Abstract
The direct causes of neurodegeneration underlying Alzheimer’s disease (AD) and many other dementias, are not known. Here we identify serum amyloid P component (SAP), a constitutive plasma protein normally excluded from the brain, as a potential drug target. After meta-analysis of three genome-wide association studies, comprising 44,288 participants, cis-Mendelian randomization showed that genes responsible for higher plasma SAP values are significantly associated with AD, Lewy body dementia and plasma tau concentration. These genetic findings are consistent with experimental evidence of SAP neurotoxicity and the strong, independent association of neocortex SAP content with dementia at death. Depletion of SAP from the blood and from the brain, as is provided by the safe, well tolerated, experimental drug, miridesap, may therefore contribute to treatment of neurodegeneration.
Keywords: Alzheimer’s disease, dementia, genome-wide association study, Lewy body dementia, Mendelian randomization, miridesap, neurodegeneration, serum amyloid P component
One Sentence Summary:
Genetic analyses of circulating serum amyloid P component (SAP) values suggest that depletion of plasma SAP may decrease the risk of Alzheimer’s disease and Lewy body dementia.
INTRODUCTION
The direct causes and mechanisms of neuronal cell death responsible for the cognitive loss in Alzheimer’s disease (AD) and many other dementias, are not known. Serum amyloid P component (SAP) is an almost invariant, constitutive, normal plasma glycoprotein produced exclusively in the liver. It circulates at a mean (SD) concentration of about 24 (8) mg/l in women and 32 (7) mg/l in men1 but it is normally rigorously excluded from the central nervous system (CNS). Cerebrospinal fluid (CSF) concentrations of SAP are one thousand-fold lower than the plasma concentration,2,3 presumably reflecting relative impermeability of the blood brain barrier (BBB). There is also evidence for an active transport mechanism exporting SAP from the CSF back into the blood4. SAP is named for its universal presence in all human amyloid deposits, which reflects the avid but reversible calcium dependent binding of SAP to all types of amyloid fibrils regardless of their protein composition5,6. Thus, although CSF and brain content of SAP are normally extremely low, SAP is nonetheless always present in the intracerebral Aβ amyloid plaques, cerebrovascular Aβ amyloid deposits and the majority of neurofibrillary tau tangles in AD. The binding of SAP stabilises amyloid fibrils7 and promotes their formation8,9, thereby contributing to amyloid deposition and persistence10. Furthermore, accumulation of SAP on intracerebral amyloid plaques, cerebrovascular amyloid deposits and neurofibrillary tangles also increases exposure of cerebral neurones to SAP.
Cerebral Aβ amyloid is a defining feature of AD, is also often present in Lewy body dementia (LBD) and is present in chronic traumatic encephalopathy. It is still not known how amyloid pathology contributes to neurodegeneration but recent reports of cognitive benefit from antibody treatments that reduce the Aβ amyloid burden in AD are encouraging11,12. Typical AD neuropathology is often seen in the brains of individuals who were cognitively normal at death, raising the possibility of other pathogenetic factors in dementia. It is therefore interesting that, unrelated to its contribution both to Aβ amyloid formation and persistence, human SAP is itself directly neurotoxic to cerebral neurones in vitro13–16 and in animal models in vivo17. Furthermore, neocortical SAP content is significantly associated with dementia at death, independently of neuropathological severity, consistent with a more direct, amyloid-independent, pathogenetic role of SAP in neurodegeneration18. Indeed, many of the risk factors for dementia, including cerebral and cerebrovascular amyloid deposition, traumatic brain injury, cerebral haemorrhage and even ‘normal’ ageing, with its associated impairment of the BBB19, are characterised by increased exposure of the brain to SAP.
In order to rigorously explore the potential causative role of human SAP in human neurodegenerative diseases, we have now sought genetic epidemiological evidence. SAP is encoded by the gene APCS (ENSG00000132703) located on chromosome 1, in close proximity to CRP (ENSG00000132693) which encodes C-reactive protein (CRP). These two proteins comprise the pentraxin family, sharing 54% strict residue for residue amino acid sequence homology, even higher genetic sequence homology and having the same secondary, tertiary and quaternary structural organisation. Despite notable phylogenetic conservation of gene and protein sequence and structure among pentraxins, there are marked biological differences between these proteins both within and between species20. Thus, human CRP is the classical acute phase protein that is among the most commonly used routine clinical chemistry analytes, whilst human SAP is a constitutive plasma protein, the assay of which has hitherto had no practical clinical significance. Human SAP is not an acute phase reactant although in chronic inflammatory conditions, in which there is sustained increased production of CRP, SAP values tend to be slightly higher, albeit within the reference range21. A few small studies in the elderly and subjects with impaired cognition have reported plasma and CSF SAP concentrations above the reference range of the healthy middle aged population2,22,23,24. Children under 10 years have circulating SAP concentrations below the adult range but reduced adult SAP values are seen only with severe hepatocellular impairment21. Unsurprisingly therefore, in contrast to CRP concentration, there have only been limited genome-wide association studies (GWAS) of plasma SAP concentration24–26. Recently, however, the SomaLogic aptamer-based proteomic platform has enabled large scale measurement of circulating SAP abundance, allowing for a growing number of GWAS identifying potential genetic instruments for plasma SAP concentration.
Cis-Mendelian randomization (MR) leverages genetic instruments associated with protein concentration to demonstrate the possible causal effects of a potential drug target and thus to anticipate safety and efficacy outcomes of specific therapeutic interventions. The random allocation of genes during gametogenesis crucially protects genetic associations against bias due to confounding and reverse causality27,28. Furthermore, through a two-sample design, MR can source aggregated data, that is point estimates and standard errors, from large scale studies, each designed to maximize the available sample size. This extensively validated MR approach can provide a precise and powerful overview of the likely causal consequences of target perturbation covering a large number of clinically relevant diseases and traits29–32.
We confirmed that the SomaLogic SAP values reliably reflect the actual plasma concentration of the protein measured by rigorously calibrated SAP immunoassay. We then conducted a meta-analysis of three GWAS of circulating SAP values, combining information from 44,288 participants, followed by a drug target MR utilizing APCS cis-variants that were strongly associated with plasma SAP values. We primarily focused on the possible causal effect of SAP in AD and LBD. Given the close proximity of APCS to CRP, and the major involvement of CRP responses with almost all inflammatory, infective, traumatic and other tissue damaging processes33, we additionally utilized MR to rule out possible effects of plasma CRP concentration acting on the SAP signal through linkage disequilibrium (LD) between variants in APCS and variants in CRP.
RESULTS
Genome-wide meta-analysis of plasma SAP values
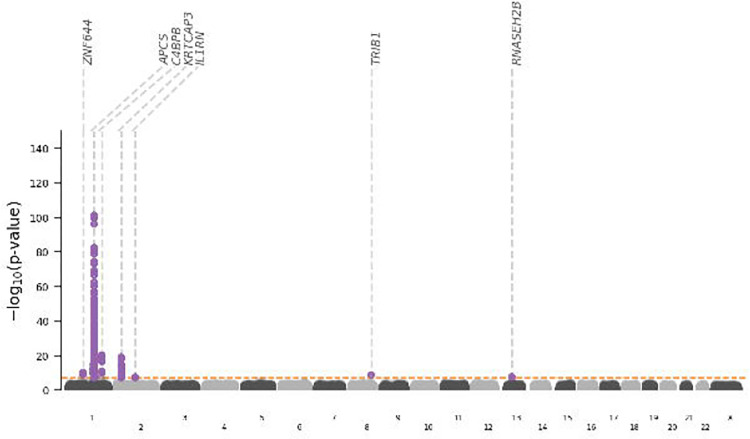
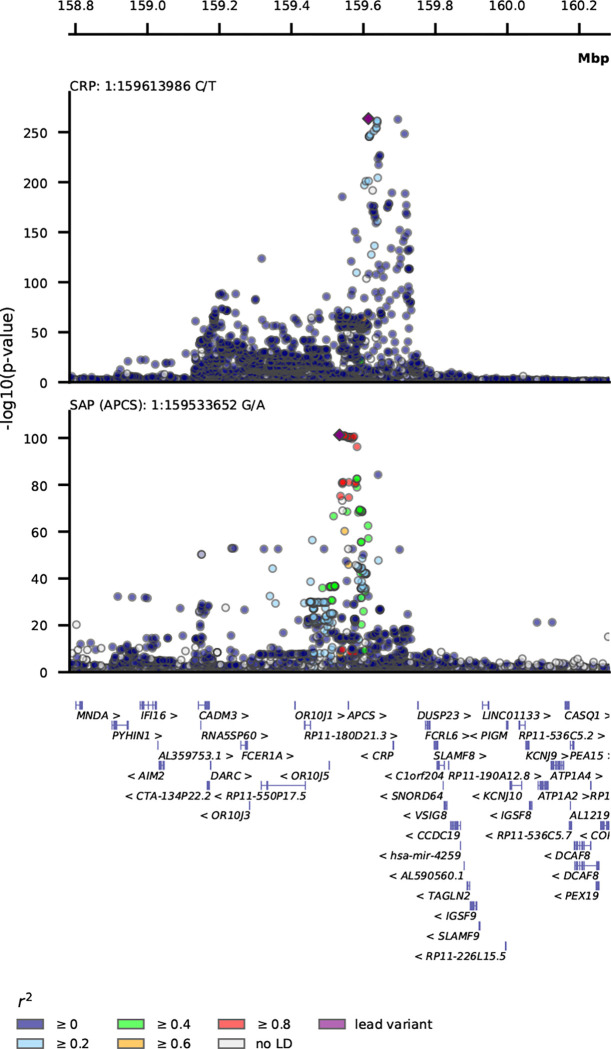
Genetic variant-specific estimates of the association with SAP values, measured by SomaLogic SomaScan assay version 4.1, were available from three independent studies: Interval, comprising 3,301 particpants24, AGES, with 5,36826, and DECODE with 35,55925. The combined data identified ten independent lead variants associating with SAP, including four cis-variants near APCS (rs140308485, rs13374652, rs1341664, rs78228389), as well as trans-variants on chromosomes 1, 2, 8 and 13 (Fig. 1–2, Table S1). Comparison of the genetic associations with plasma SAP and CRP values in the region around rs1341664, the APCS cis-variant with the strongest SAP association, indicated that these closely adjacent signals were independent (Fig. 2); the Pearson correlation coefficient comparing the −log10(p-value) for each trait was −0.06, p-value < 0.001. This was further confirmed by noting that the CRP association of the four SAP cis-variants did not reach genome-wide significance (Table S2), with the −log10(p-value) for CRP ranging between 0.04 and 6.26. Instead, the CRP signals were concordant with the SAP trans signals (Fig. S2; Table S2).
