Abstract
Career athletes, active military, and head trauma victims are at increased risk for mild repetitive traumatic brain injury (rTBI), a condition that contributes to the development of epilepsy and neurodegenerative diseases. Standard clinical imaging fails to identify rTBI-induced lesions, and novel non-invasive methods are needed. Here, we evaluated if hyperpolarized 13C magnetic resonance spectroscopic imaging (HP 13C MRSI) could detect long-lasting changes in brain metabolism 3.5 months post-injury in a rTBI mouse model. Our results show that this metabolic imaging approach can detect changes in cortical metabolism at that timepoint, whereas multimodal MR imaging did not detect any structural or contrast alterations. Using Machine Learning, we further show that HP 13C MRSI parameters can help classify rTBI vs. Sham and predict long-term rTBI-induced behavioral outcomes. Altogether, our study demonstrates the potential of metabolic imaging to improve detection, classification and outcome prediction of previously undetected rTBI.
INTRODUCTION
Individuals subject to frequent concussions such as career athletes (e.g. football players, boxers), accidental head trauma victims, domestic abuse victims, or active military are some of the population at risk for mild repetitive traumatic brain injury (rTBI). Indeed, rTBI is being steadily recognized as a risk factor for the development of epilepsy and neurodegenerative diseases, particularly chronic traumatic encephalopathy (CTE)1, 2, 3. However, to date, non-invasive diagnostic biomarkers of rTBI are lacking. In particular, clinical computed tomography (CT) and magnetic resonance imaging (MRI), the standard imaging methods for trauma patients, are unable to detect rTBI-induced pathology4, 5, 6. This lack of imaging techniques hampers proper diagnosis and appropriate clinical care, as a result new approaches are critically needed.
To further our understanding of rTBI, several models were developed over the past years7, 8. The closed-head impact model of engineered rotational acceleration (CHIMERA) device was designed to deliver multiple subconcussive mild TBI in a controlled and reproducible manner9. CHIMERA-induced rTBI has been shown to lead to reproducible pathological and behavioral changes up to several months following impacts10, mirroring the long-term effects of rTBI seen in the clinic (review by McNamara et al.11). Only a few studies have investigated the use of magnetic resonance imaging (MRI) to detect CHIMERA-induced rTBI. Diffusion MRI, which is sensitive to water diffusion in tissue and changes in tissue microstructure, detected subtle differences in rTBI animals at 7 days following injury and in the optic tract, brachium of the superior colliculus, corpus callosum and hippocampus regions12, 13; however, long-lasting changes were not studied. T2-weighted MRI, which is sensitive to changes in tissue microstructure, edema, and myelination, did not detect signs of brain injury at 7 days and 40 days post-injury10, 12. All these studies are in line with clinical findings, and further highlight the need for more sensitive approaches to detect and monitor long-term brain changes after rTBI.
Metabolic impairment following TBI has been well documented in patients and animal models in the hours following TBI using 13C-labelled substrates infusion and metabolomics approaches (reviews by Jalloh, Dermers-Marcil, and Carpenter14, 15, 16). Notably, cerebral microdialysis studies have identified that the lactate / pyruvate ratio (Lac/Pyr) parameter is associated with poor outcome17, 18. However, as cerebral microdialysis is an invasive method, its use at chronic timepoints following TBI, or in closed head injury and concussion is not feasible. Hyperpolarized 13C magnetic resonance spectroscopic imaging (HP 13C MRSI) is a unique technology that allows to measure metabolic fluxes in vivo, and to compute lactate / pyruvate ratio values as well. HP 13C MRSI applications have been extensively described in the oncology field19, 20, and its use is emerging to probe brain metabolism in health and diseases21. HP 13C MRSI of [1-13C]pyruvate enables to monitor the conversion of this key metabolite into its product(s) [1-13C]lactate and/or [13C]bicarbonate in the brain, which provides unprecedented metabolic information22, 23. Prior studies of moderate TBI have shown changes in the HP 13C lactate / pyruvate ratio (HP 13C Lac/Pyr), and HP 13C bicarbonate / lactate ratio at early timepoints (4 hours up to 7 days) following injury in preclinical models and in patients24, 25, 26, demonstrating the potential value of this technique. Furthermore, recent studies have combined injection of HP [1-13C]pyruvate with HP [13C]urea, a metabolically inactive probe, to simultaneously evaluate the pyruvate to lactate flux and tissue perfusion, respectively27, 28, 29, 30, 31. However, it remains to be determined whether HP 13C MRSI of HP [1–13C]pyruvate might be useful to the detection of rTBI at chronic timepoints, and inform on potential mechanism of metabolic injury. Furthermore, co-injection of HP [1-13C]pyruvate and HP [13C]urea has never been tested in any TBI model or patient.
Here, we questioned if advanced imaging approaches that have never been applied to the study of rTBI could detect long-lasting alterations following rTBI in the CHIMERA model. In addition to the above described HP 13C MRSI of HP [1-13C]pyruvate and HP [13C]urea, we also evaluated Susceptibility-weighted imaging (SWI) and mapping. SWI is an MRI method particularly sensitive to iron that can inform on venous deoxygenated blood and iron deposition in tissue, and which has proven very valuable to identify microbleeds in TBI, but has not yet been investigated in the CHIMERA model32, 33. Recent reports have highlighted the potential of mapping to detect oxidative stress in the rodent brain34, and thus this technique holds great potential to detect the production of reactive oxygen species that may occur following diffuse axonal injury and inflammatory processes observed in the CHIMERA model11.
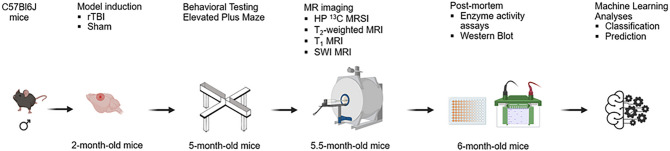
As shown in Fig. 1, we induced rTBI in two-month old male mice using the CHIMERA apparatus, tested risk-taking behavior 3 months post rTBI, and performed four MRI-based scans. We investigated the potential of 13C MRSI of HP [1-13C]pyruvate and [13C]urea to detect metabolic and tissue perfusion impairment, T2-weighted MRI to assess structural changes (as clinical standard of MR imaging), -mapping to evaluate tissue microstructure alterations and oxidative stress, and SWI MRI to detect changes in tissue microstructure, microbleed and tissue oxygenation following rTBI. Last, brain tissue was collected to evaluate changes in enzymes activity and transporter protein expression. Given the multidimensional nature of the data, we used a Machine Learning (ML) approach to identify how measured parameters could best predict changes in risk-taking behavior and HP 13C MRSI.
Figure 1. Study outline.
Experimental timeline of the study. Two-month old male mice received a rTBI using the CHIMERA device or underwent a Sham procedure (no impact). Risk-taking behavior was evaluated at 3 months post-injury using the Elevated Plus Maze. MR imaging was performed 3.5 months after Sham or rTBI, and included HP 13C MRSI, T2-weighted MRI, mapping MRI, and SWI MRI. Tissue was collected 4 months after Sham or rTBI procedures to evaluate PDH and LDH activities, and expression of MCT1 and MCT4. ML analyses methods were used to identify the best classifiers between rTBI and Sham, and the best predictors of the risk-taking behavior and HP 13C Lac/Pyr in the cortex.
RESULTS
HP 13C MRSI detects long-lasting metabolic alterations following rTBI
We investigated whether HP 13C MRSI could be used as a non-invasive tool to detect rTBI-induced long-lasting changes, and specifically to question whether metabolic alterations are present at chronic time points following injury.
