells that were immature increased (Fig. 4B). Percentages of immature NK cells rose from 5.9% (SD 3.5) at baseline to 11.1% (SD 6.6) at 28–34 days post-first PCR test (p=<0.0001) (Fig. 4B). The percentages of total NK cells that were mature CD16+ NK cells decreased following SARS-CoV-2 infection, however there were no significant differences in frequencies between baseline level and the weeks after infection (Fig. 4C). Frequencies of the CD56dim CD16− population decreased significantly in the first week after PCR positivity (20.59% SD 16.23, to 13.89% SD 8.78; p = 0.048), but overall there were no specific changes in the frequencies of mature CD16− NK cells longitudinally.
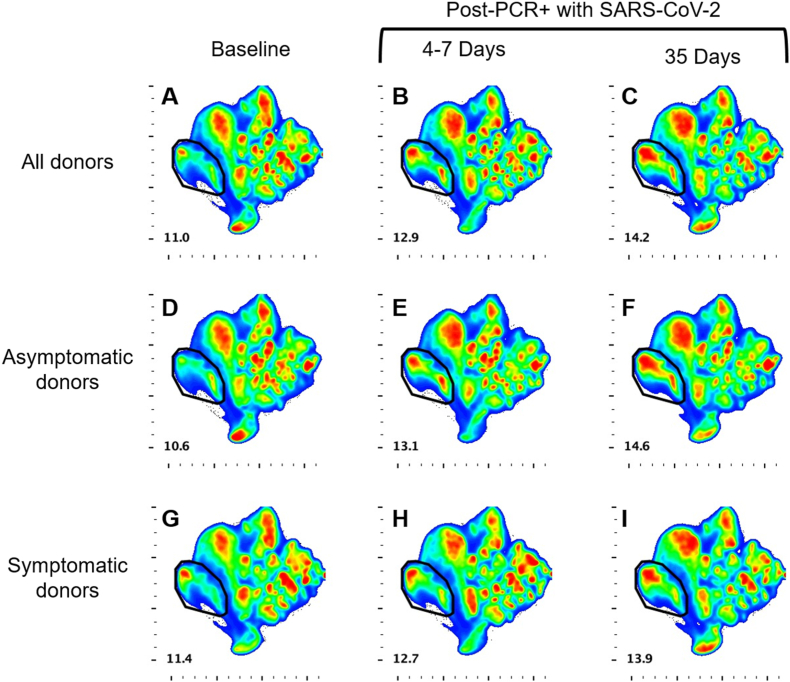
To evaluate changes in NK cell populations during the course of infection that were not defined a priori we performed multidimensional reduction analyses on longitudinal samples. UMAP analyses revealed no differences between the clustering of populations between the asymptomatic donor group (Fig. 5D–F), and the symptomatic donor group (Fig. 5G–I). The analysis did show a cluster of cells that expanded from baseline, before SARS-CoV-2 infection (Fig. 5, A, D, G; Supplemental Fig. 4), to 4–7 days post-PCR+ with SARS-CoV-2 (Fig. 5, B, E, H; Supplemental Fig. 4), with further expansion at 35 days post-infection (Fig. 5, C, F, I; Supplemental Fig. 4). The phenotype of the cells within this cluster of interest is CD56hi, CD16−, NKG2A+, CD57−, and NKG2C− (Supplemental Fig. 5). This phenotype indicates an immature population of NK cells which is expanding during infection. This directly aligns with the earlier data from Fig. 4B which demonstrated an increase in the percentage of the immature NK cell population in the weeks following infection with SARS-CoV-2.
Fig. 5.
Longitudinal UMAP analyses of NK cell clusters. UMAP analyses reveal a population of NK cells (circled) which expands from baseline (A, D, G) to 4–7 days (B, E, H) and 35 days (C, F, I) post-infection. This expansion was found when all donor samples were examined (A–C), and when examining the asymptomatic donors (D–F) and symptomatic donors (G–I) separately. Alignment with phenotypic markers reveals this population to be comprised of immature NK cells (see Supplemental Fig. 2).