Fig. 1. Manhattan plot of SAP genome-wide association study.
The −log10(p-value) of genetic variants is shown on the y-axis and GRCh37 base pair position within chromosomes on the x-axis. The horizontal dashed line is at p-value 5.8×10−8. The lead variants are labelled with the putative causal genes assigned by V2G.
Fig. 2. Stacked locus-view comparing the overlap between genetic variants for plasma SAP values and plasma CRP concentration.
The −log10(p-value) of the genetic association with SAP and CRP plotted (y-axis) against the genomic location (x-axis). The lead variant for each trait is indicated by a purple diamond. Linkage disequilibrium with the lead variant is indicated by coloured dots, with the r-squared estimated from the UKB and 1000 genomes EUR reference. Gene locations were queried from Ensembl v109 (GRCh37).
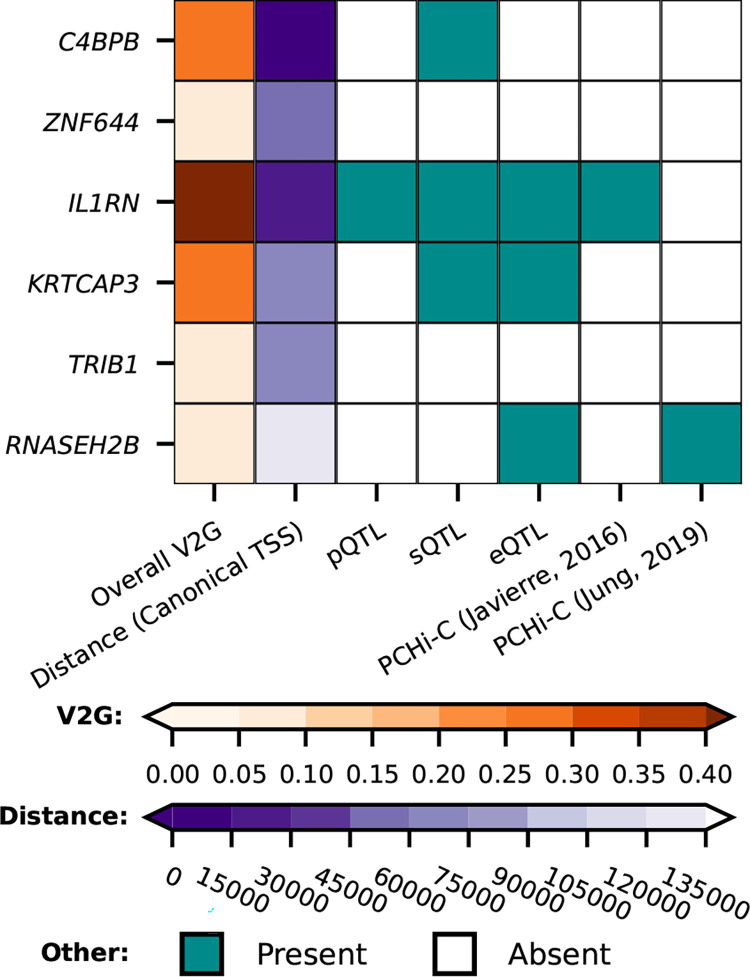
GWAS Catalog look-ups for the effects of genetic variants in APCS identified a previously reported association with white-matter microstructure34 (Fig. S3; Table S9). Using Open Target’s V2G algorithm the six SAP trans-variants were mapped to the putative causal genes: C4BPB, ZNF644, IL1RN, KRTCAP3, TRIB1, RNASEH2B (Fig. 1; 3, Tables S3–S8). Look-ups for the variants within and around the putative trans-genes for SAP, provided links with a diverse range of pathophysiology without known connections to SAP biology (Fig. S3, Table S9).
Fig. 3. Mapping of the trans signals for variants associated with plasma SAP value to putative causal genes.
First column, the overall V2G score; second column, distance in base pairs from the lead variant to the canonical gene transcription start site (TSS). The other columns show the presence or absence of x-axis criteria, specifically, whether there were quantitative trait loci (QTL) linking the gene to proteomics, transcriptomics, or splice site QTL, and PCHi-C (Promoter Capture Hi-C) chromatin interaction experiments linking the genetic variant to the indicated gene, from Jung et al.65 and Javierre et al.66. Tables S3–S8 show the whole V2G output.
Cis-MR results for plasma SAP values and dementia outcomes
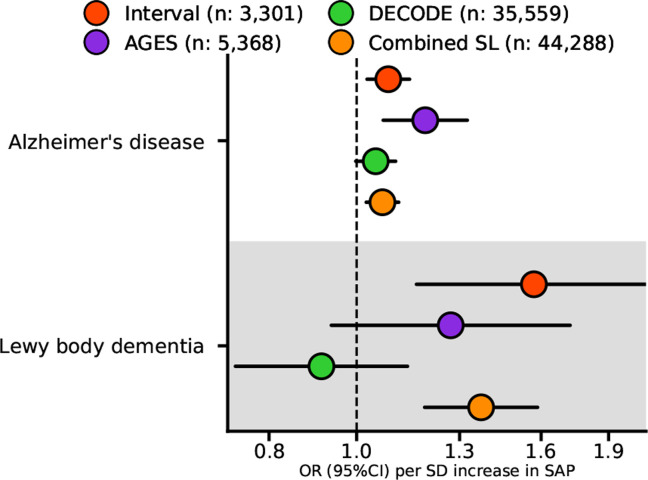
Cis-MR analysis detected significant associations of higher plasma SAP values with increased risk of dementia outcomes: AD (35,274 cases, odds ratio (OR) 1.07, 95%CI 1.02; 1.11, p=1.8×10−3), and LBD (2,981 cases, OR 1.37, 95%CI 1.19; 1.59, p=1.5×10−5) (Fig. 4, Table S10). A similar analysis for plasma CRP values did not identify links with these outcomes (Table S10).
Fig. 4. Estimates of the cis Mendelian randomization effect of plasma SAP values on Alzheimer’s disease and Lewy body dementia.
The univariable MR effects estimated from the GWAS meta-analysis of SomaLogic (SL) plasma SAP values from the Interval, AGES and DECODE studies are shown individually, together with the combined result of all three studies. We used the AD GWAS from Kunkle et al.52 which consisted of 35,274 cases and 59,163 controls, the LBD GWAS from Chia et al.53 consisted of 2,981 cases and 2,173 controls. OR, odds ratio; 95%CI, 95% confidence interval. The Figure illustrates the data in Table S10.