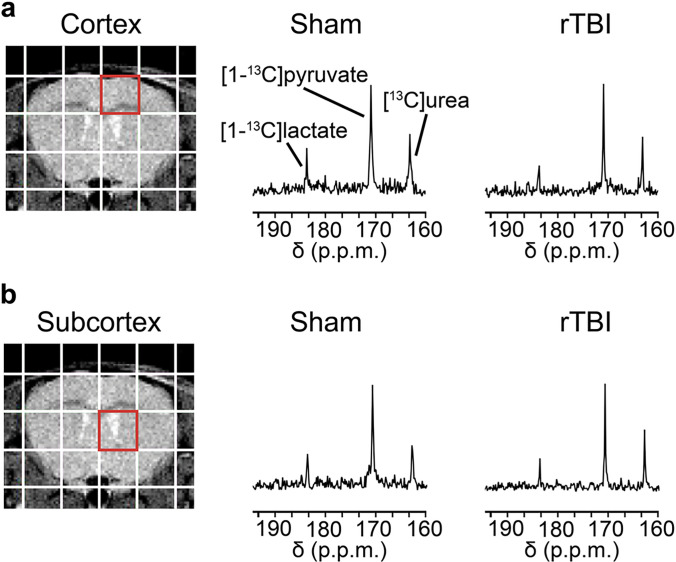
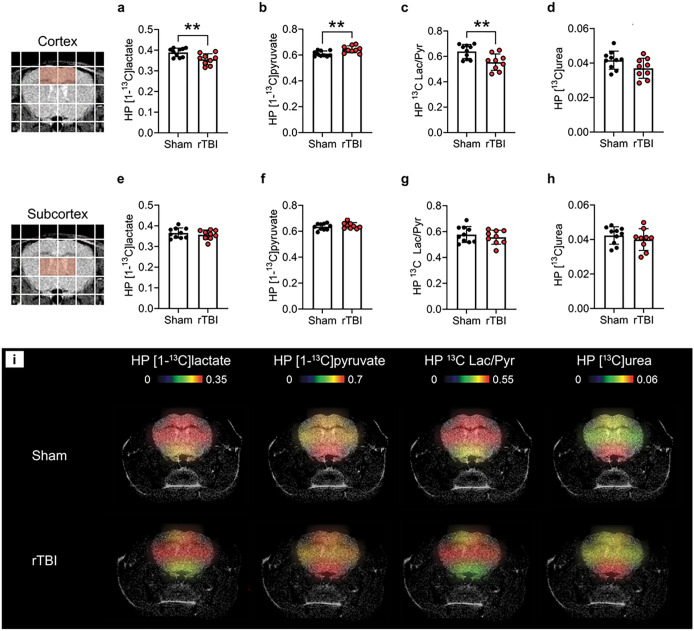
Following intravenous co-injection of HP [1-13C]pyruvate and HP [13C]urea, we observed signals from both substrates, as well as from the metabolic product HP [1-13C]lactate in the cortex (Fig. 2.a) and subcortex (Fig. 2.b) of Sham and rTBI mice at 3.5 months post-injury. Upon quantification, we observed significant metabolic differences between the cortex of rTBI compared to Sham mice: HP [1-13C]lactate levels were 1.09 fold lower in rTBI (Fig. 3.a, p = 0.0073), HP [1-13C]pyruvate levels were 1.05 fold higher (Fig. 3.b, p = 0.0073), and HP 13C Lac/Pyr was 1.15 fold lower (Fig. 3.c, p = 0.0071). No significant differences in HP [13C]urea levels were observed in the cortical area (Fig. 3.c), although a trend towards lower HP [13C]urea levels in rTBI was observed (p = 0.0794). In subcortical areas, we did not detect any differences in HP [1–13C]lactate, HP [1-13C]pyruvate, HP 13C Lac/Pyr and HP [13C]urea between rTBI and Sham mice (Fig. 3.e–h). In agreement with the metabolic quantifications, HP 13C heatmaps clearly show lower HP [1–13C]lactate, higher HP [1-13C]pyruvate and lower HP 13C Lac/Pyr in the cortex of rTBI mice (Fig. 3.i). Altogether, our results indicate that HP 13C MRSI can detect region-specific long-lasting metabolic changes following mild rTBI.
Figure 2. HP 13C spectra following co-injection of HP [1–13C]pyruvate and [13C]urea.
Representative T2-weighted MR image overlaid with the grid used for HP 13C MRSI acquisitions. Representative 13C spectra showing HP [1-13C]pyruvate, HP [13C]urea and HP [1-13C]lactate in the (a) cortex (red voxel) and (b) subcortex (red voxel) for a Sham and a rTBI mouse.
Figure 3. HP 13C MRSI detects long-lasting metabolic alterations following rTBI.
Quantitative analyses of (a) HP [1-13C]lactate levels, (b) HP [1-13C]pyruvate levels, (c) HP 13C Lac/Pyr, and (d) HP [13C]urea for the cortex (highlighted red voxels), revealed lower HP [1-13C]lactate levels (p = 0.0073), higher HP [1-13C]pyruvate levels (p = 0.0073), and lower HP 13C Lac/Pyr (p = 0.0071) in rTBI compared to Sham mice. In contrast, quantitative analyses of (e) HP [1-13C]lactate levels, (f) HP [1–13C]pyruvate levels, (g) HP 13C Lac/Pyr, and (h) HP [13C]urea for the subcortex (highlighted red voxels), did not detect differences between rTBI and Sham mice. (i) Representative HP 13C heatmaps for a Sham and a rTBI mouse, highlighting lower HP [1-13C]lactate, higher HP [1-13C]pyruvate and lower HP 13C Lac/Pyr in cortical areas in rTBI mice. N = 9 rTBI and 10 Sham mice. Unpaired t-test (**p ≤ 0.01); data are expressed as means ± SD.
Multimodal MRI does not detect long-lasting effect of injury in rTBI
We evaluated whether a comprehensive multimodal MRI approach could detect changes between rTBI and Sham mice at 3.5 months post-injury, when metabolic alterations where detected by HP 13C MRSI.
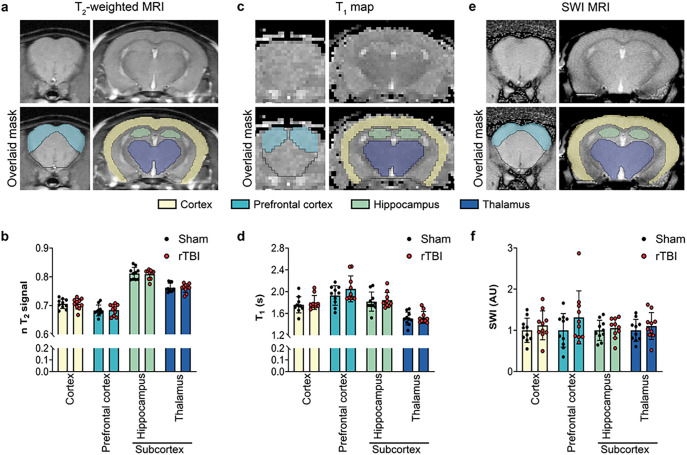
We first used T2-weighted MRI, the clinical standard of MRI approach, that is sensitive to inflammation and/or changes in myelin content. We found that T2 signal intensities were not different between rTBI and Sham mice in any of the regions studied (prefrontal cortex, cortex, hippocampus and thalamus (subcortex)) (Fig. 4.a–b). In addition, we did not detect any differences in brain region volumes between groups (Supplementary Fig. 1). Next, we used a mapping sequence that was shown to be sensitive to changes in microstructure or alterations related to oxidative stress. Similar to the T2 intensities, we did not observed any differences in the values in any of the region studied between rTBI and Sham mice (Fig. 4.c–d). Lastly, a SWI sequence was used, as it is capable of detecting microbleeds as well as potential changes in oxygenation following injury. Once again, no differences in SWI values were observed between rTBI and Sham mice in any region (Fig. 4.e–f). We did not detect any microbleed lesions in any of the studied animals.
Figure 4. Multimodal MRI does not detect long-lasting effect of injury in rTBI.
