Abstract
Genetic engineering of plants has enhanced crop productivity in the face of climate change and a growing global population by conferring desirable genetic traits to agricultural crops. Efficient genetic transformation in plants remains a challenge due to the cell wall, a barrier to exogenous biomolecule delivery. Conventional delivery methods are inefficient, damaging to tissue, or are only effective in a limited number of plant species. Nanoparticles are promising materials for biomolecule delivery, owing to their ability to traverse plant cell walls without external force and highly tunable physicochemical properties for diverse cargo conjugation and broad host range applicability. With the advent of engineered nuclease biotechnologies, we discuss the potential of nanoparticles as an optimal platform to deliver biomolecules to plants for genetic engineering.
Current Biomolecule Delivery Methods for Genetic Engineering in Plants
Food security has been threatened with decreasing crop yields and increasing food consumption in the wake of population growth, climate change, increasing shortage of arable land, and crop usage as raw materials [1,2]. Classical plant breeding to obtain plants with preferred genotypes requires crossing and selection of multiple plant generations, which disallows introduction of traits that do not currently exist in the species. A technique that enables specific horizontal gene transfer stands to greatly benefit the agricultural industry by conferring desirable traits to plants, such as increased yield, abiotic stress tolerance, and disease and pest resistance [3].
Genetic engineering has recently seen major advances in animal systems, though progress has lagged in plants. When compared to the numerous and diverse gene and protein delivery methods developed for animal systems, significantly fewer methods exist for plants (Figure 1, Key Figure). Broadly, modern genetic transformation of plants entails two major steps: genetic cargo delivery and regeneration of the transformed plant, the necessity and difficulty of the latter being highly dependent on what delivery method is used and whether stable transformation is desired. Regeneration procedures involve three parts: the induction of competent totipotent tissue, tissue culture to form calli (see Glossary), and selection and progeny segregation. Regeneration protocols are dominated by complex hormone mixtures, which are heavily species and tissue dependent, making protocol optimization the key to increasing procedure efficacy. The challenge of genetic cargo delivery to plants is attributed to the presence of the multilayered and rigid plant cell wall, otherwise absent in animal cells, which poses an additional physical barrier for intracellular delivery of biomolecules and is one of the key reasons for the slower implementation and employment of genetic engineering tools in plants [4].
Amongst conventional plant biomolecule delivery approaches, Agrobacterium-mediated and biolistic particle delivery are the two most established and preferred tools for plant genetic transformations (Box 1). Current biomolecule delivery methods to plants experience challenges that hinder their scope of use (Table 1). Methods such as electroporation, biolistics, Agrobacterium-mediated delivery, or cationic delivery typically target immature plant tissue (calli, meristems, or embryos). These methods require the regeneration of genetically modified progeny plants, which can be time-consuming and challenging, whereby efficient protocols have only been developed for a narrow range of plant species. Biolistic particle delivery circumvents the cell wall via mechanical force, but often damages portions of target tissue in the process and yields low levels of gene expression that is often sparse and sporadic. Agrobacterium-mediated delivery is subject to orthogonal challenges, the largest being that Agrobacterium displays narrow host and tissue specificity, even between specific cultivars of the same species [5]. Agrobacterium generally experiences lower transformation efficiency for both delivery and regeneration in monocotyledonous plants (monocots) over dicotyledonous plants (dicots). Additionally, Agrobacterium yields random DNA integration, which can cause disruption of important genes, or insertion into sections of the genome with poor or unstable expression [6]. Random DNA integration, however, can be prevented by utilizing magnifection with nonintegrating viruses [7], or by using a plasmid deficient in transfer DNA (T-DNA) insertion [8].
Box 1. Common Gene Delivery Methods in Plants.
Agrobacterium-Mediated Transformation
Agrobacterium tumefaciens is a soil bacterium that infects a wide range of dicots, causing crown gall disease. The formation of a gall on the host plant is achieved via the stable transfer, integration, and expression of bacterial DNA in infected plants. Engineering of the Agrobacterium plasmid by substitution of the gall-inducing virulence genes with genes of interest confers the ability of Agrobacterium to transform the host plant. For this reason, Agrobacterium has been harnessed as a tool for plant genetic transformation since the early 1980s [107].
Genetic transformation occurs through a process involving T-DNA export, targeting, and insertion into the plant nuclear genome. The export of T-DNA from the bacterium to the plant cell is facilitated by the activity of virulence genes present in the tumor inducing-plasmid of Agrobacterium, but are not themselves transferred. These virulence genes are expressed in the presence of phenolic inducers, such as acetosyringone, produced by wounded plant cells. Agrobacterium attaches to plant cells, where border sequences on either side of the T-strand (a single-stranded copy of the T-DNA sequence) are cleaved. The T-strand is then carried by a transporter with a nuclear localization sequence and integrated into the plant nuclear genome. Integration occurs at random positions in the genome via nonhomologous recombination, a repair pathway for double-stranded breaks in DNA.
Gene Gun-Mediated Transformation
A form of biolistic particle delivery (also called particle bombardment), the gene gun, is a physical method that is commonly utilized for plant genetic transformations. Developed in 1982 by Sanford and colleagues [108], the process involves gold or tungsten microparticles (or microcarriers) coated with genetic cargo that are accelerated by pressurized helium (He) gas into plant cells, rupturing cell walls and membranes. The gene gun consists of three main parts: a rupture disk, macrocarrier (holding microcarrier particles), and stopping screen. The rupture disk is a membrane designed to burst at a critical pressure of He gas. When He gas is accelerated to the desired pressure, the rupture disk bursts, creating a shock wave that propels the macrocarrier towards the plant cells. The macrocarrier’s momentum is stopped by the stopping screen, which allows genetic cargo-loaded microcarriers to pass and enter the plant cells.
Unlike Agrobacterium-mediated transformation, biolistic delivery can result in transformation of the nuclear, plastidal, or mitochondrial genomes due to the nonspecific localization of genetic cargo. Consequently, more DNA needs to be delivered with biolistic delivery than Agrobacterium-mediated delivery when targeting the nuclear genome.
Table 1.
Scope of Use Summary for Plant Biomolecule Delivery Methods
| Delivery method | Adverse effects of delivery | Target species/tissue | Cargo type and sizea | Limitations |
|---|---|---|---|---|
| Physical | ||||
| Biolistic particle-(gene gun) mediated delivery | Damage to target tissue & cargo, low penetration depth, random integration | Depends on tissue typeb/calli, embryos, leaves | DNA, siRNA, miRNA, ribonucleoproteins (RNPs), large cargo size | Targeting leaves requires detachment from plant, which limits time to observe delivery effects; targeting embryos requires laborious regeneration protocols, the effectiveness of which is highly species/cultivar-dependent |
| Electroporation | Damage to target tissue, nonspecific transport of material through pores may lead to improper cell function | Unlimited/protoplastsc, meristems, pollen grains | Nucleic acids (DNA, siRNA, miRNA) | Limited cargo-carrying capacity |
| Chemical | ||||
| Polymer-mediated delivery | High charge densities induce cytotoxicity | Species amenable to protoplast regeneration/protoplastsc | Nucleic acids (DNA, siRNA, miRNA) | Regeneration is highly inefficient for most species in transient studies and requires tissue culture |
| Biological | ||||
| Agrobacterium-mediated delivery | Can lead to apoptosis and necrosis, random integration | Narrow range of plant species, especially restricted from monocotsd/mature plants, immature tissue, protoplasts | Limited to DNA, large cargo size | Leaf-targeted delivery is transient and gene edits are not transmitted to progeny, but allow diverse biological studies; requires tissue culture (except Arabidopsis) to generate progeny; exhibits high host-specificity |
| Viral delivery | Virus integration (can be mitigated by using nonintegrating viruses) | Host plant species restrictions/mature plants, meristems | Nucleic acids (DNA, siRNA, miRNA), very limited cargo size | Highly limited cargo-carrying capacity |
While most biomolecule delivery methods to plants can deliver a variety of gene editing reagents, DNA plasmids are arguably the most common cargo of interest; DNA loading capacities are a useful metric for the upper limit for cargo sizes each method can sustain.
While biolistic particle-mediated delivery can theoretically be utilized in unlimited target species, the ability to target species depends on the target tissue (by extension, cell wall structural strength) and capability of available equipment.
The use of protoplasts as target tissue necessitates regeneration protocols and progeny segregation that are time-consuming and are challenged by the limited plant species amenable to protoplast regeneration.
Progress has been made on increasing transformation efficiency in recalcitrant monocots [9].
In sum, plant genetic engineering has lagged behind progress in animal systems; conventional methods of biomolecule delivery to plants remain challenged by intracellular transport through cell walls, and in turn limit plant genetic transformation efficacy. To date, plant biotechnology lacks a method that allows passive delivery of diverse biomolecules into a broad range of plant phenotypes and species without the aid of external force and without causing tissue damage. We posit nanotechnology as a key driver in the creation of a transformational tool to address delivery challenges and enhance the utility of plant genetic engineering.
