Abstract
Climate change and population growth are straining agricultural output. To counter these changes and meet the growing demand for food and energy, the monitoring and engineering of crops are becoming increasingly necessary. Nanoparticle-based sensors have emerged in recent years as new tools to advance agricultural practices. As these nanoparticle-based sensors enter and travel through the complex biofluids within plants, biomolecules including proteins, metabolites, lipids, and carbohydrates adsorb onto the nanoparticle surfaces, forming a coating known as the “bio-corona”. Understanding these nanoparticle–biomolecule interactions that govern nanosensor function in plants will be essential to successfully develop and translate nanoparticle-based sensors into broader agricultural practice.
Keywords: Nanosensor, biocorona, in planta, engineered nanomaterial, agricultural engineering
Graphical Abstract
As changing climates and population growth increasingly place pressure on agricultural production, the monitoring and engineering of crops are becoming essential to meet the rising demand for food and energy.1–3 To maximize the efficient use of limited resources, crops can be remotely monitored using sensors to adjust plant management strategies rapidly, minimizing losses and maximizing yields.1 In particular, innovations in nanotechnology have advanced the collection of agricultural metrics such as nutrient levels, pathogen infection, and pesticide accumulation in plants.4 Nanoparticle-based sensors have garnered much interest in agricultural applications in recent years, as reviewed elsewhere.1,5,6 The unique physical and chemical properties of nanoparticles have been leveraged to design sensing platforms that are highly portable, rapid, sensitive, and amenable to high-throughput measurements.7
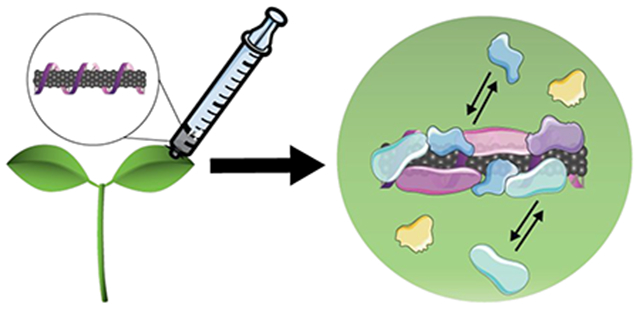
The application of nanosensors and, more broadly, of nanotechnologies in plants presents distinct obstacles that must be taken into consideration during the design phase, as the unique plant environment could drastically alter the intended nanoparticle function. Biological barriers like the waxy leaf cuticle challenge nanoparticle uptake,8 while the varying biochemical compositions in plant tissues alter nanoparticle functionality.9 When nanoparticles enter and traverse complex biological milieus in plants, biomolecules including proteins, metabolites, lipids, and carbohydrates adsorb onto the nanoparticle surfaces, forming a coating known as the “bio-corona”.9–11 As such, one major bottleneck for the seamless translation of nanosensors from in vitro validation to in planta use is in the spontaneous and as-of-yet unpredictable adsorption of biomolecules on nanosensor surfaces that attenuates their intended function (Figure 1). For the practical translation of nanotechnologies to plants, it is imperative that we understand the interactions between the nanoparticles and the biomolecules they encounter during inplant transport.
Figure 1.
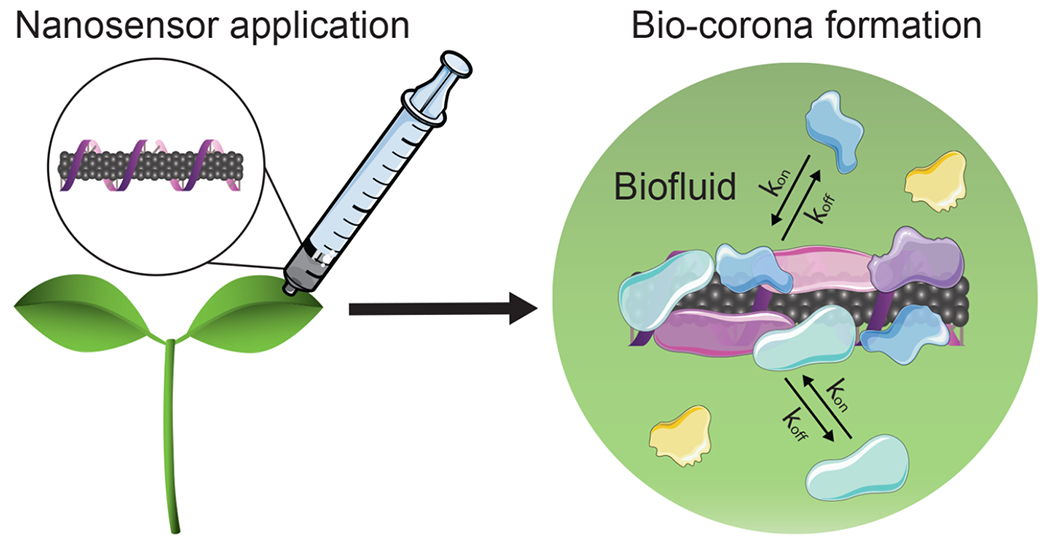
Biomolecules adsorb on the surface of nanosensors as they travel through complex plant biofluids, altering the intended nanosensor function and creating a bottleneck for the translation of nanosensors from the laboratory to applications in intact plants.
1. NANOPARTICLE-BASED SENSORS FOR AGRICULTURAL INNOVATIONS AND HOW TO SUCCESSFULLY TRANSLATE THESE TECHNOLOGIES FOR IN PLANTA USE
Sensing systems for precision agriculture have been developed to monitor water content, soil conditions, and crop health. These sensors monitor soil moisture, temperature, and nutrient levels,12–15 providing key information for crop management. Sensors for pesticides, herbicides, and insecticides in the soil or water have also been developed16–19 and are useful for improving yields and supporting food and water safety.20 Directly sensing plant health signals through wearable electronic devices and embedded sensors has more recently emerged as an exciting strategy to monitor crops.6,21,22 These sensing systems vary widely in sensing mode, from electrochemical sensors that translate chemical reactions into measured voltages, to optical sensors that measure changes in fluorescence emission for analyte quantification.
Most plant and crop sensors enable precise measurements but rely on tissue-destructive techniques that involve field- or lab-based protocols, limiting these measurements to distinct time points. We highlight a specific class of engineered nanoparticle-based sensors (<100 nm in their smallest dimension), termed “nanosensors”, that can enable continuous in planta environmental sensing and plant health monitoring in intact plants. As autosamplers and bioconcentrators of their surrounding environment,23 plants provide an exciting platform for sensing. Combining nanotechnology with the natural features of plants facilitates rapid in-field detection that circumvents expensive and time-intensive laboratory techniques. Continuous gas and fluid exchange between plants and their environment enables this mode of sensing to be used in a variety of different contexts, be it probing the soil for specific analytes or monitoring chemical signals for plant health reports in agricultural settings. Thus far, nanosensors have been used for detecting ground soil contaminants and quantifying plant defense-related biomarkers and signaling molecules (Figure 2).21,24,25 While genetically encoded plant sensors may confer similar advantages as nanosensors, they are limited by species-specific genetic transformations that require large amounts of time and effort.1 In contrast, engineered nanosensors are species-independent and thus more easily translated across plant systems. Although plants have been genetically engineered as biochemical detectors in the past, the addition of nanoparticle-based sensors to utilize plants as detectors is just emerging as a field of study.26,27 As such, the use of nanosensors in plants is promising for agricultural sustainability, with several recent examples of nanoparticle-based sensors for plant and crop monitoring outlined in Table 1 and reviewed in more detail elsewhere.4,12,17,28,29
Figure 2.
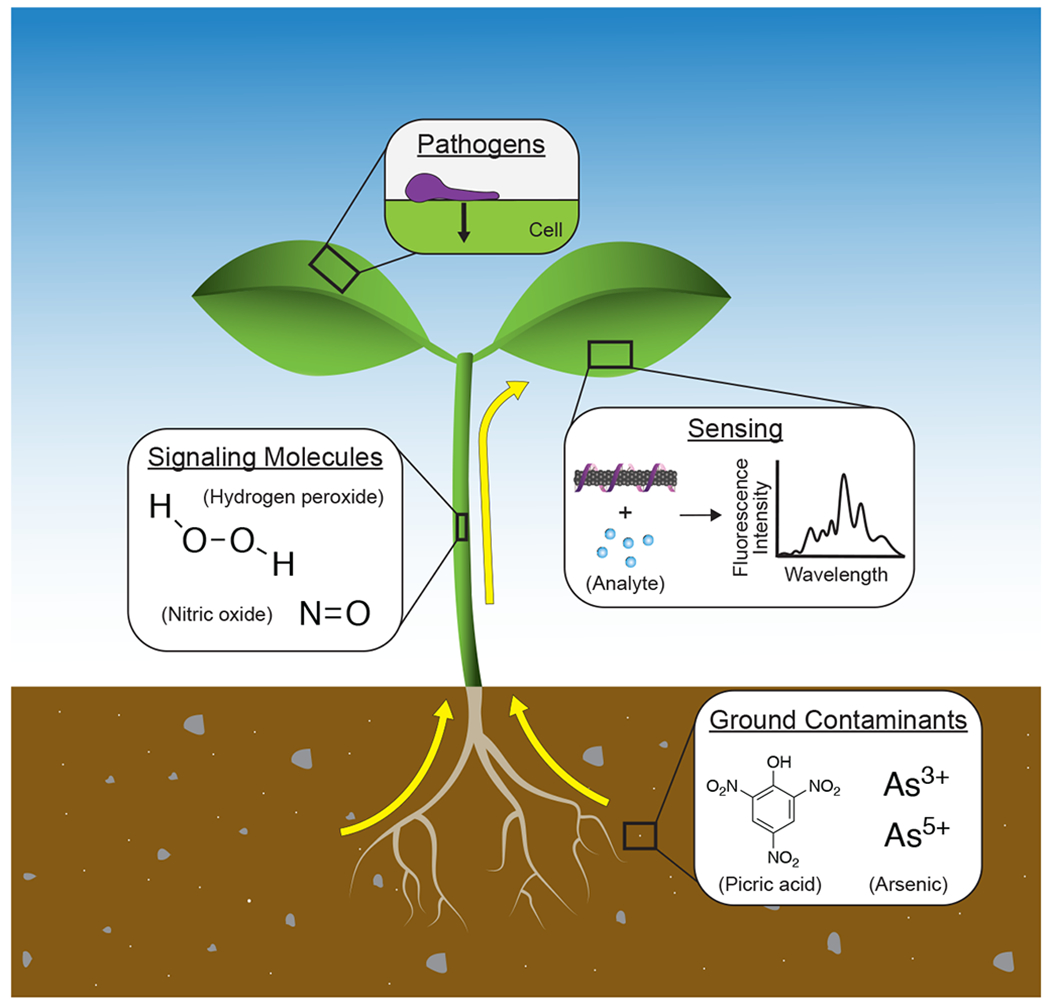
Nanosensors are used in plants for detecting soil contamination, signaling molecules, and pathogens. Early and rapid communication of plant stress through sensing innovations has the potential to improve agricultural management, while further development of plant biomarker sensors can be used to understand the complex signaling pathways within plants.
