Abstract
There was an inexplicable upsurge in the incidence of non-O1, non-O139 Vibrio cholerae among hospitalized patients admitted to the Infectious Diseases Hospital, Calcutta, India, between February and March 1996. Of the 18 strains of V. cholerae isolated during this period, 15 belonged to the non-O1, non-O139 serogroups (4 belonged to O144, 3 belonged to O11, 1 each belonged to O6, O8, O12, O19, O39, and O58, and 2 strains could not be typed), 2 belonged to the O139 serogroup, and 1 belonged to the O1 serogroup. Cell-free culture supernatants of 13 representative non-O1, non-O139 V. cholerae strains evoked a distinct cytotoxic effect on CHO and HeLa cells, and the strains examined produced the nonmembrane-damaging cytotoxin. By several PCR assays, it was determined that none of the non-O1, non-O139 strains were positive for the ctxA, zot, ace, and tcpA genes and for the genes representing the heat-labile toxin, heat-stable toxin, and verotoxin of Escherichia coli and the various variants of these genes. Studies on the clonality of non-O1, non-O139 V. cholerae strains by restriction fragment length polymorphism (RFLP) analysis of rRNA genes and of other genes (hlyA, hlyU, hlx, toxR, and attRS1) and by pulsed-field gel electrophoresis (PFGE) collectively indicate that the upsurge which occurred in February and March 1996 was caused by strains belonging to different clones. Overall, there was an excellent correlation between the results of ribotyping, RFLP analysis of various genes, and PFGE, with strains belonging to a particular serogroup showing nearly identical restriction patterns and PFGE profiles. It is clear from this study that some serogroups of V. cholerae can cause diarrhea by a mechanism quite different from that of toxigenic V. cholerae O1 and O139, and we have proposed the nomenclature of enteropathogenic V. cholerae to include these serogroups.
Vibrio cholerae strains belonging to serogroups O1 and O139 are the causative agents of cholera, while the non-O1, non-O139 serogroups of V. cholerae comprise a heterogeneous group of organisms whose clinical association with humans is inadequately understood. Clinically, apart from the O1 and O139 serogroups, the non-O1, non-O139 serogroups continue to be of negligible significance since these strains are associated with illness in only a low percentage of patients hospitalized due to acute secretory diarrhea (18). Nucleotide analysis of the asd genes of 45 strains of V. cholerae has yielded provocative evidence which indicates that the classical and El Tor biotypes and U.S. Gulf Coast strains of V. cholerae O1 evolved independently from environmental nontoxigenic, non-O1 strains (15). Therefore, it has become increasingly clear that the non-O1, non-O139 serogroups are involved in the emergence of newer variants of V. cholerae, a fact supported by the genesis of V. cholerae O139, which is believed to have evolved as a result of horizontal gene transfer between the O1 and the non-O1 serogroups (4).
These recent events have led us to fortify our cholera surveillance program in Calcutta, India, and to extend our monitoring to the non-O1, non-O139 serogroups as well. While conducting this survey, we observed an inexplicable rise in the incidence of non-O1, non-O139 V. cholerae in February and March 1996 among hospitalized patients admitted to the Infectious Diseases (ID) Hospital in Calcutta. In fact, the rate of isolation of non-O1, non-O139 strains of V. cholerae exceeded that of O1 and O139 serogroups during the period mentioned above. In this study, we report the extensive molecular characterization of the non-O1, non-O139 strains isolated between February and March 1996 from hospitalized patients admitted to the ID Hospital, Calcutta.
MATERIALS AND METHODS
Bacteriology and serogrouping.
This study was conducted among hospitalized patients admitted to the ID Hospital, Calcutta, the only hospital which admits cholera patients from metropolitan Calcutta and surrounding areas. Upon admission, a thorough clinical evaluation with particular attention to the degree of dehydration was conducted and a retrospective history was recorded in a standard proforma manner. Stool samples or rectal swabs collected in Cary Blair medium were processed in the laboratory within 2 h of collection for the isolation of V. cholerae and other enteropathogens by previously published techniques (18, 36). A multitest medium was used for the presumptive identification of V. cholerae (14, 20). All strains were subsequently examined for the oxidase reaction, and the identity of V. cholerae O1 was confirmed by serogrouping, using growth from the multitest medium, with polyvalent O1 and monospecific Inaba and Ogawa antisera raised at the National Institute of Cholera and Enteric Diseases, Calcutta. V. cholerae strains which did not agglutinate with the O1 antiserum were checked with monoclonal O139 antiserum developed at the National Institute of Cholera and Enteric Diseases (9). V. cholerae strains which did not agglutinate with either O1 or O139 antiserum were assumed to belong to the non-O1 and non-O139 serogroups, and these strains were further serogrouped by the somatic O-antigen serogrouping scheme for V. cholerae developed at the National Institute of Infectious Diseases, Tokyo, Japan (29).
Tissue culture assay.
The non-O1, non-O139 V. cholerae strains isolated during the study period were examined by tissue culture assay with CHO and HeLa cells. The strains were grown in Trypticase soy broth (TSB; Difco, Detroit, Mich.) supplemented with 0.6% yeast extract (TSB-YE) and in AKI medium (Bacto Peptone, 1.5%; yeast extract, 0.4%; NaCl, 0.5%; NaHCO3, 0.3% [pH 7.4] [13]) under shaking conditions for 18 h. The culture supernatant obtained by centrifugation at 4°C was made cell-free by passing it through a 0.22-μm-pore-size filter unit (Millex-GS; Millipore Corp., Bedford, Mass.) and collecting it in sterile test tubes which were kept at 4°C until they were used.
CHO and HeLa cells were grown as monolayers in Dulbecco’s minimum essential medium (Nissui Pharmaceutical Co. Ltd., Tokyo, Japan) supplemented with 10% (vol/vol) horse serum (Gibco Laboratories, Grand Island, N.Y.). Cell lines were maintained in 25-cm2 tissue culture flasks (NUNC, Roskilde, Denmark) at 37°C in a humidified 5% CO2 atmosphere. A confluent monolayer of CHO and HeLa cells grown for 3 to 4 days was removed from the tissue culture flasks, 200 μl of the cell suspension (ca. 6.4 × 103 cells) was added to each of the 96-well plates along with 50 μl of the cell-free culture filtrate, and the plates were incubated as described above. Morphological changes in CHO and HeLa cells were recorded at 24 h. The uninoculated culture medium was used as medium control.
Production of NMDCY.
A sensitive sandwich enzyme-linked immunosorbent assay (ELISA) for the specific detection of nonmembrane-damaging cytotoxin (NMDCY) recently purified from a strain (strain V249) of V. cholerae O26 (28) was developed (27). The use of a high-affinity monoclonal antibody belonging to the immunoglobulin G2b isotype to capture the NMDCY epitopes permitted a high-efficiency capture. By this ELISA, we sought the in vitro production of NMDCY from supernatants of the test strains grown in AKI medium (13). The culture supernatant of V. cholerae O26 (strain no. V249) served as the positive control, whereas uninoculated medium served as the negative control. Strains were considered positive for NMDCY production when the test wells yielded an absorbance of >0.1 after the absorbance of a medium control was subtracted.
