Abstract
Pneumocystis carinii f. sp. hominis isolates from 207 clinical specimens from nine countries were typed based on nucleotide sequence variations in the internal transcribed spacer regions I and II (ITS1 and ITS2, respectively) of rRNA genes. The number of ITS1 nucleotides has been revised from the previously reported 157 bp to 161 bp. Likewise, the number of ITS2 nucleotides has been changed from 177 to 192 bp. The number of ITS1 sequence types has increased from 2 to 15, and that of ITS2 has increased from 3 to 14. The 15 ITS1 sequence types are designated types A through O, and the 14 ITS2 types are named types a through n. A total of 59 types of P. carinii f. sp. hominis were found in this study.
We have previously reported that the nucleotide sequences of the internal transcribed spacer (ITS) regions of rRNA genes of different isolates of Pneumocystis carinii f. sp. hominis are variable and that this sequence variation can be used for typing (15). There are two ITS regions in the genome of P. carinii: ITS region I (ITS1) is located between the 18S and 5.8S rRNA genes, and ITS2 is located between the 5.8S and 26S rRNA genes. ITS1 was found to contain 157 bp, and ITS2 was determined to be 177 bp (15). Sequence variations in the ITS1 region that were found to be useful for typing are located at positions 6, 14, 67, 76, and 77. Based on nucleotide sequence variations at these positions, ITS1 sequences were classified into two types (A and B). Type A has a C at position 6, a T at position 14, and bases missing at positions 67, 76, and 77. Type B has a T at position 6, a base missing at position 14, a T at position 67, and AG at positions 76 and 77.
In the ITS2 region, nucleotide variations at positions 50 to 52, 59, 63 to 67, 160 and 161, and 165 were used for typing, and three types of ITS2, designated types a, b, and c, were found. Types a and b differ by two bases at positions 160 and 161, where type b has residues AT and type a lacks these two bases. Type c lacks bases at positions 50 to 52, 63 to 67, and 160 and 161. In addition, it has an A instead of a G residue at position 165.
P. carinii isolates with type A or B ITS1 may have type a, b, or c ITS2; therefore, there were six potential combination types (Aa, Ab, Ac, Ba, Bb, and Bc). We have previously reported finding four types (Ac, Ba, Bb, and Bc) (15). In this study, we have expanded the sample size and found many more new types. This paper reports a very substantial revision of ITS types of P. carinii f. sp. hominis.
MATERIALS AND METHODS
Specimens.
Specimens used for this study included bronchoalveolar lavage (BAL) fluids, BAL smears on slides, and paraffin-embedded lung biopsy specimens. A total of 207 specimens from 9 countries were used including 123 from Denmark, 44 from the United States, 10 from Ivory Coast (17, 18), 9 from Italy, 6 from France, 5 from Sweden, 4 each from The Netherlands and Portugal, and 2 from Thailand (Table 1). Specimens from Denmark and Sweden were BAL smears on slides, those from Thailand and West Africa were paraffin-embedded lung biopsy specimens, and the remaining specimens were BAL fluid specimens. The specimens from the United States included 38 from Indiana University Medical Center, 4 from the University of Medicine and Dentistry of New Jersey as described previously (15), and 2 from the University of Michigan. Fresh BAL fluid specimens and smears were processed for PCR by the procedures described previously (12, 13). Paraffin-embedded tissue sections were processed for PCR as described by Greer et al. (4).
TABLE 1.
Geographical sources of specimens used in this study
| Source | No. |
|---|---|
| Denmark | 123 |
| United States | 44 |
| Ivory Coast | 10 |
| Italy | 9 |
| France | 6 |
| The Netherlands | 4 |
| Portugal | 4 |
| Sweden | 5 |
| Thailand | 2 |
| Total | 207 |
PCR.
Nested PCR was performed on all specimens to obtain a sufficient amount of DNA fragment for analysis by the procedure of Lu et al. (15) with the following modifications. The first PCR was performed with primers 1724F2 (5′-AGTTGATCAAATTTGGTCATTTAGAG-3′) and ITS2R (5′-CTCGGACGAGGATCCTCGCC-3′) under the following conditions: (i) 3 min at 94°C; (ii) 5 cycles, with 1 cycle consisting of 1.5 min at 94°C, 1.5 min at 62°C, and 1.5 min at 72°C; and (iii) 30 cycles, with 1 cycle consisting of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C. The PCR mixture was in a total volume of 40 μl containing 50 ng of template DNA, PCR buffer, 2.5 U of Taq polymerase, 3 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, and 20 pmol of each primer. The second PCR was performed with primers FX (5′-TTCCGTAGGTGAACCTGCG-3′) and RT2 (5′-CTGATTTGAGATTAAAATTCTTG-3′) under the following conditions: (i) 3 min at 94°C; (ii) 5 cycles, with 1 cycle consisting of 1.5 min at 94°C, 1.5 min at 58°C, and 1.5 min at 72°C; and (iii) 30 cycles, with 1 cycle consisting of 1 min at 94°C, 1 min at 56°C, and 1 min at 72°C. The reaction mixture contained 2 μl of the first PCR product and the same components as those of the first PCR except that 1.5 mM MgCl2 was used.