Table 1.
Nanoparticle-Based Sensing Systems for Agricultural Engineering
| system | target | signal transduction | material | limit of detection | ref |
|---|---|---|---|---|---|
| In vitro | Carbosulfan (pesticide) | Electrochemical - voltammetry | Zinc oxide (ZnO) nanocuboids modified platinum (Pt) electrode | 0.24 nM | 16,17 |
| Soil | Atrazine (herbicide) | Electrochemical - voltammetry | Titanium dioxide (TiO2) nanotubes | 0.1 ppt | 17,19 |
| On plant | Light and humidity | Wearable optical sensor | ZnIn2S4 (ZIS) nanosheets | ~ 4 ms (light response) | 22 |
| Water | Malathion (insecticide) | Optical - Surface-Enhanced Raman Spectroscopy (SERS) | Aptamer–polymeric microsphere–gold nanoparticles (AuNPs) | 3.3 μg mL−1 | 17,18 |
| Water | Nitrite | Optical - fluorescence | Silver nanoclusters | 100 nM | 30 |
| In vitro | DNA sequence of Ganoderma boninense gene (oil palm pathogen) | Optical - fluorescence | Quantum dots (QDs) | 3.55 nM | 31 |
| In plant | Glucose | Optical - fluorescence | Thioglycolic acid-capped QDs and boronic acid-conjugated QDs | 500 μM | 25 |
| In plant | H2O2 | Optical - fluorescence | Single-walled carbon nanotubes (SWCNTs) | 10 μM | 32 |
| In plant | NO | Optical - fluorescence | SWCNTs | 100 μM | 24 |
1.1. Environmental Nanosensors to Sense Ground Analytes and Detect Plant Pathogens.
The monitoring of groundwater, soil contaminants, and early detection of plant pathogens are crucial activities in recognizing and addressing potential threats to human and plant health. By capitalizing on the innate properties of plants as microfluidic devices that sample their immediate surroundings, plant-based nanosensors have been designed for real-time biochemical sensing of soil contaminants, including explosive nitroaromatics and arsenic.23,27 In the former example, peptide-coated single-walled carbon nanotubes (SWCNTs) were embedded in spinach leaves to enable near-infrared (and thus plant-tissue transparent) optical detection of nitroaromatic compounds.27 The nitroaromatic compounds are taken up by the roots and transported through plant vasculature to the leaves where the SWCNT sensors reside. Similarly, leaf-embedded SWCNT-based sensors have been used to detect arsenic in the soil through root uptake in Spinacia oleracea (spinach), Oryza sativa (rice), and Pteris cretica (ferns).23 Of note, these SWCNT-based plant nanosensors provide both a rapid response in the presence of target analytes and stable performance over a period of months.1,32
In addition to sampling toxic ground contaminants, nanosensors can prevent disease spread among crops by nondestructively detecting the presence of plant pathogens before symptom onset. Current disease diagnostics for plants are morphology-based analyses, which occur after the disease has progressed, or sample-destructive and pathogen-specific assays such as enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR). Although robotic imaging platforms are improving detection sensitivity, such technologies often fail to detect disease before symptom onset.33 Conversely, nanoparticle-based sensors for pathogen detection have been demonstrated to function successfully in planta,5,34 though such nanoparticle-based sensors are more widely used ex vivo in rapid detection kits. A common class of nanoparticle-based sensing technologies is lateral flow immunoassays, whereby pathogens are detected with an antibody, aptamer, or DNA probe conjugated to a gold, magnetic, or fluorescent nanoparticle.35 These assays have been used to detect a variety of plant viruses such as Citrus tristeza virus from citrus leaves and fruits, Potato virus x, and the bacterial pathogen of Stewart’s wilt in sweet corn.35–38 These ex vivo sensing strategies enable convenient plant pathogen detection, yet remain limited to sampling at distinct time points. Like the ex vivo assay, a combination of antibodies, aptamers, or DNA probes and nanoparticles could be used to detect the pathogen in plant tissue. A major barrier toward translating these nanosensors in planta applications is in preserving nanosensor function in the biologically complex milieu of living plants, which would allow continuous and rapid detection during the early stages of pathogenesis. Moving forward, advancing early stage disease and pathogen detection necessitates continuous and real-time sensing capabilities with the use of in planta compatible nanosensors.
1.2. Biomarker Nanosensors to Monitor Plant Signaling and Health.
Sensing biomarkers that are indicative of plant stress and energy production provide essential insight into plant signaling and health.24,25 Abiotic stresses such as droughts and heat as well as biotic stressors like plant pathogens elicit a defense response.39 Unlike animal systems, plants have a passive immune system, and plant cells rely heavily on cell-to-cell signaling to communicate environmental threats through immunogenic signals that activate plant defenses.21,40–42 Nanosensors can be used to translate these plant chemical stress signals to electronic signals for real-time sensing, which would serve to report crop health and, thus, diagnose plant environmental stressors to enable appropriate intervention.1,43 Examples of such plant stress biomarkers include reactive oxygen species (ROS) and glucose. Fluorescent SWCNT sensors have been developed to recognize ROS such as hydrogen peroxide (H2O2), a biomarker for plant defense produced in response to stress, and nitric oxide (NO), another key signaling molecule in plants.24 Specifically, Wu et al. developed a H2O2 sensor based on SWCNTs functionalized with a DNA aptamer that binds to hemin (HeAptDNA-SWCNT). Mechanistically, this sensor takes advantage of hemin that undergoes a reaction with H2O2, producing hydroxyl radicals, resulting in SWCNT fluorescence quenching. By measuring changes in SWCNT fluorescence emission with a near-infrared camera, these H2O2 nanosensors enable continuous, real-time monitoring of plant health in response to UV–B light, high light, and pathogens, but not plant wounding in Arabidopsis thaliana which likely has H2O2 levels below the limit of detection (Figure 3).24,32 More recently, a different single-stranded DNA-functionalized SWCNT H2O2 nanosensor was developed and can sense the plant response to wounding in several common plant species including Lactuca sativa (lettuce), Eruca sativa (arugula), and Spinacia oleracea (spinach).21 SWCNT sensors have also been used for the detection of NO.24 Similarly, a quantum dot-based nanosensor has been used to ratiometrically detect glucose.25 As indicators of environmental stressors, these nanosensors for analytes like glucose and ROS have the potential to improve crop management through rapid, continuous, and nondestructive sensing.
Figure 3.
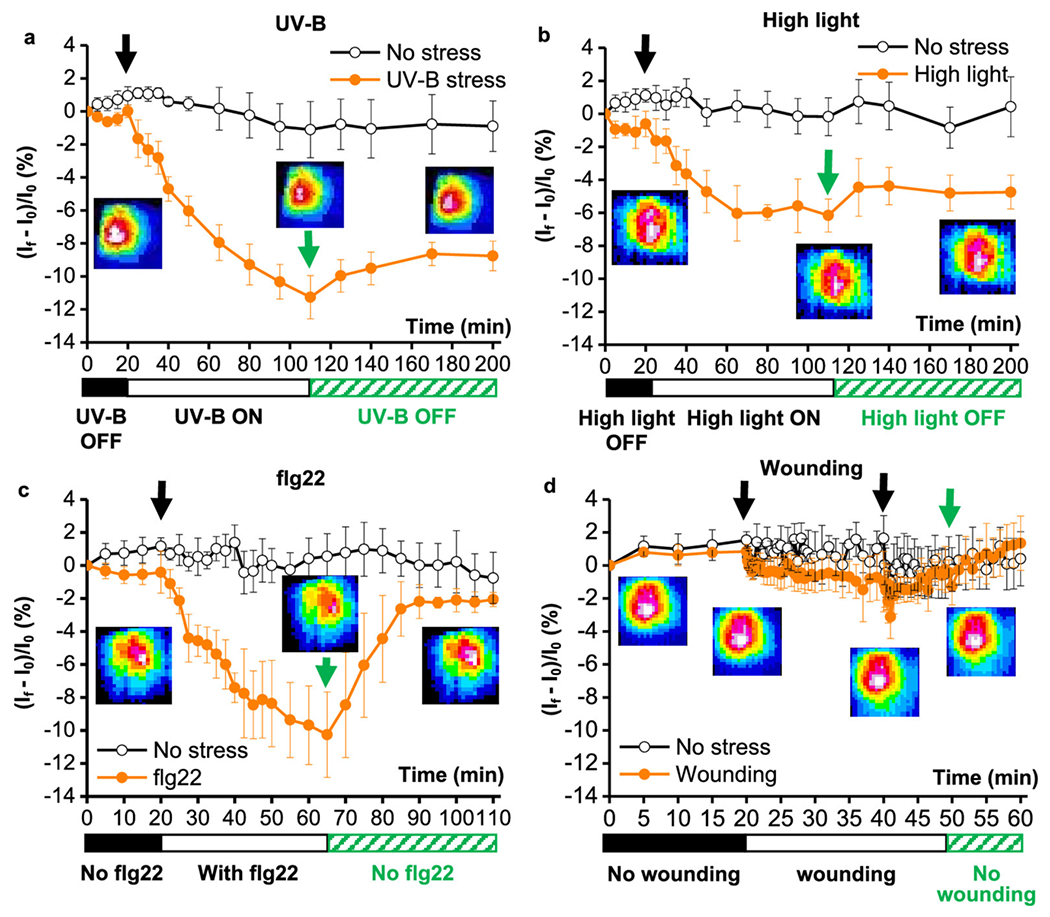
In planta monitoring of plant health signaling molecules in response to common plant stresses via a SWCNT H2O2 nanosensor. Near-infrared fluorescence intensity changes of the nanosensor embedded in leaves indicate signs of stress. The nanosensor’s near-infrared fluorescence emission decreases in the presence of (a) UV–B light, (b) high light, and (c) a pathogen-associated peptide (flg22). (d) Mechanical leaf wounding did not result in a change in the nanosensor’s near-infrared fluorescence emission, likely due to the relatively lower levels of H2O2 production. Reprinted with permission from ref 32. Copyright (2020) American Chemical Society.
The future of health and developmental monitoring in plants depends on the successful translation of these nanosensors to practical applications in the field. As these sensors are implemented in plants for crop management, the sensors will encounter challenges in less controlled environments: nanosensor biofouling is known to attenuate or abate nanosensor function, and nanosensor transport and bioaccumulation within the plant or the environment remains unpredictable. As such, nanoparticle–biomolecule interactions in plants and agricultural settings will dictate our ability to preserve nanoparticle function in planta and will need to be evaluated for environmentally conscious translation of these nanotechnologies from the lab to the field. To these ends, understanding the phenomenon of biocorona formation on nanosensors will inform designs with improved sensing capabilities and biostabilities, increasing the likelihood of success when these technologies are translated into broader applications. The process of protein corona formation on nanoparticle surfaces has been described extensively for nonplant systems elsewhere;44–46 however, it remains to be elucidated for plant and agricultural systems.
