Abstract
Surface engineering of nanoparticles has recently emerged as a promising technique for synthetic molecular recognition of biological analytes. In particular, the use of synthetic heteropolymers adsorbed onto the surface of a nanoparticle can yield selective detection of a molecular target. Synthetic molecular recognition has unique advantages in leveraging the photostability, versatility, and exceptional chemical stability of nanomaterials. In particular, single-walled carbon nanotubes (SWNT) exhibit a large Stokes shift and near infrared emission for maximum biological sample transparency. Optical biosensors with high signal transduction and molecular specificity can be synthesized with amphiphilic heteropolymers grafted to SWNT, and discovered by high-throughput screening. Herein, we describe the development and the characterization of surface-engineered nanoparticles, or “synthetic antibodies,” for protein detection.
Keywords: Protein detection, Sensors, Carbon nanotubes, DNA aptamers, Infrared fluorescence microscopy, Nanomaterials, Synthetic antibodies
1. Introduction
Sensors detect and relay information through a molecular recognition element and a signal transduction element. Together, molecular recognition and signal transduction constitute the governing principles and limitations by which a molecular sensor operates. For the detection of proteins, antibodies specific to the target analyte are traditionally used as the molecular recognition element, in conjunction with a signal transduction element that is either electrochemical, colorimetric, or optical. While most protein sensors utilize an electrochemical readout, these methods are often unable to detect an analyte with spatial and temporal resolutions which are necessary for biomolecular imaging [1–6].
Single-walled carbon nanotubes (SWNT) offer unique advantages in signal transduction. SWNT have a large Stokes shift exhibiting several hundreds of nanometers between excitation and emission wavelengths, an order of magnitude greater than that of a standard fluorophore (Fig. 1a). SWNTs also have high biological sample signal transduction lengths due to their emission in the near-infrared: an optical window beyond the photon scattering range of biological materials and before the optical absorption of photons by water (Fig. 1b).
Fig. 1.
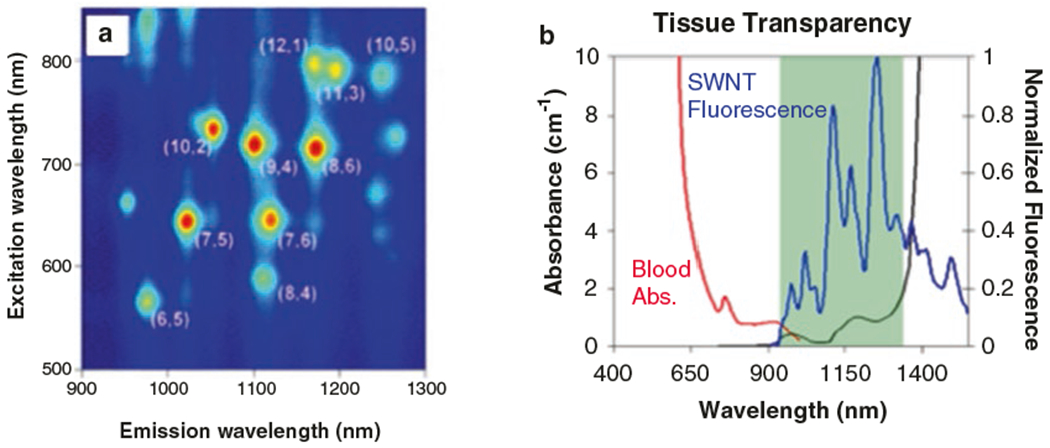
Fluorescent properties of SWNT. (a) Excitation and emission wavelengths of various SWNT chiralities (figure reproduced with permission from [7]). (b) Fluorescence of SWNT occurs in a range of wavelengths in which there is an optical window into biological tissues (figure reproduced with permission from [8])
This chapter focuses on the development of protein biosensors that serve as synthetic, non-biological antibody analogues. Heteropolymers can be engineered to adsorb onto the surface of SWNT, providing the resulting polymer-SWNT hybrid with selective molecular recognition. This approach has successfully produced synthetic sensors for analytes such as riboflavin [9], nitric oxide and hydrogen peroxide, which have gone on to be used in vivo [10, 11], and dopamine [12]. Recently, sensors using this technology have been successfully implemented to produce synthetic antibodies, such as a SWNT-based sensor for the protein fibrinogen [13].
Herein, we outline the materials and methods needed to construct and analyze surface engineered SWNT biosensors for protein detection. We first outline a three-pronged approach for the discovery of synthetic antibodies through screening-based, design-based, and ratiometric nanosensor development. Next we describe the protocols used to synthesize candidate nanosensors. Depending on the stability of the heteropolymer, adsorption to SWNT nanoparticles can be achieved using probe tip sonication, bath sonication, or dialysis. We subsequently discuss the building of essential microscopy equipment (a near-infrared spectrometer, and a near-infrared epifluorescence microscope) to identify, visualize, and utilize synthetic antibodies. Finally, we discuss the screening protocols used to validate our synthetic antibodies.
2. Materials
2.1. Candidate Synthetic Antibodies: Encapsulating Single Wall Carbon Nanotubes with a Library of Different Coronas
Raw HiPco SWNT (Unidym).
NanoPure Water (Deionized water purified to a sensitivity of 18 MΩ at 25 °C).
Bath sonicator.
Biocompatible polymers, such as DNA/RNA polymers (Integrated DNA Technologies) suspended in 100 mM NaCl or phospholipid-PEG (Avanti Polar Lipids) (see Note 1).
0.1 M sodium chloride (NaCl) solution: sodium chloride, NanoPure water. Weigh out 5.84 g of NaCl and dissolve in 1 L of H2O.
Probe tip sonicator.
3 mm probe tip (Cole-Parmer).
Cup-horn sonicator.
Surfactant, e.g., sodium dodecyl sulfate (Sigma Aldrich) (see Note 2).