4. Discussion
The specific immunological factors associated with protection against clinical disease with SARS-CoV-2 are not completely understood (Boyton and Altmann, 2021). Given that NK cells are one of the first cell types to respond to viral infections, the baseline frequency and repertoire of NK cells prior to SARS-CoV-2 infection may be an important indicator of whether an individual is at risk of developing asymptomatic or symptomatic infection. By conducting a flow cytometric study of NK cell frequencies and phenotypes in individuals enrolled in a prospective, observational study which monitored for development of SARS-CoV-2 infection by serial PCR testing, we were able to confirm the primary hypothesis that higher baseline frequencies of NK cells are associated with decreased risk of developing symptomatic disease when an individual becomes infected with SARS-CoV-2. To the best of our knowledge, this represents the first study demonstrating a relationship between baseline NK cell frequencies and the presence or absence of symptoms due to SARS-CoV-2 infection.
Our finding that NK cell frequencies are greater in asymptomatic compared to symptomatic individuals at baseline suggests that NK cells likely play a key role in controlling the severity of SARS-CoV-2 infection. This conclusion aligns with the general understanding of NK cell function in response to viral respiratory infections. In the setting of influenza infection, it is recognized that NK cells have an essential role in controlling virus titers in the lung early in the course of infection in both murine models and humans (Stein-Streilein and Guffee, 1986; Stein-Streilein et al., 1988; Nogusa et al., 2008; Ennis et al., 1981).
One of the peculiarities of the COVID-19 pandemic has been the lower disease burden in the very young compared to the elderly (Mueller et al., 2020). Most viral respiratory infections usually impact the extremes of age: the very young and the very old (Chicaiza-Ayala et al., 2018). While many young children have become severely ill from COVID-19 and hundreds have died, the percentage of young children requiring hospitalization and experiencing lethal outcomes is far lower than that observed for the elderly (Weekly Updates by Select Demographic, 2022). If NK cells play a major role in attenuating spread of SARS-CoV-2 in the body, it could account for the unbalanced impact on the elderly as NK cells become fully functional around one to five months of age but then decrease as individuals progress into older adulthood (Yabuhara et al., 1990; Hazeldine and Lord, 2013).
In addition to evaluating total NK cell frequencies, we also assessed frequencies of mature CD16+, mature CD16−, immature, and adaptive NK cell subsets before infection. As with total NK cells, at baseline individuals who developed asymptomatic infections had greater proportions of mature CD16+ NK cells, which are the main subset responsible for cytotoxic activity and killing of virally infected cells. No significant differences were observed between symptomatic and asymptomatic groups with regards to pre-infection frequencies of mature CD16−, immature, and adaptive NK cells, or in expression of various activating and inhibitory receptors. Interestingly, secondary subset analysis revealed that individuals who developed asymptomatic infection had greater frequencies of NKG2A+ immature NK cells than individuals that developed symptomatic infection. This finding is consistent with a prior study in humans that found that NKG2A+ NK cells are highly activated in humans with SARS-CoV-2 infection and strongly inhibit SARS-CoV-2 replication in vitro (Hammer et al., 2022).
In addition to evaluating NK cells at baseline, we also had the opportunity to observe longitudinal changes. Our data demonstrates that circulating NK cells decrease over the course of infection, reaching a nadir at 4 weeks post infection and then recovering to baseline levels by 8 weeks post infection. This kinetic response in NK cells aligns well with prior studies, which have noted decreased levels of NK cells in SARS-CoV-2 infected individuals compared to healthy controls (Maucourant et al., 2020; Varchetta et al., 2021; Osman et al., 2020; Bozzano et al., 2021; Zheng et al., 2020; Witkowski et al., 2021). Our results suggest that these findings are a consequence of the infection. We hypothesize that the drop in NK cells may reflect cellular efflux from the circulation to the tissues in response to virus-infected cells. Studies in mice examining the frequency and phenotype of NK cells following respiratory infection show that NK cells accumulate in the lung in the days following the initial infection, which aligns with our hypothesis that the NK cells are leaving the peripheral blood and traveling to the site of infection (Wang et al., 2012; Frank and Paust, 2020). The finding that NK cell population frequencies do not return to baseline for about two months is interesting. It may be that perturbations in NK cell homeostasis simply take several weeks to normalize, or that the host NK cell response to SARS-CoV-2 infection is prolonged even when individuals have mild outpatient infections. Interestingly, a recent autopsy study found that two individuals who had mild SARS-CoV-2 infection and died of other causes still had detectable SARS-CoV-2 RNA distributed in several tissues throughout the body weeks to months after infection (Stein et al., 2022).
