Abstract
During the origin of great apes about 14 million years ago, a series of phenotypic innovations emerged, such as the increased body size, the enlarged brain volume, the improved cognitive skill, and the diversified diet. Yet, the genomic basis of these evolutionary changes remains unclear. Utilizing the high-quality genome assemblies of great apes (including human), gibbon, and macaque, we conducted comparative genome analyses and identified 15,885 great ape-specific structural variants (GSSVs), including eight coding GSSVs resulting in the creation of novel proteins (e.g., ACAN and CMYA5). Functional annotations of the GSSV-related genes revealed the enrichment of genes involved in development and morphogenesis, especially neurogenesis and neural network formation, suggesting the potential role of GSSVs in shaping the great ape-shared traits. Further dissection of the brain-related GSSVs shows great ape-specific changes of enhancer activities and gene expression in the brain, involving a group of GSSV-regulated genes (such as NOL3) that potentially contribute to the altered brain development and function in great apes. The presented data highlight the evolutionary role of structural variants in the phenotypic innovations during the origin of the great ape lineage.
Keywords: great apes, comparative genomics, structural variant, body size, brain
Introduction
The origin of the great ape lineage (the Hominidae family) about 14 million years ago represents an evolutionary leap in primates (Hill and Ward 1988; Pozzi et al. 2014), accompanied by a series of phenotypic innovations, including the increased body size (Smith and Jungers 1997), the enlarged brain volume (MacLeod et al. 2003; Barton and Venditti 2017), the improved cognitive abilities (Alba 2010), and the diversified diet (Chivers 1998). In addition to these phenotypic changes, frequent changes in chromosome numbers have been reported in various primate lineages, including monkeys, apes, and humans (Stanyon et al. 2008). For instance, the human chromosome 2 was formed through the end-to-end fusion of two ancestral chromosomes, which remained separate in other primates. This event resulted in the 2n = 46 karyotype in humans, as opposed to the ancestral 2n = 48 karyotype typical of great apes (Kronenberg et al. 2018). This evolutionary trend eventually leads to the origin of our own species. The living Hominidae family has four major lineages, including the Homo clade (Homo sapiens), the Pan clade (Pan troglodytes and Pan paniscus), the Gorilla clade (Gorilla gorilla and Gorilla beringei), and the Pongo clade (Pongo pygmaeus, Pongo abelii, and Pongo tapanuliensis). It is of great interest to study the genetic basis of the great ape-shared evolutionary traits that have made them a successful primate lineage, which in turn is informative to delineating the genetic mechanism of human origin.
Structural variants (SVs) are large genomic alterations (≥50 bp in length), including deletions (DELs), insertions (INSs), copy number variations, inversions, and translocations. They are widely distributed in the genomes of great ape species. For example, when comparing human and chimpanzee, the divergence at single nucleotide change is only 1.23%, whereas the divergence of SVs reaches ∼ 3% (Suntsova and Buzdin 2020). Functionally, SVs are expected to have more impact than single nucleotide variants (SNVs). Previous studies have shown that in the human genome, SVs are predicted more harmful than SNVs (Abel et al. 2020) and more likely affect the expression of a gene than SNVs (Chiang et al. 2017). Specifically, SVs can affect molecular function, cellular process, regulatory function, chromatin 3D structure, and transcriptional function of the organism (Weischenfeldt et al. 2013; Spielmann et al. 2018). As a major form of genetic variations, SVs also contribute to phenotypic diversity of organisms (Stankiewicz and Lupski 2010; Patel et al. 2014). However, previously, owing to the poor quality of the great ape genome assemblies (mostly based on next-generation sequencing [NGS]), it has been difficult to systematically identify SVs and study their roles in phenotypic evolution.
Fortunately, the long-read sequencing and multiplatform scaffoldings have been widely used in constructing high-quality reference genomes. For great apes, we now have high-quality genomes covering all major great ape lineages (Gordon et al. 2016; Kronenberg et al. 2018), providing a great opportunity to analyze SVs and their phenotypic relevance during primate evolution.
Here, we used the published high-quality genomes of human, chimpanzee, gorilla, orangutan, gibbon, and macaque, and we identified 15,885 great-ape-specific SVs (GSSVs) through comparative genomic analyses. In particular, we report the potentially functional SVs that may contribute to the major phenotypic changes of great apes, especially the enlarged brains and the improved cognitive skills, which will lead to a better understanding of great ape evolution and human origin.
Results
Identification of the GSSVs
The high-quality genomes of the great ape species (human, chimpanzee, gorilla, and orangutan) were obtained from the published studies (Gordon et al. 2016; Kronenberg et al. 2018). To search for the GSSVs, we also included the high-quality genomes of two outgroup primates: white-cheeked gibbon (Nomascus leucogenys) (NCBI accession number: GCF_006542625.1) and rhesus macaque (Macaca mulatta) (Warren et al. 2020) (supplementary table S1, Supplementary Material online; fig. 1A).
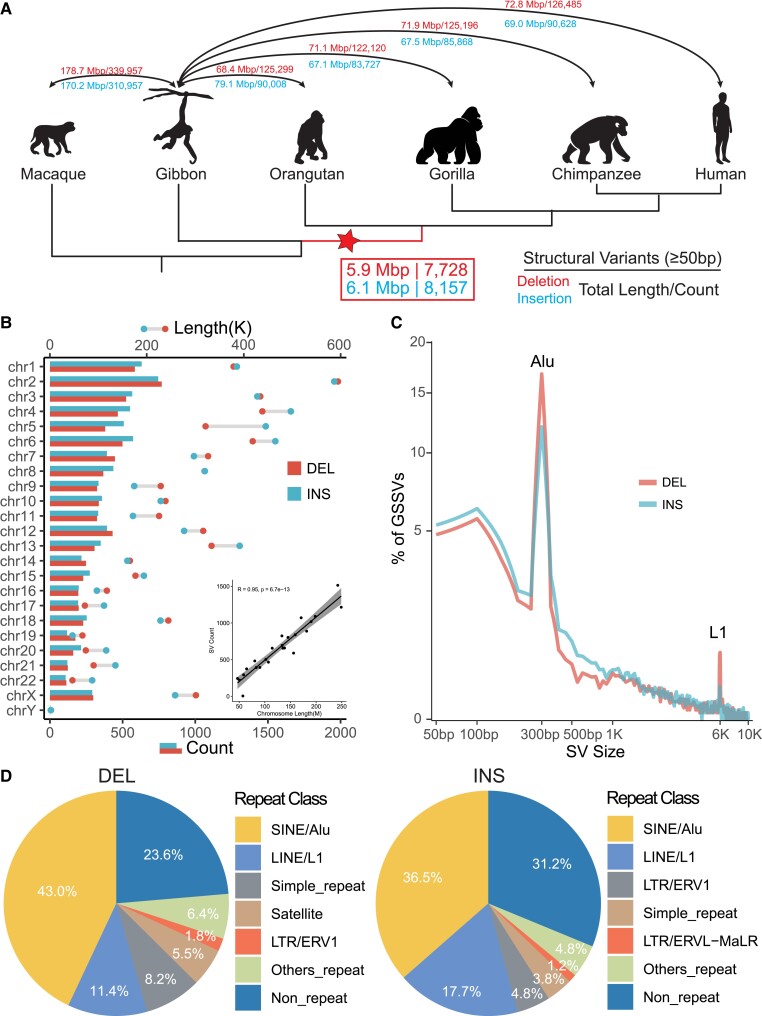
Fig. 1.
