Abstract
Here we present the karyotype analysis and genome sizing of Paracoccidioides brasiliensis, a pathogen refractory to conventional genetic analysis. We have established pulsed-field gel electrophoresis (PFGE) conditions to resolve the high-molecular-weight chromosomal bands of two clinical isolates of P. brasiliensis. Both isolates showed four megabase-sized bands, ranging from 2.0 to 10.0 Mbp. Significant differences in chromosome sizes and in the chromosomal location of genes for the gp43 antigen and chitin synthase were found. Different technical approaches were employed to estimate the DNA content and to define the ploidy of P. brasiliensis. An estimated genome size in the range of 45.7 to 60.9 Mbp was provided by the analysis of data generated by measuring the amplitude of fluorescence intensity of DAPI (4′,6-diamidino-2-phenylindole)-stained nuclei (by confocal microscopy). The nuclear genome size estimated by confocal microscopy is twice that estimated by the average sum of the molecular weight of chromosome-sized DNA molecules by PFGE, suggesting that each separated P. brasiliensis chromosomal band is diploid.
Paracoccidioides brasiliensis is the causative agent of paracoccidioidomycosis (PCM), the most prevalent systemic mycosis in South America, with areas of endemicity in Brazil, Colombia, and Venezuela (17). The disease presents multiple manifestations, and two progressive clinical forms are recognized: acute (multifocal, disseminated) and chronic (unifocal and/or multifocal) forms. The acute form (juvenile type) of PCM is serious and, if not treated, frequently culminates with the patient’s death (13).
This thermal dimorphic fungus grows in a mycelial phase at room temperature (23 to 28°C) and in a yeast phase at 35 to 37°C. A teleomorphic (sexual) stage has not been determined, greatly impairing classical genetic analysis. The hyphae are multicellular and display multinucleate structures. Budding yeasts—unicellular forms, however—were also found to be multinucleate (5, 11, 12, 26). Fungal propagules or conidia are thought to be the infective units of P. brasiliensis (27). When studying the conidia-to-yeast transformation, McEwen et al. (18) showed that >80% of conidia were uninucleate, becoming, however, binucleate or multinucleate (four to five nuclei per cell) during morphogenesis. There are few reports on the isolation and characterization of mutants. Experiments employing in vitro mutagenesis led to the selection of very few mutants with stable phenotypes, since the number of revertants was high and the multinucleate nature of the pathogen may be responsible for the instability of the in vitro-generated mutants (15). Furthermore, the genetic composition of the fungus is virtually unknown and information about the genome size and chromosome organization is scarce (20).
The development of molecular biology techniques, such as pulsed-field gel electrophoresis (PFGE), has allowed the genomic characterization, chromosomal mapping, and molecular epidemiological biotyping of microorganisms refractory to genetic analysis. Approaches applying recombinant DNA technology to the study of P. brasiliensis have recently been adopted (14, 31). The gene encoding the immunodominant antigen of the fungus, gp43 (a 43,000-Da glycoprotein) (25), which is also a laminin ligand potentially involved in the pathogenesis of PCM (36), was the first to be cloned and characterized (7).
In this report, we describe optimized conditions using PFGE for the separation of the chromosome-sized DNA molecules of two P. brasiliensis clinical isolates. By this technique we were able to separate chromosomes of up to 10 Mbp, providing evidence of chromosomal polymorphism in P. brasiliensis. Homologous DNA probes derived from the whole genomic DNA of P. brasiliensis, gp43 antigen and chitin synthase genes were used in hybridization experiments to confirm the number and polymorphism of the chromosomal bands. The genome size of the fungus was estimated by measuring the fluorescence intensity of DAPI (4′,6-diamidino-2-phenylindole)-stained nuclei by confocal microscopy and by the average sum of the molecular weights of chromosome-sized DNA molecules separated by PFGE. This study provides evidence for the diploid nature of P. brasiliensis and represents a starting point for further investigations of the genome organization of this pathogen.
MATERIALS AND METHODS
Microorganisms.
Two clinical isolates of P. brasiliensis, B-339 (ATCC 32069) and 113, were selected for this study. P. brasiliensis B-339 was originally isolated from Brazilian patients with chronic progressive PCM. Samples of isolate B-339 were kindly provided by the Mycology Division of Universidade Federal de São Paulo, São Paulo, Brazil. P. brasiliensis 113 was isolated in 1971 by Fava-Netto from a mucocutaneous lesion of a Brazilian patient. Samples of isolate 113 were obtained from the culture collection of the Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brazil. Both P. brasiliensis isolates have been largely used as a source of diagnostic antigens in Brazilian laboratories (19, 25). A Candida albicans sample was donated by the Mycology Division of Universidade Federal de São Paulo. Epimastigotes from Trypanosoma cruzi (clone CL Brener) were maintained in logarithmic growth phase at 28°C in liver infusion tryptose medium. Fungal isolates were maintained by periodic subculturing in slanted tubes of YPD medium (5 g of yeast extract liter−1, 10 g of Bacto Peptone liter−1, 15 g of dextrose liter−1, 15 g of agar liter−1) at 35 to 37°C.
