Abstract
Many research problems involving medical texts have limited amounts of annotated data available (e.g., expressions of rare diseases). Traditional supervised machine learning algorithms, particularly those based on deep neural networks, require large volumes of annotated data, and they underperform when only small amounts of labeled data are available. Few-shot learning (FSL) is a category of machine learning models that are designed with the intent of solving problems that have small annotated datasets available. However, there is no current study that compares the performances of FSL models with traditional models (e.g., conditional random fields) for medical text at different training set sizes. In this paper, we attempted to fill this gap in research by comparing multiple FSL models with traditional models for the task of named entity recognition (NER) from medical texts. Using five health-related annotated NER datasets, we benchmarked three traditional NER models based on BERT—BERT-Linear Classifier (BLC), BERT-CRF (BC) and SANER; and three FSL NER models—StructShot & NNShot, Few-Shot Slot Tagging (FS-ST) and ProtoNER. Our benchmarking results show that almost all models, whether traditional or FSL, achieve significantly lower performances compared to the state-of-the-art with small amounts of training data. For the NER experiments we executed, the F1-scores were very low with small training sets, typically below 30%. FSL models that were reported to perform well on non-medical texts significantly underperformed, compared to their reported best, on medical texts. Our experiments also suggest that FSL methods tend to perform worse on data sets from noisy sources of medical texts, such as social media (which includes misspellings and colloquial expressions), compared to less noisy sources such as medical literature. Our experiments demonstrate that the current state-of-the-art FSL systems are not yet suitable for effective NER in medical natural language processing tasks, and further research needs to be carried out to improve their performances. Creation of specialized, standardized datasets replicating real-world scenarios may help to move this category of methods forward.
Index Terms—: Natural Language Processing, Text Mining, Machine Learning, Few-shot Learning, Medical Informatics
I. Introduction
The task of named entity recognition (NER) aims to identify and locate entity names in unstructured texts, and classify them into pre-defined entity types. In medical domain specific datasets, common entity types include drug names, genes, patients, adverse drug events (ADEs), reason (for taking drugs) and symptoms, to name a few [1]–[3]. In recent years, deep neural network based methods (a.k.a., deep learning) has achieved significant success in NER tasks when large labelled datasets are available [4]–[6], especially using self-supervised pre-trained language models (PLM), such as BERT [7] and RoBERTa [8]. Typically, in such models, a linear classifier is added to the end of the encoder to classify tokens to entities, and the target domain labels are used to fine-tune the models. Recurrent neural networks have also been widely used to achieve state-of-the-art performances [9].
There are still many open challenges in automatic NER, especially for medical domain specific texts [10]. In supervised learning settings with limited training instances, the application of traditional NER methods typically leads to overfitting (i.e., the learner is incapable of generalizing the characteristics of the training data) [11], [12]. Within the medical domain, text-based datasets are often small due to the difficulty of obtaining data or of manually annotating them. For NER problems, the entities of interest may also occur sparsely (e.g., expressions of rare diseases), and there may be no possible mechanism of obtaining large training datasets. Consequently, NER systems developed for sparsely-occurring concepts often rely on manually-developed rules [?]. Few-shot learning (FSL), which aims to learn to generalize from small numbers of training instances, is one potential way to overcome this crucial problem in NER. Another challenge associated with the development of NER systems is the mismatch between entity types in different domains [14]. For example, common entity types in medical datasets, such as symptoms, treatments, drugs, and genes have are unlikely to occur frequently in datasets acquired from the News domain. In FSL-based classification tasks, a common approach to fine-tuning a system for a new domain is to use similarity measures with the representations of classes to determine the class of an unclassified instance [15], [16]. Such approaches are not very applicable for few-shot NER tasks. Consequently, FSL-based NER tasks are typically considerably more challenging than FSL-based classification tasks.
FSL for biomedical NER is an emerging research topic, and so there is a lack of benchmarks that could enable the assessment of how well different approaches perform on the same data. To the best of our knowledge, no past research attempted to benchmark different FSL-based NER approaches on medical texts, and at the same time, compared their performances to traditional NER models on the same datasets. In past related studies (e.g. [17]), only 1–2 medical text datasets were benchmarked on the same datasets. We attempt to address this gap in research in this paper. Specifically, we make the following contributions:
We benchmark several few-shot NER approaches on five medical text datasets.