Typing.
Nested PCR products of ITS regions were first probed with type-specific probes 1A, 1B, 2a, 2b, and 2c to determine whether the specimen contained a single or multiple types of P. carinii as described previously (16). The specimens which contained a single type of P. carinii were sequenced directly with the Sequenase kit (Amersham Life Science, Arlington Height, Ill.), and those which contained multiple types of P. carinii were cloned and then sequenced. Screening of recombinant clones that contained the correct insert was achieved by colony hybridization as described previously (7) using all five of the type-specific probes mentioned above. Five colonies which reacted with a given probe were sequenced. A sequence which was found to be identical in at least three of the five colonies was considered to be a correct sequence. A sequence which appeared in at least two different specimens was considered a distinct type.
Nucleotide sequence accession numbers.
All the type-specific sequences have been deposited in GenBank. The sequences and accession numbers are as follows: ITS1A, AF013806; ITS1B, AF013807; ITS1C, AF013808; ITS1D, AF013809; ITS1E, AF013810; ITS1F, AF013811; ITS1G, AF013812; ITS1H, AF013813; ITS1I, AF013814; ITS1J, AF013815; ITS1K, AF013816; ITS1L, AF013817; ITS1M, AF013818; ITS1N, AF013819; ITS1O, AF013820; ITS2a, AF013821; ITS2b, AF013822; ITS2c, AF013823; ITS2d, AF013824; ITS2e, AF013825; ITS2f, AF013826; ITS2g, AF013827; ITS2h, AF013828; ITS2i, AF013829; ITS2j, AF013830; ITS2k, AF013831; ITS2l, AF013832; ITS2m, AF013833; and ITS2n, AF013834.
RESULTS
A total of 15 ITS1 and 14 ITS2 sequence types were found from the 207 specimens that we have examined. The number of ITS1 nucleotides has been changed from 157 to 161 bp. This is due to insertions of TT between positions 22 and 23 and AT between positions 50 and 51 of the original sequence. The ITS2 sequence has been changed from 177 to 192 bp. Insertions of new bases include AA between positions 47 and 48, TAA between positions 52 and 53, T between positions 70 and 71, AT between positions 159 and 160, and AAAATAC between positions 169 and 170 of the original sequence. The nucleotides of the revised sequences have been renumbered, and new position numbers are used hereafter. The consensus nucleotide sequences of both ITS1 and ITS2 are shown in Fig. 1.
FIG. 1.
Revised nucleotide sequences of ITS1 and ITS2. The number of nucleotides has been changed from the previously reported 157 bp to 161 bp for ITS1 and from 177 to 192 bp for ITS2. The sequences shown are consensus sequences; they are not the sequences of a certain type of P. carinii f. sp. hominis.
The 15 ITS1 types are designated types A through O. Types that are close to each other are named with sequential letters. For example, types A and B differ by only one base at position 12, where type A has a T and type B lacks this base. Type A is the same type A as we have reported previously (15). The original type B has been renamed type E. All the other types are novel. Not every P. carinii isolate has the same number of nucleotides in both ITS1 and ITS2. Some isolates are missing bases at certain positions. In ITS1, sequence variations are found at nucleotide positions 6, 12, 15, 21, 23, 24, 28, 34, 42, 53, 54, 80, 81, and 115 to 118 (Fig. 2).
FIG. 2.
Nucleotide sequences of various types of ITS1 of P. carinii f. sp. hominis. A sequence which is found in two or more specimens is considered a distinct type. A total of 15 ITS1 types, designated A through O, have been found. Two sequences representing each type are aligned. Bases that are identical to those of the consensus sequence are indicated by periods, missing bases are indicated by hyphens, and bases that are different from those of the consensus sequence are given. The sequences at positions 61 to 70 and 91 to 110 are not shown, because no sequence variations are found in these areas.
