Abstract
The diversity of duckweed (Lemnaceae) associated yeasts was studied using a culture-dependent method. A total of 252 yeast strains were isolated from 53 duckweed samples out of the 72 samples collected from 16 provinces in Thailand. Yeast identification was conducted based on the D1/D2 region of the large subunit (LSU) rRNA gene sequence analysis. It revealed that 55.2% and 44.8% yeast species were Ascomycota and Basidiomycota duckweed associated yeasts, respectively. Among all, Papiliotrema laurentii, a basidiomycetous yeast, was found as the most prevalent species showing a relative of frequency and frequency of occurrence of 21.8% and 25%, respectively. In this study, high diversity index values were shown, indicated by the Shannon-Wiener index (H′), Shannon equitability index (EH) and Simpson diversity index (1-D) values of 3.48, 0.86 and 0.96, respectively. The present results revealed that the yeast community on duckweed had increased species diversity, with evenness among species. Principal coordinate analysis (PCoA) revealed no marked differences in yeast communities among duckweed genera. The species accumulation curve showed that the observed species richness was lower than expected. Investigation of the plant growth promoting traits of the isolated yeast on duckweed revealed that 178 yeast strains produced indole-3-acetic acid (IAA) at levels ranging from 0.08–688.93 mg/L. Moreover, siderophore production and phosphate solubilization were also studied. One hundred and seventy-three yeast strains produced siderophores and exhibited siderophores that showed 0.94–2.55 activity units (AU). One hundred six yeast strains showed phosphate solubilization activity, expressed as solubilization efficiency (SE) units, in the range of 0.32–2.13 SE. This work indicates that duckweed associated yeast is a potential microbial resource that can be used for plant growth promotion.
Keywords: yeast, diversity, duckweed, plant growth promotion
1. Introduction
Duckweed is a small and fast growing aquatic plant in the Lemnaceae family, which has 38 species in five genera, Landotia, Lemna, Spirodela, Wolffia, and Wolffiella [1]. They are distributed around the world and can tolerate polluted water [2]. Four of the duckweed species are found globally. However, Wolffiella is only species found in the Americas and Africa [3]. Due to their aquatic habitat, duckweed is easy to harvest and requires no arable land.
Duckweed is a well-known feed for livestock such as ducks, swine, chicken, and fish due to its high protein content alongside other nutrients such as vitamins and astaxanthin [4]. Moreover, duckweed, especially Wolffia, has long been used in traditional Asian foods in countries such as Thailand, Laos, and Cambodia [5]. The nutritional content of duckweed in terms of starch, protein, fat, minerals, vitamins, and phytosterol content, as well as amino acid and fatty acids profiles has been analyzed [6]–[8]. Results suggested that duckweed contains high value nutrients and its use is recommended by the World Health Organization (WHO). Products derived from Lemna and Wolffia species have been deemed Generally Recognized as Safe (GRAS) by the US Food and Drug Administration. Over the past decade, several companies (e.g., Parabel, Hinoman, GreenOnyx) have been established to develop duckweed as a food and protein source [9]. Additionally, the fast-growing character and chemical composition of duckweed makes it a promising energy resource.
Duckweed directly absorbs nutrients from water. Therefore, it can be used as a phytoremediation agent for waste water treatment [10]. Duckweed removes nitrogenous compounds [11] as well as heavy metals by uptake through their root fronds [12],[13]. Wetlands contaminated with hazardous chemicals can also be remediated using duckweed [14]. More recently, application of duckweed to remediate crude oil contaminants and polyester manufacturing effluents from wastewater have been reported [15],[16].
Duckweed uptakes nutrients (nitrogen and phosphorus) from waste water to support its growth and to store nutrients in its tissue. When the nutrients are completely removed from waste water, duckweed uses internally stored nutrients to support their growth for a period of time. Duckweed has the capability to accumulate starch at levels of up to 50% of its dry weight and further increases starch accumulation during waste water treatment [17]. Several treatments can induce starch accumulation in duckweed, such as abiotic stressors and nutrient limitations [18],[19]. This enables utilization of duckweed for bioenergy production [20]. Starch in duckweed can be hydrolyzed to sugars, which can consequently be fermented to alcohols such as ethanol and butanol [18]. Bioenergy production from duckweed is more feasible than that from other energy plants, such as sugarcane, since their cell walls contain less lignin. Therefore, starch can easily be pooled and converted to fermentable sugars [21],[22] prior to biofuel production. Moreover, the duckweed biomass can be used to produce biogas by anaerobic digestion [23].
Plant associated yeasts are those that colonize either inside plant tissue or on surface of host plant. This relationship between plants and yeasts is mutualistic [24]. The plant provides some nutrients and a stable environment for yeasts, while the yeasts produce metabolites that promote the plants resistance to unfavorable conditions and phytopathogens. Moreover, plant associated yeasts have capabilities to promote plant growth by production of phytohormones such as indole-3-acetic acid, siderophores, and 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase. They also solubilize phosphate and zinc, as well as present antagonistic activities against plant pathogens [25],[26].
Research on duckweed associated microorganisms has been reported [27]–[30]. However, to the best of our knowledge, a study of duckweed associated yeasts has not been previously performed. Therefore, this study aims to investigate yeast communities associated with duckweed (Lemnaceae) using culture techniques together with molecular yeast identification. A species accumulation curve was investigated and diversity indices were evaluated. The isolated yeasts were screened for plant growth promoting factors, including indole-3-acetic acid and siderophore production, as well as phosphate solubilization.
2. Materials and methods
2.1. Sample collection and yeast isolation
A total of 72 duckweed samples were collected from 28 districts in 16 provinces of Thailand between February 2021 and May 2022. The samples were collected and kept in plastic bags during transport to the laboratory. Duckweed samples were identified to genus level by eye observation of morphological characteristics [31]. Yeast isolation was carried out within three days of collection.
Yeasts were isolated directly. Approximately 1 g of duckweed was rinsed with a sterile normal saline solution (NSS, 0.85% NaCl) to remove dirt. Then, the samples were put into 250 mL Erlenmeyer flasks containing 100 mL of sterile NSS, followed by shaking at 100 rpm for 30 min. This was repeated twice for 10 min to remove the microorganisms contaminated with water from the sampling site. The effectiveness of the surface wash was verified by spreading 0.1 mL of the rinse solution onto yeast extract-malt extract (YM) agar (0.3% yeast extract, 0.3% malt extract, 0.5% peptone, 1.0% dextrose and 1.5% agar) in Petri dishes. If no microbial colonies appeared after incubation at 30 °C for seven days, the microorganisms contaminated with water from the sampling site were completely removed. After the surface wash, duckweed samples were ground using a sterile mortar and pestle with 0.3 mL of sterile NSS. Then, homogenized samples were spread onto YM agar supplemented with 0.12% sodium propionate and 0.1% chloramphenicol to prevent filamentous fungi and bacterial contamination, respectively. Plates were incubated at 30 ± 2 °C for either 2–7 days or until yeast colonies appeared. Yeast colonies with different morphologies were cross-streaked onto YM agar for purification. Purified yeast isolates were stored at −20 °C in YM broth supplemented with 30% (v/v) glycerol for long term preservation.
