Abstract
Studying the single cell protein secretome offers the opportunity to understand how a phenotypically heterogeneous population of individual cells contribute to ensemble physiology and signaling. Polarized secretion events such as neurotransmitter release and cytokine signaling necessitates spatiotemporal information to elucidate structure-function relationships. Polymer functionalized single-walled carbon nanotube protein sensor arrays allow microscopic imaging of secreted protein footprints and enable the study of the spatiotemporal heterogeneity of protein secretion at the single-cell level. The protocols for carbon nanotube sensor creation, sensor array preparation, and imaging secreted proteins in both prokaryotic and mammalian cells are presented in this chapter. Secreted RAP1 and HIV-1 integrase proteins were used as proof-of-concept examples. Additionally, we discuss potential variety of protein and nonprotein analyte effluxes that can be imaged using this platform, as well as current and future perspectives related to sensor development and deployment.
1. Introduction
Biological research primarily focuses on the ensemble behavior of cells. However, it is theorized that the inherent heterogeneity among cells of similar type in a population play a significant role in disease etiology and development. For example, tumor cell heterogeneity may give rise to the clonal evolution of drug resistance and may underlie the complex biology of cancer (Greaves & Maley, 2012). The study of such cell-to-cell variations requires the employment of single-cell omics. For instance, single-cell genome sequencing has evinced the clonal evolution of breast cancer (Wang et al., 2014). Similarly, neuronal cell type heterogeneity may underlie the diversity of signals in the brain and could convolute the underlying etiology linking disease and aberrant cellular phenotype (Iourov, Vorsanova, & Yurov, 2012). Single-cell transcriptomics have enabled cellular-level classification of neuronal subtypes based on phenotype (Darmanis et al., 2015; Lake et al., 2016). These and many other works highlight the benefits of analyzing biological phenomena at a single-cell level.
While great strides in sequencing, fluidics, and data processing have allowed common usage of single-cell techniques for exploring the cell-to-cell heterogeneity of the genome, transcriptome, and proteome, there has been limited deployment of single cell metabolomics. The metabolome is unique in that it is highly dynamic and mediates both intracellular and extracellular interactions between the cell and its environment. Furthermore, metabolites are small molecules of great diversity and varying levels of transience with examples including proteins, carbohydrates, antibodies, and other molecules which act as intermediates or end products in metabolic pathways (Zenobi, 2013). These factors combine to make the metabolome difficult to probe since analytical techniques must be both selective and sensitive, and must often function in the extracellular environment. As multiple reviews summarize, methods of analyzing the intracellular single cell metabolome are numerous and reflect the diversity of analytes (Emara et al., 2017; Zenobi, 2013). These techniques range from single-cell mass spectrometry to genetically encoded sensors (Ibáñez et al., 2013; Palmer, Jin, Reed, & Tsien, 2004). We focus herein on the detection and quantification of a subset of the metabolome that consists of peptides or proteins effluxed into the extracellular space, also known as the cellular secretome.
One such class of secreted proteins are cytokines. Cytokines are cell signaling peptides that accumulate at sites of biological trauma and can serve as biomarkers to diagnose autoimmune disorders, infections, cancers, and central nervous system disorders. The breadth of cytokines and the ubiquity with which cytokines are used as biomarkers of disease speaks to the importance of their role as signaling molecules. Most cytokine signaling starts locally via autocrine or paracrine activity. Cells secrete cytokines locally to affect neighboring cells before the signal propagates to tissues and organs—well before total blood cytokine concentrations can serve for diagnosis. Currently, cytokines are measured by ELISA, immunohistochemistry, flow cytometry, and qPCR; however, these methods yield limited temporal and no spatial information about when and where cytokines are released, despite the importance of their spatial spread intrinsic to their role as signaling molecules (Vivier, Nunès, & Vély, 2004). The secretome is especially difficult to measure because it encompasses highly variable spatiotemporal dimensions. Cellular effluxes may be directional and can widely affect near-range autocrine signaling to systems-level communication (Stanley & Lacy, 2010). Thus, probing the single cell secretome requires not only sensitive temporal detection, but also accurate spatial resolution.
Single-cell metabolic efflux measurements have been reported using a variety of tools. For redox sensitive metabolites such as the neurotransmitter dopamine, microelectrode measurements have been used successfully to measure effluxes at a single-cell level with high sensitivity and temporal resolution (Clausmeyer & Schuhmann, 2016). However, electrode measurements lack spatial resolution and have limited specificity, precluding their use in secretome sensing. As of writing, few platforms exist to sense protein efflux from cells. However, one such platform reported by Zhao et al. utilizes cell-surface tethered aptamers, oligonucleotides with protein recognition, for extracellular sensing of platelet derived growth factor (Zhao et al., 2011). Others have used various surface-immobilized and solution phase immunolabeling to detect extracellularly released proteins with spatial resolution (Huse, Lillemeier, Kuhns, Chen, & Davis, 2006; Shirasaki et al., 2014; Zhu et al., 2009). These methods are limited to sensing near the cell surface or, as in the case of immunoassays, are difficult to engineer for improved temporal response due to the tight binding as evidenced by low dissociation constants (kd) of antigen-antibody binding. Thus, we propose that nanomaterial constructs offer a highly adaptable platform for sensing protein effluxes with tunable kd (Beyene, McFarlane, Pinals, & Landry, 2017). Nanomaterials are materials with at least one dimension that is smaller than 100nm and have highly tunable physical and chemical properties. Nanomaterials can be designed for analyte recognition, binding affinity, biocompatibility, and improved stability (Holzinger, Le Goff, & Cosnier, 2014).
2. Single-walled carbon nanotubes as protein efflux sensors
A particularly interesting class of nanomaterials for metabolite imaging are single walled carbon nanotubes (SWNTs). SWNTs are graphene sheets rolled into a 1D tube that exhibit unique electrical and optical properties depending on the chirality in which they form. Certain chiralities of SWNTs act as non-photobleaching fluorescence reporters in the near-infrared (NIR) range (Heller, Baik, Eurell, & Strano, 2005). This range of optical activity is especially useful in biosensing as it potentiates deep tissue imaging due to the minimal light absorbance of tissue and scattering by water in this biological NIR window (Smith, Mancini, & Nie, 2009). Additionally, the fluorescence of single-walled carbon nanotubes is highly sensitive to changes in the local surface dielectric function, and can be modulated by changes to the local surface environment through the association of an analyte (Barone, Baik, Heller, & Strano, 2005; Zhang et al., 2013). This physico-chemical property gives rise to a class of sensors comprised of engineerable polymers non-covalently adsorbed to the surface of a fluorescent SWNT enabling molecular recognition of an analyte (Fig. 1) (Zhang et al., 2013). SWNT surface-adsorbed polymer coatings are uniquely customizable to detect a myriad of target molecules. Several sensors utilizing this technology have been developed to image or sense small molecule effluxes such as dopamine (Beyene et al., 2018; Kruss et al., 2017) or the efflux of peptides and proteins such as insulin (Bisker et al., 2018), the human immunodeficiency virus integrase (HIV-1 integrase) protein, and RAP1, a normally cytosolic protein which assists in T-cell receptor signaling (Landry et al., 2017).