Preparation of DAPI-stained cells and confocal microscopy.
Cells of 5-day-old cultures grown in YPD broth medium were harvested three times with 0.5 ml of sterile phosphate-buffered saline (PBS). The yeast pellet was resuspended in 0.5 ml of 0.01% (vol/vol) Tween 80 in PBS, and cell clusters were dispersed with a hypodermic syringe with a 28-gauge needle. This procedure was repeated until the suspension was completely homogenized. The cell suspension was then washed three times with 0.5 ml of sterile PBS. Then, cells were fixed for 30 min with 0.5 ml of 3.5% (vol/vol) formaldehyde in PBS and washed as before. The pellets were resuspended in 0.1 ml of PBS, and 10 μl of each cell suspension was applied to microwells of fluorescence slides. The slides were air dried at room temperature and stored at −20°C. Ten millimolar DAPI (Molecular Probes, Eugene, Oreg.) stock solution was diluted to 1:100 in PBS, and 0.02 ml was deposited in each well. The slides were incubated for 1 h at room temperature, washed twice with PBS, and left to dry at room temperature. The slides were mounted with buffered glycerol containing 0.5% (vol/vol) p-phenylenediamine to minimize bleaching (16). Images of DAPI-stained cells were observed on a Bio-Rad 1024-UV confocal system attached to a Zeiss Axiovert 100 microscope, using a 40× numerical aperture 1.2 Plan-Apochromatic differential interference contrast (DIC) water immersion objective. All images were collected by Kalman averaging at least 10 frames (512 by 512 pixels), using an aperture (pinhole) of 1.5 mm, a zoom set of 3.5, and a photomultiplier gain of 1200 (kept during all image acquisitions). DAPI-stained nuclei that could be clearly distinguished in different fields were then subjected to serial optical sectioning (0.14-μm steps), and the fluorescence intensity of the volume of each nucleus was estimated by using processing software (Lasersharp 1024 version 2.1A; Bio-Rad). The collected DIC images were sharpened with a minimum setting by using the same processing software. Fluorescence and DIC prints were generated by dye sublimation on a Codonics NP1600 printer.
Preparation of P. brasiliensis chromosome-sized DNA molecules.
P. brasiliensis yeast cells were subcultured three times in YPD medium, at 5-day intervals. Erlenmeyer flasks containing 50 ml of YPD broth medium were inoculated with the entire growth of two culture slants, placed in a reciprocating shaker at 120 rpm, and grown for 5 days at 35°C. Approximately 108 yeast cells of P. brasiliensis B-339 and 113 were immobilized in 2% (wt/vol) low-melting-point agarose blocks, and spheroplasts were obtained at 30°C by lysing the cell wall in PBS (pH 7.5) containing 10 U of chitinase (Sigma) ml−1 and 30 U of lyticase (Sigma) ml−1 for 1 to 2 h (22, 23, 30). The blocks were dialyzed three times against 250 mM EDTA, pH 8.0, at 37°C to inactivate the enzymes. Spheroplasts were disrupted for 24 h at 56°C by using lysis buffer (500 mM EDTA [pH 8.0], 10 mM Tris-Cl [pH 8.0], 1% [wt/vol] Sarkosyl, 10 mg of proteinase K ml−1). The blocks were washed with 500 mM EDTA and stored at 4°C in the same solution. For preparation of chromosome-sized DNA from the mycelial phase of the fungus, 100 μg (wet weight) of mycelia was also processed as described above. Whole DNAs of strains 113 and B-339 were obtained from frozen yeast cells according to the method of Cisalpino et al. (8).
PFGE separation of P. brasiliensis chromosomes.
Electrophoretic separation was performed under PFGE conditions in a Gene Navigator system (Pharmacia Biotech) with a hexagonal electrode array. DNA from spheroplasts, corresponding to approximately 107 yeast cells per well, was used, and the separations were carried out in 0.6% (wt/vol) agarose in 1.0× TAE (40 mM Tris-acetate [pH 7.5], 2 mM EDTA [pH 8.0]) kept at a constant temperature (10°C). The best separations were achieved by homogeneous pulses (north or south and east or west) with interpolation for 96 h at 42 V: phase 1, pulse time, 900 s (run time, 12 h); phase 2, pulse time, 1,800 s (run time, 12 h); phase 3, pulse time, 2,700 s (run time, 24 h); phase 4, pulse time, 3,600 s (run time, 24 h); and phase 5, pulse time, 4,500 s (run time, 24 h). After electrophoresis, gels were stained with 0.5 μg of ethidium bromide ml−1 and photographed. Chromosome-sized DNA molecules were subjected to acid depurination in the presence of 0.25 M HCl for 5 min and transferred to nylon membranes (Hybond N; Amersham), with 0.5 M Tris, pH 7.0, and 0.4 N NaOH–1.5 M NaCl as neutralization and transfer solutions, respectively (2). MegaBase IV (chromosomal DNA from Schizosaccharomyces pombe; Gibco-BRL) and MegaBase III (chromosomal DNA from Hansenula wingeii; Gibco-BRL) were used as chromosomal DNA size standards.