Identification and annotation of GSSVs. (A) The cladogram indicates the phylogenetic relationship among the analyzed primate species. We used the high-quality genomes of four great ape species, one lesser ape species, and one Cercopithecidae species. The numbers of SVs and their genomic lengths based on assembly comparisons are indicated. DELs are the numbers above the lines and INSs are those beneath the lines. The number in the box indicates the identified 15,885 GSSVs, including 7,728 DELs and 8,157 INSs. (B) Chromosomal distribution of the identified GSSVs. The number of SVs is tightly correlated with the length of the chromosome, as indicated by the Pearson correlation (R2 = 0.95, P = 6.7e−13). (C) The size distribution of GSSVs. The two peaks at 300 bp and 6 kb indicate the Alu and L1 elements, respectively. (D) Annotation of GSSVs located in the repeat regions of the genome, where SINEs (such as Alu) and LINEs (such as L1) compose of the great majority of the identified GSSVs.
Considering the methodological challenge for detecting SVs cross species with large genetic divergence (genome sequence identity < 95%), largely due to the difficulty of identifying syntenic blocks in the genomes (He et al. 2019), we assessed the two commonly used methods of detecting SVs based on an assembly-to-assembly strategy, including smartie-sv (Chaisson et al. 2015; Kronenberg et al. 2018) and minimap2 (Li 2018; Feng and Li 2021). Based on the comparison of the callsets and manual curations, we found that there were approximately 50% overlap between the results of minimap2 and smartie-sv (supplementary table S2 and fig. S1A, Supplementary Material online), and minimap2 performed significantly better than smartie-sv in detecting SVs among distantly related species (supplementary fig. S1B, Supplementary Material online). We therefore employed minimap2 in the following analyses.
We first conducted pairwise genome comparisons between gibbon and the four great ape genome assemblies (human, chimpanzee, gorilla, and orangutan) (Gordon et al. 2016; Kronenberg et al. 2018). We then took the overlapped SVs among the four SV sets as the divergent SV set between gibbon and great apes (>50% reciprocal overlapping of SV length is required for an overlapped SV). Using the genome assembly of rhesus macaque (Warren et al. 2020), we further filtered out the SVs occurred in the gibbon lineage, which eventually gave rise to the set of GSSVs (see Materials and Methods for technical details). Totally, we detected 23,300 candidate GSSVs, including 14,451 DELs and 8,849 INSs. Given the complex nature of SVs among deeply diverged species, to remove false positives, we further conducted manual curation of all the candidate GSSVs by local sequence alignments. Finally, a total of 15,885 GSSVs passed the curation, including 7,728 DELs and 8,157 INSs (fig. 1A; supplementary fig. S2A, Supplementary Material online).
These GSSVs account for 12.0 Mb genomic sequences (on average 0.44% of the great ape genomes), with fragment lengths ranging from 50 bp to 19 kb. They are randomly distributed in the genome, and the SV counts in each chromosome are positively correlated with the chromosome length (R = 0.95, P = 6.7e−13, Pearson correlation test; fig. 1B; supplementary fig. S2B and tables S6 and S7, Supplementary Material online), and 17.6% GSSVs (2,791) are longer than 1 kb. As expected, we observed two peaks around 300 bp and 6 kb in the length distribution of the GSSVs, and the majority of them are Alu and L1 elements, respectively (fig. 1C). For repeat annotation, 76.4% DELs and 68.8% INSs are composed of repeat elements, such as SINEs (short interspersed nuclear elements), LINEs (long interspersed nuclear elements), and long terminal repeats (LTRs) (fig. 1D; supplementary table S8, Supplementary Material online), suggesting that repeat elements are likely the key drivers for the generation of GSSVs.
Functional Implications of the GSSVs
To search for GSSVs with potential functional consequences, we further classified the 15,885 GSSVs into two sets based on a range of criteria used in the aforementioned manual check (see Materials and Methods), resulting in the set of 6,574 high-confident GSSVs (HC-GSSVs) (supplementary table S9, Supplementary Material online) and the set of 9,311 complex SVs (fig. 2A; see Materials and Method).
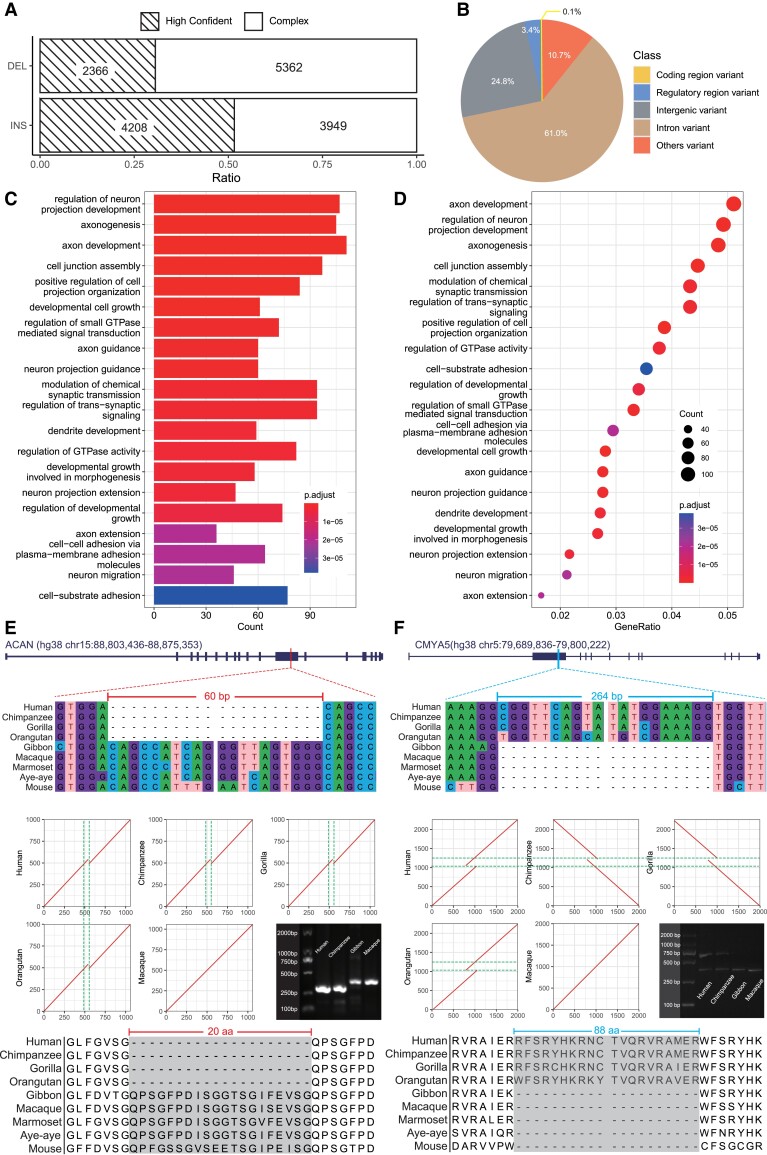
Fig. 2.
Functional annotations of the identified HC-GSSVs. (A) The ratios of the high-confident and the complex GSSVs after manual curation. (B) The pie plot shows the functional annotations of HC-GSSVs using the VEP tool. (C, D) The results of GO enrichment analysis of the 2,353 HC-GSSV-related genes. (E) A 60-bp deletion located in exon-12 of ACAN, a gene related to skeletal development. (F) A 264-bp insertion located in exon-2 of CMYA5, a well-known gene related to schizophrenia. The local sequence alignments (DNA and protein sequences) among mammalian species together with the MUMmer plots showing the locations of the GSSVs are presented.
