Abstract
The suitability of a radiometric culture medium consisting of BACTEC 12B with PANTA PLUS, mycobactin J, and egg yolk was evaluated for detection of Mycobacterium paratuberculosis in feces, mesenteric lymph nodes, and intestinal walls from cattle, sheep, and goats. In addition, a simple method that would enable the rapid identification of Mycobacterium paratuberculosis by IS900 PCR in the primary cultures was sought so that subculture to secondary egg-free radiometric medium could be avoided. An ethanol extraction followed by differential centrifugation was used to separate M. paratuberculosis from PCR inhibitors in the primary culture. PCR was then undertaken with the pellet, after boiling to lyse the mycobacteria; if this test was negative, the DNA in the lysate was purified with guanidine thiocyanate and silica. Cultures of feces, ilea, and mesenteric lymph nodes from cattle, sheep, and goats known to have or suspected of having Johne’s disease yielded positive PCR results 1 to 7 weeks after inoculation. Similar results were obtained with soil and pasture samples that had been spiked with M. paratuberculosis. The results suggested that radiometric culture was more sensitive than histopathology in detecting M. paratuberculosis infection in sheep and goats and more sensitive than culture on Herrold’s egg yolk medium for the detection of the infection in cattle. Of 259 individual PCR tests with samples from cultures with growth indices of ≥10,237 (91.5%) were positive, with only 28 (11.8%) requiring both ethanol and silica preparation to yield a positive result. Of the 22 negative PCR results for samples from cultures with growth indices of ≥10, 18 were for samples from cultures that had only just developed evidence of growth. PCR-positive cultures tended to remain PCR positive over successive weeks. Flexibility in the timing of the sampling for PCR is thus possible, facilitating batch processing of samples in large-scale disease control programs for ruminants.
Mycobacterium paratuberculosis is the etiological agent of paratuberculosis or Johne’s disease, a granulomatous enteropathy mainly affecting ruminants. Serious economic losses attributable to paratuberculosis are documented in agricultural enterprises in many countries. In Australia, a voluntary national disease control program for bovine paratuberculosis has been implemented, while similar plans for the goat, sheep, and alpaca industries are being advanced. Serology will be used in each of the disease control programs to identify putative infected herds or flocks; infection status will be confirmed by fecal culture and/or postmortem examination of seropositive animals.
One of the factors that deters farmers from participating in a voluntary paratuberculosis control program is the time taken by the laboratory to confirm a diagnosis by culture. M. paratuberculosis is a slowly growing mycobacterium, requiring up to 20 weeks to produce colonies on solid medium. Demonstration of dependence on the iron-chelating compound mycobactin following subculture onto medium with and without mycobactin has been used to identify M. paratuberculosis and requires an additional incubation of several weeks (15). Consequently, alternative methods for culture and identification of M. paratuberculosis have been investigated. Identification is now readily achieved by PCR amplification of the IS900 gene, an element unique to M. paratuberculosis (7, 10).
In studies of human tuberculosis, rapid detection of M. tuberculosis has been achieved through the use of radiometric culture systems in which a 14C-labelled substrate (often palmitic acid) in a liquid medium is metabolized to radiolabelled carbon dioxide that can be measured sensitively in the gas phase above the culture (11). This method is known as radiometric culture. Damato and Collins (6) applied radiometric culture to M. paratuberculosis and found it to be more rapid than other cultural procedures, with growth being detected as early as 9 days after inoculation of the medium. However, the time required was found to be longer with samples from animals with low-grade infections because they contained relatively few mycobacteria compared with the numbers in samples from severely affected animals (3). Radiometric culture was successfully combined with IS900 PCR analysis to obtain relatively rapid confirmation of the M. paratuberculosis status of a sample (4). The method involved inoculation of a primary radiometric culture containing egg yolk and, after a growth index (GI) was obtained, subculture to the same medium without egg yolk. A PCR assay was undertaken from the secondary culture. While the results were very encouraging, samples from only one cow and three alpacas were tested, and the necessity for subculture to avoid the PCR inhibitors present in the egg yolk added to the cost (A$5.50 per BACTEC vial) and time required to obtain a diagnosis.
In Australia, there is a need for reliable culture techniques for the strains of M. paratuberculosis that commonly infect sheep because these strains tend not to grow on conventional solid media (2). Recently, Skilbeck (12) used radiometric culture to grow M. paratuberculosis from tissues of sheep with Johne’s disease in Victoria, Australia.
The aim of the present study was to evaluate radiometric culture as a means of culturing M. paratuberculosis from feces, mesenteric lymph nodes, and intestinal walls from cattle, sheep, and goats. In addition, a simple method of enabling the specific identification of M. paratuberculosis by IS900 PCR in primary radiometric cultures containing egg yolk was sought so that the delays and cost of subculture to secondary radiometric media could be avoided.
MATERIALS AND METHODS
Collection and storage of samples.
Feces, mesenteric lymph nodes, and intestinal tissues were collected from cattle, sheep, and goats known to have or suspected of having Johne’s disease. The status of all animals was evaluated by histological examination of the ileum (four sites 2 m apart), cecum (one site), proximal colon (one site), and caudal mesenteric lymph node (one site) and/or by serological examination by a gel diffusion precipitation test (goats and sheep) and absorbed enzyme-linked immunosorbent assays (ELISAs) (cattle, goats, and sheep) (13). Feces were collected from the rectums of animals while they were on the farm or during postmortem examination of animals that were killed immediately before sample retrieval. Feces and tissues for culture were stored at 4°C overnight and then, if required, at −20 or −80°C, pending examinations.