Digestion of P. brasiliensis chromosomes with restriction endonucleases.
Agarose blocks containing P. brasiliensis chromosomes were washed three times in 10 ml of TE (10 mM Tris-HCl [pH 7.6], 1 mM EDTA [pH 8.0]) at room temperature (30 min each), followed by three washes (30 min each) with 200 μl of the specific restriction enzyme reaction buffer at 4°C. The blocks were incubated for 2 h with 200 μl of the reaction buffer containing 50 U of either SfiI or PacI restriction enzymes at 4°C, followed by incubation for 3 h at 37°C (SfiI) or 50°C (PacI). Following incubation with restriction enzymes, the blocks were washed twice in TE, loaded onto the gel, and subjected to electrophoresis. The resulting megarestriction fragments were separated under conditions used for Saccharomyces cerevisiae PFGE, modified from the method of Chu et al. (6) (phase 1: 60 s, 12 h; phase 2: 120 s, 12 h [both at 120 V]). Following electrophoresis, the gels were stained with 0.5 μg of ethidium bromide ml−1 and photographed and the DNA fragments were transferred onto nylon membranes (8). MegaBase I (chromosomal DNA from S. cerevisiae; Gibco-BRL) was used as the DNA size standard.
DNA probes.
A 630-bp BamHI/HindIII fragment which contained 70% of the coding region of the gp43 antigen gene was isolated from plasmid pUCGPb16A (7). A 600-bp fragment of the chitin synthase gene (9) was generated by PCR amplification of P. brasiliensis genomic DNA with a set of generic primers for the chitin synthase genes described by Bowen et al. (3), presenting identity with the P. brasiliensis CHS2 gene sequence (GenBank accession no. YO9231 [nucleotide 1125 to 1727]). P. brasiliensis genomic DNA was totally digested with AluI, extracted with phenol-CHCl3, and used as a probe.
Preparation of probes and Southern blot hybridization.
The DNA fragments cited above were radiolabeled by a random primer labeling system (Rad primer labeling kit; Gibco-BRL) and used as probes. Hybridizations were carried out overnight at 42°C in 50% formamide–5× SSC (1× SSC [sodium saline citrate] is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt’s solution–50 μg of yeast tRNA ml−1–100 μg of sonicated hearing sperm DNA ml−1–10 μg of poly(A) ml−1–0.1% (wt/vol) sodium dodecyl sulfate. The filters were washed twice in 0.1× SSC–0.1% (wt/vol) sodium dodecyl sulfate at 56°C.
Densitometric scanning.
Ethidium bromide-stained gels were scanned and analyzed by densitometry (550 nm) performed with a Shimadzu Dual Wavelength Flying-Spot Scanner (model C5-9000).
RESULTS
Spheroplast production from P. brasiliensis.
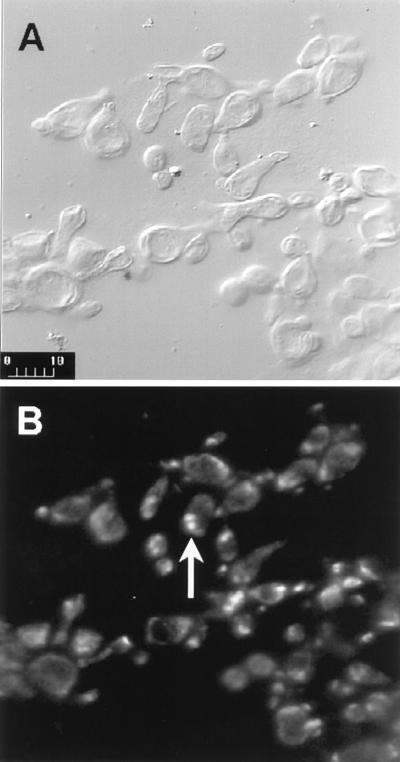
The result of digestion of yeast cell suspensions with a combination of glucanases (Novozym 234 or lyticase) and chitinase activities is shown in Fig. 1. Spheroplasts together with yeast cells were visible in the incubation mixture after 15 min. When stained with DAPI, the yeast cells and/or spheroplasts were seen to be multinucleate (Fig. 1).
FIG. 1.
Spheroplasts of P. brasiliensis yeast cells (isolate B-339) analyzed by confocal fluorescence microscopy. (A) Spheroplasts of P. brasiliensis yeast cells (Nomarski); (B) DAPI-stained spheroplasts of P. brasiliensis yeast cells analyzed under UV light. The arrow indicates a cell where two DAPI-stained nuclei are clearly seen. Bar, 10 μm.
In order to obtain intact chromosome-sized DNA molecules for PFGE studies, yeast cells were embedded in low-melting-point agarose blocks and spheroplasts were obtained at 30°C by lysing the cell wall in PBS (pH 7.5) containing chitinase and lyticase for 1 to 2 h (22, 23, 30). Chromosomal DNA prepared by this method could be stored in EDTA at 4°C for several months without apparent degradation, as assessed by conventional electrophoresis and PFGE. In our experience, the use of chitinase is critical for the efficient generation of yeast spheroplasts.
Separation of chromosome-sized DNA molecules by PFGE.