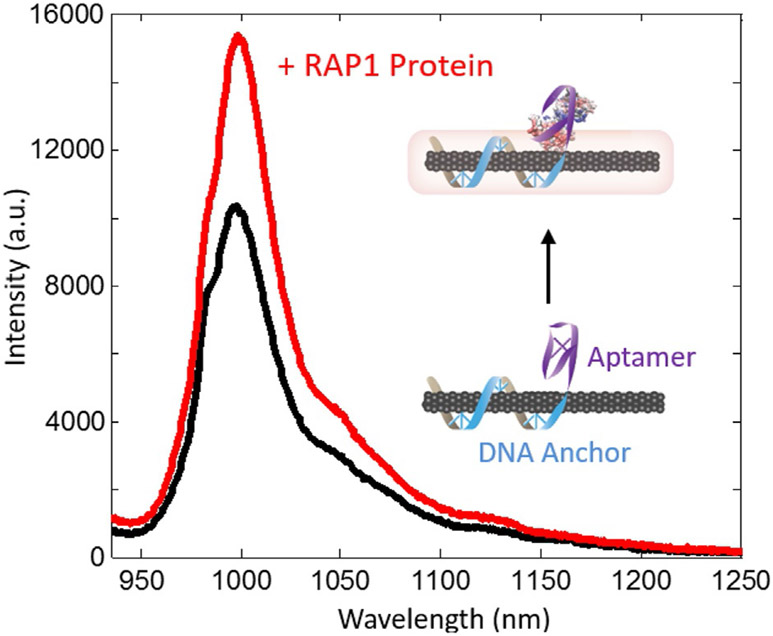
Fig. 1.
Representative fluorescence intensity change upon RAP1 protein analyte binding to DNA aptamer based SWNT sensor. Analyte binding to the polymer-wrapped SWNT induces changes in the SWNT electronic environment and fluorescence. In some SWNT sensor systems, chromatic shifts in the fluorescence maxima can also be observed (Landry et al., 2017).
2.1. Polymer selection for SWNT passivation
The diversity of analytes recognized by SWNT sensors is conferred by an amphiphilic polymer non-covalently adsorbed to the surface of the SWNT. These polymers consist of a hydrophobic and aromatic anchor domain which strongly adsorb to the hydrophobic and pi-conjugated surface of SWNTs as well as a hydrophilic component that colloidally stabilizes the SWNT in solution. Specific molecular recognition may arise from a combination of stabilizing interactions between the analyte, SWNT, and polymer. These polymers can be constructed from both abiotic and biotic monomers. For example, Zhang et al. screened a variety of abiotic polymers containing highly aromatic moieties such as rhodamine and hydrophilic domains such as polyethylene glycol (Zhang et al., 2013). Another class of polymers for SWNT-based protein sensing are peptoid polymers (Chio et al., 2019). Peptoids are synthetic peptide-mimetic polymers synthesized through solid-phase synthesis. Peptoids are resistant to proteolytic degradation and can be imparted with unique chemical attributes for both anchoring and sensing (Chio et al., 2019).
Biotic polymers include aptamers that are oligonucleotide or peptide sequences engineered to bind to target molecules. Single changes to the polymer sequence can induce great changes in binding affinities between an aptamer and its analyte. Production of specific oligonucleotide sequences is commercially feasible and cost effective, yielding a large diversity of sequence combinations and thus has potential to target many different molecules. Engineering of SWNT aptamer sensors first requires the generation of a library of oligonucleotides. These oligonucleotides should have both an anchor domain which keeps the polymer strongly attached to the SWNT as well as an aptameric capture domain which binds reversibly to the target molecule and brings the analyte close to the nanotube surface thereby perturbing the SWNT fluorescence. Typically, the anchor domain consists of strongly adhering dinucleotide repeat sequences such as (GT)15 or (AT)11 and the capture domain can be chosen from aptamer sequences available in literature. An abasic linker sequence could also be incorporated between the anchor and capture domains to improve mobility of the sensing domain, although this is not always necessary. To confirm the selectivity and sensitivity of the polymer-SWNT construct for its target analyte, selection is accomplished by screening. Screening of polymer-SWNT constructs has been well described elsewhere and extends beyond the scope of this protocol (Beyene, Demirer, & Landry, 2016; Chio, Yang, & Landry, 2017; Zhang et al., 2013). Briefly, a library of aptamer-SWNTs is synthesized and exposed to a panel of analytes in a 96-well-plate. The change in fluorescence is recorded on a NIR-spectrometer and sensors which display high signal to noise (ΔF/F) for the target molecule and also low signal for other competing molecules are good candidates for sensing applications. A titration curve can also be incorporated to determine the working concentration range and the sensitivity of the sensor. Because aptamer screening allows for tuning of kd, the end user can select a polymer that best suits the necessary limit of detection, reversibility, dynamic range, and temporal resolution. For convenience, Table 1 provides examples of polymers used to generate SWNT sensors for proteins and other small molecules.
Table 1.
Example SWNT-based sensors.
| Sensing polymer | Target | Coating | Reference |
|---|---|---|---|
| RITC-PEG-RITC | Estradiol | Synthetic polymer | Zhang et al. (2013) |
| Fmoc-Phe-PPEG8 | l-Thyroxine | Synthetic polymer | Zhang et al. (2013) |
| BA-PhO-Dex | Riboflavin | Synthetic polymer | Zhang et al. (2013) |
| (AT)11-RAP1 | RAP1 | DNA aptamer | Landry et al. (2017) |
| (AT)11-HIV-1 integrase | HIV-1 integrase | DNA aptamer | Landry et al. (2017) |
| (GT)15 and (GT)6 | Dopamine | DNA oligomer | Beyene et al. (2018) and Kruss et al. (2014) |
| C16-PEG2000-ceramide | Insulin | Phospholipid | Bisker et al. (2018) |
| ProLoop | Wheat germ agglutinin | Peptoid polymer | Chio et al. (2019) |
| CB13-DNA | Doxorubicin | DNA oligomer | Del Bonis-O’Donnell et al. (2019) |
Sensors previously screened by previous groups exhibit a wide range of sensing capability including custom-made synthetic polymers, commercially available phospholipids, and commercially procurable oligonucleotide sequences.