Dialysis cartridge with proper molecular weight cutoff for retention of SWNT and polymers, and removal of monomeric surfactant.
2.2. Synthesis of a Screening Library
To ensure selectivity, a sensor must be screened against a library of analytes that is representative of the native environment of the target analyte. For example, when screening for a synthetic antibody for fibrinogen, a protein found in blood, the analyte library will consist of the most abundant proteins in human whole blood: albumin, IgG, fibrinogen, α1-antitrypsin, transferrin, haptoglobin, α2-macroglobulin, IgA, IgM, α1-acid-glycoprotein, apolipopro-tein-A1, insulin, hCG, and CRP [13]. Other molecular constituents of the sensor’s end-working environments should also be included in the screening analyte library.
Phosphate buffered saline (PBS): 137 mM NaCl, 2.7 mM KCl, 10 mM, Na2HPO4, 1.8 mM KH2PO4, pH 7.4. Weigh out 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, and 0.24 g KH2PO4 and dissolve in 800 mL of H2O. Adjust the pH to 7.4 with HCl, and then add H2O to 1 L Sterilize by autoclaving for 20 min at 15 psion liquid cycle or by filter sterilization. Store at room temperature (see Note 3).
Fetal bovine serum (FBS): 10% vol PBS.
Protein target and other analytes.
96-well plate with transparent glass bottom.
2.3. Near-Infrared Spectrometer (1D)
Zeiss AxioVision Inverted Microscope.
785 nm photodiode laser (B&W Tek Inc.).
Princeton Instruments InGaAs 1D Detector.
PI Acton SP2500 Spectrometer.
Optical components: optical posts, post holders, post bases, optical table, broadband near-infrared optimized mirrors, mirror mounts.
Irises for beam alignment.
2.4. Near-Infrared Epifluorescence Microscope (2D)
Zeiss AxioVision Inverted Microscope.
785 nm photodiode laser (B&W Tek Inc.).
Princeton Instruments InGaAs 2D Detector. (The detector to be used is dependent on the level of signal-to-noise needed for your experiments. Several InGaAs-based 2D detectors have sub-300 e/p/s dark current with or without liquid nitrogen cooling, enabling sub-10 ms single-SWNT detection, but might not be a financially feasible option. Alternative 2D detectors are given in Table 1.)
Optical components: post, post holder, base.
Irises for beam alignment.
Near-infrared transmissive microscope objectives.
Near-infrared optimized mirrors.
Table 1.
Comparison of 2D detectors
| Manufacturer model | Photon etc. Zephir 1.7–640 | Xenics cougar-640 LN | Princeton NIRvana:640 | Photonic science SWIR VGA C | Photonic science SWIR VGAT |
|---|---|---|---|---|---|
| Material | InGaAs | InGaAs | InGaAs | InGaAs | InGaAs |
| Spectral range | 0.9–1.7 μm | 0.90–1.55 μm | 0.9–1.7 μm | 1.0–1.7 μm | 1.0–1.7 μm |
| Array size | 640 × 512 | 640 × 512 | 640 × 512 | 640 × 512 | 640 × 512 |
| Pixel size | 15 μm × 15 μm | 20 μm × 20 μm | 20 μm × 20 μm | 25 μm × 25 μm | 25 μm × 25 μm |
| Dynamic range | 13/15 bit | 24 bit | 16 bit | 16 bit | 16 bit |
| FPS | 120/350 HZ | 1.42 HZ | 110 HZ | 22 HZ | 22 HZ |
| Readout noise, gain L/H (e−) | 180/45 | 15 | 120 (high gain) | 380/150 | 380/150 |
| Dark current | 250 e/p/s | 10 e/p/s | 300 e/p/s | 6241 e/p/s | 811 e/p/s |
| Exposure time | 1 μs to minutes | 12.5 ns–53.7 s | 2 μs to minutes | – | – |
| Full well | 600 K/12 K | 400 K | 40 K/600 K | 1.9 M/39 Ke- | 1.9 M/39 Ke- |
| Cooling type | TEC | LN | TEC/water cooling | TEC air cooled | TEC water cooled |
| Operating temperature | −80 °C | −196 °C | −85 °C | −20 °C | −40 °C |
| Interface | Camera link | Camera link | GigE | GigE | GigE |
| QE | >80% | – | >85% | – | – |
| Pricing | 60,000 USD | – | 120,000 USD | 32,000 USD | 34,300 USD |
3. Methods
3.1. “Synthetic Antibodies” Development Outline
Surface engineered “synthetic antibodies” can be developed using a three-tiered approach: screening for a synthetic antibody, rational capture-anchor design of a synthetic antibody, or ratiometric detection of analytes (Fig. 2).
Fig. 2.
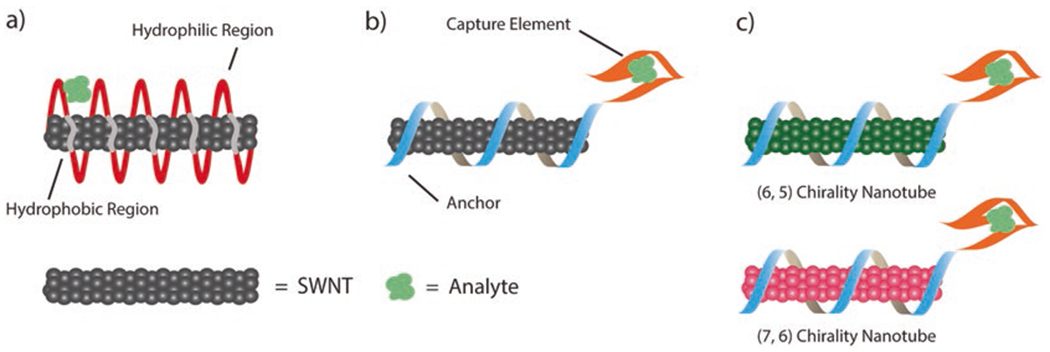
Three-tiered approach to develop synthetic antibodies. (a) In a screening approach, a large library of amphiphilic polymers is templated by the surface of the carbon nanotube for analyte capture. (b) In a design-based approach, a polymer is designed with an anchor and capture element. (c) In a ratiometric approach, two distinct carbon nanotube chiralities provide a ratiometric response to an analyte
Screening Approach:
Screening for a protein sensor requires the development of a candidate synthetic antibody library and screening candidate synthetic antibodies against a library of analytes. A candidate synthetic antibody library can comprise of SWNT surface-engineered with different polymers such as polynucleic acids, synthetic peptides and peptoids, amphiphilic heteropolymers, surfactants, and functionalized phospholipids. To create a selective and responsive sensor, a broad range of polymer-SWNT conjugates needs to be screened. The screening approach is the main focus of this chapter.