We compare the performances of six models including three traditional NER models: BERT–Linear Classifier (BLC)1, BERT–CRF (BC)2 and SANER3, and three few-shot learning NER models: StructShot & NNShot4, Few-Shot Slot Tagging (FS-ST)5 and ProtoNER6 on five medical text datasets.
We present a discussion of current research challenges for few-shot NER in the medical domain, and summarize important future research directions.
II. Materials and Methods
Common sources of biomedical NLP datasets include: (i) publicly downloadable (de-identified) data; (ii) datasets from shared tasks; and (iii) non-public datasets related to patient information from medical institutions such as hospitals. Shared tasks are competitions where the organizers release datasets publicly, and different teams train and evaluate their systems on the same datasets. As a large number of shared-tasks have been held in recent years, many datasets have bene made available to them. In this paper, we selected one dataset from category (i) above and four datasets from category (ii) to benchmark and explore the performance of six NER models under multiple different settings. Datasets from category (iii) were intentionally excluded.
A. Model Architectures
We employed the base BERT model followed by a linear classifier as our first traditional NER model, and the model architecture is shown in Figure 1(a). Taking advantage of self-supervised PLMs [7], we decided to use BERT to extract the contextualized representation of each token. Then the output of each token from BERT is passed through a fully connected neural network with a linear layer and a softmax layer at the end that projects tokens into entities. This architecture is typical for NER, and has been shown to achieve state-of-the-art performance on various datasets when sufficient training data is available [6], [18].
Fig. 1.
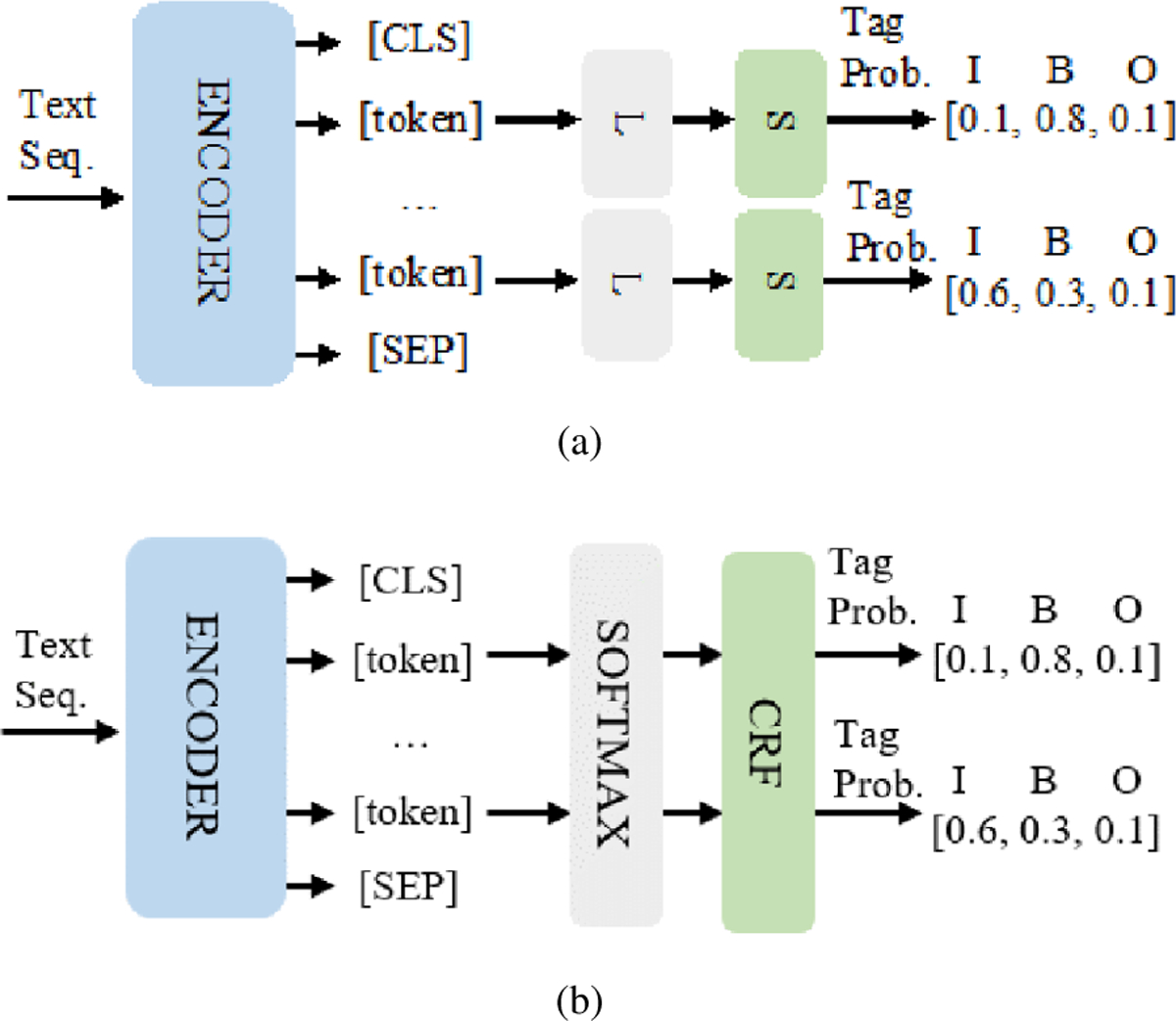
Architectures of two traditional NER models based on BERT. (a) BERT-Linear Classifier (BLC): The output of each token from BERT is passed through a fully connected neural network with a linear layer and a softmax layer at the end that project tokens into entities. (b) BERT-CRF (BC): The output of each token from BERT is fed into the CRF layer, which can explicitly model the dependencies between entities as a table with transition scores between all pairs of entity types and add some constraints to ensure that the final prediction result is valid.