2.2. SWNT chirality and sensor performance
As previously mentioned, the SWNT chirality affects SWNT optical and electronic properties. Polymer-SWNT sensors leverage a population of semiconducting SWNTs that fluoresce. The various chiralities of semiconducting SWNTs exhibit unique spectral properties that can be deconvoluted (Bachilo et al., 2002). Commercial SWNTs are typically heterogeneous mixtures of various chirality SWNTs, amorphous carbon, and catalyst byproducts. It is sufficient to use these unpurified SWNTs as sensors with no further downstream processing, as the polymer encapsulation process can enrich for SWNTs over impurities. Our laboratory routinely uses High Pressure Carbon monoxide (HiPCo) SWNTs (Nanointegris, Product #1600) for sensor development with good success. However, chiral purification can contribute to improved fluorophore brightness and signal-to-noise ratio. Since purified chiral SWNTs such as the (6,5) SWNTs used for RAP1 sensing are commercially available, and because we have found this step to not be mandatory, we have not included a procedure for chiral purification. Although the protocol described in this document will not discuss chirality separation, note that data from Landry et al. (2017) discussed in later sections does use only (6,5) chirality SWNTs. For interested readers, protocols for chirality separation of SWNTs can be found in other references (Green & Hersam, 2011; Jain, Tvrdy, Han, Ulissi, & Strano, 2014; Liu, Nishide, Tanaka, & Kataura, 2011; Turek et al., 2019; Yomogida et al., 2016).
2.3. Assembly of SWNT-polymer constructs
SWNTs are highly hydrophobic and cannot disperse in aqueous media. The process of sensor assembly not only adds a recognition element to the SWNT surface but also improves SWNT colloidal stability. One method of SWNT sensor assembly is probe-tip ultrasonication. The high energy sonication process can temporarily disperse SWNT bundles to allow for polymer adsorption. SWNTs can be directly sonicated with a target polymer to create sensors in one step (Del Bonis-O’Donnell et al., 2017). Following centrifugation, the supernatant containing a stable colloidal suspension of polymer-SWNTs can then be collected for use. The concentration of the resulting suspension is determined via IR absorbance spectroscopy where the concentration of a multichiral sample can be estimated using an extinction coefficient for the absorbance at 632nm, ε632 =0.036Lmg−1 cm−1.
In an alternate method, SWNTs can first be suspended in a surfactant such as sodium cholate, and can next undergo dialysis to exchange the surfactant layer with polymer (Giraldo et al., 2015; Kruss et al., 2014; Landry et al., 2017). Surfactant-exchange is a useful strategy following SWNT chirality purification, in which the chirally-pure SWNT product often resides in surfactant media. This surfactant-exchange method requires longer preparation time and generally produces lower sensor yields than probe-tip sonication, but protects polymers from the harsher conditions of sonication. The following protocol describes the assembly of an amphiphilic polymer-based sensor via surfactant exchange as described by Landry et al. (2017), beginning with (6,5) chirality purified SDS SWNT isolated using a previously reported protocol (Jain et al., 2014).
2.3.1. Materials and equipment: Surfactant mediated sensor assembly
Materials
(6,5) SWNT in sodium dodecyl sulfate surfactant (SDS)
Amphiphilic polymer (in this case (AT)11-RAP1)
Deionized water
Equipment
Anti-static gun
Dialysis membrane
UV/Vis spectrophotometer
2.3.2. Protocol: Surfactant mediated sensor assembly
Remove static electricity from disposable spatula, microcentrifuge tubes and SDS-SWNT stock using an anti-static gun.
Dissolve 1 wt% of (AT)11-RAP1 polymer in the SDS-SWNT solution.
Dialyze polymer-SDS-SWNT solution in a 3.5kDa dialysis membrane against 1L of deionized water or buffer for 5 days.
Note: Measure the solution absorbance using a UV/Vis spectrophotometer to determine the approximate concentration of suspended SWNTs. For multichiral samples, measure absorbance at 632nm, ε632 =0.036Lmg−1 cm−1. Appropriate absorption wavelengths for specific chiralities are provided in literature (Wei et al., 2016).
Note: Choice of dialysis membrane is dependent on the polymer’s molecular weight. A larger dialysis membrane can be used, so long as it allows removal of the surfactant of choice, but retention of both the SWNT and amphiphilic polymer. For ideal removal of surfactant and retention of polymer in the dialysis cartridge, choose a MWCO that is 50% the MW of the polymer and preferably 200% the MW of the surfactant. For reference, SWNT have MW of >300kDa.
3. Sensor array preparation
Surface fixation of metabolomic sensors on cell growth plates to form a sensor array enables spatial and temporal imaging of protein efflux. Diffusion of the target protein to the sensor array surface can be examined in real time. Furthermore, a uniform distribution of sensors prevents misrepresentation of local concentrations.
3.1. Surface passivation/sensor anchoring
Proteins can adsorb nonspecifically to many surfaces. To promote predominant surface adsorption of only SWNT sensors, the array surface must be first passivated to prevent nonspecific protein binding. This protocol describes two approaches for immobilization of SWNT sensors to create an analyte-imaging surface. The first is by sequential incubation of the microfluidic chamber with positively charged 3-aminopropyltriethoxysilane (APTES) followed by introduction of negatively charged DNA-SWNT sensors, which bind through electrostatic interactions. This protocol is rapid but does not passivate against nonspecific protein adsorption, particularly if proteins are charged. Therefore, APTES-based nanosensor immobilization and imaging should be limited to imaging in the NIR sensor emission range and not used in the visible range where significant surface coverage by proteins or debris could mar the image. The second SWNT sensor surface immobilization approach involves sequential incubation of the imaging surface, first with a dilute mixture of biotinylated polyethylene glycol succinimide carboxymethyl (BIO-PEG-SCM) in polyethylene glycol succinimide carboxymethyl (PEG-SCM). Next, addition of neutravidin protein, which binds SWNT, enables immobilization of SWNT-based sensors. This neutravidin-binding method simultaneously passivates the glass surface, and immobilizes SWNT sensors. PEG has been well documented as an effective surface passivation technique which prevents nonspecific protein-surface adsorption and is used in various forms (e.g., linear, branched, short chain, long chain) and applications (Jain, Liu, Xiang, & Ha, 2012; Zalipsky & Harris, 1997). Neutravidin binds strongly to the exposed biotin through molecular affinity, and to the negatively charged DNA-SWNT sensors electrostatically. In addition to superior surface passivation quality, this second sensor immobilization approach results in a surface coating which is more suitable for supporting cell growth as compared to the positively charged APTES surface. A schematic of the layered surface coatings is shown in Fig. 2. The use of a microfluidic chamber, as represented in Fig. 3, is beneficial for single molecule experiments as it ensures close contact between cells and the sensor array. Care should be taken to flow solutions slowly onto the imaging surface to not disrupt the sensor array, as sensors are noncovalently bonded in place. Notably, in the cell efflux experiments reported in Section 3.2.2, cells were plated onto a 35mm glass bottom culture dish (instead of microfluidic chambers) passivated with a biotin-neutravidin-sensor array.