We were also able to analyze kinetics of immature and mature subsets during SARS-CoV-2 infection. In contrast to total NK cells, we observed that immature NK cells gradually increased during the first four weeks after SARS-CoV-2 PCR positivity. The finding of increased immature NK cells was observed both with directed analyses as well as with unsupervised UMAP clustering analyses. This increase in immature NK cells may reflect a true increase in production of NK cells from the bone marrow in response to SARS-CoV-2 infection, or may be due to the egress of mature NK cells from the circulation.
Interestingly, percentages of NK cells at baseline did not correlate with inferred viral load dynamics such as duration of PCR positivity or CT value upon first PCR positivity. We suspect this may have occurred because the amount of detectable RNA from nasopharyngeal (NP) swabs does not necessarily represent the systemic or pulmonary viral load. Studies have found that CT values differ between samples obtained from NP swabs compared to samples obtained from the lower respiratory tract by endotracheal aspirate or bronchoalveolar lavage (Liu et al., 2020; Hamed et al., 2021). It has also been recognized that virus shedding time is different in various parts of the respiratory tract (Liu et al., 2020). Additionally, a study of 287 patients in France found that viral loads from nasopharyngeal swabs obtained during initial management is not predictive of subsequent COVID-19 disease severity or mortality (Le Borgne et al., 2021). Due to these challenges interpreting the results from NP swabs, it is difficult to definitively conclude that NK cells do not control viral load. We speculate that NK cells may not substantially control viral replication at the nasal mucosal surface, but likely afford protection against more invasive infection in deeper sites such as the lungs and vascular system.
There are several limitations to consider with regards to this study. First, the findings of this study may not be generalizable to the entire population. Our cohort was made up of United States Marine Corps recruits who have specific medical, physical fitness and age requirements to enlist. This limited our cohort to primarily men with an average age of 19 who are generally healthy. That stated, the homogeneity of the population may have facilitated assessment of the role of NK cells as there were few confounders. Also, as this study was carried out from May through November 2020, the participants were likely infected with wild-type D614G SARS-CoV-2 (Lizewski et al., 2022) and therefore the findings may not extend to infection with more recent variants. An additional factor to contemplate is that during boot camp the participants are undergoing extensive physical activity and research has shown that exercise does have an effect on an individual's NK cells. One study found that four weeks of moderate intensity exercise, or four weeks of high intensity exercise, results in increased numbers of NK cells in the blood compared to baseline levels from before the exercise program (Llavero et al., 2021). However, this consideration only applies to the later time point samples that were obtained once participants were infected, since the baseline samples were collected during the quarantine period before boot camp began. Nonetheless, exercise could be a possible confounding and unmeasured variable with regards to the NK cell kinetics. Another limitation is that all of the observed symptomatic infections in this study were mild in severity. The final limitation of this study is that, due to limited numbers of cryopreserved PBMCs per participant, we did not have sufficient cells to conduct in-vitro functional or transcriptional studies of NK cells.
Finally, it is interesting to note that in contrast to males, in which 46% developed symptomatic infection, all 10 females who were PCR positive for SARS-CoV-2 in this analysis had symptomatic infection. These results are in line with the CHARM cohort overall, in which infected females were more often symptomatic than infected males. In general, males have higher rates of hospitalization and mortality compared to females (Gomez et al., 2021; Nguyen et al., 2021). The CHARM study suggests that for mild infections in young and healthy adults, females may be more likely to have symptoms than males. Sensitivity analysis revealed a similar magnitude of difference in NK cell frequencies between symptomatic and asymptomatic individuals when female participants were excluded, but the differences were not statistically significant (potentially due to reduced power from analyzing a smaller number of symptomatic participants).
In conclusion, we observed that individuals who developed symptomatic COVID-19 infection had significantly fewer NK cells before they were infected compared to those who had asymptomatic infection. We also found that circulating frequencies of total and mature NK cell decrease while immature NK cells increase over the first month following initial infection. These findings suggest that NK cells may play a protective role in controlling the clinical severity of SARS-CoV-2 infection.