(a) Representative T2-weighted MRI data and corresponding manual brain masking. (b) Quantitative analyses of T2-weighted signal intensity revealed no significant differences for brain subregions between Sham and rTBI. (c) Representative map and corresponding manual brain masking. (d) Quantitative analyses of maps revealed no significant differences for brain subregions between Sham and rTBI. (e) Representative SWI data and corresponding manual brain masking. (f) Quantitative analyses of SWI intensity revealed no significant differences for brain subregions between Sham and rTBI. Brain masking color code: yellow: cortex, green: light blue: prefrontal cortex; hippocampus; dark blue: thalamus. N = 10 rTBI and 10 Sham mice. Unpaired t-test; data are expressed as means ± SD.
Altogether, our results indicate that a comprehensive multimodal MRI approach combining T2-weighted MRI, mapping and SWI was not able to detect any signs of injury in rTBI mice at 3.5 months post-injury, unlike HP 13C MRSI.
Disrupted enzymatic activity, but not transporter expression, is observed at chronic time points after rTBI
To further investigate potential underlying mechanisms responsible for the observed changes in HP 13C MRSI readouts, we evaluated the activity of enzymes responsible for pyruvate conversion into its downstream metabolites and the expression of transporters that control the entry of pyruvate into cells and the efflux of metabolites outside of the cells.
In the brain, lactate dehydrogenase (LDH) converts pyruvate into lactate, and pyruvate dehydrogenase (PDH) controls pyruvate entry into the tricarboxylic cycle and its conversion into acetyl-coA. We observed that PDH was 1.6 fold lower in the prefrontal cortex and 1.7 fold lower in the cortex of rTBI compared to Sham mice (Table 1, p = 0.0044 and p = 0.0375, respectively). No differences in PDH were observed in subcortical areas that include the hippocampus and thalamus. The activity of LDH was not significantly different between rTBI and Sham mice for cortical and subcortical areas.
Table 1.
PDH and LDH enzyme activity.
| PDH activity | LDH activity | ||||
|---|---|---|---|---|---|
| mean ± SD | P-value | mean ± SD | P-value | ||
| Prefrontal cortex | rTBI | 0.0007 ± 0.0001 | 0.0044 ( ** ) | 0.0070 ± 0.0007 | 0.3345 |
| Sham | 0.0011 ± 0.0003 | 0.0078 ± 0.0020 | |||
| Cortex | rTBI | 0.0012 ± 0.0005 | 0.0375 ( * ) | 0.0022 ± 0.0004 | 0.5837 |
| Sham | 0.002 ± 0.0008 | 0.0023 ± 0.0003 | |||
| Hippocampus | rTBI | 0.0074 ± 0.003 | 0.4171 | 0.0020 ± 0.0002 | 0.1602 |
| Sham | 0.0091 ± 0.005 | 0.0023 ± 0.0005 | |||
| Thalamus | rTBI | 0.00013 ± 0.00003 | 0.6509 | 0.0023 ± 0.0004 | 0.1282 |
| Sham | 0.00014 ± 0.00006 | 0.0021 ± 0.0002 |
Unpaired t-test.
p ≤ 0.05
p ≤ 0.01.
As levels of HP metabolites and HP 13C Lac/Pyr also depend on HP [1-13C]pyruvate intake by cells and on the efflux of HP [1-13C]lactate, we evaluated the expression of monocarboxylate transporters (MCTs). MCT1 is primarily responsible for pyruvate entry into the cell, and MCT4 is principaly involved in the efflux of lactate outside of the cell. Protein quantification of MCT1 and MCT4 performed for the prefrontal cortex, cortex, hippocampus and thalamus did not show any differences between rTBI and Sham mice (Table 2), indicating that the differences observed with HP 13C MRSI are likely not due to MCT1 and MCT4 expression.
Table 2.
MCT1 and MCT4 protein expression.
| MCT1 | MCT4 | ||||
|---|---|---|---|---|---|
| mean ± SD | P-value | mean ± SD | P-value | ||
| Prefrontal cortex | rTBI | 1.3 ± 0.31 | 0.0685 | 1.59 ± 0.72 | 0.1109 |
| Sham | 1 ± 0.32 | 1 ± 0.54 | |||
| Cortex | rTBI | 1 ± 0.14 | 0.6399 | 0.72 ± 0.35 | 0.1502 |
| Sham | 1 ± 0.07 | 1 ± 0.33 | |||
| Hippocampus | rTBI | 0.91 ± 0.31 | 0.5090 | 1 ± 0.77 | 0.9147 |
| Sham | 1 ± 0.17 | 1 ± 0.58 | |||
| Thalamus | rTBI | 0.89 ± 0.34 | 0.4891 | 0.75 ± 0.59 | 0.3361 |
| Sham | 1 ± 0.24 | 1 ± 0.27 |
Unpaired t-test.
Altogether, our results indicate that PDH activity is decreased in the cortical areas following rTBI, while LDH activity, MCT1 and MCT4 expression are not significantly different compared to Sham at 3.5 months post-injury.
Machine learning identifies rTBI/Sham classifiers, and predictors of behavior and HP 13C readouts
The machine learning (ML) analysis included all the data described above, as well as behavioral data we previously reported in Krukowski et al.35, which showed that mild rTBI leads to increased risk-taking behavior in male mice at 100 days post-injury.
Given n = 20 (10 for rTBI and 10 for Sham) mice along with 44 measured variables (see Table 3 for list of variables and abbreviations), we used ML to perform two types of analyses. First, we wanted to identify the best classifying variables allowing for separation of the two groups (rTBI vs Sham). Second, we aimed to find the best predictors of changes in risk-taking behavior, as it recapitulates a key behavioral component observed in rTBI patients, and of cortical HP 13C Lac/Pyr, due to its potential to serve as a novel biomarker for long-lasting consequences of rTBI.
Table 3.
List of variables used for ML analyses.