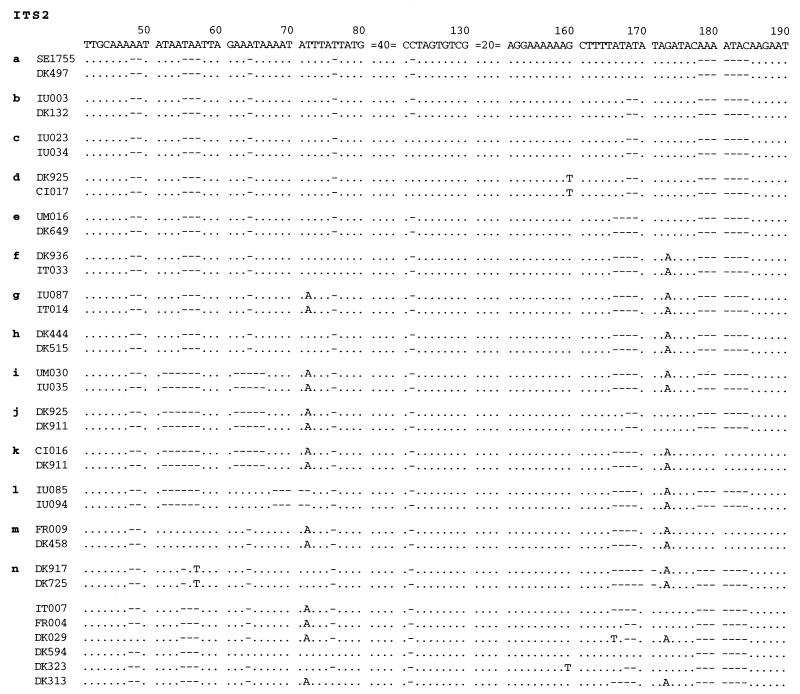
Nucleotide sequence variations in ITS2 are much more extensive. They are found at positions 48 and 49, 52 and 57, 62 to 70, 72, 76, 122, 160, 166 to 171, 173, and 178 to 184 (Fig. 3). There are 14 types of ITS2 sequences, designated types a through n. Similar to ITS1 types, those that are close to each other are named with sequential letters. The original type b remains type b, whereas the original types a and c have been renamed types e and m, respectively.
FIG. 3.
Nucleotide sequences of various types of ITS2 of P. carinii f. sp. hominis. A sequence which is found in two or more specimens is considered a distinct type, and 14 sequences (designated a through n) met this requirement. Two sequences representing each type are aligned. Bases that are identical to those of the consensus sequence are indicated by periods, missing bases are indicated by hyphens, and bases that are different from those of the consensus sequence are given. The sequences at positions 1 to 40, 81 to 120, and 131 to 150, are not shown, because no sequence variations are found in these areas. Six sequences from specimens (IT007, FR004, DK029, DK594, DK323, and DK313) are also listed but are not considered distinct types, because they are found in only one specimen each.
There are six sequences from specimens IT007, FR004, DK029, DK594, DK323, and DK313 (Fig. 3) that have variations at variable positions of ITS2 but are not considered distinct types, because only one example of each has been found so far. The sequence found in specimen IT007 differs from that of type g at position 173, where IT007 has a G and type g has an A. The sequence of specimen FR004 differs from that of type b by only one base at position 72 where type b has a T and FR004 has an A. The sequence found in specimen DK029 is unique. It has no base deletions at positions 48 and 49. It also lacks bases at positions 55 to 57, 64, 76, 122, 168 and 169, and 178 to 184 and has an A residue at position 72, a T at position 166, and an A at position 173. The sequence found in specimen DK594 has an A at position 64 and a T at position 76; otherwise, it is identical to type b. The sequence of DK323 differs from that of type d by only one base at position 122, where it is missing the base C. The sequence of DK313 differs from type f by one base at position 72, where DK313 has an A instead of a T.
As previously described, P. carinii sequence types are designated with a two-letter code (15). The first letter is the ITS1 type, and the second one is the ITS2 type. Among the 207 specimens that have been typed, there are a total of 59 combination types of P. carinii f. sp. hominis (Table 2). Type Eg is the most prevalent; 59 of 217 specimens contain this type. The second most prevalent type is Ne (43 specimens), which is followed by Eb (25 specimens); Ai (12 specimens); Bi, Ec, and Ee (11 specimens each); Al and Me (10 specimens each); Eh (7 specimens); Gg, Jf, and Kf (6 specimens each); Bl, Ei, and Og (4 specimens each); Bb, Bg, and Li (3 specimens each); Ad, Bk, De, Di, Ea, Ej, Em, Fg, Gb, Ih, Ng, Oe, and On (2 specimens each); and Ab, Ba, Be, Bh, Bm, Cg, Dh, Ed, Ef, El, Gf, Gh, Gi, He, Hg, Hh, Ie, Ii, In, Jg, Na, Nb, Nc, Nl, Nn, Of, and Oi (1 specimen each). Among the 207 specimens examined, 155 (75%) are found to contain a single type of P. carinii and 52 (25%) had more than one type, including 33 specimens with two types, 8 specimens with three types, 6 specimens with four types, and two specimens with 6 types (Table 3).
TABLE 2.