1.3. Understanding Biocorona Formation to Increase the Translational Value of Plant Nanosensors.
Although nanosensors have the potential to revolutionize agriculture, the parameters governing nanosensor performance in planta have been understudied in plant systems. Successful in vivo translation of nanosensor technologies requires a more thorough understanding of the interactions between the nanosensor and its local plant environment. Despite the prevalence of protein corona characterization in nanomedicine toward human health applications, biocorona formation in plant systems has only received limited acknowledgment and research.47 Plant-based nanosensors are developed and implemented without taking into account the inevitable changes in physicochemical properties as the nanosensor is progressed from in vitro development to in vivo use. We propose that more informed plant nanosensor designs can be developed and predictably translated into practical applications, guided by fundamental studies of plant nanoparticle biocoronas.
The field of nanomedicine has largely motivated the study of nanoparticle–protein interactions to improve nanoparticle function in applications including drug delivery, disease diagnostics, treatment, and prevention.48–52 Nanoparticle-based sensors have been used to detect metal ions, small molecules, and proteins including biomarkers for early cancer and kidney disease.53–55 As nanotechnologies become more widely used in biological settings, it is increasingly important to understand and predict nanoparticle function and fate in vivo; despite the successes of some, many nanomaterials produce unsatisfactory results or off-target effects during clinical trials.56–60 This translation of nanomedicines from laboratory development to clinical practice is limited by our lack of control over interactions between the nanoparticle and its surrounding bioenvironment.44,60–63 Although the intrinsic physicochemical characteristics of the nanoparticle determine in vitro functionality, the environment around the nanoparticle in the applied setting, such as blood plasma for intravenous delivery, will play a dominant role in determining the ultimate nanoparticle fate and function.
Likewise, to develop a fundamental understanding of biocorona formation on nanoparticles in plants, the governing parameters must be considered. Biocorona formation is determined by the interplay of intrinsic properties of nanoparticles, including electrostatic charge, hydrophobicity, and surface structure, and characteristics of the environment surrounding the nanoparticle, such as biomolecule composition and solution conditions.64 As nanoparticles are introduced to a bioenvironment, an inner layer of more tightly bound, higher-affinity biomolecules forms, referred to as the “hard corona”, and a rapidly exchanging outer layer of biomolecules, the “soft corona”, more loosely associates.65–67 This coating of biomolecules alters the lab-engineered properties of the nanoparticle, since in many cases, it is this biocorona that ultimately interacts with the biological environment.11,68,69 Accordingly, these biomolecular interactions can lead to loss of nanoparticle targeting ability,61 impact nanoparticle uptake in vivo by influencing membrane adhesion and internalization pathways,62,69,70 and even eliminate nanoparticle efficacy.64,71 More specifically, nanoparticle–protein interactions often lead to protein denaturation on the nanoparticle surface and colloidal aggregation of the complexes.64,72,73 By preemptively considering this phenomenon, we can reduce biofouling and preserve nanoparticle functionality in planta.
2. CURRENT STATUS OF PROBING BIOCORONA FORMATION ON NANOPARTICLES IN PLANTS
Proteomic and metabolomic studies detailing biocorona composition are necessary to understand the biological identity that nanoparticles acquire in plants. In turn, knowledge of biocorona constituents will inform improved sensor design strategies to reduce biofouling and even tune biocorona formation to enhance nanosensor localization and function. These studies are crucial, but, in contrast to animal systems, the literature on biocorona formation and the corresponding impact on nanoparticle behavior in plant systems is limited.
2.1. Proteomic and Metabolomic Analyses of Biocorona Formation in Plant Biofluids.
To-date, plant-based biocorona formation has been investigated with three types of nanoparticles: titanium dioxide nanoparticles (TiO2), magnetic nanoparticles, and gold nanoparticles. These studies have focused on individual assessments of either the protein or the metabolite corona in plant biofluids.
In vitro combinations of nanoparticles and specific plant proteins demonstrate the significance of protein identity in the resulting biocorona formation, as anticipated from analogous studies in animal systems.67 Bing et al. studied the effects of incubating common plant proteins including glutenin, gliadin, zein, and soy protein with TiO2 nanoparticles and found that the corona formed in the presence of each protein produced varied effects on TiO2 surface potential and morphology, with 4–60 nm thickness of the adsorbed protein layers.74 Separately, a recent study probed the evolution of the protein corona on 16 nm gold nanoparticles in crude protein extracts and nuclear fractions of Brassica juncea and observed differences in protein corona components between the two biofluids, with more than a quarter of the hard corona proteins in the crude protein fraction involved in energy generation pathways.75 Given that gold nanoparticles have been shown to induce a change in Brassica juncea overall growth and seed yield,76 Prakash and Deswal suggest that the protein corona could be implicated in system-level effects observed from nanomaterial–plant interactions.
Although corona formation has system-wide implications, the phenomenon of biocorona formation can instead be harnessed for molecularly specific applications. The Smalle group has pioneered an approach coined “nano-harvesting” in which nanoparticles preferentially bind and extract catechol-containing flavonoids from plants. Within this body of work, Kurepa et al. conducted multiple studies centered around the metabolite corona formed on TiO2 nanoparticles in Arabidopsis thaliana, Ocimum sanctum, and Rubia tinctorum.77–79 TiO2 nanoparticles were incubated with mature leaves, and after subsequent metabolomic analysis, the nanoparticle surface was found to be enriched in lipids and in particular flavonoids, which are polyphenolic small molecules involved in secondary metabolism.78 These results also revealed that lipids and flavonoids compete for nanoparticle surface sites during biocorona formation. In a separate study, Qing and co-workers used human serum albumin-functionalized magnetic nanoparticles to extract bioactive molecules from Dioscorea panthaica via preferential corona binding, greatly expediting the isolation of four saponin compounds.80 In combination with Kurepa’s work, these findings show that biocorona formation can be leveraged to achieve desirable molecule enrichment. The tuning of the nanoparticle surface properties to control biocorona formation while retaining targeting and delivery functions has been achieved in nanomedicine81,82 and provides a roadmap in harnessing knowledge of the biocorona to design nanosensors that maintain their utility in planta.
2.2. Challenges Associated with Nanoparticle-Based Proteomic and Metabolomic Studies in Plants.
Technological advances have led to more rigorous characterization of the biocorona formed on nanoparticles, yet several challenges persist before the process of studying plant-based protein and metabolite coronae is rapid, comprehensive, and efficient. These obstacles are briefly summarized here and detailed more comprehensively in other reviews.83–86
The main challenge in advancing biocorona studies in plants is the lack of sufficient proteomic and metabolomic information on the plants alone, in contrast to the extensive-omics characterization of human and many model animal systems. Obtaining this data for plants requires a meticulously designed experimental process and in-depth data analytics, which are both time- and energy-intensive. Additional experimental considerations apply in data collection because plant physiology and biochemistry vary across different strata: (i) spatial resolution, accounting for plant organs, subcellular and extracellular spaces, and (ii) temporal distribution, spanning growth stages, photoperiodism, and seasonal variations.85 These distinctive protein abundance patterns in plant tissues87 and biochemical changes over time that represent challenges to proteomic and metabolomics studies will further influence biocorona formation. Thus, it is essential to address the gap in proteomic and metabolomic plant literature to accurately characterize these nanoparticle–biomolecule interactions.
The plant metabolome poses significant difficulties in compound identification and quantification: between 100,000–1,000,000 metabolites are estimated to belong to the plant kingdom,88 while in contrast the number of detected and quantified human metabolites is only 18,609.89 The metabolomic compositional complexity also differs vastly across plant species,85 and the dynamic range of abundances (up to 12 orders of magnitude) and identity of sampled metabolites are heavily dependent on biotic and abiotic factors during plant growth.86,90 Proteomic analysis encounters similar challenges, requiring careful and unbiased sample preparation.91 Protein identification and quantification must also consider the high dynamic range and harness genomes, which for plants like barley (5000 Mbp genome) are larger than the human genome (~3300 Mbp),92 and expressed sequence tag (EST) data,93,94 presenting a challenge in itself since there is no central curated database.95
Advances in proteomic and metabolomic data acquisition and analysis remain key to furthering the study of the biocorona in plants (Figure 4). Such advances will subsequently enable the correlation of proteins and metabolites in the nanoparticle biocorona with biological effects of nanoparticles on plants. The prospect of these advances motivates the synthesis of genomics, proteomics, and metabolomics information to uncover biochemical pathways. Excitingly, the rise of integrative approaches to -omics,96 decreasing costs of high-throughput sequencing and mass spectrometry, as well as developments in bioinformatics promise to accelerate the development of plant -omics.97–99 Through these innovations, knowledge of plant -omics is quickly expanding,87 enabling biocorona characterization on nanoparticles in plant tissues with distinct biochemical compositions. As these technologies, tools, and databases become more widely developed and utilized, compositional knowledge of the biocorona can be employed to better inform plant-based nanosensor design strategies.
Figure 4.
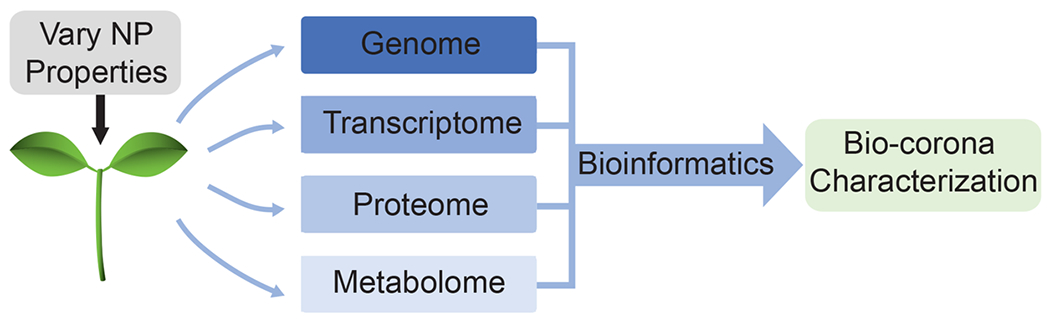
Innovations in -omics research may enable improved nanoparticle (NP)-based sensor design through biocorona characterization with the high-throughput combination of genomics, transcriptomics, proteomics, and metabolomics with bioinformatics.
Previous biocorona studies have broadly shown preferential enrichment of biomolecules but fail to consider the multiple unique bioenvironments that a nanosensor encounters within the plant. We must carefully consider the physiological and biochemical compositions in which nanoparticles travel through and/or localize at to understand the full picture of nanoparticle–biomolecule interactions. For instance, to study the efficiency of a nanosensor that traverses through plant vasculature would require nanoparticle incubation within phloem or xylem sap. Conversely, investigating nanosensor biofouling for a nanosensor embedded within a leaf might necessitate considering the different localization end points such as the cytosol or cell wall. In future research, contextualizing studies of biocorona formation may provide more realistic insight into nanoparticle fate.