Author contributions
EKG: Conceived and designed the study/experiments, Acquired data/performed experiments, Created detailed Formal analysis plan and/or analyzed the data, Interpreted findings, Contributed Resources; reagents/materials/specimens, Composed first draft of manuscript, Provided critical revisions and edits to provisional drafts, Reviewed and approved final version for submission. AGL: Conceived and designed the study/experiments, Created detailed Formal analysis plan and/or analyzed the data, Interpreted findings, Contributed Resources; reagents/materials/specimens, Provided critical revisions and edits to provisional drafts, Reviewed and approved final version for submission. EM: Conceived and designed the study/experiments, Created detailed Formal analysis plan and/or analyzed the data, Interpreted findings, Composed first draft of manuscript, Provided critical revisions and edits to provisional drafts, Reviewed and approved final version for submission. PS: Acquired data/performed experiments, Reviewed and approved final version for submission. SL: Acquired data/performed experiments, Contributed resources; reagents/materials/specimens, Reviewed and approved final version for submission. RL: Acquired data/performed experiments, Contributed Resources; reagents/materials/specimens, Reviewed and approved final version for submission. DLW: Acquired data/performed experiments, Contributed Resources; reagents/materials/specimens, Reviewed and approved final version for submission. CWG: Acquired data/performed experiments, Contributed Resources; reagents/materials/specimens, Reviewed and approved final version for submission. AM: Created detailed Formal analysis plan and/or analyzed the data, Reviewed and approved final version for submission. KM: Created detailed Formal analysis plan and/or analyzed the data, Interpreted findings, Reviewed and approved final version for submission. DP-P: Created detailed Formal analysis plan and/or analyzed the data, Interpreted findings, Reviewed and approved final version for submission. SA: Created detailed Formal analysis plan and/or analyzed the data, Provided critical revisions and edits to provisional drafts, Reviewed and approved final version for submission.
Funding
This work was supported by grant 9700130 from the Defense Health Agency through the Naval Medical Research Center (AGL). This work was supported by a cooperative agreement (WW81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DOD). This research was supported in part by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research, under contract HHSN261200800001E.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Uniformed Services University of the Health Sciences, Department of the Navy, Department of Defense, the U.S. Government, nor the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. AMWM, PS, SL, RL, DLW, CWG, AGL, and EM are military Service members or employees of the U.S. Government. This work was prepared as part of their official duties. Title 17, U.S.C., §105 provides that copyright protection under this title is not available for any work of the U.S. Government. Title 17, U.S.C., §101 defines a U.S. Government work as a work prepared by a military Service member or employee of the U.S. Government as part of that person’s official duties.
Acknowledgements
The many U.S. Navy corpsmen who assisted in the logistics and sample acquisition and the Marine Corps recruits who volunteered for this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crimmu.2023.100064.
Appendix A. Supplementary data