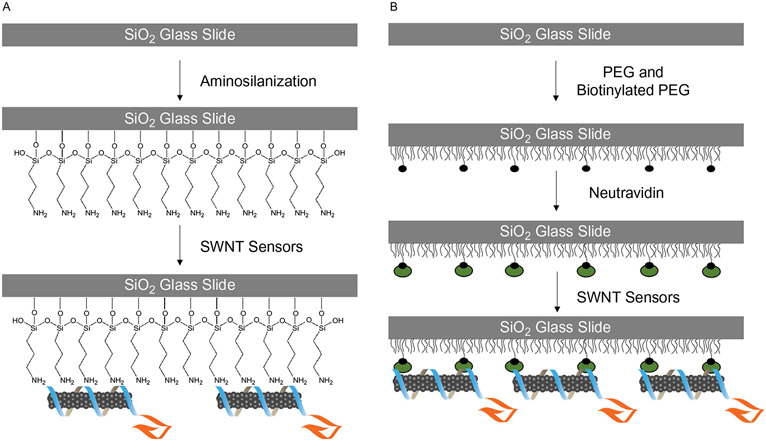
Fig. 2.
Schematic of surface passivation and immobilization. Glass surfaces can be passivated both to prevent nonspecific protein adsorption, and permit sensor immobilization (Lamichhane et al., 2015). (A) APTES provides a positive charge onto which negatively charged DNA-SWNT sensors can be adsorbed. (B) PEG surface passivation prevents nonspecific adsorption. A small fraction of biotinylated-PEG sequentially binds to neutravidin, which electrostatically binds DNA-SWNT sensors.
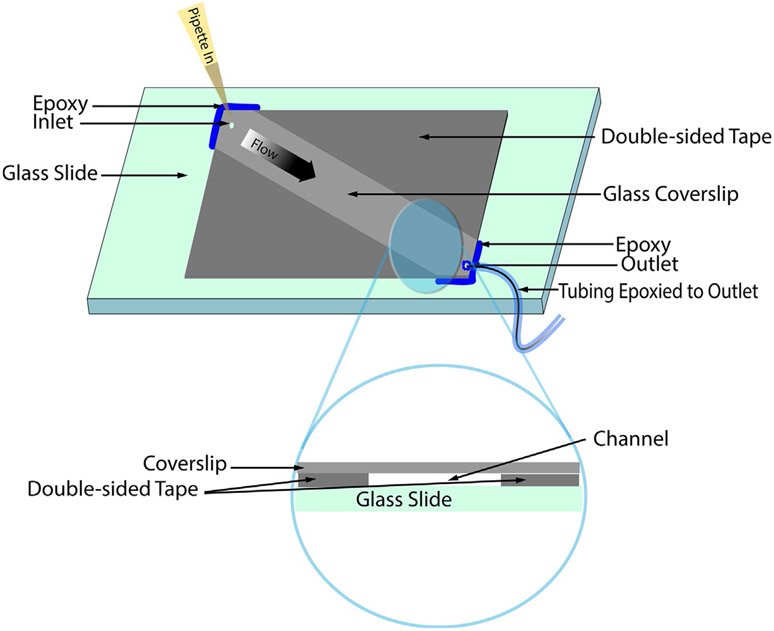
Fig. 3.
Schematic of a microfluidic chamber. Fabrication of a microfluidic chamber with surfaces that function as sensor arrays can be constructed using a glass slide, glass coverslip, double sided tape, and epoxy (Roy, Hohng, & Ha, 2008). Negative pressure can be applied to the outlet tubing using a syringe in order to flow the added liquid through the channel.
3.1.1. Materials: Assembly of microfluidic chamber
Materials
Microscope slide and glass coverslip with precut inlet and outlet holes (Ibidi Glass Coverslip No. 1.5)
Deionized water
Methanol
Acetone
Double sided tape (Scotch™, 3M©)
Epoxy (5 Minute®, ITW Devcon)
Tubing (0.51mm I.D., Dow Corning™ Silastic™ Laboratory Tubing)
3.1.2. Protocol: Assembly of microfluidic chamber
This protocol describes a microfluidic chamber constructed to hold ~30 μL sample volume (Fig. 3). A coverslip with inlet and outlet holes as described by other groups (Roy et al., 2008) facilitates fluid exchange, which is necessary for surface passivation, sensor immobilization, analyte introduction, and cell culture media regeneration. A conventional coverslip can also be used for channel construction as detailed by Del Bonis-O’Donnell et al. (2017). Alternatively, commercial microscope slides with microfluidic channels can also be used (Ibidi μ-Slide Luer, Cat. No. 80171).
Using a diamond-tip drill bit, drill holes into the desired flow inlet and outlet locations of a microscope slide, as described by Zhao and Rueda (2009).
Clean the microscope slide and long glass coverslip sequentially with deionized water, methanol, acetone, and again with deionized water.
Press two pieces of double-sided tape firmly onto the glass slide approximately 5mm apart. Each piece forms the long border to the microfluidic channel.
Insert laboratory tubing through the outlet hole of the microscope slide. Apply epoxy glue on the side of the coverslip that will form the exterior of the chamber to fix the tubing in place. Follow manufacturer’s instructions for how long to dry epoxy (typically 30 min).
Carefully cut the protruding segment of the tube so that it is flush with the coverslip.
Press the coverslip tightly onto the glass slide, making sure the inlet and outlet holes are not covered by tape.
Apply a small amount of epoxy to seal the coverslip perimeter. Allow it to harden fully following the manufacturer’s instructions (typically 30 min).