Nanoparticle-Mediated Biomolecule Delivery in Animal Systems
Nanoparticles as Molecular Transporters in Living Systems
Nanotechnology has advanced a variety of fields, including manufacturing, energy, and medicine. Of particular interest is the use of nanoparticles (NPs) (Box 2) as molecular transporters in cells, an area that has largely focused on molecular delivery in animal systems. NPs allow manipulation on a subcellular level, giving rise to a previously unattainable degree of control over exogenous interactions with biological systems. Therefore, the impact of NPs as drug and gene delivery vehicles in animals has been nothing short of revolutionary.
Box 2. Nanoparticles.
Nanoparticles (1–100 nm in at least one dimension) can be engineered with varied compositions, morphologies, sizes, and charges, enabling tunable physical and chemical properties. Ranging from zero to three dimensional, NPs are novel tools that have a wide range of applications, including but not limited to energy storage, sensing devices, and biomedical applications [109,110].
In addition to their high degree of tunability, NPs possess several advantages that validate their recent widespread use, with particular emphasis in the biomedical industry. Most NPs can be prepared with consistent properties for low batch-to-batch variability, and can be designed to target biological systems, tissues, cells, or subcellular structures with high specificity [52]. Moreover, NP-mediated gene and drug delivery can overcome common issues faced with viral vectors; NPs are often less immunogenic and oncogenic and can carry diverse and larger cargo, although the increased NP sizes when biomolecules are surface-loaded raise the challenge of bypassing biological barriers [111]. Furthermore, the effects of NP use have yet to be thoroughly studied, though existing research points to nanoparticle chemistry, size, and dose as tunable parameters to control cytotoxicity[112,113].
NPs are typically classified based on morphology and chemical properties. The most common categories include polymeric [114], lipid [115], magnetic [116], metallic [117], and carbon-based NPs [118]. NPs can be synthesized with either a top-down or bottom-up approach using techniques such as lithography [119], deposition [120], and self-assembly [121].
In NP-based delivery, a variety of strategies are employed to load NPs with the desired cargo. Physical techniques such as encapsulation or entrapment are commonly used in drug delivery to ensure the progressive release of drugs. Chemical techniques where the NP surface is modified for cargo grafting are in development, including noncovalent conjugation (electrostatic interaction [122], π-π stacking [123]) and covalent conjugation [23].
The small size of NPs and their highly tunable chemical and physical properties have enabled NP engineering for NPs to bypass biological barriers and even localize NPs in subcellular domains of CHO and HeLa cells, among others [10–13]. NPs serve as nonviral, biocompatible, and noncytotoxic vectors that can transport a range of biomolecules [small molecules, DNA, siRNA, miRNA, proteins, and ribonucleoproteins (RNPs)] [14–19] to biological cells. To this end, various features of NPs, including size, shape, functionalization, tensile strength, aspect ratio, and charge, have been tuned for efficient intracellular biomolecule delivery to animal systems. Furthermore, ‘smart’ NPs have been developed to achieve responsive release of cargo for increased control of site-specificity [20]. Various NPs have been manufactured and are responsive to a range of stimuli, including temperature [21], pH [22], redox [23], and the presence of enzymes [24].
Outlook and Implications for Nanocarriers in Plant Science
In contrast to the proliferate studies demonstrating NP-mediated delivery in animals, analogous research in plants is relatively sparse (Figure 1), owing to the transport challenge imposed by the plant cell wall, which renders biomolecule delivery more challenging than for most mammalian systems.
Nevertheless, knowledge gained from biomolecule delivery to animals provides a blueprint for translation to plant systems, and could accelerate advancements in NP-mediated plant biomolecule delivery. NP-mediated delivery may overcome the three foremost limitations of current delivery techniques in plant systems by controlling NP size to traverse the cell wall, tuning charge and surface properties to carry diverse cargo, and greater breadth in utility across plant species.
NP-mediated delivery in animals has successfully carried many types of cargo indiscriminately, whereby certain methods for plants, such as Agrobacterium, can only deliver DNA. For instance, Wang and colleagues report NP-mediated RNP delivery to mammalian cells via lipid encapsulation [25]. Additionally, plastid engineering is not achievable with Agrobacterium, which only targets the plant nuclear genome and cannot target the chloroplast or mitochondrial genomes. Conversely, targeting moieties can be attached to NPs to obtain subcellular localization and modification of the desired genome. Hoshino and coworkers demonstrate the delivery of quantum dots to the nucleus and mitochondria of Vero kidney cells using respective localizing signal peptides [26]. Active targeting and controlled release is not achievable with conventional plant biomolecule delivery methods, but has been demonstrated in animal systems with NP-based delivery. Davis and colleagues designed a polymeric NP with a human transferrin protein-targeting ligand and polyethylene-glycol (PEG) on the NP exterior to deliver siRNA to human melanoma tumor cells, specifically [15]. Additionally, Lai and coworkers accomplished stimuli-responsive controlled release of drug molecules and neurotransmitters encapsulated within mesoporous silica NPs (MSNs) to neuroglial cells [27]. Drawing inspiration from progress in NP-mediated delivery for animal systems, NP-mediated controlled delivery and release of biomolecules without species limitations in plants is a forthcoming goal.
NP-Mediated Biomolecule Delivery to Plants
NP–Plant Interactions
To date, most literature on NP–plant systems focuses on plant-based metallic nanomaterial synthesis [28], agrochemical delivery [29], and NP uptake, showing both valuable and deleterious effects on plant growth [30,31]. Dicot and monocot plants exhibit variable degrees of direct uptake of many NP types, including MSNs [32], carbon nanotubes (CNTs) [33], quantum dots [34], and metal/metal oxide NPs [35–37]. Once uptaken, certain types of NPs exhibit phytotoxicity via vascular blockage, oxidative stress, or DNA structural damage [30]. Conversely, NPs have been shown to improve root and leaf growth, and chloroplast production [31]. Tradeoffs between phytotoxicity and growth enhancement as a function of species, growth conditions, NP properties, and dosage are not well understood and call for more studies with a focus on NP physical and chemical properties. Closing the knowledge gap in plant physiological response to NP uptake is important and should be pursued in parallel with the enhancement of plant science using engineered nanomaterials, as the ‘nanorevolution’ in targeted delivery to animals suggests tremendous potential for analogous progress in plants.
Heuristics for Nanocarrier Design
While a complete structure–function landscape of physical and chemical NP properties that drive cargo loading and cellular internalization remains elusive, a heuristic approach to nanocarrier design is a useful starting point. NP uptake and transport throughout plant tissue is limited by pore diameters, setting size exclusion limits (SELs) for various tissues and organs that are discussed extensively in the literature [30,38–43]. The cell wall is commonly thought to exclude particles >5–20 nm, although recently NPs up to 50 nm in diameter have been reported as cell wall-permeable through unclear mechanisms [38,41]. For genetic engineering applications, where cytosolic or nuclear localization is necessary to affect gene function, the plasma and nuclear membranes pose additional barriers to delivery. In practice, the cell wall (SEL <50 nm) plays a dominant role in NP size internalization limitations, as the cell membrane SEL is much larger (>500 nm) [38]. NP charge and shape greatly influence cell membrane translocation and thus these properties are central to nanocarrier optimization [44]. Plant cellular uptake can occur through energy-dependent (endocytosis) and energy-independent (direct penetration) pathways that are not well understood. It is commonly reported that internalization is faster and more efficient for cationic NPs versus anionic NPs, due to cationic NP binding with the negatively charged cell membrane [44]. This charge preference has been demonstrated in protoplasts and walled plant cells [45,46].
Endosomal escape is critical for subcellular delivery, as vesicle-entrapped NPs can be trafficked for degradation or exocytosis, and remain inaccessible for downstream processing if trapped in the endosome. Subcellular localization of NPs in plants is not well understood but will depend on the uptake pathway, as endocytic proteins and vesicle cargo play a role in endosome fate [47], whereby direct cell penetration bypasses endosomal vesicle formation entirely. Serag and colleagues report CNT internalization in protoplasts through both direct penetration and endocytosis, supporting prior demonstrations in mammalian cells that high aspect ratio NPs undergo vesicle-free internalization [48,49]. However, for Serag and colleagues, direct penetration was only observed for cell wall-impermeable multiwalled CNTs in protoplasts [48,49], motivating further studies for plant cell wall internalization by high aspect ratio NPs. Wong and colleagues have demonstrated passive internalization of single-walled CNTs in extracted chloroplasts [129] through a mechanism dependent on NP size and zeta potential [130]. Cationic, pH-buffering polymers are well-known endosome disruption agents [50] that can function as ligands to improve endosomal escape. Chang and colleagues report energy-independent internalization to walled root cells by organically functionalized spherical MSNs [51]. Notably, endocytosed single-walled CNTs in plants are trafficked to vacuoles but localize in the cytosol when loaded with DNA [33,48].
