Abstract
The present study describes the identification of inhibitors of a Mycobacterium tuberculosis-specific gap ligase chain reaction (LCR) DNA amplification assay as well as a method for their removal. A major contributor to inhibition was deduced to be a calcium phosphate precipitate, CaHPO4. The precipitate forms during N-acetyl-l-cysteine–sodium hydroxide (NALC-NaOH) decontamination, digestion, and concentration of respiratory specimens. The solubility product of CaHPO4 precipitate at pH 7.8, the pH at which gap LCR is optimized, indicates that the precipitate releases an amount of phosphate ions sufficient to inhibit amplification. A method for removal of the precipitate was identified. The precipitate is dissociated by exposing it to a mildly acidic (pH 4.1) buffer during the first of two centrifugation steps; the inhibitory phosphate ions are removed by the centrifugation steps. When 100 NALC-NaOH respiratory sediments were tested by gap LCR, none of the sediments were inhibitory when the acidic buffer was used while 24 samples were inhibitory when TE buffer, pH 7.8, was used. In another study, when the acidic buffer wash was applied to 1,440 NALC-NaOH respiratory sediments, only 10 sediments were found to be inhibitory. None of the inhibited sediments were culture positive for M. tuberculosis. This work demonstrates that when inhibition mechanisms are identified, relatively simple protocols can be used to obtain low inhibition rates and to allow the use of larger volume equivalents in amplification reactions.
Many groups have developed Mycobacterium tuberculosis-specific nucleic acid amplification assays for rapid detection of active tuberculosis (16, 22). The utility of these assays, however, has been limited by specimen-associated inhibitors. Inhibitors reduce the activity of the enzyme(s) responsible for nucleic acid amplification, which can result in false-negative assay results (6, 16, 22). Inhibition rates varying from 3 to 52% have been reported when M. tuberculosis-specific PCR assays have been used to test respiratory specimens decontaminated, digested, and concentrated by N-acetyl-l-cysteine–sodium hydroxide (NALC-NaOH) (1, 12, 20, 21, 24), NaOH (2, 14, 15, 25), sputolysin-NaOH (26), and sodium lauryl sulfate-NaOH (9) protocols. In these studies, lower inhibition rates (5% or less) were obtained when relatively small volume equivalents (≤25 μl) of NALC respiratory sediment were tested per amplification reaction or when the sediments underwent relatively complex specimen processing protocols prior to amplification.
Ligase chain reaction (LCR) is a DNA amplification method which uses thermostable DNA ligase for the detection of infectious agents, as well as point mutations associated with cancer, and genetic disease (5, 8). A more sensitive version of LCR, called gap LCR, requires thermostable DNA polymerase in addition to thermostable DNA ligase (4, 8). Gap LCR assays have been developed on the automated LCx system for the specific detection of Chlamydia trachomatis, Neisseria gonorrhoeae, and Mycobacterium tuberculosis (7, 10, 11, 13, 17).
The purpose of this study was to identify the major inhibitor(s) of gap LCR in NALC-NaOH respiratory sediments and to develop simple methods to alleviate inhibition, so that a relatively large volume equivalent (100 μl) of sediment could be tested per assay. The present study demonstrates that phosphate ions are potent inhibitors of gap LCR amplification. It also suggests that the NALC-NaOH decontamination, digestion, and concentration process, in which phosphate buffer is added, produces a calcium phosphate precipitate which acts as a source of inhibitory phosphate ions. Finally, methods for alleviation of the inhibitory effects of these substances are presented.
MATERIALS AND METHODS
Specimen collection, transport, and storage.
Respiratory specimens were decontaminated, digested, and concentrated by the NALC-NaOH protocol (18). Following microscopy for detection of acid-fast bacteria and the initiation of liquid and solid cultures, the remainder of each NALC-NaOH respiratory sediment was transported to Abbott Laboratories and stored at 2 to 8 or −20°C.
Preparation of calcium phosphate precipitate.
Ten milliliters of water, 5 or 10 mM calcium chloride solutions were incubated individually with 10 ml of 3% NaOH for 15 min at room temperature, after which 30 ml of 67 mM phosphate buffer (pH 6.8) was added. After the addition of the phosphate buffer, the tubes were centrifuged at 2,000 × g for 30 min. The supernatants were removed, and each pellet was resuspended in 5 ml of 67 mM phosphate buffer, pH 6.8.
LCR.