Antimicrobial susceptibility.
The non-O1, non-O139 V. cholerae strains under analysis were examined for resistance to ampicillin (10 μg), chloramphenicol (30 μg), co-trimoxazole (25 μg), ciprofloxacin (5 μg), furazolidone (100 μg), gentamicin (10 μg), neomycin (30 μg), nalidixic acid (30 μg), norfloxacin (10 μg), streptomycin (10 μg), and tetracycline (30 μg) with commercial discs (Hi Media, Bombay, India). A 4-h culture of each of the test strains in TSB (Difco) was spread plated onto well-dried Mueller-Hinton agar (Difco). The plates were incubated for 24 h at 37°C. Characterization of the strains as sensitive, intermediate, or resistant was based on the size of the inhibition zones around each disc according to the manufacturer’s instruction. Strains showing intermediate zones of inhibition were interpreted as resistant to that drug.
PCR assay.
A PCR-based assay was used as described elsewhere (16, 18) to determine whether the ctxA, zot, ace, and tcpA genes were present. Additionally, all the strains were also examined for the presence of heat-labile toxin (LT), heat-stable toxin (ST), and verotoxin (VT) of Escherichia coli and its variants with primers common to the toxin and the variants. The primers used for this assay and the expected amplicon sizes are listed in Table 1. The following were added to each 100 μl of PCR mixture: 10 μl of Mg-free 10× amplification buffer (500 mM KCl, 100 mM Tris HCl [pH 9.0], 0.1% Triton X-100); 8 μl of 25 mM MgCl2; 2 μl each of 2 mM dATP, dTTP, dGTP, and dCTP; 50 pmol each of the primers; and 2.5 U of Taq DNA polymerase (Takara Shuzo, Otsu, Japan). PCR was carried out in 0.5-ml microcentrifuge tubes with 43.5 μl of the PCR mixture described above and 6.5 μl of a Luria broth (Difco) culture of the test strains heated at 94°C for 5 min. The solution was overlaid with a drop of sterile mineral oil (Sigma), and PCR was performed in an automated thermocycler (UNO-Thermoblock; Biometra, Göttingen, Germany) for 30 cycles, and the cycling conditions were as follows: denaturation at 94°C for 1.5 min, annealing at 60°C (but in the case of zot and ace the annealing temperature was 55°C and for LTc, VTc, and STc the annealing temperature was 50°C), and extension at 72°C for 1.5 min. A reagent blank (containing all the components of the reaction mixture and water instead of broth containing template DNA) and strains VC20 (V. cholerae O1 El Tor Ogawa), 569B (V. cholerae O1 classical Inaba), B2C (LT- and ST-positive strain of E. coli), and T17 (VT-positive strain of E. coli) were run as controls. Amplified products from the PCR were electrophoresed in 2.5% agarose gels and were stained with ethidium bromide. A 1-kb molecular size ladder (Gibco BRL, Gaithersburg, Md.) was run with each gel.
TABLE 1.
Sequences of primers used for detection of different genes by the PCR assay
| Gene | Primer sequence | Ampli- con size (bp) | Refer- ence |
|---|---|---|---|
| ctxA | CTC AGA CGG GAT TTG TTA GGC ACG | 301 | 16 |
| TCT ATC TCT GTA GCC CCT ATT ACG | |||
| zot | CAC TGT TGG TGA GCG TTA TCG | 243 | 6 |
| CAA GCG CTG TGG GTA GAA GTG AAA | |||
| ace | GCT TAT GAT GGA CAC CCT TTA | 284 | 6 |
| TTT GCC CTG CGA GCG TTA AAC | |||
| tcpA (El Tor) | GAA GAA GTT TGT AAA AGA AGA ACA C | 471 | 16 |
| GAA AGG ACC TTC TTT CAC GTT G | |||
| tcpA (classical) | CAC GAT AAG AAA ACC GGT CAA GAG | 617 | 16 |
| ACC AAA TGC AAC GCC GAA TGG AG | |||
| LTc | GCG ACA AAT TAT ACC GTG CT | 708 | 33 |
| CCG AAT TCT GTT ATA TAT GT | |||
| STc | ATT TTT C/ATT TCT GTA TTA/G TC | 105 | 30 |
| GC/TA CAG/A GCA GGA TTA CAA AA | |||
| VTc | GAA CGA AAT AAT TTA TAT GTG | 521 | 32 |
| TGA TGA TGG CAA TTG AGT AT |
Preparation of DNA probes, Southern blotting, and DNA hybridization.
The DNA probes for hlyU, hlyA, and toxR were a 0.769-kb EcoRI fragment, a 1.7-kb EcoRI fragment, and a 0.9-kb XbaI-SalI fragment, respectively, from plasmids pHU2, pGT89, and pToxR II, respectively. These probes were amplified by PCR with V. cholerae 569B O1 chromosomal DNA as the template and with various oligonucleotide primers, which are listed in Table 2. The heat-stable enterotoxin of V. cholerae non-O1 (NAG-ST) probe used was a 0.271-kb EcoRI-BamHI fragment from pAO111 (31). A 0.8-kb SmaI fragment and a 1.5-kb SstI-PvuII fragment of plasmid pKK 3535 containing the 16S and 23S rRNA genes of E. coli, respectively, were used as the probes (5). The hemolysin X (hlx) probe was a 29-base oligonucleotide (5′-GTT CTG CTC ACT GGC TGA GCG CAA GA-3′) located in the middle of the hlx-coding region (19). The attRS1 probe was an 18-base oligonucleotide (5′-CCT TAG TGC GTA TTA TGT-3′) corresponding to the 17-bp target sequence termed attRS1, in which the RS1 of the CTX genetic element integrates into the chromosome of V. cholerae strains (22). The 13 non-O1, non-O139 V. cholerae strains listed in Table 3 were analyzed for restriction fragment length polymorphisms (RFLPs) of hlyA, hlyU, hlx, attRS1, and toxR genes. Chromosomal DNA was prepared as reported earlier (21). Samples of 2 μg of the DNA preparations were digested with a variety of restriction endonucleases (Promega, Madison, Wis.) according to the manufacturer’s instruction. The restricted fragments were separated by electrophoresis through 0.7% (wt/vol) gels, and Southern hybridization was performed as described before (21). Probes were labeled with [32P]dATP either by nick translation or by 5′ end labeling with polynucleotide kinase (25).
TABLE 2.