P. carinii f. sp. hominis types found in various countries
| Type no. | Type | No. of specimens found in countrya:
|
Total no.b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DK | US | CI | IT | FR | NE | PT | SE | TH | |||
| 1 | Ab | 1 | 1 | ||||||||
| 2 | Ad | 1 | 1 | 2 | |||||||
| 3 | Ai | 5 | 2 | 2 | 3 | 12 | |||||
| 4 | Al | 4 | 5 | 1 | 10 | ||||||
| 5 | Ba | 1 | 1 | ||||||||
| 6 | Bb | 1 | 1 | 1 | 3 | ||||||
| 7 | Be | 1 | 1 | ||||||||
| 8 | Bg | 3 | 3 | ||||||||
| 9 | Bh | 1 | 1 | ||||||||
| 10 | Bi | 6 | 2 | 2 | 1 | 11 | |||||
| 11 | Bk | 1 | 1 | 2 | |||||||
| 12 | Bl | 2 | 2 | 4 | |||||||
| 13 | Bm | 1 | 1 | ||||||||
| 14 | Cg | 1 | 1 | ||||||||
| 15 | De | 1 | 1 | 2 | |||||||
| 16 | Dh | 1 | 1 | ||||||||
| 17 | Di | 1 | 1 | 2 | |||||||
| 18 | Ea | 2 | 2 | ||||||||
| 19 | Eb | 15 | 6 | 3 | 1 | 25 | |||||
| 20 | Ec | 5 | 5 | 1 | 11 | ||||||
| 21 | Ed | 1 | 1 | ||||||||
| 22 | Ee | 8 | 2 | 1 | 11 | ||||||
| 23 | Ef | 1 | 1 | ||||||||
| 24 | Eg | 38 | 11 | 3 | 1 | 2 | 3 | 1 | 59 | ||
| 25 | Eh | 6 | 1 | 7 | |||||||
| 26 | Ei | 3 | 1 | 4 | |||||||
| 27 | Ej | 2 | 2 | ||||||||
| 28 | El | 1 | 1 | ||||||||
| 29 | Em | 1 | 1 | 2 | |||||||
| 30 | Fg | 1 | 1 | 2 | |||||||
| 31 | Gb | 1 | 1 | 2 | |||||||
| 32 | Gf | 1 | 1 | ||||||||
| 33 | Gg | 3 | 2 | 1 | 6 | ||||||
| 34 | Gh | 1 | 1 | ||||||||
| 35 | Gi | 1 | 1 | ||||||||
| 36 | He | 1 | 1 | ||||||||
| 37 | Hg | 1 | 1 | ||||||||
| 38 | Hh | 1 | 1 | ||||||||
| 39 | Ie | 1 | 1 | ||||||||
| 40 | Ih | 1 | 1 | 2 | |||||||
| 41 | Ii | 1 | 1 | ||||||||
| 42 | In | 1 | 1 | ||||||||
| 43 | Jf | 5 | 1 | 6 | |||||||
| 44 | Jg | 1 | 1 | ||||||||
| 45 | Kf | 1 | 1 | 1 | 3 | 6 | |||||
| 46 | Li | 1 | 2 | 3 | |||||||
| 47 | Me | 4 | 2 | 4 | 10 | ||||||
| 48 | Na | 1 | 1 | ||||||||
| 49 | Nb | 1 | 1 | ||||||||
| 50 | Nc | 1 | 1 | ||||||||
| 51 | Ne | 26 | 13 | 1 | 1 | 1 | 1 | 43 | |||
| 52 | Ng | 1 | 1 | 2 | |||||||
| 53 | Nl | 1 | 1 | ||||||||
| 54 | Nn | 1 | 1 | ||||||||
| 55 | Oe | 1 | 1 | 2 | |||||||
| 56 | Of | 1 | 1 | ||||||||
| 57 | Og | 1 | 1 | 1 | 1 | 4 | |||||
| 58 | Oi | 1 | 1 | ||||||||
| 59 | On | 2 | 2 | ||||||||
| Totalc | 45 | 21 | 18 | 8 | 8 | 3 | 3 | 4 | 3 | ||
Abbreviations: DK, Denmark; US, United States; CI, Ivory Coast; IT, Italy; FR, France; NE, The Netherlands; PT, Portugal; SE, Sweden; TH, Thailand.
Total number of each type found in this study.
Total numbers of types in each country.
TABLE 3.