Cis-MR results for plasma SAP values and other outcomes
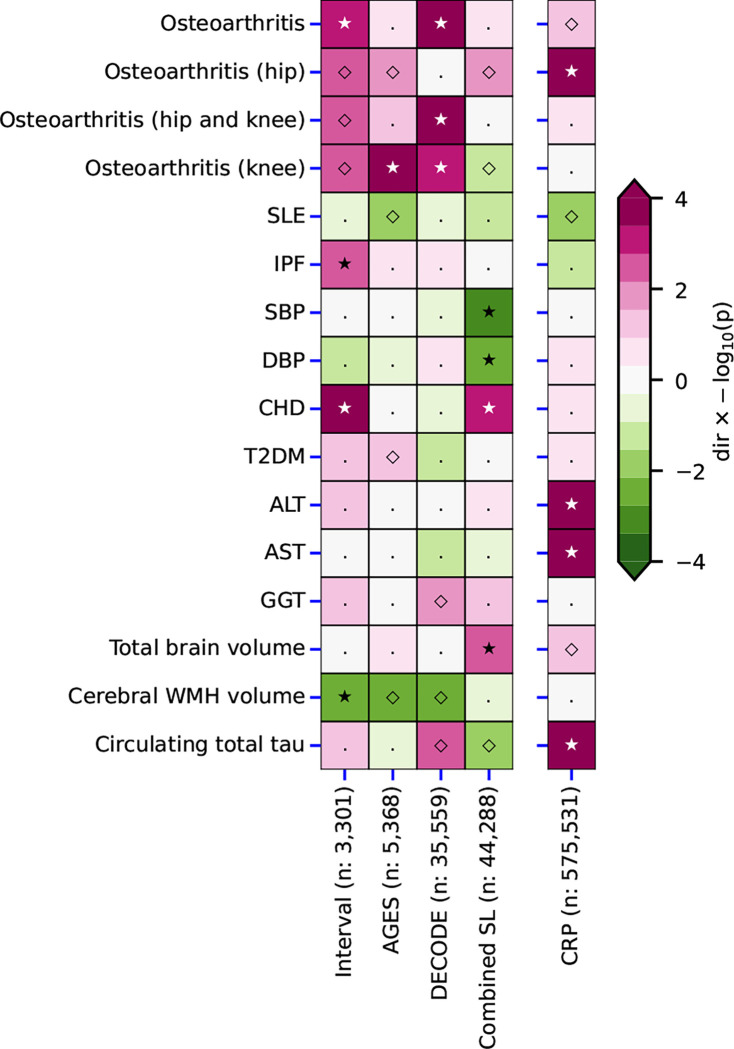
Cis-MR analysis suggested that higher plasma SAP value was associated with increased coronary heart disease (CHD) risk (OR 1.03, 95%CI 1.01; 1.05), greater total brain volume (0.06 SD, 95%CI 0.02; 0.10), lower systolic blood pressure (SBP) (−0.16 mm Hg, 95%CI −0.26; −0.07) and lower diastolic blood pressure (DBP) (Fig. 5, Table S11). In contrast, the MR analysis of plasma CRP values showed a distinct effects profile in which higher CRP concentrations were associated with serum concentrations of hepatocellular enzymes, with osteoarthritis, and with total plasma tau concentration (Fig. 5, Table S12).
Fig. 5. Comparison of the cis Mendelian randomization effect estimates of plasma SAP values and CRP concentration.
Effect direction, magenta for positive and green for negative, is shown by the −log10(p-value) to 4 significant figures. Open diamonds, p<0,05; closed stars, p<2.38 × 10−5.. Left block, SAP; right column, CRP. Abbreviations: ALT, alanine transaminase; AST, aspartate transaminase; CHD, coronary heart disease; DBP, diastolic blood pressure; GGT, gamma-glutamyl transferase; IPF, idiopathic pulmonary fibrosis; SBP, systolic blood pressure; SLE, systemic lupus erythematosus; T2DM, type 2 diabetes; WMH, white matter hyperintensities. The Figure illustrates the data in Tables S11–S12.
Multivariable MR results for outcomes linked to both SAP and CRP
APCS and CRP are closely co-located on chromosome 1, potentially challenging the recognition of, and discrimination between, more subtle apparent independent effects. We therefore identified outcomes significant at the more liberal value of p<0.05 for both plasma SAP and CRP values, specifically, total brain volume, plasma total tau concentration and osteoarthritis. Application of multivariable MR (MVMR) to identify the mutually independent effects of both proteins, and accounting for potential influence of LD, then showed that higher values of each increased circulating total tau concentration: (0.06 log2(ng/L), 95%CI 0.03; 0.08) for higher plasma SAP value, and (0.20 log2(ng/L), 95%CI 0.14; 0.25) for higher plasma concentration of CRP. The MVMR analysis did not confirm significant associations for osteoarthritis, and total brain volume (Table S13).
DISCUSSION
We report here a large-scale meta-analysis of GWAS of SomaLogic values for plasma SAP comprising 44,288 participants. We confirmed that the SomaLogic intensity scores for plasma SAP agree with immunoassay of actual SAP concentrations. The GWAS results then enabled MR analysis sourcing cis-acting variants within and around APCS, the gene encoding SAP, to explore potential involvement of SAP in pathogenesis of dementia. We found that higher plasma SAP values increased the risk of AD (OR 1.07, 95%CI 1.02; 1.11, p=1.8×10−3) and LBD (OR 1.37, 95%CI 1.19; 1.59, p=1.5×10−5), implying that pharmaceutical depletion of SAP might reduce the risk of both diseases. Furthermore, using multivariable MR to account for possible horizontal pleiotropy by plasma CRP concentration, we also detected a significant association of higher plasma SAP values with higher total plasma tau concentration (0.06 log2(ng/L), 95%CI 0.03; 0.08), which is itself also associated with AD dementia35.
The presence of SAP in all cerebral Aβ plaques and on most neurofibrillary tangles in AD, long known from immunohistochemical studies, has recently been shown to strongly discriminate between AD brains and cognitively normal brains36. However, the present finding of significant associations between genetically determined higher plasma SAP values and increased risk of AD and LBD, is more specifically consistent with the association between neocortex SAP content and cognitive status at death that was recently observed in the Cognitive Function and Ageing Study18. In this unselected, population-representative, elderly brain donor population, the OR for dementia at death, between the top tertile and the lowest tertile of neocortical SAP content, was 5.24 (95%CI 1.79; 15.29). Furthermore the association of dementia with SAP content was independent of Braak stage, Thal phase and all other classical neuropathological hallmarks of dementia18. It was thus specific for abundance of SAP itself in the neocortex rather than SAP content just being a surrogate for the Aβ amyloid and neurofibrillary tau tangle pathology which is always present in AD and also frequently found in LBD. This SAP-dementia association, which is consistent with a possible pathogenetic role of SAP in neurodegeneration, is now directly supported by the present MR results.
In contrast to a previously reported MR study, which did not detect an association between SAP values and AD37, our analysis was improved in multiple ways. First and foremost, instead of simply taking a single SAP GWAS, we meta-analysed combined data from three independent studies to produce the largest GWAS of SAP values to date. This resulted in an AD MR analysis using 53 SAP variants instead of the 14 used by Yueng et al37. The larger number of variants allowed us to consider outlier and leverage statistics, identifying and removing variants with possible horizontal pleiotropic effects, further ensuring the robustness of the present findings. Furthermore, we performed confirmatory analyses to refute possible bias due to the location of the CRP gene closely adjacent to APCS, and we found no meaningful overlap between the effect profiles of plasma SAP and CRP values. Similar to the analysis by Yueng et al37, we conducted a two-sample MR analysis, where the exposure GWAS did not, or only partially, overlapped with the outcome GWAS. Any potential weak instrument bias will thus, on average, act towards a null effect, hence our results are conservative. This, however, also implies that non-significant findings should not be over-interpreted as providing proof of absence38. Finally, we note that, under the null hypothesis and accounting for the number of evaluated outcomes, the probability of finding an effect of plasma SAP value on AD as well as on LBD is equal to the square of the alpha: (2.78×10−3)2 =7.73×10−6. The concordant findings thus strongly imply that SAP contributes to pathogenesis of dementia.
Increased duration and/or intensity of brain exposure to SAP may be pathogenic through its direct cytotoxicity for some cerebral neurones, as has been demonstrated experimentally in vitro and in vivo13–16, and potentially also by promoting the formation and the persistence of Aβ amyloid fibrils and neurofibrillary tau tangles. However, SAP is produced only by the liver; it is not in the brain transcriptome39. Normally the brain is strongly protected against exposure to SAP by the BBB and by active transport back to the plasma of any SAP that leaks through4, so that the CSF SAP concentration is about one thousandth of that in the plasma2,3. It is therefore striking that even modestly higher plasma SAP concentrations are associated with dementia risk. There have been very few studies of SAP concentration in paired samples of plasma or serum and CSF but in addition to the circulating SAP concentration, BBB integrity and the efficiency of the SAP active export mechanism must affect brain exposure to SAP. Nevertheless, across the large populations studied here, there was a significant effect of higher plasma SAP concentration on clinical dementia outcomes.
In addition to the four cis-loci, our GWAS results identified variants mapping to genes outside APCS, including C4BPB, ZNF644, IL1RN, KRTCAP3, TRIB1 and RNASEH2B. SAP binds specifically to C4-binding protein, encoded by C4BPB, under particular experimental conditions in vitro40, though no functional effect of the interaction has been reported. IL1RN, which encodes the IL-1 receptor antagonist (IL-1RA) might have a functional effect on SAP via the acute phase response, which is mediated by IL-1 both directly and via other pro-inflammatory cytokines. Even though human SAP is not an acute phase reactant, its concentration does tend to rise modestly within the reference range in chronic inflammatory diseases with a sustained acute phase response21. TRIB1 has diverse, wide ranging effects across many different physiological systems.