3. PHYSIOLOGICAL CHARACTERISTICS OF PLANTS RELEVANT TO BIOCORONA FORMATION
The broad concepts and underlying physical phenomena driving biocorona formation can be readily translated to nanoparticles in plant bioenvironments. Yet, we emphasize that additional factors must be considered in terms of the distinct biological characteristics and obstacles that nanoparticles encounter while moving through plants in comparison to those more widely studied in animal systems. Such plant-specific aspects include (i) the modes of nanoparticle transport, (ii) unique biological barriers, such as the presence of a multilayered cellulosic cell wall, and (iii) markedly different biofluid conditions and constituents.
3.1. Transport Phenomena of Nanoparticles in Plants and How the Biocorona May Modulate Movement.
Modes of nanoparticle transport through plants predominantly consist of nanoparticle uptake, translocation, internalization, and accumulation, as reviewed elsewhere.8,100–102 In addition to more commonly considered nanoparticle characteristics such as size and charge, biocorona formation on nanoparticles is expected to influence and be influenced by each of these modes of movement. Yet, the effect of nanoparticle corona formation has not been studied in relation to nanoparticle transport in plants. As such, this molecular phenomenon of corona formation will propagate effects through the macro- and microscales of nanosensor outcomes in plant bioenvironments. Macroscale transport through plant vasculature occurs by movement of water and ions through the xylem with pore diameters ranging from 40–340 nm and movement of photosynthetic products via the phloem with pore diameters ranging from 200–1500 nm.8 Conversely, microscale transport through the plant cell wall and membrane is reserved for smaller nanosensors developed for intracellular measurements that must measure below the plant cell wall size exclusion limit of approximately 5–20 nm.103
Nanoparticle transport through plant vasculature begins with uptake, as governed by the method of delivery, generally including foliar application (onto leaves), root application, or direct injection into other plant tissues. While the latter method is considered the most efficient for nanoparticle delivery,8 foliar and root application are ideal for certain nanosensing applications, whereby numerous studies have demonstrated that leaves are the main sinks of airborne contaminants and roots serve to uptake organic compounds in the soil.27 For example, the nitroaromatic, arsenic, and hydrogen peroxide nanosensors, as discussed in earlier sections, involve embedding nanosensors directly in the spongy leaf mesophyll.21,23,27 In this case, nanoparticle transport needs end upon uptake, as the analytes localize to the leaf-residing nanosensors. However, even this seemingly straightforward journey requires interaction with the leaf cuticle, stomata, and parenchyma of leaf lamina, all of which may result in dynamic biocorona formation that impacts nanosensor function.
Beyond direct foliar embedding, nanosensors requiring longer distance transport will translocate through the plant vascular system. Typically, nanosensors are directly embedded in the target tissue and do not translocate through the vasculature, but as these technologies are scaled from the laboratory to the field, feeding nanoparticles directly to crops is emerging as a delivery method.8 Considering how nanoparticle transport through vascular tissue will impact biocorona formation is necessary for developing these technologies. Colloidal interactions of nanoparticles with vascular walls are expected to play a role in ease of translocation. It has been demonstrated with classic colloid modeling using a DLVO framework,8 with experimental validation,104,105 that approximately neutral (+5 mV) and positively charged (+35 mV) nanoparticles that lack steric stabilization will resist transport by depositing on negatively charged vascular walls. In contrast, negatively charged (−35 mV) nanoparticles can more freely translocate through plant vessels. As the adsorption of biomolecules onto nanoparticles from the surrounding plant medium will modulate nanoparticle size, effective surface charge, steric character, and other nanoparticle surface properties, biocorona formation becomes imperative to understand in optimizing efficient nanoparticle transport phenomena in plants.
3.2. Biological Barriers That Nanoparticles Encounter in Plants and How the Biocorona May Impact and Be Modified by Traversal.
Nanoparticles encounter distinct biological barriers during transport into and through plants. Additionally, an adsorbed biocorona can either hinder nanoparticle passage or be influenced by the different biological conditions that the nanoparticles are exposed to during passage, as studied in animal systems.106 Nanoparticles encounter barriers in plants during (i) initial uptake through roots and leaves and (ii) eventual entry into the plant cell, across the cell wall, cell membrane, and potentially organelle membranes (Figure 5). Even if nanoparticles are expected to pass through these structures based on characterization of the pristine nanoparticle hydrodynamic size, unintended biocorona formation and nanoparticle aggregation may prevent nanoparticle transport through such barriers. For context, the protein corona can add approximately 10–30 nm to the nanoparticle diameter in animal circulation environments,46 increasing the hydrodynamic size by 50% in the case of 60-nm-diameter gold nanoparticles in blood serum.107
Figure 5.
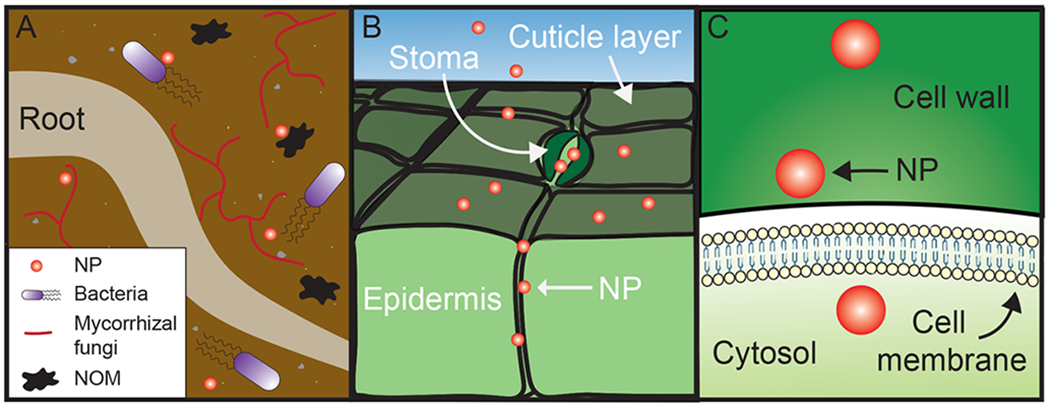
Nanoparticles (NPs) encounter barriers and interact with a variety of biomolecules in plant bioenvironments. (A) During uptake through roots, nanoparticles interact with microorganisms, such as bacteria and mycorrhizal fungi, and natural organic matter (NOM). (B) In foliar application, NPs encounter the waxy cuticle coating leaf surfaces, and stomatal pores for gas exchange. (C) As NPs are internalized by the cell, they must cross the cell wall, with a size exclusion limit of 5–20 nm, and the cell membrane.
Nanoparticle uptake by roots is often reported to have low efficiencies, presumably occurring through passage cells of intact roots or at sites of new or damaged roots, bypassing the root cuticle.8,108 An added complexity expected to influence root uptake of nanoparticles is that of the rhizosphere. The rhizosphere presents a rich environment of root exudates and mucilage originating from plants, in addition to the surrounding population of soil-based microorganisms including bacteria and mycorrhizal fungi. Nanoparticle interaction with these secreted substances and organisms of the rhizosphere is a key, yet underexplored, consideration anticipated to affect the biocorona formed on nanoparticles, and thus nanoparticle surface properties, stability, and bioavailability.101,108,109 Accordingly, this rhizosphere-imparted corona on nanoparticles may at least partly explain the frequent observation of little-to-no nanoparticle uptake by root application.108 As an illustrative example, flavonoid signaling in the rhizosphere has been well established, and nanoparticles have previously been shown to form a biocorona rich with flavonoid compounds.78 These results suggest that the rhizosphere will impact nanoparticle corona composition and subsequent plant uptake and that nanoparticles can interfere with native signaling functions necessary for healthy organism maintenance.78,108,110
After the rhizosphere has been navigated and nanoparticles engage directly with the roots, multiple studies have found that positively charged nanoparticles are more readily adsorbed onto and in through root surfaces.101,104,111,112 However, once inside the plant, the opposite trend occurs, whereby negatively charged nanoparticles promote higher translocation efficiency. This presents an opportunity to harness an engineered biocorona, promoting either shedding or adsorption of an outer layer to enable charge reversal upon internalization, such as the adsorption of negatively charged root mucilage upon root traversal. Similar scenarios exist in animal systems, such as with the mucosal and intestinal barriers that nanoparticles encounter during oral delivery, and designer corona approaches have displayed success in allowing a changing surface charge to mediate effective biological barrier passage.113
For foliar application, nanoparticles first come into contact with the leaf cuticle and stomata openings. The cuticle is a waxy coating designed to protect the plant from water loss and nonselective molecular entry and consists of lipids and hydrocarbons that cover most of the leaf exterior. The cuticular pathway comprises modes of access for lipophilic molecules via diffusion and permeation or for polar and ionic solutes via pores, with pore diameters <5 nm.108 Although nanoparticle passage through the cuticle is typically not observed unless surfactants are employed,8,102 Avellan et al. found that PVP-coated gold nanoparticles at least partly enter leaves via disruption and/or diffusion through the leaf cuticle.47 This cuticular uptake route or mere interaction with the surface could bestow nanoparticles with a hydrophobic surface coating that fundamentally modifies subsequent nanoparticle interactions with the internal biofluid. However, nanoparticle entry through leaves is generally found to occur through stomata, despite this uptake mechanism remaining unclear.8,114 Stomata are the leaf pores for gas exchange, displaying pore openings of tens of microns (although the actual size exclusion limit is found to be a few orders of magnitude smaller108) that also facilitate nanoparticle entry and access to the phloem for transport through plants.8,102 Akin to the rhizosphere for roots and similarly understudied in the context of nanoparticles, leaves support a phyllosphere of microorganisms that secrete extracellular polymeric substances that are expected to modulate nanoparticle surface properties through the introduction of a biocorona.108
The plant cell wall, otherwise absent in animal systems, presents a barrier that hinders nanoparticle movement and targeted localization. Even for nanosensors that do not explicitly require cell internalization, nanoparticles must cross cell walls simply to reach the vasculature for translocation to other plant organs. As such, the unique parameters facilitating cell wall traversal must be considered. Cell walls exhibit a small size exclusion limit of approximately 5–20 nm.103 Additionally, stiffer nanoparticle constructs have exhibited higher plant cell internalization.115 Biocorona formation is expected to play a role in prohibiting cell uptake due to the increase in hydrodynamic nanoparticle size and the reduction of inherent nanoparticle stiffness, with the adsorption of a soft biomolecular shell. More broadly, analogous cellular internalization studies in animal systems demonstrate that this process is both governed by the extracellular corona and further imparts an intracellular corona on nanoparticles, potentially disrupting nanotechnology function.116–118 However, fundamental understanding of the interaction of nanoparticles with cell walls and how the corona would impact internalization is lacking due to the difficulties of both measuring and modeling such systems, where more focus has been placed on coronacoated nanoparticle interactions with cell membranes.119,120 For cell membrane passage, the biocorona is expected to govern uptake mechanisms, as seen in mammalian systems.121 Intracellular nanosensors may also traverse the lipid bilayers of organelle membranes, such as for carbon nanotube-based sensors used in chloroplasts.21,122 Toward this latter point, a model termed Lipid Exchange Envelope Penetration (LEEP) has been developed to describe nanoparticle internalization into chloroplasts as a function of effective nanoparticle surface charge (zeta potential) and smallest dimension.123 Interestingly, this study reveals that the magnitude, not the sign, of the zeta potential governs spontaneous nanoparticle uptake and trapping in chloroplasts. Biocorona formation is expected to modulate both of these nanoparticle parameters, such as adsorbed ligands reducing electrostatic stabilization to potentially prohibit effective chloroplast localization.