2.2. Yeast identification
The DNA of purified yeasts was extracted according to Ruiz-Barba et al. [32] with slight modifications. Yeast cells grown on YM agar for 18–24 h were suspended in 100 µL of sterile deionized water in a sterile 1.5 mL plastic microtube. Then, an aliquot (100 µL) of chloroform/isoamyl alcohol (24:1) was added to the suspension prior to vortexing for 5 min. The mixtures were centrifuged at 14,000 × g for 5 min. An aliquot of the upper aqueous phase was used as a DNA template for amplification. The D1/D2 domain of the large subunit (LSU) rRNA gene was amplified using a polymerase chain reaction (PCR) with NL1 and NL4 primers [33]. The PCR products were examined using agarose gel electrophoresis under blue light and compared with DNA markers. Then, the PCR products were purified with a FavorPrep™ GEL/PCR Purification Mini kit (Favorgen, Austria). The purified PCR products were sent for DNA sequencing at First BASE Laboratories, Malaysia. The sequences were compared with those in the GenBank (http://www.ncbi.nlm.nih.gov/) database using a nucleotide BLASTN search [34]. The criteria of yeast species identification using similarity of the D1/D2 region of the LSU rRNA gene sequence were 99.41% and 99.51% for ascomycetous and basidiomycetous yeasts, respectively. The criterion for distinguish yeast genera using the similarity of D1/D2 region of LSU rRNA gene sequence was 97.11% [35].
2.3. Phylogenetic analysis
Phylogenetic analysis was performed based on the sequence of the D1/D2 region of the LSU rRNA gene to confirm the yeast identification using the MEGA (Version 7.0.26) program. The sequence of a representative yeast from an individual species was subjected to alignment with the type strain sequences from GenBank. Then, the alignment was used for phylogenetic tree construction. A phylogenetic tree was built from the evolutionary distance using a GTR evolutionary model and the maximum-likelihood method. Bootstrap values were calculated from 1000 replicates.
2.4. Biodiversity analysis
Yeast isolates that showed identical DNA sequences were excluded from the collection for analysis of biodiversity indices, species richness, and for principal coordinate analysis (PCoA). Yeast diversity was analyzed using the Shannon Wiener index (H′). Yeast community evenness was analyzed with Shannon equitability index (EH) [36],[37], which assumes a value between 0 and 1; a value approaching 1 indicates complete evenness among the species, while a value approaching 0 indicates no evenness. The equations are as follows:
| (1) |
| (2) |
where, Pi is the proportion of each species in the sample and S is the total number of species in the total sample.
The Simpson diversity index (1-D) is a measure of diversity that considers both richness and evenness. This value is between 0 and 1, where 1 represents infinite diversity and 0 is no diversity [38]. Its equation is as follows:
| (3) |
where, ni is the number of strains of each species and N is the total number of strains of all species.
The frequency of occurrence (%) was calculated as the number of samples in which a particular species was observed divided by the total number of samples. The relative frequency (%) was calculated as the number of strains of an individual species as a proportion of the total number of strains.
The species richness was estimated using the EstimateS software, Version 9.1 which calculated species richness from the sampling effort of the Chao 1, Jack 1, and bootstrap estimators with sample-based abundance data (i.e., the classic EstimateS input) [39].
The similarity of yeast communities associated with duckweed was measured using PCoA based on Jaccard similarity indices. Computational analysis was performed using PAST software, Version 4.0 [40].
2.5. Plant growth promoting factors
2.5.1. Indole-3-acetic acid production
Yeast was inoculated into 3 mL of yeast extract peptone dextrose medium (YPD) containing 1% yeast extract, 2% peptone, and 2% dextrose and incubated at 30 °C and 200 rpm for 16–18 h. Then, the yeast inoculum was transferred to a 125 mL Erlenmeyer flask containing 25 mL of YPD medium supplemented with 0.1% L-tryptophan. The initial OD600 was adjusted to 0.2 prior to incubation on a rotary shaker at 30 °C and 170 rpm for 3 days. Final OD600 values were spectrophotometrically determined. The samples were centrifuged at 14,000 × g for 5 min and the supernatants were collected for IAA analysis using high performance liquid chromatography (HPLC; Nexera LC-40 series, Shimadzu, Japan) with a Cosmosil SC18-MS-II column (Nacalai Tesque, Japan) and UV detector at 280 nm. A mixture of ethanol, acetic acid, and water (60:1.5:40 v/v/v) was used as a mobile phase with a flow rate of 0.3 mL/min, as described by Nutaratat et al. [41]. Authentic IAA (Sigma, USA) was used as a standard.
2.5.2. Siderophore production and phosphate solubilization
A yeast inoculum was prepared on a YPD medium supplement with 1.5% agar at 30 °C for 24 h. Then, the yeast inoculum was point inoculated on Chrome-Azurol S (CAS) agar [42] and Pikovskaya's agar [43] to determine siderophore production and phosphate solubilization, respectively, prior to incubation at 30 °C for seven days. Siderophore activity unit (AU) values were calculated as a ratio between the diameter of an orange halo zone and that of its associated colony. The phosphate solubilization efficiency (SE) was calculated as a ratio of the diameter of a clear zone to that of its associated colony.
3. Results
3.1. Sample collection and yeast isolation
Two-hundred and fifty-two yeast strains were isolated from 53 duckweed samples out of a total 72 samples collected from 16 Thai provinces in 2021–2022. The results indicated that 183, 36, 23, and 10 yeast strains were isolated from 33 Lemna, 9 Spirodela, 8 Landotia, and 3 Wolffia samples, corresponding to 45.83%, 12.5%, 11.11%, and 4.17% of individual duckweed genera associated with yeast, respectively (Table S1). The effectiveness of the sample surface wash procedure was tested by spreading the final rinse water onto YM agar plates. A few fungal colonies were found in the final rinse water of 3 out of 72 samples after seven days of incubation. However, no yeast or bacterial colonies were found in the final rinse water for any of the samples.
3.2. Yeast identification
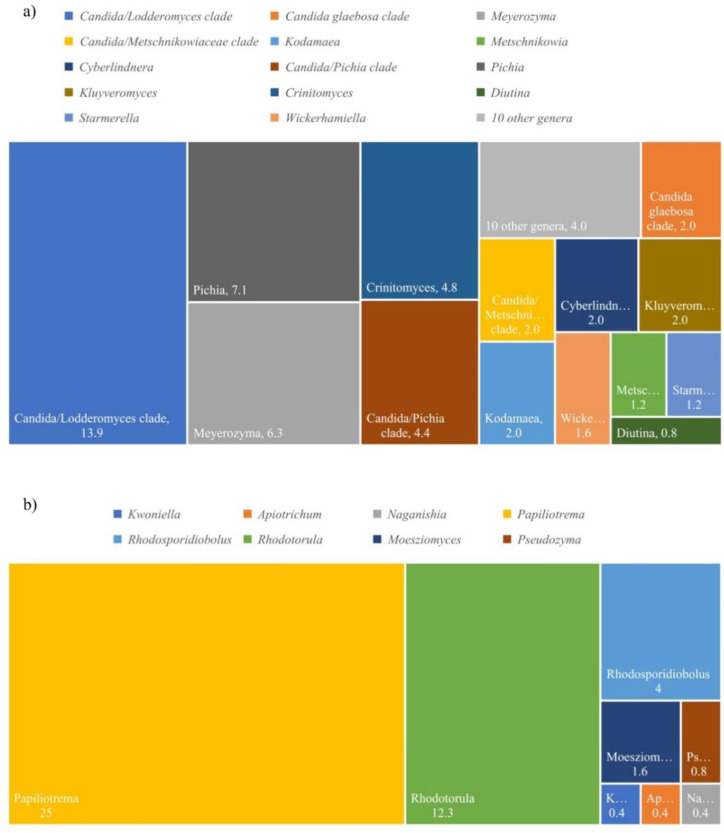
Two-hundred and fifty-two yeast strains were identified based on the D1/D2 region of the LSU rRNA gene sequence. According to Vu et al. [35], there is an increased number of yeast in the phylum Ascomycota (55.2%) when compared to Basidiomycota (44.8%). The proportion of yeast genera in duckweed samples is shown in Figure 1, whereas the relative of frequency data of each species is shown in Table S2.
Figure 1. Percentage of yeast genera found in duckweed samples, a) yeast in the phylum Ascomycota, b) yeast in the phylum Basidiomycota.