Our downstream analyses were focused on the HC-GSSV set containing 2,366 DELs and 4,208 INSs. We first conducted PCR (polymerase chain reaction) validation of ten DELs and ten INSs randomly selected from the HC-GSSV set, and 19 of them were validated as true GSSVs, and only one failed because of nonspecific amplification of the target region (supplementary fig. S3, Supplementary Material online). Accordingly, the HC-GSSV set can be considered a conservative set of GSSVs.
We annotated the identified HC-GSSVs using the Variant Effect Predictor (VEP) tool according to the human GRCh38 coordinates (see Materials and Methods). The majority of HC-GSSVs (85.8%) are located in the intronic (61.0%) or the intergenic regions (24.8%), and 10.7% of them are located in the other regions (e.g., the 3′ or 5′ UTR regions). Specifically, among the 222 HC-GSSVs located in the known regulatory regions, 53.6% (119/222) of them are located in enhancers, 19.8% (44/222) in the CTCF binding site regions, 19.4% (43/222) in the open chromatin regions, 0.5% (1/222) in the promoter regions, and 6.8% (15/222) in the transcription factor (TF) binding site regions (fig. 2B; supplementary table S10, Supplementary Material online).
Next, we conducted functional enrichment analysis for the 2,353 HC-GSSV-related genes (those genes overlapped with the HC-GSSVs, see Materials and Methods for details) (supplementary table S11, Supplementary Material online). The enriched functional terms are related to the known traits of the great ape lineage, such as developmental growth involved in morphogenesis (body size) and axonogenesis (brain). Remarkably, more than half of the enriched terms are related to brain development and function. The top terms include axon development, neurogenesis, neural projection, and neuronal differentiation (fig. 2C and D). These results indicate a potential role of HC-GSSVs in shaping the great-ape-specific phenotypic traits, especially the central nervous system.
In particular, we found eight coding HC-GSSVs (two DELs and six INSs) (table 1; supplementary table S12, Supplementary Material online; fig. 2B; supplementary fig. S4, Supplementary Material online). Although several of them are located in exons and were predicted causing protein sequence changes, none of them actually result in truncated proteins (table 1). Among these GSSV-related genes, POU5F1B is a great-ape-specific novel gene caused by the retroposition of its mother copy (OCT4) on chromosome 8 (a 1,412-bp INS specifically occurred in the great ape lineage), confirming the previous report (Simó-Riudalbas et al. 2022). Initially, it was thought to be a pseudogene of OCT4 with 95% sequence similarity. However, there have been reports suggesting its susceptibility in tumors (Zanke et al. 2007; Rafnar et al. 2014), and it likely acts as a transcriptional activator to promote cancer cell proliferation (Panagopoulos et al. 2008; Pan et al. 2018). In addition, POU5F1B is implicated in the regulation of OCT4 activity (Suo et al. 2005). The functional role of POU5F1B in the evolution of great apes is yet to be understood.
Table 1.
The Eight HC-GSSVs Located in the Gene-Coding Regions.
| Positiona | Length (bp) | Type | Gene | Consequenceb | Gene function or diseasec |
|---|---|---|---|---|---|
| Chr2:232,330,767–232,330,767 | 186 | DEL | DIS3L2 | c, d | RNA degradation |
| Chr15:88,857,790–88,857,790 | 60 | DEL | ACAN | d | Skeletal development |
| Chr5:79,736,810–79,736,810 | 264 | INS | CMYA5 | e | Schizophrenia, skeletal muscle development |
| Chr15:81,135,350–81,135,983 | 633 | INS | CFAP161 | a, f | Ciliary motion |
| Chr11:71,961,971–71,962,270 | 299 | INS | RNF121 | a, g, h | Regulation of cell cycle, signal transduction |
| Chr8:127,415,807–127,417,219 | 1,412 | INS | POU5F1B | b, h | Weak transcriptional activator |
| Chr10:77,994,516–77,999,310 | 4,794 | INS | POLR3A | a, f, g, h | Spastic ataxia |
| ChrX:14,915,424–14,915,588 | 164 | INS | MOSPD2 | a, g, h | Intracellular exchanges and communication |
The coordinates are based on human GRCh38.
The consequence contains the following: (a) coding_sequence; (b) start_lost; (c) stop_gained; (d) inframe_insertion; (e) inframe_deletion; (f) splice_donor; (g) splice_acceptor; and (h) UTR.
The gene functions and disease are collected from GeneCard, MGI database, and literatures (Zanke et al. 2007; Panagopoulos et al. 2008; Chen et al. 2011; Austin-Tse et al. 2013; Morris et al. 2013; Rafnar et al. 2014; Gao et al. 2017; Hu et al. 2017; Di Mattia et al. 2018; Hoya et al. 2018; Lee et al. 2018; Pan et al. 2018; Infante et al. 2020; Wei et al. 2021).
Interestingly, one coding HC-GSSV, a 60-bp DEL, is located in exon-12 of ACAN. This gene is involved in skeletal development (fig. 2E). It is one of the major components of cartilage and binds to hyaluronan and links protein to form huge aggregates, a hydrated gel-like structure with resistibility to compression and deformation in joints (Watanabe et al. 1998). Many studies have shown that mutations in ACAN can lead to short stature and poor bone development (Hu et al. 2017; Wei et al. 2021). Besides, as the main component of the neural extracellular matrix, ACAN is also expressed in the brain (Morawski et al. 2012). We speculate that the deletion of 20 amino acids (due to the 60-bp DEL) in the binding domain of the free sugar chains of ACAN may affect bone development in great apes, which might be related to the increased body size during the origin of the great ape lineage.
The other potentially important case is a 264-bp INS located in exon-2 of CMYA5, resulting in an 88 amino-acid insertion (fig. 2F). Previous GWAS (genome-wide association study) have found that variants in CMYA5 are associated with schizophrenia (Chen et al. 2011; Hoya et al. 2018), implying that this HC-GSSV in CMYA5 may play a role in the brain evolution of great ape species. Another interesting case is the 186-bp DEL in DIS3L2. This gene is mainly related to RNA degradation, and mutations in this gene can cause Perlman syndrome, characterized by a larger head size and developmental delay (Morris et al. 2013), providing a hint of this GSSV in the great-ape-specific pattern of brain/head development.
The remaining four HC-GSSVs are not associated with the known great ape–specific traits based on current knowledge. CFAP161 is associated with cilia movement (Austin-Tse et al. 2013), whereas RNF121 is involved in the regulation of cell cycle, signal transduction, genomic integrity under hypoxia, and metastasis of cancer cells (Gao et al. 2017; Lee et al. 2018). POLR3A is associated with spastic ataxia, and the best recognized phenotype is cerebellar ataxia (Infante et al. 2020), and MOSPD2 is involved in intracellular exchange and communication (Di Mattia et al. 2018). Whether and how these HC-GSSVs contribute to the origin and evolution of great apes are yet to be explored (supplementary fig. S4, Supplementary Material online).
The Regulatory HC-GSSVs with Potential Functions in the Brain
Compared with gibbons and Cercopithecidae, the most crucial changes that occurred during the origin of the great apes include the increase in body size, the expansion of brain volume, and the improvement of cognitive ability. As aforementioned, the GO enrichment analysis shows that most of the GSSV-associated genes are involved in central nervous system (64.0% GSSV-associated genes [233/364] of the top ten GO terms), and the majority of the HC-GSSVs were mapped to the intronic or intergenic regions, suggesting that these HC-GSSVs would be more likely on the level of gene regulation, if they have functional impact. It has been postulated a few decades ago that the differences between human and chimpanzee are mostly caused by gene regulation changes rather than by alterations in their protein-coding sequences (King and Wilson 1975), and a recent study also reached the same conclusion (Suntsova and Buzdin 2020).