Types of P. carinii f. sp. hominis found in each specimen
| Sp. IDa | Type(s) | Sp. ID | Type(s) | Sp. IDa | Type(s) | Sp. IDa | Type(s) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| DK004 | Ne | DK412 | Bg, Bi, Ne | DK713 | Bl, Eg | IU085 | Al | |||
| DK005 | Eg | DK414 | Jf | DK714 | Bi, Ea, Eg | IU086 | Al | |||
| DK006 | Ee | DK415 | Eg, Jf | DK720 | Eg | IU087 | Eg | |||
| DK008 | Eg | DK439 | Eb | DK725 | Ii, In, On | IU088 | Ne | |||
| DK010 | Ne | DK441 | Ng, Eh | DK735 | Bg | IU089 | Eg, Eh | |||
| DK013 | Eg | DK444 | Bi, Bg, Bh | DK911 | Ai, Bi, Bk, Ed, Ej, Em | NJ114 | Ne | |||
| DK015 | Eg | DK451 | Al, Nn | DK912 | Eg | NJ00A | Ec | |||
| DK017 | He, Me | DK458 | Bi, Bm | DK913 | Gg | NJ00W | Eg | |||
| DK027 | Eg, Ef, Kf | DK464 | Eb, Eg | DK918 | Ee, Ei | NJ00Z | Eb | |||
| DK033 | Ne | DK465 | Eb, Eg | DK921 | Eb | UM016 | Eb, Nb, Ne | |||
| DK035 | Oh | DK472 | Ee | DK922 | Ne | UM030 | Ai | |||
| DK038 | Ee, Me | DK478 | Ai, Gg | DK923 | Ne | CI006 | Bi, Dh, Li | |||
| DK057 | Eb, Ih | DK482 | Eg | DK925 | Ab, Ad, Eb, Ei, Ej, Gb, On | CI010 | Ai, De, Gb, Oe | |||
| DK058 | Eg | DK484 | Eb | DK931 | Ne | CI011 | Ai | |||
| DK071 | Og | DK488 | Eg | DK937 | Jf | CI012 | Me, Ne | |||
| DK072 | Ne | DK494 | Ec, Eh | DK939 | Ne | CI014 | Bi, Ec, Eg, Li | |||
| DK073 | Me | DK495 | Cg, Eg | DK941 | Ai | CI015 | Ee, Gg, Me, Og | |||
| DK132 | Eb | DK497 | Ea | DK942 | Ai | CI016 | Bk | |||
| DK140 | Ec | DK507 | Eh | DK944 | Eb | CI017 | Ad | |||
| DK237 | Bl | DK512 | Eh, Ne | IU001 | Ne | CI018 | Eg | |||
| DK241 | Nl | DK515 | Eh | IU002 | Bi | CI019 | Eg, Fg, Kf | |||
| DK246 | Eg | DK516 | Eg, Ei | IU003 | Bb, Bi, Eb | IT007 | Eo | |||
| DK254 | Eg | DK521 | Ne | IU004 | Eg | IT014 | Eg | |||
| DK255 | Ne | DK530 | Eg | IU023 | Ec, Nc, Ne, Oe | IT022 | Hg | |||
| DK258 | Jf | DK543 | Ne | IU031 | Ne | IT029 | Kf | |||
| DK272 | Ec | DK544 | Eg | IU033 | Ai | IT033 | Gf, Of | |||
| DK275 | Eg | DK545 | Eg | IU034 | Ec | IT040 | Kf | |||
| DK284 | Eb | DK547 | Eg | IU035 | Be, Eg, Li | IT046 | Kf | |||
| DK288 | Al | DK561 | Ee | IU036 | Ne | IT057 | Ih | |||
| DK295 | Ne | DK563 | Ec | IU037 | Ne | IT062 | Jf, Jg | |||
| DK296 | Eg | DK567 | Ne | IU038 | Ne | FR001 | Eb | |||
| DK300 | Eg | DK574 | Ie | IU039 | Eg | FR002 | Ne | |||
| DK303 | Ne | DK601 | Eg | IU040 | Kf | FR004 | Eb, Eg, Eo | |||
| DK310 | Ee | DK622 | Ne, De | IU041 | Hh | FR006 | Bb, Eb, Eg, Ei | |||
| DK313 | Eg | DK630 | Eb | IU043 | Og | FR009 | Em | |||
| DK315 | Eg | DK631 | Eg | IU044 | Eg, Gg | FR010 | Bi, Gi | |||
| DK319 | Eg | DK641 | Eg | IU047 | Ee | NL091 | Ne | |||
| DK321 | Gg | DK647 | Eg | IU050 | Eg | NL092 | Ai | |||
| DK330 | Eb | DK649 | Ee | IU051 | Ee, Eg, Ne, Ng | NL093 | Ai | |||
| DK332 | Jf | DK650 | Eg | IU052 | Gg | NL097 | Ai, Oi | |||
| DK336 | Al | DK651 | Ba | IU058 | Ec | PT115 | Bl | |||
| DK338 | Eb | DK652 | Fg | IU059 | Eg | PT153 | Al | |||
| DK340 | Eb | DK654 | Ne | IU070 | Ne | PT154 | Bl | |||
| DK341 | Ne | DK654-B | Ne | IU071 | Ne | PT167 | Eb | |||
| DK345 | Ec | DK665 | Ai, Di | IU077 | Eb | SE911 | Eg | |||
| DK346 | Ne | DK669 | Ne | IU078 | Al, Eb | SE1337 | Eg | |||
| DK347 | Eh | DK671 | Ne | IU079 | Al, Me | SE1632 | Ne, Og | |||
| DK375 | Al | DK674 | Bb | IU080 | Me | SE1755 | Na | |||
| DK399 | Eg | DK676 | El, Me | IU081 | Ne | SE1969 | Eg | |||
| DK402 | Eg, Ne | DK681 | Ne | IU082 | Al | TH001 | Di | |||
| DK403 | Bi, Eg | DK698 | Eb | IU083 | Ec | TH002 | Eg, Gh | |||
| DK406 | Ne | DK702 | Ee, Eg | IU084 | Eb |
Sp. ID, specimen identification.