In our first attempts to establish the molecular karyotype of P. brasiliensis, we used different separation programs and electrophoretic systems. The electrophoresis performed in a contour-clamped homogeneous electric field apparatus, using the program for separation of S. cerevisiae chromosomes, showed that P. brasiliensis chromosomes migrated as two 1.90-Mbp compressed bands (data not shown). Separation of P. brasiliensis chromosomes in a Gene Navigator apparatus using the PFGE conditions previously described for T. cruzi (4) also showed two 3.3-Mbp compressed bands, corresponding to the size of the largest chromosome of H. wingeii (data not shown). These results indicated that P. brasiliensis chromosomes were larger than those of S. cerevisiae and H. wingeii.
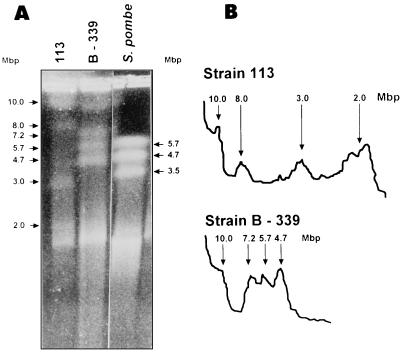
To achieve satisfactory separation of the largest chromosomes of P. brasiliensis, we tested different PFGE conditions, selecting one which resolved molecules larger than 5.7 Mbp and allowed the detection of chromosomal polymorphisms between the two isolates. The results of PFGE and respective densitometric tracings for P. brasiliensis 113 and B-339 isolates are shown in Fig. 2. The karyotype of isolate 113 shows four chromosomal bands, with sizes of approximately 2.0, 3.0, 8.0, and 10.0 Mbp. The sizes of chromosomal bands of isolate B-339 are approximately 4.7, 5.7, 7.2, and 10.0 Mbp. Under the conditions adopted, the electrophoretic karyotypes and staining intensity of the chromosomal bands were reproducible from one preparation to another. The stability of the karyotypes of isolates B-339 and 113 was confirmed by the fact that no changes were detected after more than 10 electrophoretic runnings in three independent chromosomal preparations obtained from both fungal isolates subcultured for 2 years in YPD medium.
FIG. 2.
Electrophoretic karyotype of P. brasiliensis. (A) Ethidium bromide-stained 0.6% (wt/vol) agarose gel of chromosomal preparations of isolates B-339 and 113. The separation of chromosomal bands was carried out on a Gene Navigator apparatus using the electrophoretic conditions described in Materials and Methods. The sizes of P. brasiliensis and S. pombe chromosomal bands (arrows) are indicated to the left and right of the gel. Chromosomal DNA size standard: S. pombe (MegaBase IV; Gibco-BRL). (B) Densitometric-scanning profiles of the ethidium bromide-stained chromosomal bands of isolates 113 and B-339. Peaks corresponding to chromosomal bands (arrows) and their respective sizes are indicated on the figure.
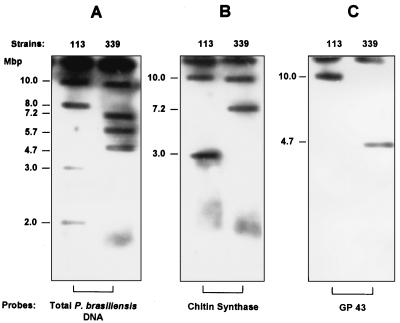
For a better characterization of the distinct chromosomal profiles observed among the isolates, Southern blots carrying intact chromosomes were hybridized with radiolabeled AluI-digested genomic DNA of P. brasiliensis. The probe hybridized with all chromosomal bands of both isolates, confirming the number of ethidium bromide-stained molecules separated by PFGE (Fig. 3).
FIG. 3.
Southern blot hybridization of P. brasiliensis chromosome-sized DNA molecules with homologous probes. Chromosomal bands of isolates 113 and B-339 were separated on a Gene Navigator apparatus using the electrophoretic conditions described in Materials and Methods. Chromosomal DNA was transferred onto nylon filters and hybridized with the following radiolabeled probes: total genomic DNA of P. brasiliensis digested with AluI (A), a 600-bp fragment of the chitin synthase gene (GenBank accession no. YO9231) generated by PCR amplification of P. brasiliensis genomic DNA (B), and a 630-bp BamHI/HindIII fragment from the coding region of the gp43 antigen gene of P. brasiliensis (GenBank accession no. U26160) (C).
Our PFGE analysis shows that the fungus has a genome comprising four large chromosome-sized DNA bands of approximately 2 to 10 Mbp, resulting in distinct karyotypes for the two isolates studied. The genome size, calculated by the addition of the average molecular weights of chromosomal bands, was approximately 23.0 Mbp for isolate 113 and 27.6 Mbp for isolate B-339. The summation of the average molecular sizes of these individual chromosomes demonstrates that the genome sizes of isolates B-339 and 113 are very similar.
Chromosomal mapping of genes encoding gp43 and chitin synthase.