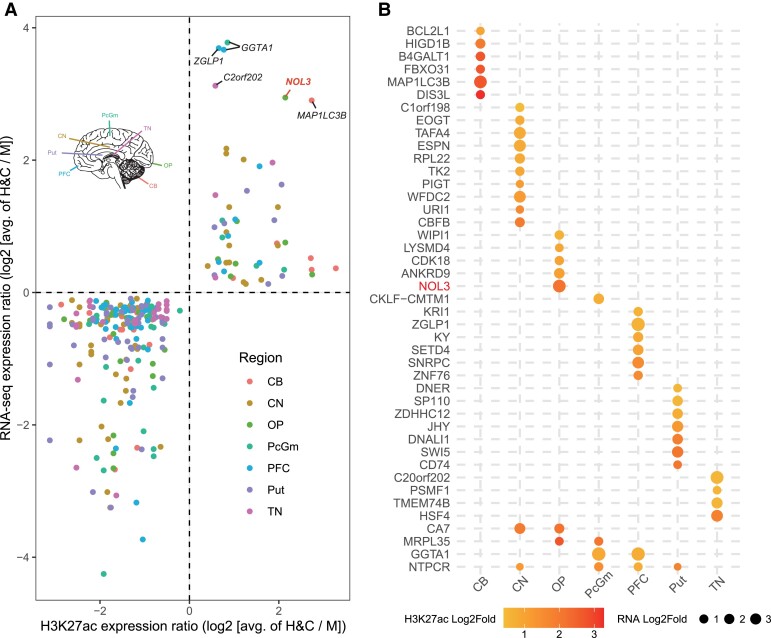
With the use of the published brain ChIP-seq (H3K27ac) and RNA-seq data from human, chimpanzee, and rhesus macaque (Vermunt et al. 2016; Sousa et al. 2017), we identified 105 HC-GSSVs that overlapped with 105 human-chimpanzee-specific cis-regulatory elements (CREs). Among the 105 CREs, there are 186 associated genes (genes within the 500 kb downstream and upstream of CRE) showing human-chimpanzee-specific expression changes compared with rhesus macaque, and the change direction of gene expression is consistent with the H3K27ac signals (indication of CRE activity). These 105 CREs include 36 DEL-containing CREs associated with 62 nearby genes and 69 INS-containing CREs associated with 126 genes (two genes are present in both gene sets) (supplementary tables S13–S15, Supplementary Material online).
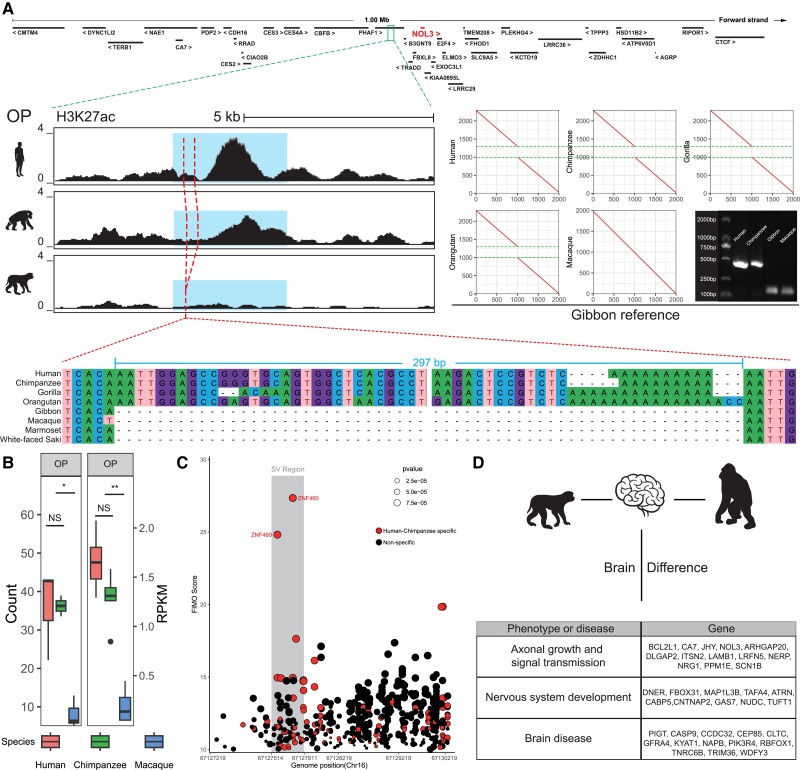
We firstly ranked the 43 upregulated genes (human-chimp vs. macaque) by log2 fold changes of gene expression. The top five upregulated genes include GGTA1, ZGLP1, C20orf202, NOL3, and MAP1LC3B (fig. 3A). Of these genes, NOL3 is closely associated with neurodevelopment. Markedly, NOL3 also ranks the top gene in the occipital pole (OP) of the cortex (fig. 3B). Previous studies have shown that NOL3 is related to abnormal neural potential, and the absence of NOL3 causes excessive excitation (Russell et al. 2012). NOL3 is 42.9 kb away from the nearest human-chimpanzee-specific CRE, and a 297-bp INS is located in this CRE. The H3K27ac peak map shows that human and chimpanzee have higher peaks than that of macaque (P < 0.05, Welch's two-tailed unpaired t-test, fig. 4A) and the comparison of epigenome and transcriptome data also shows human and chimpanzee have higher expression (fig. 4B). The multiple sequence alignment shows that the genomic region containing this INS is conserved in primates (supplementary fig. S5A, Supplementary Material online), and the 297-bp INS likely causes a CRE activity change, creating a human-chimpanzee-specific enhancer that regulates NOL3 expression.
Fig. 3.
The HC-GSSVs located in the CREs of the brain. (A) The HC-GSSV-related genes showing human–chimp-specific CRE activity (by the H3K27ac signals) and gene expression changes (by RNA-seq) compared with macaque across seven brain regions, including CB, CN, OP, PcGm, PFC, Put, and TN. (B) Ranking of the 43 upregulated genes based on gene expression and the H3K27ac signals.
Fig. 4.
Overview of GSSV occurred in NOL3. (A) A 297-bp INS located in a human-chimpanzee-specific CRE (blue region) in which the nearest human–chimpanzee-specific gene is NOL3. The left plot shows the difference of H3K27ac signal in OP region among human, chimpanzee, and rhesus monkey; the right plot shows the MUMmer plot of the 297-bp INS between great apes and gibbon; and bottom plot shows multiple sequence alignments of representative primate species. (B) The comparisons of normalized read count of H3K27ac signal of CRE (left) and expression (right) of NOL3. NS, not significant; *P < 0.05; **P < 0.01. (C) TF enrichment of CREs. The CREs with human-chimpanzee-specific TF binding sites are denoted in red scatters, and two ZNF460 binding sites are labeled, which have the highest FIMO score and located in SV region (gray region). (D) Summary of brain-related genes with human–chimp-specific CREs and overlapped to identified GSSVs.
In addition, using the FIMO tool (Grant et al. 2011), we performed TF enrichment analysis of the homologous CRE regions, and we identified 78 human–chimpanzee-specific TFs (supplementary fig. S5B, Supplementary Material online) with 27 of them located in the HC-GSSV regions. The strongest signal (by the FIMO score) is ZNF460 (fig. 4C; supplementary table S16, Supplementary Material online), a TF mainly expressed in the brain (supplementary fig. S5D, Supplementary Material online), though its function is largely unknown.