Two-hundred and thirty-seven yeasts out of 252 strains were identified as yeast species and 15 strains were identified to the genus level. There were four genera and four groups of Candida, including one strain of Candida sp. Group 1 (closely related to Candida tropicalis in the Candida/Lodderomyces clade), two strains of Candida sp. Group 2 (closely related to C. suratensis in the Candida/Metschnikowiaceae clade), one strain of Candida sp. Group 3 (closely related to C. yuanshanica in the Candida/Wickerhamomyces clade), one strain of Candida sp. Group 4 (closely related to C. pseudolambica in the Pichia/Candida clade), three strains of a Starmerella sp. (closely related to Starmerella caucasica), one strain of a Zygoascus sp. (closely related to Zygoascus polysorbophila), five strains of Papiliotrema sp. (closely related to Papiliotrema laurentii), and one strain of Rhodotorula sp. (closely related to Rhodotorula toruloides) (Table S3). These 15 yeast strains showed nucleotide sequence similarities that ranged between 97.11% and 99.41% to their closest species in the GenBank database.
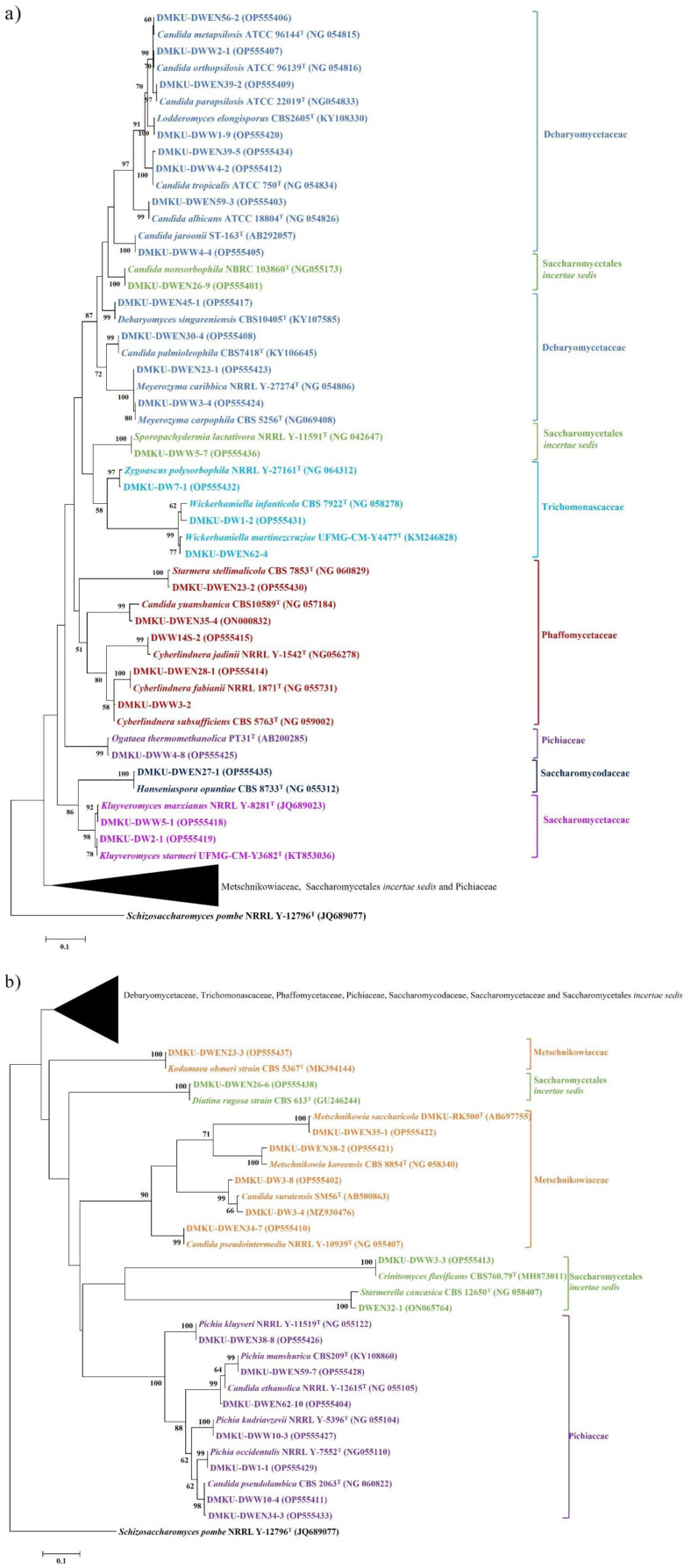
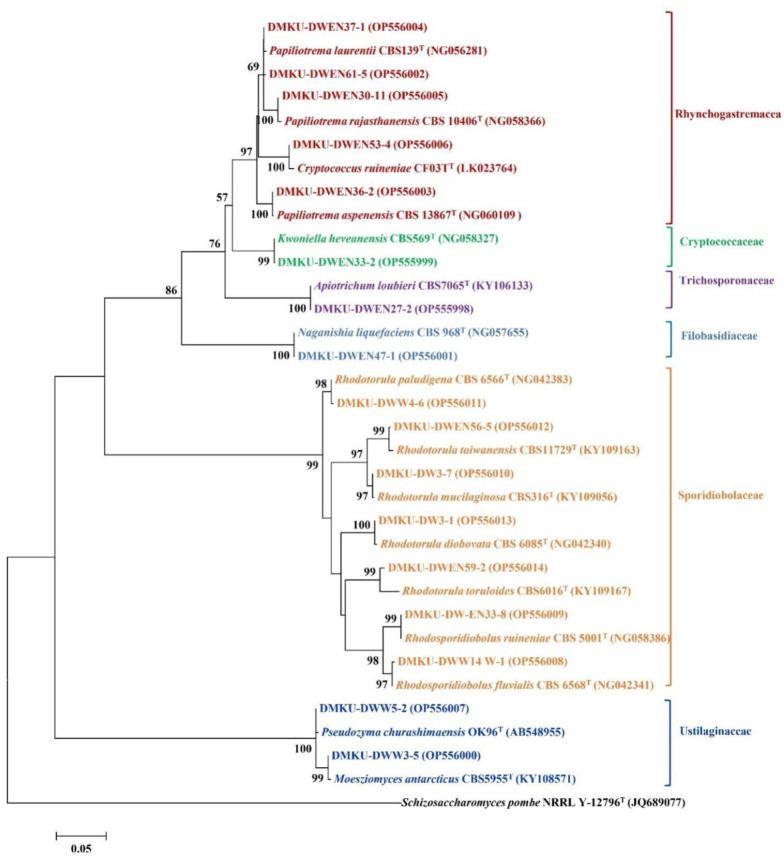
Among the 237 strains of known yeast species, 130 strains were identified as 35 known members of eight families in the phylum Ascomycota, including Debaryomycetaceae (11 species, 58 strains), Metschnikowiaceae (4 species, 11 strains), Phaffomycetaceae (4 species, 6 strains), Pichiaceae (7 species, 29 strains), Saccharomycetaceae (2 species, 5 strains), Saccharomycodaceae (1 species, 1 strain), Trichomonascaceae (2 species, 4 strains), and Saccharomycetales incertae sedis (4 species, 16 strains). However, 107 strains were identified as 15 known members of six families and three subphyla in the phylum Basidiomycota, including Filobasidiaceae (1 species, 1 strain), Cryptococcaceae (1 species, 1 strain), Rhynchogastremaceae (4 species, 58 strains), Trichosporonaceae (1 species, 1 strain), Sporidiobolaceae (6 species, 40 strains), and Ustilaginaceae (2 species, 6 strains). The phylogenetic placement of representative yeast isolated from duckweed is shown in Figure 2 (phylum Ascomycota) and Figure 3 (phylum Basidiomycota). The representative sequence data of those yeasts showing the highest similarity of the D1/D2 region of the LSU rRNA gene to the corresponding type strains were submitted to the GenBank database under the following accession numbers: ON065764, ON000832, MZ930476, OP609955, OP555401-OP555438, and OP555998-OP556014.