The second traditional NER scheme we used is also based on BERT but with a conditional random fields (CRF) layer after the softmax layer instead of a linear layer like the previous system. Figure 1(b) shows the basic architecture of this model. The linear classifier leads to the conditional independence of each classification decision, and thus, it is necessary to design a transition matrix with context relevance. The CRF layer can explicitly model the dependencies between entities as a table with transition scores between all pairs of entity types and add some constraints to ensure that the final prediction result is valid. These constraints can be automatically learned by the CRF layer during training phase. For NER, texts are usually encoded in BIO or IO format (B represents “the beginning of the entity”, I represents “the inside of the entity”, and O represents “outside”). CRFs are effective in capturing dependencies between entities (e.g., I-Drug cannot follow O, it must always follow B-Drug). In addition to transition matrix, a CRF learner also includes an emission matrix. This emission matrix can be trained with Bi-LSTM, or it can be initialized randomly, but the performance is typically not as good as BERT. Consequently, we chose the BERT-CRF model as one of the traditional NER models in our experiments.
The last traditional NER model we employed is SANER[19]. SANER is a neural-based NER method for social media text that utilizes augmented semantics to improve the performance. The system contains a semantic enhancement module and a gate module to encode and aggregate these information separately, so as to solve the problems of data sparsity when dealing with short and informal texts. We included this model since medical texts invariably contain domain-specific formal or informal language and abbreviations.
The first two FSL NER models we explored were StructShot and NNShot [20]. StructShot uses contextual representations for each token in the support (training) set, and then uses a nearest neighbor (NN) and a Viterbi decoder to capture label dependencies, since the mismatch between the tags in the source domain and the target domain. The authors of this model use a standard and reproducible evaluation setup for the few-shot NER task by using standard test sets and development sets from several domains. NNShot is a simpler variant of StructShot, which computes a similarity score between a token in the test example and all tokens in the support set without using the Viterbi decoder. The performance of StructShot was shown to be better than that of NNShot.
The second model we included is the few-shot slot tagging model [21], which also includes a CRF layer. Since CRF considers both the transition score and the emission score to find the global optimal label sequence for each input, the framework in this paper includes two components: Transition Scorer and Emission Scorer. The transition scorer component captures the dependencies between labels. So the authors introduce a collapsed dependency transfer mechanism into the CRF to transfer abstract label dependency patterns as transition scores. Specifically, they collapse specific labels into three abstract labels: O, B and I and modeled transition from B and I to the same B (sB), a different B (dB), the same I (sI) and a different I (dI). To calculate the label transition probability for a new domain, the authors evenly distribute the abstract transition probabilities into corresponding target transitions. Then, the similarity between the word and each entity type is used as the CRF emission score. In order to calculate this similarity, the paper proposes a label-enhanced task-adaptive projection network (L-TapNet) based on the few-shot classification model TapNet, which represents labels by using label name semantics. Unlike StructShot model, FS-ST model uses the more popular meta-learning (aka., “learning to learn” [22]) framework for training and evaluation.