Southern blots carrying intact chromosomes were hybridized with a fragment of the gene coding for the gp43 antigen (7) and with a chitin synthase amplimer (0.6-kb genomic fragment) carrying sequences of the chitin synthase gene (GenBank accession no. YO9231). The gene coding for the gp43 antigen mapped onto the upper chromosomal band (10 Mbp) of isolate 113 and onto the 4.7-Mbp band of isolate B-339. The chitin synthase probe hybridized with two chromosomal bands of 10.0 and 3.0 Mbp of isolate 113 and with two chromosomal bands of 10 and 7.2 Mbp of isolate B-339 (Fig. 3).
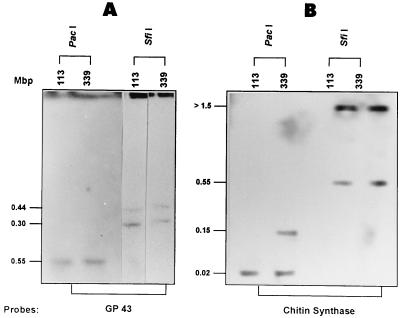
Chromosome-sized DNA molecules from isolates B-339 and 113 were digested with enzymes that infrequently cleave DNA (SfiI and PacI), separated by PFGE, and hybridized with the fragment of the gene coding for gp43 and with the chitin synthase probe. The hybridization profiles obtained with these probes were very similar for isolates B-339 and 113. Figure 4 shows that the gp43 probe hybridized with two SfiI fragments of approximately 440 and 300 kbp and with a single PacI fragment of 50 kbp in both isolates. On the other hand, the chitin synthase probe also hybridized with two SfiI fragments of 550 and 1,500 kbp in both isolates. The only difference was observed with PacI digests. The chitin synthase probe hybridized with two 20-kbp PacI fragments in both isolates, whereas a 150-kbp PacI fragment was only present in isolate B-339.
FIG. 4.
Hybridization patterns of P. brasiliensis chromosome-sized DNA molecules after digestion with rare-cutting site restriction enzymes and separation by PFGE. The chromosomal DNA of isolates 113 and B-339 embedded in agarose were digested with PacI or SfiI restriction endonucleases, subjected to PFGE, and transferred onto nylon filters. Southern blots were hybridized with the gp43 (A) and chitin synthase (B) probes described in the legend to Fig. 3. Sizes of the genomic fragments are shown to the left of each panel.
Estimation of P. brasiliensis genome size.
In this study we used two independent methods, i.e., PFGE and confocal fluorescence microscopy, to estimate the genome size of P. brasiliensis. Using confocal microscopy of DAPI-stained nuclei, we selected on different fields nuclei that could be clearly distinguished (Fig. 1). These were optically sectioned, and the fluorescence intensity of the volume of each nucleus was estimated by using the processing software described in Materials and Methods. After averaging the fluorescence intensity values of at least 12 nuclei, we compared these values with those obtained for C. albicans and T. cruzi, which have known genome sizes (1, 4).
The results summarized in Table 1 indicated that the genome sizes (excluding those of mitochondrial DNA) correlated well with the measurements obtained by confocal microscopy for C. albicans and T. cruzi. Also, the fluorescence intensity measurements were precise and reliable and could be used to accurately determine the size of the P. brasiliensis genome (Table 1). A single P. brasiliensis nucleus contains approximately 45.7 to 60.9 Mbp. The microscopic observation of DAPI-stained P. brasiliensis yeast cells confirmed the presence of 4 to 8 nuclei per cell (Fig. 1) (26). However, it is not possible to claim that each nucleus is an independent genomic entity or that it might be considered part of the total genome of the organism. On the other hand, if we base our calculations on the summation of the average molecular sizes of individual chromosome DNA molecules separated by PFGE, the nuclear genome sizes of P. brasiliensis B-339 and 113 are approximately 27.6 and 23 Mbp, respectively. Thus, by this approach the size of the nuclear genome of the fungus is shown to be about half of that estimated by fluorescence intensity measurements (45.7 to 60.9 Mbp). The present results suggest that the nuclei of P. brasiliensis yeast forms are diploid.
TABLE 1.
Relative amounts of nuclear and mitochondrial DNAs, based on confocal fluorescence microscopy and PFGE
| Organism | No. of nuclei/ cell | No. of chromo- somes/ nucleus | Genome sizea (Mbp) | Fluores- cence amplitudeb | Genome sizec (Mbp) | Mito- chondrial DNA (Mbp) |
|---|---|---|---|---|---|---|
| T. cruzi | 1 | 64d | 87d | 1.944 × 104 | 61–86 | 25–30 |
| P. brasiliensis | 4–8 | 4–5 | 23–27.6 | 1.365 × 104 | 45.7–60.9 | Primitive |
| C. albicans | 1 | 8–12e | 27–36e | 0.806 × 104 | 24–34.8 | 0.04 |
Based on the average sum of the chromosome-sized bands.
Average sum of 12 DNA histograms. The fluorescence amplitude of each nucleus was measured with a Bio-Rad model MCR 1000 confocal microscope (zoom, 3.5; iris 1.5 mm; gain, 200).
Based on fluorescence amplitude.
Cano et al. (4).
Altboum (1).