In this study, substantial numbers of specimens were collected from Denmark and the United States. A total of 45 types were found in the 123 specimens from Denmark. The three most prevalent types in Denmark are Eg (38 specimens), Ne (26 specimens), and Eb (15 specimens). Twenty-one different types were found in 44 specimens from the United States, and the three most prevalent types are Ne (13 specimens), Eg (11 specimens), and Eb (6 specimens). The numbers of specimens from other countries are not sufficient to make a significant conclusion on the prevalence of different types.
DISCUSSION
We have previously described the finding of four types of P. carinii based on nucleotide sequence variations in ITS regions, but there were a number of sporadic sequence variations which were not considered distinct types because less than three isolates had the same sequence (15). By expanding the sample size in this study, we have found more isolates that have the same sequence as those previously considered as sporadic variations. Most of those sporadic variations are now considered new types. We have also discovered many more new sequences.
Although a single nucleotide variation may result in a distinct type, sequence variation in one area within ITS1 was not used for typing. This area is located at positions 62 to 71, where there are 10 T’s in a row (Fig. 1). It has been reported that the number of T’s determined in a poly(T) tract varied when the same sample was sequenced repeatedly (6, 20). In this study, the number of T’s was found to vary from 8 to 12 in different specimens. Although there are several other poly(T) and poly(A) tracts in both ITS1 and ITS2, we did not encounter the same problem in these areas.
Tsolaki et al. (19) has recently examined 24 specimens and found five types of ITS1 (designated A2, B1, B2, B3, and C) and seven types of ITS2 sequences (designated a1, a2, a3, b1, b2, c, and d). Types B1, B2, and C of Tsolaki et al. are the same as our new ITS1 types E, N, and F, respectively. Type A2 can be type A or B and type B3 can be type C or D, depending on the presence (types A and D) or absence (types B and C) of a T residue at position 12 of the ITS1 sequence. ITS2 types a1, a3, b1, c, and d of Tsolaki et al. are the same as our new types e, g, b, l, and a, respectively. Type a2 has the same sequence as that of IT007 (Fig. 3), which is not considered a distinct type in this study, because only one sample has this sequence. Type b2 can be type c or d, depending on whether there is a G (type c) or T (type d) residue at position 160 of the ITS2 sequence.
Similar results have been reported by Latouche et al. (10, 11). They have examined 20 specimens in one study (10) and 36 in another (11) from AIDS patients and found eight types of ITS1 (designated A1, A2, B1, B2, B3, B4, B5, and B6) and seven types of ITS2 (designated a1, a2, a3, a4, b1, b5, and c1). ITS1 types A1 and A2 of Latouche et al. (11) are identical to our new ITS1 types B and A, respectively. Types B2 and B5 differ in the numbers of T’s in the poly(T) tract, which is not used for typing in our study. These two types should be considered the same and are equivalent to our type N. Type B3 is the same as our type J, except that position 21 is an A instead of a T. Similarly, type B4 is the same as our type I, except that position 21 is an A instead of a T. Types B1 and B6 of Latouche et al. (11) also differ in the numbers of T’s in the poly(T) tract. These two types can be our type E, F, L, or M, depending on the nucleotides at positions 34, 42, and 115 to 118 (Fig. 2). Since the identities of nucleotides of types B1 and B6 at these positions are not reported, we are unable to identify these two types. In the ITS2 region, types a1, a2, a3, a4, b5, and c1 of Latouche et al. (11) are identical to our new types e, h, n, f, a, and l, respectively. Their type b1 can be our new type b, c, or d, depending on the nucleotides at positions 122 and 160. If the C residue at position 122 is missing, it is a type b. At position 160, type c has a G and type d has a T. Since there is no information as to the nucleotides at these two positions, it is impossible to determine whether their type b1 belongs to our type b, c, or d.