Specimens (either phosphate buffer [pH 6.8], calcium phosphate precipitate, or NALC-NaOH respiratory sediments) were vortexed for 2 to 5 s, after which 500-μl aliquots were pipetted into 1.7-ml screw-cap tubes which contained 50 mg of 150- to 212-μm acid-washed glass beads and 900 μl of buffer. The buffer used was either 10 mM Tris-HCl (pH 7.8)–1 mM EDTA (TE) buffer, 0.5 M potassium acetate buffer (pH 4.1), or 1.0 M potassium acetate buffer (pH 4.1). After the specimens were added, the tubes were recapped and vortexed for 2 to 5 s before being centrifuged at 1,500 × g for 10 min in an aerosol-contained clinical centrifuge. The supernatant was removed from every tube, and each pellet was resuspended in 1.0 ml of resuspension buffer [30 mM N-(2-hydroxyethyl) piperazine-N-(3-propanesulfonic acid) (EPPS) (pH 7.8), 75 mM magnesium chloride, 0.001% acetylated bovine serum albumin, 0.0003% amaranth dye, and 1% sodium azide]. The tubes were recapped, vortexed for 2 to 5 s, and centrifuged (1,500 × g for 10 min). The supernatants were removed, and each pellet was resuspended in 0.5 ml of resuspension buffer. The recapped tubes were vortexed for 2 to 5 s and incubated at 95°C for 20 min in an LCx covered dry bath. After being heated, the specimens were allowed to cool to room temperature and then were sonicated for a defined time (approximately 10 min) in an LCx Lysor. Sonication was performed to increase the efficiency with which mycobacteria were lysed. After sonication, the specimen tubes were microfuged for 2 min before gap LCR amplification was initiated.
One hundred microliters of processed specimens was added to an amplification vial which contained 100 μl of prealiquoted amplification mixture. After the specimens were added, each gap LCR assay mixture contained 18,000 U of recombinant Thermus thermophilus thermostable DNA ligase, 2 U of native Thermus flavus thermostable DNA polymerase, 5 μM NAD, 37.5 mM magnesium chloride, 1 mM spermidine, 0.001% acetylated bovine serum albumin, 1.0 μM dATP, 1.0 μM dCTP, 10 mM potassium as either potassium chloride or potassium hydroxide, 40 mM EPPS buffer (pH 7.8), 0.00015% amaranth dye, 0.6% sodium azide, and 1012 of each of the four probes which constitute the LCR probe set. The probe set was designed to detect nucleotides 347 to 390 of the single-copy chromosomal gene which encodes protein antigen b of M. tuberculosis (3). The PAB gene appears to be specific to the four subspecies of the M. tuberculosis complex (11, 14, 23, 26). Gap LCR assay mixtures were thermocycled (37 cycles of 94°C for 1 s, 64°C for 1 s, and 69°C for 40 s) in an LCx thermal cycler. Two calibrators (25 M. tuberculosis DNA genomes in 1 μg of salmon testes DNA per gap LCR amplification mixture) and two negative controls (1 μg of salmon testes DNA per gap LCR amplification reaction mixture) were subjected to thermocycling along with each batch of 20 processed specimens. After thermocycling, double-hapten-labeled amplification product, if produced, was detected by microparticle enzyme immunoassay on an LCx analyzer, a 24-position automated batch analyzer, with the following run order: two negative controls, two calibrators, 20 specimens. Detectable fluorescence was expressed as counts per second per second. The assay cutoff was 0.3 times the mean fluorescence rate of the two calibrators. Specimens were considered gap LCR positive if their sample-to-cutoff (S/CO) ratios were ≥1.0. After detection, the LCx analyzer chemically inactivated the amplification product (17).
Detection of inhibitors.
Each specimen processed for gap LCR amplification was amplified in the presence of 25 M. tuberculosis DNA genomes, which were added as a 10-μl spike-in. Specimens were considered inhibitory if their S/CO ratio was <1.0.
RESULTS
Phosphate buffer inhibition.
Phosphate buffer (67 mM; pH 6.8) was diluted so that the final concentrations in gap LCR mixtures were 2.16, 1.08, 0.54, and 0.27 mM. Each dilution, as well as a water control, was tested in triplicate for the presence of inhibition. Table 1 shows that two of three gap LCR assays were inhibited when each mixture contained 2.16 mM phosphate buffer, pH 6.8. No inhibition was observed when the concentration of phosphate buffer, pH 6.8, in gap LCR amplification mixtures was reduced to 1.08 mM. Consequently, the inhibitory threshold of gap LCR amplification for phosphate buffer, pH 6.8, lies between 1.08 mM and 2.16 mM. A double-centrifugation protocol was instituted to assure that phosphate buffer was diluted consistently below the inhibitory threshold for gap LCR amplification.