Figure 2. Phylogenetic placement of known species of the representative yeast species from duckweed (phylum Ascomycota) based on sequences of the D1/D2 region of the LSU rRNA gene. Reference sequences retrieved from the GenBank database are included. The tree was constructed with the maximum-likelihood method and the GTR evolutionary model. Numbers on the branches represent the bootstrap values (>50%) from 1000 random replicates. The scale bar corresponds to a genetic distance of 0.1 substitutions per position. Schizosaccharomyces pombe NRRL Y-12796T (JQ689077) was used as an outgroup in this analysis. a) A part of the tree showing the phylogenetic relationships of a partial taxa within Debaryomycetaceae, Saccharomycetales incertae sedis, Trichomonascaceae, Phaffomycetaceae, Pichiaceae, Saccharomycodaceae and Saccharomycetaceae b) Part of the tree that shows the phylogenetic relationships of another partial taxa within Metschnikowiaceae, Saccharomycetales incertae sedis and Pichiaceae.
Figure 3. Phylogenetic placement of known species of the representative yeast species from duckweed (phylum Basidiomycota) based on the sequence of the D1/D2 region of the LSU rRNA gene. Reference sequences retrieved from the GenBank database are included. The tree was constructed with the maximum-likelihood method and the GTR evolutionary model. Numbers on the branches represent the bootstrap values (>50%) from 1000 random replicates. The scale bar corresponds to a genetic distance of 0.05 substitutions per position. Schizosaccharomyces pombe NRRL Y-12796T (JQ689077) was used as an outgroup in this analysis.
3.3. Yeast diversity
The results shown in Figures 2 and 3 indicate a slightly higher number of yeast strains in the phylum Ascomycota than in Basidiomycota. However, the most abundant species in this study was Papiliotrema laurentii (55 strains out of 252 strains, which is equivalent to a 21.8% relative frequency). Additionally, the highest occurring species was Pa. laurentii, found in 18 out of 72 samples (25% frequency of occurrence; FO), followed by Crinitomyces flavificans (16.7% FO), Candida tropicalis (15.3% FO), and Rhodotorula mucilaginosa (13.9% FO). The other species had frequency of occurrence values between 1.4–9.7% (Table S2). Moreover, two yeast species, Pa. laurentii and Cr. flavificans, were found in all four genera of duckweed investigated in the present study.
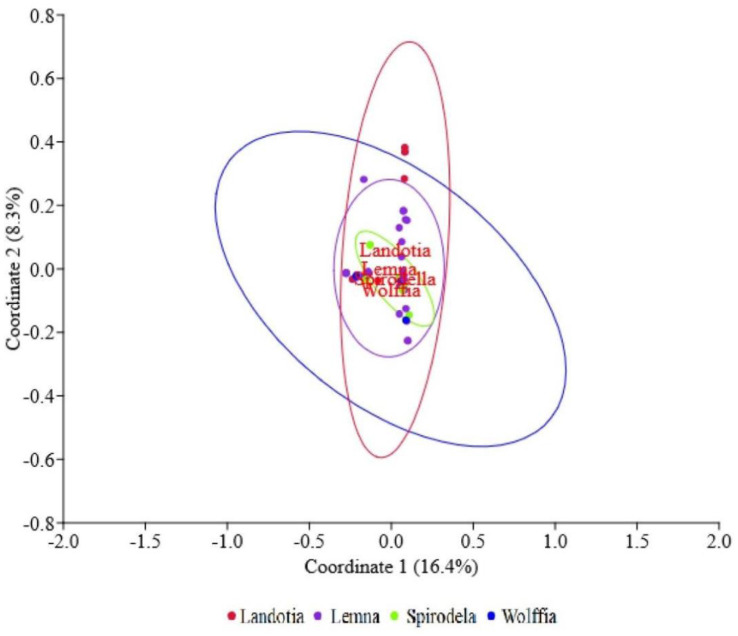
From 8 samples of Landotia duckweeds, 11 yeast species were found (members of 9 yeast genera in 6 families), whereas from 33 samples of Lemna duckweeds, 46 yeast species were found (members of 29 yeast genera in 13 families). In the case of Spirodella duckweeds, 20 yeast species (members of 14 yeast genera in 9 families) were found from 9 duckweed samples, while 5 yeast species (members of 5 yeast genera in 5 families) were found from 3 samples of Wolffia duckweeds. In order to determine difference in yeast diversity, the principal coordinate analysis (PCoA) was performed. As a result, no marked differences could be found in yeast species among duckweed genera (Figure 4). However, we observed that two yeast species, namely Pa. laurentii and Cr. Flavificans, can be isolated from all four duckweed genera.
Figure 4. Principal Coordinate Analysis (PCoA) plots of yeast communities on duckweed samples using Jaccard similarity coefficient.
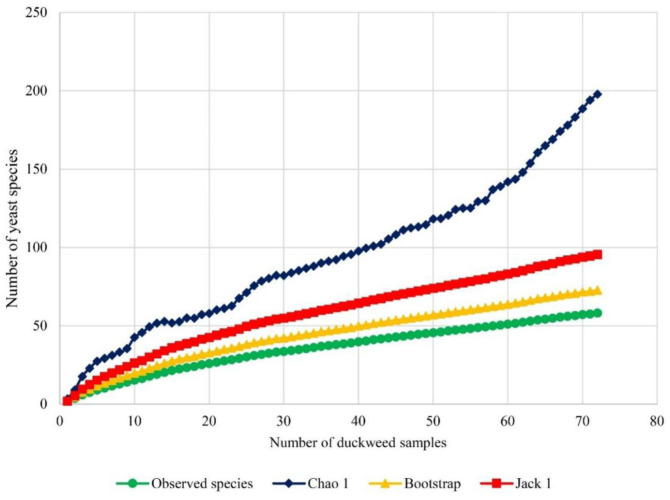
Diversity index values were calculated to evaluate the diversity of duckweed (Lemnaceae) associated yeasts. The Shannon-Wiener index (H′), Shannon equitability index (EH), and Simpson diversity index (1-D) values were 3.48, 0.86, and 0.96, respectively (Table 1). As the Shannon-Wiener index (H′) indicates diversity of yeast species, a higher value signifies an increased diversity of species in the habitat. If the index value is 0, only one species is present in the community. An H′ value is usually between 1.5 and 3.5 [44]. The H′ value obtained in the current study was 3.48. This value suggests that the yeast species from duckweed were highly diverse. The Shannon equitability index (EH) explains the evenness of species in a community. The term “evenness” simply refers to similarity of the abundance of different species in the community. An EH value is calculated between 0 and 1, and a value close to 1 indicates that the community has full evenness, while a value close to 0 means that there is no evenness among the species. The EH value of the present study was 0.86, revealing that yeasts species from duckweed in this community were highly even. The Simpson diversity index (1-D) is a measurement of diversity that is calculated from the number of species present, as well as the relative abundance of each species. When species richness and evenness increase, the diversity is greater. The value of the Simpson's diversity index in the present study was 0.96, which was close to 1, indicating that the yeast species from duckweed were highly rich and even. Estimation of the expected species richness demonstrated that the observed species richness was lower than the expected species richness (Figure 5). This result reveals that some yeast species may not have been recovered in this study.