| Variable abbreviation | Variable description |
|---|---|
| EPM frequency extreme | Number of entries into the extreme zones on the EPM |
| EPM duration extreme | Time (sec) in the extreme zones on the EPM |
| EPM frequency openandcenter | Number of entries in the Open + Center on EPM |
| EPM duration openandcenter | Time (sec) in the open + Center on the EPM |
| EPM totaldistance centerpoint | Total distance traveled on EPM, measured by centerpoint, units = cm |
| EPM averagevelocity centerpoint | Average animal velocity on the EPM, units = cm/sec |
| HP 13C Urea Ctx | Hyperpolarized [13C]urea level in cortical area |
| HP 13C Lac/Pyr Ctx | Hyperpolarized lactate/pyruvate ratio in cortical area |
| HP 13C Urea Subctx | Hyperpolarized [13C]urea level in subcortical area |
| HP 13C Lac/Pyr Subctx | Hyperpolarized lactate/pyruvate ratio in subcortical area |
| PDH Pfc | PDH activity in prefrontal cortex |
| PDH Ctx | PDH activity in cortex |
| PDH Hp | PDH activity in hippocampus |
| PDH Thai | PDH activity in thalamus |
| nT2 Ctx | Normalized T2 intensity value in cortex |
| nT2 Hp | Normalized T2 intensity value in hippocampus |
| nT2 Pfc | Normalized T2 intensity value in prefrontal cortex |
| nT2 Subctx | Normalized T2 intensity value in subcortical areas |
| Volume Ctx | Volume of cortex calculted from T2w MRI |
| Volume Hp | Volume of hippocampus calculated from T2w MRI |
| Volume Pfc | Volume of prefrontal cortex calculated from T2w MRI |
| Volume Subctx | Volume of subcortical areas calculated from T2w MRI |
| Volume Ventricle | Volume of ventricles calculated from T2w MRI |
| Volume Brain | Volume of whole brain calculated from T2w MRI |
| T1 Ctx | T1 value in cortex |
| T1 Hp | T1 value in hippcampus |
| T1 Pfc | T1 value in prefrontal cortex |
| T1 Subctx | T1 value in subcortex |
| MCT1 Thal | MCT1 expression in thalamus |
| MCT1 Hp | MCT1 expression in hippocampus |
| MCT1 Ctx | MCT1 expression in cortex |
| MCT1 Pfc | MCT1 expression in prefrontal cortex |
| MCT4 Thal | MCT4 expression in thalamus |
| MCT4 Hp | MCT4 expression in hippocampus |
| MCT4 Ctx | MCT4 expression in cortex |
| MCT4 Pfc | MCT4 expression in prefrontal cortex |
| LDH Thal | LDH activity in thalamus |
| LDH Hp | LDH activity in hippocampus |
| LDH Ctx | LDH activity in cortex |
| LDH Pfc | LDH activity in prefrontal cortex |
| SWI Ctx | SWI value in cortex |
| SWI Hp | SWI value in hippcampus |
| SWI Pfc | SWI value in prefrontal cortex |
| SWI Subctx | SWI value in subcortex |
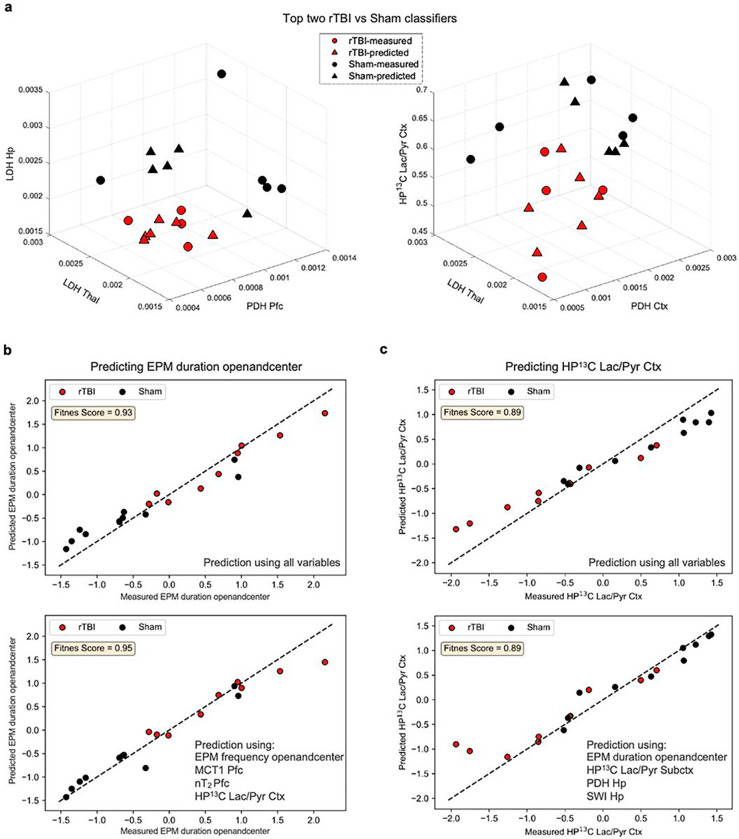
Various classification and feature extraction methods were implemented to identify the best classifying variables between Sham and rTBI mice. Consequently, we identified five triplets of variables that could accurately distinguish between either group and ranked them based on their feature importance scores computed using various feature extraction algorithms (see methods). The top two triplets with high feature scores are presented in Fig. 5.a and the others are shown in Supplementary Fig. 2. The top two triplets are: 1) PDH Pfc, LDH Thal, and LDH Hp, and 2) PDH Ctx, LDH Thal, and HP 13C Lac/Pyr Ctx. These findings suggest that although one single feature is not sufficient to identify the difference between rTBI and Sham, the combined PDH and LDH activity, as well as HP 13C imaging readouts can help distinguish differences between the two conditions. The three lower-tier triplets consisted of the subsequent variables: 3) PDH Pfc, EPM duration openandcenter, and LDH Thal, 4) PDH Pfc, EPM duration openandcenter, and HP 13C Lac/Pyr Ctx, and 5) PDH Pfc, MCT1 Pfc, and HP 13C Lac/Pyr Ctx. These three last triplets further idenfy the risk-taking behavior and MCT1 expression in the prefrontal cortex as important variables to classify Sham and rTBI.
Figure 5. ML analyses identify best rTBI and Sham classifiers and best predictors of changes in risk-taking behavior and HP 13C Lac/Pyr Ctx.
(a) Top two triplets that can classify rTBI (red) and Sham (black) mice. Here, circles represent the mice for which all three variables are measured whereas triangles represent mice for which at least one of the three variables were missing and predicted by ML algorithms (see Methods). (b-c) Prediction performance of the best predictors of risk-taking behavior (b, bottom panel), and HP 13C Lac/Pyr Ctx (c, bottom panel) compared to the prediction performance of the case in which all variables are used (b-c top panel). N = 10 rTBI and 10 Sham mice.
Next, we identified the best predictors of the changes in risk-taking behavior and HP 13C Lac/Pyr Ctx presented in Fig. 5.b and 5.c, respectively. Interestingly, four variables, namely EPM frequency openandcenter, MCT1 Pfc, nT2 Pfc, and HP 13C Lac/Pyr Ctx, were sufficient to predict risk-taking behavior (Fig. 5.b bottom panel) with similar accuracy than when all variables were used for prediction (Fig. 5.b top panel). This result shows that a systems approach comprising behavioral, transporter expression, structural and HP 13C imaging measures yields a tangible prediction for changes in risk-taking behavior. Similarly, four variables, namely EPM duration openandcenter, HP 13C Lac/Pyr Subctx, PDH Hp, and SWI Hp, were enough to predict the HP 13C Lac/Pyr Ctx (Fig. 5.c bottom panel) with similar accuracy than when all the variables were used for prediction (Fig. 5.c top panel). This result demonstrates that using a systemic combination of behavioral, enzyme activity, and advanced MRI measures best predicts the HP 13C Lac/Pyr Ctx.
DISCUSSION
In this study, we demonstrated that HP 13C MRSI of [1-13C]pyruvate can detect metabolic changes 3.5 months following rTBI when structural T2 MRI, mapping, SWI MRI and HP 13C MRSI of [13C]urea did not. Specifically, we measured a significantly lower HP 13C Lac/Pyr in the mouse cortex 3.5 months post-injury, which was associated with lower PDH activity. Using a ML approach, we further validated that the HP 13C Lac/Pyr is among the best classifiers of rTBI and Sham groups, and is a predictor of the risk-taking behavior observed in this rTBI model. Our findings demonstrate the ability of HP 13C MRSI of [1–13C]pyruvate to detect rTBI-induced damages and highlight promising potential to improve diagnosis and monitoring of rTBI patients at chronic time points when other imaging techniques are insufficient.
Cerebral metabolism has been probed using HP [1-13C]pyruvate both in preclinical and clinical settings in healthy and diseased brain (see Le Page & al. for recent literature review21). Prior studies performed in moderate TBI preclinical models of contusion injury have reported increased HP 13C Lac/Pyr at early timepoints following injury (4 hours up to 7 days following injury), but no significant changes at chronic timepoints (28 days post-injury)24, 25. In contrast, in this study we observed a decreased HP 13C Lac/Pyr at chronic timepoints, suggesting different underlying pathological changes between contusion injury and rTBI. Studies performed using positron emission tomography (PET) imaging with the glucose analogue 18F-fluorodeoxyglucose (18F-FDG) have detected long-term brain hypometabolism following TBI4, 5, 36, which is in line with the lower HP 13C Lac/Pyr measured here at chronic timepoints. To the best of our knowledge, 18F-FDG PET imaging has never been applied to CHIMERA rTBI model. Furthermore, the use of this method for TBI is limited by ionizing radiations, and the high background of 18F-FDG PET signal in the brain tissue, hampering the detection of small changes in glucose uptake.