3.3. Biological Conditions That Nanoparticles Encounter in Plants, with Posited Effects on Biocorona Formation.
Beyond the modes of nanoparticle transport and barriers to such movement, the molecular entities and conditions uniquely present in plants at each of these points must be considered in the context of biocorona composition. Broadly, such constituents include biomolecules (proteins, sugars, lipids, etc.), inorganic ions, and natural organic matter (NOM) in the surrounding environment, and conditions such as ionic strength, pH, and sap flow rates. Plant organs each express similar proteins at different abundance levels, with a dynamic range of over 6 orders of magnitude for the case of Arabidopsis thaliana,87 and plant saps exhibit distinct constituents dependent on function, such as xylem versus phloem sap.8,124
To reiterate from section 3.2, soil-administered nanoparticles are expected to possess an adsorbed biocorona prior to contact with plant roots, likely entailing NOM. Nanoparticle-bound organic macromolecules including humic acid, fulvic acid, and citric acid and soluble extracellular polymeric substances have been demonstrated to enhance nanoparticle stability against high-ionic-strength-induced nanoparticle aggregation,125,126 particularly as driven by divalent cations (Ca2+ and Mg2+).127 Increased stability is presumably by means of both electrostatic and steric stabilization, yet polymer bridging effects could also bring about nanoparticle flocculation. NOM adsorption has the potential to displace pre-existing surface moieties and form highly heterogeneous surface coatings,128 potentially rendering the nanosensor construct nonfunctional.
In the vasculature, nanoparticles can interact with the distinct sap constituents of the xylem versus the phloem. Studies delving into the interactions of sap components with nanoparticles, and the subsequent impact on nanoparticle fate in plants, remain rare in the literature.8 Both the xylem and phloem vascular bundles transport water, nutrients, and metabolites, and the phloem additionally plays a role in transporting signaling molecules including proteins and small signaling molecules such as hormones and mRNAs.124 To briefly summarize the differing compositions, the phloem consists of appreciable amounts of potassium, calcium, magnesium, sodium, chlorine, phosphorus, nitrogen, sulfur, sugars, amino acids, organic acids, and proteins, while the xylem consists of similar inorganic ions and proteins at lower concentrations (approximately an order of magnitude for the former) and no sugars or organic acids.8 As such, nanoparticles entering by roots and traveling by xylem may be challenged with adsorption of far fewer biomolecules, in contrast to nanoparticles entering by leaves and traveling by phloem.
Plant biofluids can be further distinguished on the microscale in terms of the apoplastic and symplastic fluids as well as organelles. The apoplast is the space outside of plant cell membranes, encompassing the cell wall matrix and intercellular spaces. As the apoplastic fluid acts as the interface between the xylem and phloem, the composition correspondingly reflects exchange between the vascular bundles, while specifically, leaf apoplastic fluid consists mainly of proteins for metabolic processes.124 The symplast comprises the intracellular region, facilitating cell-to-cell transport of biomacromolecules that is rarely observed for nanoparticles.108 Although the apoplastic space has been posited as a nanoparticle translocation pathway, there is no general agreement in the literature as to whether or not nanoparticles primarily move through the apoplastic or symplastic pathway.100 Within organelles, prior work has taken advantage of the large pool of flavonoids inside vacuoles,78 yet such highly abundant metabolites may interfere with intended nanosensor outcomes and must be taken into account a priori. For example, the highly abundant protein RuBisCO that composes nearly half of the stroma protein content of chloroplasts may be relevant for the biocorona expected on nanosensors localizing to the chloroplasts, important for sensing tasks such as monitoring photosynthesis.129 It is important to also note that morphological and physiological characteristics of plant biofluids and tissues vary as functions of plant species, growth stage, and external conditions including weather, time of day, and nutrients.8,108 All such factors lead to variability in experiments that must be evaluated on a case-by-case basis.
CONCLUSION
The translation of nanoparticle-based sensors for widespread agricultural applications could greatly advance plant monitoring through continuous, nondestructive sensing of crop health. Although nanoparticle interactions and transformations in the context of these plant bioenvironments have been reported, our understanding of biocorona formation in plant systems and its impact on nanoparticle function remain fairly limited. To further develop our knowledge of biocorona formation on nanosensors, studies in animal systems provide a template to guide our inquiry into plant systems, with key challenges for working in plants that include proteomic and metabolomic considerations, plant transport, biological barriers, and biofluid constituents. By considering these biological features in plant systems in the context of engineered nanosensor properties, we can tune and improve nanosensor design for the seamless translation of these nanotechnologies in agricultural practice.
ACKNOWLEDGMENTS
We acknowledge support of the IGI LGR ERA, GlaxoSmithKline, and Citris/Banatao Seed Funding. We acknowledge support of a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI) (to M.P.L.), a Dreyfus foundation award (to M.P.L.), a Stanley Fahn PDF Junior Faculty Grant with Award # PF-JFA-1760 (to M.P.L.), a Beckman Foundation Young Investigator Award (to M.P.L.), an NIH MIRA award (to M.P.L.), an NSF CAREER award (to M.P.L), an NSF CBET award (to M.P.L.), an NSF CGEM award (to M.P.L.), a FFAR Young Investigator award (to M.P.L.), a CZI investigator award (to M.P.L), a Sloan Foundation Award (to M.P.L.), a USDA BBT EAGER award (to M.P.L), a USDA NIFA Award (to M.P.L), a Moore Foundation Award (to M.P.L.), a Cisco Research Center grant (to M.P.L), and a DARPA Young Investigator Award (to M.P.L.). M.P.L. is a Chan Zuckerberg Biohub investigator, a Hellen Wills Neuroscience Institute Investigator, and an IGI Investigator. R.L.P. acknowledges financial support from the National Science Foundation Graduate Research Fellowship (NSF DGE 1752814). N.S.G. acknowledges financial support from the Foundation for Food and Agriculture Research (FFAR) Fellows program. We would like to acknowledge the use of medical clipart from Servier Medical Art by Servier (http://smart.servier.com), licensed under a Creative Commons Attribution 3.0 Unported License.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acssensors.1c01159
The authors declare no competing financial interest.
Contributor Information
Elizabeth Voke, Department of Chemical and Biomolecular Engineering, University of California, Berkeley, California 94720, United States.
Rebecca L. Pinals, Department of Chemical and Biomolecular Engineering, University of California, Berkeley, California 94720, United States.
Natalie S. Goh, Department of Chemical and Biomolecular Engineering University of California, Berkeley, California 94720, United States
Markita P. Landry, Department of Chemical and Biomolecular Engineering and California Institute for Quantitative Biosciences, QB3, University of California, Berkeley, California 94720, United States; Innovative Genomics Institute (IGI), Berkeley, California 94720, United States; Chan-Zuckerberg Biohub, San Francisco, California 94158, United States.
REFERENCES
- (1).Giraldo JP; Wu H; Newkirk GM; Kruss S Nano-biotechnology Approaches for Engineering Smart Plant Sensors. Nat. Nanotechnol 2019, 14 (6), 541–553. [DOI] [PubMed] [Google Scholar]
- (2).Tilman D; Balzer C; Hill J; Befort BL Global Food Demand and the Sustainable Intensification of Agriculture. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (50), 20260–20264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Reid WV; Ali MK; Field CB The Future of Bioenergy. Glob. Change Biol 2020, 26 (1), 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Antonacci A; Arduini F; Moscone D; Palleschi G; Scognamiglio V Nanostructured (Bio)Sensors for Smart Agriculture. TrAC, Trends Anal. Chem 2018, 98, 95–103. [Google Scholar]
- (5).Kwak S-Y; Wong MH; Lew TTS; Bisker G; Lee MA; Kaplan A; Dong J; Liu AT; Koman VB; Sinclair R; Hamann C; Strano MS Nanosensor Technology Applied to Living Plant Systems. Annu. Rev. Anal. Chem 2017, 10 (1), 113–140. [DOI] [PubMed] [Google Scholar]
- (6).Roper JM; Garcia JF; Tsutsui H Emerging Technologies for Monitoring Plant Health in Vivo. ACS Omega 2021, 6, 5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Arduini F; Cinti S; Scognamiglio V; Moscone D; Palleschi G How Cutting-Edge Technologies Impact the Design of Electrochemical (Bio)Sensors for Environmental Analysis. A Review. Anal. Chim. Acta 2017, 959, 15–42. [DOI] [PubMed] [Google Scholar]