DISCUSSION
Chromosome-sized DNA molecules of P. brasiliensis were successfully prepared from yeast-derived, multinucleate spheroplasts and resolved by PFGE. The electrophoretic conditions established in this study permitted the clear separation of four chromosomal bands in the range of 2.0 to 10.0 Mbp, and the karyotypes obtained under these conditions were very reproducible. Our results suggest that the chromosomes from P. brasiliensis are comparable in size and number to Coccidioides immitis, Neurospora crassa, and Dictyostelium discoideum chromosomes (10, 22, 23). We did not observe chromosomes smaller than 2.0 Mbp under a variety of electrophoretic conditions. Moreover, when chromoblots were hybridized with P. brasiliensis DNA probe no additional band was detected. Thus, it is unlikely that chromosomes went undetected because they were too small to form visible bands under the electrophoretic conditions employed in this work.
The chromosome number of P. brasiliensis was estimated from analysis of the number and intensity of the chromosomal bands separated by PFGE. The results are consistent with P. brasiliensis having a haploid number of 4 if each band represents one chromosome. The total DNA content derived from addition of the average molecular weights of chromosome-sized bands indicates that the haploid genome size of P. brasiliensis could be in the range of 23 to 27.6 Mbp.
To determine the DNA content and to examine the ploidy of other fungi, optical and photometric methods (flow microfluorometry, fluorescence microscopy, photometry, and cytofluorimetry) have been employed (21, 23, 28, 34, 35). Our results of microfluorometry with P. brasiliensis DAPI-stained nuclei by confocal fluorescence microscopy indicated a DNA content in the range of 45.7 to 60.9 Mbp. The calculated value is twice that estimated by the addition of the molecular weights of chromosomal bands separated by PFGE. This means the DNA content is almost equal to that of two genomes of C. albicans (27 to 36 Mbp) or C. immitis (29 Mbp) (1, 21, 23). The genome complexity of P. brasiliensis is equivalent to that reported for N. crassa (45 to 47 Mbp) or D. discoideum (52 to 56 Mbp) (10, 22). Therefore, we suggest that each separated P. brasiliensis chromosomal band may correspond to two comigrating chromosome molecules of the same size, if the value estimated by PFGE is compared with the more accurate measurements of the fluorescence intensity following optical sectioning of individual DAPI-stained nuclei by confocal microscopy.
Hybridization of selected gene probes (gp43 and chitin synthase genes) on chromoblots was used to further characterize the P. brasiliensis molecular karyotype. The lack of smearing below the hybridization signals indicated that the chromosome-sized DNA molecules were intact and bands had undergone little or no degradation. It is interesting that the gp43 gene mapped onto chromosomal bands with different sizes of each isolate (10.0 Mbp for isolate 113 and 4.7 Mbp for isolate B-339). However, the profiles generated by Southern blot hybridization of megarestriction fragments with the gp43 probe were very similar for both isolates, showing two SfiI fragments of approximately 440 and 300 kbp and a single PacI fragment of 50 kbp. No restriction sites for SfiI or PacI exist on the known sequence of the gene coding for the gp43 antigen, and previous work showed that it is present in a few copies per genome (7). Our results could be interpreted as indicative of the existence of, at least, two copies of the gp43 gene on the same chromosomal band or as evidence of the existence of two allelic forms of the gene, mapping onto two closely comigrating chromosomes.
The chitin synthase probe hybridized with two chromosomal bands on each strain. The profiles obtained by Southern hybridization of the chitin synthase probe with megarestriction fragments could be explained by the existence of several copies of the chitin synthase gene, as has been described for other fungi. It is also noteworthy that in isolate 113, the gp43 and chitin synthase sequences may constitute a genetic linkage group.
Differences in the electrophoretic mobilities of bands of the isolates of P. brasiliensis examined in the present study suggest that chromosome polymorphisms exist and make it difficult to correlate the banding patterns among strains and isolates. In a preliminary report, Montoya et al. (20) presented the molecular karyotype of five clinical P. brasiliensis Colombian isolates, all of which exhibited five chromosome-sized molecules with a unique banding pattern (three bands within the same size range of the S. pombe chromosome and two other bands larger than 5.7 Mbp). Although their results are slightly different from ours, they suggest the existence of a third karyotype which could be explained by the use of different fungal isolates and/or technical approaches. Variation in band mobility and chromosome number is a common feature among strains and isolates of several other pathogenic fungi, including Histoplasma capsulatum (33), C. immitis (23), C. albicans (1), and Cryptococcus neoformans (24). The results of the molecular karyotypes of pathogenic fungi overwhelmingly demonstrate the fluidity of chromosome organization among eukaryotes with small genomes. The plasticity of these genomes could have implications for the maintenance of genome functionality and for the control of gene expression in these organisms (29, 32, 37).
In the present study, we have taken preliminary steps in karyotyping and mapping of species-specific genes of P. brasiliensis and estimated fungus total genome size by two methods, PFGE and confocal fluorescence microscopy of DAPI-stained nuclei, providing strong evidence for the diploid nature of P. brasiliensis. There is little or no genetic data on this fungus. A molecular kayotype combined with physical mapping studies should aid in the identification and isolation of genes of interest, facilitating gene targeting and enabling the construction of physical and genetic maps of this pathogen. The results presented in this work could provide the basis for future genetic, taxonomic, and epidemiological research on P. brasiliensis.