Decreased HP 13C bicarbonate / lactate ratio was found in moderate TBI models25, and decreased HP 13C bicarbonate levels were found in TBI patients26, highlighting possible changes in mitochondrial function and aerobic versus anaerobic respiration following trauma. In agreement with these findings, a decreased of PDH activity after injury has been previously described24, 37, including in our current findings in rTBI. In this study, we were not able to detect 13C bicarbonate, likely due to the low signal to noise and the fast relaxation rate of this metabolite at ultra-high field (14.1 Tesla). HP 13C Lac/Pyr can be influenced by the activity of LDH and/or PDH, the availability of their cofactors38, 39, 40, as well as by MCTs expression, where increased MCT1 expression leads to increased HP 13C Lac/Pyr in selected cell lines41. In this study, we did not find any significant differences in MCT1 and MCT4 between rTBI and Sham mice, suggesting that they do not play a prominent role in the changes observed in the HP 13C Lac/Pyr at this late time post injury.
It has previously been shown that the blood-brain-barrier (BBB) limits the entry of HP probes, which could in turn influence the measured HP 13C Lac/Pyr42, 43. To evaluate potential changes in perfusion and delivery, we co-injected [1-13C]pyruvate with HP [13C]urea, a metabolically inactive probe. Although decreased cerebral blood flow alteration has been reported following rTBI44, 45, we did not observe any differences in HP [13C]urea levels between Sham and rTBI, indicating that perfusion was likely unaltered at 3.5 months post-injury. To the best of our knowledge, this is the first study reporting the use of HP [13C]urea in a rTBI model. Additional studies at earlier timepoints following injury, and in other TBI models, are needed to evaluate the full potential of HP [13C]urea in detecting BBB alterations and/or vasculature changes following brain injury.
Conventional and advanced anatomical MRI did not detect any differences between Sham and rTBI at 3.5 months post-injury. In agreement with these findings, Haber et al. and our group previously reported no differences using T2 MRI at 7 days and 40 days post-injury, respectively10, 12. These results suggest that conventional T2 MRI may not be able to detect rTBI patholology-induced using the CHIMERA device, either at early or late timepoints following injury. Using mapping we investigated whether we could detect changes in tissue microstructure and reactive oxygen species production, but found no differences between Sham and rTBI34. Oxidative stress and reactive oxygen species have been shown to play an important role in TBI pathogenesis and in mediating axonal degeneration46, 47, 48. However it remains unclear if these events may predominantly occur at early timepoints following injury and would have resolved by the time we performed our imaging study (3.5 months injury), or whether mapping was not able to detect these events in this rTBI model. HP [1-13C]dehydroascorbic acid (DHA) and HP [1-13C]N-acetyl cysteine (NAC) have been shown to be sensitive probes to investigate redox changes in vivo49, 50, and therefore represent attractive probes to further interrogate the involvement of oxidative stress using HP 13C MRSI. We also included SWI MRI exams as this method has been shown to improve the detection of microbleeds and hemorrhagic diffusive axonal injury after TBI, which was associated with neurologic deficits and long-term outcome in human TBI51, 52. However, we did not detect any differences in SWI MRI between Sham and rTBI. As for mapping, it remains to be determined whether no microbleeds or oxygenation changes occurs in this model at early timepoints, or whether potential changes have resolved by the time we conducted our imaging exams.
We took a systems approach to measure behavior outcomes using ML analyses. We found that sets of variables were able to classify rTBI and Sham mice. These variables are linked to cognitive abilities (risk-taking behavior), metabolism and molecule transport (PDH and LDH activity, HP 13C Lac/Pyr, and MCT1), highlighting the importance of long-term metabolic impairment in rTBI and suggesting their potential as injury biomarkers. Interestingly, ML was able to identify that enzymatic changes in the thalamus and hippocampus regions, which were not statistically significant using conventional unpaired t-test in isolation, became important variables to classify rTBI and Sham when considered together. This is in agreement with changes in hippocampal function that have been previously reported in rTBI up to 6 months post-injury53. Future HP 13C imaging studies will aim to also include the thalamus and hippocampus to determine whether in vivo metabolic changes can be detected in these regions. ML was also used to determine which variables are best predictors of the risk-taking behavior. We found that the changes in risk-taking behavior were best predicted by variables from the cortical areas, including HP 13C Lac/Pyr, MCT1 expression, structural MRI, and behavioral parameters. Similarly, cerebral microdialysis studies have shown that the Lac/Pyr is an important variable associated to clinical outcome. High Lac/Pyr within the first days after injury was associated to poor clinical outcome 6 months later18, while here we show that a lower HP 13C Lac/Pyr is associated to higher risk-taking behavior 3.5 months after injury. This discrepancy might be explained by different timing of the Lac/Pyr measurement (days versus months after injury), and the injury severity (severe TBI versus mild rTBI). Nonetheless, our study provides further evidence that the Lac/Pyr is a useful marker to predict behavioral outcome, which can now be measured in a non-invasive manner using HP 13C MRSI, thus opening new avenues to evaluate metabolic alterations months after trauma. Last, ML was used to determine the best predictors of the HP 13C Lac/Pyr in the cortex, and identified subcortical variables, including PDH activity, SWI MRI and metabolic MRSI, as well as risk-taking behavior. Altogether, these results demonstrate the importance of multimodal approaches to detect rTBI pathology and associated long-lasting changes.
Thanks to the recent efforts of the community to enable easy data sharing though data repository54, future studies with higher sample size will become possible, which in turn can lead to improvement of our understanding of biological and functional pathway involved in rTBI, and help identify novel biomarkers.
In summary, our findings demonstrate the potential of HP [1-13C]pyruvate to detect long-lasting metabolic alterations in a mouse model of rTBI. In addition, ML identified HP 13C MRSI as a key parameter to predict long-term rTBI-induced behavioral outcomes. Over the past few years, the use of HP 13C MRSI in clinical trials worldwide has been rapidly expanding, and the injection of HP [1-13C]pyruvate has proven feasible and safe, with no reported side effects55. In this study, we were able to measure changes in HP 13C MRS parameters from two regions, the cortex which is closest to the impact, and the subcortex, which is more remote. We were not able to differentiate between smaller brain regions (e.g. prefrontal cortex, hippocampus, thalamus) due to the large voxel size used in this study relative to the size of the mouse brain. Current sequences available on clinical scanners can achieve up to 1 cm3 spatial resolution and cover the entire human brain, thus providing metabolic information from brain areas close and remote to the site of injury. With the growing availability of the HP 13C MRS technology, our findings provide a strong rationale to translate its use in patients suffering from rTBI, with the aim to improve the detection of rTBI-induced damages, help in understanding metabolic pathways involved in rTBI pathogenesis, and eventually aid the development of treatment strategies.
METHODS
Animals and rTBI model induction
All animal research was approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco. Mice were given one week of acclimation and housed with a reversed 12-h light/12-h dark cycle and provided food and water ad libitum. At 8 weeks of age, mice were randomly assigned to the rTBI or sham control group. Animals were anesthetized using isoflurane (2–3%) in oxygen 1 L/min during the procedure. rTBI animals were subjected to multiple, mild, closed-head injuries using the CHIMERA device as previously reported10, 35. Briefly, rTBI animals were placed supinely into an angled holding platform without any shaving of the head or incision into the skin so that the head was level with the piston target hole while aligning the eyes, ears, and nose such that the impact was centered on the dorsal convexity of the skull, targeting a 5-mm area surrounding bregma. A nose cone delivering isoflurane was removed just prior to the impact. Impact was initiated using RealTerm software, which was connected to a system including air tank, pressure regulator, digital pressure gauge, two-way solenoid valve, and piston. The impact was administered with a velocity range of 3.9–4.5 m/sec, resulting in an impact energy of 0.5 J from the 5 mm, 50 g piston10, 35. Animals were moved to an incubator immediately after the impact and monitored until fully recovered. rTBI animals received an injury once per day for 5 days with a 24 h interval in between impacts. Five repeated hits were chosen to specifically focus on the effects of repeated exposure to TBI, as athletes, veterans and sometimes trauma victims are exposed to constant and repeated blows, even without experiencing concussive symptoms. Sham mice were exposed to the same isoflurane anesthesia paradigm without sustaining an impact. Skull fractures, seizures, apnea, or mortality were not observed in any animals, and no animals were excluded from the study due to injury parameters.