Design-based Approach:
Design-based engineering of synthetic antibodies involves the tethering of polymers with an anchor domain and a capture element onto the surface of a SWNT [14]. Anchor domains must noncovalently and strongly adsorb to the surface of the SWNT often through hydrophobic or pi-stacking interactions. A known strong anchor domain is a C30 nucleic acid sequence [15]. Capture elements have predetermined specificity to the target protein. Unwanted interactions between the capture domain and the SWNT surface could lead to unforeseeable loss of detection. This method is advantageous over a screening approach in that it leverages specific polymer sequences with predetermined affinities for protein analytes, and as such requires a smaller polymer and analyte library at the onset.
Ratiometric Approach:
If both or either of the above methods provides a sensitive, but not selective, response for the protein of interest, a ratiometric detection strategy can be developed. A SWNT sensor can be created using a ratio of two distinct intensities from SWNT chiralities with different emission wavelengths. For example, the [5, 6] SWNT chirality emits at 1000 nm and the [6, 7] SWNT chirality emits at 1150 nm. A ratiometric relationship between the signal of different chiralities can provide additional information about the system by monitoring one signal as invariant to the analyte and the second signal as responsive to the analyte. Ratiometric monitoring can also eliminate baseline noise of different and often biologically complex sensing environments. While chirality-purified SWNT are available commercially, more information about isolating distinct SWNT chiralities can be found in ref. [16].
3.2. Protocol to Conjugate SWNT with Polymer Corona Phases
The method used to conjugate polymers to the surface of the carbon nanotubes depends on two parameters: the robustness of the polymer, and the affinity of the polymer for the SWNT. Probe tip sonication results in the highest SWNT-polymer conjugate yield, is completed in a few minutes, but exposes the polymer to harsh high-power conditions. This method is suitable for structurally robust polymers such as short ssDNA oligonucleotides. Bath sonication gives lower yield than probe-tip sonication, is completed in a few hours, and uses gentler sonication power conditions. Dialysis of a surfactant-suspended SWNT solution with polymer is completed in a few days’ time, and involves the gentle exchange of surfactant with polymer on SWNT surface for structurally fragile polymers in which shearing is to be avoided. The three methods are compared in Table 2. Before conjugation, raw carbon nanotube samples require processing to remove impurities often found in SWNT materials (Subheading 3.2.1).
Table 2.
Comparison of the three methods for synthetic antibody development
| Method | Probe tip sonication | Bath sonication | Dialysis |
|---|---|---|---|
| Conditions | High-power exposure, significant heating | High-power exposure (gentler than probe tip) | Gradual exchange of surfactant for polymer, on SWNT surface |
| Time scale | Minutes | Hours | Days |
| Subheading | 3.2.2 | 3.2.3 | 3.2.4 |
| Yield | High | Low | High |
| Recommendations | For robust polymers (DNA and RNA) | For robust polymers prone to shearing, with high affinity for SWNT (peptide capture elements) | For fragile polymers, protein, and antibodies, with low affinity for SWNT (large dsDNA, proteins) |
3.2.1. Processing Raw Single-Walled Carbon Nanotubes (SWNT) for Surface Engineering
Raw SWNT must be washed and processed before surface engineering. Washing the carbon nanotubes removes the organic binder, typically methanol, and residual catalyst from carbon nanotube synthesis. Initially 0.8–1.2 nm in diameter and 100 nm-1 μm in length, carbon nanotubes have a mean length of 550 nm after processing. Carbon nanotubes above ~100 nm in length retain their near-infrared fluorescence (see Note 4).
Add 200–300 mg of raw SWNT to a 50 mL Falcon tube with 45 mL of deionized water.
Vortex the tube at the highest setting for 2 min.
Place the tube in a bath sonicator for 5 min to disperse the nanoparticles.
Centrifuge for 20 min at 2000 × g. Then discard the supernatant.
Add 45 mL of fresh deionized water and repeat the vortexing and centrifugation steps (steps 2 and 4) 7 additional times.
Place the wet SWNT in a drying oven at 120 °C until completely dried (typically 24–48 h).
Grind SWNT with a mortar and pestle to homogenize SWNT length.
3.2.2. Probe Tip Sonication to Conjugate SWNT with Polymer Corona Phases
In a fume hood, add 1 mg of SWNT to 2 mg of polymer in 1 mL of 0.1 M sodium chloride solution in a 2 mL centrifuge tube.
In an ice bath, sonicate the solution with a 3 mm probe tip for 5 min at a power of 4 W (see Notes 5 and 6).
Benchtop-centrifuge the solution for 90 min at 16,100 × g (see Notes 7 and 8).
Pipette to collect the top 80–90% of the supernatant for further experimentation being careful not to disrupt the pellet. The pellet can be discarded (see Note 9).
3.2.3. Bath Sonication to Conjugate SWNT with Polymer Corona Phases
In a fume hood, add 1 mg of SWNT to 2 mg of polymer in 1 mL of 0.1 M sodium chloride solution in a 2 mL tube.
Bath-sonicate the solution for 1 h (see Note 5).
Benchtop-centrifuge the solution for 90 min at 16,100 × g (see Notes 7 and 8).