Table 1. Diversity indices of yeast from duckweed.
| Diversity indices | Values/Yeast name |
| Number of total samples | 72 |
| Number of total yeast strains | 252 |
| Total number of yeast species (S) | 58 |
| Shannon Weiner index (H′) | 3.48 |
| Shannon Equitability index (EH) | 0.86 |
| Simpson diversity index (1-D) | 0.96 |
| The most prevalent yeast species | Papiliotrema laurentii |
Figure 5. Species accumulation curves showing the relationship between the number of duckweed samples and the number of observed species. Chao 1, Jack 1 and bootstrap species richness estimators were plotted.
3.4. Study of plant growth promoting factors
The obtained yeasts were studied to determine their plant growth promoting factors, including production of indole-3-acetic acid (IAA) and siderophores, as well as phosphate solubilization. The results are shown in Table 2.
Table 2. Production of IAA, siderophores and phosphate solubilization of the yeast isolated from duckweed.
| Yeast species | Yeast strain code | IAA production (mg/L) | Siderophore production (AU) | Phosphate solubilization (SE) |
| Candida albicans (Candida/Lodderomyces clade) | DWEN59-3 | 117.07 ± 1.67 | 1.10 ± 0 | 1.06 ± 0.03 |
| Candida jaroonii (Yamadazyma clade) | DWW4-4 | 151.35 ± 2.76 | 0 | 0 |
| Candida metapsilosis (Candida/Lodderomyces clade) | DWEN56-2 | 0 | 0 | 0 |
| Candida orthopsilosis (Candida/Lodderomyces clade) | DWW2-1 | 25.62 ± 3.21 | 1.04 ± 0.03 | 1.11 ± 0.13 |
| DWW2-2 | 24.07 ± 1.08 | 1.41 ± 0.31 | 1.08 ± 0 | |
| DWW2-3 | 19.88 ± 8.13 | 1.08 ± 0.01 | 1.10 ± 0.09 | |
| DWW2-4 | 26.59 ± 3.11 | 1.16 ± 0.07 | 0 | |
| Candida palmioleophila (Candida glaebosa clade) | DWEN30-4 | nd | 1.09 ± 0 | 0 |
| DWEN30-5 | 24.73 ± 1.17 | 1.20 ± 0 | 0 | |
| DWEN30-6 | 52.68 ± 32.04 | 1.23 ± 0.06 | 0 | |
| DWEN30-13 | 16.28 ± 0.46 | 1.31 ± 0.17 | 0 | |
| DWEN56-10 | 225.28 ± 4.46 | 0 | 0 | |
| Candida parapsilosis (Candida/Lodderomyces clade) | DW8-4 | 3.39 ± 0.13 | 1.50 ± 0 | 0 |
| DWEN30-3 | 2.05 ± 2.90 | 1.67 ± 0 | 0.97 ± 0.05 | |
| DWEN30-7 | nd | 2.00 ± 0 | 0.43 ± 0.37 | |
| DWEN30-9 | 0.08 ± 0.12 | 2.00 ± 0 | 0.94 ± 0.05 | |
| DWEN30-10 | 82.66 ± 3.27 | 1.83 ± 0.29 | 0.97 ± 0.05 | |
| DWEN30-15 | 27.15 ± 0.54 | - | 0.63 ± 0.15 | |
| DWEN39-1 | 76.5 ± 1.69 | 1.43 ± 0.06 | 0.44 ± 0.08 | |
| DWEN39-2 | 128.05 ± 1.64 | 1.7 ± 0.26 | 0.65 ± 0.09 | |
| DWEN62-9 | 0 | 1.31 ± 0.05 | 0.67 ± 0.12 | |
| Candida tropicalis (Candida/Lodderomyces clade) | DWW4-2 | 166.44 ± 5.43 | 0 | 0 |
| DW18-1 | 168.6 ± 2.05 | 1.44 ± 0.19 | 0.52 ± 0.06 | |
| DW18-2 | 176.89 ± 0.63 | 1.75 ± 0 | 0.84 ± 0.07 | |
| DWEN23-4 | nd | 1.28 ± 0.05 | 0.50 ± 0.04 | |
| DWEN23-6 | 50.13 ± 4.8 | 1.50 ± 0 | 0.71 ± 0.08 | |
| DWEN23-7 | 88.15 ± 43.97 | 1.49 ± 0.15 | 0.71 ± 0.06 | |
| DWEN23-8 | 60.2 ± 3.54 | 1.50 ± 0 | 0.73 ± 0.04 | |
| DWEN29-1 | 0 | - | 1.19 ± 0.02 | |
| DWEN30-2 | 64.31 ± 0.18 | 2.00 ± 0 | 1.06 ± 0.05 | |
| DWEN30-8 | 81.39 ± 1.17 | 1.67 ± 0.29 | 0.57 ± 0.1 | |
| DWEN30-14 | nd | 1.50 ± 0 | 0.62 ± 0.06 | |
| DWEN30-16 | 235.37 ± 8.6 | 1.83 ± 0.29 | 0.61 ± 0.05 | |
| DWEN33-3 | nd | 1.33 ± 0 | 0.76 ± 0.07 | |
| DWEN38-1 | 408.03 ± 37.79 | - | 0.67 ± 0 | |
| DWEN49-1 | 133.84 ± 5.36 | 1.50 ± 0 | 0.86 ± 0.02 | |
| DWEN54-2 | 127.48 ± 1.05 | 1.33 ± 0 | 0.77 ± 0.02 | |
| DWEN56-1 | 174.64 ± 8.57 | 1.42 ± 0.14 | 0.77 ± 0.07 | |
| DWEN62-1 | 115.49 ± 3.55 | 1.28 ± 0.10 | 0.55 ± 0.14 | |
| DWEN62-6 | 122.75 ± 1.78 | 1.5 ± 0 | 0.67 ± 0.04 | |
| Candida sp. group 1 (closely related to C. tropicalis in Candida/Lodderomyces clade) | DWEN39-5 | 527.77 ± 5.41 | 1.50 ± 0 | 0.47 ± 0.05 |
| Debaryomyces singareniensis | DWEN45-1 | 179.76 ± 30.68 | 0 | 0.78 ± 0.14 |
| Lodderomyces elongisporus | DWW1-9 | 13.83 ± 0.27 | 1.5 ± 0.29 | 0 |
| Meyerozyma caribbica | DWEN23-1 | 76.45 ± 11.95 | 1.53 ± 0.06 | 0.86 ± 0.04 |
| DWEN23-9 | 17.2 ± 0.11 | 1.28 ± 0.05 | 0.56 ± 0.05 | |
| DWEN34-2 | 254.34 ± 7.87 | 1.5 ± 0 | 0 | |
| DWEN34-4 | 45.14 ± 5.14 | 1.19 ± 0.02 | 0.51 ± 0.11 | |
| DWEN34-5 | 39.86 ± 1.36 | 1.23 ± 0.03 | 0 | |
| DWEN34-6 | 40.61 ± 0.75 | 1.19 ± 0.02 | 0.32 ± 0.01 | |
| DWEN43-1 | 153.72 ± 16.31 | 0 | 1.05 ± 0.04 | |
| DWEN53-2 | 494.8 ± 40.30 | 0 | 0 | |
| DWEN60-3 | 128.34 ± 1.40 | 1.13 ± 0.02 | 0.75 ± 0.11 | |
| DWEN61-2 | 0 | 1.4 ± 0 | 1.19 ± 0.17 | |
| DWEN62-11 | 37.23 ± 3.17 | 1.17 ± 0 | 0.74 ± 0.03 | |
| Meyerozyma carpophila | DWW3-4 | 101.93 ± 5 | 0 | 0.67 ± 0.6 |
| DWW4-1 | 122.84 ± 3.12 | 0 | 0 | |
| DWW4-9 | 125.61 ± 5.33 | 0 | 0 | |
| DW3-2 | 173.89 ± 5.81 | 1.38 ± 0.11 | 0 | |