Risk-taking behavioral test
For all behavioral assays, the experimenters were blinded to surgery. Before behavioral analysis, animals were inspected for gross motor impairments. Animals were inspected for whisker loss, limb immobility (including grip strength), and eye occlusions. If animals displayed any of these impairments, they were eliminated from the study. Behavioral tests were recorded and scored using a video tracking and analysis setup (Ethovision XT 8.5, Noldus Information Technology). If tracking was unsuccessful, videos were scored by two individuals blinded to surgery. Risk-taking behavioral phenotype was evaluated using the Elevated Plus Maze (EPM) at ~ 100 days (3 months) post-injury (counted from the day of the first injury) as described previously10, 35. The EPM consists of two exposed, open arms (35 cm) opposite each other and two enclosed arms (30.5 cm) also across from each other. The four arms are attached to a center platform (4.5 cm square), and the entire maze is elevated 40 cm off the floor. Bright white lights illuminated both ends of the open arm. Mice were placed individually onto the center of the maze and allowed to explore the maze for 5 min, and their activity was recorded. The maze was cleaned with 70% ethanol between animals. Risk-taking behavior was measured by changes in time spent in the open arms + center of the EPM. We also measured the number of entries into the extremes zones on the EPM, the time spent in the extreme zons on the EPM, the number of entries in the open arms + center on the EPM, the total distance traveled on EPM measured by centerpoint, and the average animal velocity on the EPM. These behavioral data were previously reported in Krukowski et al.35.
Magnetic resonance imaging
Mice were anesthetized using isoflurane (1.5–2% in O2) and a 27-gauge catheter was placed in the tail vein to allow for intravenous (i.v.) injection. Next, animals were placed in a dedicated cylindrical cradle allowing for reproducible positioning of the mouse head; which was subsequently inserted inside a dual tune 1H-13C volume coil (ØI = 40 mm) or a single tuned 1H proton coil (ØI = 40 mm) in a 14.1 T vertical MR system (Agilent Technologies). Respiration rate was continuously monitored through the PC-sam software interface (SA Instrument, NY, USA).
First, T2-weighted images from the entire brain were acquired for adequate positioning of the grid used for hyperpolarized 13C acquisitions using the following parameters: repetition time (TR) = 1200 ms, echo time (TE) = 20 ms, slice thickness 1.8 mm, 2 averages, matrix 256 × 256, field of view (FOV) 30 × 30 mm². For HP 13C MRSI acquisitions, 24 μL of [1-13C]pyruvate and 55 μL [13C]urea preparation were co-polarized using a Hypersense DNP polarizer (Oxford Instruments) for one hour. After dissolution, the HP [1–13C]pyruvate and [1-13C]urea preparation was rapidly dissolved in isotonic buffer (pH ~ 7) to a final concentration of 80 mM and 78 mM, respectively. A final volume of 300 μL of the HP [1-13C]pyruvate and [13C]urea solution was then injected i.v. through the tail vein catheter. 2D dynamic chemical shift imaging 13C data were acquired 16 seconds post i.v. injection of the HP [1-13C]pyruvate and [13C]urea solution using the following parameters: TR = 67 ms, TE = 0.58 ms, spectral width 5000 Hz, 256 points, flip angle 10°; matrix = 16 ×16, field of view (FOV) = 32 × 32 mm²; slice thickness 4 mm.
Next, for T2-weighted MRI, MRI and SWI acquistions, the dual tune 1H-13C volume coil (ØI = 40 mm) was removed and replaced by a 1H volume only coil (ØI = 40 mm). T2-weighted MRI was acquired using a 2D fast spin-echo, with effective echo time (TEeff)/TR = 11.80/2006 ms, FOV = 25 × 25 mm2, in 256 × 256 array and 0.5 mm slice thickness. -mapping data were acquired using fast spin-echo with inversion recovery: TEeff/ TR = 7.44/10000 ms, 8 inversion times (TI): 100, 170, 310, 530, 940, 1640, 2900, 5000 ms, FOV = 30 × 30 mm2, in 128 × 128 array and 1 mm slice thickness. SWI acquisitions were acquired using TEeff/TR = 4.64/111.29 ms, FOV = 20 × 20 mm2, in a 256 × 256 array and 0.4 mm slice thickness.
Magnetic resonance imaging data analysis
Brain regions were manually delineated on maps, T2-weighted and SWI magnitude images for each mouse based on the Allen Adult Mouse Brain atlas (Allen Institute) using the Aedes region of interest package for MATLAB (Mathworks). For each region, brain volumes were calculated with the T2-weighted data, as well as the mean T2-weighted values which were normalized to the mean of the cerebrospinal fluid signal from the ventricles as signal value standard. The mean relaxation times was calculated from maps generated in VNMRj by pixel-wise fitting according to (Eq. 1).
| (eq. 1) |
Where is the measured signal from fast spin echo with multiple inversion recovery, and the three fit parameters: relaxation time , equilibrium longitudinal magnetization , and pre-inversion recovery longitudinal relaxation . The TI list is used as input for time .
SWI data were processed as previously described32 with phase images unwrapped by PRELUDE (FSL), High pass Gaussian filtered with pixel size 32 × 32, and positive phase map scaling used (Eq. 2).
| (eq. 2) |
Normalized positive phase map where is the filtered, unwrapped phase at location , and is the maximum phase of the slice of interest. The positive phase map is a spatial map varying between zero and one, with higher phase approaching zero and thus increasing contrast on the final merged SWI data. The phase map is multiplied with the magnitude image four times to create final SWI data56. The mean SWI intensity was calculated for each mouse for each brain region and normalized to the mean of the Sham for each region, which corresponds to 1.
HP 13C MRS imaging data was analyzed using the in-house SIVIC software (http://sourceforge.net/apps/trac/sivic/) and custom-built programs written in MATLAB (MATLAB R2011b, The MathWorks Inc.). The area under the curve (AUC) of HP [1-13C]pyruvate, AUC of HP [1–13C]lactate and AUC of HP [13C]urea were measured for each voxel and normalized to the noise level. To account for variations in polarization levels and delivery, HP [1-13C]pyruvate signal and HP [1-13C]lactate signal were calculated by dividing each value by the sum of HP [1-13C]pyruvate and HP [1-13C]lactate for each voxel. HP [13C]urea signal was obtained by normalizing HP [13C]urea signal from each brain voxel to the sum of [13C]urea signal from tissue surrounding the brain. HP 13C Lac/Pyr was calculated as the ratio of the AUC. Next, average from voxels containing cortex or subcortex was calculated, and the obtained mean values were used to evaluate statistical significance between sham and rTBI groups. Color heatmaps of HP [1-13C]pyruvate, HP [1-13C]lactate, HP [13C]urea and HP 13C Lac/Pyr were generated using a linear-based interpolation of the 13C 2D CSI data to the resolution of the anatomical images using custom-built programs written in MATLAB and SIVIC.
Ex vivo analyses of brain samples
Following the imaging session, mice were transcardially perfused with ice-cold phosphate buffered saline, and brains were rapidly dissected. Next, prefrontal cortex, cortex, thalamus and hippocampus were isolated and snap-frozen in liquid nitrogen. Samples were stored at − 80°C until further processing for activity assays and western blots.
LDH and PDH activities were evaluated using spectrophotometric activity assay kits according to manufacturer’s guidelines (ab102526 and ab109902; Abcam, respectively), and normalized to protein concentration determined by the Bradford protein assay method.