Among the 59 types of P. carinii f. sp. hominis found in this study, types Eg, Ne, and Eb are present in most countries from which we have obtained specimens. There are types that are found only in certain countries in this study. For example, types Be, Hh, Nb, and Nc are found only in the United States. However, it is premature to conclude that these types are unique to certain regions, since the sample size from each country was relatively small in this study. A considerably greater number of specimens from each country will have to be typed to make a significant conclusion. This study focused on finding new types rather than studying the distribution of different types.
As reported previously (2, 8, 9, 11, 15, 19), a portion of specimens examined in this study contained more than one type of P. carinii. Although it is likely that these specimens represent mixed infections, the following possibilities remain to be ruled out. The mixed types may be derived from different copies of rRNA genes in the same P. carinii genome, since multiple copies of rRNA genes are commonly found in eukaryotic organisms. It is unknown whether P. carinii f. sp. hominis has more than one copy of the rRNA gene, although rat-derived P. carinii has been shown to contain less than two copies (3). The other possibility is that they are due to heterozygosity of the organism. There is also the possibility of contamination during specimen collection or processing. In this study, two specimens were found to contain six or more types of P. carinii. It is quite likely that these two specimens were contaminated, since it is unusual to see a host infected by so many different types of the same organism.
Typing of P. carinii based on nucleotide sequence variations has been conducted by several investigators on several different loci (1, 5, 8–10, 12, 19, 20). We have examined the sequences of mitochondrial rRNA gene (12) and the ITS regions (7, 15, 16). In addition to these two loci, Latouche et al. (10) have examined the 5S rRNA gene and the thymidylate synthase gene. Sequence variations were not found in the thymidylate synthase gene. The mitochondrial rRNA gene has only 2 or 3 nucleotides that are variable. This limited sequence variation may not have sufficient discriminatory power for typing. Isolates that have the same mitochondrial rRNA gene type may have different ITS types (9). The 5S rRNA gene also has limited sequence variation; variations are found only in five positions. Until other genetic loci of P. carinii are found that are more variable, we consider ITS sequence variation the method of choice for typing P. carinii isolates. The development of P. carinii typing has enabled preliminary epidemiological studies (14) and investigation of potential air-borne transmission of P. carinii infections (2). It has also made it possible to determine whether recurrent pneumocystosis is due to a relapse of a previous infection or a new infection (8, 19).
ACKNOWLEDGMENTS
This study was supported by NIH grant RO1 AI 34304.
In the collection of the autopsy samples from patients in Ivory Coast, we acknowledge the assistance of A. Kadio, J. Andoh, and M. Hondé of the University Hospital of Treichville in Abidjan. We also thank S. H. Vermund for assistance with specimen collection and valuable advice on the project and S. Meshnick, M. J. Lebowitz, and B. Sathapatayavongs for providing specimens.
REFERENCES
- 1.Banerji S, Lugli E B, Miller R F, Wakefield A E. Analysis of genetic diversity at the arom locus in isolates of Pneumocystis carinii. J Eukaryot Microbiol. 1995;42:675–679. doi: 10.1111/j.1550-7408.1995.tb01614.x. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett M S, Vermund S H, Jacobs R, Durant P J, Shaw M M, Smith J W, Tang X, Lu J J, Li B, Jin S, Lee C H. Detection of Pneumocystis carinii DNA in air samples: likely environmental risk to susceptible persons. J Clin Microbiol. 1997;35:2511–2513. doi: 10.1128/jcm.35.10.2511-2513.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giuntoli D, Stringer S L, Stringer J R. Extraordinary low number of ribosomal RNA genes in P. carinii. J Eukaryot Microbiol. 1994;41:88S. [PubMed] [Google Scholar]
- 4.Greer C E, Wheeler M, Manos M M. Sample preparation and PCR amplification from paraffin-embedded tissues. PCR Methods Appl. 1994;3:S113–S122. doi: 10.1101/gr.3.6.s113. [DOI] [PubMed] [Google Scholar]
- 5.Hunter J A, Wakefield A E. Genetic divergence at the mitochondrial small subunit ribosomal RNA gene among isolates of Pneumocystis carinii from five mammalian host species. J Eukaryot Microbiol. 1996;43:24S–25S. doi: 10.1111/j.1550-7408.1996.tb04962.x. [DOI] [PubMed] [Google Scholar]