Acid-fast-stained smears and histopathology.
Smears were prepared from feces and scrapings of intestinal mucosa, dried in an oven at 65°C, and stained by a Ziehl-Neelsen technique (5). Tissues were fixed in 10% buffered neutral formalin, embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin and by a Ziehl-Neelsen method (9).
Culture of M. paratuberculosis.
The double incubation method of Whitlock and Rosenberger (16) was used to prepare feces. Briefly, a fecal sample (2 to 5 g) was placed in a 15-ml polypropylene tube containing a swab stick, which was used to break up the feces in 10 to 12 ml of sterile normal saline. After mixing, the tube was allowed to stand for 30 min at room temperature. Five milliliters of the surface fluid was transferred to a fresh tube containing 25 ml of 0.9% hexadecylpyridinium chloride (HPC; Sigma Chemical Co., St. Louis, Mo.) in half-strength brain heart infusion broth (Oxoid, Basingstoke, England), and the contents were allowed to stand at 37°C for 24 h. After centrifugation at 900 × g for 30 min, the pellet was collected and resuspended in 1 ml of half-strength brain heart infusion broth with vancomycin (100 μg/ml), nalidixic acid (100 μg/ml), and amphotericin B (50 μg/ml) (all reagents were from Sigma), and the mixture was incubated for 48 to 72 h at 37°C.
A lymph node or terminal ileum sample (tested separately) of approximately 5 g was trimmed of fat and fibrous tissue, cut into small pieces, and homogenized for 30 s in 2 ml of sterile normal saline in a blender. After adding 25 ml of HPC, the homogenates were left standing at room temperature for 48 to 72 h. The sediment from the base of the tube was collected.
For radiometric culture, 0.1 ml of the prepared fecal or tissue sediment was inoculated into each culture vial. The radiometric medium was based on those of Collins et al. (3) and Cousins et al. (4) and consisted of Middlebrook 7H12 medium (BACTEC 12B; Becton Dickinson, Sparks, Md.) with 200 μl of PANTA PLUS (Becton Dickinson), 1 ml of egg yolk, 5 μg of mycobactin J (Allied Monitor Inc., Fayette, Mo.), and 0.7 ml of water. The vials were incubated at 37°C for 8 weeks. The GI was determined weekly with an automatic ion chamber (BACTEC 460; Johnston Laboratories, Towson, Md.).
For culture on solid medium, the prepared fecal sediment (250 μl) or tissue sediment (50 μl) was inoculated onto each of three (feces) or two (tissue) 35-ml screw-cap polystyrene Macartney tubes containing 10 ml of Herrold’s egg yolk medium (HEYM) (1) containing 8 egg yolks/liter, 0.4% (wt/vol) sodium pyruvate (Sigma), and 2 μg of mycobactin J (Allied Monitor Inc.) per ml and a single tube of the same medium without pyruvate. The tubes were incubated at 37°C for 20 weeks. Growth was determined visually at weeks 1, 2, 4, 6, 8, 9, 10, 12, 16, and 20. Identification of M. paratuberculosis was achieved by demonstration of mycobactin dependency. A colony from a primary culture was streaked onto HEYM with and without mycobactin and incubated for 1 month before the result was read. Mycobactin dependency on HEYM was also undertaken for radiometric cultures in trial 3 by using an inoculum of 50 μl after the cultures had been incubated for 10 weeks (see below).
Preparation of radiometric culture samples for PCR analysis.
BACTEC 12B medium containing egg is inhibitory to PCR assays (4). Several simple methods for removing the egg yolk and overcoming the PCR inhibitor were investigated. These included a variety of differential centrifugation protocols, heating followed by differential centrifugation protocols, and alcohol treatments followed by differential centrifugation. Only a method with alcohol subsequently resulted in the successful amplification of IS900 from cultures that were known to contain M. paratuberculosis (see Results). All work was conducted in a class II biosafety cabinet by using precautions for the containment of radioactivity and the protection of personnel.
Removal of residual inhibitors of PCR.
Lysates (45 μl) were thawed, and DNA was purified from the entire lysate by binding to silica in a column with 6 M guanidine thiocyanate according to the manufacturer’s instructions (Wizard PCR Preps DNA Purification System; Promega Corporation, Madison, Wis.) and with the elution of purified DNA in 50 μl of sterile distilled water. A volume of 5 μl of purified DNA solution was added to each PCR mixture.
PCR.