The third few-shot NER model is the ProtoNER model[23]. Metric learning methods, such as prototypical networks[15], which use prototypes (the average embeddings of support instances of each class) as the representations of each class, then compare the similarities between query instances and prototypes of each class based on certain distance metrics, showed state-of-the-art results in FSL for image classification tasks. Despite its success in image processing, metric learning has not been widely used in NLP tasks. Instead, in FSL settings for NLP, transfer learning is a more popular approach. Therefore, trying to adapt prototype-based methods such as prototypical networks for few-shot NER task naturally becomes another way of solving this problem. The ProtoNER model explored this possibility.
B. Datasets
We used five medical text datasets for comparing traditional NER models and FSL NER models. The datasets included MIMIC III (Medical Information Mart for Intensive Care) dataset [1], which contains information related to patients and is publicly available, and four additional datasets from different shared tasks. These four shared tasks are: (i) the N2C2 2018 shared task track 2 [24], which focuses on adverse drug event (ADE) and medication mention extraction; (ii) the I2B2 2014 shared task [2], which focuses on de-identification of longitudinal medical records; (iii) the BioNLP 2016 shared task [25], which focuses on extracting descriptions of genetic and molecular mechanisms from scientific articles; and (iv) the SMM4H (Social Media Mining for Health Applications) 2021 shared task 1b [26], which focuses on ADE mention extraction in social media data. Table I presents relevant statistics for all datasets—contains the source and aim of each NER task, training and test set sizes, the number of entity types and the number of entities in each dataset. The datasets combined included a total of 134,562 manually-annotated instances, with 90,776 (67.5%) instances for training and 43,786 (32.5%) instances for evaluation.
Table I.
Statistics of five biomedical datasets, including the source and aim of their tasks, training and test sizes (number of tokens), the number of entity types and the number of entities in each dataset.
| Datasets | Training Size | Test Size | Entity Types | Entities |
|---|---|---|---|---|
| N2C2 2018 track 2 (adverse drug events and medication extraction) | 60.3k | 37.7k | 9 | 7.6k |
| I2B2 2014 (de-identification of longitudinal medical records) | 47.2k | 18.1k | 23 | 2.1k |
| MIMIC III (information relating to patients) | 3.7k | 0.7k | 12 | 0.9k |
| BioNLP 2016 (Genetic and molecular mechanisms) | 51.5k | 14.9k | 1 | 2.8k |
| SMM4H 2021 task 1b (distinguishing adverse effect mentions) | 30.6k | 15.9k | 1 | 0.1k |
C. Data Collection and Preparation
Datasets are often reconstructed from existing ones to fit FSL scenarios. The most common way to represent data is K-Shot-N-Way, where “-shot” applies to the number of examples per category, and the suffix “-way” refers to the number of possible categories. Therefore, K-Shot-N-Way means that each of N classes or entities contains K labeled samples. Among these, 1-shot and 5-shot are the most common ‘shot’ settings. Therefore, in this paper, we conducted four sets of experiments using various proportions of the training data for the three traditional NER modes: 1-shot, 5-shot, 10% and 100%. For the FSL models, (StructShot NNShot and FS-ST), we conducted experiments using 1-shot, 5-shot and 15-shot settings. For the 10% setting, we randomly sampled 10% of the training set, and for 100% setting, we used the full training set.
For few-shot NER tasks, reconstructing datasets can be complex, as each text segment may contain more than one entity, often making it difficult to ensure that the reconstructed datasets include only K labeled samples for each entity type. Therefore, we followed the construction method proposed by Yang et al. [20], and used the greedy sampling strategy to construct the training sets (support sets). In particular, we sampled entity sentences in increasing order relative to their frequency. Take 5-shot setting (K=5) as an example. We first extracted the entity type with the lowest frequencies, and after collecting 5 text segments containing this entity, we considered the entity type with the second lowest frequency and checked whether it appeared in the support set for less than 5 times, as selected text segments may contain multiple entities. If it did occur less than 5 times, then we added more segments until it occurred 5 times. We followed these steps until all the entity types were included. The result of the greedy sampling strategy is to ensure that all entity types appear in the support set at least K times. And If any (instance, entity) pair is deleted from the support set, at least one entity type appears in the support set less than K times.
D. Experimental Setup
Due to the structural differences in the included models, we could not use uniform parameter settings for all of them. Thus, we implemented experiments according to the parameter settings described in the publication associated with each model, including the number of training epochs, batch size, learning rate, and random seed numbers.