Pipette to collect and keep the top 80–90% of the supernatant, being careful not to disrupt the pellet. The pellet can be discarded (see Note 9).
3.2.4. Dialysis to Conjugate SWNT with Polymer Corona Phases
For this protocol, the carbon nanotube is first suspended with sodium dodecyl sulfate. Another surfactant may be chosen as long as it is compatible with the polymer being adsorbed, and smaller in molecular mass than the dialysis membrane chosen.
Add 4 g of sodium dodecyl sulfate (SDS) to 150 mL of water (see Note 10).
Add 60 mg of single-walled nanotubes to the aqueous solution (see Note 4).
Add 50 mL of water to the solution.
Homogenize the sample for 1 h using a homogenizer on the lowest setting. This will disperse the sample without breaking the SWNT.
Sonicate the sample for 10 min using a cup-horn sonicator with an amplitude of 90%.
Properly weigh out the sample in separate centrifuge tubes to ensure the centrifuge is balanced.
Ultracentrifuge the sample for 4 h at 47,000 × g.
Pipette to collect the top 80–90% of the supernatant for further experimentation being careful not to disrupt the pellet. The pellet can be discarded (see Note 9).
Add the polymer to the SDS-SWNT suspension to a final polymer concentration of 2 mg/mL.
Dialyze the mixture using an appropriate molecular weight cutoff dialysis cartridge against 2 L of water for 4–5 days. Run a concurrent control of a SDS-SWNT only suspension.
Change the dialysis buffer every 12 h to completely remove SDS from the SWNT surface. Dialysis is complete when the SWNT from the control precipitates out of solution (see Note 11).
3.3. Building Near-Infrared Screening and Imaging Microscopy
3.3.1. 1D InGaAs Infrared Screening Spectrograph
Fix the Zeiss AxioVision Inverted Microscope and 785 nm laser to the optical table.
To align the beam, first draw the desired beam path on the optical table. Screw in post holders for the laser and shutter using Fig. 3 as a guide. Fix the laser on the corresponding post loosely so that its position may be adjusted.
Align the laser beam using two irises, a set distance apart. Adjust the irises to be at the same height as the microscope port through which the beam will enter. First, adjust the tilt of the laser so that it passes through the center of the first iris. Next, adjust its height on the stability post so that the beam passes through both irises.
Attach post holders to the table at the corners of the beam path for placement of the mirrors. Place the mirrors on the post so that the laser hits the center of the mirror (see Notes 12 and 13).
Align the mirrors two at a time using the irises. Place the irises along the desired beam path ahead of two mirrors. The irises should be at the same height as before and a set distance apart from one another. Adjust the angle of the first mirror until the laser beam passes through the center of the first iris. Next, turn the second mirror until the beam is aligned with the center of the second iris. Repeat these two steps iteratively until the beam is perfectly aligned through the center of both irises.
Align the laser with the front and back focal plane of the objective of the inverted microscope. Similar to the previous step. Iteratively align the second mirror from the objective with the crosshair on the front focal plane, and the first mirror with the center of the eyepiece.
Fix the appropriate filter cube to the filter wheel of the microscope. For our setup, we use a Chroma RT—Raman 785 nm longpass set (see Note 14).
Ensure the laser line is collimated by reflecting the laser beam from the filter set and allowing the beam to reflect upwards toward the ceiling unperturbed. Laser beam should be collimated, or slightly divergent within a several meter-long path. Laser beam should also be approximately 1 cm in diameter as it enters the microscope port, to overfill the back of the objective (see Notes 15 and 16).
Place a 20× objective with near-infrared transmission capabilities in the microscope nosepiece.
Place the spectrometer (IsoPlane, Princeton Instruments) in line with the microscope side port and place a lens in the beam path such that the emitted light from the sample plane is directed into the entrance slit of the spectrometer. Attach a 1D InGaAs array (PyLoN-IR, Princeton Instruments) to the camera port of the IsoPlane spectrometer.
Fig. 3.
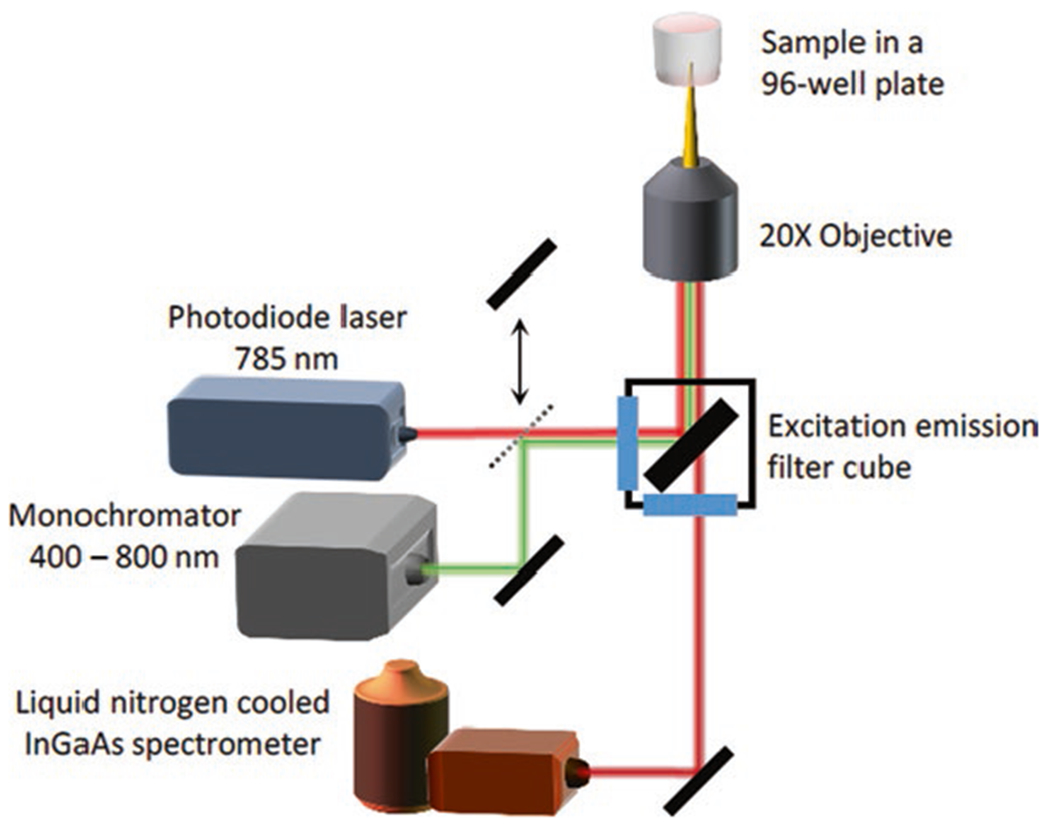
Schematic of the 1D InGaAs Infrared Spectrometer for screening of synthetic antibody fluorescence response to analyte libraries
3.3.2. 2D InGaAs Infrared Imaging Microscope
Fix the Zeiss AxioVision Inverted Microscope and 785 nm laser to the optical table.