| DWEN39-4 | 70.29 ± 1.03 | 1.37 ± 0.11 | 0.34 ± 0.06 | |
| Candida pseudointermedia (Candida/Metschnikowiaceae clade) | DWEN30-1 | 130.51 ± 6.9 | 1.13 ± 0 | 0.63 ± 0.1 |
| DWEN34-7 | 366.45 ± 0.04 | 1.37 ± 0.05 | 1.08 ± 0 | |
| DWEN62-8 | 419.48 ± 15.87 | 1.33 ± 0.07 | 0.79 ± 0.04 | |
| Candida sp. group 2 (closely related to Candida suratensis in Candida/Metschnikowiaceae clade) | DW3-4 | 19.89 ± 0.56 | 1.17 ± 0 | 0 |
| DW3-8 | 85.62 ± 16.9 | 1.33 ± 0 | 0 | |
| Kodamaea ohmeri | DWEN23-3 | nd | 1.38 ± 0 | 1.00 ± 0 |
| DWEN23-5 | nd | 1.46 ± 0.07 | 0.65 ± 0.06 | |
| DWEN38-6 | 5.12 ± 6.47 | 1.40 ± 0.05 | 0.77 ± 0.05 | |
| DWEN38-9 | 0 | 1.67 ± 0 | 0.47 ± 0.15 | |
| DWEN50-1 | 337.68 ± 3.46 | 1.53 ± 0.12 | 0.81 ± 0.04 | |
| Metschnikowia koreensis | DWEN38-2 | 67.65 ± 2.38 | 1.67 ± 0 | 0.65 ± 0.3 |
| Metschnikowia saccharicola | DWEN35-1 | 14.52 ± 0.01 | - | 1.18 ± 0 |
| DWEN35-3 | 24.54 ± 0.49 | 2.00 ± 0 | 1.06 ± 0.19 | |
| Candida sp. group 3 (closely related to Candida yuanshanica in Candida/Wickerhamomyces clade) | DWEN35-4 | 47.90 ± 0.33 | 0 | 0.95 ± 0.08 |
| Cyberlindnera fabianii | DWEN28-1 | 0 | - | 1.15 ± 0.14 |
| DWEN28-2 | 0 | - | 1.24 ± 0.13 | |
| Cyberlindnera jadinii | DWW14 S-2 | 0 | 0 | 1.41 ± 0.17 |
| Cyberlindnera subsufficiens | DWW3-2 | 0 | 0 | 1.00 ± 0.14 |
| DWW3-8 | 10.86 ± 0.45 | 0 | 0 | |
| Starmera stellimalicola | DWEN23-2 | 0 | - | 1.13 ± 0.06 |
| Candida ethanolica (Candida/Pichia clade) | DWEN62-10 | 43.16 ± 0.69 | 1.50 ± 0 | 0 |
| DWEN62-12 | nd | 0 | 0 | |
| DWEN62-13 | 0 | 1.36 ± 0.13 | 1.00 ± 0 | |
| Candida pseudolambica (Candida/Pichia clade) | DWW3-1 | 40.39 ± 0.68 | 0 | 0 |
| DWW10-2 | nd | nd | nd | |
| DWW10-4 | 97.06 ± 15.43 | 0 | 1.00 ± 0 | |
| DWEN26-3 | nd | 1.17 ± 0 | 0 | |
| DWEN35-2 | 52.30 ± 2.30 | 0 | 0 | |
| DWEN39-3 | 137.94 ± 4.00 | 1.19 ± 0.12 | 0.60 ± 0.04 | |
| DWEN49-2 | 0 | - | 0 | |
| Candida sp. group 4 (closely related to Candida pseudolambica in Candida/Pichia clade) | DWEN34-3 | 44.43 ± 3.80 | 1.58 ± 0.14 | 0.62 ± 0.08 |
| Ogataea thermomethanolica | DWW4-8 | 92.56 ± 17.61 | 0 | 0 |
| Pichia kluyveri | DWEN38-8 | 322.46 ± 17.87 | - | 2.13 ± 0.05 |
| Pichia kudriavzevii | DWW10-3 | 51.28 ± 2.77 | 0 | 0 |
| DWW11-1 | 124.38 ± 6.63 | 0 | 0.72 ± 0.05 | |
| DWEN26-7 | 17.79 ± 1.02 | 2.00 ± 0 | 1.00 ± 0 | |
| DWEN54-3 | 82.89 ± 3.64 | 1.25 ± 0 | 0.93 ± 0.03 | |
| DWEN56-7 | 46.82 ± 1.15 | 1.67 ± 0 | 1.03 ± 0.09 | |
| DWEN56-11 | 46.78 ± 0.36 | 1.42 ± 0.14 | 0.94 ± 0.04 | |
| DWEN59-4 | 45.76 ± 1.20 | 1.25 ± 0 | 0.88 ± 0.03 | |
| DWEN59-5 | 56.25 ± 1.51 | 1.25 ± 0 | 0.86 ± 0.12 | |
| DWEN59-6 | 155.65 ± 133.22 | 1.39 ± 0.10 | 0.84 ± 0.17 | |
| DWEN59-8 | 13.39 ± 0.01 | 1.22 ± 0.03 | 0.94 ± 0.07 | |
| DWEN60-1 | 0 | 1.25 ± 0 | 0.91 ± 0.08 | |
| DWEN61-1 | 0 | 1.56 ± 0.19 | 0.99 ± 0.09 | |
| DWEN62-2 | 30.38 ± 0.34 | 1.67 ± 0.14 | 1.02 ± 0.03 | |
| DWEN62-5 | 31.28 ± 1.00 | 0.94 ± 0.82 | 0.97 ± 0.02 | |
| DWEN62-7 | 49.54 ± 2.35 | 1.31 ± 0.05 | 0.82 ± 0.12 | |
| Pichia manshurica | DWEN59-7 | 11.75 ± 5.41 | 1.17 ± 0 | 0.68 ± 0.30 |
| Pichia occidentalis | DW1-1 | 688.93 ± 24.76 | 1.50 ± 0 | 0 |
| Kluyveromyces marxianus | DWW5-1 | 20.25 ± 0.68 | 0 | 0 |
| DWW5-11 | 30.32 ± 0.32 | 0 | 0 | |
| Kluyveromyces starmeri | DW2-1 | 69.63 ± 0.55 | 1.00 ± 0.87 | 1.19 ± 0.01 |
| DWEN26-4 | 181.24 ± 52.91 | 2.00 ± 0 | 1.37 ± 0.15 | |
| DWEN26-5 | 5.63 ± 0.52 | 2.00 ± 0 | 1.91 ± 0.37 | |
| Candida nonsorbophila (Candida/Saccharomycetales clade) | DWEN26-9 | 274.15 ± 2.01 | 1.00 ± 0 | 1.00 ± 0 |
| Crinitomyces flavificans | DWW3-3 | 36.77 ± 1.3 | 0 | 1.07 ± 0.39 |
| DWW3-6 | nd | 0 | 1.06 ± 0.33 | |
| DWW4-7 | 38.34 ± 1.57 | 0 | 1.04 ± 0 | |
| DWW5-3 | nd | - | 0.88 ± 0.03 | |
| DWW5-6 | nd | nd | nd | |
| DWW5-8 | 78.69 ± 62.34 | - | 0.98 ± 0.39 | |
| DWW5-9 | 105.03 ± 32.6 | - | 1.14 ± 0.34 | |
| DWW5-12 | 86.58 ± 65.39 | - | 1.15 ± 0.09 | |
| DWW7-1 | 114.76 ± 11.59 | 0 | 1.15 ± 0.09 | |
| DWW8-1 | nd | 0 | 1.01 ± 0.15 | |
| DWW11-2 | 121.04 ± 3.19 | - | 0.98 ± 0.06 | |
| DW1-3 | 133.29 ± 2.4 | - | 0.83 ± 0.04 | |
| Diutina rugosa | DWEN26-6 | 14.61 ± 0.17 | 2.00 ± 0 | 0 |
| DWEN53-3 | 15.12 ± 0.09 | 1.39 ± 0.10 | 0 | |
| Sporopachydermia lactativora | DWW5-7 | 71.47 ± 3.8 | 0 | 0 |
| Starmerella sp. (closely related to Starmerella caucasica) | DWEN32-1 | 41.62 ± 1.13 | 1.47 ± 0.12 | 0.84 ± 0.01 |
| DWEN32-2 | 43.85 ± 1.12 | 1.50 ± 0 | 0.81 ± 0.08 | |
| DWEN32-3 | 18.98 ± 9.18 | 1.42 ± 0.22 | 0.89 ± 0.1 | |
| Hanseniaspora opuntiae | DWEN27-1 | 0 | nd | nd |