III. Results
Table II presents the F1-scores of six NER models on each medical dataset. The table shows that all traditional NER models have relatively good performances when using full training data during training phases, especially on the relatively high-quality datasets (i.e., datasets which were collected and analyzed using a strict set of guidelines that ensure consistency and accuracy (low ambiguity), such as the N2C2 2018 and I2B2 2014 datasets). Even on the relatively noisy SMM4H dataset (social media), which has a small training set size, only one entity type, and relatively ambiguous annotations, their performances have been shown to quite good[27]. SANER outperformed the other two traditional NER models on most datasets and most settings. In the few-shot scenarios, however, the F1-scores for the BLC and BC models are mostly 0.0, suggesting that it is difficult for these models to generalize the characteristics from such a small training data.
Table II.
F1-scores of six NER models on five medical dataset. The best performance in 5-shot settings and 1-shot settings has been highlighted in bold and underlined.
| Models | Training Size | N2C2 2018 | I2B2 2014 | MIMIC III | BioNLP 2016 | SMM4H 2021 |
|---|---|---|---|---|---|---|
| SANER (traditional NER model) | Whole training data | 80.63 | 90.62 | 66.57 | 81.78 | 44.56 |
| 10% training data | 79.27 | 80.67 | 46.68 | 70.50 | 23.4 | |
| 5-shot | 10.27 | 36.38 | 21.25 | 23.14 | 0.00 | |
| 1-shot | 7.92 | 31.14 | 7.07 | 4.32 | 0.00 | |
| BERT + Classifier (traditional NER model) | Whole training data | 59.47 | 76.47 | 65.71 | 81.23 | 47.30 |
| 10% training data | 42.32 | 34.69 | 30.29 | 58.44 | 25.83 | |
| 5-shot | 3.27 | 0.00 | 0.00 | 1.71 | 0.00 | |
| 1-shot | 0.00 | 0.21 | 0.57 | 0.15 | 0.00 | |
| BERT + CRF (traditional NER model) | Whole training data | 82.79 | 80.63 | 59.58 | 77.62 | 45.45 |
| 10% training data | 64.09 | 27.84 | 20.5 | 61.35 | 2.74 | |
| 5-shot | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| 1-shot | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| StructShot (few-shot model) | 5-shot | 25.44 | 20.30 | 3.18 | 0.03 | 0.00 |
| 1-shot | 17.59 | 20.26 | 0.63 | 0.00 | 0.00 | |
| NNShot (few-shot model) | 5-shot | 25.29 | 19.73 | 19.71 | 28.88 | 0.00 |
| 1-shot | 16.70 | 16.35 | 15.37 | 6.42 | 0.00 | |
| FewShot-Tagging (few-shot model) | 5-shot | 0.94 | 0.27 | 0.60 | 3.32 | 0.32 |
| 1-shot | 4.59 | 0.14 | 5.17 | 6.81 | 0.35 |
From the table, we can also see that NNShot outperforms most other models in few-shot settings. This finding contrasts the performances reported previously in the literature. We suspect that compared with StructShot, NNShot might have better ability to extract and generalize features from medical texts. Another somewhat surprising result comes from the FS-ST model. In their original work, FS-ST model used the Ontonotes 5.0 dataset [28], WikiGold dataset [29], and several domains of the SNIPS dataset [30] for training, and the remaining domains of SNIPS were used as the test set for evaluating. The reported performances of the FS-ST model on these datasets are far better than those for the medical domain data that we used. This suggests that both the similarity of texts and the overlap of entity types between these three datasets are higher than those of the medical datasets.
Table III shows the results of ProtoNER model. This model does not reconstruct datasets for satisfying few-shot settings. Instead, it randomly selected N sentences for training from the original dataset whose entity types are not evenly distributed. Then, it conducts separate experiments for each entity type. Therefore, we were unable to obtain the F1-score for the entire datasets, and thus did not compare its results with other models. However, the values in table III show that, when the number of instances of an entity is obviously insufficient, the F1-score is also very low, even going down to zero on occasions. In contrast to the success of the prototypical network in the field of image classification, its performance on few-shot NER tasks is not as competitive. The performances shown in the table are not high enough for application in real-life settings.
Table III.