3.1.3. Materials: Surface passivation and sensor attachment using APTES
Materials
Tris base (Trizma base) (Sigma Aldrich No. 93362)
Sodium chloride (NaCl)
Concentrated hydrochloric acid (HCl)
MilliQ water
3-Aminopropyltriethoxysilane (APTES) (Sigma Aldrich No. 440140)
Ethanol
DNA-SWNT
Microfluidic chamber
3.1.4. Protocol: Surface passivation and sensor adhesion using APTES
This protocol allows for SWNT-sensor immobilization on a surface using 3-aminopropyltriethoxysilane (APTES). This does not passivate the surface against non-specific protein surface adsorption and therefore should be used for surfaces that will only undergo NIR imaging. This protocol was adapted from Del Bonis-O’Donnell et al. (2017).
Prepare Tris imaging buffer by dissolving 22.23 g of Tris base and 58.44 g of NaCl in 500mL of MilliQ water. Carefully add concentrated HCl until a pH of 8.1 is reached. Add MilliQ water to reach a final volume of 1L of 185mM Tris buffered 1M saline.
Dilute 3-aminopropyltriethoxysilane (APTES) to form a 10% (w/v) solution of APTES in ethanol
Flow 100 μL 1 × Tris buffer through microfluidic chamber.
Flush the channel with APTES solution (50 μL) and incubate for 5 min.
Wash five times with 1 × Tris buffer (250 μL of buffer).
Dilute stock solutions of suspended DNA-SWNTs in Tris buffer to a concentration of 1–10 mg/L and flow 50 μL of the solution into the channel. Incubate for 5 min.
Gently rinse away excess and unbound SWNTs using 50 μL of Tris buffer.
Note: Volume of the microfluidic chamber is ~30 μL but can vary depending on the positioning of the double-sided tape. Do not allow the assembled microfluidic chamber to dry out. Slowly perform buffer exchanges to avoid introducing bubbles into the microfluidic chambers, and to avoid detaching SWNT sensors from the imaging surface.
3.1.5. Materials: Surface passivation and sensor attachment using biotinylated PEG and neutravidin
Materials
Biotin polyethylene glycol succinimidyl carboxymethyl (BIO-PEG-SCM, MW 3400/5000, Laysan Bio. Inc.)
Methoxy polyethylene glycol succinimidyl carboxymethyl (m-PEG-SCM, MW 5000, Laysan Bio. Inc.)
Sodium bicarbonate
Neutravidin (Thermo Scientific No. 31000)
1 × phosphate buffered saline (PBS)
Sodium chloride (NaCl)
Microfluidic chamber
Tris base
DNA-SWNT
3.1.6. Protocol: Surface passivation and sensor attachment using biotinylated PEG and neutravidin
For cell efflux experiments described in Section 3.2.2 of this protocol, a 35mm glass-bottom culture dish should form the basis of the sensor array instead of a microfluidic chamber. The same passivation steps apply, simply incubate the dish surface in each solution as described below, ensuring to rinse with buffer between steps.
Mix 1:100 solution of biotin polyethylene glycol succinimidyl carboxymethyl (BIO-PEG-SCM) and methoxy polyethylene glycol succinimidyl carboxymethyl (m-PEG-SCM) in 100mM Tris buffer (pH8.5).
Flush the channel of the microfluidic chamber with PEG solution and incubate covered for 3–4h.
Rinse three to five times with MilliQ water.
Prepare a 33nM neutravidin (NA) solution in 20 μL of 1 × PBS buffer (pH 7.5).
Flush the channel with this NA solution and incubate for 10 min.
Rinse three to five times with 0.1 M NaCl.
Dilute stock solutions of suspended DNA-SWNTs in Tris buffer (preparation described in Section 3.1.4) to a concentration of 1–10mg/L and flow 50 μL of the solution into the channel and incubate for 5 min.
Gently rinse away excess DNA-SWNTs using 50 μL of Tris buffer.
Note: 20 μL of NA solution should be sufficient to fill one channel—scale accordingly if multiple channels are in use. See Section 3.1.4 for additional recommendations on optimal fabrication of the microfluidic chamber.
3.2. Cell growth and inducible secretion
To date, secreted protein imaging using SWNT sensors has been accomplished in mammalian, bacterial, and yeast cells. SWNT sensors have also been used to visualize small molecule efflux such as dopamine from PC-12 neuronal cells (Kruss et al., 2017), and from neurons in intact brain tissue (Beyene et al., 2018). Proof-of-principle protein efflux imaging experiments have been performed by using inducible heterologous protein expression, with RAP1 protein imaging accomplished using an inducible riboregulation schema. Constitutive HIV-1 integrase protein secretion was also imaged in HEK293 cells on SWNT sensor arrays. Although a concern might be that strong promoter driven protein expression would not reflect physiological levels of protein release, the working concentration range of these SWNT sensors extends well into the picomolar and single-molecule regime, thus spanning the range of expected physiological extracellular protein concentrations. Inducible constructs are also ideal for proof-of-principle experiments, as the analyte secretion can be temporally controlled for proper validation. Additionally, as demonstrated by Kruss et al., the sensor array can be washed with fresh biological buffer to restore baseline signal after several minutes or halfmax-intensity after several seconds (Kruss et al., 2014). The sensor recovery time is dependent on the sensor kd and the diffusion of the analyte.
Previously, heterologous protein expression was achieved using a tetracycline (aTc) inducible promoter in Escherichia coli and a constitutive CMV promoter in mammalian HEK293 cells (Landry et al., 2017). A peptide fragment of human RAP1 GTP-ase (UniProt: Q9NYB0) fused to the N-terminal secretory peptide sequence of a secreted bacterial protein, csgA (UniProt: P28307), was expressed in E. coli using an aTc driven riboregulator expression system. For mammalian cells, HIV-1 integrase, a vital HIV protein (UniProt: Q76353) N-terminally fused to a secretory signal derived from Gaussia luciferase was constitutively expressed. We do not describe here the construction of these vectors or the transfection process as they are routine procedures. We leave the choice of expression system and cell type to the end user, noting that both heterologous and endogenous secretory proteins can be imaged using the appropriate SWNT sensor. The following protocol describes the growth, seeding, and induction conditions to achieve imaging of the secreted protein ‘footprint’ from immobilized cells (Fig. 4). Fig. 4 demonstrates the unique spatial data that is possible to acquire using immobilized sensor arrays. Individual cell secretion of RAP1 protein can be tracked relative to the position of the cell.