TABLE 1.
Inhibitory effect of phosphate buffer, pH 6.8, on gap LCR amplification
| LCR reaction mixture | S/CO ratio for indicated concentration (mM) of phosphate buffer
|
||||
|---|---|---|---|---|---|
| 0 (water) | 0.271 | 0.542 | 1.08 | 2.16 | |
| Replicate 1 | 3.41 | 3.49 | 3.52 | 2.69 | 0.02 |
| Replicate 2 | 3.36 | 3.45 | 3.29 | 3.17 | 0.70 |
| Replicate 3 | 3.57 | 3.43 | 2.89 | 3.01 | 1.30 |
| Mean | 3.45 | 3.46 | 3.23 | 2.96 | 0.67 |
Calcium phosphate precipitate formation and removal of its inhibitory effect by using potassium acetate buffer (pH 4.1).
Visible precipitate was formed when mock specimens, consisting of either 5 or 10 mM CaCl2, were subjected to conditions similar to those found under NALC-NaOH decontamination, digestion, and concentration. No precipitate was formed when water was used as the mock specimen. Even though the precipitate was not chemically identified, it was believed to be calcium phosphate. The calcium phosphate precipitate was inhibitory to gap LCR amplification when the first centrifugation was performed with 10 mM TE buffer, pH 7.8 (Table 2). By contrast, the inhibitory effect of the calcium phosphate precipitate was partially removed when the first centrifugation was performed with 0.5 M potassium acetate buffer, pH 4.1, and completely removed when 1.0 M potassium acetate buffer, pH 4.1, was used in the first centrifugation.
TABLE 2.
Inhibitory effect of calcium phosphate precipitate on gap LCR amplification and removal of inhibition by potassium acetate buffer, pH 4.1
| Centrifugation buffer | Mean (n = 2) S/CO ratio with:
|
||
|---|---|---|---|
| Water | 5 mM CaCl2 | 10 mM CaCl2 | |
| TE, pH 7.8 | 3.33 | 0.05 | 0.08 |
| 0.5 M potassium acetate, pH 4.1 | 3.13 | 2.33 | 0.03 |
| 1.0 M potassium acetate, pH 4.1 | 2.81 | 2.78 | 2.19 |
Comparison of TE buffer (pH 7.8) with potassium acetate buffer (pH 4.1) for removal of inhibition.
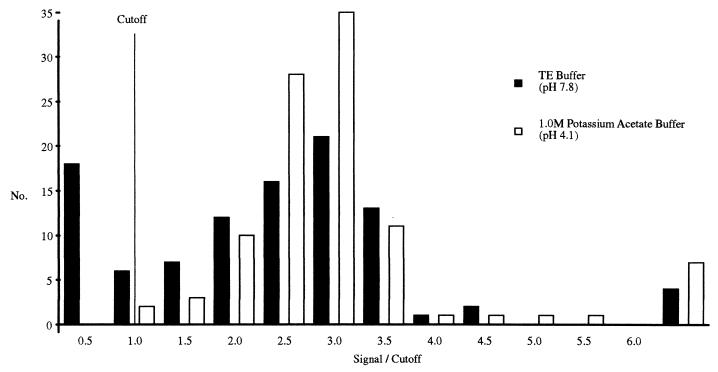
Separate aliquots from 100 NALC-NaOH respiratory sediments were processed with either 10 mM TE buffer (pH 7.8) or 1.0 M potassium acetate buffer (pH 4.1). Inhibition testing determined that 24 of the TE-processed specimens inhibited gap LCR amplification (Fig. 1). By contrast, none of the same NALC-NaOH respiratory sediments were inhibitory to gap LCR amplification when processed with 1.0 M potassium acetate buffer, pH 4.1. Figure 1 also shows that some sediments produced considerably higher S/CO ratios than others. This is likely the result of these sediments containing endogenous M. tuberculosis DNA in addition to the M. tuberculosis DNA added to test for inhibition.
FIG. 1.