Most NPs are amenable to surface adsorption (physisorption) of biomolecules as a simple conjugation strategy. However, physisorption may be unstable depending on the specific NP and cargo, and thus electrostatic interactions are preferable for noncovalent cargo loading [52]. Cationic surface chemistry not only enhances endocytic uptake and escape, but is also amenable to electrostatic loading of genetic cargo via attraction with negatively charged DNA and RNA. Covalent NP surface functionalization is typically achieved by one of many of ‘click’ chemistries [53]. Notably, covalent attachment of thiolated DNA and proteins to gold NPs has shown recent success [54] but the field remains open to new strategies for covalent bioconjugation, especially for applications in plants. As an alternative to surface functionalization, porous NPs such as MSNs can be internally loaded with macromolecules or small chemicals alike, for controlled intracellular release [55].
NPs with some or all of the properties mentioned above have demonstrated successful biomolecule delivery in plants and are good starting points for choosing the appropriate NP, ligand, and cargo for a given application. However, it should be noted that nanocarrier design is a complex, multivariable optimization process, such that success will likely require tweaking of these heuristics for different systems until a complete NP structure–function relationship is established for plant systems.
Nanomaterials for Plant Genetic Engineering
NPs are valuable materials for intracellular biomolecule delivery, owing to their ability to cross biological membranes, protect and release diverse cargoes, and achieve multifaceted targeting via chemical and physical tunability. Such properties have enabled NPs to revolutionize targeted delivery and controlled release in mammalian systems. However, nanocarrier delivery in plants remains largely underexplored due to the cell wall, which is typically overcome by chemical or mechanical aid (Figure 1). Passive biomolecule delivery to plants is promising for minimally invasive, species-independent, in vivo genetic engineering of plants, especially for transient expression in somatic tissue (Table 2). The potential of NP-based plant delivery methods is underscored by the limitations of in vitro plant studies in general, wherein regeneration capacity varies widely across species, genotype, and even within a single plant depending on developmental age of source tissue [56]. Currently, stable transformation requires progeny regeneration from embryogenic calli regardless of the delivery method (Table 2). Thus, parallel optimization of delivery and regeneration is necessary to improve efficiency and expand stable transformation capabilities to all plant species.
Table 2.
Challenges in Plant Genetic Engineering and Proposed Advantages of NP Delivery
| Desired outcome | Nonheritablea (somatic/transient expression) | Heritable (germline/stable transformation) | |||
|---|---|---|---|---|---|
| Targeted tissue | Leaves | Roots | Protoplasts | Zygotic embryo | Somatic embryogenic calli |
| Tissue-specific biological and experimental challenges |
|
|
|
|
|
| Proposed advantages of NP delivery |
|
|
|
|
|
While these somatic tissues (leaves, roots, protoplasts) are most commonly targeted for transient expression experiments, heritable outcomes may be derived through somatic embryogenesis (dedifferentiation of somatic tissue).
Key Figure
Nanoparticle (NP)-Mediated Genetic Cargo Delivery to Animals and Plants
Figure 1.
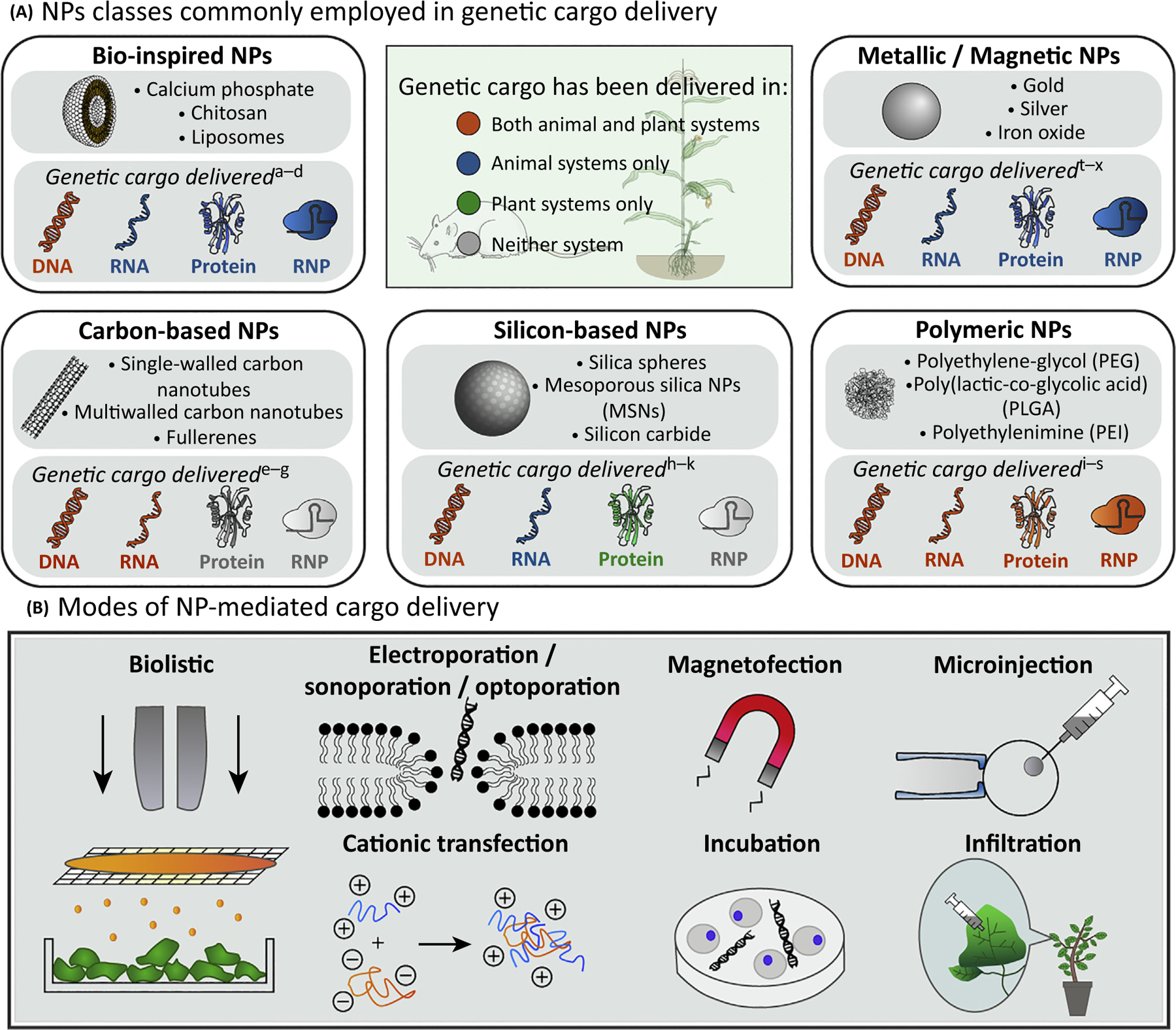
(A) NPs commonly used for biomolecule delivery in both animal and plant systems cover five major categories: bio-inspired, carbon-based, silicon-based, polymeric, and metallic/magnetic. We provide a visual comparison of delivery of various genetic cargo [DNA, RNA, proteins (site-specific recombinases or nucleases), and ribonucleoprotein (RNP)] with each of the five NP types across animal and plant systems. It is evident that NP-mediated delivery has been utilized with a greater variety of genetic cargo in animals than in plants. (b) NP-mediated cargo delivery is conducted via several means. Physical methods include creating transient pores in the cell membrane with electric fields, sound waves, or light, magnetofection, microinjection, and biolistic particle delivery. Nonphysical methods include the use of cationic carriers, incubation, and infiltration. a[64], b[86], c[87], d[88], e[89], f[68], g[90], h[91], i[45], j[92], k[58], l[93], m[94], n[95], o[96], p[97], q[98], r[99], s[81], t[100], u[63], v[101], w[102], x[54].
In 2007, Torney and colleagues were the first to demonstrate NP co-delivery of DNA and chemicals to Nicotiana tabacum plants via biolistic delivery of 100–200-nm gold-capped MSNs [45]. In this study, a chemical expression inducer was loaded into MSN pores (~3 nm) that were subsequently covalently capped with gold NPs. The capped MSNs were then coated with GFP plasmids and delivered by gene gun to N. tabacum cotyledons, wherein GFP expression was triggered upon uncapping and release of the expression inducer [45]. This seminal paper demonstrated proof of concept that strategies common for NP delivery of DNA to mammalian systems can be adapted to plants. Notably, gold MSNs were also used for biolistic co-delivery of DNA and proteins, namely GFP and Cre-recombinase, demonstrating the ability of MSNs to deliver proteins for gene editing [58]. Many delivery strategies still require a gene gun, electromagnetic field, or protoplast PEG-transfection [58–63] as NP structure–function parameters have not yet been fully optimized to passively bypass the cell wall (Table 3). However, for systems where mechanical or chemical aid is necessary for NP internalization, the small size and high surface area of nanocarriers still offers superior performance over conventional methods. For instance, Torney and colleagues’ MSN study achieved transgene expression with 1000 × less DNA than the tens to hundreds of micrograms of DNA typically required for conventional PEG-transfection in protoplasts [45].
Table 3.