For western blot analyses, frozen brain samples were then homogenized with RIPA buffer (Pierce, 89900) and protease inhibitor Halt ™ Protease Inhibitor Cocktail (Thermo Scientific, 1862209) using a TissueLyser II (Qiagen). Lysate were incubated on ice for 15 minutes and then centrifuged at 14,000 rpm for 10 min at 4°C. Protein concentration in supernatants was determined using the Bradford protein assay method. Equal amount of proteins was loaded on Mini-Protean® TGX TM Precast gels 12% (BioRad, 456–1043) for 34 minutes at 200 V and 400 mA in Tris/Glycine/SDS Buffer, (BioRad, 1610732). Proteins were transferred onto 0.2 μm PVDF membranes (BioRad, 1704157) using Trans-Blot Turbo (BioRad) for 7 minutes using the following settings: 1.3 A, 25 V. Next, the membranes were blocked for 1 hour using Tris-buffered saline supplemented with 0.1% Tween 20% and 5% milk (Research Product International, M17200–500). Membranes were incubated with the primary antibodies: rabbit anti-MCT1/SLC16A1 pAb (Novus, NBP1–59656, lot C, 1:400), rabbit anti-MCT4 (Bethyl, A304–439A-M, 1:1000), rabbit mAb anti-beta-tubulin (9F3) (Cell Signaling, 2128S, 1:2000) diluted in Tris-buffered saline supplemented with 0.1% Tween 20% and 5% milk overnight. An anti-rabbit IgG HRP-linked secondary antibody (Cell Signaling, 7074S, 1:3000) was used to detect immune-reactive bands using enhanced chemiluminescence (ECL Western Blotting Substrate, Pierce, 32209) according to the manufacturer instructions. Quantification of protein bands was done by measuring band intensities using ImageJ software. MCT1 and MCT4 levels were normalized to beta-tubulin expression and expressed as the levels relative to the expression of Sham mice, which corresponds to 1.
Machine Learning analyses
The ML pipeline is summarized in Supplementary Fig. 3. Preprocessing of the raw experimental data was performed before ML analyses to (i) scale the measurements for fair comparison and (ii) predict the missing measurements. The raw measurements were rescaled using Scikit-learn python library’s standard scaler57 (see Supplementary Fig. 4 for an example of original versus scaled data distributions). For a few mice, we were not able to measure some of the variables due to tissue isolation (n = 2 rTBI and n = 2 Sham missing for PDH activity, n = 3–4 rTBI and n = 3 Sham missing for LDH activity, MCT1 and MCT4 expression) and low signal to noise of the HP [13C] imaging data (n = 1 rTBI mouse excluded). To impute the missing measurements we separated the data into rTBI and Sham groups, and within each group the data was split as training and testing. Mice with all measured variables were used to train a Random Forrest Regressor (RFR). The trained regressor was then used to predict missing measurements associated with the testing data.58 To identify the variables that are the best classifiers of Sham and rTBI groups, and to eliminate sensitivity of the results to different methods, we used multiple Scikit-learn classifiers and feature selection algorithims that are neural network, logistic regression, recursive feature elimination (RFE), SelectKBest, feature importance with ExtraTreesClassifier, and LASSO. Using each method, all the variables were scored based on their importance in the classification of Sham versus rTBI and a pool of variables with high importance scores was created. From this pool, we created groups of best classifiers with the minimum number of variables, that is three in this case, and ranked those triplets of variables based on their individual importance scores across different methods. Similarly, to identify the best predictors of the risk-taking behavior and HP 13C Lac/Pyr in the cortex, we trained multiple regression algorithms of Scikit-learn library that are LASSO, RFR, Ridge Regression, Support Vector Regressor, using the measurements of variable to be predicted as the y-values and all the remaining variables as the x-values (predictors). Then, the variables with high absolute coefficients were identified as the most important features that contribute to the prediction of the regressor. After pooling and grouping the variables with high feature scores across different algorithms, we observed the best predictor group with the minimum number of variables.
Statistical analysis
Results are expressed as mean ± standard deviation (SD). Statistical analyses of MRI, behavioral, and ex vivo parameters was performed using unpaired t-test (GraphPad Prism (v 9.1.2), (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
Acknowledgements
This work was supported by research grants: NIH R01NS102156 (MC), NIH NIA grants R01AG056770 (SR) and NIH R21NS096718 (SR, MC), Dana Foundation: The David Mahoney Neuroimaging program (MC) and the NIH Hyperpolarized MRI Technology Resource Center P41EB013598. K.K. was supported by an NRSA post-doctoral fellowship from the NIA F32AG054126. AN was supported by the NINDS K08 NS114170.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Myriam Chaumeil, University of California, San Francisco.
Caroline Guglielmetti, University of California, San Francisco.
Kai Qiao, University of California, San Francisco.
Brice Tiret, University of California, San Francisco.
Mustafa Ozen, Bay Area Institute of Science, Altos Labs.
Karen Krukowski, Bay Area Institute of Science, Altos Labs.
Amber Nolan, University of Washington.
Maria Serena Paladini, Bay Area Institute of Science, Altos Labs.
Carlos Lopez, Bay Area Institute of Science, Altos Labs.
Susanna Rosi, Bay Area Institute of Science, Altos Labs.
References
- 1.Brett BL, Gardner RC, Godbout J, Dams-O’Connor K, Keene CD. Traumatic Brain Injury and Risk of Neurodegenerative Disorder. Biol Psychiatry 91, 498–507 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding K, Gupta PK, Diaz-Arrastia R. Epilepsy after Traumatic Brain Injury. In: Translational Research in Traumatic Brain Injury (eds Laskowitz D, Grant G) (2016). [PubMed] [Google Scholar]
- 3.Hay J, Johnson VE, Smith DH, Stewart W. Chronic Traumatic Encephalopathy: The Neuropathological Legacy of Traumatic Brain Injury. Annu Rev Pathol 11, 21–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng TS, et al. Neuroimaging in repetitive brain trauma. Alzheimers Res Ther 6, 10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asken BM, Rabinovici GD. Identifying degenerative effects of repetitive head trauma with neuroimaging: a clinically-oriented review. Acta Neuropathol Commun 9, 96 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toth A. Magnetic Resonance Imaging Application in the Area of Mild and Acute Traumatic Brain Injury: Implications for Diagnostic Markers? In: Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects (ed Kobeissy FH) (2015). [PubMed] [Google Scholar]
- 7.McAteer KM, Turner RJ, Corrigan F. Animal models of chronic traumatic encephalopathy. Concussion 2, CNC32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoogenboom WS, Branch CA, Lipton ML. Animal models of closed-skull, repetitive mild traumatic brain injury. Pharmacol Ther 198, 109–122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Namjoshi DR, et al. Merging pathology with biomechanics using CHIMERA (Closed-Head Impact Model of Engineered Rotational Acceleration): a novel, surgery-free model of traumatic brain injury. Mol Neurodegener 9, 55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolan A, et al. Repeated Mild Head Injury Leads to Wide-Ranging Deficits in Higher-Order Cognitive Functions Associated with the Prefrontal Cortex. J Neurotrauma 35, 2425–2434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNamara EH, Grillakis AA, Tucker LB, McCabe JT. The closed-head impact model of engineered rotational acceleration (CHIMERA) as an application for traumatic brain injury pre-clinical research: A status report. Exp Neurol 333, 113409 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Haber M, et al. Defining an Analytic Framework to Evaluate Quantitative MRI Markers of Traumatic Axonal Injury: Preliminary Results in a Mouse Closed Head Injury Model. eNeuro 4, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komlosh ME, et al. Using double pulsed-field gradient MRI to study tissue microstructure in traumatic brain injury (TBI). Microporous Mesoporous Mater 269, 156–159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jalloh I, Carpenter KL, Helmy A, Carpenter TA, Menon DK, Hutchinson PJ. Glucose metabolism following human traumatic brain injury: methods of assessment and pathophysiological findings. Metab Brain Dis 30, 615–632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demers-Marcil S, Coles JP. Cerebral metabolic derangements following traumatic brain injury. Curr Opin Anaesthesiol 35, 562–569 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter KL, et al. (13)C-labelled microdialysis studies of