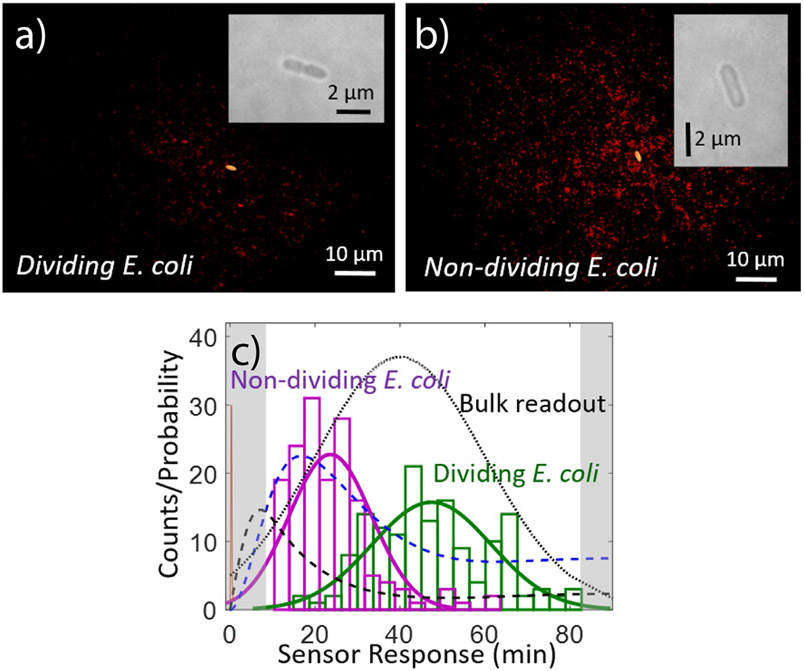
Fig. 4.
Protein secretion footprint imaging. Typical NIR protein footprint (red), as measured with a RAP1 SWNT nanosensor array 1h post-induction of E. coli cells with aTc for (A) a dividing E. coli cell and (B) a non-dividing cell (bright-field insets). Dividing cells show a noticeably smaller protein secretion footprint than non-dividing cells (154.0 ± 69.2 versus 486.2 ± 113.0 protein-responding sensors, respectively). (C) Quantification of first-response times for each nanosensor under each cell for N = 22 individual protein secretion events for dividing (green) or non-dividing (magenta) cells. As measured by the sensor array, a faster first-protein-secretion events are measured for non-dividing cells. Kinetic model for the protein secretion pathway for RAP1 secretion: aTc diffusion to the cell (red spike), protein transcription (mRNA concentration, dashed black), and protein translation (protein concentration, dashed blue). The black dotted line models the expected protein content in the cell supernatant if measured using an SDS-PAGE gel to monitor for protein band intensity (Landry et al., 2017).
3.2.1. Materials and equipment: Cell seeding and inducible secretion of RAP1 in E. coli
Materials
Transformed BL21 E. coli cells
Temperature controlled incubator-shaker or warm room
1000 × 50 μM anhydrotetracycline (aTc) stock solution
M63 minimal media
LB media
Bacto-Agar
35mm glass bottom culture dish prepared with a sensor array described in Section 3.1
Equipment
Centrifuge
UV-Vis-NIR absorbance spectrophotometer
NIR fluorescence microscope with heated stage, InGaAs detector, and 785nm excitation laser
3.2.2. Protocol: Cell seeding and inducible secretion of RAP1 in E. coli
Grow 3mL cultures of BL21 cells (DE3) (NEB) transformed with secretory signal-RAP1 fusion inducible expression vector overnight at 37 °C with shaking in liquid LB broth.
Pellet the cultures by centrifugation at 3–5000rpm for 5 min and discard the supernatant.
Resuspend the pellet in 37 °C M63 minimal media with 0.2 % (w/v) agar to a density of 1 × 109–1.5 × 109 cfumL−1 for bulk protein imaging and ~1.5 × 107 cfumL−1 for single cell imaging. Care should be taken that the cells are not cast in overly-hot media to ensure viability. A low percentage of agar is added to immobilize the E. coli for imaging while still allowing diffusion of the inducer and resulting analyte.
Slowly cast a small ~1–200 μL amount of the bacteria-agar media solution into the bottom of a 35mm glass-bottom dish prepared with a sensor array and allow to set. Avoid incorporating bubbles into the media.
Image the cell sample in the microscope. Identify a cell of interest with brightfield microscopy, and switch to NIR imaging of sensors.
Add fresh liquid M63 media with 50nM aTc inducer and no added agar to cover the entire bottom of the 35mm dish, around 1mL. Record the time at which the inducer is added to the cells.
Collect images using a 100 × oil objective lens on a custom-built inverted - near-IR microscope equipped with a 785nm 450mW excitation laser. Images can be collected on either time-lapse or snapshot mode. More information about the necessary filters and optical elements for NIR microscopy can be found in Section 3.3.
Notes: At ~1.5 × 107 cfUmL−1, one E. coli cell on average is visible in a 60 × 80 μM field of view. The concentration of bacteria can be estimated by an optical density measurement and a corresponding colony-forming unit calibration curve. The aTc should be added following initiation of NIR imaging. Using M63 minimal media allows for expression of the protein while retarding cell division, which would disallow measuring efflux from a single cell.
3.2.3. Materials and equipment: Constitutive HIV-1 integrase secretion imaging in HEK293
Materials
Transfected HEK 293 cells
Dulbecco’s Minimum Essential Media (DMEM)
100 × Penicillin-Streptomycin Stock (PS stock)
Fetal Bovine Serum (FBS)
Equipment
Centrifuge
Cell culture biosafety cabinet
Temperature controlled incubator with CO2 control
Corning cell-culture treated T25 flask
Hemocytometer
Wide-field phase contrast light microscope with 20 × air objective
NIR fluorescence microscope with heated stage, InGaAs detector, and 785nm excitation laser
3.2.4. Protocol: Constitutive HIV-1 integrase secretion imaging in HEK293
Culture HEK293 cells transiently transfected to constitutively express a Gaussia luciferase secretion signal fused HIV-1 integrase protein in a 25 cm2 flask in DMEM supplemented with 10 (v/v)% FBS and 1 (v/v)% penicillin-streptomycin (PS) stock solution at 37 °C in 5% CO2 until 90% confluence.
Pellet the cells at 1000 × g for 5 min and aspirate the old media prior to imaging.
Resuspend the cells in 5mL of fresh, pre-warmed DMEM supplemented with 10% FBS and 1% PS antibiotic. Upon suspension, the pellet should be pipetted up and down to mix to get singly suspended cells for best imaging results.
Adjust the cell concentration to 5 × 104–2 × 105 cells/mL using a hemocytometer.
Load ~100 μL of the adjusted cell suspension into the prepared microfluidic chamber.
Immediately place the microfluidic channel on a heated microscope stage at 37 °C for image collection. For longer imaging times, a stage equipped with an additional CO2 chamber is recommended.
For imaging, see step 6 from prior Section 3.2.2 for imaging E. coli protein secretion.