For the 143 downregulated genes (supplementary fig. S6, Supplementary Material online), there are also worth-noting cases, such as LRFN5, 21.4 kb away from the nearest human-chimpanzee-specific CRE, and a 508-bp INS is located in this CRE (supplementary fig. S7, Supplementary Material online). As a synaptic adhesion molecule implicated in autism, LRFN5 can induce presynaptic differentiation through binding to the LAR family receptor protein tyrosine phosphatases (LAR-RPTPs) that have been highlighted as presynaptic hubs for synapse formation (Goto-Ito et al. 2018; Lin et al. 2018).
The brains of great apes are structurally and functionally more complex than those of gibbons and macaques. We summarized the annotated functions of all the identified HC-GSSV-related genes showing human-chimpanzee-specific expression changes in the brain. They are mainly involved in three functional aspects, including synapse formation and signal transmission, nervous system development (such as neural migration and neural differentiation), and various brain diseases caused by gene mutations (such as intellectual disability, microcephaly, schizophrenia, and Parkinson's) (fig. 4D).
Taken together, we identified a set of genes related to HC-GSSVs that are associated with great ape brain function. These HC-GSSVs could serve as a resource to study the genetic basis of brain structure/function change that emerged during the origin of the great ape lineage. We listed the GWAS-based functional annotations of the 194 HC-GSSV-related genes of interest (including the eight coding GSSVs) in supplementary table S17, Supplementary Material online. However, owing to the lack of ENCODE data of the correspondent tissues in most of the nonhuman primate species, the speculated regulatory roles of these HC-GSSVs are yet to be validated.
Discussion
A number of previous studies have addressed SVs in great ape species. They found significant increases in DELs and duplications, as well as segmental duplications, after the emergence of the great ape ancestor, especially in the chimpanzee lineage. Presumably, these mutation events altered the genome structure of the great ape ancestor and might have an important impact on subsequent evolution (Marques-Bonet et al. 2009; Sudmant et al. 2013). In addition, some studies have compared the Neanderthals genome with other primates and explored the evolution of small INSs and DELs in modern humans (Chintalapati et al. 2017). However, given the prior knowledge on SVs in hominid evolution, our current understanding of SVs in the great ape ancestor still remains elusive due to the limitations in genome quality and analytic tools.
Based on the high-quality great ape assemblies, we identified 15,885 great-ape-specific SV by cross-species comparison. Through further manual curation with a stringent filtering, we report 6,574 high-confidence GSSVs that overlap with 2,353 genes. These HC-GSSV-related genes show functional connections with the great-ape-specific traits, such as body size and brain. Markedly, many of the enriched functional terms of these genes are related to brain development and function, implying an important role of SVs in shaping the central nervous system of great apes during evolution. In particular, we report eight coding GSSVs that lead to the generation of novel proteins during the origin of the great ape lineage, and some of these great-ape-specific proteins (such as ACAN and CMYA5) are involved in bone development and brain function, providing clues to the genetic basis of the great-ape-shared phenotypic innovations.
Previous studies had suggested that the body size or weight of great apes is significantly larger than lesser apes and Cercopithecidae (Wheatley 1987; Smith and Jungers 1997; Zihlman and McFarland 2000; Zihlman and Bolter 2015). In the HC-GSSVs set, we found a 60-bp DEL leading to a 20 amino-acid deletion of ACAN, a gene related to bone development. Previous genetic studies in humans reported that the ACAN mutations could cause short stature and spinal disease (Hu et al. 2017; Wei et al. 2021). Hence, this SV-induced novel ACAN protein may play a potential role in body size evolution of great apes. Another important phenotypic innovation in the great ape lineage involves the brain. Accordingly, we identified many HC-GSSVs associated with brain development and function. For example, a 264-bp INS changes the coding sequence of CMYA5, and previous human studies suggest its involvement in schizophrenia (Chen et al. 2011; Hoya et al. 2018).
Besides the eight coding HC-GSSVs, the great majority of the identified HC-GSSVs are located in the noncoding regions of the genome, which presumably contribute to gene expression regulation. One notable example is a 297-bp INS that influences the enhancer of NOL3, a gene related with abnormal neural potential.
During primate evolution, there have been two leaps of brain volume enlargement, one in the common ancestor of the great ape lineage and the other in the human lineage (Holloway et al. 2009). Consequently, the brain volume of great apes is much larger than lesser apes. The larger brain means an improved cognition, because of the higher relative cortex volume and neuron packing density (NPD). Likewise, information processing capacity becomes higher due to short interneuronal distance and high axonal conduction velocity (Roth and Dicke 2012). Thanks to the published brain epigenomic data that include human, chimpanzee, and macaque (Vermunt et al. 2016; Sousa et al. 2017), we were able to search for the HC-GSSVs located in the CREs and to infer their potential influences on gene expression regulation. We found that many genes affected by HC-GSSVs are related with brain size, such as WDFY3, CCDC32, and CLTC, all of which are involved in microcephaly. More importantly, these genes are also associated with intellectual disability and developmental delay (DeMari et al. 2016; Le Duc et al. 2019; Nabais Sá et al. 2020; Abdalla et al. 2022). Given the more complex cortex structure of great apes, we found several HC-GSSV-related genes (FBXO31 and NUDC) acting on neuronal differentiation and migration, which may help form the complex neural network in the brain. Consistently, several GSSV-related genes (BCL2L1, CA7, and LRFN5) are involved in axonal growth and signal transmission, highlighting their potential roles in the higher information processing capacity of great apes. Together, the enriched HC-GSSVs and the target genes for brain development and function indicate the evolutionary significance of SVs that may contribute to the larger brain and higher cognitive abilities of great apes. Future functional experiments are warranted to reveal the molecular and developmental mechanisms underlying the bigger and smarter brains of great apes.
In conclusion, given the challenge of studying SVs among distantly related species, in this study, we identified the GSSVs, providing a useful resource for understanding the genetic basis of phenotypic innovations in the evolution of great apes.
Materials and Methods
Data Information
All the genomes we used can be downloaded from NCBI, including human (GRCH38.P13, accession number: GCF_000001405.39), chimpanzee (Clint_PTRv2, accession number: GCF_002880755.1), gorilla (Kamilah_GGO_v0, accession number: GCF_008122165.1), orangutan (Susie_PABv2, accession number: GCF_002880775.1), gibbon (Asia_NLE_v1, accession number: GCF_006542625.1), and macaque (Mmul_10, accession number: GCF_003339765.1), and the details are listed in supplementary table S1, Supplementary Material online. The epigenome data of human, chimpanzee, and macaque can be downloaded from the published study (Vermunt et al. 2016), as well as the online database of PsychENCODE (http://evolution.psychencode.org/#).
Comparing Minimap2 and Smartie-sv
We mapped each long-read assembled genome of the four great ape species to the gibbon assembly and called SVs by Minimap2 + paftools and Blasr + printgaps (Smartie-sv), respectively. For the reverse calling, we called the SVs by mapping the gibbon genome back to each of the great ape genomes. We calculated the overlap between the two methods (supplementary fig. S1A and table S2, Supplementary Material online) and used the command “shuf –n 100 SV_list.bed” to randomly select 100 SVs from the gibbon–human SV set generated separately by the two methods and further validated the ratio of true SVs by manual check. The analysis was repeated ten times (resulting in a total of 1,000 SVs), and the results were summarized (supplementary fig. S1B, Supplementary Material online).