Select Summary of NP-Mediated Genetic Engineering in Plants
| NP type | Cargo | Plant species; cell/tissue type | Delivery method | Comments | Year | Refs | |
|---|---|---|---|---|---|---|---|
| With external aid | Gold capped MSNs | GFP plasmid; chemical expression inducer | N. tabacum cotyledons; Z. mays embryos | Biolistic | Co-delivery and controlled release of DNA and chemicals | 2007 | [45] |
| Poly-L-lysine coated starch NPs | GFP plasmid | Dioscorea zingiberensis C.H. Wright calli suspension | Sonoporation | 5% transient expression efficiency; some integration occurs | 2008 | [60] | |
| Gold-plated MSNs | GFP and mCherry plasmids; GFP protein | Allium cepa epidermis tissue | Biolistic | DNA and protein co-delivery | 2012 | [59] | |
| Magnetic gold NPs | β-glucuronidase (GUS) plasmid | Brassica napus protoplasts and walled cell suspension | Magnetic field | Transient GUS expression | 2013 | [61] | |
| Gold-plated MSNs | AmCyan1 and DsRed2 plasmids; Cre protein | Z. mays embryos | Biolistic | DNA and protein co-delivery; both transient and stable expression | 2014 | [58] | |
| Dimethylaminoethyl methacrylate (DMAEM) polymer NPs | Yellow fluorescent protein (YFP) and GFP plasmids | N. tabacum and Ceratodon purpureus protoplasts | PEG transfection | Both transient and stable expression | 2017 | [62] | |
| Magnetic Fe3O4 NPs | Selectable marker gene plasmids | Gossypium hirsutum pollen | Magnetic field | ~1% efficiency for generating stable transgenic seeds | 2017 | [63] | |
| In vitro without external aid | Polyamidoamine (PAMAM) dendrimer NPs | GFP plasmid | Agrostis stolonifera L. calli | Passive | 48.5% cells showed transient expression | 2008 | [65] |
| Calcium phosphate NPs (CaPNPs) | GUS plasmid | Brassica juncea hypocotyl explants | Passive | 80.7% stable transformation efficiency | 2012 | [64] | |
| Organically functionalized CNTs | YFP plasmid | N. tabacum protoplasts and leaf explants | Passive | Both transient and stable expression | 2015 | [66] | |
| In vivo without external aid | Organically functionalized MSNs | mCherry plasmid | A. thaliana roots | Passive | 46.5% transient expression efficiency | 2013 | [51] |
| PAMAM dendrimer NPs | Double-stranded DNA for RNA interference | A. thaliana roots | Passive | Developmental gene silencing led to systemic phenotypes | 2014 | [67] | |
| Polymer functionalized CNTs | GFP plasmid; siRNA for transgenic GFP silencing | E. sativa, N. benthamiana, and T. aestivum leaves | Passive | 95% transient silencing efficiency; transient expression in mature leaves | [68] |
A few recent examples show promise for NP-mediated passive delivery to plants in vitro [64–66] and in vivo [51,67] in, for example, N. tabacum protoplasts [66] and Arabidopsis thaliana roots [51,67], respectively (Table 3). Demirer and colleagues have recently achieved passive delivery of DNA plasmids and protected siRNA using functionalized CNT NPs for transient GFP expression in Eruca sativa (arugula) leaves and transient silencing of constitutively expressed GFP in transgenic Nicotiana benthamiana leaves [68]. This study also demonstrates CNT-mediated transient GFP expression in Triticum aestivum (wheat), indicating the potential for passive NP delivery in both model and crop species with high efficiency and low toxicity. While many more studies are needed to optimize NP properties and functionalization, these early results are promising for further exploration of NPs as a plant biomolecule delivery platform that addresses the shortcomings of conventional methods. Furthermore, with the advent of nuclease-based gene editing technologies (Box 3), it is of great interest to optimize the delivery of these revolutionary genome engineering tools by exploring NP-based delivery strategies for diverse biomolecular cargoes.
Box 3. Traditional Genetic Engineering versus Nuclease-Enabled Genome Editing.
Genetic engineering refers broadly to manipulating a cell’s genome and gene expression profile. Techniques for genetic engineering may cause recombinant protein expression, up/downregulation of a gene, permanent gene knockout, targeted mutations in the host gene, or insertion of large foreign DNA segments into the host genome. Genome modifications may be transient, permanent, or heritable and involve many types of biomolecules (most commonly RNA, DNA, and proteins) which are sometimes taken up passively by cells but often require enhanced delivery techniques, such as gene guns, microinjection, electroporation, sonoporation, nanoparticle-assisted delivery, and engineered bacteria or viruses. In plants, genetic engineering is hindered by the cell wall, requiring delivery methods that are highly host-specific or limited by challenges in plant regeneration.
Nuclease-enabled genome editing refers to techniques where genes are removed or changed with engineered nucleases, a class of enzymes that perform targeted double-stranded breaks (DSBs) at specific locations in the host genome. When nucleases perform DSBs, the cell undergoes homology-directed repair (HDR) or nonhomologous end-joining (NHEJ) to repair the cut. NHEJ is a random, error-prone repair process that involves realignment of a few bases, such that the high error frequency provides a simplistic pathway for gene knockout. HDR is a nonrandom repair process requiring large stretches of sequence homology, allowing for precise edits by introducing customized homologous recombination sequences for gene knockout, knock-in, and targeted mutations. Prominent tools in genome editing are zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR (clustered regularly interspaced short palindromic repeat)-Cas (CRISPR associated) systems. In the 1990s, ZFNs became the first nuclease system engineered for selectable genome editing in bacteria [124]. TALEN and CRISPR-Cas genome editing systems were developed for bacteria and eukaryotes more recently, around 2009 and 2012, respectively [125–128]. Composed of protein complexes containing a DNA-binding domain and a DNA-cleaving domain, ZFNs and TALENs rely on protein/DNA recognition to induce endogenous DNA repair. CRISPR-Cas systems are composed of a nuclease protein (Cas) and a guide RNA (gRNA) with sequence homology to the genomic target, and therefore rely on the formation of a ribonucleoprotein (RNP) complex to induce HDR or NHEJ. While all three systems have their drawbacks, CRISPR-Cas has revolutionized the field of genome editing owing to its relatively superior simplicity, efficiency, and multiplexing ability (i.e., simultaneous editing of different genes) over ZFNs and TALENs.
Genome Editing has Enabled a New Era of Plant Science
Engineered Nucleases for Plant Genome Editing
Engineered nuclease systems, namely ZFNs, TALENs, and CRISPR-Cas, have emerged as breakthrough genome editing tools owing to their high genetic engineering specificity and efficiency (Box 3), whereby CRISPR-Cas has demonstrated increased simplicity, affordability, and multiplexing capabilities over TALENs and ZFNs in plants [69,70]. Since 2012, CRISPR-Cas has shown success for genome editing in both model and crop species, including A. thaliana, N. benthamiana, N. tabacum (tobacco), Oryza sativa (rice), T. aestivum (wheat), Zea mays (corn), Solanum lycopersicum (tomato), and Sorghum bicolor, among others [71,72]. Notably, CRISPR-Cas mutations as small as 1 bp have been conserved through three plant generations [73,74], which is promising for stable transgene-free modified crops. As with traditional genetic engineering of plants, many of the limitations for implementing gene editing tools in plants (low editing efficiency, tissue damage, species limitations, cargo-type limitations) originate in biomolecular transport into plant cells. As such, NP-based biomolecule delivery to plants stands to enable higher-throughput plant genome editing via DNA, single guide RNA (sgRNA), and RNP delivery, and thus warrants a discussion on the state of the plant genome editing field.
Global Landscape of Regulatory Uncertainty towards Genetically Engineered Crops
Genetic engineering of crops has evolved to overcome limitations in traditional breeding, as breeding is slow, laborious, and lacks precise control over plant genotype and phenotype generation. Modern biotechnology enables rapid development of crop variants with disease and pest resistance, stress tolerance, higher yield, and enhanced nutritional value. Since 1996, global genetically modified organism (GMO) cultivation has increased 110-fold to 185 mega-hectares in 2016 [75] (Figure 2). The US is a leader in GMO production but highly regulates production of modified crops, which poses, among other challenges, significant financial barriers to commercialization of new crop variants [76]. The US GMO pipeline is product-based but sensitive to plant pests, such that Agrobacterium automatically triggers regulation, while other methods of gene delivery are often deregulated if the product is nontransgenic [76,77]. European Union GMO regulation is process-based and affects any organism whose genome has been modified other than by mating or natural recombination [78], but includes exceptions for certain types of mutagenesis that will likely exempt modern gene editing [79]. The advent of nuclease-based gene editing (Box 3) has set forth a global reevaluation of the legislation surrounding genetically engineered crops, wherein several leading GMO cultivators have exempted nontransgenic genome-edited plants from regulation (Figure 2). Recently, the USDA officially stated that there are no future plans to include genome-edited plants under the current US regulatory umbrella for GMOs [131]. However, due to differences in regulatory philosophy and public opinion, several countries oppose deregulation of nontransgenic genome-edited plants and it remains unclear how enforcement of GMO status will proceed worldwide in the future [80]. Despite the heterogenous and dynamic global regulatory landscape, nuclease-based genome editing currently plays a critical role in overcoming regulatory restrictions and ensuring scientific progress, as well as commercial implementation of engineered crop variants.