A reaction volume of 50 μl containing 5 μl of the DNA sample, 250 ng of each of the M. paratuberculosis IS900 primers P90 (5′-GAAGGGTGTTCGGGGCCGTCGCTTAGG) and P91 (5′-GGCGTTGAGGTCGATCGCCCACGTGAC) (10), 200 μM (each) the nucleotides dATP, dTTP, dGTP, and dCTP, 66.8 mM Tris-HCl, 16.6 mM (NH4)2SO4, 2.5 mM MgCl2, 1.65 mg of bovine serum albumin per ml, 10 mM β-mercaptoethanol, and 2 U of Taq polymerase in buffer (10 mM Tris-HCl, 0.1 mM EDTA [pH 8.8]) was used. Amplification was undertaken in 200-μl tubes in a 96-place thermal cycler (Corbett Research, Sydney, Australia) with the following conditions: 1 cycle of denaturation at 94°C for 2 min followed by 37 cycles of denaturation at 94°C for 30 s, annealing at 62°C for 15 s, and extension at 72°C for 1 min. Products of approximately 400 bp were predicted, and the samples were evaluated for 400-bp products by electrophoresis at 94 V for 45 min in 2% agarose gels stained with ethidium bromide. The specificity of the reaction for IS900 was confirmed during optimization experiments by Southern hybridization with an internal probe (10).
Experimental design.
A preincubation control sample was taken immediately after inoculation of each radiometric culture vial. A sample was again collected from the culture vial when its GI was ≥10; thereafter, samples were collected from the vial at 7-day intervals for up to 8 weeks. All samples were prepared with ethanol as described above. Samples that had GIs of ≥10 but a negative result in the PCR assay were then purified with silica and retested by PCR.
Four trials were undertaken. The first trial was with frozen-stored samples from cattle and sheep that were known to contain M. paratuberculosis on the basis of the results of conventional culture on solid medium (cattle) or histopathology and examination of tissue or fecal smears stained with Ziehl-Neelsen stain (sheep). In the second trial, samples from goats and sheep with unknown infection status were selected for culture on the basis of the results of serological testing, to mimic the approach taken in a disease control program; the animals were known to have been exposed to M. paratuberculosis on a farm where Johne’s disease is endemic, and the samples were processed for culture after overnight storage at 4°C. In the third trial, feces from seropositive cattle from a herd known to be infected were cultured to mimic the approach taken in a disease control program. In the fourth trial, the suitability of the culture method for environmental samples was evaluated, with the main considerations being sensitivity, contamination with rapidly growing environmental organisms, and inhibition of the PCR. Feces from two sheep excreting either high or low numbers of M. paratuberculosis were mixed with soil and pasture samples to achieve 3 log10 dilutions and were then cultured.
RESULTS
Sample preparation and purification methods.
The ethanol extraction method described below was a simple procedure. Briefly, the rubber stopper lid of the radiometric culture vial was wetted with 70% ethanol, left for 20 s, and then dried. The vial was inverted several times to mix the contents, and 200 μl of medium was removed with a sterile syringe and needle and transferred to a screw-cap 1.5-ml polypropylene microcentrifuge tube. Five hundred microliters of absolute ethanol was added, and the tube was left to stand for 2 min before mixing vigorously on a vortex mixer for 5 s and centrifuging at 8 × g for 10 min at 22°C. Partially flocculated egg yolk accumulated at the base and sides of the tube. The supernatant was transferred to a clean microcentrifuge tube, and the tube was then centrifuged at 18,000 × g for 5 min. The resulting pellet was washed twice in 200 μl of sterile phosphate-buffered saline by resuspension and centrifugation and was then resuspended in 50 μl of sterile distilled water. The tube was placed in a dry-heating block at 100°C for 20 min to lyse the mycobacteria. A volume of 5 μl of the lysate was added to each PCR mixture. The lysate was then stored at −20°C.
To evaluate the samples for the presence of residual inhibitors of PCR and excess target DNA, five lysates from samples that had GIs of ≥10 but negative PCR results and one PCR-positive lysate were diluted 10- and 100-fold and retested by PCR, with negative results. DNA was then purified from the same lysates with silica, diluted 10- and 100-fold, and tested by PCR. Four samples that were previously PCR negative then gave positive PCR results. Dilution of the purified DNA samples was detrimental, with the PCR results then being negative.
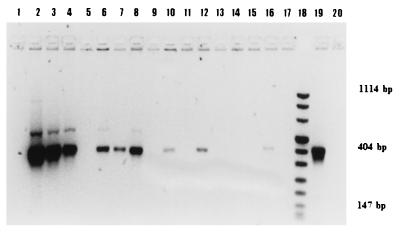
To test whether there were significant losses of target DNA during silica purification, three PCR-positive lysates with graded responses were tested again to ensure the consistency of the response and were then purified with silica and tested again. They remained PCR positive. The intensity of the PCR product from a strongly positive sample was slightly diminished by the silica purification. The intensities of the PCR products from a moderately positive sample and a weakly positive sample were enhanced, while a sample that had appeared initially negative gave a detectable product after silica purification (Fig. 1). These results correlated with the GI. Unless treated with ethanol, all samples were inhibitory to the PCR assay (Fig. 1).
FIG. 1.
Effect of sample treatment on the results of PCR analysis. The PCR product (20 μl) was resolved on a 2% agarose gel for 45 min at 94 V and was stained with ethidium bromide. The following samples were tested: A, GI of 999, lanes 1 to 4; B, GI of 91, lanes 5 to 8; C, GI of 39, lanes 9 to 12; D, GI of 7, lanes 13 to 16. The treatments were as follows: nil, lanes 1, 5, 9, and 13; ethanol, first test, lanes 2, 6, 10, and 14; ethanol, second test, lanes 3, 7, 11, and 15; ethanol plus silica, lanes 4, 8, 12, and 16. Lane 18, DNA size markers; lane 19, positive control with IS900; lane 20, negative control.