The following is the supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abel A.M., Yang C., Thakar M.S., Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béziat V., Liu L.L., Malmberg J.A., Ivarsson M.A., Sohlberg E., Björklund A.T., Retière C., Sverremark-Ekström E., Traherne J., Ljungman P., Schaffer M., Price D.A., Trowsdale J., Michaëlsson J., Ljunggren H.G., Malmberg K.J. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 2013;121:2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkström N.K., Strunz B., Ljunggren H.-G. Natural killer cells in antiviral immunity. Nat. Rev. Immunol. 2022;22:112–123. doi: 10.1038/s41577-021-00558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyton R.J., Altmann D.M. The immunology of asymptomatic SARS-CoV-2 infection: what are the key questions? Nat. Rev. Immunol. 2021;21:762–768. doi: 10.1038/s41577-021-00631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzano F., Dentone C., Perrone C., Di Biagio A., Fenoglio D., Parodi A., Mikulska M., Bruzzone B., Giacobbe D.R., Vena A., Taramasso L., Nicolini L., Patroniti N., Pelosi P., Gratarola A., De Palma R., Filaci G., Bassetti M., De Maria A. Extensive activation, tissue trafficking, turnover and functional impairment of NK cells in COVID-19 patients at disease onset associates with subsequent disease severity. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski J.F., Woda B.A., Habu S., Okumura K., Welsh R.M. Natural killer cell depletion enhances virus synthesis and virus-induced hepatitis in vivo. J. Immunol. 1983;131:1531–1538. [PubMed] [Google Scholar]
- Chicaiza-Ayala W., Henríquez-Trujillo A.R., Ortiz-Prado E., Douce R.W., Coral-Almeida M. The burden of acute respiratory infections in Ecuador 2011-2015. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis F., Beare A.S., Riley D., Schild G., Meager A., Yi-Hua Q., Schwarz G., Rook A. Interferon induction and increased natural killer-cell activity in influenza infections in man. Lancet. 1981;318:891–893. doi: 10.1016/s0140-6736(81)91390-8. [DOI] [PubMed] [Google Scholar]
- Frank K., Paust S. Dynamic natural killer cell and T cell responses to influenza infection. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freud A.G., Mundy-Bosse B.L., Yu J., Caligiuri M.A. The broad spectrum of human natural killer cell diversity. Immunity. 2017;47:820–833. doi: 10.1016/j.immuni.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J.M.D., Du-Fay-de-Lavallaz J.M., Fugar S., Sarau A., Simmons J.A., Clark B., Sanghani R.M., Aggarwal N.T., Williams K.A., Doukky R., Volgman A.S. Sex differences in COVID-19 hospitalization and mortality. J. Womens Health (Larchmt) 2021;30:646–653. doi: 10.1089/jwh.2020.8948. [DOI] [PubMed] [Google Scholar]
- Gondois-Rey F., Chéret A., Granjeaud S., Mallet F., Bidaut G., Lécuroux C., Ploquin M., Müller-Trutwin M., Rouzioux C., Avettand-Fenoël V., Moretta A., Pialoux G., Goujard C., Meyer L., Olive D. NKG2C(+) memory-like NK cells contribute to the control of HIV viremia during primary infection: optiprim-ANRS 147. Clin. Transl. Immunol. 2017;6 doi: 10.1038/cti.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumá M., Angulo A., Vilches C., Gómez-Lozano N., Malats N., López-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- Hamed I., Shaban N., Nassar M., Cayir D., Love S., Curran M.D., Webb S., Yang H., Watson K., Rostron A., Navapurkar V., Mahroof R., Conway Morris A. Paired nasopharyngeal and deep lung testing for severe acute respiratory syndrome coronavirus-2 reveals a viral gradient in critically ill patients: a multicenter study. Chest. 2021;159:1387–1390. doi: 10.1016/j.chest.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Q., Dunst J., Christ W., Picarazzi F., Wendorff M., Momayyezi P., Huhn O., Netskar H.K., Maleki K.T., García M., Sekine T., Sohlberg E., Azzimato V., Aouadi M., Degenhardt F., Franke A., Spallotta F., Mori M., Michaëlsson J., Björkström N.K., Rückert T., Romagnani C., Horowitz A., Klingström J., Ljunggren H.-G., Malmberg K.-J. SARS-CoV-2 Nsp13 encodes for an HLA-E-stabilizing peptide that abrogates inhibition of NKG2A-expressing NK cells. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazeldine J., Lord J.M. The impact of ageing on natural killer cell function and potential consequences for health in older adults. Ageing Res. Rev. 2013;12:1069–1078. doi: 10.1016/j.arr.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. Orange, Human natural killer cell deficiencies and susceptibility to infection. Microb. Infect. 2002;4:1545–1558. doi: 10.1016/s1286-4579(02)00038-2. [DOI] [PubMed] [Google Scholar]