To align the beam, first draw the desired beam path on the optical table. Screw in post holders for the laser and shutter using Fig. 4 as a guide. Fix the laser on the laser mounting post loosely so that its position may be adjusted.
Align the laser beam using two irises, a set distance apart. First, adjust the irises to be at the same height as the microscope port through which the beam will enter. Adjust the tilt of the laser so that it passes through the center of the first iris. Next, adjust its height on the stability post so that the beam passes through both irises.
Attach post holders to the table at the corners of the beam path for placement of the mirrors. Place the mirrors on the post so that the laser hits the center of the mirror (see Notes 12 and 13).
Align the mirrors two at a time using the irises. Place the irises along the desired beam path ahead of two mirrors. The irises should be at the same height as before and a distance apart from one another. Adjust the angle of the first mirror until the laser beam passes through the center of the first iris. Next, turn the second mirror until the beam is aligned with the center of the second iris. Repeat these two steps iteratively until the beam passes through the center of both irises.
Align the laser with the front and back focal plane of the objective of the inverted microscope. Similar to the previous step, iteratively align the second mirror from the objective with the crosshair on the front focal plane, and the first mirror with the center of the eyepiece.
Fix the appropriate filter cube to the filter wheel of the microscope. For our setup, we use a Chroma RT—Raman 785 nm longpass set (see Note 14).
Ensure the laser line is collimated by reflecting the laser beam from the filter set and allowing the beam to reflect upwards unperturbed. Laser beam should be collimated, or slightly divergent within a several meter-long path. Laser beam should also be approximately 1 cm in diameter to overfill the back of the objective (see Notes 15 and 16).
Place a 100× objective in the microscope nosepiece, ensuring the objective is near-infrared transmissive.
Put a 2-camera adapter at the exit port of the microscope Place the Princeton Instruments InGaAs 2D Detector (NIRvana 640ST) on the optical table. Mount the camera using a C-mount camera adapter to fit the exit port of the microscope and the 1-in. threaded mount of the 2D detector.
If a brightfield image is desired, mount a brightfield camera such as a Zeiss axiocam on the top line of the 2-camera adapter.
Fig. 4.
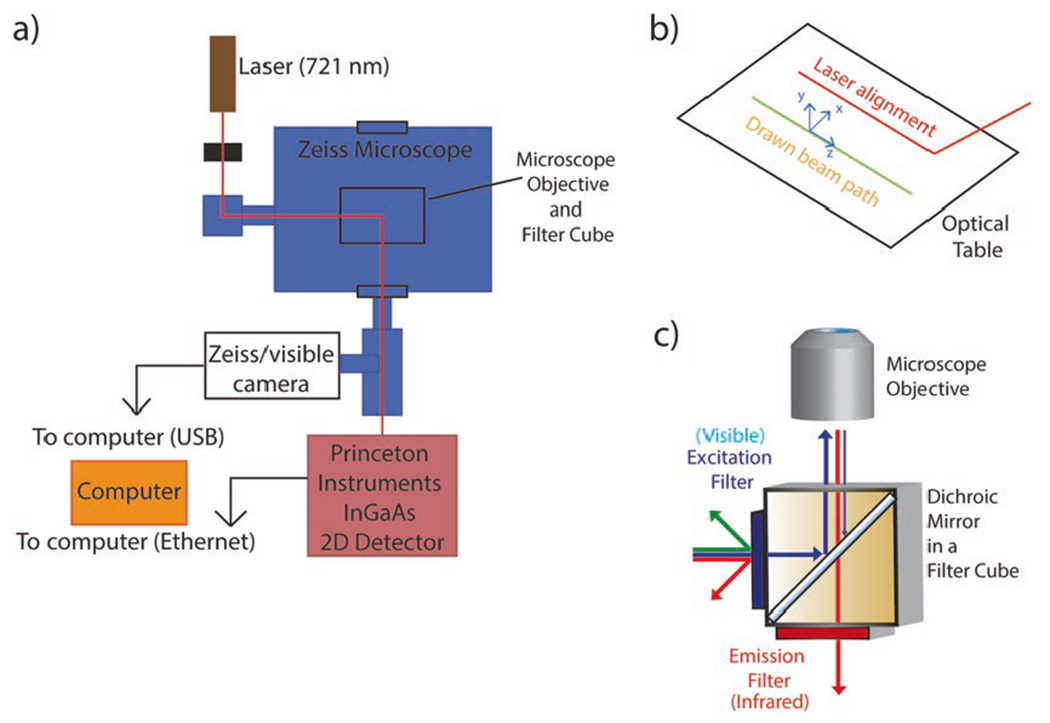
(a) Schematic of the set up for a 2D InGaAs Infrared Screening Spectrograph to screen for synthetic antibodies. (b) The beam path is drawn before laser alignment. (c) An appropriate filter cube enables excitation with visible wavelengths and enables emission of near infrared wavelengths should be chosen for imaging of SWNT-based synthetic antibodies
3.4. Spectroscopic Screening for Synthetic Antibodies
Fill the nIR detector (PyLoN-IR, Princeton Instruments) coolant tank with liquid N2. Wait approximately 3 h for the detector to cool to −100 °C before collecting absorbance spectra (see Note 17).