Trial 1.
In the first trial, ovine and bovine samples that were known to contain M. paratuberculosis were used. M. paratuberculosis was cultured in radiometric medium and was identified by IS900 PCR from each sample within 6 weeks of inoculation (Table 1). Tissues (ileum or lymph node) yielded positive results in 1 to 4 weeks, while feces yielded positive results in 2 to 6 weeks. When a GI of ≥10 was first noted in the cultures, 5 of 12 samples prepared with ethanol alone yielded positive PCR results. These cultures had GIs in the range of 236 to 999. Three cultures that had lower GIs (range, 10 to 23), and the M. paratuberculosis control culture (GI, 26) required ethanol and silica preparation in order to yield a positive PCR result on the first occasion that it was tested. Samples from three of the remaining four cultures (GIs, 29, 99, 157, and 755) were PCR negative when they were first tested, regardless of the method of sample preparation, but they were positive 1 week later. Samples from three cultures were PCR positive immediately after inoculation (Table 1, animals 4, 5, and 8), indicating the presence of large numbers of M. paratuberculosis cells in the inoculum, but a GI was not detected for 3 weeks in one of these cultures, suggesting the presence of few viable M. paratuberculosis cells in the original inoculum. Samples from the majority of cultures were PCR negative immediately after inoculation, confirming that replication of M. paratuberculosis had occurred in the medium; this was a significant finding for the samples from sheep. In each of the weeks following the first confirmation of growth by PCR, samples from each of the cultures, except one in 1 week, were PCR positive after ethanol preparation alone. The GIs of these samples ranged from 329 to 999. Overall, 80 of 85 samples examined by PCR between 1 and 8 weeks after inoculation of the culture medium were positive; 5 of the 80 positive samples had required preparation with ethanol and silica. None of the samples from sheep produced visible growth on HEYM after 20 weeks, while each of the bovine samples yielded colonies visible at 10 weeks.
TABLE 1.
Culture of ovine and bovine clinical samples known to contain M. paratuberculosis by using radiometric medium, HEYM, and identification of M. paratuberculosis by IS900 PCR with a sample from the primary culturea
| Animal no. | Species | Sample | Time to positive PCR result (wk) | GI when the culture PCR was positive | Result of culture on HEYM |
|---|---|---|---|---|---|
| 1 | Ovine | Ileum | 1 | 236 | Negative |
| 2 | Ovine | Lymph node | 2 | 10 | Negative |
| 3 | Ovine | Lymph node | 2 | 421 | Negative |
| 4 | Ovine | Ileum | 1 | 999 | Negative |
| 5 | Ovine | Lymph node | 4b | 999 | Negative |
| 6 | Ovine | Feces | 2 | 512 | Negative |
| 7 | Ovine | Feces | 2 | 664 | Negative |
| 8 | Ovine | Feces | 4c | 999 | Negative |
| 9 | Bovine | Feces | 5c | 913 | Positive, 10 wk |
| 10 | Bovine | Feces | 4d | 20 | Positive, 10 wk |
| 11 | Bovine | Feces | 6c | 566 | Positive, 10 wk |
| 12 | Bovine | Feces | 3 | 23 | Positive, 10 wk |
| Control | M. avium | Laboratory culture | —e | 999 | Not tested |
| Control | M. paratuberculosis | Laboratory culture | 1 | 26 | Not tested |
Radiometric cultures were incubated for 8 weeks, and HEYM cultures were incubated for 20 weeks.
Samples from culture were PCR negative for the first 2 weeks that they were tested but were PCR positive in all later weeks.
A sample from culture was PCR negative when the first sample was obtained but PCR positive in all later weeks.
A sample from culture was PCR negative in week 5.
—, did not test positive by IS900 PCR.
Trial 2.
In the second trial, samples were taken from goats and sheep from a property on which livestock were known to have Johne’s disease. The animals were chosen on the basis of a reaction in a gel diffusion precipitation assay or absorbed ELISA for antibodies against M. paratuberculosis. A mesenteric lymph node and feces from each animal were cultured. Four of 5 goats and 4 of 11 sheep were found by radiometric culture followed by PCR to be infected with M. paratuberculosis (Table 2). Seven lymph node samples and six fecal samples were culture positive. Of these 13 culture-positive samples, 8 were PCR positive when a GI was first detected (GI range, 10 to 480) following ethanol preparation alone, and an additional 2 samples (GIs, 28 and 35) were PCR positive at this time after ethanol and silica preparation. The remaining three samples were PCR negative when a GI was first detected (GIs, 15, 107, and 207), despite ethanol and silica preparation, but were PCR positive a week later (GI range, 586 to 999) after ethanol preparation alone (Table 2). After first becoming PCR positive, samples collected weekly from all but one culture in 1 week remained PCR positive for the duration of the 8-week incubation. Two cultures were PCR positive immediately after inoculation. Overall, 63 of 67 samples examined by PCR between 1 and 8 weeks after inoculation of the radiometric medium were positive; 2 of the 63 positive samples had required preparation with ethanol and silica. M. paratuberculosis was not recovered from any of the samples cultured on HEYM during an incubation of 20 weeks.
TABLE 2.