The putatively mapped SAP trans-genes have previously been variously linked to a broad range of different metabolic, cardiac and haematological traits and to increased plasma concentrations of liver enzymes. Inclusion of these traits in our cis-MR analysis identified an association of higher SAP values with increased CHD and decreased SBP and DBP, but, since there is no known functional connection between these cardiovascular features and SAP, the protein itself is unlikely to be directly involved. Potential pathogenetic connections have been suggested between SAP and two different, unrelated diseases, osteoarthritis41 and systemic lupus erythematosus42. Plasma SAP concentrations were not strongly associated with lupus but there were apparent associations between increased SAP values and some osteoarthritis outcomes (Fig. 5), perhaps reflecting our conservative analyses. We also found a significant positive association between SAP and idiopathic pulmonary fibrosis, suggesting that SAP may also have a pathogenetic role in this condition (Fig. 5). In contrast, osteoarthritis43 and lupus44, respectively, have well established positive and negative clinical links with CRP, and, interestingly, both were associated with circulating CRP concentrations in the corresponding directions in the current analysis (Fig. 5).
Potential limitations to our study comprise, firstly, our use of SomaLogic values for plasma SAP, which are only relative intensities not actual SAP concentrations. SomaLogic assays alone therefore cannot enable precise determination of the effect magnitude relevant for potential pharmaceutical intervention, even though we rigorously demonstrated that the SomaLogic values are in acceptably close agreement with the actual SAP concentrations measured by precise, rigorously standardised electroimmunoassay.
Secondly, previous GWAS studies of AD and other types of dementia have not reported APCS as a potential gene for disease onset. However, GWAS is deliberately designed to limit detection of false positive results and may accordingly leave additional signals undiscovered27. It is therefore important to emphasize that drug target MR does not require the GWAS data used for the outcome trait to reach GWAS significance28.
Thirdly, while the present results imply that SAP depletion might reduce dementia risk, they do not indicate optimal timing for the intervention. Neuropathological changes are well known to long precede clinically detectable cognitive loss in AD and other dementias, so SAP depletion might be most effectively introduced prophylactically. Nevertheless, in view of the present evidence for a causal relationship between increased circulating SAP and risk of dementia, prompt SAP depletion may protect residual cognition at any stage.
Fortunately, the experimental drug, miridesap, (CPHPC; hexanoyl bis-D-proline; (R)-1-[6-[(R)-2-carboxy-pyrrolidin-1-yl]-6-oxo-hexanoyl]pyrrolidine-2-carboxylic acid))45, safely provides extremely effective SAP depletion. It reduces plasma SAP concentration by more than 95% for as long as the drug is administered46 and thereby removes all detectable SAP from the CSF in patients with AD3 and from the brain in human SAP transgenic AD model mice47. DESPIAD, a small, academic, phase 2b clinical trial of SAP depletion by miridesap in established AD, is now in progress (EudraCT number 2016–003284-19) and will report in 2025. Meanwhile, our present genetic analysis indicates that depletion of plasma SAP is expected to decrease the risk of AD and LBD.
MATERIALS AND METHODS
Validation of SomaLogic SAP assay
The read out from the SomaLogic aptamer-based mass spectrometric method is relative reagent intensity, a proxy for SAP concentration rather than actual mass per volume. We therefore used the robust electroimmunoassay method, rigorously standardised with isolated, pure human SAP1, to measure the actual concentration of SAP in 100 human plasma samples from a random sub-cohort of the EPIC-Norfolk study (https://www.epic-norfolk.org.uk/) in which SAP had been quantified by the SomaLogic method. The electroimmunoassay confirmed that the SomaLogic results accurately reflected plasma SAP concentrations: for the two sets of results, the Pearson correlation coefficient was 0.86 (p< 0.001) and Spearman correlation was 0.84 (p<0.001) (Fig. S1).
GWAS of SAP plasma values
Three independent studies with plasma SAP values determined by the SomaLogic method: Interval (n: 3,301)24, AGES (n: 5,368)26, and DECODE (n: 35,559)25, were used to provide aggregate genetic data. To account for potential heterogeneity in genetic associations, due to difference in participants and/or environment, we performed a DerSimonian-Laird random effects meta-analysis using METAL48.
Independent lead genetic variants were identified by filtering associations on a genome-wide significant p-value of 5.8×10−8 and clumping to an R-squared of 0.01 based on LD reference data from a random 5,000 participant subset of the UK Biobank (UKB). The nearest protein coding genes were identified by querying the GRCh37 assembly via Ensembl REST API49. Lead variants within 2 megabase pairs (MB) of APCS, the gene encoding SAP, were assigned to this gene; trans-variants (outside ±2 MB of APCS) were mapped to putative causal genes using the V2G algorithm offered by Open Targets50. The V2G algorithm ranks putative causal genes based on integrated information on molecular traits, such as, information on splice-sites, mRNA expression, chromatin interaction, functional predictions and distance from the canonical transcription start site. Potential pleiotropic associations of these putative causal genes were explored by querying the author-assigned gene in GWAS Catalog, which comprises the largest source of gene to phenotype information51.
Drug target Mendelian randomization
Drug target MR was employed to ascertain the possible causal effects that a unit increase in standard deviation (SD) of mean plasma SAP concentration had on clinically relevant traits, with a primary focus on AD52 and LBD53.
To limit the potential for bias due to pre-translational horizontal pleiotropy, variants were extracted from within and around APCS, applying a ±1 MB pairs flank54. Variants were filtered to a minor allele frequency of 0.01 or larger, and clumped to an R-squared of 0.40. Residual LD was modelled using generalized least square (GLS) solutions55 and a 5,000 random sample of UKB participants. To reduce the risk of weak-instrument bias56, we selected genetic variants with an F-statistic of 15 or higher. Furthermore, due to the absence of sample overlap between the SAP GWAS dataset and the GWAS used for many of the outcome traits, any potential weak-instrument bias would act towards a null effect, reducing power rather than increasing type 1 errors56,57.
Estimates of the potential causal effect of higher plasma SAP value were obtained using the GLS implementation of the inverse-variance weighted (IVW) estimator and the MR-Egger estimator, the latter being unbiased in the presence of horizontal pleiotropy at the cost of lower precision58. To minimize the potential influence of horizontal pleiotropy, variants beyond 3 times the mean leverage or an outlier Chi-square statistic larger than 10.83, were pruned59. Finally, a model selection framework was applied to select the most appropriate estimator, IVW or MR-Egger59,60. This model selection framework61 utilizes the difference in heterogeneity between the IVW Q-statistic and the Egger Q-statistic to decide which method provides the best model to describe the available data and hence optimizes the bias-variance trade-off.
Given the close proximity of CRP to APCS we additionally conducted an MR analysis of CRP concentration, taking advantage of availability of the largest CRP GWAS conducted to date62. The MR effect estimates of SAP and CRP were compared to identify outcomes which seemingly were affected by both proteins using a p-value of 0.05. For the subset of outcomes which were affected by both SAP and CRP, we additionally conducted MVMR to analytically control any influence of CRP on the SAP signal and vice versa. MVMR is similar to standard multiple regression, where multiple variables, in our case two, are included in the same model, resulting in estimates that are mutually independent of one another63. Importantly, MVMR allowed us to account for any horizontal pleiotropy that might act through CRP concentration63. In addition, to correct for any potential remaining horizontal pleiotropy acting through non-CRP pathways, we applied the same model selection framework to decide between MVMR with and without Egger correction. Where relevant, we differentiate between MVMR and regular MR results by referring to the latter as univariable MR.
Effect estimates and multiple testing
Unless otherwise specified, all point estimates, that is OR or mean differences, refer to a unit change of the independent variable, typically one standard deviation in plasma protein value for MR results or an increase in number of risk alleles for GWAS results, respectively. Results are provided with 95% confidence intervals (CI) and p-values. Significance in the GWAS analysis was evaluated using the standard multiplicity corrected alpha, that is the false positive rate, of 5.8×10−8, accounting for the estimated number of independent genetic variants in the genome64. The MR results were tested against a Bonferroni corrected alpha of 2.78×10−3, accounting for the 18 evaluated traits (Data availability section).
Supplementary Material
Figures
S1 Correlation between SomaLogic values and immunoassay for actual plasma SAP concentration 2
S2 Locus view plots 6
S3 GWAS Catalog look-ups 9
Tables
S1 Lead variants for serum amyloid P component (SAP) plasma value 11
S2 SAP lead variant associations with plasma C-reactive protein (CRP) concentration 12
S3 Open target v2g results mapping trans-variant rs2808467 to a putative causal gene 13
S4 Open target v2g results mapping trans-variant rs165316 to a putative causal gene 14
S5 Open target v2g results mapping trans-variant rs10188292 to a putative causal gene 15
S6 Open target v2g results mapping trans-variant rs4665972 to a putative causal gene 16
S7 Open target v2g results mapping trans-variant rs112875651 to a putative causal gene 17
S8 Open target v2g results mapping trans-variant rs9591359 to a putative causal gene 18
S9 GWAS Catalog look-ups for putative causal genes for plasma SAP value 19
S10 The cis-Mendelian randomization results for the effects of one standard deviation higher plasma SAP value or plasma CRP concentration on dementia outcomes 20
S11 The cis-Mendelian randomization results for the effects of one standard deviation higher plasma SAP value on secondary outcomes 21
S12 The cis-Mendelian randomization results for the effects of one standard deviation higher plasma CRP concentration on secondary outcomes 23
S13 The cis-multivariable Mendelian randomization results for the effects of one standard deviation higher plasma SAP value or plasma CRP concentration 24
Acknowledgments:
A preprint version of this manuscript has been deposited at medRxiv. We thank Professor Nick Wareham and the EPIC-Norfolk team for generous provision of the plasma samples we used to validate the SomaLogic method for quantification of SAP concentration.