- Le Borgne P., Solis M., Severac F., Merdji H., Ruch Y., Alamé Intern K., Bayle E., Hansmann Y., Bilbault P., Fafi-Kremer S., Meziani F. SARS-CoV-2 viral load in nasopharyngeal swabs in the emergency department does not predict COVID-19 severity and mortality. Acad. Emerg. Med. 2021;28:306–313. doi: 10.1111/acem.14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letizia A.G., Ge Y., Goforth C.W., Weir D.L., Lizewski R., Lizewski S., Soares-Schanoski A., Vangeti S., Marjanovic N., Sealfon S.C., Ramos I. SARS-CoV-2 seropositivity among US marine recruits attending basic training, United States, spring-fall 2020. Emerg. Infect. Dis. 2021;27:1188–1192. doi: 10.3201/eid2704.204732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Yi S., Zhang J., Lv Z., Zhu C., Zhang Y. Viral load dynamics in sputum and nasopharyngeal swab in patients with COVID-19. J. Dent. Res. 2020;99:1239–1244. doi: 10.1177/0022034520946251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizewski R.A., Sealfon R.S.G., Park S.W., Smith G.R., Porter C.K., Gonzalez-Reiche A.S., Ge Y., Miller C.M., Goforth C.W., Pincas H., Termini M.S., Ramos I., Nair V.D., Lizewski S.E., Alshammary H., Cer R.Z., Chen H.W., George M.-C., Arnold C.E., Glang L.A., Long K.A., Malagon F., Marayag J.J., Nunez E., Rice G.K., Santa Ana E., Schilling M.A., Smith D.R., Sugiharto V.A., Sun P., van de Guchte A., Khan Z., Dutta J., Vangeti S., Voegtly L.J., Weir D.L., Metcalf C.J.E., Troyanskaya O.G., Bishop-Lilly K.A., Grenfell B.T., van Bakel H., Letizia A.G., Sealfon S.C. SARS-CoV-2 outbreak dynamics in an isolated US military recruit training center with rigorous prevention measures. Epidemiology. 2022;33:797–807. doi: 10.1097/EDE.0000000000001523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llavero F., Alejo L.B., Fiuza-Luces C., López Soto A., Valenzuela P.L., Castillo-García A., Morales J.S., Fernández D., Aldazabal I.P., Ramírez M., Santos-Lozano A., Zugaza J.L., Lucia A. Exercise training effects on natural killer cells: a preliminary proteomics and systems biology approach. Exerc. Immunol. Rev. 2021;27:125–141. [PubMed] [Google Scholar]
- Lopez-Vergès S., Milush J.M., Schwartz B.S., Pando M.J., Jarjoura J., York V.A., Houchins J.P., Miller S., Kang S.M., Norris P.J., Nixon D.F., Lanier L.L. Expansion of a unique CD57⁺NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14725–14732. doi: 10.1073/pnas.1110900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., Strunz B., Lentini A., Reinius B., Brownlie D., Cuapio A., Ask E.H., Hull R.M., Haroun-Izquierdo A., Schaffer M., Klingström J., Folkesson E., Buggert M., Sandberg J.K., Eriksson L.I., Rooyackers O., Ljunggren H.-G., Malmberg K.-J., Michaëlsson J., Marquardt N., Hammer Q., Strålin K., Björkström N.K. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A.L., McNamara M.S., Sinclair D.A. Why does COVID-19 disproportionately affect older people? Aging (Albany NY) 2020;12:9959–9981. doi: 10.18632/aging.103344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N.T., Chinn J., De Ferrante M., Kirby K.A., Hohmann S.F., Amin A. Male gender is a predictor of higher mortality in hospitalized adults with COVID-19. PLoS One. 2021;16 doi: 10.1371/journal.pone.0254066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogusa S., Ritz B.W., Kassim S.H., Jennings S.R., Gardner E.M. Characterization of age-related changes in natural killer cells during primary influenza infection in mice. Mech. Ageing Dev. 2008;129:223–230. doi: 10.1016/j.mad.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Osman M., Faridi R.M., Sligl W., Shabani-Rad M.-T., Dharmani-Khan P., Parker A., Kalra A., Tripathi M.B., Storek J., Cohen Tervaert J.W., Khan F.M. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Adv. 2020;4:5035–5039. doi: 10.1182/bloodadvances.2020002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralainirina N., Poli A., Michel T., Poos L., Andrès E., Hentges F., Zimmer J. Control of NK cell functions by CD4+CD25+ regulatory T cells. J. Leukoc. Biol. 2007;81:144–153. doi: 10.1189/jlb.0606409. [DOI] [PubMed] [Google Scholar]