Figure 2.
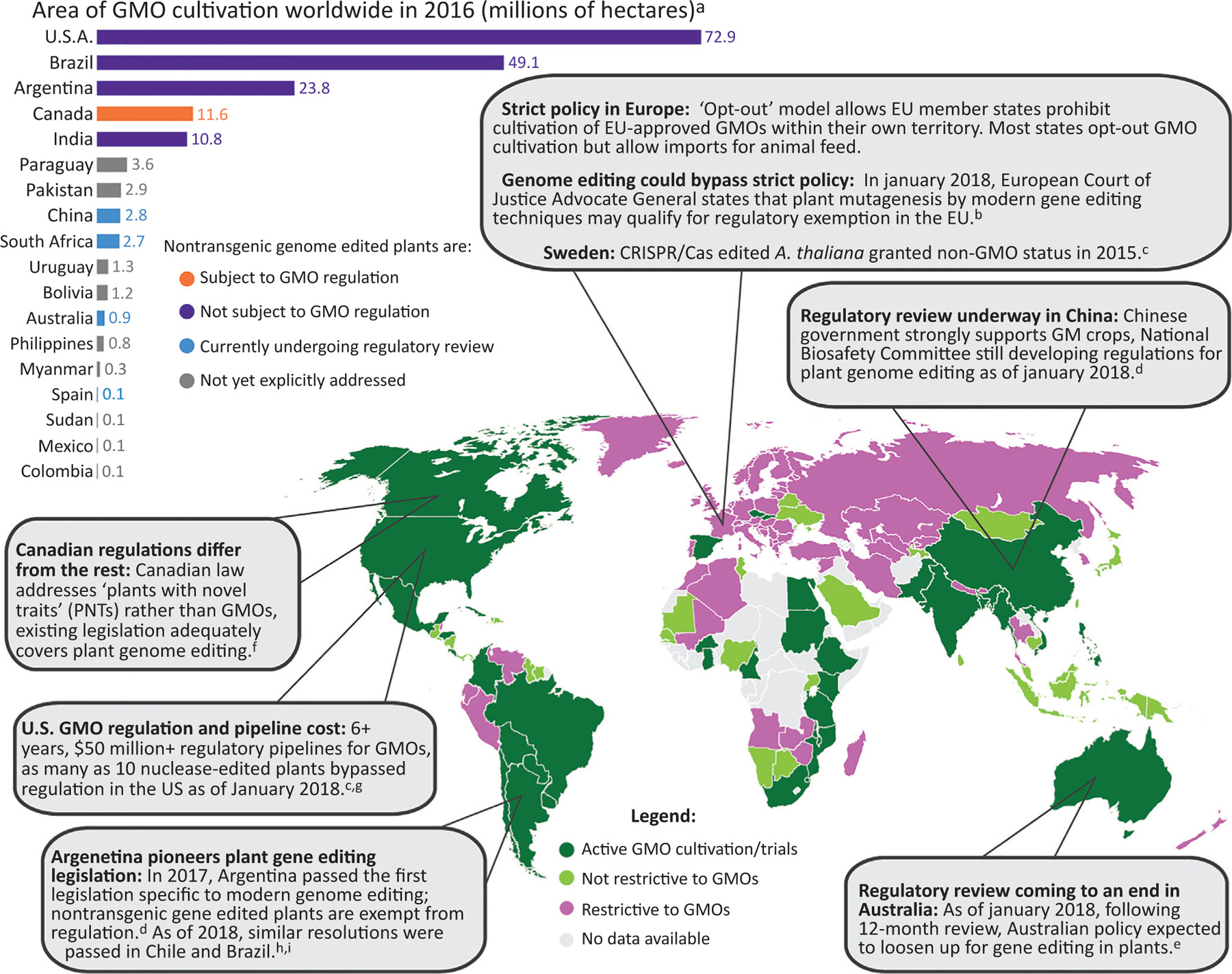
Genetically Modified Organism (GMO) Cultivation and Regulatory Attitudes Worldwide. Despite a long, expensive regulatory pipeline, the US is a leader for GMO cultivation worldwide, followed by Brazil and Argentina, with Argentina being the first to directly address modern genome editing techniques in GMO legislation. European and Australian regulatory attitudes are strict but have recently evolved as of January 2018, suggesting that regulations for genome-edited plants will soon be relaxed in these regions. Nuclease-based edits without transgene integration escape regulation, even in countries with large agricultural GMO industries and complex regulatory systems. Globally, GMO regulation and commercial use is heterogenous and uncertain due to economic, ecological, and sociopolitical complexities. This map is a simplification of the convoluted global landscape regarding genetically engineered crops. ‘Restrictive to GMOs’ indicates a complete or partial ban on GMOs and GMO-derived products for commercial or research purposes. a[75], b[79], c[80], d[103], e[104], f[105], g[106], h[132], i[133].
Nanocarriers Hold Promise for Nuclease-Based Plant Genome Editing
Genome editing tools may increase the throughput of plant molecular biology and genetic studies, and as such could shift the paradigm in regulatory oversight of transgenic plants. Species, amenable tissue, expression strategy (DNA, RNA, or protein), and delivery method contribute to the efficacy of transgene expression or modification and to the propensity of transgene integration into the host genome. ‘DNA-free’ genome editing techniques are increasingly attractive, especially from a regulatory perspective, to eliminate all risk of transgene integration. Recently, RNP delivery has been demonstrated in A. thaliana and O. sativa protoplasts via PEG-transfection [81] and Z. mays embryos via gene gun delivery [82]; the methods used in both of these studies are primarily throughput-limited by challenges in progeny regeneration. The challenge to realizing efficient, stable gene editing in plants is twofold. First, plant germline cells cannot be transformed by any current method (with the exception of Arabidopsis floral dip [83]) and therefore progeny must be regenerated from embryo genic calli. Second, the cell wall imposes a rigid transport barrier to biomolecule delivery, such that conventional delivery in plants is either destructive and inefficient, or host-specific. Thus, the foremost limitation for broad-scale implementation of plant genome editing originates from an inability to target germline cells, and the absence of an efficient and species-independent bio-cargo delivery strategy. While engineered nuclease systems have begun to reveal remarkable potential for the future of plant genome engineering, novel carriers are required to overcome the restrictions of conventional delivery methods, but could also begin to pave the way for efficient progeny regeneration or direct germline editing in plants.
NPs have begun to facilitate and enhance genome editing through efficient and targeted delivery of plasmids, RNA, and RNPs [84]. In mammalian cells, NPs are routinely used for efficient, direct cytosolic/nuclear delivery of Cas-RNPs in many cell types [85], and RNP delivery has been shown to greatly reduce off-target effects in comparison with plasmid-based CRISPR systems [84]. However, in plants, the cell wall has hindered the development of an analogous system that can passively deliver genome editing cargo to mature plants and across species. Thus, there remains much potential for designing NP carriers with diverse cargo loading capabilities (DNA, RNA, proteins) and optimal geometry/chemistry to efficiently bypass the cell wall and membranes in dense plant tissues without external aid. Previous work [51,67,68] shows that some NP formulations are capable of passive internalization in planta with DNA, RNA, or protein cargo. These NP scaffolds, namely CNTs, MSNs, and polymeric NPs, should be further explored for delivering engineered nuclease systems to plants.
Concluding Remarks and Future Perspectives
Genetic engineering of plants has greatly accelerated scientific progress and paved the way for crop variants with improved growth characteristics, disease and pest resistance, environmental stress tolerance, and enhanced nutritional value. In parallel, advances in site-specific genome editing technologies have optimized the precision with which genetic engineering of organisms can be accomplished. However, conventional methods of plant genetic engineering and genome editing are limited in scope. This is primarily due to the cell wall that imposes a barrier to efficient delivery of biomolecules, which could potentially be overcome by NPs. Agrobacterium is a preferred method for plant genetic transformation, but is only effective in a limited range of host species and is an automatic trigger for regulatory oversight in the United States. Biolistic particle delivery and PEG-transfection are effective, host-independent transformation methods, but difficulties in regenerating healthy plant tissue and low-efficiency editing are severe drawbacks to their broad-scale and high-throughput implementation. NPs have recently emerged as a novel method of targeted biomolecule delivery in mammalian cells, especially for clinical applications. However, exploration of nanocarriers for biomolecule delivery in plants remains a nascent field, with much potential for the future of plant biotechnology and genome editing (see Outstanding Questions). Preliminary studies show that NPs with proper surface chemistry and physical properties analogous to those developed for animal systems are capable of delivering biomolecules to plants in vivo and in vitro with improvements over conventional methods. However, as of yet, most nanocarriers in plants still require assistance from conventional methods (i.e., gene gun), or are limited to in vitro studies. To our knowledge, the field of plant bioengineering has yet to fully demonstrate a reliable strategy for NP-mediated passive biomolecule delivery to plants. To realize the full scientific and humanitarian potential lingenetic engineering of both model and crop species, especially with the advent of nuclease-based genome editing, a promising focus will be to optimize NPs as efficient and ubiquitous delivery vessels of diverse biomolecules, tunable across cargo types, species, and tissues, for both transient and stable genetic engineering. However, because germline transformation is currently limited to only one model plant species (Arabidopsis), even a ubiquitous delivery strategy for precise genome editing would be limited by the success of regenerating progeny from somatic tissue. A remarkable, yet conceivable, future accomplishment of NP delivery in plants could be enablement of unprecedented, highly parallel genetic studies that elucidate the precedents for success in tissue regeneration, and the direct manipulation of germline plant cells.