Culture of caprine and ovine clinical samples with radiometric medium and identification of M. paratuberculosisa by IS900 PCR
| Animal no. | Species | Sample | Status of animal based on histology and Ziehl-Neelsen smear of feces or tissuesb | Time to positive PCR result (wk) | GI when the culture was PCR positive |
|---|---|---|---|---|---|
| 1 | Goat | Lymph node | I | 2 | 411 |
| Feces | I | 3 | 269 | ||
| 2 | Goat | Lymph node | U | 4 | 438 |
| Feces | U | 5 | 28 | ||
| 3 | Goat | Lymph node | I | 2 | 480 |
| Feces | I | 5c | 999 | ||
| 4 | Goat | Lymph node | U | −d | − |
| Feces | U | 5 | 10 | ||
| 5 | Goat | Lymph node | U | − | − |
| Feces | U | − | − | ||
| 6 | Sheep | Lymph node | I | 4 | 35 |
| Feces | I | − | − | ||
| 7 | Sheep | Lymph node | U | − | − |
| Feces | U | − | − | ||
| 8 | Sheep | Lymph node | U | − | − |
| Feces | U | − | − | ||
| 9 | Sheep | Lymph node | U | 4 | 190 |
| Feces | U | 5 | 194 | ||
| 10 | Sheep | Lymph node | U | − | − |
| Feces | U | − | − | ||
| 11 | Sheep | Lymph node | U | − | − |
| Feces | U | − | − | ||
| 12 | Sheep | Lymph node | I | 5c | 586 |
| Feces | I | − | − | ||
| 13 | Sheep | Lymph node | U | − | − |
| Feces | U | − | − | ||
| 14 | Sheep | Lymph node | I | 2e | 331 |
| Feces | I | 5c | 999 | ||
| 15 | Sheep | Lymph node | U | − | − |
| Feces | U | − | − | ||
| 16 | Sheep | Lymph node | U | − | − |
| Feces | U | − | − |
The animals were from a farm where livestock were known to have been infected with M. paratuberculosis and were selected on the basis of serological results. Radiometric cultures were incubated for 8 weeks. The results of culture on HEYM were negative at 20 weeks in each case.
I, infected; U, uninfected.
Samples from culture were PCR negative when they were first tested but were PCR positive in later weeks.
−, negative.
The sample from culture was PCR negative in week 4.
Culture detected a greater number of infected goats and sheep than did the other tests. Only two of the four infected goats and three of the four infected sheep were shown to be infected with M. paratuberculosis by examination of fecal or tissue smears stained with Ziehl-Neelsen stain and/or histopathological examination of the ileum, cecum, colon, and mediastinal lymph node (Table 2).
Trial 3.
In a third trial, fecal samples were cultured from 21 cows that were suspected of being infected with M. paratuberculosis on the basis of the results of serological tests, as is done during routine surveillance for Johne’s disease. Twelve cultures were PCR positive within 7 weeks (Table 3). Of these, five were positive when first tested (GIs, 38, 57, 460, 638, and 835), with the two with the highest GIs requiring ethanol and silica preparation, while 7 were negative (GI range, 11 to 121) even after silica purification. Samples from the latter cultures were all PCR positive 1 week later (GIs, 472 to 999). Samples from two cultures were PCR negative in week 8, even though samples from these cultures were PCR positive over the previous 2 to 4 weeks. None of the samples were PCR positive immediately after inoculation. Overall, 50 of 59 samples examined by PCR between 1 and 8 weeks after inoculation were positive. Of the 50 positive samples, 18 had required ethanol and silica preparation. Of the 27 samples tested by PCR after ethanol and silica preparation because of an earlier negative PCR result, 18 were positive. M. paratuberculosis was recovered on HEYM from 8 of the 12 samples that were culture positive in radiometric medium (Table 3). A mycobactin dependency test on HEYM was used to identify M. paratuberculosis in the 12 PCR-positive radiometric cultures, but the results for 3 of the 12 cultures could not be confirmed with this method because of overgrowth of fast-growing bacterial species (Table 3).
TABLE 3.
Culture of bovine fecal samples with radiometric medium and HEYMa
| Animal no. | Time to positive PCR result (wk) | GI when the culture was PCR positive | Result of mycobactin dependency test on HEYM | Result of primary culture on HEYM |
|---|---|---|---|---|
| 1 | −b | − | Not tested | Negative |
| 2 | − | − | Not tested | Negative |
| 3 | 5c | 472 | Positive | Positive, 10 wk |
| 4 | − | − | Not tested | Negative |
| 5 | − | − | Not tested | Negative |
| 6 | 4c | 999 | Positive | Positive, 9 wk |
| 7 | 4c | 999 | Positive | Positive, 8 wk |
| 8 | 4c | 999 | Negatived | Contaminated |
| 9 | 7c | 644 | Negatived | Negative |
| 10 | 4 | 38 | Positive | Positive, 12 wk |
| 11 | − | − | Not tested | Negative |
| 12 | 4 | 638 | Positive | Positive, 9 wk |
| 13 | − | − | Not tested | Negative |
| 14 | − | − | Not tested | Negative |
| 15 | 4e | 835 | Negatived | Negative |
| 16 | 6e | 460 | Positive | Negative |
| 17 | 5c | 561 | Positive | Positive, 10 wk |
| 18 | − | − | Not tested | Negative |
| 19 | − | − | Not tested | Negative |
| 20 | 5c | 999 | Positive | Positive, 9 wk |
| 21 | 4 | 57 | Positive | Positive, 9 wk |
M. paratuberculosis from radiometric cultures was identified by IS900 PCR analysis and the mycobactin dependency test on HEYM. Cattle were from a farm where livestock were known to have been infected with M. paratuberculosis and were selected on the basis of serological results. Radiometric cultures were incubated for 8 weeks, and primary HEYM cultures were incubated for 20 weeks.