- Romee R., Foley B., Lenvik T., Wang Y., Zhang B., Ankarlo D., Luo X., Cooley S., Verneris M., Walcheck B., Miller J. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17) Blood. 2013;121:3599–3608. doi: 10.1182/blood-2012-04-425397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S.R., Ramelli S.C., Grazioli A., Chung J.-Y., Singh M., Yinda C.K., Winkler C.W., Sun J., Dickey J.M., Ylaya K., Ko S.H., Platt A.P., Burbelo P.D., Quezado M., Pittaluga S., Purcell M., Munster V.J., Belinky F., Ramos-Benitez M.J., Boritz E.A., Lach I.A., Herr D.L., Rabin J., Saharia K.K., Madathil R.J., Tabatabai A., Soherwardi S., McCurdy M.T., Babyak A.L., Perez Valencia L.J., Curran S.J., Richert M.E., Young W.J., Young S.P., Gasmi B., Sampaio De Melo M., Desar S., Tadros S., Nasir N., Jin X., Rajan S., Dikoglu E., Ozkaya N., Smith G., Emanuel E.R., Kelsall B.L., Olivera J.A., Blawas M., Star R.A., Hays N., Singireddy S., Wu J., Raja K., Curto R., Chung J.E., Borth A.J., Bowers K.A., Weichold A.M., Minor P.A., Moshref M.A.N., Kelly E.E., Sajadi M.M., Scalea T.M., Tran D., Dahi S., Deatrick K.B., Krause E.M., Herrold J.A., Hochberg E.S., Cornachione C.R., Levine A.R., Richards J.E., Elder J., Burke A.P., Mazzeffi M.A., Christenson R.H., Chancer Z.A., Abdulmahdi M., Sopha S., Goldberg T., Sangwan Y., Sudano K., Blume D., Radin B., Arnouk M., Eagan J.W., Palermo R., Harris A.D., Pohida T., Garmendia-Cedillos M., Dold G., Saglio E., Pham P., Peterson K.E., Cohen J.I., de Wit E., Vannella K.M., Hewitt S.M., Kleiner D.E., Chertow D.S., Consortium N.C.-A. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature. 2022;612:758–763. doi: 10.1038/s41586-022-05542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-Streilein J., Guffee J. In vivo treatment of mice and hamsters with antibodies to asialo GM1 increases morbidity and mortality to pulmonary influenza infection. J. Immunol. 1986;136:1435–1441. [PubMed] [Google Scholar]
- Stein-Streilein J., Guffee J., Fan W. Locally and systemically derived natural killer cells participate in defense against intranasally inoculated influenza virus. Reg. Immunol. 1988;1:100–105. [PubMed] [Google Scholar]
- Varchetta S., Mele D., Oliviero B., Mantovani S., Ludovisi S., Cerino A., Bruno R., Castelli A., Mosconi M., Vecchia M., Roda S., Sachs M., Klersy C., Mondelli M.U. Unique immunological profile in patients with COVID-19. Cell. Mol. Immunol. 2021;18:604–612. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner S.N., Reighard S.D., Gyurova I.E., Cranert S.A., Mahl S.E., Karmele E.P., McNally J.P., Moran M.T., Brooks T.R., Yaqoob F., Rydyznski C.E. Roles of natural killer cells in antiviral immunity. Curr. Opin. Virol. 2016;16:15–23. doi: 10.1016/j.coviro.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li F., Zheng M., Sun R., Wei H., Tian Z. Lung natural killer cells in mice: phenotype and response to respiratory infection. Immunology. 2012;137:37–47. doi: 10.1111/j.1365-2567.2012.03607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekly Updates by Select Demographic and Geographic Characteristics. National Center for Health Statistics; 2022. [Google Scholar]
- Witkowski M., Tizian C., Ferreira-Gomes M., Niemeyer D., Jones T.C., Heinrich F., Frischbutter S., Angermair S., Hohnstein T., Mattiola I., Nawrath P., McEwen S., Zocche S., Viviano E., Heinz G.A., Maurer M., Kölsch U., Chua R.L., Aschman T., Meisel C., Radke J., Sawitzki B., Roehmel J., Allers K., Moos V., Schneider T., Hanitsch L., Mall M.A., Conrad C., Radbruch H., Duerr C.U., Trapani J.A., Marcenaro E., Kallinich T., Corman V.M., Kurth F., Sander L.E., Drosten C., Treskatsch S., Durek P., Kruglov A., Radbruch A., Mashreghi M.-F., Diefenbach A. Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells. Nature. 2021;600:295–301. doi: 10.1038/s41586-021-04142-6. [DOI] [PubMed] [Google Scholar]
- Yabuhara A., Kawai H., Komiyama A. Development of natural killer cytotoxicity during childhood: marked increases in number of natural killer cells with adequate cytotoxic abilities during infancy to early childhood. Pediatr. Res. 1990;28:316–322. doi: 10.1203/00006450-199010000-00002. [DOI] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.