Sequences of primers used for preparation of DNA probes for detection of indicated genes
| Gene | Primer sequence |
|---|---|
| hlyU | 5′-TTC TGA TCA CTA CCC AAT GAT AAG TG-3′ |
| 5′-TTC GCT AGC CGT CGT TCT ATG ATG-3′ | |
| hlyA | 5′-TTC ACA GAG TCA GTG AGG TTT ATA TCG C-3′ |
| 5′-TTC GCT GTA GAC ATT GGT CAA TTC ATC-3′ | |
| toxR | 5′-TTA CTA CTC ACA CAC TTT GAT GGC ATC GTT-3′ |
| 5′-TTA ATG TTC GGA TTA GGA CAC AAC TCA AAA G-3′ |
TABLE 3.
Characteristics of non-O1, non-O139 V. cholerae strains isolated during February and March 1996 in Calcuttaa
| Strain no. | Date of isolation (day.mo.yr) | Sero- group | Cleavage
pattern of the following genes:
|
Ribo- type | NMDCY A492 | Tissue culture
activity
|
Antibiogram | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hlyU | hlx | attRS1 | toxR | CHO
|
HeLa
|
||||||||
| TSB-YE | AKI medium | TSB-YE | AKI medium | ||||||||||
| AM107 | 29.2.1996 | O144 | HUII | HLII | A1 | TR1 | R1 | 1.43 | Ct | Ct | Ct | Ct | A Cf Co Fz N Na Nx S |
| AM108 | 29.2.1996 | O144 | HUII | HLII | A1 | TR1 | R1 | 2.15 | Ct | Ct | Ct | Ct | A C Co Fz N Na Nx S |
| AS60 | 29.2.1996 | O144 | HUI | HLII | A1 | TR1 | R1 | 1.59 | Ct | Ct | Ct | Ct | A Cf Co Fz N Na S |
| AS61 | 29.2.1996 | O144 | HUI | HLII | A1 | TR1 | R1 | 1.51 | Ct | Ct | Ct | Ct | A Cf Co Fz N Na Nx S |
| AM109 | 14.3.1996 | O19 | HUI | HLI | UP | TR3 | R3 | 1.82 | Ct | Ct | Ct | Ct | A Fz N S |
| AM111 | 18.3.1996 | O12 | HUI | HLI | UP | TR3 | R4 | ND | Ct | Ct | Ct | Ct | A Cf Fz N Na |
| AM112 | 19.3.1996 | O39 | HUI | HLI | UP | TR2 | R5 | 2.13 | Ct | Ct | Ct | Ct | A Fz N S |
| AM113 | 20.3.1996 | O6 | HUI | HLI | UP | TR2 | R6 | 2.46 | Ct | Ct | Ct | Ct | A Fz N S |
| AS64 | 21.3.1996 | O11 | HUI | HLI | A2 | TR3 | R2 | 1.21 | Ct | Ct | Ct | Ct | A Fz N S |
| AS65 | 21.3.1996 | O11 | HUI | HLI | A2 | TR3 | R2 | 0.89 | Ct | Ct | Ct | Cr | A Fz N S |
| AS66 | 21.3.1996 | O11 | HUII | HLI | — | TR3 | R2 | 1.15 | Ct | Ct | Ct | Cr | A Fz N S |
| AS67 | 25.3.1996 | OUT | HUI | HLI | A2 | TR3 | R2 | 1.20 | Cr | Cr | Ct | Cr | A Fz N S |
| AS68 | 29.3.1996 | OUT | HUI | HLI | A2 | TR3 | R2 | 1.04 | Ct | Ct | Ct | Cr | A Fz N S |
Abbreviations: —, partial digestion; UP, unique pattern; Ct, cytotoxic; Cr, cell rounding; A, ampicillin; C, chloramphenicol, Co, co-trimoxazole; Cf, ciprofloxacine; Fz, furazolidone; N, neomycin; Na, nalidixic acid; Nx, norfloxacin; S, streptomycin; ND, Not done.
PFGE.
For the pulsed-field gel electrophoresis (PFGE) study, 16 strains of V. cholerae were analyzed. These included nine of the non-O1, non-O139 V. cholerae strains associated with the upsurge in Calcutta (the remaining six strains could not be analyzed by PFGE due to endonuclease activity), 3 strains of non-O1, non-O139 V. cholerae belonging to serogroups O60 (strain SG2), O2 (strain SG12), and O37 (strain SG9) associated with sporadic infections and isolated in 1992 from hospitalized patients, one strain of Vibrio fluvialis (strain AS63) representing a species other than V. cholerae isolated from a patient with diarrhea in the same time frame as the non-O1, non-O139 upsurge, and three reference strains of toxigenic V. cholerae representing the O1 classical biotype (strain 569B), the O1 El Tor biotype (strain V5), and the O139 serogroup (strain SG26), respectively.
To perform PFGE, the genomic DNAs of the various strains were prepared in agarose plugs as described previously (17). Agarose blocks containing genomic DNA were equilibrated in restriction enzyme buffer for 1 h at room temperature and were cleaved in fresh buffer at the appropriate incubation temperature. For complete digestion of the DNAs, 50 U of NotI was used. PFGE of the inserts was performed by the contour-clamped homogeneous electric field method on a CHEF DR-II apparatus (Bio-Rad, Richmond, Calif.) in 0.4× TBE buffer (44.5 mM Tris-HCl, 44.5 mM boric acid, 1.0 mM EDTA [pH 8.0]). The gels were electrophoresed for 19.5 h with pulse times of 5 to 35 s, and electrophoresis was continued for 23.5 h with pulse times of 6.72 to 17.3 s. A DNA size standard (bacteriophage λ ladder; Bio-Rad) was used as the molecular mass standard, and a model 1000 mini-chiller (Bio-Rad) was used to maintain the temperature of the buffer at 14°C. The gels were stained in distilled water containing 1.0 μg of ethidium bromide per ml for 30 min, rinsed several times in tap water, and photographed under UV light.
Interpretation of PFGE patterns.
The picture obtained from PFGE was directly scanned on a model 420 optically enhanced densitometer-scanner interfaced with a computer in which the program Diversity One (version 1.6) was installed. Comparisons of differences in the patterns of NotI-digested DNA were made to ascertain the phylogenetic relationship between strains by using the software that runs on a Sun workstation (pdi Inc., Huntington Station, N.Y.). A phylogenetic tree was made with the average percentage of matched bands ranging from 97 to 485 kb; the tree was unrooted to summarize the relationships among the strains compared.