F1-scores of ProtoNER model. The results obtained according to each entity type of each dataset. the entity types with less than 10 instances and their performance have been highlighted in bold and underlined.
| Datasets | Labels | Instances of label | F1-score |
|---|---|---|---|
| N2C2 2018 | Drug | 12510 | 63.92 |
| Strength | 5519 | 83.45 | |
| Form | 5398 | 87.93 | |
| Frequency | 4062 | 63.57 | |
| Dosage | 3280 | 75.12 | |
| Route | 4672 | 85.72 | |
| Duration | 461 | 0.00 | |
| Reason | 2962 | 40.81 | |
| ADE | 692 | 24.24 | |
| I2B2 2014 | PATIENT | 903 | 61.81 |
| DOCTOR | 1986 | 63.37 | |
| USERNAME | 60 | 93.88 | |
| PROFESSION | 161 | 66.67 | |
| HOSPITAL | 945 | 55.03 | |
| ORGANIZATION | 88 | 19.99 | |
| STREET | 162 | 96.87 | |
| CITY | 293 | 69.99 | |
| STATE | 250 | 76.71 | |
| COUNTRY | 61 | 85.71 | |
| ZIP | 164 | 90.14 | |
| LOCATION-OTHER | 4 | 0.00 | |
| AGE | 874 | 79.99 | |
| DATE | 5087 | 69.31 | |
| PHONE | 175 | 81.97 | |
| FAX | 5 | 0.00 | |
| 2 | 0.00 | ||
| URL | 6 | 0.00 | |
| HEALTHPLAN | 1 | 0.00 | |
| MEDICALRECORD | 337 | 78.57 | |
| IDNUM | 78 | 54.05 | |
| DEVICE | 7 | 0.00 | |
| BIOID | 1 | 0.00 | |
| MIMIC III | CONDITION/SYMPTOM | 2365 | 40.01 |
| DRUG | 690 | 65.24 | |
| AMOUNT | 403 | 50.01 | |
| TIME | 326 | 40.63 | |
| MEASUREMENT | 665 | 49.85 | |
| LOCATION | 618 | 47.31 | |
| EVENT | 757 | 36.86 | |
| FREQUENCY | 62 | 0.00 | |
| ORGANIZATION | 114 | 28.62 | |
| DATE | 2 | 0.00 | |
| AGE | 44 | 95.25 | |
| GENDER | 36 | 99.98 | |
| BioNLP 2016 | GENE | 18258 | 27.87 |
| SMM4H 2021 | ADE | 1124 | 7.84 |
IV. Discussion
In the benchmarking results shown in Table II and Table III, the most important observation is perhaps that in few-shot medical NLP settings, all the models perform relatively poorly. The F1-scores almost invariably fall below 30%, which renders them unsuitable for practical applications. More research is clearly required to develop FSL methods that are applicable in practical settings. This is particularly true for NER tasks involving medical data. Some of their performances in low-shot settings were, however, higher than the performances of traditional NER systems, which suggests that there is some promise for FSL NER methods.
We found that the quality of (e.g., in terms of ambiguity) or the amount of noise in the datasets also plays a very important role in the performance of models on them. Although it is not shown in the tables, the performances of many models on the high-quality Conll2003 dataset [31] are much better than that on other datasets (StructShot 1-shot on Conll2003: 74.82%, FS-ST 1-shot on conll2003: 43.25%). It can also be seen in the table that almost all models have 0 F1-scores on the SMM4H dataset when the labeled data is very few. The SMM4H dataset is the only one that involves data from social media. Past research has shown that social media based medical NLP datasets are more difficult to obtain high performances on compared to medical datasets from other sources [32]. This is because social media data has specific characteristics that make NLP challenging, such as the presence of misspellings, colloquial expressions and noise. For example, “nosleep”, as a symptom after taking drugs is marked as “adverse drug event” in one tweet, but not in another tweet, which might be due to the subtle differences in the contexts in which they are mentioned (i.e., it can be an adverse event in some contexts and symptom in others, and it is not a standard medical term for either and therefore is unlikely to occur in other medical datasets).
The overarching aim of FSL is to enable systems to learn from few examples, as humans are often capable of doing[33]. Improving the intrinsic evaluation performances of FSL methods, especially on the datasets that have been explored, will still be one of the most important work in the future. Perhaps the other most influential work can be the creation of standardized publicly available datasets that will replicate real-world scenarios, and present actual FSL challenges. Currently, there is a paucity of such datasets, resulting in the need to reconstruct existing datasets to represent few-shot settings. Re-constructed datasets often do not accurately capture real-world scenarios. Specialized datasets representing few-shot scenarios will facilitate the thorough comparison of different FSL NER strategies, as well as the comparison of FSL NER methods with traditional NER methods. Furthermore, there is currently no specialized dataset for FSL-based biomedical NLP, and contributions in this space are necessary to move the state-of-the-art in FSL-based NER for medical text forward. Future shared tasks should consider designing problems relevant for FSL-based NLP approaches.