- (8).Su Y; Ashworth V; Kim C; Adeleye AS; Rolshausen P; Roper C; White J; Jassby D Delivery, Uptake, Fate, and Transport of Engineered Nanoparticles in Plants: A Critical Review and Data Analysis. Environ. Sci.: Nano 2019, 6 (8), 2311–2331. [Google Scholar]
- (9).Nel AE; Mädler L; Velegol D; Xia T; Hoek EMV; Somasundaran P; Klaessig F; Castranova V; Thompson M Understanding Biophysicochemical Interactions at the Nano–Bio Interface. Nat. Mater 2009, 8 (7), 543–557. [DOI] [PubMed] [Google Scholar]
- (10).Chetwynd AJ; Lynch I The Rise of the Nanomaterial Metabolite Corona, and Emergence of the Complete Corona. Environ. Sci.: Nano 2020, 7 (4), 1041–1060. [Google Scholar]
- (11).Monopoli MP; Åberg C; Salvati A; Dawson KA Biomolecular Coronas Provide the Biological Identity of Nanosized Materials. Nat. Nanotechnol 2012, 7 (12), 779–786. [DOI] [PubMed] [Google Scholar]
- (12).Yin H; Cao Y; Marelli B; Zeng X; Mason AJ; Cao C Soil Sensors and Plant Wearables for Smart and Precision Agriculture. Adv. Mater 2021, 33, 2007764. [DOI] [PubMed] [Google Scholar]
- (13).AndradeSánchez P; Upadhyaya SK; AgüeraVega J; Jenkins BM EVALUATION OF A CAPACITANCE-BASED SOIL MOISTURE SENSOR FOR REAL-TIME APPLICATIONS. Trans. ASAE 2004, 47 (4), 1281–1287. [Google Scholar]
- (14).McGee TD Principles and Methods of Temperature Measurement; Wiley: New York, 1988. [Google Scholar]
- (15).Ali MA; Wang X; Chen Y; Jiao Y; Mahal NK; Moru S; Castellano MJ; Schnable JC; Schnable PS; Dong L Continuous Monitoring of Soil Nitrate Using a Miniature Sensor with Poly(3-Octyl-Thiophene) and Molybdenum Disulfide Nanocomposite. ACS Appl. Mater. Interfaces 2019, 11 (32), 29195–29206. [DOI] [PubMed] [Google Scholar]
- (16).Nesakumar N; Sethuraman S; Krishnan UM; Rayappan JBB Electrochemical Acetylcholinesterase Biosensor Based on ZnO Nanocuboids Modified Platinum Electrode for the Detection of Carbosulfan in Rice. Biosens. Bioelectron 2016, 77, 1070–1077. [DOI] [PubMed] [Google Scholar]
- (17).Srivastava AK; Dev A; Karmakar S Nanosensors and Nanobiosensors in Food and Agriculture. Environ. Chem. Lett 2018, 16 (1), 161–182. [Google Scholar]
- (18).Barahona F; Bardliving CL; Phifer A; Bruno JG; Batt CA An Aptasensor Based on Polymer-Gold Nanoparticle Composite Microspheres for the Detection of Malathion Using Surface-Enhanced Raman Spectroscopy. Ind. Biotechnol 2013, 9, 42. [Google Scholar]
- (19).Yu Z; Zhao G; Liu M; Lei Y; Li M Fabrication of a Novel Atrazine Biosensor and Its Subpart-per-Trillion Levels Sensitive Performance. Environ. Sci. Technol 2010, 44 (20), 7878–7883. [DOI] [PubMed] [Google Scholar]
- (20).Yang T; Duncan TV Challenges and Potential Solutions for Nanosensors Intended for Use with Foods. Nat. Nanotechnol 2021, 16 (3), 251–265. [DOI] [PubMed] [Google Scholar]
- (21).Lew TTS; Koman VB; Silmore KS; Seo JS; Gordiichuk P; Kwak S-Y; Park M; Ang MC-Y; Khong DT; Lee MA; Chan-Park MB; Chua N-H; Strano MS Real-Time Detection of Wound-Induced H2O2 Signalling Waves in Plants with Optical Nanosensors. Nat. Plants 2020, 6 (4), 404–415. [DOI] [PubMed] [Google Scholar]
- (22).Lu Y; Xu K; Zhang L; Deguchi M; Shishido H; Arie T; Pan R; Hayashi A; Shen L; Akita S; Takei K Multimodal Plant Healthcare Flexible Sensor System. ACS Nano 2020, 14, 10966. [DOI] [PubMed] [Google Scholar]
- (23).Lew TTS; Park M; Cui J; Strano MS Plant Nanobionic Sensors for Arsenic Detection. Adv. Mater 2021, 33 (1), 2005683. [DOI] [PubMed] [Google Scholar]
- (24).Giraldo JP; Landry MP; Kwak S-Y; Jain RM; Wong MH; Iverson NM; Ben-Naim M; Strano MS A Ratiometric Sensor Using Single Chirality Near-Infrared Fluorescent Carbon Nanotubes: Application to In Vivo Monitoring. Small 2015, 11 (32), 3973–3984. [DOI] [PubMed] [Google Scholar]
- (25).Li J; Wu H; Santana I; Fahlgren M; Giraldo JP Standoff Optical Glucose Sensing in Photosynthetic Organisms by a Quantum Dot Fluorescent Probe. ACS Appl. Mater. Interfaces 2018, 10 (34), 28279–28289. [DOI] [PubMed] [Google Scholar]
- (26).Nagata T; Nakamura A; Akizawa T; Pan-Hou H Genetic Engineering of Transgenic Tobacco for Enhanced Uptake and Bioaccumulation of Mercury. Biol. Pharm. Bull 2009, 32 (9), 1491–1495. [DOI] [PubMed] [Google Scholar]
- (27).Wong MH; Giraldo JP; Kwak S-Y; Koman VB; Sinclair R; Lew TTS; Bisker G; Liu P; Strano MS Nitroaromatic Detection and Infrared Communication from Wild-Type Plants Using Plant Nanobionics. Nat. Mater 2017, 16 (2), 264–272. [DOI] [PubMed] [Google Scholar]
- (28).Kundu M; Krishnan P; Kotnala RK; Sumana G Recent Developments in Biosensors to Combat Agricultural Challenges and Their Future Prospects. Trends Food Sci. Technol 2019, 88, 157–178. [Google Scholar]
- (29).Sarma H; Joshi SJ; Prasad R; Jampilek J Biobased Nanotechnology for Green Applications; Springer Nature, 2021. [Google Scholar]
- (30).Chen C; Yuan Z; Chang H-T; Lu F; Li Z; Lu C Silver Nanoclusters as Fluorescent Nanosensors for Selective and Sensitive Nitrite Detection. Anal. Methods 2016, 8 (12), 2628–2633. [Google Scholar]
- (31).Bakhori NM; Yusof NA; Abdullah AH; Hussein MZ Development of a Fluorescence Resonance Energy Transfer (FRET)-Based DNA Biosensor for Detection of Synthetic Oligonucleotide of Ganoderma Boninense. Biosensors 2013, 3 (4), 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wu H; Nißler R; Morris V; Herrmann N; Hu P; Jeon SJ; Kruss S; Giraldo JP Monitoring Plant Health with Near-Infrared Fluorescent H2O2 Nanosensors. Nano Lett. 2020, 20 (4), 2432–2442. [DOI] [PubMed] [Google Scholar]
- (33).Mishra P; Polder G; Vilfan N Close Range Spectral Imaging for Disease Detection in Plants Using Autonomous Platforms: A Review on Recent Studies. Curr. Robot. Rep 2020, 1 (2), 43–48. [Google Scholar]
- (34).Li SJ; Hsu JJ; Lin ZP; Cheng TC; Chiu CY; Liao CY; Chang CY; Yao KS; Tzeng KC Fluorescence Silica Nanoprobe as a Biomarker for Rapid Detection of Plant Pathogens. Adv. Mater. Res 2009, 79–82, 513. [Google Scholar]
- (35).Khater M; de la Escosura-Muñiz A; Merkoçi A Biosensors for Plant Pathogen Detection. Biosens. Bioelectron 2017, 93, 72–86. [DOI] [PubMed] [Google Scholar]
- (36).Salomone A; Mongelli M; Roggero P; Boscia D; Delle S Reliability of Detection of Citrus Tristeza Virus by an Immunochromatographic Lateral Flow Assay in Comparison with ELISA. J. Plant Pathol 2004, 86, 1. [Google Scholar]
- (37).Drygin YF; Blintsov AN; Grigorenko VG; Andreeva IP; Osipov AP; Varitzev YA; Uskov AI; Kravchenko DV; Atabekov JG Highly Sensitive Field Test Lateral Flow Immunodiagnostics of PVX Infection. Appl. Microbiol. Biotechnol 2012, 93 (1), 179–189. [DOI] [PubMed] [Google Scholar]
- (38).Zhang F; Zou M; Chen Y; Li J; Wang Y; Qi X; Xue Q Lanthanide-Labeled Immunochromatographic Strips for the Rapid Detection of Pantoea Stewartii Subsp. Stewartii. Biosens. Bioelectron 2014, 51, 29–35. [DOI] [PubMed] [Google Scholar]
- (39).Pandey P; Irulappan V; Bagavathiannan MV; Senthil-Kumar M Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-Morphological Traits. Front. Plant Sci 2017, 8, 1 DOI: 10.3389/fpls.2017.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Savatin DV; Gramegna G; Modesti V; Cervone F Wounding in the Plant Tissue: The Defense of a Dangerous Passage. Front. Plant Sci 2014, 5, 1 DOI: 10.3389/fpls.2014.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hedrich R; Salvador-Recatalà V; Dreyer I Electrical Wiring and Long-Distance Plant Communication. Trends Plant Sci. 2016, 21 (5), 376–387. [DOI] [PubMed] [Google Scholar]
- (42).Zhou J-M; Zhang Y Plant Immunity: Danger Perception and Signaling. Cell 2020, 181 (5), 978–989. [DOI] [PubMed] [Google Scholar]
- (43).Lew TTS; Koman VB; Gordiichuk P; Park M; Strano MS The Emergence of Plant Nanobionics and Living Plants as Technology. Adv. Mater. Technol 2020, 5 (3), 1900657. [Google Scholar]
- (44).Pinals RL; Yang D; Rosenberg DJ; Chaudhary T; Crothers AR; Iavarone AT; Hammel M; Landry MP Quantitative Protein Corona Composition and Dynamics on Carbon Nanotubes in Biological Environments. Angew. Chem., Int. Ed 2020, 59 (52), 23668–23677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lundqvist M; Stigler J; Cedervall T; Berggård T; Flanagan MB; Lynch I; Elia G; Dawson K The Evolution of the Protein Corona around Nanoparticles: A Test Study. ACS Nano 2011, 5 (9), 7503–7509. [DOI] [PubMed] [Google Scholar]
- (46).Monopoli MP; Walczyk D; Campbell A; Elia G; Lynch I; Baldelli Bombelli F; Dawson KA Physical-Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc 2011, 133 (8), 2525–2534. [DOI] [PubMed] [Google Scholar]
- (47).Avellan A; Yun J; Zhang Y; Spielman-Sun E; Unrine JM; Thieme J; Li J; Lombi E; Bland G; Lowry GV Nanoparticle Size and Coating Chemistry Control Foliar Uptake Pathways, Translocation, and Leaf-to-Rhizosphere Transport in Wheat. ACS Nano 2019, 13 (5), 5291–5305. [DOI] [PubMed] [Google Scholar]
- (48).Martínez-Negro M; González-Rubio G; Aicart E; Landfester K; Guerrero-Martínez A; Junquera E Insights into Colloidal Nanoparticle-Protein Corona Interactions for Nanomedicine Applications. Adv. Colloid Interface Sci 2021, 289, 102366. [DOI] [PubMed] [Google Scholar]
- (49).Baetke SC; Lammers T; Kiessling F Applications of Nanoparticles for Diagnosis and Therapy of Cancer. Br. J. Radiol 2015, 88 (1054), 20150207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Han X; Xu K; Taratula O; Farsad K Applications of Nanoparticles in Biomedical Imaging. Nanoscale 2019, 11 (3), 799–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Cherukuri P; Glazer ES; Curley SA Targeted Hyperthermia Using Metal Nanoparticles. Adv. Drug Delivery Rev 2010, 62 (3), 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Fasoli E Protein Corona: Dr. Jekyll and Mr. Hyde of Nanomedicine. Biotechnol. Appl. Biochem 2020, 1 DOI: 10.1002/bab.2035. [DOI] [PubMed] [Google Scholar]