RESULTS
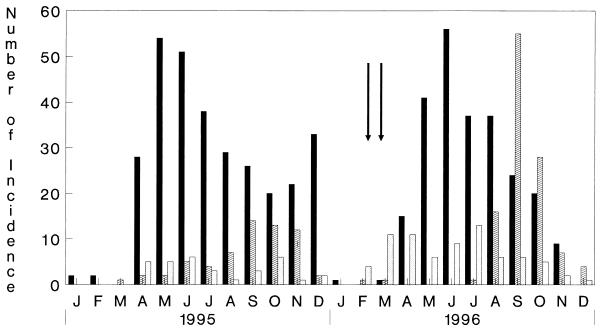
In 1996, an unusual event occurred in the beginning of the cholera season in Calcutta. From the end of February 1996, there was an inexplicable upsurge in the incidence of non-O1, non-O139 V. cholerae infections among hospitalized patients admitted to the ID Hospital (Fig. 1). The incidence of the non-O1, non-O139 serogroup exceeded the incidence of the O1 or the O139 serogroup of V. cholerae during this period for the first time since we initiated the focused cholera surveillance program in Calcutta 141 months earlier. A total of 18 strains of V. cholerae were isolated from 18 of the 232 patients examined during February and March 1996. For all 18 patients, V. cholerae was the sole pathogen isolated. Of these 18 isolates, 15 belonged to non-O1, non-O139 serogroups, 2 belonged to the O139 serogroup, and 1 belonged to the O1 serogroup. Serotyping of the 15 non-O1, non-O139 strains revealed that 4 belonged to O144, 3 belonged to O11, 1 each belonged to O6, O8, O12, O19, O39, and O58, and 2 strains could not be typed. The antibiotic susceptibilities of the 13 non-O1, non-O139 V. cholerae strains isolated in this study are presented in Table 3. All strains were resistant to multiple antibiotics with the V. cholerae strains belonging to serogroup O144 being resistant to more than seven of the antibiotics used in this study.
FIG. 1.
Monthly isolation of various serogroups of V. cholerae from patients hospitalized because of secretory diarrhea at the ID Hospital, Calcutta, from January 1995 to December 1996. The arrows denote the months when non-O1, non-O139 V. cholerae dominated over V. cholerae O1 and O139. ▪, O1; ▨, O139; ░⃞, non-O1, non-O139.
All the patients excreting the non-O1, non-O139 V. cholerae strains developed severe dehydration with hypovolemic shock after a short period of diarrhea (mean preadmission duration of less than 12 h) and vomiting. The patients required large amounts of intravenous fluids for correction of the initial dehydration and hypovolemic shock. The postadmission duration of purging was about 35 h. All the patients were successfully rehydrated with intravenous and oral fluids, and there was no mortality in this series.
The 13 non-O1, non-O139 V. cholerae strains associated with cholera-like disease were further characterized in extensive detail to obtain an understanding of the virulence traits which might have contributed to the disease. As indicated in Table 3, cell-free culture supernatants of all of the strains grown in TSB-YE and AKI medium evoked a distinct cytotoxic effect on CHO and HeLa cells, while culture supernatants of the strains examined also produced NMDCY when they were cultured in AKI medium. In the PCR assay, all the strains yielded negative results for the ctxA, zot, ace, and tcpA genes. Furthermore, all non-O1, non-O139 strains were negative by PCR for genes representing LT, ST, and VT of E. coli and its various variants and also did not hybridize with a DNA probe specific for NAG-ST. Further analysis of 13 of the non-O1, non-O139 V. cholerae strains showed that all strains hybridized with DNA probes specific for the El Tor hemolysin (hlyA), for the regulatory gene hlyU which upregulates expression of the El Tor hemolysin (34), and with the oligonucleotide probe specific for the recently described hlx gene. In addition, all the strains hybridized with the DNA probe specific for toxR and with an oligonucleotide probe for the specific 17-bp target sequence termed attRS1.
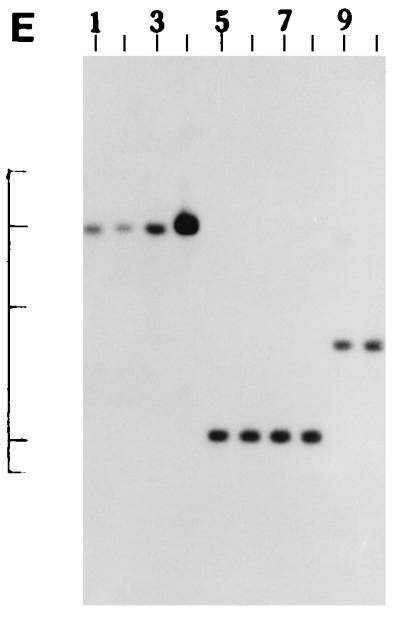
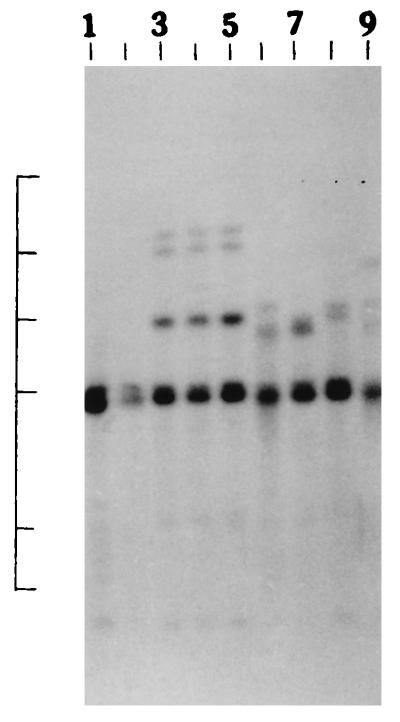
We further examined the genetic relatedness between the various strains of non-O1, non-O139 V. cholerae isolated between February and March 1996 by investigating the RFLPs of the hlyA, hlyU, hlx, toxR, attRS1, and rRNA genes. A single PstI fragment of 6 kb hybridized with the hlyA gene probe for all strains examined, as indicated in Fig. 2A. RFLP analysis of the hlyU gene with XbaI-BglII showed two patterns designated HUI and HUII (Fig. 2B; Table 3) among the strains examined. Pattern HUI consisted of only one band of 3.3 kb, while pattern HUII consisted of four bands of 6.3, 5.0, 4.3, and 3.3 kb. RFLP analysis of the hlx gene with Sau3AI displayed two profiles designated HLI and HLII (Fig. 2C; Table 3), with profile HLI consisting of a band of 0.568 kb and HLII consisting of a band of 0.7 kb. RFLP analysis of the attRS1 target sequence with HindIII displayed two main profiles designated A1 and A2. Strains AM109, AM111, AM112, and AM113 displayed unique RFLP patterns, while strain AS66 showed partial digestion, and therefore, multiple bands were seen (Fig. 2D; Table 3). Finally, RFLP analysis of the toxR gene with PstI showed three patterns (TR1, TR2, and TR3) among the strains examined (Fig. 2E). Ribotyping of the 13 non-O1, non-O139 V. cholerae strains examined with BglI produced six patterns which were designated R1 to R6 (Fig. 3). All strains belonging to serogroup O144 were found to have pattern R1, while the strains belonging to O11 and OUT (O untypeable) had pattern R2. The remaining four strains showed polymorphisms in their ribotype patterns. None of the six ribotypes matched any of the patterns documented for the standardized ribotypes documented for V. cholerae by Popovic et al. (23).
FIG. 2.