| Wickerhamiella infanticola | DW1-2 | 16.88 ± 18.84 | 1.50 ± 0 | 0 |
| DW3-3 | 20.51 ± 1.44 | 1.50 ± 0 | 0 | |
| DWEN30-12 | 196.65 ± 281.78 | 0 | 0 | |
| Wickerhamiella martinezcruziae | DWEN62-4 | 0 | 1.39 ± 0.10 | 0 |
| Zygoascus sp. (closely related to Zygoascus polysorbophila) | DW7-1 | 0 | 1.60 ± 0 | 1.26 ± 0.15 |
| Naganishia liquefaciens | DWEN47-1 | 32.74 ± 39.54 | 0 | 0 |
| Kwoniella heveanensis | DWEN33-2 | 15.15 ± 1.00 | - | 0 |
| Papiliotrema aspenensis | DWEN36-2 | 22.68 ± 0.89 | - | 0 |
| Papiliotrema laurentii | DWW1-8 | 14.23 ± 0.33 | 1.41 ± 0.16 | 1.24 ± 0.04 |
| DWW9-1 | nd | 1.45 ± 0.18 | 0 | |
| DWW14 S-1 | 5.45 ± 0.77 | 1.17 ± 0 | 0 | |
| DWW14 S-3 | 5.09 ± 6.18 | 1.19 ± 0.05 | 0 | |
| DWW14 S-8 | 0 | 1.15 ± 0.02 | 0 | |
| DWW14 W-2 | 0 | 1.00 ± 0.88 | 0 | |
| DWW14 W-3 | 0 | 1.44 ± 0.1 | 0 | |
| DW8-3 | nd | nd | nd | |
| DW15-1 | 0 | 1.50 ± 0 | 0 | |
| DW15-2 | 0 | 1.39 ± 0.19 | 0 | |
| DWEN21-1 | 1.12 ± 0.11 | 2.17 ± 0.29 | 0 | |
| DWEN21-2 | nd | nd | nd | |
| DWEN22-3 | nd | 1.89 ± 0.19 | 0 | |
| DWEN22-4 | 1.51 ± 0.81 | 2.00 ± 0 | 0 | |
| DWEN22-5 | 0 | 1.83 ± 0.29 | 0 | |
| DWEN22-6 | 0 | 1.47 ± 0.21 | 0 | |
| DWEN22-7 | 0 | 0 | 0 | |
| DWEN22-8 | 0.97 ± 1.36 | 1.50 ± 0 | 0 | |
| DWEN29-2 | 0 | 1.33 ± 0 | 0 | |
| DWEN29-4 | 0 | 2.00 ± 0 | 0 | |
| DWEN29-5 | 6.53 ± 0.05 | 2.00 ± 0 | 0 | |
| DWEN29-6 | nd | 2.00 ± 0 | 0 | |
| DWEN29-7 | 5.42 ± 1.07 | 1.83 ± 0.14 | 0 | |
| DWEN29-8 | 6.64 ± 1.09 | 2.00 ± 0 | 0 | |
| DWEN29-9 | nd | 2.00 ± 0 | 0 | |
| DWEN29-10 | 0 | 2.00 ± 0 | 0 | |
| DWEN29-11 | nd | 2.00 ± 0 | 0 | |
| DWEN29-12 | nd | 2.00 ± 0 | 0 | |
| DWEN31-1 | 26.45 ± 2.25 | 1.83 ± 0.14 | 0 | |
| DWEN31-2 | 17.21 ± 0.56 | 1.75 ± 0 | 0 | |
| DWEN31-3 | 24.92 ± 0.51 | 1.50 ± 0 | 0 | |
| DWEN36-1 | 0 | 2.00 ± 0 | 0 | |
| DWEN36-3 | 0 | 1.44 ± 0.1 | 0 | |
| DWEN36-4 | nd | 1.50 ± 0 | 0 | |
| DWEN36-5 | nd | 1.25 ± 0 | 0 | |
| DWEN36-6 | 0 | 0 | 0 | |
| DWEN36-7 | 0 | 1.50 ± 0 | 1.09 ± 0.01 | |
| DWEN36-8 | 4.85 ± 6.86 | 2.00 ± 0 | 0 | |
| DWEN37-1 | 14.99 ± 1.29 | 0 | 0.67 ± 0.21 | |
| DWEN37-4 | 13.79 ± 0.97 | 0 | 0 | |
| DWEN37-7 | 109.63 ± 8.05 | 0 | 0 | |
| DWEN37-8 | 54.08 ± 0.45 | 2.00 ± 0 | 0.05 ± 0.08 | |
| DWEN37-9 | 33.54 ± 5.14 | 2.00 ± 0 | 0 | |
| DWEN37-10 | 79.38 ± 18.56 | 2.00 ± 0 | 0 | |
| DWEN38-4 | 42.91 ± 22.87 | 1.44 ± 0.19 | 0 | |
| DWEN38-5 | 2.31 ± 0.76 | 1.33 ± 0 | 0.56 ± 0.21 | |
| DWEN38-7 | 49.09 ± 2.15 | 1.33 ± 0.33 | 0 | |
| DWEN41-1 | 0 | nd | nd | |
| DWEN41-2 | 0 | 2.08 ± 0.38 | 0 | |
| DWEN41-4 | 47.77 ± 0.25 | 2.17 ± 0.29 | 0 | |
| DWEN54-6 | 0 | 1.00 ± 0.87 | 0 | |
| DWEN55-1 | 0 | 1.67 ± 0.29 | 0 | |
| DWEN60-2 | 108.04 ± 0.45 | 1.31 ± 0.05 | 0 | |
| DWEN60-4 | 78.43 ± 23.85 | 1.16 ± 0.15 | 0 | |
| DWEN61-3 | 23.81 ± 19.01 | 1.19 ± 0.08 | 0 | |
| Papiliotrema rajasthanensis | DWEN30-11 | 74.25 ± 0.65 | 1.39 ± 0.1 | 0 |
| Papiliotrema ruineniae | DWEN53-4 | 5.89 ± 0.30 | 0 | 0 |
| Papiliotrema sp. (closely related to Papiliotrema laurentii) | DWW14 S-6 | 0 | 1.19 ± 0.05 | 0 |
| DWEN29-3 | 0 | 2.00 ± 0 | 0 | |
| DWEN55-2 | 0 | 1.50 ± 0 | 0 | |
| DWEN60-5 | 0 | 1.3 ± 0.09 | 0 | |
| DWEN61-5 | 0 | 1.5 ± 0 | 0 | |
| Apiotrichum loubieri | DWEN27-2 | 0 | 0 | 0 |
| Rhodosporidiobolus fluvialis | DWW14 S-4 | 277.08 ± 4.74 | - | 0 |
| DWW14 S-7 | 606.28 ± 92.19 | 0 | 0 | |
| DWW14 W-1 | 562.11 ± 14.29 | 1.50 ± 0 | 0 | |
| DWW14 W-4 | 663.10 ± 17.5 | 1.19 ± 0.13 | 0 | |
| Rhodosporidiobolus ruineniae | DW8-2 | 0 | nd | nd |
| DWEN26-10 | 50.63 ± 16.08 | 2.33 ± 0.29 | 1.22 ± 0.03 | |
| DWEN33-1 | nd | nd | nd | |
| DWEN33-5 | 73.73 ± 13.04 | 1.67 ± 0 | 0 | |
| DWEN33-6 | 42.78 ± 38.98 | 1.61 ± 0.10 | 0 | |
| DWEN33-8 | 50.75 ± 7.91 | 2.55 ± 0.45 | 0 | |
| Rhodotorula mucilaginosa | DW3-5 | 99.93 ± 3.01 | - | 0 |
| DW3-7 | 164.46 ± 10.19 | - | 0 | |
| DW4-1 | 96.41 ± 0.47 | - | 0 | |
| DW4-2 | 91.34 ± 0.19 | 1.50 ± 0.17 | 0 | |
| DWEN22-1 | 24.46 ± 0.07 | 1.58 ± 0.07 | 0 | |
| DWEN22-2 | 30.06 ± 2.24 | 2.00 ± 0 | 0 | |
| DWEN39-6 | 386.8 ± 188.83 | 2.11 ± 0.19 | 0 | |
| DWEN42-1 | 405.24 ± 130.15 | 2.22 ± 0.19 | 0 | |
| DWEN43-2 | 307.03 ± 11.12 | 1.92 ± 0.14 | 0 | |
| DWEN53-1 | 122.83 ± 2.46 | 1.94 ± 0.59 | 0 | |
| DWEN53-5 | 102.44 ± 0.49 | 1.92 ± 0.14 | 0 | |
| DWEN56-3 | 94.57 ± 0.94 | 1.78 ± 0.19 | 0 | |
| DWEN56-6 | 109.11 ± 2.09 | 1.39 ± 0.10 | 0 | |
| DWEN56-9 | 97.17 ± 15.38 | 0 | 0 | |
| DWEN59-1 | 143.32 ± 10.22 | 1.61 ± 0.34 | 0 | |
| DWEN62-3 | 0 | 1.89 ± 0.10 | 0 | |
| Rhodotorula paludigena | DWW4-6 | nd | nd | nd |
