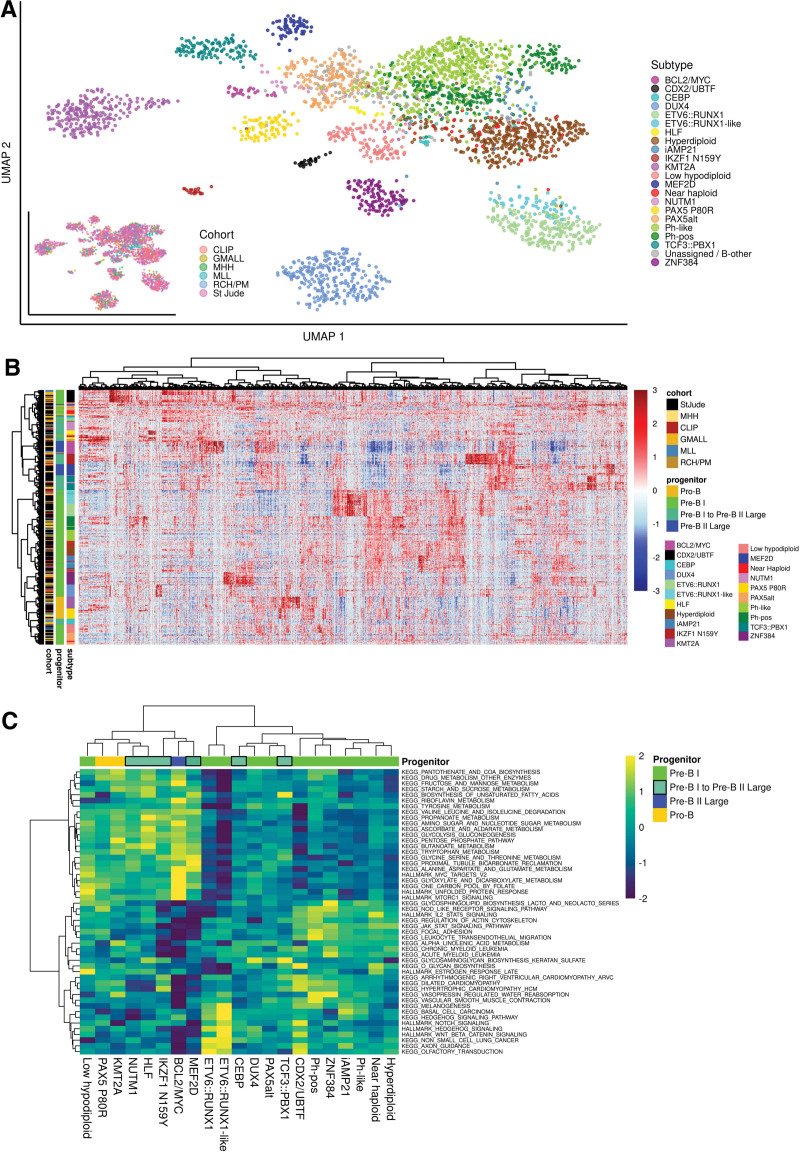
Figure 6.
The gene expression landscape in BCP-ALL. (A) UMAP plot showing all n = 2998 samples used in this study. Count data from the 6 data sets was batch corrected using the sva package29 and TPM values were calculated. The plot is based on 2802 genes selected by LASSO for training of ALLCatchR. Cohorts are highlighted on the bottom left plot. The expression data before batch correction is shown in Suppl. Figure S3A. (B) ALLCatchR predictions were used to define samples that best represented their respective molecular subtype. A total of n = 20 top ranking samples per subtype (exceptions with lesser samples available: HLF n = 14, CEBP n = 16, NUTM1 n = 17, IKZF1 N159Y n = 18) were used to obtain a homogenous data set representing all 21 BCP-ALL subtypes (n = 405). Differential gene expression analyses for each subtype versus the remaining cohort using DESeq230 revealed 5110 differentially expressed genes (cutoff: 1.5-log2-fold change, FDR: 0.001) used for unsupervised clustering. Suppl. Figure S12 and Suppl. Tables S9-S16, S17-S22, and S23-S29 provide detailed information on the derived gene sets. (C) Canonical signaling pathways (KEGG, HALLMARK gene sets; MSigDB) were used for single-sample gene set enrichment analysis using the BCP-ALL subcohort from (B) for balanced representation of all subtypes. Enrichment scores for top variable enriched pathways are shown. BCP-ALL = B-cell precursor acute lymphoblastic leukemia; UMAP = uniform manifold approximation plot.