−, negative.
Samples from culture were PCR negative when they were first tested but were PCR positive in all later weeks.
The culture was overgrown with fast-growing bacteria.
The sample from culture was PCR negative in week 8.
Trial 4.
In the fourth trial, an ovine fecal sample in which large numbers of acid-fast rods were visualized and a second ovine fecal sample in which very small numbers of acid-fast rods were visualized were diluted and thoroughly mixed with soil or grass clippings so as to achieve dilutions of the feces of 1:10, 1:100, and 1:1,000. The resulting specimens were treated as described above for fecal samples. Samples from each of the cultures were PCR positive after 2 to 6 weeks of incubation in the radiometric medium, and observations were discontinued at 6 weeks (Table 4). Samples from four cultures were PCR negative on the first occasion that a GI was noted (GI range, 24 to 242) but were PCR positive a week later. Samples collected from two cultures in week 6 required ethanol and silica preparation in order to yield a positive PCR result. Overall, 44 of 48 samples examined by PCR between 1 and 6 weeks after inoculation were positive; 3 of the 44 positive samples had required both ethanol and silica preparation. None of the cultures was discarded because of contamination.
TABLE 4.
Radiometric culture of soil and pasture samples inoculated with ovine feces containing M. paratuberculosisa
| Sample | Level of M. paratuberculosis in feces | Dilution of feces in sample | Time to positive PCR result (wk) | GI when the culture was PCR positive |
|---|---|---|---|---|
| Soil | High | 1:10 | 2b | 58 |
| High | 1:100 | 5 | 11 | |
| High | 1:1,000 | 5c | 778 | |
| Low | 1:10 | 3b | 434 | |
| Low | 1:100 | 6c | 899 | |
| Low | 1:1,000 | 4 | 10 | |
| Pasture | High | 1:10 | 4c | 999 |
| High | 1:100 | 4 | 373 | |
| High | 1:1,000 | 4 | 18 | |
| Low | 1:10 | 4c | 612 | |
| Low | 1:100 | 4 | 11 | |
| Low | 1:1,000 | 4 | 14 |
IS900 PCR was used to identify M. paratuberculosis. Cultures were incubated for 6 weeks.
The sample collected from culture in week 6 was PCR positive only after ethanol and silica preparation.
The samples from culture were PCR negative when they were first tested but were PCR positive in all later weeks.
In each of the trials, the GIs of the cultures increased rapidly after becoming detectable so that 1 to 2 weeks later most had GIs equal to or greater than the maximum level (GI, 999). Toward the end of the incubation period, the GIs of some of the cultures declined, presumably in association with repeated removal of the gas phase and depletion of the labelled substrate in the liquid phase (8).
DISCUSSION
This study, like several others (3, 4), has confirmed a useful role for radiometric culture in the detection of M. paratuberculosis, with the principal advantage over conventional culture on solid medium being the relatively short time required before being able to report a result. This is particularly so if PCR amplification of the IS900 gene rather than demonstration of mycobactin dependency is used to confirm the identities of isolates. However, a disadvantage of this approach has been the need to subculture samples from the primary radiometric culture vial to radiometric medium without egg yolk in order to obtain a sample that is noninhibitory for the PCR assay (4). The need for subculture of samples into a secondary radiometric culture vial adds greatly to the costs of labor and materials associated with the detection of M. paratuberculosis. To gain a saving in time but to avoid additional costs associated with the identification of M. paratuberculosis by PCR after subculture, we have routinely used primary radiometric culture and then subcultured samples onto HEYM to demonstrate mycobactin dependency. This approach has not been possible for ovine samples, which have generally failed to grow on HEYM. The method that we report here enables culture in radiometric medium followed by PCR with samples from the primary culture for confirmation of the presence of M. paratuberculosis. In addition to savings in labor and materials, there is a further saving in time because a second incubation period is avoided. Our results indicated that simple treatment of samples removed from primary radiometric cultures effectively removed substances that are inhibitory to PCR and permitted testing for the presence of the M. paratuberculosis-specific IS900 gene.
During this study 259 individual samples from cultures obtained between 1 and 8 weeks after inoculation were examined by PCR following ethanol or ethanol plus silica preparation, and 237 (91.5%) were positive. Of the 22 PCR-negative samples, 18 were taken on the first occasion that a GI was noted, 17 had GIs of <250, and 11 had GIs of <100. All these cultures were PCR positive 1 week later, which strongly suggests that there were insufficient M. paratuberculosis cells in the cultures when they were first sampled. The reasons why some cultures with relatively high GIs were PCR negative are unclear but may include the presence of residual inhibitors of the PCR in the sample after the treatment steps the presence in the culture of organisms other than M. paratuberculosis which contributed to the GI, clumped growth which resulted in the failure to include sufficient numbers of M. paratuberculosis cells in the sample for PCR, and excessive numbers of M. paratuberculosis or other cells which resulted in an excess of target DNA or irrelevant DNA in the PCR. The last possibility seems remote given the fact that dilution of the sample for PCR failed to result in a positive result. The chance of residual PCR inhibition is real and perhaps is overcome only when the amount of target DNA reaches a certain level. Because inhibitors may be contributed by the inoculum as well as the culture medium, it is likely that the level of residual inhibition will vary from sample to sample. There was evidence of this in the present study, in which in trial 3 a relatively high proportion (36%) of the PCR-positive samples required silica purification. However, overall only 28 of 237 (11.8%) PCR-positive samples in this study required silica purification.