Outstanding Questions.
Are there nanoparticle varieties yet to be discovered for efficient biomolecule delivery in plants, or do we lack knowledge of, or control over, optimal nanoparticle modifications for applications in plant systems?
Can we narrow the current design space to a single nanoparticle type with tunable functionalization for passive delivery in plants, regardless of cargo type, plant species, and tissue variety?
How might we gain a better mechanistic understanding of nanoparticle internalization into plant cells, and how can we harness this knowledge towards rational design of nanoparticles for a range of biological delivery applications?
Will challenges in biomolecule delivery and progeny regeneration always remain decoupled, or will nanoparticle delivery enable significant increase in throughput and efficiency of genetic studies on plant regenerative biology and stable transformation?
While genome editing by induced nonhomologous end-joining does not invoke regulatory oversight in many countries, how will genome edits introduced by homology-directed repair (where integration of a repair template is necessary) be classified from a legislative standpoint?
How can scientists, the public, and regulatory bodies create a space for open communication to address the risks of introducing crop variants to the environment, while continuing to enable scientific progress and commercialization of sustainable and resilient crop variants?
Highlights.
Plant biotechnology is key to ensuring food and energy security; however, biomolecule delivery and progeny regeneration continue to be key challenges in plant genetic engineering.
Conventional biomolecule delivery methods in plants have critical drawbacks, such as low efficiency, narrow species range, limited cargo types, and tissue damage.
Advances in nanotechnology have created opportunities to overcome limitations in conventional methods: nanoparticles are promising for species-independent passive delivery of DNA, RNA, and proteins.
The advent of nuclease-based genome editing (e.g., CRISPR-Cas9) has ushered in a new era of precise genetic engineering that, among other impacts, has enabled the development of genetically engineered crops without harsh regulatory restrictions.
The potential of nanoparticles to overcome limitations in conventional delivery makes them excellent candidates for delivery of nuclease-based genome editing cargo, thus making nanoparticle delivery a critical technology for the advancement of plant genetic engineering.
Glossary
- Callus
a mass of undifferentiated cells that can be used to regenerate plants.
- Cultivars
short for cultivated varieties, a group of plants with desired characteristics that have been selected from a naturally occurring species and are passed through propagation.
- Dicotyledonous plants
one of the two major groups of flowering plants. The eponymous term originates from the presence of two embryonic leaves upon germination. Additionally, dicots can be distinguished from monocots by a number of characteristics that include leaf veins, vascular bundles, root development, floral bundles, and pollen. See monocotyledonous plants.
- Electroporation
a physical transfection method where an electric field is applied to create temporary pores in cell membranes for the uptake of genetic cargo into a cell.
- Explant
any segment of a plant that is removed to initiate a culture.
- In planta
a transformation paradigm involving the genetic transformation of any segment of a plant without the need for tissue culture and regeneration.
- Magnifection
delivering virus vectors using Agrobacterium T-DNA transfer.
- Meristems
regions of tissue containing undifferentiated cells.
- Monocotyledonous plants
one of the two major groups of flowering plants that have one embryonic leaf upon germination. Monocots include crops that make up the majority of a balanced diet, such as rice, wheat, and barley. See dicotyledonous plants.
- Passive delivery
transport of cargo across cell wall and membrane to an intracellular location without the use of mechanical force.
- Protoplasts
plant cells with their cell walls removed, typically through either mechanical or enzymatic means.
- Recalcitrant
a species of plant that is difficult to genetically transform and regenerate into mature plants. Often used in the context of Agrobacterium-mediated transformation.
- Transgene
a gene taken from an organism and transferred into the genome of another. Consequently, transgene integration results in transgenic plants.
References
- 1.Ray DK et al. (2013) Yield trends are insufficient to double global crop production by 2050. PLoS One 8, e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao C et al. (2017) Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. U. S. A 114, 9326–9331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdallah NA et al. (2015) Genome editing for crop improvement: challenges and opportunities. GM Crops Food 6, 183–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azencott HR et al. (2007) Influence of the cell wall on intracellular delivery to algal cells by electroporation and sonication. Ultrasound Med. Biol 33, 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nyaboga E et al. (2014) Agrobacterium-mediated genetic transformation of yam (Dioscorea rotundata): an important tool for functional study of genes and crop improvement. Front. Plant Sci 5, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelvin SB (2017) Integration of Agrobacterium T-DNA into the plant genome. Annu. Rev. Genet 51, 195–217 [DOI] [PubMed] [Google Scholar]
- 7.Gleba Y et al. (2005) Magnifection—a new platform for expressing recombinant vaccines in plants. Vaccine 23, 2042–2048 [DOI] [PubMed] [Google Scholar]
- 8.Stoddard T et al. Cellectis. Agrobacterium-mediated genome modification without t-dna integration, WO2016125078A1 [Google Scholar]
- 9.Lowe K et al. (2016) Morphogenic regulators baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng X et al. (2014) Nanoparticle-directed sub-cellular localization of doxorubicin and the sensitization breast cancer cells by circumventing GST-mediated drug resistance. Biomaterials 35, 1227–1239 [DOI] [PubMed] [Google Scholar]
- 11.Dekiwadia CD et al. (2012) Peptide-mediated cell penetration and targeted delivery of gold nanoparticles into lysosomes. J. Pept. Sci 18, 527–534 [DOI] [PubMed] [Google Scholar]
- 12.Mout R et al. (2017) Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 11, 2452–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong J et al. (2015) A smart polymeric platform for multistage nucleus-targeted anticancer drug delivery. Biomaterials 65, 43–55 [DOI] [PubMed] [Google Scholar]
- 14.Mao H-Q et al. (2001) Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J. Control Release 70, 399–421 [DOI] [PubMed] [Google Scholar]
- 15.Davis ME et al. (2010) Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan M et al. (2010) A novel intracellular protein delivery platform based on single-protein nanocapsules. Nat. Nanotechnol 5, 48–53 [DOI] [PubMed] [Google Scholar]
- 17.Sengupta S et al. (2005) Temporal targeting of tumour cells and neovasculature with a nanoscale delivery system. Nature 436, 568–572 [DOI] [PubMed] [Google Scholar]
- 18.Swyer T et al. (2014) Nanoparticle oxygen delivery to the ischemic heart. Perfusion 29, 539–543 [DOI] [PubMed] [Google Scholar]
- 19.Haham M et al. (2012) Stability and bioavailability of vitamin D nanoencapsulated in casein micelles. Food Funct. 3, 737. [DOI] [PubMed] [Google Scholar]
- 20.Karimi M et al. (2016) Smart micro/nanoparticles in stimulus-responsive drug/gene delivery systems. Chem. Soc. Rev 45, 1457–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ta T et al. (2014) Localized delivery of doxorubicin in vivo from polymer-modified thermosensitive liposomes with MR-guided focused ultrasound-mediated heating. J. Control. Release 194, 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mu Q et al. (2015) Stable and efficient paclitaxel nanoparticles for targeted glioblastoma therapy. Adv. Healthc. Mater 4, 1236–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svenson S et al. (2016) Tumor selective silencing using an RNAi-conjugated polymeric nanopharmaceutical. Mol. Pharm 13, 737–747 [DOI] [PubMed] [Google Scholar]
- 24.Hou X-F et al. (2015) Enzyme-responsive protein/polysaccharide supramolecular nanoparticles. Soft Matter 11, 2488–2493 [DOI] [PubMed] [Google Scholar]
- 25.Wang M et al. (2016) Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. U. S. A 113, 2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoshino A et al. (2004) Quantum dots targeted to the assigned organelle in living cells. Microbiol. Immunol 48, 985–994 [DOI] [PubMed] [Google Scholar]
- 27.Lai CY et al. (2003) A mesoporous silica nanosphere-based carrier system with chemically removable CdS nanoparticle caps for stimuli-responsive controlled release of neurotransmitters and drug molecules. J. Am. Chem. Soc 125, 4451–4459 [DOI] [PubMed] [Google Scholar]