cerebral metabolism in TBI patients. Eur J Pharm Sci 57, 87–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guilfoyle MR, et al. Characterising the dynamics of cerebral metabolic dysfunction following traumatic brain injury: A microdialysis study in 619 patients. PLoS One 16, e0260291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timofeev I, et al. Cerebral extracellular chemistry and outcome following traumatic brain injury: a microdialysis study of 223 patients. Brain 134, 484–494 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Nelson SJ, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(1) (3)C]pyruvate. Sci Transl Med 5, 198ra108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang ZJ, et al. Hyperpolarized (13)C MRI: State of the Art and Future Directions. Radiology 291, 273–284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Page LM, Guglielmetti C, Taglang C, Chaumeil MM. Imaging Brain Metabolism Using Hyperpolarized (13)C Magnetic Resonance Spectroscopy. Trends Neurosci 43, 343–354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Vigneron DB, Xu D. Current human brain applications and challenges of dynamic hyperpolarized carbon-13 labeled pyruvate MR metabolic imaging. Eur J Nucl Med Mol Imaging 48, 4225–4235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurd RE, Yen YF, Tropp J, Pfefferbaum A, Spielman DM, Mayer D. Cerebral dynamics and metabolism of hyperpolarized [1-(13)C]pyruvate using time-resolved MR spectroscopic imaging. J Cereb Blood Flow Metab 30, 1734–1741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guglielmetti C, et al. In vivo metabolic imaging of Traumatic Brain Injury. Sci Rep 7, 17525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeVience SJ, et al. Metabolic imaging of energy metabolism in traumatic brain injury using hyperpolarized [1-(13)C]pyruvate. Sci Rep 7, 1907 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hackett EP, et al. Imaging Acute Metabolic Changes in Patients with Mild Traumatic Brain Injury Using Hyperpolarized [1-(13)C]Pyruvate. iScience 23, 101885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau AZ, Miller JJ, Robson MD, Tyler DJ. Simultaneous assessment of cardiac metabolism and perfusion using copolarized [1-(13) C]pyruvate and (13) C-urea. Magn Reson Med 77, 151–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin H, et al. Simultaneous Metabolic and Perfusion Imaging Using Hyperpolarized (13)C MRI Can Evaluate Early and Dose-Dependent Response to Radiation Therapy in a Prostate Cancer Mouse Model. Int J Radiat Oncol Biol Phys 107, 887–896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin H, et al. Clinical translation of hyperpolarized (13) C pyruvate and urea MRI for simultaneous metabolic and perfusion imaging. Magn Reson Med 87, 138–149 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Morze C, et al. Investigating tumor perfusion and metabolism using multiple hyperpolarized (13)C compounds: HP001, pyruvate and urea. Magn Reson Imaging 30, 305–311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bok R, et al. The Role of Lactate Metabolism in Prostate Cancer Progression and Metastases Revealed by Dual-Agent Hyperpolarized (13)C MRSI. Cancers (Basel) 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haacke EM, Xu Y, Cheng YC, Reichenbach JR. Susceptibility weighted imaging (SWI). Magn Reson Med 52, 612–618 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Haller S, Vernooij MW, Kuijer JPA, Larsson EM, Jager HR, Barkhof F. Cerebral Microbleeds: Imaging and Clinical Significance. Radiology 287, 11–28 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Tain RW, Scotti AM, Li W, Zhou XJ, Cai K. Imaging short-lived reactive oxygen species (ROS) with endogenous contrast MRI. J Magn Reson Imaging 47, 222–229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krukowski K, et al. Integrated Stress Response Inhibitor Reverses Sex-Dependent Behavioral and Cell-Specific Deficits after Mild Repetitive Head Trauma. J Neurotrauma 37, 1370–1380 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koerte IK, et al. A review of neuroimaging findings in repetitive brain trauma. Brain Pathol 25, 318–349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma P, Benford B, Li ZZ, Ling GS. Role of pyruvate dehydrogenase complex in traumatic brain injury and Measurement of pyruvate dehydrogenase enzyme by dipstick test. J Emerg Trauma Shock 2, 67–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brindle KM. Imaging metabolism with hyperpolarized (13)C-labeled cell substrates. J Am Chem Soc 137, 6418–6427 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Day SE, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med 13, 1382–1387 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Witney TH, et al. A comparison between radiolabeled fluorodeoxyglucose uptake and hyperpolarized (13)C-labeled pyruvate utilization as methods for detecting tumor response to treatment. Neoplasia 11, 574–582, 571 p following 582 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao Y, et al. Hyperpolarized [1-(13)C]pyruvate-to-[1-(13)C]lactate conversion is rate-limited by monocarboxylate transporter-1 in the plasma membrane. Proc Natl Acad Sci U S A 117, 22378–22389 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller JJ, et al. (13)C Pyruvate Transport Across the Blood-Brain Barrier in Preclinical Hyperpolarised MRI. Sci Rep 8, 15082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hackett EP, et al. Probing Cerebral Metabolism with Hyperpolarized (13)C Imaging after Opening the Blood-Brain Barrier with Focused Ultrasound. ACS Chem Neurosci 12, 2820–2828 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark AL, Weigand AJ, Bangen KJ, Merritt VC, Bondi MW, Delano-Wood L. Repetitive mTBI is associated with age-related reductions in cerebral blood flow but not cortical thickness. J Cereb Blood Flow Metab 41, 431–444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ojo JO, et al. Chronic Repetitive Mild Traumatic Brain Injury Results in Reduced Cerebral Blood Flow, Axonal Injury, Gliosis, and Increased T-Tau and Tau Oligomers. J Neuropathol Exp Neurol 75, 636–655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frati A, et al. Diffuse Axonal Injury and Oxidative Stress: A Comprehensive Review. Int J Mol Sci 18, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delage C, Taib T, Mamma C, Lerouet D, Besson VC. Traumatic Brain Injury: An Age-Dependent View of Post-Traumatic Neuroinflammation and Its Treatment. Pharmaceutics 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ismail H, et al. Traumatic Brain Injury: Oxidative Stress and Novel Anti-Oxidants Such as Mitoquinone and Edaravone. Antioxidants (Basel) 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keshari KR, Kurhanewicz J, Bok R, Larson PE, Vigneron DB, Wilson DM. Hyperpolarized 13C dehydroascorbate as an endogenous redox sensor for in vivo metabolic imaging. Proc Natl Acad Sci U S A 108, 18606–18611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto K, et al. Real-Time insight into in vivo Real-Time insight into in vivo redox status utilizing hyperpolarized [1-(13)C] N-acetyl cysteine. Sci Rep 11, 12155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JH, Park SW, Kang SH, Nam TK, Min BK, Hwang SN. Detection of traumatic cerebral microbleeds by susceptibility-weighted image of MRI. J Korean Neurosurg Soc 46, 365–369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tong KA, et al. Diffuse axonal injury in children: clinical correlation with hemorrhagic lesions. Ann Neurol 56, 36–50 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Chen H, Desai A, Kim HY. Repetitive Closed-Head Impact Model of Engineered Rotational Acceleration Induces Long-Term Cognitive Impairments with Persistent Astrogliosis and Microgliosis in Mice. J Neurotrauma 34, 2291–2302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chou A, et al. Empowering Data Sharing and Analytics through the Open Data Commons for Traumatic Brain Injury Research. Neurotrauma Rep 3, 139–157 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaeggemose M, FS R, Laustsen C. Comprehensive Literature Review of Hyperpolarized Carbon-13 MRI: The Road to Clinical Application. Metabolites 11, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haacke EM, Mittal S, Wu Z, Neelavalli J, Cheng YC. Susceptibility-weighted imaging: technical aspects and clinical applications, part 1. AJNR Am J Neuroradiol 30, 19–30 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pedregosa. F, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research 12, 2825–2830 (2011). [Google Scholar]
- 58.Emmanuel T, Maupong T, Mpoeleng D, Semong T, Mphago B, Tabona O. A survey on missing data in machine learning. J Big Data 8, 140 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]