Funding:
AFS is supported by BHF grant PG/18/5033837, PG/22/10989, and the UCL BHF Research Accelerator AA/18/6/34223. This work was supported by the National Institutes of Health (USA) [R01 LM010098], as well as by the UKRI/NIHR Multimorbidity Fund Mechanism and Therapeutics Research Collaborative MR/V033867/1. Core support for the work of MBP was provided by the UK National Institute for Health Research (NIHR) Biomedical Research Centre and Unit Funding Scheme via the University College London Hospitals/University College London Biomedical Research Centre. MNR is supported by the National Institute for Health Research, (NIHR) University College London Hospitals (UCLH)/ University College London (UCL) Biomedical Research Centre (BRC).
Footnotes
Competing interests: AFS and CF have received funding from NewAmsterdam Pharma for unrelated work. MBP is the inventor on expired patents on SAP depletion by miridesap (CPHPC). GlaxoSmithKline’s abandoned patents on an experimental miridesap prodrug are assigned to UCL spinout company, Pentraxin Therapeutics Ltd, founded and directed by MBP. The other authors have no competing interests.
Ethics declaration:
The study exclusively uses information from aggregate data resources. As such this study is exempt from approval by an Ethics Committee or Institutional Review Board.
Data and materials availability:
All of the source data for this study are publicly available: SAP plasma value from DECODE (https://download.decode.is/form/folder/proteomics), AGES (https://doi.org/10.5281/zenodo.5711426), and Interval (https://www.ebi.ac.uk/gwas/studies/GCST90242796). Data on CRP concentration were obtained from: https://www.ebi.ac.uk/gwas/studies/GCST90029070. GWAS data were accessed for the following traits: Alzheimer’s disease (https://www.niagads.org/datasets/ng00075), Lewy body dementia (https://www.ebi.ac.uk/gwas/publications/33589841), osteoarthritis (https://www.ebi.ac.uk/gwas/publications/30664745), systemic lupus erythematosus (https://www.ebi.ac.uk/gwas/publications/26502338), idiopathic pulmonary fibrosis, (https://www.ebi.ac.uk/gwas/publications/33197388), systolic/diastolic blood pressure (https://www.ebi.ac.uk/gwas/publications/30224653), coronary heart disease (https://www.ebi.ac.uk/gwas/publications/36474045), type 2 diabetes (http://diagram-consortium.org/downloads.html, from Mahajan et al.), liver enzymes (https://www.ebi.ac.uk/gwas/publications/33972514 and https://www.ebi.ac.uk/gwas/publications/33547301), brain volume (https://ctg.cncr.nl/software/summary_statistics, from Jansen et al.), cerebral white matter hyperintensities (https://www.ebi.ac.uk/gwas/publications/26674333), circulating total tau values (https://www.ebi.ac.uk/gwas/publications/35396452). All data needed to evaluate the conclusions in the paper are present in the paper and the Supplementary Materials.
References and Notes
- 1.Nelson SR, Tennent GA, Sethi D, Gower PE, Ballardie FW, Amatayakul-Chantler S, Pepys MB. Serum amyloid P component in chronic renal failure and dialysis. Clin Chim Acta 1991;200:191–199. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins PN, Rossor MN, Gallimore JR, Miller B, Moore EG, Pepys MB. Concentration of serum amyloid P component in the CSF as a possible marker of cerebral amyloid deposits in Alzheimer’s disease. Biochem Biophys Res Commun 1994;201:722–726. [DOI] [PubMed] [Google Scholar]
- 3.Kolstoe SE, Ridha BH, Bellotti V, Wang N, Robinson CV, Crutch SJ, Keir G, Kukkastenvehmas R, Gallimore JR, Hutchinson WL, Hawkins PN, Wood SP, Rossor MN, Pepys MB. Molecular dissection of Alzheimer’s disease neuropathology by depletion of serum amyloid P component. Proc Natl Acad Sci U S A 2009;106:7619–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veszelka S, Laszy J, Pázmány T, Németh L, Obál I, Fábián L, Szabó G, Abrahám CS, Deli MA, Urbányi Z. Efflux transport of serum amyloid P component at the blood-brain barrier. Eur J Microbiol Immunol (Bp) 2013;3:281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pepys MB, Booth DR, Hutchinson WL, Gallimore JR, Collins IM, Hohenester E. Amyloid P component. A critical review. Amyloid 1997;4:274–295. [Google Scholar]
- 6.Pepys MB, Dyck RF, Beer FC de, Skinner M, Cohen AS. Binding of serum amyloid P-component (SAP) by amyloid fibrils. Clin Exp Immunol 1979;38:284–293. [PMC free article] [PubMed] [Google Scholar]
- 7.Tennent GA, Lovat LB, Pepys MB. Serum amyloid P component prevents proteolysis of the amyloid fibrils of Alzheimer disease and systemic amyloidosis. Proc Natl Acad Sci U S A 1995;92:4299–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamazaki H. Amyloid P component promotes aggregation of Alzheimer’s beta-amyloid peptide. Biochem Biophys Res Commun 1995;211:349–353. [DOI] [PubMed] [Google Scholar]
- 9.Mold M, Shrive AK, Exley C. Serum amyloid P component accelerates the formation and enhances the stability of amyloid fibrils in a physiologically significant under-saturated solution of amyloid-β42. J Alzheimers Dis 2012;29:875–881. [DOI] [PubMed] [Google Scholar]
- 10.Botto M, Hawkins PN, Bickerstaff MC, Herbert J, Bygrave AE, McBride A, Hutchinson WL, Tennent GA, Walport MJ, Pepys MB. Amyloid deposition is delayed in mice with targeted deletion of the serum amyloid P component gene. Nat Med 1997;3:855–859. [DOI] [PubMed] [Google Scholar]
- 11.Lilly’s Donanemab Significantly Slowed Cognitive and Functional Decline in Phase 3 Study of Early Alzheimer’s Disease | Eli Lilly and Company https://investor.lilly.com/news-releases/news-release-details/lillys-donanemab-significantly-slowed-cognitive-and-functional (26 May 2023)
- 12.FDA approves new antibody to slow Alzheimer’s disease, even as safety concerns linger | Science | AAAS https://www.science.org/content/article/fda-approves-new-antibody-slow-alzheimer-s-disease-even-safety-concerns-linger (15 June 2023)
- 13.Urbányi Z, Lakics V, Erdö SL. Serum amyloid P component-induced cell death in primary cultures of rat cerebral cortex. Eur J Pharmacol 1994;270:375–378. [DOI] [PubMed] [Google Scholar]
- 14.Duong T, Acton PJ, Johnson RA. The in vitro neuronal toxicity of pentraxins associated with Alzheimer’s disease brain lesions. Brain Res 1998;813:303–312. [DOI] [PubMed] [Google Scholar]
- 15.Urbányi Z, László L, Tomasi TB, Tóth E, Mekes E, Sass M, Pázmány T. Serum amyloid P component induces neuronal apoptosis and beta-amyloid immunoreactivity. Brain Res 2003;988:69–77. [DOI] [PubMed] [Google Scholar]
- 16.Pisalyaput K, Tenner AJ. Complement component C1q inhibits beta-amyloid- and serum amyloid P-induced neurotoxicity via caspase- and calpain-independent mechanisms. J Neurochem 2008;104:696–707. [DOI] [PubMed] [Google Scholar]
- 17.Urbányi Z, Sass M, Laszy J, Takács V, Gyertyán I, Pázmány T. Serum amyloid P component induces TUNEL-positive nuclei in rat brain after intrahippocampal administration. Brain Res 2007;1145:221–226. [DOI] [PubMed] [Google Scholar]
- 18.Ellmerich S, Taylor GW, Richardson CD, Minett T, Schmidt AF, Brayne C, Matthews FE, Ince PG, Wharton SB, Pepys MB, Cognitive Function and Ageing Study. Dementia in the older population is associated with neocortex content of serum amyloid P component. Brain Commun 2021;3:fcab225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montagne A, Barnes SR, Sweeney MD, Halliday MR, Sagare AP, Zhao Z, Toga AW, Jacobs RE, Liu CY, Amezcua L, Harrington MG, Chui HC, Law M, Zlokovic BV. Blood-brain barrier breakdown in the aging human hippocampus. Neuron 2015;85:296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pepys MB. The Pentraxins 1975–2018: Serendipity, Diagnostics and Drugs. Front Immunol 2018;9:2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepys MB, Dash AC, Markham RE, Thomas HC, Williams BD, Petrie A. Comparative clinical study of protein SAP (amyloid P component) and C-reactive protein in serum. Clin Exp Immunol 1978;32:119–124. [PMC free article] [PubMed] [Google Scholar]
- 22.Jensson O, Bjornsson A, Arnason A, Birgisdottir B, Pepys MB Serum amyloid P-component and C-reactive protein in serum of healthy Icelanders and members of an Icelandic family with macroglobulinaemia. Acta medica Scandinavica 1982;211. [DOI] [PubMed] [Google Scholar]