V. Conclusion
We benchmarked the performances of six NER models on five biomedical text datasets. We found that achieving good results on few-shot learning NER tasks by using biomedical data is still a big challenge for all models we tested, which implies the progress on this field has been limited. In addition, we found that transferring systems to low ambiguity datasets such as the I2B2 2014 dataset is more effective than to noisy text datasets such as the SMM4H 2021 dataset. Broadly speaking, our experiments suggest that improving the performance of internal evaluations, creating new and specialized dataset, and proposing benchmarks for this field are the most important directions for future research.
Acknowledgment
Research reported in this publication was supported by the National Institute on Drug Abuse (NIDA) of the National Institutes of Health (NIH) under award number R01DA046619. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Contributor Information
Yao Ge, Department of Biomedical Informatics School of Medicine, Emory University Atlanta, GA.
Yuting Guo, Department of Biomedical Informatics School of Medicine, Emory University Atlanta, GA.
Yuan-Chi Yang, Department of Biomedical Informatics School of Medicine, Emory University Atlanta, GA.
Mohammed Ali Al-Garadi, Department of Biomedical Informatics School of Medicine, Emory University Atlanta, GA.
Abeed Sarker, Department of Biomedical Informatics School of Medicine, Emory University Atlanta, GA.
References
- [1].Johnson A, Pollard T, Shen L et al. “Mimic-iii, a freely accessible critical care database.” Scientific data 3, pp. 1–9, doi: 10.1038/sdata.2016.35, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stubbs A and Uzuner Ö “Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/uthealth corpus” Journal of Biomedical Informatics, vol. 58, pp. S20–S29, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen Y, Zhou C, Li T, Wu H, Zhao X, Ye K and Liao J. “Named entity recognition from Chinese adverse drug event reports with lexical feature based BiLSTM-CRF and tri-training.” Journal of Biomedical Informatics, vol. 96, pp. 103252, doi: 10.1016/j.jbi.2019.103252, 2019. [DOI] [PubMed] [Google Scholar]
- [4].Cetoli A, Bragaglia S, O’Harney A and Sloan M “Graph convolutional networks for named entity recognition.” arXiv preprint arXiv:1709.10053, 2017. [Google Scholar]
- [5].Li Y, Shetty P, Liu L, Zhang C and Song L “Bertifying the hidden markov model for multi-source weakly supervised named entity recognition.” arXiv preprint arXiv:2105.12848, 2021. [Google Scholar]
- [6].Labusch K, Neudeckery C and Zellhofer D “Bert for named entity recognition in contemporary and historical German,” In Proceedings of the 15th Conference on Natural Language Processing, Erlangen, Germany, pp. 8–11, 2019. [Google Scholar]
- [7].Devlin J, Chang MW, Lee K and Toutanova K “Bert: Pretraining of deep bidirectional transformers for language understanding.” In Proceedings of the 2019 North American Chapter of the Association for Computational Linguistics: Human Language Technologies, 2019. [Google Scholar]
- [8].Liu Y et al. “Roberta: A robustly optimized bert pretraining approach.” arXiv preprint arXiv:1907.11692, 2019. [Google Scholar]
- [9].Magge A, Weissenbacher D, Sarker A, Scotch M, Gonzalez-Hernandez G “Deep neural networks and distant supervision for geographic location mention extraction” Bioinformatics, vol. 58, issue 13, pp. i565–i573, PMID: 29950020, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hofer M, Kormilitzin A, Goldberg P and Nevado-Holgado A “Few-shot learning for named entity recognition in medical text.” arXiv preprint arXiv:1811.05468, 2018. [Google Scholar]
- [11].Dong N, Xing EP “Few-shot semantic segmentation with prototype learning.” BMVC, vol. 3. No. 4, 2018. [Google Scholar]
- [12].Li W, Wang L, Xu J, Huo J, Gao Y, and Luo J “Revisiting local descriptor based image-to-class measure for few-shot learning.” In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 7260–7268, 2019. [Google Scholar]