Identification of GSSV
Genome comparisons were performed using Minimap2 (Li 2018). We mapped each long-read assembled genome of the four great ape species to the gibbon assembly (NLE), including human-GRCh38.p13 (V38), Chimpanzee-Clint_PTRv2 (CCP), Gorilla-Kamilah_v0 (GGO), Orangutan-Susie_PABv2 (PAB), and rhesus-Mmul_10 (RM10). Using gibbon genome as the reference genome, we mapped the genomes of great apes to the gibbon genome for SV calling, referred as the forward calling. For the reverse calling, we called the SVs by mapping the gibbon genome back to each of the great ape genomes. Then, we filtered the SVs by intersecting the two SV sets and obtained the SVs for each pair (supplementary table S3, Supplementary Material online). GSSVs were identified by taking the intersection of all the gibbon–great ape SV sets. Furthermore, to exclude the gibbon lineage–specific SVs, we used the published rhesus macaque genome to repeat the forward and reverse calling, and the intersected GSSV set was taken as the final set of GSSVs (supplementary table S3, Supplementary Material online-S5).
GSSV Manual Check and PCR Validation
The sequences of the GSSV regions were extracted by the “bedtools getfasta” command, and sequence comparison was performed and plotted by MUMmer (NUCmer-3.1 and MUMmerplot-3.5). The candidate GSSVs with 1 kb upstream/downstream sequences were aligned to the gibbon genome using NUCmer. The GSSVs were classified into three categories based on manual check: 1) If the GSSV region and its 1 kb flanking sequences of macaque could be completely aligned to the corresponding gibbon GSSV region using NUCmer and MEGA-7.0.26 (MUSCLE under the default parameters), they were considered as orthologous between gibbon and macaque. These GSSVs were defined as HC-GSSVs. 2) If the GSSV region with 1 kb flanking sequences of macaque could only be partially aligned to the corresponding gibbon GSSV sequence, we classified these GSSVs as “complex” GSSVs. And 3) if the potential GSSV region with 1 kb flanking sequence of macaque could be completely aligned to the corresponding gibbon GSSV sequences except for the SV regions, we inferred that these SVs are present in macaque, and we defined these GSSVs as “false” GSSVs. The detailed list is shown in supplementary figure S9, Supplementary Material online. For PCR and Sanger sequencing validation, we randomly selected ten DELs and ten INSs by command “shuf –n 10,” and the tested DNA samples included one rhesus macaque, one white-cheeked gibbon, one chimpanzee, and one human. The primers were designed by Primer Premier5 (supplementary table S18, Supplementary Material online). The PCR products were visualized by agarose gel electrophoresis to verify the lengths of GSSVs (supplementary fig. S3, Supplementary Material online), and for Sanger sequencing, supplementary figure S10, Supplementary Material online displays the ACAN sequencing results and the identification of SV breakpoints based on the sequencing sequences. All the presented SVs in this study were validated by both PCR and Sanger sequencing, and the data were submitted to https://github.com/kizzb/Great-apes-specific-SVs/blob/main/All_sanger_sequence_results.zip.
Repeat Analysis
Transposable elements (TEs) were identified by using RepeatMasker (v4.0.9) to search against the known Repbase TE library (Repbase21.08). Since the DEL coordinates are present only in the gibbon genome (the great apes have only one point of the matching coordinates), we could only obtain DEL sequences from the gibbon genome, and the same reasoning applies to INS. Therefore, we analyzed the TE ratios of INS in the human genome and of DEL in the gibbon genome, respectively (supplementary table S8, Supplementary Material online).
HC-GSSV Function Prediction by VEP
After identifying 6,574 HC-GSSVs (2,366 DELs and 4,208 INSs), the functional effects of DEL and INS were predicted separately by VEP (http://www.ensembl.org/info/docs/tools/vep/index.html); in the parameter section, “Species” select Homo (GRCh38.p13), “Restrict results” select “show most severe consequence per variant,” and the others were the default parameters (supplementary table S10, Supplementary Material online).
GO Enrichment Analysis
For GO analysis of 6,574 HC-GSSVs, we first obtained the matched coordinate of human and then associate with the human genes that intersect with SVs by command “bedtools intersect –a SV_coord.bed –b Gene_coord.bed –wa –wb.” We next performed GO enrichment analysis by ClusterProfiler-4.6.0 (Wu et al. 2021), and the selected background database was “org.Hs.eg.db,” which is a human database. The selected enrichment class was “BP,” and the P value cutoff was 0.05 (supplementary table S11, Supplementary Material online).
Identification of HC-GSSVs Located in the Great Ape–Specific CREs
The published ChIP-seq data were normalized and used to identify the great ape–specific CREs in seven brain regions that have correspondent H3K27ac and RNA-seq data (Vermunt et al. 2016; Sousa et al. 2017). We downloaded the 60,702 genomic regions with H3K27ac signals, which were generated from three humans, two chimpanzees, and three rhesus monkeys in each of the seven brain regions, including prefrontal cortex (PFC), precentral gyrus (PcGm), OP, caudate nucleus (CN), putamen (Put), cerebellum (CB), and thalamic nuclei (TN). We performed pairwise comparison of the enhancer signals (the H3K27ac peaks) among the three species (macaque–human, macaque–chimpanzee, and human–chimpanzee), and we used the Welch two-tailed unpaired t-test for statistical assessment. We defined an enhancer as a great ape–specific CRE if this enhancer showed significant difference (P < 0.05) of the same direction in both the macaque–human and the macaque–chimpanzee comparisons while showing no difference (P > 0.05) in the human–chimpanzee comparison. According to this criterion, we obtained 18,105 human–chimpanzee-specific CREs (supplementary table S13, Supplementary Material online). Next, we identified those genes located in the 500k flanking (upstream 500k and downstream 500k, total 1M) regions of these human–chimpanzee-specific CREs, and we further overlap HC-GSSVs with these CREs (by command “bedtools intersect -a SV_hg38.bed -b CRE_coord.bed -wa -wb |sort -k1,1 -s -V -k2n,2 |uniq”) to link HC-GSSVs with human–chimpanzee-specific CREs. In the end, 105 CREs overlapped with the HC-GSSVs were identified, involving 186 genes. To quantify the activities of these great ape–specific CREs, we calculated the log2(fold change) between great apes (the average of human and chimpanzee) and rhesus macaque (supplementary table S15, Supplementary Material online). The overlapping threshold is ≥1 bp intersection between HC-GSSV and CRE.
Comparative Gene Expression Analysis of Seven Brain Regions
The gene expression data of adult brains of human, chimpanzee, and macaque were obtained from the published study, and these data are already-normalized expression data (Sousa et al. 2017), which contain samples from six humans, five chimpanzees, and five macaques of 16 brain regions. To match the expression data with the ChIP-seq data, we used the data of nine brain regions, including primary motor cortex (M1C), mediodorsal nucleus of the thalamus (MD), primary visual cortex (V1C), cerebellar cortex (CBC), striatum (STR), ventrolateral prefrontal cortex (VFC), orbital prefrontal cortex (OFC), dorsolateral prefrontal cortex (DFC), and medial prefrontal cortex (MFC), and the matching relationships between the ChIP-seq and the gene expression data are shown in supplementary figure S8, Supplementary Material online. The Welch two-tailed paired t-test was used in accessing expression differences between great apes and macaques and obtained 1,714 human-chimpanzee-specific genes (supplementary table S14, Supplementary Material online). In the end, we linked HC-GSSVs with human–chimpanzee-specific CREs and genes (supplementary table S15, Supplementary Material online).