- 23.Nybo M, Olsen H, Jeune B, Andersen-Ranberg K, Holm Nielsen E, Svehag SE. Increased plasma concentration of serum amyloid P component in centenarians with impaired cognitive performance. Dement Geriatr Cogn Disord 1998;9:126–129. [DOI] [PubMed] [Google Scholar]
- 24.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Genomic atlas of the human plasma proteome. Nature 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, Gunnarsdottir K, Helgason A, Oddsson A, Halldorsson BV, Jensson BO, Zink F, Halldorsson GH, Masson G, Arnadottir GA, Katrinardottir H, Juliusson K, Magnusson MK, Magnusson OT, Fridriksdottir R, Saevarsdottir S, Gudjonsson SA, Stacey SN, Rognvaldsson S, Eiriksdottir T, Olafsdottir TA, Steinthorsdottir V, Tragante V, Ulfarsson MO, Stefansson H, Jonsdottir I, Holm H, Rafnar T, Melsted P, Saemundsdottir J, Norddahl GL, Lund SH, Gudbjartsson DF, Thorsteinsdottir U, Stefansson K. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet 2021;53:1712–1721. [DOI] [PubMed] [Google Scholar]
- 26.Gudjonsson A, Gudmundsdottir V, Axelsson GT, Gudmundsson EF, Jonsson BG, Launer LJ, Lamb JR, Jennings LL, Aspelund T, Emilsson V, Gudnason V. A genome-wide association study of serum proteins reveals shared loci with common diseases. Nat Commun 2022;13:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt AF, Hingorani AD, Finan C. Human Genomics and Drug Development. Cold Spring Harb Perspect Med 2021:a039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt AF, Finan C, Gordillo-Marañón M, Asselbergs FW, Freitag DF, Patel RS, Tyl B, Chopade S, Faraway R, Zwierzyna M, Hingorani AD. Genetic drug target validation using Mendelian randomisation. Nat Commun 2020;11:3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swerdlow DI, Hingorani AD, Casas JP, Consortium IMR, The Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium, Swerdlow DI, Holmes MV, Kuchenbaecker KB, Engmann JEL, Shah T, Sofat R, Guo Y, Chung C, Peasey A, Pfister R, Mooijaart SP, Ireland HA, Leusink M, Langenberg C, Li K, Palmen J, Howard P, Cooper JA, Drenos F, Hardy J, Nalls MA, Li YR, Lowe G, Stewart M, Bielinski SJ, Peto J, Timpson NJ, Gallacher J, Dunlop M, Houlston R, Tomlinson I, Tzoulaki I, Luan J, Boer JMA, Forouhi NG, Onland-Moret NC, Van Der Schouw YT, Schnabel R, Hubacek JA, Kubinova R, Baceviciene M, Tamosiunas A, Pajak A, Topor-Madry R, Malyutina S, Baldassarre D, Sennblad B, Tremoli E, De Faire U, Ferrucci L, Bandenelli S, Tanaka T, Meschia JF, Singleton A, Navis G, Leach IM, Bakker SJL, Gansevoort RT, Ford I, Epstein SE, Burnett MS, Devaney JM, Jukema JW, Westendorp RGJ, De Borst GJ, Van Der Graaf Y, De Jong PA, Maitland-Van Der Zee AH, Klungel OH, De Boer A, Doevendans PA, Stephens JW, Eaton CB, Robinson JG, Manson JE, Fowkes FGR, Frayling TM, Price J, Whincup PH, Morris RW, Lawlor DA, Smith GD, Ben-Shlomo Y, Redline S, Lange LA, Kumari M, Wareham NJ, Verschuren WMM, Benjamin EJ, Whittaker JC, Hamsten A, Dudbridge F, Delaney JAC, Wong A, Kuh D, Hardy R, Castillo BA, Connolly JJ, Van Der Harst P, Brunner EJ, Marmot MG, Wassel CL, Humphries SE, Talmud PJ, Kivimaki M, Asselbergs FW, Voevoda M, Bobak M, Pikhart H, Wilson JG, Hakonarson H, Reiner AP, Keating BJ, Sattar N, Hingorani AD, Casas JP, Consortium IMR, The Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium, Swerdlow DI, Holmes MV, Kuchenbaecker KB, Engmann JEL, Shah T, Sofat R, Guo Y, Chung C, Peasey A, Pfister R, Mooijaart SP, Ireland HA, Leusink M, Langenberg C, Li K, Palmen J, Howard P, Cooper JA, Drenos F, Hardy J, Nalls MA, Li YR, Lowe G, Stewart M, Bielinski SJ, Peto J, Timpson NJ, Gallacher J, Dunlop M, Houlston R, Tomlinson I, Tzoulaki I, Luan J, Boer JMA, Forouhi NG, Onland-Moret NC, Van Der Schouw YT, Schnabel R, Hubacek JA, Kubinova R, Baceviciene M, Tamosiunas A, Pajak A, Topor-Madry R, Malyutina S, Baldassarre D, Sennblad B, Tremoli E, De Faire U, Ferrucci L, Bandenelli S, Tanaka T, Meschia JF, Singleton A, Navis G, Leach IM, Bakker SJL, Gansevoort RT, Ford I, Epstein SE, Burnett MS, Devaney JM, Jukema JW, Westendorp RGJ, De Borst GJ, Van Der Graaf Y, De Jong PA, Maitland-Van Der Zee AH, Klungel OH, De Boer A, Doevendans PA, Stephens JW, Eaton CB, Robinson JG, Manson JE, Fowkes FGR, Frayling TM, Price J, Whincup PH, Morris RW, Lawlor DA, Smith GD, Ben-Shlomo Y, Redline S, Lange LA, Kumari M, Wareham NJ, Verschuren WMM, Benjamin EJ, Whittaker JC, Hamsten A, Dudbridge F, Delaney JAC, Wong A, Kuh D, Hardy R, Castillo BA, Connolly JJ, Van Der Harst P, Brunner EJ, Marmot MG, Wassel CL, Humphries SE, Talmud PJ, Kivimaki M, Asselbergs FW, Voevoda M, Bobak M, Pikhart H, Wilson JG, Hakonarson H, Reiner AP, Keating BJ, Sattar N, Hingorani AD, Casas JP. The interleukin-6 receptor as a target for prevention of coronary heart disease: A mendelian randomisation analysis. The Lancet 2012;379:1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cupido AJ, Reeskamp LF, Hingorani AD, Finan C, Asselbergs FW, Hovingh GK, Schmidt AF. Joint Genetic Inhibition of PCSK9 and CETP and the Association With Coronary Artery Disease: A Factorial Mendelian Randomization Study. JAMA Cardiol 2022;7:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt AF, Hunt NB, Gordillo-Marañón M, Charoen P, Drenos F, Kivimaki M, Lawlor DA, Giambartolomei C, Papacosta O, Chaturvedi N, Bis JC, O’Donnell CJ, Wannamethee G, Wong A, Price JF, Hughes AD, Gaunt TR, Franceschini N, Mook-Kanamori DO, Zwierzyna M, Sofat R, Hingorani AD, Finan C. Cholesteryl ester transfer protein (CETP) as a drug target for cardiovascular disease. Nat Commun 2021;12:5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng J, Haberland V, Baird D, Walker V, Haycock PC, Hurle MR, Gutteridge A, Erola P, Liu Y, Luo S, Robinson J, Richardson TG, Staley JR, Elsworth B, Burgess S, Sun BB, Danesh J, Runz H, Maranville JC, Martin HM, Yarmolinsky J, Laurin C, Holmes MV, Liu JZ, Estrada K, Santos R, McCarthy L, Waterworth D, Nelson MR, Smith GD, Butterworth AS, Hemani G, Scott RA, Gaunt TR. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet 2020;52:1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest 2003;111:1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Xia K, Ahn M, Jha SC, Blanchett R, Crowley JJ, Szatkiewicz JP, Zou F, Zhu H, Styner M, Gilmore JH, Knickmeyer RC. Genome-Wide Association Analysis of Neonatal White Matter Microstructure. Cereb Cortex 2021;31:933–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattsson N, Zetterberg H, Janelidze S, Insel PS, Andreasson U, Stomrud E, Palmqvist S, Baker D, Tan Hehir CA, Jeromin A, Hanlon D, Song L, Shaw LM, Trojanowski JQ, Weiner MW, Hansson O, Blennow K. Plasma tau in Alzheimer disease. Neurology 2016;87:1827–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts JA, Varma VR, Candia J, Tanaka T, Ferrucci L, Bennett DA, Thambisetty M. Unbiased proteomics and multivariable regularized regression techniques identify SMOC1, NOG, APCS, and NTN1 in an Alzheimer’s disease brain proteomic signature. npj Aging 2023;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeung CHC, Lau KWD, Au Yeung SL, Schooling CM. Amyloid, tau and risk of Alzheimer’s disease: a Mendelian randomization study. Eur J Epidemiol 2021;36:81–88. [DOI] [PubMed] [Google Scholar]
- 38.Altman DG, Bland JM. Statistics notes: Absence of evidence is not evidence of absence. BMJ 1995;311:485–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, Lagemaat LN van de, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, David Daly B, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SGN, Jones AR. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012;489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beer FC de Baltz ML, Holford S, Feinstein A, Pepys MB. Fibronectin and C4-binding protein are selectively bound by aggregated amyloid P component. J Exp Med 1981;154:1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pepys MB, Hawkins PN. Compounds inhibiting the binding of SAP for treating osteoarthritis (U.S. Patent No. 7659299B2) 2010.