- 6.Jennings M P, Hood D W, Peak I R A, Virji M, Moxon E R. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 7.Jiang B, Lu J J, Li B, Tang X, Bartlett M S, Smith J W, Lee C H. Development of type-specific PCR for typing Pneumocystis carinii f. sp. hominis based on nucleotide sequence variations of the internal transcribed spacer regions of ribosomal RNA genes. J Clin Microbiol. 1996;34:3245–3248. doi: 10.1128/jcm.34.12.3245-3248.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keely S P, Stringer J R, Baughman R P, Linke M J, Walzer P D, Smulian A G. Genetic variation among Pneumocystis carinii hominis isolates in recurrent pneumocystosis. J Infect Dis. 1995;172:595–598. doi: 10.1093/infdis/172.2.595. [DOI] [PubMed] [Google Scholar]
- 9.Keely S P, Stringer J R. Sequences of Pneumocystis carinii f. sp. hominis strains associated with recurrent pneumonia vary at multiple loci. J Clin Microbiol. 1997;35:2745–2747. doi: 10.1128/jcm.35.11.2745-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latouche S, Ortona E, Masers E, Margutti P, Tamburrini E, Siracusano A, Guyot K, Nigou M, Roux P. Biodiversity of Pneumocystis carinii hominis: typing with different DNA regions. J Clin Microbiol. 1997;35:383–387. doi: 10.1128/jcm.35.2.383-387.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latouche S, Poirot J-L, Bernard C, Roux P. Study of internal transcribed spacer and mitochondrial large-subunit genes of Pneumocystis carinii hominis isolated by repeated bronchoalveolar lavage from human immunodeficiency virus-infected patients during one or several episodes of pneumonia. J Clin Microbiol. 1997;35:1687–1690. doi: 10.1128/jcm.35.7.1687-1690.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C H, Lu J J, Bartlett M S, Durkin M M, Liu T H, Wang J, Jiang B, Smith J W. Nucleotide sequence variation in Pneumocystis carinii strains that infect humans. J Clin Microbiol. 1993;31:754–757. doi: 10.1128/jcm.31.3.754-757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C H, Wang J, Durkin M M, Brady S L, Bartlett M S, Smith J W. Amplification of Pneumocystis carinii DNA on specimens scraped from slides. Diagn Microbiol Infect Dis. 1994;18:197–199. doi: 10.1016/0732-8893(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 14.Lee C H, Lu J J, Tang X, Jiang B, Li B, Jin S, Bartlett M S, Lundgren B, Atzori C, Orlando G, Cargnel A, Smith J W. Prevalence of various Pneumocystis carinii f. sp. hominis types in different geographical locations. J Eukaryot Microbiol. 1996;43:37S. doi: 10.1111/j.1550-7408.1996.tb04973.x. [DOI] [PubMed] [Google Scholar]
- 15.Lu J J, Bartlett M S, Shaw M M, Smith J W, Ortiz-Rivera M, Leibowitz M J, Lee C H. Typing of Pneumocystis carinii strains that infect humans based on nucleotide sequence variations of internal transcribed spacers of rRNA genes. J Clin Microbiol. 1994;32:2904–2912. doi: 10.1128/jcm.32.12.2904-2912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J J, Bartlett M S, Smith J W, Lee C H. Typing of Pneumocystis carinii strains with type-specific oligonucleotide probes derived from nucleotide sequences of internal transcribed spacers of rRNA genes. J Clin Microbiol. 1995;33:2973–2977. doi: 10.1128/jcm.33.11.2973-2977.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas S B, Hounnou A, Peacock C S, Beaumel A, Djomand G, N’Gbichi J-M, Yeboue K, Hondé M, Diomandé M, Giordano C, Doorly R, Brattegaard K, Kestens L, Smithwick R W, Kadio A, Ezani N, Yapi A, De Cock K M. The mortality and pathology of HIV disease in a West African city. AIDS. 1993;7:1569–1579. doi: 10.1097/00002030-199312000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Lucas S B, Peacock C S, Hounnou A, Brattegaard K, Koffi K, Hondé M, Andoh J, Bell J E, De Cock K M. Disease in children infected with HIV in Abidjan, Côte d’Ivoire. Br Med J. 1996;312:335–338. doi: 10.1136/bmj.312.7027.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsolaki A G, Miller R F, Underwood A P, Banerji S, Wakefield A E. Genetic diversity at the internal transcribed spacer regions of the rRNA operon among isolates of Pneumocystis carinii from AIDS patients with recurrent pneumonia. J Infect Dis. 1996;174:141–156. doi: 10.1093/infdis/174.1.141. [DOI] [PubMed] [Google Scholar]
- 20.Wakefield A E, Fritscher C C, Malin A S, Gwanzura L, Hughes W T, Miller R F. Genetic diversity in human-derived Pneumocystis carinii isolates from four geographical locations shown by analysis of mitochondrial rRNA gene sequences. J Clin Microbiol. 1994;32:2959–2961. doi: 10.1128/jcm.32.12.2959-2961.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]