- (53).Singh S; Gill AAS; Nlooto M; Karpoormath R Prostate Cancer Biomarkers Detection Using Nanoparticles Based Electrochemical Biosensors. Biosens. Bioelectron 2019, 137, 213–221. [DOI] [PubMed] [Google Scholar]
- (54).Chinen AB; Guan CM; Ferrer JR; Barnaby SN; Merkel TJ; Mirkin CA Nanoparticle Probes for the Detection of Cancer Biomarkers, Cells, and Tissues by Fluorescence. Chem. Rev 2015, 115 (19), 10530–10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Du B; Yu M; Zheng J Transport and Interactions of Nanoparticles in the Kidneys. Nat. Rev. Mater 2018, 3 (10), 358–374. [Google Scholar]
- (56).Lima T; Bernfur K; Vilanova M; Cedervall T Understanding the Lipid and Protein Corona Formation on Different Sized Polymeric Nanoparticles. Sci. Rep 2020, 10 (1), 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Getts DR; Shea LD; Miller SD; King NJC Harnessing Nanoparticles for Immune Modulation. Trends Immunol. 2015, 36 (7), 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Tang J; Pérez-Medina C; Zhao Y; Sadique A; Mulder WJM; Reiner T A Comprehensive Procedure to Evaluate the In Vivo Performance of Cancer Nanomedicines. J. Visualized Exp 2017, e55271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Ledford H Bankruptcy Filing Worries Developers of Nanoparticle Cancer Drugs. Nature 2016, 533 (7603), 304. [DOI] [PubMed] [Google Scholar]
- (60).He H; Liu L; Morin EE; Liu M; Schwendeman A Survey of Clinical Translation of Cancer Nanomedicines—Lessons Learned from Successes and Failures. Acc. Chem. Res 2019, 52 (9), 2445–2461. [DOI] [PubMed] [Google Scholar]
- (61).Salvati A; Pitek AS; Monopoli MP; Prapainop K; Bombelli FB; Hristov DR; Kelly PM; Åberg C; Mahon E; Dawson KA Transferrin-Functionalized Nanoparticles Lose Their Targeting Capabilities When a Biomolecule Corona Adsorbs on the Surface. Nat. Nanotechnol 2013, 8 (2), 137–143. [DOI] [PubMed] [Google Scholar]
- (62).Tenzer S; Docter D; Kuharev J; Musyanovych A; Fetz V; Hecht R; Schlenk F; Fischer D; Kiouptsi K; Reinhardt C; Landfester K; Schild H; Maskos M; Knauer SK; Stauber RH Rapid Formation of Plasma Protein Corona Critically Affects Nanoparticle Pathophysiology. Nat. Nanotechnol 2013, 8 (10), 772–781. [DOI] [PubMed] [Google Scholar]
- (63).Mahmoudi M Debugging Nano-Bio Interfaces: Systematic Strategies to Accelerate Clinical Translation of Nanotechnologies. Trends Biotechnol. 2018, 36 (8), 755–769. [DOI] [PubMed] [Google Scholar]
- (64).Pinals RL; Chio L; Ledesma F; Landry MP Engineering at the Nano-Bio Interface: Harnessing the Protein Corona towards Nanoparticle Design and Function. Analyst 2020, 145 (15), 5090–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Weiss ACG; Kempe K; Förster S; Caruso F Microfluidic Examination of the “Hard” Biomolecular Corona Formed on Engineered Particles in Different Biological Milieu. Biomacromolecules 2018, 19 (7), 2580–2594. [DOI] [PubMed] [Google Scholar]
- (66).Docter D; Westmeier D; Markiewicz M; Stolte S; Knauer SK; Stauber RH The Nanoparticle Biomolecule Corona: Lessons Learned - Challenge Accepted? Chem. Soc. Rev 2015, 44 (17), 6094–6121. [DOI] [PubMed] [Google Scholar]
- (67).Lundqvist M; Stigler J; Elia G; Lynch I; Cedervall T; Dawson KA Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (38), 14265–14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Walkey CD; Chan WCW Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem. Soc. Rev 2012, 41 (7), 2780–2799. [DOI] [PubMed] [Google Scholar]
- (69).García KP; Zarschler K; Barbaro L; Barreto JA; O’Malley W; Spiccia L; Stephan H; Graham B Zwitterionic-Coated “Stealth” Nanoparticles for Biomedical Applications: Recent Advances in Countering Biomolecular Corona Formation and Uptake by the Mononuclear Phagocyte System. Small 2014, 10 (13), 2516–2529. [DOI] [PubMed] [Google Scholar]
- (70).Lesniak A; Fenaroli F; Monopoli MP; Åberg C; Dawson KA; Salvati A Effects of the Presence or Absence of a Protein Corona on Silica Nanoparticle Uptake and Impact on Cells. ACS Nano 2012, 6 (7), 5845–5857. [DOI] [PubMed] [Google Scholar]
- (71).Naidu PSR; Gavriel N; Gray CGG; Bartlett CA; Toomey LM; Kretzmann JA; Patalwala D; McGonigle T; Denham E; Hee C; Ho D; Taylor NL; Norret M; Smith NM; Dunlop SA; Iyer KS; Fitzgerald M Elucidating the Inability of Functionalized Nanoparticles to Cross the Blood-Brain Barrier and Target Specific Cells in Vivo. ACS Appl. Mater. Interfaces 2019, 11 (25), 22085–22095. [DOI] [PubMed] [Google Scholar]
- (72).Raoufi M; Hajipour MJ; Kamali Shahri SM; Schoen I; Linn U; Mahmoudi M Probing Fibronectin Conformation on a Protein Corona Layer around Nanoparticles. Nanoscale 2018, 10 (3), 1228–1233. [DOI] [PubMed] [Google Scholar]
- (73).Warning LA; Zhang Q; Baiyasi R; Landes CF; Link S Nanoscale Surface-Induced Unfolding of Single Fibronectin Is Restricted by Serum Albumin Crowding. J. Phys. Chem. Lett 2020, 11, 1170. [DOI] [PubMed] [Google Scholar]
- (74).Bing J; Xiao X; McClements DJ; Biao Y; Chongjiang C Protein Corona Formation around Inorganic Nanoparticles: Food Plant Proteins-TiO2 Nanoparticle Interactions. Food Hydrocolloids 2021, 115, 106594. [Google Scholar]
- (75).Prakash S; Deswal R Analysis of Temporally Evolved Nanoparticle-Protein Corona Highlighted the Potential Ability of Gold Nanoparticles to Stably Interact with Proteins and Influence the Major Biochemical Pathways in Brassica Juncea. Plant Physiol. Biochem 2020, 146, 143–156. [DOI] [PubMed] [Google Scholar]
- (76).Arora S; Sharma P; Kumar S; Nayan R; Khanna PK; Zaidi MGH Gold-Nanoparticle Induced Enhancement in Growth and Seed Yield of Brassica Juncea. Plant Growth Regul. 2012, 66 (3), 303–310. [Google Scholar]
- (77).Kurepa J; Nakabayashi R; Paunesku T; Suzuki M; Saito K; Woloschak GE; Smalle JA Direct Isolation of Flavonoids from Plants Using Ultra-small Anatase TIO 2 Nanoparticles. Plant J. 2014, 77 (3), 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Kurepa J; Shull TE; Smalle JA Metabolomic Analyses of the Bio-Corona Formed on TiO2 Nanoparticles Incubated with Plant Leaf Tissues. J. Nanobiotechnol 2020, 18 (1), 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Kurepa J; Smalle JA Composition of the Metabolomic BioCoronas Isolated from Ocimum Sanctum and Rubia Tinctorum. BMC Res. Notes 2021, 14, 1 DOI: 10.1186/s13104-020-05420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Qing L-S; Shan X-Q; Xu X-M; Xue Y; Deng W-L; Li B-G; Wang X-L; Liao X Rapid probe and isolation of bioactive compounds from Dioscorea panthaica using human serum albumin functionalized magnetic nano-particles (HSA-MNPs)-based ligand fishing coupled with electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom 2010, 24 (22), 3335–3339. [DOI] [PubMed] [Google Scholar]
- (81).Wang C; Feng N; Chang F; Wang J; Yuan B; Cheng Y; Liu H; Yu J; Zou J; Ding J; Chen X Injectable Cholesterol-Enhanced Stereocomplex Polylactide Thermogel Loading Chondrocytes for Optimized Cartilage Regeneration. Adv. Healthcare Mater 2019, 8, No. 1900312. [DOI] [PubMed] [Google Scholar]
- (82).Oh JY; Kim HS; Palanikumar L; Go EM; Jana B; Park SA; Kim HY; Kim K; Seo JK; Kwak SK; Kim C; Kang S; Ryu J-H Cloaking Nanoparticles with Protein Corona Shield for Targeted Drug Delivery. Nat. Commun 2018, 9 (1), 4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Heazlewood JL; Millar AH Plant Proteomics: Challenges and Resources. Annual Plant Reviews Online 2018, 1–31. [Google Scholar]
- (84).Shannahan J The Biocorona: A Challenge for the Biomedical Application of Nanoparticles. Nanotechnol. Rev 2017, 6 (4), 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Hegeman AD Plant Metabolomics—Meeting the Analytical Challenges of Comprehensive Metabolite Analysis. Briefings Funct. Genomics 2010, 9 (2), 139–148. [DOI] [PubMed] [Google Scholar]
- (86).Peters K; Worrich A; Weinhold A; Alka O; Balcke G; Birkemeyer C; Bruelheide H; Calf OW; Dietz S; Dührkop K; Gaquerel E; Heinig U; Kücklich M; Macel M; Müller C; Poeschl Y; Pohnert G; Ristok C; Rodríguez VM; Ruttkies C; Schuman M; Schweiger R; Shahaf N; Steinbeck C; Tortosa M; Treutler H; Ueberschaar N; Velasco P; Weiß BM; Widdig A; Neumann S; Dam N. M. van. Current Challenges in Plant Eco-Metabolomics. Int. J. Mol. Sci 2018, 19 (5), 1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Mergner J; Frejno M; List M; Papacek M; Chen X; Chaudhary A; Samaras P; Richter S; Shikata H; Messerer M; Lang D; Altmann S; Cyprys P; Zolg DP; Mathieson T; Bantscheff M; Hazarika RR; Schmidt T; Dawid C; Dunkel A; Hofmann T; Sprunck S; Falter-Braun P; Johannes F; Mayer KFX; Jürgens G; Wilhelm M; Baumbach J; Grill E; Schneitz K; Schwechheimer C; Kuster B Mass-Spectrometry-Based Draft of the Arabidopsis Proteome. Nature 2020, 579 (7799), 409–414. [DOI] [PubMed] [Google Scholar]
- (88).Fang C; Fernie AR; Luo J Exploring the Diversity of Plant Metabolism. Trends Plant Sci. 2019, 24 (1), 83–98. [DOI] [PubMed] [Google Scholar]
- (89).Human Metabolome Database: Database Statistics; http://hmdb.ca/statistics (accessed Mar 2, 2021).