Prepare a library of polymer-SWNT samples in PBS at a concentration of 1 mg/L using methods detailed in Subheading 3.1. Remove excess polymer in solution through filtration with a spin desalting column, or dialysis.
In a 96 well plate, add 198 μL of polymer-SWNT solution at a concentration of 1 mg/L in PBS to wells B2–11, C2–11, D2–11, and E2–8 using a 200 μL micropipette with each well corresponding to a particular polymer coating (see Note 18).
To a separate 96 well plate, add 150 μL of PBS solution to well B2. Label this as the background sample and set it aside for now.
Screen for successful suspension using fluorescence absorbance/emission spectroscopy. Turn on the custom epifluorescence microscope (Subheading 3.4) and the 785 nm laser with the shutter closed. Switch to the 20× objective and select filter cube to reflect 785 nm excitation light, and transmit light above 900 nm (see Note 19).
Place a well plate containing 15 mg/L SWNT-based candidate sensor on the motorized translation stage of the microscope with the first well, B2, positioned over the 20× objective. The objective should be positioned far from the bottom of the well plate.
Collect several sequential test spectra using an exposure time of 1000 ms. Move the objective upwards toward the bottom of the well plate slowly until maximum emission is observed. The software used for data acquisition is camera specific; however, for Princeton Instruments detectors, LightField software interfaces well with the detector. Once you achieve maximum fluorescence, keep the objective at that working distance (see Note 20).
Move to an empty well within your well plate. Collect a background spectrum using the exposure time from the previous step, with the objective positioned at its optimal distance from the objective.
Place the original sample well back onto the microscope objective. Run the automation script to collect fluorescence spectra for all wells using the same optimal exposure time. Verify that the background has been subtracted from all sample spectra. This is the initial intensity spectrum for each of your candidate sensors.
Remove the plate from the microscope. Using a 10 μL micropipette, add 2 μL of each analyte from the 10 mM stock solution in PBS to the corresponding polymer conjugated SWNT in the well plate. Gently stir the well plate in a circular motion while pushing the plunger in and out to thoroughly mix the solution. The final concentration of analyte in each well is 100 μM (see Note 21).
Place the well plate back onto the microscope. Run the automated script to scan and collect fluorescence spectra for all wells using the same exposure time as in step 8. These fluorescence spectra represent the fluorescence modulation incurred by each analyte.
Cover the well plate using a 5 cm × 5 cm sheet of Parafilm and incubate on a tabletop orbital shaker at room temperature for 1 h.
Remove the Parafilm and place the plate on the optical table and repeat the screening protocol to collect fluorescence spectra for each sample. These fluorescence spectra represent any time-dependent fluorescence modulation incurred on the SWNT sensor by each analyte.
For each well, calculate the change in fluorescence intensity at a particular wavelength upon addition of analyte, and 1-h post analyte addition (Fig. 5a). Plot these results in a heat map to determine which of your sensor-analyte pairs exhibit a significant and selective response (Fig. 5b). Candidate sensors responding selectively and strongly to the addition of a protein analyte can be used as synthetic antibodies.
Fig. 5.
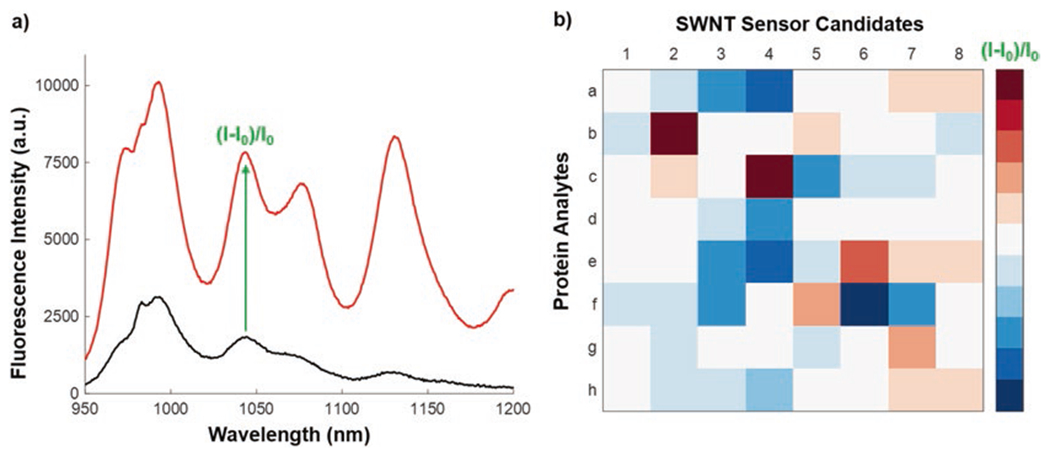
Result of spectroscopic screening for a sample analyte-SWNT candidate nanosensor. (a) Fluorescence spectrum for a representative SWNT-polymer conjugate before (black) and after (red) addition of an analyte shown to induce fluorescence modulation. The peak at 1044 nm, for instance, can be used to calculate the sensor signal, (I-I0)/I0. (b) Simulated heat map of sensor response to the library of protein analytes. Candidate sensors with a significant turn-off or turn-on response to a single analyte are deemed selective synthetic antibodies, such as Protein B + candidate SWNT sensor 2, or Protein C + candidate SWNT sensor 4
3.5. Fluorescence Imaging of “Synthetic Antibodies” Using Near-Infrared Microscopy
Dilute polymer-SWNT conjugate solutions to a final SWNT concentration of 2 mg/L (see Note 22).