3.3. Imaging
A distinct advantage to SWNT based fluorescent sensors is that their excitation and emission window lies outside the visible range and thereby avoids issues of scattering in biological media. Spatial resolution can be a challenge when using NIR fluorescence as the long emission wavelength dictates a relatively large wavelength diffraction limit of ~λ/2. Our laboratory uses a Zeiss AxioVision Inverted Microscope, 785nm photodiode laser (B&W Tek Inc.), Princeton Instruments InGaAs 1D Detector, and PI Acton SP2500 Spectrometer and is described in detail in Chio et al. (2017) and Beyene et al. (2016). Here we briefly describe the imaging protocol.
3.3.1. NIR fluorescence microscopy
Using a 100 × oil immersion objective (1.4 NA), apply fluorescence-free immersion oil and place the immobilized SWNT sample onto the microscope stage.
Raise the objective until the oil contacts the bottom of the cover glass (#1.5, 170μm thickness). Make adjustments to the objective collar, if necessary, to optimize imaging conditions, e.g., temperature, glass thickness, etc.
Slowly raise the objective with the excitation source on and monitor the fluorescence signal of the SWNT sensors on the InGaAs camera. Fluorescence intensity should gradually increase as the focal plane approaches the surface and the immobilized sensors come into focus.
Acquire the NIR movie with a ~1 min baseline prior to experimentation, for example before the induction of metabolite secretion.
4. Conclusion
SWNT sensors have great potential for imaging cellular efflux and broadening our understanding of the cell secretome. While other methods of probing the metabolome with single molecule measurements do exist, examples of these such as redox readings using electrodes or surface-tethered aptamers restrict analyte detection either spatially or temporally. SWNT NIR fluorescence is particularly advantageous for sensing in biological environments, where the NIR fluorescence is least attenuated by scattering or absorption by biological molecules or water, respectively. Aptamer sequences on SWNT nanoparticles offer a robust and customizable platform for label-free single molecule imaging. For an aptamer-free approach, molecular selectivity can also be conferred by implementing high throughput screens for target molecules with a library of amphiphilic polymers adsorbed on SWNT. The generation of SWNT-based sensor arrays and subsequent NIR imaging of effluxed cellular proteins have been detailed in our protocol.
Using an array of polymer-SWNT sensors to image cellular efflux, we can gain a better understanding of cellular metabolism. We described the uniform coating of a microfluidic chamber with DNA-SWNT sensors upon which cells can be deposited and protein efflux can be imaged using NIR microscopy. Previously, such sensor arrays were used by Landry et al. to detect RAP1 protein efflux from E. coli and HIV-1 integrase protein from HEK293 and Pichia pastoris cells at the single-cell level, and similar cell sensor arrays could be implemented to measure other cellular metabolites. By imaging cellular efflux at a single molecule level, we are able to obtain spatiotemporal optical visualization of real-time cellular activity. For example, the spatial and temporal characteristics of phenomena such as polar cytokine release and neurotransmitter secretion are processes of interest that can be studied using this nanosensor array platform (Beyene et al., 2018; Kruss et al., 2017; Stanley & Lacy, 2010; Thurley, Gerecht, Friedmann, & Höfer, 2015). With different molecular-recognition conferring polymers adsorbed to SWNT, this platform can be adapted for imaging a wide variety of extracellular proteins and metabolites.
References
- Bachilo SM, Strano MS, Kittrell C, Hauge RH, Smalley RE, & Weisman RB (2002). Structure-assigned optical spectra of single-walled carbon nanotubes. Science, 298(5602), 2361–2366. [DOI] [PubMed] [Google Scholar]
- Barone PW, Baik S, Heller DA, & Strano MS (2005). Near-infrared optical sensors based on single-walled carbon nanotubes. Nature Materials, 4(1), 86–92. [DOI] [PubMed] [Google Scholar]
- Beyene AG, Delevich K, Del Bonis O’Donnell JT, Piekarski DJ, Lin WC, Thomas AW, et al. (2018). Imaging striatal dopamine release using a non-genetically encoded near-infrared fluorescent catecholamine nanosensor. bioRxiv, 356543. https://www.biorxiv.org/content/biorxiv/early/2018/10/19/356543.full.pdf. 10.1101/356543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene AG, Demirer GS, & Landry MP (2016). Nanoparticle-templated molecular recognition platforms for detection of biological analytes. Current Protocols in Chemical Biology, 8(3), 197–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene AG, McFarlane IR, Pinals RL, &Landry MP (2017). Stochastic simulation of dopamine neuromodulation for implementation of fluorescent neurochemical probes in the striatal extracellular space. ACS Chemical Neuroscience, 8(10), 2275–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisker G,Bakh NA, Lee MA, Ahn J, Park M, O’Connell EB,et al. (2018). Insulin detection using a corona phase molecular recognition site on single-walled carbon nanotubes. ACS Sensors, 3(2), 367–377. [DOI] [PubMed] [Google Scholar]
- Chio L, Del Bonis-O’Donnell JT, Kline M, Kim JH, McFarlane IR, Zuckermann RN, et al. (2019). Electrostatic-assemblies of single-walled carbon nanotubes and sequence-tunable peptoid polymers detect a lectin protein and its target sugars. Nano Letters. 10.1021/acs.nanolett.8b04955. [DOI] [PubMed] [Google Scholar]
- Chio L, Yang D,& Landry M (2017). Surface engineering of nanoparticles to create synthetic antibodies. In Tiller T (Ed.), Synthetic antibodies: Methods and protocols (pp. 363–380). New York, NY: Springer New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausmeyer J, & Schuhmann W (2016). Nanoelectrodes: Applications in electrocatalysis, single-cell analysis and high-resolution electrochemical imaging. TrAC Trends in Analytical Chemistry, 79, 46–59. [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. (2015). A survey of human brain transcriptome diversity at the single cell level. Proceedings of the National Academy of Sciences of the United States of America, 112(23), 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bonis-O’Donnell JT, Pinals RL, Jeong S, Thakrar A, Wolfinger RD, & Landry MP (2019). Chemometric approaches for developing infrared nanosensors to image anthracyclines. Biochemistry, 58(1), 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bonis-O’Donnell JT, Beyene A, Chio L, Demirer G, Yang D, & Landry MP (2017). Engineering molecular recognition with bio-mimetic polymers on single walled carbon nanotubes. Journal of Visualized Experiments: JoVE, 119, 55030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara S, Amer S, Ali A, Abouleila Y, Oga A, & Masujima T, (2017). Single-cell metabolomics. In Sussulini A (Ed.), Metabolomics: From fundamentals to clinical applications (pp. 323–343). Cham: Springer International Publishing. [Google Scholar]