- (90).Jorge TF; Mata AT; António C Mass Spectrometry as a Quantitative Tool in Plant Metabolomics. Philos. Trans. R. Soc., A 2016, 374 (2079), No. 20150370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Liu Y; Lu S; Liu K; Wang S; Huang L; Guo L Proteomics: A Powerful Tool to Study Plant Responses to Biotic Stress. Plant Methods 2019, 15 (1), 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Bindschedler LV; Cramer R Quantitative Plant Proteomics. Proteomics 2011, 11 (4), 756–775. [DOI] [PubMed] [Google Scholar]
- (93).Chandramouli K; Qian P-Y Proteomics: Challenges, Techniques and Possibilities to Overcome Biological Sample Complexity. Hum. Genomics Proteomics 2009, 1, 1 DOI: 10.4061/2009/239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).van Wijk KJ Challenges and Prospects of Plant Proteomics. Plant Physiol. 2001, 126 (2), 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Subba P; Narayana Kotimoole C; Prasad TSK Plant Proteome Databases and Bioinformatic Tools: An Expert Review and Comparative Insights. OMICS J. Integr. Biol 2019, 23 (4), 190–206. [DOI] [PubMed] [Google Scholar]
- (96).Karczewski KJ; Snyder MP Integrative Omics for Health and Disease. Nat. Rev. Genet 2018, 19 (5), 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Pérez-Alonso M-M; Carrasco-Loba V; Pollmann S Advances in Plant Metabolomics. Annual Plant Reviews Online 2018, 557–588. [Google Scholar]
- (98).Timp W; Timp G Beyond Mass Spectrometry, the next Step in Proteomics. Sci. Adv 2020, 6 (2), eaax8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Tsugawa H Advances in Computational Metabolomics and Databases Deepen the Understanding of Metabolisms. Curr. Opin. Biotechnol 2018, 54, 10–17. [DOI] [PubMed] [Google Scholar]
- (100).Hubbard JD; Lui A; Landry MP Multiscale and Multidisciplinary Approach to Understanding Nanoparticle Transport in Plants. Curr. Opin. Chem. Eng 2020, 30, 135–143. [Google Scholar]
- (101).Ma C; White JC; Zhao J; Zhao Q; Xing B Uptake of Engineered Nanoparticles by Food Crops: Characterization, Mechanisms, and Implications. Annu. Rev. Food Sci. Technol 2018, 9 (1), 129–153. [DOI] [PubMed] [Google Scholar]
- (102).Schwab F; Zhai G; Kern M; Turner A; Schnoor JL; Wiesner MR Barriers, Pathways and Processes for Uptake, Translocation and Accumulation of Nanomaterials in Plants - Critical Review. Nanotoxicology 2016, 10 (3), 257–278. [DOI] [PubMed] [Google Scholar]
- (103).Navarro E; Baun A; Behra R; Hartmann NB; Filser J; Miao A-J; Quigg A; Santschi PH; Sigg L Environmental Behavior and Ecotoxicity of Engineered Nanoparticles to Algae, Plants, and Fungi. Ecotoxicology 2008, 17 (5), 372–386. [DOI] [PubMed] [Google Scholar]
- (104).Zhu Z-J; Wang H; Yan B; Zheng H; Jiang Y; Miranda OR; Rotello VM; Xing B; Vachet RW Effect of Surface Charge on the Uptake and Distribution of Gold Nanoparticles in Four Plant Species. Environ. Sci. Technol 2012, 46 (22), 12391–12398. [DOI] [PubMed] [Google Scholar]
- (105).Spielman-Sun E; Avellan A; Bland GD; Tappero RV; Acerbo AS; Unrine JM; Giraldo JP; Lowry GV Nanoparticle Surface Charge Influences Translocation and Leaf Distribution in Vascular Plants with Contrasting Anatomy. Environ. Sci.: Nano 2019, 6 (8), 2508–2519. [Google Scholar]
- (106).Cox A; Andreozzi P; Dal Magro R; Fiordaliso F; Corbelli A; Talamini L; Chinello C; Raimondo F; Magni F; Tringali M; Krol S; Jacob Silva P; Stellacci F; Masserini M; Re F Evolution of Nanoparticle Protein Corona across the Blood–Brain Barrier. ACS Nano 2018, 12 (7), 7292–7300. [DOI] [PubMed] [Google Scholar]
- (107).Zhang Y; Wu JLY; Lazarovits J; Chan WCW An Analysis of the Binding Function and Structural Organization of the Protein Corona. J. Am. Chem. Soc 2020, 142 (19), 8827–8836. [DOI] [PubMed] [Google Scholar]
- (108).Lv J; Christie P; Zhang S Uptake, Translocation, and Transformation of Metal-Based Nanoparticles in Plants: Recent Advances and Methodological Challenges. Environ. Sci.: Nano 2019, 6 (1), 41–59. [Google Scholar]
- (109).Avellan A; Schwab F; Masion A; Chaurand P; Borschneck D; Vidal V; Rose J; Santaella C; Levard C Nanoparticle Uptake in Plants: Gold Nanomaterial Localized in Roots of Arabidopsis Thaliana by X-Ray Computed Nanotomography and Hyperspectral Imaging. Environ. Sci. Technol 2017, 51 (15), 8682–8691. [DOI] [PubMed] [Google Scholar]
- (110).Dai Y; Chen F; Yue L; Li T; Jiang Z; Xu Z; Wang Z; Xing B Uptake, Transport, and Transformation of CeO2 Nanoparticles by Strawberry and Their Impact on the Rhizosphere Bacterial Community. ACS Sustainable Chem. Eng 2020, 8 (12), 4792–4800. [Google Scholar]
- (111).Li J; Tappero RV; Acerbo AS; Yan H; Chu Y; Lowry GV; Unrine JM Effect of CeO 2 Nanomaterial Surface Functional Groups on Tissue and Subcellular Distribution of Ce in Tomato (Solanum Lycopersicum). Environ. Sci.: Nano 2019, 6 (1), 273–285. [Google Scholar]
- (112).Hu P; An J; Faulkner MM; Wu H; Li Z; Tian X; Giraldo JP Nanoparticle Charge and Size Control Foliar Delivery Efficiency to Plant Cells and Organelles. ACS Nano 2020, 14 (7), 7970–7986. [DOI] [PubMed] [Google Scholar]
- (113).Wang A; Yang T; Fan W; Yang Y; Zhu Q; Guo S; Zhu C; Yuan Y; Zhang T; Gan Y Protein Corona Liposomes Achieve Efficient Oral Insulin Delivery by Overcoming Mucus and Epithelial Barriers. Adv. Healthcare Mater 2019, 8 (12), 1801123. [DOI] [PubMed] [Google Scholar]
- (114).El-Shetehy M; Moradi A; Maceroni M; Reinhardt D; Petri-Fink A; Rothen-Rutishauser B; Mauch F; Schwab F Silica Nanoparticles Enhance Disease Resistance in Arabidopsis Plants. Nat. Nanotechnol 2021, 16, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Zhang H; Demirer GS; Zhang H; Ye T; Goh NS; Aditham AJ; Cunningham FJ; Fan C; Landry MP DNA Nanostructures Coordinate Gene Silencing in Mature Plants. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (15), 7543–7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Wang C; Chen B; He M; Hu B Composition of Intracellular Protein Corona around Nanoparticles during Internalization. ACS Nano 2021, 15 (2), 3108–3122. [DOI] [PubMed] [Google Scholar]
- (117).Gravely M; Safaee MM; Roxbury D Biomolecular Functionalization of a Nanomaterial To Control Stability and Retention within Live Cells. Nano Lett. 2019, 19 (9), 6203–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Bertoli F; Garry D; Monopoli MP; Salvati A; Dawson KA The Intracellular Destiny of the Protein Corona: A Study on Its Cellular Internalization and Evolution. ACS Nano 2016, 10 (11), 10471–10479. [DOI] [PubMed] [Google Scholar]
- (119).Shadmani P; Mehrafrooz B; Montazeri A; Naghdabadi R Protein Corona Impact on Nanoparticle-Cell Interactions: Toward an Energy-Based Model of Endocytosis. J. Phys.: Condens. Matter 2020, 32 (11), 115101. [DOI] [PubMed] [Google Scholar]
- (120).Forest V; Pourchez J Preferential Binding of Positive Nanoparticles on Cell Membranes Is Due to Electrostatic Interactions: A Too Simplistic Explanation That Does Not Take into Account the Nanoparticle Protein Corona. Mater. Sci. Eng. C 2017, 70, 889–896. [DOI] [PubMed] [Google Scholar]
- (121).Digiacomo L; Cardarelli F; Pozzi D; Palchetti S; Digman MA; Gratton E; Capriotti AL; Mahmoudi M; Caracciolo G An Apolipoprotein-Enriched Biomolecular Corona Switches the Cellular Uptake Mechanism and Trafficking Pathway of Lipid Nanoparticles. Nanoscale 2017, 9 (44), 17254–17262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Giraldo JP; Landry MP; Faltermeier SM; McNicholas TP; Iverson NM; Boghossian AA; Reuel NF; Hilmer AJ; Sen F; Brew JA; Strano MS Plant Nanobionics Approach to Augment Photosynthesis and Biochemical Sensing. Nat. Mater 2014, 13 (4), 400–408. [DOI] [PubMed] [Google Scholar]
- (123).Wong MH; Misra RP; Giraldo JP; Kwak S-Y; Son Y; Landry MP; Swan JW; Blankschtein D; Strano MS Lipid Exchange Envelope Penetration (LEEP) of Nanoparticles for Plant Engineering: A Universal Localization Mechanism. Nano Lett. 2016, 16 (2), 1161–1172. [DOI] [PubMed] [Google Scholar]
- (124).Rodríguez-Celma J; Ceballos-Laita L; Grusak MA; Abadía J; López-Millán A-F Plant Fluid Proteomics: Delving into the Xylem Sap, Phloem Sap and Apoplastic Fluid Proteomes. Biochim. Biophys. Acta, Proteins Proteomics 2016, 1864 (8), 991–1002. [DOI] [PubMed] [Google Scholar]
- (125).Adeleye AS; Keller AA Interactions between Algal Extracellular Polymeric Substances and Commercial TiO2 Nanoparticles in Aqueous Media. Environ. Sci. Technol 2016, 50 (22), 12258–12265. [DOI] [PubMed] [Google Scholar]
- (126).Yang X; Wang Q; Qu X; Jiang W Bound and Unbound Humic Acids Perform Different Roles in the Aggregation and Deposition of Multi-Walled Carbon Nanotubes. Sci. Total Environ 2017, 586, 738–745. [DOI] [PubMed] [Google Scholar]
- (127).Barbero F; Mayall C; Drobne D; Saiz-Poseu J; Bastús NG; Puntes V Formation and Evolution of the Nanoparticle Environmental Corona: The Case of Au and Humic Acid. Sci. Total Environ 2021, 768, 144792. [DOI] [PubMed] [Google Scholar]
- (128).Lead JR; Batley GE; Alvarez PJJ; Croteau M-N; Handy RD; McLaughlin MJ; Judy JD; Schirmer K Nanomaterials in the Environment: Behavior, Fate, Bioavailability, and Effects—An Updated Review. Environ. Toxicol. Chem 2018, 37 (8), 2029–2063. [DOI] [PubMed] [Google Scholar]
- (129).Ferro M; Brugière S; Salvi D; Seigneurin-Berny D; Court M; Moyet L; Ramus C; Miras S; Mellal M; Le Gall S; Kieffer-Jaquinod S; Bruley C; Garin J; Joyard J; Masselon C; Rolland N AT_CHLORO, a Comprehensive Chloroplast Proteome Database with Subplastidial Localization and Curated Information on Envelope Proteins*. Mol. Cell. Proteomics 2010, 9 (6), 1063–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]