Frequency distribution of a comparison between TE, pH 7.8, and 1.0 M potassium acetate buffer, pH 4.1, for the removal of inhibition. A portion of each NALC-NaOH respiratory sediment was processed with either TE, pH 7.8, or 1.0 M potassium acetate buffer, pH 4.1, before being tested by gap LCR amplification in the presence of 25 purified M. tuberculosis DNA genomes. A NALC-NaOH respiratory sediment was considered inhibitory if its gap LCR amplification produced an S/CO ratio of <1.0.
Effectiveness of 1.0 M potassium acetate buffer (pH 4.1) for removing inhibition from NALC-NaOH respiratory sediments.
A total of 1,440 NALC-NaOH respiratory sediments were processed with 1.0 M potassium acetate buffer, pH 4.1, and then each specimen was tested twice by gap LCR. The first gap LCR assay was performed to determine if the specimen contained endogenous M. tuberculosis DNA, while the second was done to determine if the sample inhibited gap LCR. Of the 1,440 specimens, 197 contained endogenous M. tuberculosis DNA. Gap LCR detected 116 of 128 specimens that were culture positive for M. tuberculosis (61 of 62 microscopy-positive and 55 of 66 microscopy-negative specimens were gap LCR positive). Ten of the 1,440 specimens inhibited gap LCR. However, none of the inhibitory specimens were culture positive for M. tuberculosis.
DISCUSSION
The development of relatively simple specimen preparation protocols that reduce the inhibitory potential of NALC-NaOH respiratory sediments would increase the utility of DNA amplification reactions for the detection of M. tuberculosis. When inhibition is substantially reduced, assay sensitivity can be increased by permitting greater volume equivalents of NALC-NaOH respiratory sediment to be tested per amplification reaction. Fulfillment of this goal requires that the most important inhibitors be identified and effectively removed.
Phosphate ions at concentrations as low as 1 to 2 mM were found to inhibit gap LCR amplification. This is an issue because 67 mM phosphate buffer, pH 6.8, is added to respiratory specimens during the NALC-NaOH decontamination, digestion, and concentration process. A double-centrifugation protocol was therefore instituted to reduce the concentration of phosphate ions to below 1.0 mM, when 100-μl equivalents of NALC-NaOH sediment were tested per gap LCR amplification assay. Despite the use of the double-centrifugation protocol, a substantial proportion of NALC-NaOH respiratory sediments inhibited gap LCR amplification. An explanation for the remaining inhibition is the fact that some inhibitors are not removed by the centrifugation steps but instead centrifuge down with the M. tuberculosis bacilli.
One such substance was deduced to be calcium phosphate precipitate, Ca3(PO4)2. This highly insoluble precipitate (solubility product, 0.06 mM) (19) formed when calcium ion-containing specimens were treated with the alkaline NALC-NaOH solution, followed by the addition of phosphate buffer. The precipitate would centrifuge down with M. tuberculosis bacilli and would not be removed even when two centrifugation steps were performed. If the precipitate was subjected to a near-neutral pH, such as pH 7.8, which is found in gap LCR amplification assays, it would convert to a more soluble (solubility product, 1.84 mM) type of precipitate (CaHPO4) (19). The higher concentration of phosphate ions would inhibit gap LCR amplification. If a calcium phosphate precipitate was the culprit, it follows that inhibition could be reduced if the precipitate was dissociated by exposure to acidic conditions and then had its ionic constituents removed by double centrifugation. This conclusion was supported by the observations that calcium phosphate precipitate was formed in a process similar to NALC-NaOH decontamination, digestion, and concentration; that this precipitate inhibited gap LCR when sediments were processed with TE buffer, pH 7.8; and that inhibition was reduced significantly when mildly acidic buffer, pH 4.1, was used in place of TE buffer.
The utility of the double-centrifugation protocol, in which the first centrifugation was performed in the presence of a mildly acidic buffer, was demonstrated when 1,440 NALC-NaOH respiratory sediments were processed and tested for the presence of inhibitors, as well as for endogenous M. tuberculosis DNA. Only 10 sediments were found to be inhibitory to gap LCR. In addition, none of 128 specimens that were culture positive for M. tuberculosis showed inhibition. Gap LCR results indicated that 197 sediments contained endogenous M. tuberculosis DNA, and they were not included in the inhibition rate calculations. This is due to our definition of inhibition, which is based on a S/CO ratio of <1.0 when 25 M. tuberculosis genomes are added to negative specimens. Sediments containing endogenous M. tuberculosis DNA will produce higher gap LCR signals, giving rise to S/CO ratios of >1, even though partial inhibition may be present. The gap LCR inhibition rate was thus calculated to be 0.8% (10 of 1,243).