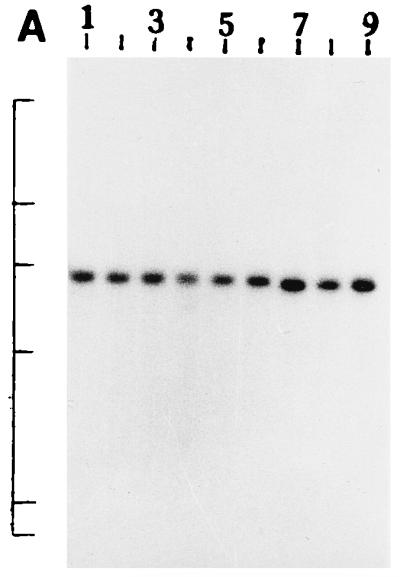
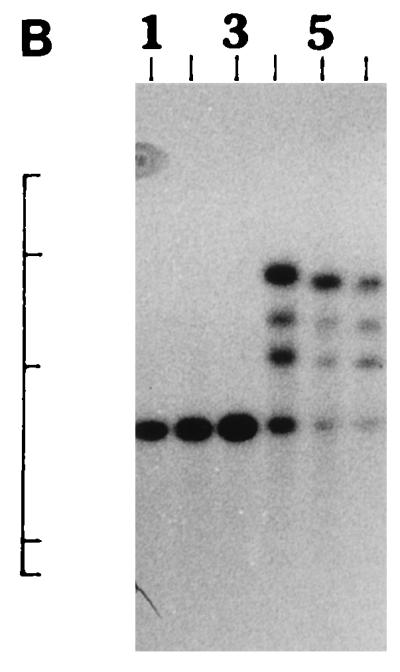
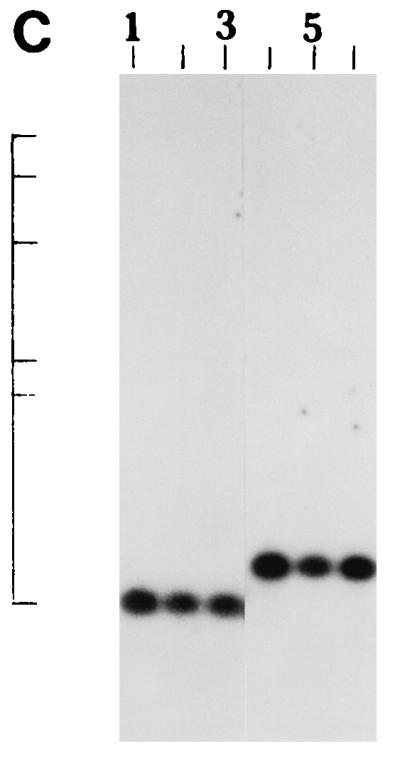
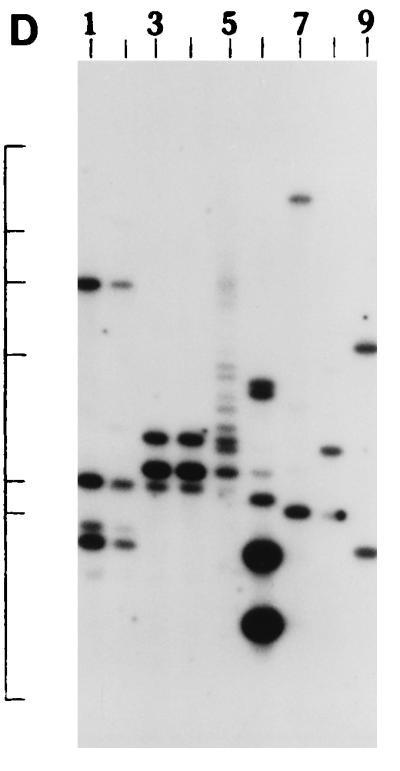
(A) Southern blot hybridization of PstI-digested non-O1, non-O139 V. cholerae chromosomal DNA with the hlyA probe. Lanes 1 to 9, strains AM107, AS61, AM109, AM111, AM112, AM113, AS64, AS67, and AS68, respectively. The positions of bacteriophage λ HindIII molecular size markers run on the same gel are indicated by bars from top to bottom (23.13, 9.41, 6.55, 4.36, 2.32, and 2.0 kb). The patterns for only representative strains are shown here. (B) Southern blot hybridization of XbaI-BglII-digested non-O1, non-O139 V. cholerae chromosomal DNA with the hlyU probe. Lanes 1 to 3, strains AS61, AM112, and AS64 (strains showing the HUI pattern), respectively; lanes 4 to 6, strains AM107, AM108, and AS66 (strains showing the HUII pattern), respectively. The positions of bacteriophage λ HindIII molecular size markers run on the same gel are indicated by bars from top to bottom (9.41, 6.55, 4.36, 2.32, and 2.0 kb). The patterns for only representative strains are shown here. (C) Southern blot hybridization of Sau3AI-digested non-O1, non-O139 V. cholerae chromosomal DNA with the hlx probe. Lanes 1 to 3, strains AM109, AS64, and AS68 (strains showing the HLI pattern), respectively; lanes 4 to 6, strains AM107, AS60, and AS61 (strains showing the HLII pattern), respectively. The positions of bacteriophage λ HindIII molecular size markers run on the same gel are indicated by bars from top to bottom (9.4, 6.55, 4.36, 2.32, 2.0 and 0.56 kb). The patterns for only a few representative strains are shown. (D) Southern blot hybridization of HindIII-digested non-O1, non-O139 V. cholerae chromosomal DNA with the attRS1 probe. Lanes 1 and 2, strains AM107 and AS61 (strains the showing A1 pattern), respectively; lanes 3 and 4, strains AS64 and AS68 (strains showing the A2 pattern), respectively; lane 5, strain AS66 (showing a partial digest); lanes 6 to 9, strains showing unique patterns (strains AM109, AM111, AM112, and AM113, respectively). The positions of bacteriophage λ HindIII molecular size markers run on the same gel are indicated by bars from top to bottom (23.13, 9.41, 6.55, 4.36, 2.32, 2.0, and 0.5 kb). The patterns for representative strains are shown. (E) Southern blot hybridization of PstI-digested non-O1, non-O139 V. cholerae chromosomal DNA with the toxR probe. Lanes 1 to 4, strains AM107, AM108, AS60, and AS61 (strains showing the TR1 pattern), respectively; lanes 5 to 8, strains AM109, AM113, AS64, and AS68 (strains showing the TR3 pattern), respectively; lanes 9 and 10, strains AM111 and AM112 (strains showing the TR2 pattern), respectively. The positions of bacteriophage λ HindIII molecular size markers run on the same gel are indicated by bars from top to bottom (9.41, 6.55, 4.36, 2.32, and 2.0 kb). The patterns for only representative strains are shown.
FIG. 3.