TF Enrichment Analysis
We used liftover to obtain the homologous coordinates of the great ape–specific CREs (chr16_67128753_hg38) in human, chimpanzee, and macaque and then extracted the DNA sequences. Next, using FIMO, we predicted the enriched TFs on the sequences of each species (by command “fimo –o TF_result JASPAR_TF_data/Vertebrates/all.meme species.fa”), and we screened out the human–chimpanzee-specific TFs (supplementary table S16, Supplementary Material online).
Supplementary Material
Acknowledgments
This work has been supported by grants from the National Natural Science Foundation of China (U2002207 to B.S.), the National Key Research and Development Program of China (2021ZD0200100 to B.S.), and the Youth Innovation Promotion Association of CAS (to Y.H.).
Contributor Information
Bin Zhou, State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China; National Resource Center for Non-Human Primates, Kunming Primate Research Center, and National Research Facility for Phenotypic & Genetic Analysis of Model Animals (Primate Facility), Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Beijing, China.
Yaoxi He, State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China; National Resource Center for Non-Human Primates, Kunming Primate Research Center, and National Research Facility for Phenotypic & Genetic Analysis of Model Animals (Primate Facility), Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China.
Yongjie Chen, State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China; National Resource Center for Non-Human Primates, Kunming Primate Research Center, and National Research Facility for Phenotypic & Genetic Analysis of Model Animals (Primate Facility), Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Beijing, China.
Bing Su, State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China; National Resource Center for Non-Human Primates, Kunming Primate Research Center, and National Research Facility for Phenotypic & Genetic Analysis of Model Animals (Primate Facility), Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China; Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, Yunnan, China.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
B.S. designed the project; B.Z. and Y.H. performed bioinformatics analyses, genotyping, and sequencing experiments; B.S., Y.H., and B.Z. wrote the manuscript. All authors have discussed the results and read the manuscript.
Data Availability
All genome, epigenome, and transcriptome data could be downloaded in the NCBI database, and the code used in this study had been submitted to github (https://github.com/kizzb/Great-apes-specific-SVs).
References
- Abdalla E, Alawi M, Meinecke P, Kutsche K, Harms FL. 2022. Cardiofacioneurodevelopmental syndrome: report of a novel patient and expansion of the phenotype. Am J Med Genet A. 188:2448–2453. [DOI] [PubMed] [Google Scholar]
- Abel HJ, Larson DE, Regier AA, Chiang C, Das I, Kanchi KL, Layer RM, Neale BM, Salerno WJ, Reeves C, et al. 2020. Mapping and characterization of structural variation in 17,795 human genomes. Nature 583:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba DM. 2010. Cognitive inferences in fossil apes (Primates, Hominoidea): does encephalization reflect intelligence? J Anthropol Sci. 88:11–48. [PubMed] [Google Scholar]
- Austin-Tse C, Halbritter J, Zariwala MA, Gilberti RM, Gee HY, Hellman N, Pathak N, Liu Y, Panizzi JR, Patel-King RS, et al. 2013. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am J Hum Genet. 93:672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Venditti C. 2017. Rapid evolution of the cerebellum in humans and other great apes. Curr Biol. 27:1249–1250. [DOI] [PubMed] [Google Scholar]
- Chaisson MJP, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, Antonacci F, Surti U, Sandstrom R, Boitano M, et al. 2015. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lee G, Maher BS, Fanous AH, Chen J, Zhao Z, Guo A, van den Oord E, Sullivan PF, Shi J, et al. 2011. GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol Psychiatry. 16:1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Scott AJ, Davis JR, Tsang EK, Li X, Kim Y, Hadzic T, Damani FN, Ganel L, GTEx Consortium, et al. 2017. The impact of structural variation on human gene expression. Nat Genet. 49:692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintalapati M, Dannemann M, Prüfer K. 2017. Using the Neandertal genome to study the evolution of small insertions and deletions in modern humans. BMC Evol Biol. 17:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers DJ. 1998. Measuring food intake in wild animals: primates. Proc Nutr Soc. 57:321–332. [DOI] [PubMed] [Google Scholar]
- DeMari J, Mroske C, Tang S, Nimeh J, Miller R, Lebel RR. 2016. CLTC as a clinically novel gene associated with multiple malformations and developmental delay. Am J Med Genet A. 170:958–966. [DOI] [PubMed] [Google Scholar]
- Di Mattia T, Wilhelm LP, Ikhlef S, Wendling C, Spehner D, Nominé Y, Giordano F, Mathelin C, Drin G, Tomasetto C, et al. 2018. Identification of MOSPD2, a novel scaffold for endoplasmic reticulum membrane contact sites. EMBO Rep. 19:e45453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Li H. 2021. Higher rates of processed pseudogene acquisition in humans and three great apes revealed by long-read assemblies. Mol Biol Evol. 38:2958–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Cai A, Xi H, Li J, Xu W, Zhang Y, Zhang K, Cui J, Wu X, Wei B, et al. 2017. Ring finger protein 43 associates with gastric cancer progression and attenuates the stemness of gastric cancer stem-like cells via the Wnt-β/catenin signaling pathway. Stem Cell Res Ther. 8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Huddleston J, Chaisson MJP, Hill CM, Kronenberg ZN, Munson KM, Malig M, Raja A, Fiddes I, Hillier LW, et al. 2016. Long-read sequence assembly of the gorilla genome. Science 352:aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto-Ito S, Yamagata A, Sato Y, Uemura T, Shiroshima T, Maeda A, Imai A, Mori H, Yoshida T, Fukai S. 2018. Structural basis of trans-synaptic interactions between PTPδ and SALMs for inducing synapse formation. Nat Commun. 9:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Luo X, Zhou B, Hu T, Meng X, Audano PA, Kronenberg ZN, Eichler EE, Jin J, Guo Y, et al. 2019. Long-read assembly of the Chinese rhesus macaque genome and identification of ape-specific structural variants. Nat Commun. 10:4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Ward S. 1988. Origin of the Hominidae: the record of African large hominoid evolution between 14 My and 4 My. Am J Phys Anthropol. 31:49–83. [Google Scholar]
- Holloway RL, Sherwood CC, Hof PR, Rilling JK. 2009. Evolution of the brain in humans—paleoneurology. In: Binder MD Hirokawa N and Windhorst U, editors. Encyclopedia of neuroscience. Berlin, Heidelberg: Springer Berlin Heidelberg. p. 1326–1334. [Google Scholar]
- Hoya S, Watanabe Y, Shibuya M, Someya T. 2018. Updated meta-analysis of CMYA5 rs3828611 and rs4704591 with schizophrenia in Asian populations. Early Interv Psychiatry. 12:938–941. [DOI] [PubMed] [Google Scholar]
- Hu X, Gui B, Su J, Li H, Li N, Yu T, Zhang Q, Xu Y, Li G, Chen Y, et al. 2017. Novel pathogenic ACAN variants in non-syndromic short stature patients. Clin Chim Acta. 469:126–129. [DOI] [PubMed] [Google Scholar]
- Infante J, Serrano-Cárdenas KM, Corral-Juan M, Farré X, Sánchez I, de Lucas EM, García A, Martín-Gurpegui JL, Berciano J, Matilla-Dueñas A. 2020. POLR3A-related spastic ataxia: new mutations and a look into the phenotype. J Neurol. 267:324–330. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188:107–116. [DOI] [PubMed] [Google Scholar]