ACKNOWLEDGMENTS
This work was supported by grants from FAPESP, FINEP/BID (66/96/0792/00), PADCT/CNPq, and CYTED (Subprograma III, Biotecnologia).
We thank Roberto Tedesco for help with confocal microscopy and Irvane L. S. Tersariol for his assistance with scanning.
REFERENCES
- 1.Altboum Z. Genetic studies in Candida albicans. In: Segal E, Baum G, editors. Pathogenic yeasts and yeast infections. Boca Raton, Fla: CRC Press; 1994. pp. 33–48. [Google Scholar]
- 2.Birren B, Lai E. Pulsed field gel electrophoresis, a practical guide. San Diego, Calif: Academic Press; 1993. pp. 141–147. [Google Scholar]
- 3.Bowen A R, Chen-Wu J C, Momany M, Young R, Szanislo P J, Robbins P W. Classification of fungal chitin synthases. Proc Natl Acad Sci USA. 1992;89:519–523. doi: 10.1073/pnas.89.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cano M I, Gruber A, Vasquez M, Cortes A, Levin M J, Gonzalez A, Degrave W, Rondinelli E, Zingales B, Ramirez J I, Alonso C, Requema J M, Franco da Silveira J. Trypanosoma cruzi genome project: molecular karyotype of clone CL Brener. Mol Biochem Parasitol. 1995;71:273–278. doi: 10.1016/0166-6851(95)00066-a. [DOI] [PubMed] [Google Scholar]
- 5.Carbonell L M, Gil F. Ultraestrutura del Paracoccidioides brasiliensis. In: del Negro G, Lacaz C S, Fiorillo A M, editors. Paracoccidioidomicose (Blastomicose sul americana). São Paulo, Brazil: Sarvier; 1982. pp. 23–34. [Google Scholar]
- 6.Chu G, Vollrath D, Davis R W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 7.Cisalpino P, Puccia R, Yamauchi L, Cano M I, Franco da Silveira J, Travassos L R. Cloning, characterization and epitope expression of the major diagnostic antigen of Paracoccidioides brasiliensis. J Biol Chem. 1996;271:4553–4560. doi: 10.1074/jbc.271.8.4553. [DOI] [PubMed] [Google Scholar]
- 8.Cisalpino P, Franco da Silveira J, Travassos L R. RNA and DNA isolation from Paracoccidioides brasiliensis yeast cells: establishment of cDNA and genomic libraries, and PCR amplification. In: Maresca B, Kobayashi G, editors. Molecular biology of pathogenic fungi, a laboratory manual. New York, N.Y: Telos Press; 1994. pp. 461–467. [Google Scholar]
- 9.Cisalpino, P. S., L. R. Travassos, and J. Franco da Silveira. Unpublished data.
- 10.Cox E C, Vocke C E, Walter S, Gregg K Y, Bain E S. Electrophoretic karyotype for Dictyostelium discoideum. Proc Natl Acad Sci USA. 1990;87:8247–8251. doi: 10.1073/pnas.87.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drouet E, Zapater R C. Phase levure et phase filamenteuse de Paracoccidioides brasiliensis: étude des noyaux. Ann Inst Pasteur. 1954;87:396–403. [PubMed] [Google Scholar]
- 12.Emmons C W. Fungus nuclei in the diagnosis of mycoses. Mycologia. 1959;51:227–236. [Google Scholar]
- 13.Franco M F, Montenegro M R, Mendes R P, Marques S A, Dillon N L, Mota N G S. Paracoccidioidomycosis: a recent proposed classification of clinical forms. Rev Soc Bras Med Trop. 1987;20:129–131. doi: 10.1590/s0037-86821987000200012. [DOI] [PubMed] [Google Scholar]
- 14.Goldani L Z, Maia A L, Sugar A M. Cloning and nucleotide sequence of a specific DNA fragment from Paracoccidioides brasiliensis. J Clin Microbiol. 1995;33:1652–1654. doi: 10.1128/jcm.33.6.1652-1654.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallack J, San-Blas F, San-Blas G. Isolation and wall analysis of dimorphic mutants of Paracoccidioides brasiliensis. Sabouraudia. 1982;20:51–62. doi: 10.1080/00362178285380081. [DOI] [PubMed] [Google Scholar]
- 16.Koch G L E, Macer D R J, Smith M J. Visualization of the intact endoplasmic reticulum by immunofluorescence with antibodies to the major ER glycoprotein, endoplasmin. J Cell Sci. 1987;87:535–542. doi: 10.1242/jcs.87.4.535. [DOI] [PubMed] [Google Scholar]
- 17.Lacaz C S. Paracoccidioides brasiliensis: morphology, evolutionary cycle, maintenance during saprophytic life, biology, virulence, taxonomy. In: Franco M, Lacaz C S, Restrepo-Moreno A, Del Negro G, editors. Paracoccidioidomycosis. Boca Raton, Fla: CRC Press; 1994. pp. 13–22. [Google Scholar]
- 18.McEwen J G, Restrepo B I, Salazar M E, Restrepo A. Nuclear staining of P. brasiliensis conidia. J Med Vet Mycol. 1987;25:343–345. [PubMed] [Google Scholar]