Ribotypes of non-O1, non-O139 V. cholerae strains. Lanes 1 and 2, strains AM107 and AS61 (strains showing the R1 pattern), respectively; lanes 3 to 5, strains AS66, AS67, and AS68 (strains showing the R2 pattern), respectively; lanes 6, strain AM109 (a strain showing the R3 pattern); lane 7, strain AM111 (a strain showing the R4 pattern); lane 8, strain AM112 (a strain showing the R5 pattern); lane 9, strain AM113 (a strain showing the R6 pattern). The positions of bacteriophage λ HindIII molecular size markers are indicated by bars from top to bottom (23.13, 9.41, 6.55, 4.36, 2.32, and 2.0 kb). The patterns for only representative strains are shown.
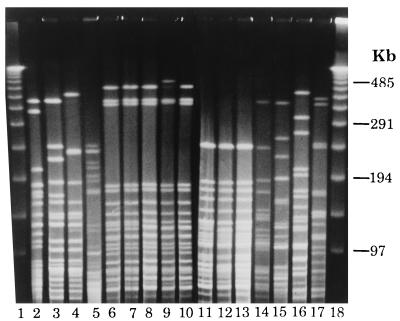
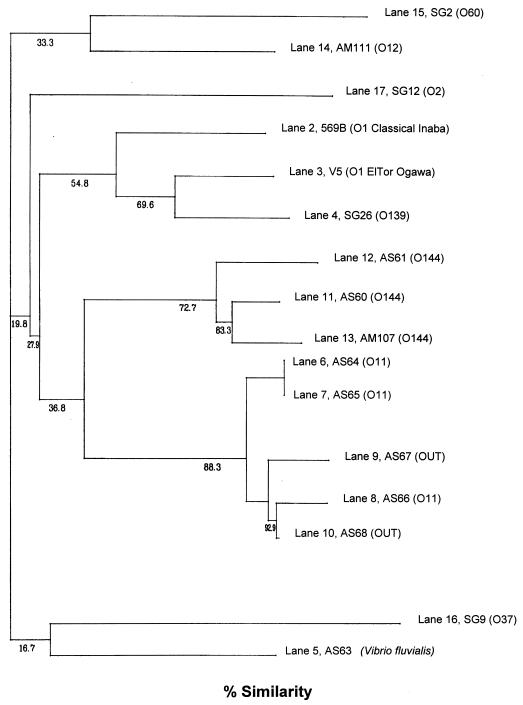
The PFGE profiles of the V. cholerae strains obtained with NotI showed a variety of patterns which correlated well with the profiles obtained by ribotyping and serogrouping. For example, all O144 strains had identical patterns (Fig. 4, lanes 11, 12, and 13). Likewise, strains belonging to O11 and OUT also displayed nearly similar profiles (Fig. 4, lanes 6 to 10). The PFGE profiles of strains belonging to serogroups O144 and O11 and OUT differed from each other and from the patterns displayed by the reference strains belonging to toxigenic V. cholerae serogroups O1, O139 and those belonging to other non-O1, non-O139 serogroups. The genomic restriction patterns of individual strains were compared quantitatively, and the percent similarity in restriction patterns between the strains was estimated and is represented as a dendrogram (Fig. 5). It can be seen from the dendrogram that strains belonging to serogroups O144, O11, and OUT exhibited similar restriction patterns to form two different clusters and showed a 36.8% similarity to each other. The other interesting finding was that the O144 cluster and the O11 and OUT cluster were more closely related to the toxigenic V. cholerae O1, O139 cluster than to the clusters containing non-O1, non-O139 V. cholerae strains associated with sporadic infections in 1992.
FIG. 4.
PFGE profiles of non-O1, non-O139 V. cholerae strains obtained with the NotI enzyme. Lanes 1 and 18, bacteriophage λ ladder; lane 2, 569B O1 classical Inaba; lane 3, V5 O1 El Tor Ogawa; lane 4, SG26 (O139); lane 5, V. fluvialis AS63; lane 6, AS64 (O11); lane 7, AS65 (O11); lane 8, AS66 (O11); lane 9, AS67 (OUT); lane 10, AS68 (OUT); lane 11, AS60 (O144); lane 12, AS61 (O144); lane 13, AM107 (O144); lane 14, AM111 (O12); lane 15, SG2 (O60); lane 16, SG9 (O37); lane 17, SG12 (O2).
FIG. 5.
Dendrogram generated by the average percentage of matched bands summarizing the degree of similarity of the NotI restriction patterns of genomic DNAs of non-O1, non-O139 V. cholerae strains.
DISCUSSION
The mechanism of pathogenesis and the factors involved in the virulence of non-O1, non-O139 V. cholerae remain enigmas. It is now clear beyond a reasonable doubt that some strains of non-O1, non-O139 V. cholerae have the capacity to precipitate a cholera-like syndrome and the capacity to flare into a localized outbreak. There has been an escalation in interest in the non-O1, non-O139 serogroups following the discovery of the O139 serogroup (1, 24) and following the finding that environmental nontoxigenic, non-O1 strains play an important role in the evolution of toxigenic V. cholerae (15). At least three localized outbreaks of diarrhea caused by non-O1, non-O139 serogroups have been described in the recent literature. These include an outbreak caused by V. cholerae O10 and O12 in February 1994 in Lima, Peru (7), another caused by O10 in East Delhi, India (26), and an epidemic caused by non-O1 V. cholerae that produced ST among Khmers in a camp in Thailand (3).
The strains of non-O1, non-O139 V. cholerae characterized in this study belonged to different O serogroups and lacked all the known virulence traits associated with toxigenic V. cholerae O1, O139 and those associated with toxigenic E. coli. In particular, all the non-O1, non-O139 strains lacked the genes comprising the core part of the CTX genetic element and also the tcpA gene, both of which are recognized as important components contributing to the pathogenicity of toxigenic V. cholerae O1, O139. However, all the non-O1, non-O139 strains examined in this study possessed the gene encoding the regulatory protein ToxR, which controls the coordinate expression of genes associated with pathogenicity in toxigenic V. cholerae O1/O139. In addition, all the strains also possessed the 17-bp target sequence termed attRS1.
In this context, the most significant finding from the PFGE study was that strains of V. cholerae belonging to serogroups O144, O11, and OUT were more closely related to the toxigenic V. cholerae O1/O139 strains than to strains of non-O1, non-O139 V. cholerae associated with sporadic infections. The fact that the non-O1, non-O139 strains isolated in this study possess the basic regulatory and insertional elements necessary to receive the CTX genetic element coupled with the proximity of some of these serogroups to the O1, O139 cluster in the dendrogram suggests that the non-O1, non-O139 serogroups could be a proto-cholera agent. If these non-O1, non-O139 serogroups were to acquire the TCP gene, for example, by horizontal transfer, they could acquire the CTX element through exposure to the toxinoferous phage CTXφ (34). However, given the rare occurrence of strains of the non-O1, non-O139 serogroups from clinical sources possessing the tcpA and the CTX genetic elements (18), it would appear that when and if such an event occurs there would possibly be attendant changes in the somatic antigen. Molecular biology-based studies have provided evidence of the horizontal transfer of the O antigen, since isolates with nearly identical asd gene sequences had different O antigens and isolates with the O1 antigen did not cluster together but were found in different lineages (15).