- 42.Breathnach SM, Kofler H, Sepp N, Ashworth J, Woodrow D, Pepys MB, Hintner H. Serum amyloid P component binds to cell nuclei in vitro and to in vivo deposits of extracellular chromatin in systemic lupus erythematosus. J Exp Med 1989;170:1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spector TD, Hart DJ, Nandra D, Doyle DV, Mackillop N, Gallimore JR, Pepys MB. Low-level increases in serum C-reactive protein are present in early osteoarthritis of the knee and predict progressive disease. Arthritis Rheum 1997;40:723–727. [DOI] [PubMed] [Google Scholar]
- 44.Pepys MB, Lanham JG, Beer FC de. C-reactive Protein in SLE. Clin in Rheum Dis 1982;8:91–103. [PubMed] [Google Scholar]
- 45.Pepys MB, Herbert J, Hutchinson WL, Tennent GA, Lachmann HJ, Gallimore JR, Lovat LB, Bartfai T, Alanine A, Hertel C, Hoffmann T, Jakob-Roetne R, Norcross RD, Kemp JA, Yamamura K, Suzuki M, Taylor GW, Murray S, Thompson D, Purvis A, Kolstoe S, Wood SP, Hawkins PN. Targeted pharmacological depletion of serum amyloid P component for treatment of human amyloidosis. Nature 2002;417:254–259. [DOI] [PubMed] [Google Scholar]
- 46.Gillmore JD, Tennent GA, Hutchinson WL, Gallimore JR, Lachmann HJ, Goodman HJB, Offer M, Millar DJ, Petrie A, Hawkins PN, Pepys MB. Sustained pharmacological depletion of serum amyloid P component in patients with systemic amyloidosis. Br J Haematol 2010;148:760–767. [DOI] [PubMed] [Google Scholar]
- 47.Al-Shawi R, Tennent GA, Millar DJ, Richard-Londt A, Brandner S, Werring DJ, Simons JP, Pepys MB. Pharmacological removal of serum amyloid P component from intracerebral plaques and cerebrovascular Aβ amyloid deposits in vivo. Open Biol 2016;6:150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yates A, Beal K, Keenan S, McLaren W, Pignatelli M, Ritchie GRS, Ruffier M, Taylor K, Vullo A, Flicek P. The Ensembl REST API: Ensembl Data for Any Language. Bioinformatics 2015;31:143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mountjoy E, Schmidt EM, Carmona M, Schwartzentruber J, Peat G, Miranda A, Fumis L, Hayhurst J, Buniello A, Karim MA, Wright D, Hercules A, Papa E, Fauman EB, Barrett JC, Todd JA, Ochoa D, Dunham I, Ghoussaini M. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat Genet 2021;53:1527–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, Lee SJ van der, Amlie-Wolf A, Bellenguez C, Frizatti A, Chouraki V, Martin ER, Sleegers K, Badarinarayan N, Jakobsdottir J, Hamilton-Nelson KL, Moreno-Grau S, Olaso R, Raybould R, Chen Y, Kuzma AB, Hiltunen M, Morgan T, Ahmad S, Vardarajan BN, Epelbaum J, Hoffmann P, Boada M, Beecham GW, Garnier J-G, Harold D, Fitzpatrick AL, Valladares O, Moutet M-L, Gerrish A, Smith AV, Qu L, Bacq D, Denning N, Jian X, Zhao Y, Del Zompo M, Fox NC, Choi S-H, Mateo I, Hughes JT, Adams HH, Malamon J, Sanchez-Garcia F, Patel Y, Brody JA, Dombroski BA, Naranjo MCD, Daniilidou M, Eiriksdottir G, Mukherjee S, Wallon D, Uphill J, Aspelund T, Cantwell LB, Garzia F, Galimberti D, Hofer E, Butkiewicz M, Fin B, Scarpini E, Sarnowski C, Bush WS, Meslage S, Kornhuber J, White CC, Song Y, Barber RC, Engelborghs S, Sordon S, Voijnovic D, Adams PM, Vandenberghe R, Mayhaus M, Cupples LA, Albert MS, De Deyn PP, Gu W, Himali JJ, Beekly D, Squassina A, Hartmann AM, Orellana A, Blacker D, Rodriguez-Rodriguez E, Lovestone S, Garcia ME, Doody RS, Munoz-Fernadez C, Sussams R, Lin H, Fairchild TJ, Benito YA, Holmes C, Karamujić-Čomić H, Frosch MP, Thonberg H, Maier W, Roshchupkin G, Ghetti B, Giedraitis V, Kawalia A, Li S, Huebinger RM, Kilander L, Moebus S, Hernández I, Kamboh MI, Brundin R, Turton J, Yang Q, Katz MJ, Concari L, Lord J, Beiser AS, Keene CD, Helisalmi S, Kloszewska I, Kukull WA, Koivisto AM, Lynch A, Tarraga L, Larson EB, Haapasalo A, Lawlor B, Mosley TH, Lipton RB, Solfrizzi V, Gill M, Longstreth WT, Montine TJ, Frisardi V, Diez-Fairen M, Rivadeneira F, Petersen RC, Deramecourt V, Alvarez I, Salani F, Ciaramella A, Boerwinkle E, Reiman EM, Fievet N, Rotter JI, Reisch JS, Hanon O, Cupidi C, Andre Uitterlinden AG, Royall DR, Dufouil C, Maletta RG, Rojas I de, Sano M, Brice A, Cecchetti R, George-Hyslop PS, Ritchie K, Tsolaki M, Tsuang DW, Dubois B, Craig D, Wu C-K, Soininen H, Avramidou D, Albin RL, Fratiglioni L, Germanou A, Apostolova LG, Keller L, Koutroumani M, Arnold SE, Panza F, Gkatzima O, Asthana S, Hannequin D, Whitehead P, Atwood CS, Caffarra P, Hampel H, Quintela I, Carracedo Á, Lannfelt L, Rubinsztein DC, Barnes LL, Pasquier F, Frölich L, Barral S, McGuinness B, Beach TG, Johnston JA, Becker JT, Passmore P, Bigio EH, Schott JM, Bird TD, Warren JD, Boeve BF, Lupton MK, Bowen JD, Proitsi P, Boxer A, Powell JF, Burke JR, Kauwe JSK, Burns JM, Mancuso M, Buxbaum JD, Bonuccelli U, Cairns NJ, McQuillin A, Cao C, Livingston G, Carlson CS, Bass NJ, Carlsson CM, Hardy J, Carney RM, Bras J, Carrasquillo MM, Guerreiro R, Allen M, Chui HC, Fisher E, Masullo C, Crocco EA, DeCarli C, Bisceglio G, Dick M, Ma L, Duara R, Graff-Radford NR, Evans DA, Hodges A, Faber KM, Scherer M, Fallon KB, Riemenschneider M, Fardo DW, Heun R, Farlow MR, Kölsch H, Ferris S, Leber M, Foroud TM, Heuser I, Galasko DR, Giegling I, Gearing M, Hüll M, Geschwind DH, Gilbert JR, Morris J, Green RC, Mayo K, Growdon JH, Feulner T, Hamilton RL, Harrell LE, Drichel D, Honig LS, Cushion TD, Huentelman MJ, Hollingworth P, Hulette CM, Hyman BT, Marshall R, Jarvik GP, Meggy A, Abner E, Menzies GE, Jin L-W, Leonenko G, Real LM, Jun GR, Baldwin CT, Grozeva D, Karydas A, Russo G, Kaye JA, Kim R, Jessen F, Kowall NW, Vellas B, Kramer JH, Vardy E, LaFerla FM, Jöckel K-H, Lah JJ, Dichgans M, Leverenz JB, Mann D, Levey AI, Pickering-Brown S, Lieberman AP, Klopp N, Lunetta KL, Wichmann H-E, Lyketsos CG, Morgan K, Marson DC, Brown K, Martiniuk F, Medway C, Mash DC, Nöthen MM, Masliah E, Hooper NM, McCormick WC, Daniele A, McCurry SM, Bayer A, McDavid AN, Gallacher J, McKee AC, Bussche H van den, Mesulam M, Brayne C, Miller BL, Riedel-Heller S, Miller CA, Miller JW, Al-Chalabi A, Morris JC, Shaw CE, Myers AJ, Wiltfang J, O’Bryant S, Olichney JM, Alvarez V, Parisi JE, Singleton AB, Paulson HL, Collinge J, Perry WR, Mead S, Peskind E, Cribbs DH, Rossor M, Pierce A, Ryan NS, Poon WW, Nacmias B, Potter H, Sorbi S, Quinn JF, Sacchinelli E, Raj A, Spalletta G, Raskind M, Caltagirone C, Bossù P, Orfei MD, Reisberg B, Clarke R, Reitz C, Smith AD, Ringman JM, Warden D, Roberson ED, Wilcock G, Rogaeva E, Bruni AC, Rosen HJ, Gallo M, Rosenberg RN, Ben-Shlomo Y, Sager MA, Mecocci P, Saykin AJ, Pastor P, Cuccaro ML, Vance JM, Schneider JA, Schneider LS, Slifer S, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tang M, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu C-E, Yu L, Saba Y, Pilotto A, Bullido MJ, Peters O, Crane PK, Bennett D, Bosco P, Coto E, Boccardi V, De Jager PL, Lleo A, Warner N, Lopez OL, Ingelsson M, Deloukas P, Cruchaga C, Graff C, Gwilliam R, Fornage M, Goate AM, Sanchez-Juan P, Kehoe PG, Amin N, Ertekin-Taner N, Berr C, Debette S, Love S, Launer LJ, Younkin SG, Dartigues J-F, Corcoran C, Ikram MA, Dickson DW, Nicolas G, Campion D, Tschanz J, Schmidt H, Hakonarson H, Clarimon J, Munger R, Schmidt R, Farrer LA, Van Broeckhoven C, C O’Donovan M, DeStefano AL, Jones L, Haines JL, Deleuze J-F, Owen MJ, Gudnason V, Mayeux R, Escott-Price V, Psaty BM, Ramirez A, Wang L-S, Ruiz A, Duijn CM van, Holmans PA, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Lambert J-C, Pericak-Vance MA, Alzheimer Disease Genetics Consortium (ADGC), European Alzheimer’s Disease Initiative (EADI), Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE ),, Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES),. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet 2019;51:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chia R, Sabir MS, Bandres-Ciga S, Saez-Atienzar S, Reynolds RH, Gustavsson E, Walton RL, Ahmed S, Viollet C, Ding J, Makarious MB, Diez-Fairen M, Portley MK, Shah Z, Abramzon Y, Hernandez DG, Blauwendraat C, Stone DJ, Eicher J, Parkkinen L, Ansorge O, Clark L, Honig LS, Marder K, Lemstra A, St George-Hyslop P, Londos E, Morgan K, Lashley T, Warner TT, Jaunmuktane Z, Galasko D, Santana I, Tienari PJ, Myllykangas L, Oinas M, Cairns NJ, Morris JC, Halliday GM, Van Deerlin VM, Trojanowski JQ, Grassano M, Calvo A, Mora G, Canosa A, Floris G, Bohannan RC, Brett F, Gan-Or Z, Geiger JT, Moore A, May P, Krüger R, Goldstein DS, Lopez G, Tayebi N, Sidransky E, American Genome Center, Norcliffe-Kaufmann L, Palma J-A, Kaufmann H, Shakkottai VG, Perkins M, Newell KL, Gasser T, Schulte C, Landi F, Salvi E, Cusi D, Masliah E, Kim RC, Caraway CA, Monuki ES, Brunetti M, Dawson TM, Rosenthal LS, Albert MS, Pletnikova O, Troncoso JC, Flanagan ME, Mao Q, Bigio EH, Rodríguez-Rodríguez E, Infante J, Lage C, González-Aramburu I, Sanchez-Juan P, Ghetti B, Keith J, Black SE, Masellis M, Rogaeva E, Duyckaerts C, Brice A, Lesage S, Xiromerisiou G, Barrett MJ, Tilley BS, Gentleman S, Logroscino G, Serrano GE, Beach TG, McKeith IG, Thomas AJ, Attems J, Morris CM, Palmer L, Love S, Troakes C, Al-Sarraj S, Hodges AK, Aarsland D, Klein G, Kaiser SM, Woltjer R, Pastor P, Bekris LM, Leverenz JB, Besser LM, Kuzma A, Renton AE, Goate A, Bennett DA, Scherzer CR, Morris HR, Ferrari R, Albani D, Pickering-Brown S, Faber K, Kukull WA, Morenas-Rodriguez E, Lleó A, Fortea J, Alcolea D, Clarimon J, Nalls MA, Ferrucci L, Resnick SM, Tanaka T, Foroud TM, Graff-Radford NR, Wszolek ZK, Ferman T, Boeve BF, Hardy JA, Topol EJ, Torkamani A, Singleton AB, Ryten M, Dickson DW, Chiò A, Ross OA, Gibbs JR, Dalgard CL, Traynor BJ, Scholz SW. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet 2021;53:294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fauman EB, Hyde C. An optimal variant to gene distance window derived from an empirical definition of cis and trans protein QTLs. BMC Bioinformatics 2022;23:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burgess S, Zuber V, Valdes-Marquez E, Sun BB, Hopewell JC. Mendelian randomization with fine-mapped genetic data: Choosing from large numbers of correlated instrumental variables. Genetic Epidemiology 2017;41:714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgess S, Thompson SG. Avoiding bias from weak instruments in mendelian randomization studies. Int J Epidemiol 2011;40:755–764. [DOI] [PubMed] [Google Scholar]
- 57.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. 2016;40:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bowden J, Smith GD, Burgess S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowden J, Del Greco M F, Minelli C, Davey Smith G, Sheehan N, Thompson J, Del Greco M F, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Statistics in medicine 2017;36:1783–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bowden J, Spiller W, Del Greco M F, Sheehan N, Thompson J, Minelli C, Davey Smith G . Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol 2018;47:1264–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rücker G, Schwarzer G, Carpenter JR, Binder H, Schumacher M. Treatment-effect estimates adjusted for small-study effects via a limit meta-analysis. Biostatistics 2011;12:122–142. [DOI] [PubMed] [Google Scholar]
- 62.Said S, Pazoki R, Karhunen V, Võsa U, Ligthart S, Bodinier B, Koskeridis F, Welsh P, Alizadeh BZ, Chasman DI, Sattar N, Chadeau-Hyam M, Evangelou E, Jarvelin M-R, Elliott P, Tzoulaki I, Dehghan A. Genetic analysis of over half a million people characterises C-reactive protein loci. Nat Commun 2022;13:2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burgess S, Dudbridge F, Thompson SG. Re: “Multivariable Mendelian Randomization: The Use of Pleiotropic Genetic Variants to Estimate Causal Effects”. Am J Epidemiol 2015;181:290–291. [DOI] [PubMed] [Google Scholar]
- 64.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genetic Epidemiology 2008;32:227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jung I, Schmitt A, Diao Y, Lee AJ, Liu T, Yang D, Tan C, Eom J, Chan M, Chee S, Chiang Z, Kim C, Masliah E, Barr CL, Li B, Kuan S, Kim D, Ren B. A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat Genet 2019;51:1442–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Várnai C, Thiecke MJ, Burden F, Farrow S, Cutler AJ, Rehnström K, Downes K, Grassi L, Kostadima M, Freire-Pritchett P, Wang F, BLUEPRINT Consortium, Stunnenberg HG, Todd JA, Zerbino DR, Stegle O, Ouwehand WH, Frontini M, Wallace C, Spivakov M, Fraser P. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 2016;167:1369–1384.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures
S1 Correlation between SomaLogic values and immunoassay for actual plasma SAP concentration 2
S2 Locus view plots 6
S3 GWAS Catalog look-ups 9
Tables
S1 Lead variants for serum amyloid P component (SAP) plasma value 11
S2 SAP lead variant associations with plasma C-reactive protein (CRP) concentration 12
S3 Open target v2g results mapping trans-variant rs2808467 to a putative causal gene 13
S4 Open target v2g results mapping trans-variant rs165316 to a putative causal gene 14
S5 Open target v2g results mapping trans-variant rs10188292 to a putative causal gene 15
S6 Open target v2g results mapping trans-variant rs4665972 to a putative causal gene 16
S7 Open target v2g results mapping trans-variant rs112875651 to a putative causal gene 17
S8 Open target v2g results mapping trans-variant rs9591359 to a putative causal gene 18
S9 GWAS Catalog look-ups for putative causal genes for plasma SAP value 19
S10 The cis-Mendelian randomization results for the effects of one standard deviation higher plasma SAP value or plasma CRP concentration on dementia outcomes 20
S11 The cis-Mendelian randomization results for the effects of one standard deviation higher plasma SAP value on secondary outcomes 21
S12 The cis-Mendelian randomization results for the effects of one standard deviation higher plasma CRP concentration on secondary outcomes 23
S13 The cis-multivariable Mendelian randomization results for the effects of one standard deviation higher plasma SAP value or plasma CRP concentration 24