- 28.Vijayaraghavan K and Ashokkumar T (2017) Plant-mediated biosynthesis of metallic nanoparticles: a review of literature, factors affecting synthesis, characterization techniques and applications. J. Environ. Chem. Eng 5, 4866–4883 [Google Scholar]
- 29.Baker S et al. (2016) Nanoagroparticles emerging trends and future prospect in modern agriculture system. Environ. Toxicol. Pharmacol 53, 10–17 [DOI] [PubMed] [Google Scholar]
- 30.Tripathi DK et al. (2017) An overview on manufactured nanoparticles in plants: uptake, translocation, accumulation and phytotoxicity. Plant Physiol. Biochem 110, 2–12 [DOI] [PubMed] [Google Scholar]
- 31.Zuverza-Mena N et al. (2017) Exposure of engineered nanomaterials to plants: insights into the physiological and biochemical responses-a review. Plant Physiol. Biochem 110, 236–264 [DOI] [PubMed] [Google Scholar]
- 32.Hussain HI et al. (2013) Mesoporous silica nanoparticles as a biomolecule delivery vehicle in plants. J. Nanoparticle Res 15, 1676 [Google Scholar]
- 33.Liu Q et al. (2009) Carbon nanotubes as molecular transporters for walled plant cells. Nano Lett. 9, 1007–1010 [DOI] [PubMed] [Google Scholar]
- 34.Koo Y et al. (2015) Fluorescence reports intact quantum dot uptake into roots and translocation to leaves of Arabidopsis thaliana and subsequent ingestion by insect herbivores. Environ. Sci. Technol 49, 626–632 [DOI] [PubMed] [Google Scholar]
- 35.Kurepa J et al. (2010) Uptake and distribution of ultrasmall anatase TiO2 alizarin red S nanoconjugates in Arabidopsis thaliana. Nano Lett. 10, 2296–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.González-Melendi P et al. (2008) Nanoparticles as smart treatment-delivery systems in plants: assessment of different techniques of microscopy for their visualization in plant tissues. Ann. Bot 101, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larue C et al. (2012) Accumulation, translocation and impact of TiO2 nanoparticles in wheat (Triticum aestivum spp.): influence of diameter and crystal phase. Sci. Total Environ 431, 197–208 [DOI] [PubMed] [Google Scholar]
- 38.Wang P et al. (2016) Nanotechnology: a new opportunity in plant sciences. Trends Plant Sci. 21, 699–712 [DOI] [PubMed] [Google Scholar]
- 39.Eichert T and Goldbach HE (2008) Equivalent pore radii of hydrophilic foliar uptake routes in stomatous and astomatous leaf surfaces – further evidence for a stomatal pathway. Physiol. Plant 132, 491–502 [DOI] [PubMed] [Google Scholar]
- 40.Eichert T et al. (2008) Size exclusion limits and lateral heterogeneity of the stomatal foliar uptake pathway for aqueous solutes and water-suspended nanoparticles. Physiol. Plant 134, 151–160 [DOI] [PubMed] [Google Scholar]
- 41.Schwab F et al. (2015) Barriers, pathways and processes for uptake, translocation and accumulation of nanomaterials in plants – critical review. Nanotoxicology 10, 1–22 [DOI] [PubMed] [Google Scholar]
- 42.Ma X et al. (2010) Interactions between engineered nanoparticles (ENPs) and plants: phytotoxicity, uptake and accumulation. Sci. Total Environ 408, 3053–3061 [DOI] [PubMed] [Google Scholar]
- 43.Larue C et al. (2014) Foliar exposure of the crop Lactuca sativa to silver nanoparticles: evidence for internalization and changes in Ag speciation. J. Hazard. Mater 264, 98–106 [DOI] [PubMed] [Google Scholar]
- 44.Albanese A et al. (2012) The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng 14, 1–16 [DOI] [PubMed] [Google Scholar]
- 45.Torney F et al. (2007) Mesoporous silica nanoparticles deliver DNA and chemicals into plants. Nat. Nanotechnol 2, 295–300 [DOI] [PubMed] [Google Scholar]
- 46.Zhu Z-J et al. (2012) Effect of surface charge on the uptake and distribution of gold nanoparticles in four plant species. Environ. Sci. Technol 46, 12391–12398 [DOI] [PubMed] [Google Scholar]
- 47.Fan L et al. (2015) Endocytosis and its regulation in plants. Trends Plant Sci. 20, 388–397 [DOI] [PubMed] [Google Scholar]
- 48.Serag MF et al. (2013) Nanobiotechnology meets plant cell biology: carbon nanotubes as organelle targeting nanocarriers. RSC Adv. 3, 4856 [Google Scholar]
- 49.Serag MF et al. (2011) Trafficking and subcellular localization of multiwalled carbon nanotubes in plant cells. ACS Nano 5, 493–499 [DOI] [PubMed] [Google Scholar]
- 50.Selby LI et al. (2017) Nanoescapology: progress toward understanding the endosomal escape of polymeric nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 9, e1452. [DOI] [PubMed] [Google Scholar]
- 51.Chang F-P et al. (2013) A simple plant gene delivery system using mesoporous silica nanoparticles as carriers. J. Mater. Chem. B 1, 5279. [DOI] [PubMed] [Google Scholar]
- 52.Zhao H et al. (2016) Polymer-based nanoparticles for protein delivery: design, strategies and applications. J. Mater. Chem. B 4060, 4060–4071 [DOI] [PubMed] [Google Scholar]
- 53.Lallana E et al. (2012) Click chemistry for drug delivery nanosystems. Pharm. Res 29, 1–34 [DOI] [PubMed] [Google Scholar]
- 54.Lee K et al. (2017) Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng 1, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slowing II et al. (2008) Mesoporous silica nanoparticles as controlled release drug delivery and gene transfection carriers. Adv. Drug Deliv. Rev 60, 1278–1288 [DOI] [PubMed] [Google Scholar]
- 56.Birnbaum KD and Sánchez Alvarado A (2008) Slicing across kingdoms: regeneration in plants and animals. Cell 132, 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raliya R et al. (2016) Quantitative understanding of nanoparticle uptake in watermelon plants. Front. Plant Sci 7, 1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin-Ortigosa S et al. (2014) Mesoporous silica nanoparticle-mediated intracellular Cre protein delivery for maize genome editing via loxP site excision. Plant Physiol. 164, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin-Ortigosa S et al. (2012) Gold functionalized mesoporous silica nanoparticle mediated protein and DNA codelivery to plant cells via the biolistic method. Adv. Funct. Mater 22, 3576–3582 [Google Scholar]
- 60.Liu J et al. (2008) Preparation of fluorescence starch-nanoparticle and its application as plant transgenic vehicle. J. Cent. South Univ. Technol. (Engl. Ed.) 15, 768–773 [Google Scholar]
- 61.Hao Y et al. (2013) Magnetic gold nanoparticles as a vehicle for fluorescein isothiocyanate and DNA delivery into plant cells. Botany 91, 457–466 [Google Scholar]
- 62.Finiuk N et al. (2017) Investigation of novel oligoelectrolyte polymer carriers for their capacity of DNA delivery into plant cells. Plant Cell. Tissue Organ Cult 131, 27–39 [Google Scholar]
- 63.Zhao X et al. (2017) Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 3, 956–964 [DOI] [PubMed] [Google Scholar]
- 64.Naqvi S et al. (2012) Calcium phosphate nanoparticle mediated genetic transformation in plants. J. Mater. Chem 22, 3500 [Google Scholar]
- 65.Pasupathy K et al. (2008) Direct plant gene delivery with a poly (amidoamine) dendrimer. Biotechnol. J 3, 1078–1082 [DOI] [PubMed] [Google Scholar]
- 66.Burlaka OM et al. (2015) Plant genetic transformation using carbon nanotubes for DNA delivery. Cytol. Genet 49, 349–357 [PubMed] [Google Scholar]
- 67.Jiang L et al. (2014) Systemic gene silencing in plants triggered by fluorescent nanoparticle-delivered double-stranded RNA. Nanoscale 6, 9965. [DOI] [PubMed] [Google Scholar]
- 68.Demirer GS et al. (2018) High aspect ratio nanomaterials enable biomolecule delivery and transgene expression or silencing in mature plants. bioRxiv Published online January 30, 2018. 10.1101/179549 [DOI] [Google Scholar]
- 69.Ma X et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284 [DOI] [PubMed] [Google Scholar]
- 70.Osakabe Y and Osakabe K (2015) Genome editing with engineered nucleases in plants. Plant Cell Physiol. 56, 389–400 [DOI] [PubMed] [Google Scholar]
- 71.Bortesi L and Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv 33, 41–52 [DOI] [PubMed] [Google Scholar]
- 72.Arora L and Narula A (2017) Gene editing and crop improvement using CRISPR-Cas9 system. Front. Plant Sci 8, 1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feng Z et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 111, 4632–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang W et al. (2014) Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS One 9, e99225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.James C (2016) Global Status of Commercialized Biotech/GM Crops: 2016. ISAAA Brief No. 52, ISAAA [Google Scholar]
- 76.Camacho A et al. (2014) Genetically engineered crops that fly under the US regulatory radar. Nat. Biotechnol 32, 1087–1091 [DOI] [PubMed] [Google Scholar]
- 77.Jones HD (2015) Regulatory uncertainty over genome editing. Nat. Plants 1, 14011. [DOI] [PubMed] [Google Scholar]
- 78.Parliament EU (2001) Directive 2001/18/EC of the European Parliament and of the Council of 12 March 2001 on the deliberate release into the environment of genetically modified organisms and repealing Council Directive. Off. J. Eur. Comm L. 106, 1–39 [Google Scholar]