General recommendations can be made regarding the timing of the PCR examination of radiometric cultures that have evidence of growth. We did not seek to determine the threshold GI required to yield a positive result in PCR assays because GIs tend to increase rapidly and we wished to monitor cultures only weekly. However, because of the factors mentioned above that might result in negative PCR results even with high GIs, positive PCR results probably should not be expected to be related to a narrow threshold. This is illustrated in the present study, in which most of the negative PCR results were in samples from cultures with GIs of <250 but in which 17 of the 50 PCR-positive cultures were detected when the GI was <200. For routine batch processing of diagnostic samples it would be appropriate to wait 1 to 2 weeks after first recording a GI of >10 before undertaking PCR examination or to undertake PCR examination of all cultures with GIs of >200 at 6 to 8 weeks after inoculation. By either of these approaches, most samples would be expected to yield a positive PCR result after ethanol preparation alone. If it is desired that results be reported after a minimum incubation period, samples from cultures with GIs should be purified with silica before PCR examination, and if negative, a sample should be taken and tested by PCR after a further 7 days of incubation. Certain batches of clinical samples may be troublesome and may require the additional silica purification, but rather than doing this routinely, it would be easier and cheaper to apply silica purification only to those samples that gave a negative PCR result in the first instance.
An unexpected finding from this study was that BACTEC 12B medium containing egg yolk, mycobactin J, and PANTA PLUS appeared to be satisfactory for culture of M. paratuberculosis strains from sheep. In Australia, these ovine strains have proven to be very difficult to culture on conventional solid or liquid media and have been regarded as noncultivable for practical diagnostic purposes. Although it has been recognized since 1996 that culture of these strains from tissues may be possible in BACTEC 12B medium (12), an inference from the unpublished findings was that positive PCR results for samples from BACTEC 12B cultures may have been due simply to the continued presence of M. paratuberculosis organisms that were in the inoculum. We have shown this inference to be incorrect. Replication of M. paratuberculosis determined by a change in the PCR status of the culture from negative to positive was shown to have occurred in most instances. Primary culture of Australian ovine strains of M. paratuberculosis on solid medium or successful subculture from radiometric medium to solid medium has not yet been achieved reliably. Furthermore, repeated subculture of these strains in the same radiometric medium results in the loss of viability (unpublished observations). Therefore, an effort is required to elucidate the cultural requirements of M. paratuberculosis strains from sheep in Australia.
The results of this study support and extend those of Cousins et al. (4) and suggest that culture of clinical samples in BACTEC 12B medium with egg yolk, mycobactin J, and PANTA PLUS can enable the relatively rapid detection of the principal types of M. paratuberculosis that are endemic in Australia. Johne’s disease is known to occur in cattle, goats, sheep, and alpacas in Australia and is associated with infection with several distinct genotypes of M. paratuberculosis. Although we did not include samples from infected alpacas, isolates of M. paratuberculosis from alpacas have been found to be similar to isolates from cattle and can be grown in modified BACTEC 12B medium (4).
The fact that several culture-positive sheep and goats were classified as uninfected on the basis of histopathology and examination of fecal or tissue smears suggests that radiometric culture followed by IS900 PCR may be more sensitive than the other tests and may be able to detect relatively few bacteria in tissue or fecal samples. It is reasonable to speculate on the numbers of M. paratuberculosis cells that might have been present in the spiked soil and pasture samples used in this study. On the basis of a likely detection limit of 103 to 104 organisms per gram by microscopic examination of smears stained with Ziehl-Neelsen stain (14), the soil and pasture samples prepared with the highest final dilution of feces with low numbers of organisms (Table 4) would have contained 1 to 10 to 10 to 100 cells per g. Unfortunately, we were unable to obtain samples from cattle with very low numbers of M. paratuberculosis organisms in their tissues or feces, as was a problem in a previous study (4). Consequently, we are still unsure of the sensitivity of radiometric culture compared to that of conventional culture on solid medium for the detection of animals in the early stages of infection with strains of M. paratuberculosis that can be grown on solid media. For this reason we plan to compare radiometric culture and conventional culture over a longer period to establish the relative sensitivities of these methods. However, the preliminary results (Table 3) suggest that radiometric culture may be more sensitive than culture on HEYM.
The radiometric culture and PCR preparation methods described here appeared to be suitable for culture and identification of M. paratuberculosis from soil and pasture samples that had been spiked with the organism. There did not appear to be contamination of cultures with rapidly growing environmental actinomycetes and fungi, and PCR inhibition appeared to be minimal.