Clean and oxidize a glass #1.5 coverslip using a series of rinses: methanol, ethanol, acetone, and water.
Fix the coverslip onto the adhesive surface of a microfluidics channel slide (Ibidi, Martinsried, Germany). Only the nanotubes immobilized on the surface of the coverslip will be imaged so cleaning of this slide is not necessary.
Wash the channel by flowing through 50 μL of PBS solution using a micropipette.
Coat the surface of the coverslip with 50 μL of biotin labeled BSA (1 mg/mL) taking care not to introduce any air bubbles into the channel. Incubate for 5 min at room temperature.
Rinse with 100 μL PBS solution to remove any unbound BSA-Biotin.
Add 50 μL of 0.2 mg/mL neutravidin in PBS and let incubate for 5 min. This step sparsely populates the coverslip with SWNT binding sites.
Rinse again with 100 μL PBS.
Add 50 μL of the synthetic antibody solution to the coverslip and incubate for 5 min at room temperature. Rinse with only 50 μL PBS to prevent washing away any immobilized nanotubes (see Note 23).
Rinse a final time with 50 μL PBS to remove any remaining unbound synthetic antibodies. The slide is now ready to be imaged which should be done as soon as possible after completion of this step. If this is not possible, the sample can be kept refrigerated for 1–2 days.
Place the microscope slide on a 100× near-infrared transmissive objective with oil immersion, onto the 2D InGaAs imaging microscope with 500 ms exposure time. Slowly move the objective toward the microscope slide until surface-immobilized synthetic antibodies come into the field of view in focus (see Fig. 6b) (see Note 24).
Begin acquiring data by acquiring a sequence of frames. Make note of the frame at which you add 50 μL of protein analyte at a concentration of 10 nM. Your synthetic antibodies should respond similarly to the response recorded in the screening heat map (Fig. 5b).
Calculate the relative change in synthetic antibody intensity upon protein analyte addition, ((I-I0)/I0). This value will likely be higher than the value determined in Fig. 5b.
Fig. 6.
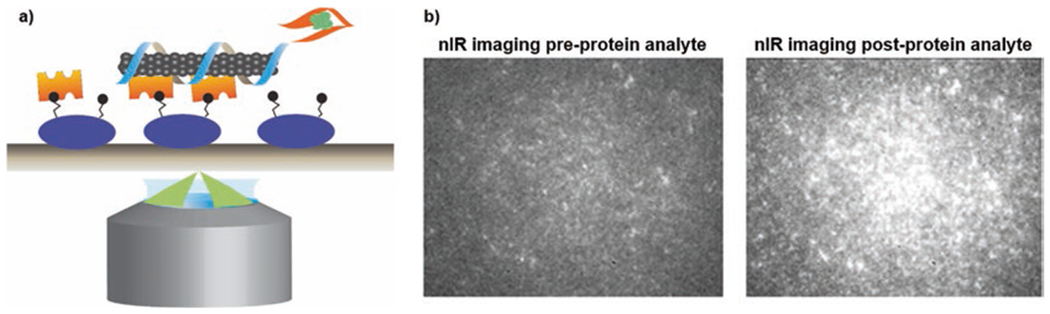
Imaging of immobilized synthetic antibodies. (a) Schematic of excitation and emission of polymer-SWNT fixed to a coverslip. (b) The addition of protein analyte causes a detectable increase in the fluorescence intensity of the synthetic sensors. This can be used for the localization of analytes on a single molecule level on hour-long timescales, enabled by the theoretically infinite fluorescence photobleaching lifetime of SWNT
This surface can also be utilized to detect protein analytes from biological samples. For instance, one might introduce biological cells into the microscope slide and monitor the infrared response of surface-immobilized sensors as a measure of protein secretion from the cell [14]. Analyte binding can be verified by incorporating a visible single-molecule fluorescence line into the near-infrared fluorescence microscope setup as described in [17].
Acknowledgments
This work was supported by Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), a Simons Foundation grant, and a Brain and Behavior Research foundation young investigator grant. D.Y. acknowledges an NSF Graduate Research Fellowship and L.C. acknowledges a LAM research fellowship. L. C. wrote the introduction, Subheadings 3.1 and 3.2. D.Y. wrote Subheadings 3.3–3.5. L.C. and D.Y. collaboratively wrote support commentary and edited the manuscript with guidance from M. L. We thank Roger Chang for insightful feedback on this work.
4 Notes
Polymers used generally will not exhibit affinity for a target protein prior to conjugation to SWNT.
Dry surfactants should be handled in a ventilated hood in case of aerosolization. Solid sodium dodecyl sulfate is flammable and should be handled in a fume hood away from sparks and flames.
Concentrated HCl (12 N) can be used at first to narrow the pH gap from the starting pH to the required pH. When pH is within a pH unit of the required final pH, use a series of HCl stock solutions (e.g., 6 N and 1 N) with lower ionic strengths, added dropwise, to avoid a sudden drop in pH below the required pH. Observe caution when handling caustic materials such as highly concentrated HCl.
Always work in a properly ventilated space, such as a fume hood or a glove box, when handling nanoparticles to avoid inhalation.
The SWNT initially forms small clumps in solution. If SWNT is successfully suspended by the polymer, the solution should have higher optical density and greater homogeneity.
Be sure that the sonicator tip is not touching the bottom or sides of the tube. Exercise caution after sonication; sonicator tip will be hot to the touch.
Always balance the centrifuge before operation so that the tube masses are within 0.05 g of each other.
If the supernatant appears clear with all SWNT in the pellet after a full 90 min of sonication, repeat the experiment and centrifuge in 5 min intervals to check stability of the polymer-SWNT suspension.
As soon as centrifugation ends, the pellet will begin to dissolve into the supernatant. Therefore, quick decanting is required to obtain solution of only single nanotubes, rather than tube bundles.