- Kronenberg ZN, Fiddes IT, Gordon D, Murali S, Cantsilieris S, Meyerson OS, Underwood JG, Nelson BJ, Chaisson MJP, Dougherty ML, et al. 2018. High-resolution comparative analysis of great ape genomes. Science 360:eaar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Duc D, Giulivi C, Hiatt SM, Napoli E, Panoutsopoulos A, Harlan De Crescenzo A, Kotzaeridou U, Syrbe S, Anagnostou E, Azage M, et al. 2019. Pathogenic WDFY3 variants cause neurodevelopmental disorders and opposing effects on brain size. Brain 142:2617–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NS, Chang HR, Kim S, Ji J-H, Lee J, Lee HJ, Seo Y, Kang M, Han JS, Myung K, et al. 2018. Ring finger protein 126 (RNF126) suppresses ionizing radiation-induced p53-binding protein 1 (53BP1) focus formation. J Biol Chem. 293:588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Liu J, Ding H, Xu F, Liu H. 2018. Structural basis of SALM5-induced PTPδ dimerization for synaptic differentiation. Nat Commun. 9:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod CE, Zilles K, Schleicher A, Rilling JK, Gibson KR. 2003. Expansion of the neocerebellum in Hominoidea. J Hum Evol. 44:401–429. [DOI] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. 2009. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457:877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawski M, Brückner G, Arendt T, Matthews RT. 2012. Aggrecan: beyond cartilage and into the brain. Int J Biochem Cell Biol. 44:690–693. [DOI] [PubMed] [Google Scholar]
- Morris MR, Astuti D, Maher ER. 2013. Perlman syndrome: overgrowth, Wilms tumor predisposition and DIS3L2. Am J Med Genet C Semin Med Genet. 163:106–113. [DOI] [PubMed] [Google Scholar]
- Nabais Sá MJ, Venselaar H, Wiel L, Trimouille A, Lasseaux E, Naudion S, Lacombe D, Piton A, Vincent-Delorme C, Zweier C, et al. 2020. De novo CLTC variants are associated with a variable phenotype from mild to severe intellectual disability, microcephaly, hypoplasia of the corpus callosum, and epilepsy. Genet Med. 22:797–802. [DOI] [PubMed] [Google Scholar]
- Pan Y, Zhan L, Chen L, Zhang H, Sun C, Xing C. 2018. POU5F1B promotes hepatocellular carcinoma proliferation by activating AKT. Biomed Pharmacother. 100:374–380. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Möller E, Collin A, Mertens F. 2008. The POU5F1P1 pseudogene encodes a putative protein similar to POU5F1 isoform 1. Oncol Rep. 20:1029–1033. [PubMed] [Google Scholar]
- Patel A, Schwab R, Liu Y-T, Bafna V. 2014. Amplification and thrifty single-molecule sequencing of recurrent somatic structural variations. Genome Res. 24:318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi L, Hodgson JA, Burrell AS, Sterner KN, Raaum RL, Disotell TR. 2014. Primate phylogenetic relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 75:165–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafnar T, Sulem P, Thorleifsson G, Vermeulen SH, Helgason H, Saemundsdottir J, Gudjonsson SA, Sigurdsson A, Stacey SN, Gudmundsson J, et al. 2014. Genome-wide association study yields variants at 20p12.2 that associate with urinary bladder cancer. Hum Mol Genet. 23:5545–5557. [DOI] [PubMed] [Google Scholar]
- Roth G, Dicke U. 2012. Evolution of the brain and intelligence in primates. Prog Brain Res. 195:413–430. [DOI] [PubMed] [Google Scholar]
- Russell JF, Steckley JL, Coppola G, Hahn AFG, Howard MA, Kornberg Z, Huang A, Mirsattari SM, Merriman B, Klein E, et al. 2012. Familial cortical myoclonus with a mutation in NOL3. Ann Neurol. 72:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simó-Riudalbas L, Offner S, Planet E, Duc J, Abrami L, Dind S, Coudray A, Coto-Llerena M, Ercan C, Piscuoglio S, et al. 2022. Transposon-activated POU5F1B promotes colorectal cancer growth and metastasis. Nat Commun. 13:4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Jungers WL. 1997. Body mass in comparative primatology. J Hum Evol. 32:523–559. [DOI] [PubMed] [Google Scholar]
- Sousa AMM, Zhu Y, Raghanti MA, Kitchen RR, Onorati M, Tebbenkamp ATN, Stutz B, Meyer KA, Li M, Kawasawa YI, et al. 2017. Molecular and cellular reorganization of neural circuits in the human lineage. Science 358:1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann M, Lupiáñez DG, Mundlos S. 2018. Structural variation in the 3D genome. Nat Rev Genet. 19:453–467. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. 2010. Structural variation in the human genome and its role in disease. Annu Rev Med. 61:437–455. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Rocchi M, Capozzi O, Roberto R, Misceo D, Ventura M, Cardone MF, Bigoni F, Archidiacono N. 2008. Primate chromosome evolution: ancestral karyotypes, marker order and neocentromeres. Chromosome Res. 16:17–39. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Huddleston J, Catacchio CR, Malig M, Hillier LW, Baker C, Mohajeri K, Kondova I, Bontrop RE, Persengiev S, et al. 2013. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 23:1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntsova MV, Buzdin AA. 2020. Differences between human and chimpanzee genomes and their implications in gene expression, protein functions and biochemical properties of the two species. BMC Genomics 21:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo G, Han J, Wang X, Zhang J, Yannan Z, Yanhong Z, Dai J. 2005. Oct4 pseudogenes are transcribed in cancers. Biochem Biophys Res Commun. 337:1047–1051. [DOI] [PubMed] [Google Scholar]
- Vermunt MW, Tan SC, Castelijns B, Geeven G, Reinink P, de Bruijn E, Kondova I, Persengiev S, Bank NB, Bontrop R, et al. 2016. Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat Neurosci. 19:494–503. [DOI] [PubMed] [Google Scholar]
- Warren WC, Harris RA, Haukness M, Fiddes IT, Murali SC, Fernandes J, Dishuck PC, Storer JM, Raveendran M, Hillier LW, et al. 2020. Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science 370:eabc6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Yamada Y, Kimata K. 1998. Roles of aggrecan, a large chondroitin sulfate proteoglycan, in cartilage structure and function. J Biochem. 124:687–693. [DOI] [PubMed] [Google Scholar]
- Wei M, Ying Y, Li Z, Weng Y, Luo X. 2021. Identification of novel ACAN mutations in two Chinese families and genotype-phenotype correlation in patients with 74 pathogenic ACAN variations. Mol Genet Genomic Med. 9:e1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO. 2013. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 14:125–138. [DOI] [PubMed] [Google Scholar]
- Wheatley BP. 1987. The evolution of large body size in orangutans: a model for hominoid divergence. Am J Primatol. 13:313–324. [DOI] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. 2021. Clusterprofiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb). 2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanke BW, Greenwood CMT, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, et al. 2007. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 39:989–994. [DOI] [PubMed] [Google Scholar]
- Zihlman AL, Bolter DR. 2015. Body composition in Pan paniscus compared with Homo sapiens has implications for changes during human evolution. Proc Natl Acad Sci U S A. 112:7466–7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zihlman AL, McFarland RK. 2000. Body mass in lowland gorillas: a quantitative analysis. Am J Phys Anthropol. 113:61–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genome, epigenome, and transcriptome data could be downloaded in the NCBI database, and the code used in this study had been submitted to github (https://github.com/kizzb/Great-apes-specific-SVs).