The inhibition rates achieved here are substantially lower than those reported by other workers who also tested a relatively large volume equivalent of NALC-NaOH respiratory sediment per amplification assay, following centrifugation to remove inhibitors. Nolte et al. (20) observed an inhibition rate of 13.6% when using a PCR internal control, after processing NALC-NaOH respiratory sediments with a single centrifugation step and then testing 50- to 100-μl equivalents of NALC-NaOH respiratory sediment per PCR amplification. By contrast, Clarridge et al. (12) centrifuged each specimen twice before testing 166-μl equivalents of NALC-NaOH respiratory sediment per PCR assay; they observed that 14.3% (4 of 28 sputa or bronchial washes) of PCR-negative specimens which were culture positive for M. tuberculosis contained inhibitors.
Low inhibition rates have been achieved by other workers by testing small volume equivalents (≤25 μl) of respiratory sediment per amplification reaction after performing a simple specimen preparation protocol or by testing >25-μl equivalents per amplification reaction after performing relatively complicated, multistep specimen preparation protocols (1, 2, 9, 21, 24, 26). Both of these strategies have disadvantages. Testing small volume equivalents of respiratory sediment per amplification reaction reduces sensitivity compared to that of BACTEC liquid culture, which uses 500 μl of respiratory sediment per culture. By contrast, the more complicated the specimen preparation method, the more it is prone to cross-contamination and loss of analyte (25).
This work demonstrates the value of identifying the most potent inhibitors of nucleic acid amplification reactions. It permits simple protocols to be designed which effectively remove the most troublesome inhibitors and also promotes increased sensitivity by permitting relatively large volume equivalents of specimen to be tested per amplification reaction.
REFERENCES
- 1.Amicosante M, Richeldi L, Trenti G, Paone G, Campa M, Bisetti A, Saltini C. Inactivation of polymerase inhibitors for Mycobacterium tuberculosis DNA amplification in sputum by using capture resin. J Clin Microbiol. 1995;33:629–630. doi: 10.1128/jcm.33.3.629-630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen A B, Thybo S, Godfrey-Faussett P, Stoker N G. Polymerase chain reaction for detection of Mycobacterium tuberculosis in sputum. Eur J Clin Microbiol Infect Dis. 1993;12:922–927. doi: 10.1007/BF01992166. [DOI] [PubMed] [Google Scholar]
- 3.Andersen Å B, Hansen E B. Structure and mapping of antigenic domains of protein antigen b, a 38,000-molecular-weight protein of Mycobacterium tuberculosis. Infect Immun. 1989;57:2481–2488. doi: 10.1128/iai.57.8.2481-2488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backman K. Diagnostic technology for the 1990s and beyond. Clin Chem. 1992;38:457–458. [Google Scholar]
- 5.Barany F. Ligase chain reaction in a PCR world. PCR Methods Applic. 1991;5:5–16. doi: 10.1101/gr.1.1.5. [DOI] [PubMed] [Google Scholar]
- 6.Betsch D F. Nucleic acid amplification-based diagnostics: barriers to commercialization. Med Rev Diagn Ind. 1995;17:22–28. [Google Scholar]
- 7.Birkenmeyer L, Armstrong A S. Preliminary evaluation of the ligase chain reaction for specific detection of Neisseria gonorrhoeae. J Clin Microbiol. 1992;30:3089–3094. doi: 10.1128/jcm.30.12.3089-3094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkenmeyer L G, Mushahwar I K. DNA probe amplification methods. J Virol Methods. 1991;35:117–126. doi: 10.1016/0166-0934(91)90127-l. [DOI] [PubMed] [Google Scholar]
- 9.Brisson-Noel A, Aznar C, Chureau C, Nguyen S, Pierre C, Bartoli M, Bonette R, Pialoux G, Gicquel B, Garrigue G. Diagnosis of tuberculosis by DNA amplification in clinical practice evaluation. Lancet. 1991;338:364–366. doi: 10.1016/0140-6736(91)90492-8. [DOI] [PubMed] [Google Scholar]
- 10.Burczak J D, Ching S F, Hu H Y, Lee H. Ligase chain reaction for the detection of infectious agents. In: Wiedbrauk D, Farkas D H, editors. Molecular methods for virus detection. New York, N.Y: Academic Press, Inc.; 1995. pp. 315–327. [Google Scholar]