For best purity, use NanoPure or Milli-Q filtered water.
The complete removal of SDS from the SWNT surface allows for the adsorption of the polymer. The resulting polymer suspensions should be clear to the eye and free of SWNT aggregates.
When aligning the mirrors, keep in mind that the actual position of the mirror will be offset by the post.
Use mirrors with extended wavelength ranges that allow the passage of high wavelength beams (for the near-infrared you will need reflection of wavelengths of 700 nm and above). Some companies’ standard mirrors are only optimized to manipulate light in the visible range, up to 700 nm.
Many filter sets have not been optimized for infrared emission. Make sure to check that the emission filter is capable of transmitting light beyond 1000 nm, specifically for the 950–1300 nm emission range of most SWNT chiralities. Check the product page of the filter cube for %Transmission using a wavelength range between 750 and 1200 nm.
If laser beam is smaller than 1 cm, place a beam expander in the laser alignment path along the optical table and repeat the process of iterative alignment.
If beam is divergent, convergent, or if the beam profile is not Gaussian, check the quality of the laser output beam, the mirrors and dichroics used for beam steering, any accidental beam path occlusions, or the beam expander lenses.
Thermal gloves and eye protection should be worn while handling liquid nitrogen.
For experiments in serum, dilute polymer-SWNT to a concentration of 5 mg/L in FBS.
785 nm light is damaging to eyes. Whenever the laser is on, laser eye protection certified for this wavelength should be worn.
The lights in the room should be turned off while the microscope is running.
Protein analytes should be frozen and thawed before use. Solutions should be prepared fresh immediately before experimentation.
To determine polymer-SWNT concentration, take absorption spectrum of supernatant with a UV-Vis spectrometer. Concentration is roughly equal to (absorbance at 632 nm)/(0.036 L mg−1 cm−1).
Longer incubation times can be used to increase SWNT adsorption. If this is required, the channel openings should be capped to prevent evaporation of the solution and formation of air bubbles.
If there is insufficient SWNT binding to the surface, increase the concentration of neutravidin in step 7.
References
- 1.Cao Y, Zhu S, Yu J, Zhu X, Yin Y, Li G (2012) Protein detection based on small molecule-linked DNA. Anal Chem 84:4314–4320 [DOI] [PubMed] [Google Scholar]
- 2.Luo X, Freeman C, James T, Davis JJ (2013) Ultrasensitive label free electrical detection of insulin in neat blood serum. Anal Chem 85:4129–4134 [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Liu B, Tang D, Niessner R, Chen G, Knopp D (2012) DNA-based hybridization chain reaction for amplified bioelectronics signal and ultrasensitive detection of proteins. Anal Chem 84:5392–5399 [DOI] [PubMed] [Google Scholar]
- 4.Taleat Z, Cristea C, Marrazza G, Mazloum-Ardakani M, Sandulescu R (2014) Electrochemical immunoassay based on aptamer-protein interaction and functionalized polymer for cancer biomarker detection. J Electrochem Soc 717(718):119–124 [Google Scholar]
- 5.Shi H, He X, Wang K, Wu X, Ye X, Guo Q, Tan W, Qing Z, Yang X, Zhou B (2011) Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc Natl Acad Sci 108(10):3900–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sriwichai S, Baba A, Phanichphant S, Shinbo K, Kato K, Kaneko F (2010) Electrochemically controlled surface plasmon resonance immunosensor for the detection of human immunoglobulin G on poly(3-aminobenzoic acid) ultrathin films. Sens Actuators B 147:322–329 [Google Scholar]
- 7.Miyauchi Y, Chiashi S, Murakami Y, Hayashida Y, Maruyama S (2004) Fluorescence spectroscopy of single-walled carbon nanotubes synthesized from alcohol. Chem Phys Lett 387(1–3):198–203 [Google Scholar]
- 8.Boghossian AA, Zhang J et al. (2011) Near-infrared fluorescent sensors based on single-walled carbon nanotubes for life sciences application. Chem Sus Chem 4:848–863 [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Landry MP, Barone PW, Kim JH et al. (2013) Molecular recognition using corona phase complexes made of synthetic polymers adsorbed on carbon nanotubes. Nat Nanotechnol 8:959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraldo JP, Landry MP et al. (2015) A ratiometric sensor using single chirality near-infrared fluorescent carbon nanotubes: application to in vivo monitoring. Small 11(32):3973–3984 [DOI] [PubMed] [Google Scholar]
- 11.Giraldo JP et al. (2014) Plant nanobionics approach to augment photosynthesis and biochemical sensing. Nat Mater 13:400–408 [DOI] [PubMed] [Google Scholar]
- 12.Kruss S, Landry MP et al. (2014) Neurotransmitter detection using corona phase molecular recognition on fluorescent single-walled carbon nanotube sensors. J Am Chem Soc 136:713–724 [DOI] [PubMed] [Google Scholar]
- 13.Bisker G et al. (2016) Protein targeted corona phase molecular recognition. Nat Commn 7:10241. doi: 10.1038/ncomms10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landry MP et al. (2017) Single-molecule detection of protein efflux from microorganisms using fluorescent single walled carbon nanotube sensor arrays. Nat Nanotechnol. doi: 10.1038/nnano.2016284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landry MP et al. (2015) Comparative dynamics and sequence dependence of DNA and RNA binding to single walled carbon nanotubes. J Phys Chem C Nanomater Interfaces 119(18):10048–10058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H et al. (2015) Large-scale single-chirality separation of single-wall carbon nanotubes by simple gel chromatography. Nat Commn 2:309. doi: 10.1038/ncomms1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beyene AG et al. (2016) Nanoparticle-templated molecular recognition platforms for detection of biological analytes. Curr Protoc Chem Biol 8:197–223. doi: 10.1002/cpch.10 [DOI] [PMC free article] [PubMed] [Google Scholar]