- Giraldo JP, Landry MP, Kwak S-Y, Jain RM, Wong MH, Iverson NM, et al. (2015). A ratiometric sensor using single chirality near-infrared fluorescent carbon nanotubes: Application to In vivo monitoring. Small, 11(32), 3973–3984. [DOI] [PubMed] [Google Scholar]
- Greaves M, & Maley CC (2012). Clonal evolution in cancer. Nature, 481, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AA, & Hersam MC (2011). Nearly single-chirality single-walled carbon nanotubes produced via orthogonal iterative density gradient ultracentrifugation. Advanced Materials, 23(19), 2185–2190. [DOI] [PubMed] [Google Scholar]
- Heller DA, Baik S, Eurell TE, & Strano MS (2005). Single-walled carbon nanotube spectroscopy in live cells: Towards long-term labels and optical sensors. Advanced Materials, 17(23), 2793–2799. [Google Scholar]
- Holzinger M, Le Goff A, & Cosnier S (2014). Nanomaterials for biosensing applications: A review. Frontiers in Chemistry, 2, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Lillemeier BF, Kuhns MS, Chen DS, & Davis MM (2006). T cells use two directionally distinct pathways for cytokine secretion. Nature Immunology, 7, 247. [DOI] [PubMed] [Google Scholar]
- Ibáñez AJ, Fagerer SR, Schmidt AM, Urban PL, Jefimovs K, Geiger P, et al. (2013). Mass spectrometry-based metabolomics of single yeast cells. Proceedings of the National Academy of Sciences of the United States of America, 110(22), 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iourov IY, Vorsanova SG, & Yurov YB (2012). Single cell genomics of the brain: Focus on neuronal diversity and neuropsychiatric diseases. Current Genomics, 13(6), 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Liu R, Xiang YK, & Ha T (2012). Single-molecule pull-down for studying protein interactions. Nature Protocols, 7, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RM, Tvrdy K, Han R, Ulissi Z, & Strano MS (2014). Quantitative theory of adsorptive separation for the electronic sorting of single-walled carbon nanotubes. ACS Nano, 8(4), 3367–3379. [DOI] [PubMed] [Google Scholar]
- Kruss S, Landry MP, Vander Ende E, Lima BMA, Reuel NF, Zhang J, et al. (2014). Neurotransmitter detection using corona phase molecular recognition on fluorescent single-walled carbon nanotube sensors. Journal of the American Chemical Society, 136(2), 713–724. [DOI] [PubMed] [Google Scholar]
- Kruss S, Salem DP, Vuković L, Lima B, Vander Ende E, Boyden ES, et al. (2017). High-resolution imaging of cellular dopamine efflux using a fluorescent nanosensor array. Proceedings of the National Academy of Sciences, 114(8), 1789–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, et al. (2016). Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science, 352(6293), 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane R, Liu JJ, Pljevaljcic G, White KL, van der Schans E, Katritch V, et al. (2015). Single-molecule view of basal activity and activation mechanisms of the G protein-coupled receptor β2AR. Proceedings of the National Academy of Sciences of the United States of America, 112(46), 14254–14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry MP, Ando H, Chen AY, Cao J, Kottadiel VI, Chio L, et al. (2017). Single-molecule detection of protein efflux from microorganisms using fluorescent single-walled carbon nanotube sensor arrays. Nature Nanotechnology, 12, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Nishide D, Tanaka T, & Kataura H (2011). Large-scale single-chirality separation of single-wall carbon nanotubes by simple gel chromatography. Nature Communications, 2, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AE, Jin C, Reed JC, & Tsien RY (2004). Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proceedings of the National Academy of Sciences of the United States of America, 101(50), 17404–17409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, & Ha T (2008). A practical guide to single-molecule FRET. Nature Methods, 5, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki Y, Yamagishi M, Suzuki N, Izawa K, Nakahara A, Mizuno J, et al. (2014). Real-time single-cell imaging of protein secretion. Scientific Reports, 4, 4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Mancini MC, & Nie S (2009). Bioimaging: Second window for in vivo imaging. Nature Nanotechnology, 4(11), 710–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley AC, & Lacy P (2010). Pathways for cytokine secretion. Physiology, 25(4), 218–229. [DOI] [PubMed] [Google Scholar]
- Thurley K, Gerecht D, Friedmann E, & Höfer T (2015). Three-dimensional gradients of cytokine signaling between T cells. PLoS Computational Biology, 11(4), e1004206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek E, Shiraki T, Shiraishi T, Shiga T, Fujigaya T, & Janas D (2019). Single-step isolation of carbon nanotubes with narrow-band light emission characteristics. Scientific Reports, 9(1), 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Nunès JA, & Vély F (2004). Natural killer cell signaling pathways. Science, 306(5701), 1517–1519. [DOI] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, et al. (2014). Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature, 512, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Tanaka T, Yomogida Y, Sato N, Saito R, &Kataura H (2016). Experimental determination of excitonic band structures of single-walled carbon nanotubes using circular dichroism spectra. Nature Communications, 7, 12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yomogida Y, Tanaka T, Zhang M, Yudasaka M,Wei X,& Kataura H (2016). Industrial-scale separation of high-purity single-chirality single-wall carbon nanotubes for biological imaging. Nature Communications, 7, 12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalipsky S, & Harris JM (1997). Introduction to chemistry and biological applications of poly(ethylene glycol). In Vol. 680. Poly(ethylene glycol) (pp. 1–13): American Chemical Society. [Google Scholar]
- Zenobi R (2013). Single-cell metabolomics: Analytical and biological perspectives. Science, 342(6163), 1243259. [DOI] [PubMed] [Google Scholar]
- Zhang J, Landry MP, Barone PW, Kim J-H, Lin S, Ulissi ZW, et al. (2013). Molecular recognition using corona phase complexes made of synthetic polymers adsorbed on carbon nanotubes. Nature Nanotechnology, 8, 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, & Rueda D (2009). RNA folding dynamics by single-molecule fluorescence resonance energy transfer. Methods, 49(2), 112–117. 10.1016/j.ymeth.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Zhao W, Schafer S, Choi J, Yamanaka YJ, Lombardi ML, Bose S, et al. (2011). Cell-surface sensors for real-time probing of cellular environments. Nature Nanotechnology, 6, 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Stybayeva G, Silangcruz J, Yan J, Ramanculov E, Dandekar S, et al. (2009). Detecting cytokine release from single T-cells. Analytical Chemistry, 81(19), 8150–8156. 10.1021/ac901390j. [DOI] [PMC free article] [PubMed] [Google Scholar]