- 19.Mendes-Giannini M J S, Toscano E, Del Negro G B, Assis C M, Garcia N M. Immunochemical study of a Paracoccidioides brasiliensis polysaccharide-like antigen. J Med Vet Mycol. 1995;33:379–383. [PubMed] [Google Scholar]
- 20.Montoya A E, Moreno M N A, McEwen J G O, Restrepo A M. Resumenes del VI Encuentro Internacional sobre Paracoccidioidomicosis, Montevideo, Uruguay. 1996. Cariotipo electroforetico del Paracoccidioides brasiliensis; p. 156. [Google Scholar]
- 21.Olaiya A F, Sogin S J. Ploidy determination of Candida albicans. J Bacteriol. 1979;140:1043–1049. doi: 10.1128/jb.140.3.1043-1049.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orbach M J, Vollrath D, Davis R W, Yanofsky C. An electrophoretic karyotype of Neurospora crassa. Mol Cell Biol. 1988;8:1469–1473. doi: 10.1128/mcb.8.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan S, Cole G T. Electrophoretic karyotype of clinical isolates of Coccidioides immitis. Infect Immun. 1992;60:4872–4880. doi: 10.1128/iai.60.11.4872-4880.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perfect J R, Magee B B, Magee P T. Separation of chromosomes of Cryptococcus neoformans by pulsed field gel electrophoresis. Infect Immun. 1989;57:2624–2627. doi: 10.1128/iai.57.9.2624-2627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puccia R, Schenkman S, Gorin P A J, Travassos L R. Exocellular components of Paracoccidioides brasiliensis: identification of a specific antigen. Infect Immun. 1986;53:199–206. doi: 10.1128/iai.53.1.199-206.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Queiroz-Telles F. Paracoccidioides brasiliensis ultrastructural findings. In: Franco M, Lacaz C S, Restrepo-Moreno A, Del Negro G, editors. Paracoccidioidomycosis. Boca Raton, Fla: CRC Press; 1994. pp. 27–47. [Google Scholar]
- 27.Restrepo B I, McEwen J G, Salazar M E, Restrepo A. Morphological development of the conidia produced by P. brasiliensis mycelial form. J Med Vet Mycol. 1986;24:337–339. [PubMed] [Google Scholar]
- 28.Riggsby W S, Torres-Bauza L J, Wills J W, Townes T M. DNA content, kinetic complexity, and the question of ploidy in Candida albicans. Mol Cell Biol. 1982;2:853–862. doi: 10.1128/mcb.2.7.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rustchenko E P, Howard D H, Sherman F. Variation in assimilating functions occurs in spontaneous Candida albicans mutants having chromosomal alterations. Microbiology. 1997;143:1765–1778. doi: 10.1099/00221287-143-5-1765. [DOI] [PubMed] [Google Scholar]
- 30.San-Blas F, San-Blas G. Protoplast production from the yeast phase of Paracoccidioides brasiliensis. In: Maresca B, Kobayashi G S, editors. Molecular biology of pathogenic fungi, a laboratory manual. New York, N.Y: Telos Press; 1994. pp. 469–478. [Google Scholar]
- 31.Soares C M, Mollinari-Madlun E, Silva S, Pereira M, Felipe S. Characterization of Paracoccidioides brasiliensis isolates by random amplified polymorphic DNA analysis. J Clin Microbiol. 1995;33:505–507. doi: 10.1128/jcm.33.2.505-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soll D R. Gene regulation during high-frequency switching in Candida albicans. Microbiology. 1997;143:279–288. doi: 10.1099/00221287-143-2-279. [DOI] [PubMed] [Google Scholar]
- 33.Steele P E, Carle G, Kobayashi G, Medoff G. Electrophoretic analysis of Histoplasma capsulatum chromosomal DNA. Mol Cell Biol. 1989;9:983–987. doi: 10.1128/mcb.9.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki T, Kanbe T, Kuroiwa T, Tanaka K. Occurrence of ploidy shift in a strain of the imperfect yeast Candida albicans. J Gen Microbiol. 1986;132:443–453. doi: 10.1099/00221287-132-2-443. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki T, Nishibayashi S, Kuroiwa T, Kanbe T, Tanaka K. Variance of ploidy in Candida albicans. J Bacteriol. 1982;152:893–896. doi: 10.1128/jb.152.2.893-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vicentini A P, Gesztesi J-L, Franco M F, Souza W, Moraes J Z, Travassos L R, Lopes J D. Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect Immun. 1994;62:1465–1469. doi: 10.1128/iai.62.4.1465-1469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickes B L, Golin J E, Weber E, Kwon-Chung K J. Chromosomal rearrangement in Candida stellatoidea results in a positive effect on phenotype. Infect Immun. 1991;59:1762–1771. doi: 10.1128/iai.59.5.1762-1771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]