- 79.Bobek M (2018) Opinion of Advocate General Bobek in Case C-528/16, Court of Justice of the European Union [Google Scholar]
- 80.Ishii T and Araki M (2017) A future scenario of the global regulatory landscape regarding genome-edited crops. GM Crops Food 8, 44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Woo JW et al. (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol 33, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 82.Svitashev S et al. (2016) Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun 7, 13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X et al. (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc 1, 641. [DOI] [PubMed] [Google Scholar]
- 84.Liu C et al. (2017) Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release 266, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mout R et al. (2017) Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 11, 2452–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Antonelli NM and Stadler J (1990) Genomic DNA can be used with cationic methods for highly efficient transformation of maize protoplasts. Theor. Appl. Genet 80, 395–401 [DOI] [PubMed] [Google Scholar]
- 87.Ragelle H et al. (2014) Chitosan nanoparticles for siRNA delivery: optimizing formulation to increase stability and efficiency. J. Control. Release 176, 54–63 [DOI] [PubMed] [Google Scholar]
- 88.Zuris JA et al. (2015) Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol 33, 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pantarotto D et al. (2004) Functionalized carbon nanotubes for plasmid DNA gene delivery. Angew. Chem. Int. Ed. Engl 43, 5242–5246 [DOI] [PubMed] [Google Scholar]
- 90.Kam NW et al. (2005) Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of siRNA and potent gene silencing. J. Am. Chem. Soc 127, 12492–12493 [DOI] [PubMed] [Google Scholar]
- 91.Kneuer C et al. (2000) A nonviral DNA delivery system based on surface modified silica-nanoparticles can efficiently transfect cells in vitro. Bioconjug. Chem 11, 926–932 [DOI] [PubMed] [Google Scholar]
- 92.Chen AM et al. (2009) Co-delivery of doxorubicin and Bcl-2 siRNA by mesoporous silica nanoparticles enhances the efficacy of chemotherapy in multidrug-resistant cancer cells. Small 5, 2673–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boussif O et al. (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Biochemistry 92, 7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Negrutiu I et al. (1987) Hybrid genes in the analysis of transformation conditions. Plant Mol. Biol 8, 363–373 [DOI] [PubMed] [Google Scholar]
- 95.Aigner A et al. (2002) Delivery of unmodified bioactive ribozymes by an RNA-stabilizing polyethylenimine (LMW-PEI) efficiently down-regulates gene expression. Gene Ther. 9, 1700–1707 [DOI] [PubMed] [Google Scholar]
- 96.Silva AT et al. (2010) Conjugated polymer nanoparticles for effective siRNA delivery to tobacco BY-2 protoplasts. BMC Plant Biol. 10, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sgolastra F et al. (2017) Sequence segregation improves noncovalent protein delivery. J. Control. Release 254, 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mizutani O et al. (2012) Modified Cre-loxP recombination in Aspergillus oryzae by direct introduction of Cre recombinase for marker gene rescue. Appl. Environ. Microbiol 78, 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun W et al. (2015) Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed. Engl 54, 12029–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sandhu KK et al. (2002) Gold nanoparticle-mediated transfection of mammalian cells. Bioconjug. Chem 13, 3–6 [DOI] [PubMed] [Google Scholar]
- 101.Lee J-H et al. (2009) All-in-one target-cell-specific magnetic nanoparticles for simultaneous molecular imaging and siRNA delivery. Angew. Chem. Int. Ed. Engl 48, 4174–4179 [DOI] [PubMed] [Google Scholar]
- 102.Mout R et al. (2017) General strategy for direct cytosolic protein delivery via protein-nanoparticle co-engineering. ACS Nano 11, 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gao W et al. (2018) Risk analysis for genome editing-derived food safety in China. Food Control 84, 128–137 [Google Scholar]
- 104.McCarthy M (2018) Genetic modification laws set for shake-up, with health and agriculture research industries to benefit (interview with Raj Bhula). ABC Australia. http://www.abc.net.au/news/rural/2018-01-19/gene-tech-regulator-proposes-shakeup-for-genetic-modification/9341354 [Google Scholar]
- 105.Schuttelaar & Partners (2015) The regulatory status of New Breeding Techniques in countries outside the European Union. http://www.nbtplatform.org/background-documents/rep-regulatory-status-of-nbts-oustide-the-eu-june-2015.pdf [Google Scholar]
- 106.Waltz E (2018) With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol 36, 6–7 [DOI] [PubMed] [Google Scholar]
- 107.Bevan M (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klein TM et al. (1987) High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 327, 70–73 [PubMed] [Google Scholar]
- 109.Tiwari JN et al. (2012) Zero-dimensional, one-dimensional, two-dimensional and three-dimensional nanostructured materials for advanced electrochemical energy devices. Prog. Mater. Sci 57, 724–803 [Google Scholar]
- 110.Khan I et al. (2017) Nanoparticles: properties, applications and toxicities. Arab. J. Chem Published online May 18, 2017. 10.1016/J.ARABJC.2017.05.011 [DOI] [Google Scholar]
- 111.Wang L et al. (2016) In vivo delivery systems for therapeutic genome editing. Int. J. Mol. Sci 17, E626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim I-Y et al. (2015) Toxicity of silica nanoparticles depends on size, dose, and cell type. Nanomedicine 11, 1407–1416 [DOI] [PubMed] [Google Scholar]
- 113.Shang L et al. (2014) Engineered nanoparticles interacting with cells: size matters. J. Nanobiotechnol 12, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Petersen MA et al. (2013) Bioresorbable polymersomes for targeted delivery of cisplatin. Bioconjug. Chem 24, 533–543 [DOI] [PubMed] [Google Scholar]
- 115.Tekedereli I et al. (2013) Therapeutic silencing of Bcl-2 by systemically administered siRNA nanotherapeutics inhibits tumor growth by autophagy and apoptosis and enhances the efficacy of chemotherapy in orthotopic xenograft models of ER (−) and ER (+) breast cancer. Mol. Ther. Nucleic Acids 2, e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yin PT et al. (2014) Combined magnetic nanoparticle-based microRNA and hyperthermia therapy to enhance apoptosis in brain cancer cells. Small 10, 4106–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang K et al. (2012) Antibody-linked spherical nucleic acids for cellular targeting. J. Am. Chem. Soc 134, 16488–16491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smith BR et al. (2014) Selective uptake of single-walled carbon nanotubes by circulating monocytes for enhanced tumour delivery. Nat. Nanotechnol 9, 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen P-C et al. (2016) Polyelemental nanoparticle libraries. Science 352, 1565–1569 [DOI] [PubMed] [Google Scholar]
- 120.Lu J et al. (2014) Toward atomically-precise synthesis of supported bimetallic nanoparticles using atomic layer deposition. Nat. Commun 5, 3264. [DOI] [PubMed] [Google Scholar]
- 121.Hu X et al. (2013) Polyprodrug amphiphiles: hierarchical assemblies for shape-regulated cellular internalization, trafficking, and drug delivery. J. Am. Chem. Soc 135, 17617–17629 [DOI] [PubMed] [Google Scholar]
- 122.Agudelo D et al. (2016) tRNA conjugation with chitosan nanoparticles: an AFM imaging study. Int. J. Biol. Macromol 85, 150–156 [DOI] [PubMed] [Google Scholar]
- 123.Shi Y et al. (2015) Complete regression of xenograft tumors upon targeted delivery of paclitaxel via π-π stacking stabilized polymeric micelles. ACS Nano 9, 3740–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim YG et al. (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U. S. A 93, 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jinek M et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cong L et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boch J et al. (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 128.Moscou MJ and Bogdanove AJ (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501. [DOI] [PubMed] [Google Scholar]
- 129.Wong MH et al. (2016) Lipid exchange envelope penetration (LEEP) of nanoparticles for plant engineering: a universal localization mechanism. Nano Lett. 16, 1161–1172 [DOI] [PubMed] [Google Scholar]
- 130.Giraldo JP et al. (2014) Plant nanobionics approach to augment photosynthesis and biochemical sensing. Nat. Mater 13, 400–408 [DOI] [PubMed] [Google Scholar]
- 131.USDA (2018) Secretary Perdue Issues Usda Statement on Plant Breeding Innovation, Press Release No. 0070.18 [Google Scholar]
- 132.National Technical Biosafety (2018) Normative Resolution No. 16/2018, CTNBio [Google Scholar]
- 133.Servicio Agrícola y Ganadero (2017) Applicability of Resolution No. 1523/2001 on Propagation Material Developed by New Plant Breeding Techniques, SAG [Google Scholar]