- 11.Cao J, Davis A, Erickson D, Facey I, Halaka F, He Q, Hu H, Kawa D, Lin B-C, Winter G, Leckie G. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Direct detection of Mycobacterium tuberculosis by the LCxRMycobacterium tuberculosis assay, abstr. U-10. [Google Scholar]
- 12.Clarridge J E, Shawar R M, Shinnick T M, Plikaytis R B. Large-scale use of polymerase chain reaction for detection of Mycobacterium tuberculosis in a routine mycobacteriology laboratory. J Clin Microbiol. 1993;31:2049–2056. doi: 10.1128/jcm.31.8.2049-2056.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dille B J, Butzen C C, Birkenmeyer L G. Amplification of Chlamydia trachomatis DNA by ligase chain reaction. J Clin Microbiol. 1992;31:729–731. doi: 10.1128/jcm.31.3.729-731.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes B A, Hicks K E S. Direct detection of Mycobacterium tuberculosis in respiratory specimens using a polymerase chain reaction. J Clin Microbiol. 1993;31:1688–1694. doi: 10.1128/jcm.31.7.1688-1694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forbes B A, Hicks K E. Substances interfering with direct detection of Mycobacterium tuberculosis in clinical specimens by PCR: effects of bovine serum albumin. J Clin Microbiol. 1996;34:2125–2128. doi: 10.1128/jcm.34.9.2125-2128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forbes B A. Critical assessment of gene amplification approaches on the diagnosis of tuberculosis. Immunol Investig. 1997;26:105–116. doi: 10.3109/08820139709048919. [DOI] [PubMed] [Google Scholar]
- 17.Hu H Y, Burczak J D, Leckie G W, Ray K A, Muldoon S, Lee H H. Analytic performance and contamination control methods of a ligase chain reaction DNA amplification assay for detection of Chlamydia trachomatis in urogenital specimens. Diagn Microbiol Infect Dis. 1996;24:71–76. doi: 10.1016/0732-8893(95)00272-3. [DOI] [PubMed] [Google Scholar]
- 18.Kent P T, Kubica G P. Public health mycobacteriology. A guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 19.Lide D R, editor. CRC handbook of chemistry and physics, 1993–1994. Boca Raton, Fla: CRC Press Inc.; 1994. pp. 4–49. [Google Scholar]
- 20.Nolte F S, Metchock B, McGowan J E, Edwards A, Okwumabua O, Thurmond C, Mitchell P S, Plikaytis B, Shinnick T. Direct detection of Mycobacterium tuberculosis in sputum by polymerase chain reaction and DNA hybridization. J Clin Microbiol. 1993;31:1777–1782. doi: 10.1128/jcm.31.7.1777-1782.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noordhoek G T, Kaan J A, Mulder S, Wilke H, Kolk A H J. Routine application of the polymerase chain reaction for detection of Mycobacterium tuberculosis in clinical samples. J Clin Pathol. 1995;48:810–814. doi: 10.1136/jcp.48.9.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfyffer G E. Amplification techniques: hope or illusion in the direct detection of tuberculosis? Med Microbiol Lett. 1994;3:335–347. [Google Scholar]
- 23.Sjobring U, Mecklenburg M, Andersen A B, Miorner H. Polymerase chain reaction for detection of Mycobacterium tuberculosis. J Clin Microbiol. 1990;28:2200–2204. doi: 10.1128/jcm.28.10.2200-2204.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soini H, Skurnik M, Liippo K, Tala E, Viljanen M K. Detection and identification of mycobacteria by amplification of a segment of the gene coding for the 32-kilodalton protein. J Clin Microbiol. 1992;30:2025–2028. doi: 10.1128/jcm.30.8.2025-2028.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson S M, McNerney R, Nye P M, Godfrey-Faussett P D, Stoker N G, Voller A. Progress toward a simplified polymerase chain reaction and its application to diagnosis of tuberculosis. J Clin Microbiol. 1993;31:776–782. doi: 10.1128/jcm.31.4.776-782.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuen K Y, Chan K S, Chan C M, Ho B S W, Dai L K, Chau P Y, Ng M H. Use of PCR in routine diagnosis of treated and untreated pulmonary tuberculosis. J Clin Pathol. 1993;46:318–322. doi: 10.1136/jcp.46.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]