In the absence of CT and TCP proteins, what is clear from this study is that some non-O1, non-O139 strains precipitate diarrhea by a mechanism which is entirely different from that used by toxigenic V. cholerae O1, O139. Studies with the O10 and O12 strains of V. cholerae isolated from the outbreak in Peru showed that none of the strains produced enterotoxin but the majority of the strains produced a cytotoxin, as assessed in Y1 and HeLa cells (7). Interestingly, all the strains of V. cholerae isolated in the study also produced a cytotoxin, while all strains examined produced NMDCY. We have previously reported that the purified NMDCY has impressive enterotoxic activity (28) and is widely distributed among strains of V. cholerae, irrespective of serogroup (27). Studies on the clonality of non-O1, non-O139 V. cholerae strains by RFLP analysis of rRNA genes and of the other genes examined and by PFGE collectively indicate that the upsurge which occurred in February and March 1996 was caused by strains belonging to different clones. Overall there was an excellent correlation between the results of ribotyping, RFLP analysis of various genes, and PFGE, with strains belonging to a particular serogroup (e.g., O144, O11, and OUT) showing nearly identical restriction patterns and PFGE profiles. This would indicate that a discrete set of V. cholerae serogroups had, for reasons that are unclear, a competitive advantage at that point in time.
Although controversial, the El Tor hemolysin has been implicated as a virulence factor for toxigenic V. cholerae. All the strains of V. cholerae examined in this study had the genetic potential to produce the El Tor hemolysin and the novel hemolysin recently described by Nagamune et al. (19). The El Tor hemolysin is reportedly a potent toxin with both enterotoxic and cytotoxic activities (2, 11, 12). The V. cholerae O10 and O12 strains associated with the outbreak in Peru were all positive for the El Tor hemolysin, cytotoxin, invasiveness, and mannose-sensitive hemagglutinin, leading Dalsgaard et al. (7) to conclude that a combination of these factors and perhaps an unknown factor may have caused the diarrhea.
The pathogenic mechanism of non-O1, non-O139 V. cholerae in a way resembles that of enteropathogenic E. coli which is known to be a complex and multifaceted process that manifests as severe secretory diarrhea and which is caused by antigenically similar populations (35). Among non-O1, non-O139 V. cholerae strains, some serogroups (e.g., O10, O11, O12, and O144) seem to be more often associated with disease, despite the absence of the virulence package (ctxA, ctxB, zot, ace, and other genes), indicating that these serogroups have a mode of pathogenesis different from that of toxigenic V. cholerae. The inability to identify such serogroups (clones) earlier was related to the fact that serotyping of non-O1, non-O139 V. cholerae was not systematically pursued, and therefore, such pathogenic clones remained unidentified. There have been earlier instances in which clones of non-O1 V. cholerae have caused fairly large outbreaks, like the one caused by non-O1 V. cholerae in the Kumbh Fair at Allahabad in India in early 1954 (10). Likewise, the predilection of a serogroup was also observed among people with infections caused by non-O1 V. cholerae in Cancun, Mexico, with Smith serotype 12 accounting for 46% of the infections (8). Therefore, as for enteropathogenic E. coli, we would like to propose the nomenclature of enteropathogenic V. cholerae (EPVC) to include serogroups of non-O1, non-O139 V. cholerae that do not possess the CTX genetic element but that can cause diarrhea in humans by a hitherto unknown mechanism. It must be emphasized that at present the proposal of EPVC is based on data for a relatively small number of patients. Apart from serotyping, there are no other good markers to identify such clones of non-O1, non-O139 V. cholerae associated with diarrhea compared to their innocuous environmental counterparts. On the basis of the comparison of the asd gene sequences of a diverse collection of V. cholerae, Karaolis et al. (15) opined that it is also probable that some O antigens of V. cholerae are compatible with pathogenesis. As a beginning, isolates of serogroups such as O10, O11, O12, and O144 should be included under the category of EPVC. Future epidemiological studies backed with systematic serogrouping is likely to contribute to the rapid expansion of members constituting EPVC.
ACKNOWLEDGMENT
We thank Pradip Kumar Saha for help in performing the ELISA for determination of NMDCY production.
REFERENCES
- 1.Albert M J, Siddique A K, Islam M S, Faruque A S G, Ansaruzzaman M, Faruque S M, Sack R B. Large outbreak of clinical cholera due to V. choleraenon-O1 in Bangladesh. Lancet. 1993;341:704. doi: 10.1016/0140-6736(93)90481-u. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Mayrhofer G, Kotlarski I, Manning P A. Amino-terminal domain of the ElTor haemolysin of Vibrio choleraeO1 is expressed in classical strains and is cytotoxic. Vaccine. 1991;9:588–594. doi: 10.1016/0264-410x(91)90247-4. [DOI] [PubMed] [Google Scholar]
- 3.Bagchi K, Echeverria P, Arthur J D, Sethabutr O, Serichantalergs O, Hoge C W. Epidemic diarrhoea caused by Vibrio choleraenon-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J Clin Microbiol. 1993;31:1315–1317. doi: 10.1128/jcm.31.5.1315-1317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio choleraeO139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Ullrich A, Raker M A, et al. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 6.Colombo M M, Mastrandrea S, Santona A, de Amdrade A P, Uzzau S, Rabino S, Cappuccinelli P. Distribution of the ace, zot, and ctxA toxin genes in the clinical and environmental Vibrio cholerae. J Infect Dis. 1994;170:750–751. doi: 10.1093/infdis/170.3.750. [DOI] [PubMed] [Google Scholar]
- 7.Dalsgaard A, Albert M J, Taylor D N, Shimada T, Meza R, Serichantalergs O, Echeverria P. Characterization of Vibrio choleraenon-O1 serogroup obtained from an outbreak of diarrhoea in Lima, Peru. J Clin Microbiol. 1995;33:2715–2722. doi: 10.1128/jcm.33.10.2715-2722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finch M J, Valdespino J L, Wells J G, Perez-Perez G, Arjona F, Sepulveda A, Bessudo D, Blake P A. Non-O1 Vibrio choleraeinfections in Cancun, Mexico. Am J Trop Med Hyg. 1987;36:393–397. doi: 10.4269/ajtmh.1987.36.393. [DOI] [PubMed] [Google Scholar]