Radiometric cultures are incubated for only 8 weeks before a negative result is reported, whereas conventional cultures require 20 weeks of incubation on solid medium. M. paratuberculosis was detected within 7 weeks in all culture-positive samples evaluated in this study. The time saved through the use of radiometric culture followed directly by IS900 PCR may remove a significant impediment to the adoption of disease control programs for Johne’s disease in ruminants. These programs are based on herd screening by serology, which may yield some seropositive individuals that must be examined by culture. Farmers are sometimes reluctant to enter into a program that creates uncertainty about infection status when clarification of infection status cannot be made for 5 to 6 months. Radiometric culture with the identification of M. paratuberculosis by IS900 PCR offers a 3- to 4-month respite from this uncertainty.
ACKNOWLEDGMENTS
We gratefully acknowledge the financial assistance of the McGarvie Smith Trust, without which this study would not have been possible.
Andrew Berneutz provided valuable technical assistance in the early stages of this study.
REFERENCES
- 1.Anonymous. Laboratory methods for veterinary mycobacteriology for the isolation and identification of mycobacteria. Ames, Iowa: National Animal Disease Laboratory; 1972. [Google Scholar]
- 2.Carrigan M J, Seaman J T. The pathology of Johne’s disease in sheep. Aust Vet J. 1990;67:47–50. doi: 10.1111/j.1751-0813.1990.tb07693.x. [DOI] [PubMed] [Google Scholar]
- 3.Collins M T, Kenefick K B, Sockett D C, Lambrecht S R, McDonald J, Jorgensen J B. Enhanced radiometric detection of Mycobacterium paratuberculosis by using filter-concentrated bovine fecal specimens. J Clin Microbiol. 1990;28:2514–2519. doi: 10.1128/jcm.28.11.2514-2519.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cousins D V, Evans R J, Francis B R. Use of BACTEC radiometric culture method and polymerase chain reaction for the rapid screening of faeces and tissues for Mycobacterium paratuberculosis. Aust Vet J. 1995;72:458–462. doi: 10.1111/j.1751-0813.1995.tb03489.x. [DOI] [PubMed] [Google Scholar]
- 5.Cruickshank R, Duguid J P, Marmion B P, Swain R H A. Medical microbiology. 12th ed. II. The practice of medical microbiology. Edinburgh, United Kingdom: Churchill Livingstone, Ltd.; 1973. [Google Scholar]
- 6.Damato J J, Collins M T. Growth of Mycobacterium paratuberculosis in radiometric, Middlebrook and egg-based media. Vet Microbiol. 1990;22:31–42. doi: 10.1016/0378-1135(90)90122-c. [DOI] [PubMed] [Google Scholar]
- 7.Green E P, Tizard M L V, Moss M T, Thompson J, Winterbourne D J, McFadden J J, Hermon-Taylor J. Sequence and characteristics of IS900, an insertion element identified in a human Crohn’s disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 1989;17:9063–9073. doi: 10.1093/nar/17.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambrecht R S, Carriere J F, Collins M T. A model for analyzing growth kinetics of a slowly growing Mycobacterium sp. Appl Environ Microbiol. 1988;54:910–916. doi: 10.1128/aem.54.4.910-916.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luna L G. Manual of histologic staining methods of the Armed Forces Institute of Pathology. 3rd ed. New York, N.Y: McGraw-Hill Book Company; 1968. [Google Scholar]
- 10.Millar D S, Withey S J, Tizard M L V, Ford J G, Hermon-Taylor J. Solid-phase hybridization capture of low-abundance target DNA sequences: application to the polymerase chain reaction detection of Mycobacterium paratuberculosis and Mycobacterium avium subsp. sylvaticum. Anal Biochem. 1995;226:325–330. doi: 10.1006/abio.1995.1232. [DOI] [PubMed] [Google Scholar]
- 11.Reggiardo Z, Tigertt W D. Automatable radiometric detection of growth of Mycobacterium tuberculosis in selective media. Am Rev Respir Dis. 1977;115:1066–1069. doi: 10.1164/arrd.1977.115.6.1066. [DOI] [PubMed] [Google Scholar]
- 12.Skilbeck, N. W. 1996. Personal communication.
- 13.Stephens L R. Johne’s disease (paratuberculosis) In: Corner L A, Bagust T J, editors. Australian Standard Diagnostic Techniques for Animal Diseases. East Melbourne, Australia: Standing Committee on Agriculture and Resource Management, Commonwealth Scientific and Industrial Research Organisation Publications; 1987. [Google Scholar]
- 14.Vaneechoutte M, Van Eldere J. The possibilities and limitations of nucleic acid amplification technology in diagnostic microbiology. J Med Microbiol. 1997;46:188–194. doi: 10.1099/00222615-46-3-188. [DOI] [PubMed] [Google Scholar]
- 15.Whipple D L, Callihan D R, Jarnagin J L. Cultivation of Mycobacterium paratuberculosis from bovine fecal specimens and a suggested standardized procedure. J Vet Diagn Invest. 1991;3:368–373. doi: 10.1177/104063879100300424. [DOI] [PubMed] [Google Scholar]
- 16.Whitlock R H, Rosenberger A E. Proceedings of the 94th Annual Meeting of the U.S. Animal Health Association; 1990. Fecal culture protocol for Mycobacterium paratuberculosis. A recommended protocol; pp. 280–285. [Google Scholar]