- [13].Klein AZ, Sarker A, Cai H, Weissenbacher D, and Gonzalez-Hernandez G “Social media mining for birth defects research: A rule-based, bootstrapping approach to collecting data for rare health-related events on Twitter” Journal of Biomedical Informatics, vol. 87, pp. 68–78. doi: 10.1016/j.jbi.2018.10.001, PMID: 30292855, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hou Y et al. “Fewjoint: A few-shot learning benchmark for joint language understanding.” arXiv preprint arXiv:2009.08138, 2020. [Google Scholar]
- [15].Snell J, Swersky K and Zemel RS “Prototypical networks for few-shot learning.” Advances in neural information processing systems, vol. 30, 2017. [Google Scholar]
- [16].Vinyals O, Blundell C, Lillicrap T and Wierstra D “Matching networks for one shot learning.” Advances in neural information processing systems, vol. 29, pp. 3630–3638, doi: https://dl.acm.org/doi/abs/10.5555/3157382.3157504, 2016. [Google Scholar]
- [17].Huang J et al. “Few-shot named entity recognition: A comprehensive study.” arXiv preprint arXiv:2012.14978, 2020. [Google Scholar]
- [18].Souza F, Nogueira R and Lotufo R “Portuguese named entity recognition using bert-crf.” arXiv preprint arXiv:1909.10649, 2019. [Google Scholar]
- [19].Nie Y, Tian Y, Wan X, Song Y and Dai B “Named entity recognition for social media texts with semantic augmentation.” arXiv preprint arXiv:2010.15458, 2020. [Google Scholar]
- [20].Yang Y and Katiyar A “Simple and effective few-shot named entity recognition with structured nearest neighbor learning.” arXiv preprint arXiv:2010.02405, 2020. [Google Scholar]
- [21].Hou Y et al. “Few-shot slot tagging with collapsed dependency transfer and label-enhanced task-adaptive projection network.” arXiv preprint arXiv:2006.05702, 2020. [Google Scholar]
- [22].Hospedales T, Antoniou A, Micaelli P, Storkey A “Meta-learning in neural networks: A survey.” arXiv preprint arXiv:2004.05439, 2020. [DOI] [PubMed] [Google Scholar]
- [23].Fritzler A, Logacheva V and Kretov M “Few-shot classification in named entity recognition task.” Proceedings of the 34th ACM/SIGAPP Symposium on Applied Computing, pp. 993–1000, doi: 10.1145/3297280.3297378, 2019. [DOI] [Google Scholar]
- [24].Henry S, Buchan K, Filannino M, Stubbs A and Uzuner Ö “2018 n2c2 shared task on adverse drug eventsand medication extraction in electronic health records.” Journal of the American Medical Informatics Association, vol. 27, pp. 3–12, doi: 10.1093/jamia/ocz166, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chaix E et al. “Overview of the regulatory network of plant seed development (seedev) task at the bionlp shared task 2016.” The Association for Computational Linguistics, pp. 113, 2016. [Google Scholar]
- [26].Weissenbacher D et al. “Overview of the fourth social media mining for health (smm4h) shared tasks at acl 2019.” Proceedings of the fourth social media mining for health applications (SMM4H) workshop shared task, pp. 21–30, 2019. [Google Scholar]
- [27].Guo Y, Ge Y, Al-Garadi MA, et al. “Pre-trained Transformer-based Classification and Span Detection Models for Social Media Health Applications” Proceedings of the Sixth Social Media Mining for Health (SMM4H) Workshop and Shared Task, pp. 52–57, 2021. [Google Scholar]
- [28].Weischedel R et al. “Ontonotes release 5.0.” Penn.: Linguistic Data Consortium, 2013. [Google Scholar]
- [29].Balasuriya D, Ringland N, Nothman J, Murphy T and Curran JR “Named entity recognition in wikipedia.” In Proceedings of the 2009 Workshop on The People’s Web Meets NLP: Collaboratively Constructed Semantic Resources (People’s Web), pp. 10–18, 2009. [Google Scholar]
- [30].Coucke A et al. “Snips voice platform: an embedded spoken language understanding system for private-by-design voice interfaces.” arXiv preprint arXiv:1805.10190, 2018. [Google Scholar]
- [31].Sang Erik F., and Fien De Meulder. “Introduction to the conll-2003 shared task: Language-independent namedentity recognition.” arXiv preprint cs/0306050, 2003. [Google Scholar]
- [32].Sarker A et al. “Utilizing social media data for pharmacovigilance: A review.” Journal of Biomedical Informatics, vol. 54, pp. 202–212, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Brown TB et al. “Language models are few-shot learners.” Advances in neural information processing systems, vol. 33, pp. 1877–1901, 2020. [Google Scholar]