- 9.Garg S, Ramamurthy T, Mukhopadhyay A K, Deb B C, Nair G B, Shimada T, Takeda T, Huq A, Colwell R R, Takeda Y. Production and cross reactivity patterns of a panel of high affinity monoclonal antibodies to Vibrio choleraeO139 Bengal. FEMS Immunol Med Microbiol. 1994;8:293–298. doi: 10.1111/j.1574-695X.1994.tb00455.x. [DOI] [PubMed] [Google Scholar]
- 10.Gupta N P, Gupta S P, Mangalik V S, Prasad B G, Yajnik B S. Investigations into the nature of the Vibriostrains isolated from the epidemic of gastroenteritis in Kumbh fair at Allahabad in 1954. Indian J Med Sci. 1956;X:781–791. [Google Scholar]
- 11.Honda T, Finkelstein R A. Purification and characterization of a hemolysin produced by Vibrio choleraebiotype El Tor: another toxic substance produced by cholera vibrios. Infect Immun. 1979;26:1020–1027. doi: 10.1128/iai.26.3.1020-1027.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichinose Y, Yamamoto K, Nakasone N, Tanabe M J, Takeda T, Miwatani T, Iwanaga M. Enterotoxicity of El Tor-like hemolysin of non-O1 Vibrio cholerae. Infect Immun. 1987;55:1090–1093. doi: 10.1128/iai.55.5.1090-1093.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwanaga M, Kuyyakanond T. New medium for the production of cholera toxin by Vibrio choleraeO1 biotype El Tor. J Clin Microbiol. 1985;22:405–408. doi: 10.1128/jcm.22.3.405-408.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaper J B. Isolation, ecology and taxonomy of human pathogens in an estuary. Ph.D. thesis. College Park: University of Maryland; 1979. [Google Scholar]
- 15.Karaolis D K R, Lan R, Reeves P R. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keasler S P, Hall R H. Detecting and biotyping V. choleraeO1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 17.Kurazono H, Okuda J, Nair G B, Albert M J, Sack R B, Chong-nguan M, Chaicumpa W, Takeda Y. Vibrio choleraeO139 Bengal isolated from India, Bangladesh and Thailand are clonal as determined by pulsed-field gel electrophoresis. J Infect. 1994;29:109–110. doi: 10.1016/s0163-4453(94)95357-0. [DOI] [PubMed] [Google Scholar]
- 18.Mukhopadhyay A K, Garg S, Mitra R, Basu A, Dutta D, Bhattacharya S K, Shimada T, Takeda T, Takeda Y, Nair G B. Temporal shifts in traits of Vibrio choleraestrains isolated from hospitalized patients in Calcutta: a 3-year (1993 to 1995) analysis. J Clin Microbiol. 1996;34:2537–2543. doi: 10.1128/jcm.34.10.2537-2543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagamune K, Yamamoto K, Honda T. Cloning and sequencing of a novel hemolysis gene of Vibrio cholerae. FEMS Microbiol Lett. 1995;128:265–269. doi: 10.1111/j.1574-6968.1995.tb07534.x. [DOI] [PubMed] [Google Scholar]
- 20.Nair G B, Misra S, Bhadra R K, Pal S C. Evaluation of the multitest medium for rapid presumptive identification of Vibrio choleraefrom environmental sources. Appl Environ Microbiol. 1987;53:1203–1205. doi: 10.1128/aem.53.5.1203-1205.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pajni S, Sharma C, Bhasin N, Ghosh A, Ramamurthy T, Nair G B, Ramjayam S, Das B, Kar S, Roychowdhury S, Ghosh R K. Studies on the genesis of Vibrio choleraeO139: identification of probable progenitor strains. J Med Microbiol. 1994;42:20–25. doi: 10.1099/00222615-42-1-20. [DOI] [PubMed] [Google Scholar]
- 22.Pearson G D N, Woods A, Chiang S, Mekalanos J J. CTX genetic element encodes a site specific recombination system and an intestinal colonization. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popovic T, Bopp C, Olsvik O, Wachsmuth K. Epidemiologic application of a standardized ribotype scheme for Vibrio choleraeO1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazono H, Pal A, Takeda Y. Emergence of novel strain of Vibrio choleraewith epidemic potential in Southern and Eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 25.Rigby P W J, Dieckmann M, Rhodes C, Berg P. Labelling deoxyribonucleic acid to high specific activity in vitroby nick translation with DNA polymerase I. J Mol Biol. 1977;113:237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- 26.Rudra S, Mahajan R, Mathur M, Kathuria K, Talwar V. Cluster of cases of clinical cholera due to Vibrio choleraeO10 in east Delhi. Indian J Med Res. 1996;103:71–73. [PubMed] [Google Scholar]
- 27.Saha P K, Nair G B. Production of monoclonal antibodies to the non-membrane-damaging cytotoxin (NMDCY) purified from Vibrio cholerae O26 and distribution of NMDCY among strains of Vibrio choleraeand other enteric bacteria determined by monoclonal-polyclonal sandwich enzyme-linked immunosorbent assay. Infect Immun. 1997;65:801–805. doi: 10.1128/iai.65.2.801-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha P K, Koley H, Nair G B. Purification and characterization of an extracellular secretogenic non-membrane-damaging cytotoxin produced by clinical strains of Vibrio choleraenon-O1. Infect Immun. 1996;64:3101–3108. doi: 10.1128/iai.64.8.3101-3108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimada T, Arakawa E, Itoh K, Okitsu T, Matsushima A, Asai Y, Yamai S, Nakazato T, Nair G B, Albert M J, Takeda Y. Extended serotyping scheme for Vibrio cholerae. Curr Microbiol. 1994;28:175–178. [Google Scholar]
- 30.Takeda, T. Personal communication.
- 31.Takeda T, Peina Y, Ogawa A, Dohi S, Abe H, Nair G B, Pal S C. Detection of heat-stable enterotoxin in a cholera toxin gene-positive strain of Vibrio choleraeO1. FEMS Microbiol Lett. 1991;80:23–28. doi: 10.1016/0378-1097(91)90203-m. [DOI] [PubMed] [Google Scholar]
- 32.Takeda Y, Kurazono H, Yamasaki S. Verotoxins (Shiga-like toxins) produced by enterohaemorrhagic Escherichia coli (Verocytotoxin producing E. coli) Microbiol Immunol. 1993;37:591–599. doi: 10.1111/j.1348-0421.1993.tb01681.x. [DOI] [PubMed] [Google Scholar]
- 33.Tornieporth N G, John J, Salgado K, Jesus P D, Latham E, Melo M C N, Gunzburz S T, Riley L W. Differentiation of pathogenic Escherichia colistrains in Brazilian children by PCR. J Clin Microbiol. 1995;33:1371–1374. doi: 10.1128/jcm.33.5.1371-1374.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 35.Williams P H, Baldwin T J, Knutton S. Enteropathogenic Escherichia coli. In: Sussman M, editor. Escherichia coli: mechanisms of virulence. Cambridge, United Kingdom: Cambridge University Press; 1997. pp. 403–420. [Google Scholar]
- 36.World Health Organization. Manual for laboratory investigation of acute enteric infection. CDD/83.3. Geneva, Switzerland: Program for Control of Diarrhoeal Diseases, World Health Organization; 1983. [Google Scholar]