| Rhodotorula taiwanensis | DWW5-5 | nd | nd | nd |
| DWW14 S-5 | 14.21 ± 5.07 | 0 | 0 | |
| DWEN37-2 | 9.51 ± 4.54 | 0 | 0 | |
| DWEN37-5 | 105.59 ± 31.58 | 0 | 0 | |
| DWEN37-6 | 49.18 ± 0.19 | 0 | 0 | |
| DWEN37-11 | 87.49 ± 16.72 | 2.00 ± 0 | 0 | |
| DWEN41-3 | 73.29 ± 0.05 | 2.19 ± 0.17 | 0.36 ± 0.63 | |
| DWEN55-3 | 11.14 ± 1.73 | 1.89 ± 0.19 | 0 | |
| DWEN55-4 | 4.47 ± 0.06 | 1.67 ± 0.58 | 0 | |
| DWEN55-6 | 24.62 ± 25.33 | 2.00 ± 0 | 0 | |
| DWEN56-5 | 2.00 ± 0.16 | 1.50 ± 0.07 | 0 | |
| Rhodotorula diobovata | DW3-1 | 4.25 ± 3.18 | 1.89 ± 0.19 | 0 |
| DW3-9 | 9.84 ± 13.92 | 2.00 ± 0 | 0 | |
| Rhodotorula sp. (closely related to Rhodotorula toruloides) | DWEN59-2 | 29.57 ± 27.64 | 1.14 ± 0.01 | 0 |
| Moesziomyces antarcticus | DWW3-5 | 101.93 ± 5 | nd | nd |
| DWW3-7 | nd | nd | nd | |
| DWW4-3 | nd | nd | nd | |
| DWW6-1 | 0 | 1.60 ± 0.17 | 0 | |
| Pseudozyma churashimaensis | DWW5-2 | 33.91 ± 33.4 | 1.61 ± 0.11 | 0 |
| DWW5-4 | 127.45 ± 3.19 | 0 | 0.93 ± 0.06 |
The results are expressed as mean ± standard deviation of duplicate of IAA production result, triplicate phosphate solubilization and siderophore production results. SE; The ratio of diameter of a clear zone and diameter of an associated colony, AU; The ratio of diameter of an orange halo zone and diameter of an associated colony.
nd; The results are not available, -; Yeast that did not grow on CAS agar
One hundred seventy-eight yeast produced IAA at levels ranging from 0.08–688.93 mg/L. The highest IAA producing yeast in the present study was Pichia occidentalis DW1-1. Among these, 58, 36, and 85 yeast strains produced IAA in the ranges of 0.08–49.54, 50.13–99.93, and 100–688 mg/L, respectively. It is notable that five strains were found to produce higher than 500 mg/L of IAA in YPD broth with tryptophan supplementation.
One hundred seventy-three yeast strains produced siderophores, evaluated as siderophore activity units (AU), in the range of 0.94–2.55. Rhodosporidiobolus ruineniae DWEN33-8 showed the highest siderophore producing capability. From a total of 173 strains, 138 and 35 exhibited siderophore AU units in ranges of 0.94–1.94 and 2.00–2.55, respectively. However, 21 yeast strains did not grow on CAS agar after seven days of incubation.
The results revealed that 106 yeast strains exhibited phosphate solubilization activity, expressed as solubilization efficiency (SE) units, in the range of 0.32–2.13. Among 106 strains of phosphate solubilizing yeasts, 68 and 38 strains exhibited activities in range of 0.32–0.99 and 1–2.13 SE units, respectively. The highest phosphate solubilizing yeast in this study was Pichia kluyveri DWEN38-8.
4. Discussion
The Lemnaceae family consists of five genera, Landotia, Lemna, Spirodella, Wolffia, and Wolffiella. In the present study, four duckweed genera, Landotia, Lemna, Spirodella, and Wolffia were collected from 16 provinces in Thailand. It was not possible to obtain Wolffiella in Thailand since this duckweed genus is only found in the Americas and Africa [3]. Two-hundred and fifty-two yeasts were isolated from 53 samples of duckweed, but 19 of these samples contained no yeast species. The results of yeast identification and the proportion of yeast genera found in this study, shown in Figure 1, suggest a slightly higher number of yeasts in the phylum Ascomycota (55.2%) than those in Basidiomycota (44.8%). In accordance with this study, the ascomycetous yeast isolated from the inside of apple fruits (Malus domestica) and pear fruits (Pyrus communis) were more diverse than basidiomycetous yeast [45]. It was reported that the ascomycetous yeast belonging to the genera Hanseniaspora and Metschnikowia were predominant in the tissues of fleshly fruits such as chokeberry, hawthorn, pumpkin, euonymus, gooseberry, sea-buckthorn, honeysuckle, tomato, apple, plum, pear, oak, currant, brier, ash berries, and black haw [46]. Additionally, out of 98 yeast strains of 114 yeasts, the study of the phylloplane yeast of diverse plants in Thailand also showed that the majority were ascomycetous yeast [47]. In contrast, Khunnamwong et al. [48] reported increased numbers of basidiomycetous yeast when compared to ascomycetous yeast in the leaf tissue of the three crops, rice, corn, and sugarcane. The plant associated yeast species can either be ascomycetes or basidiomycetes. Relationships between yeast and plant species cannot yet be ruled out.
Thirty-five known yeast species in the phylum Ascomycota and 15 known species in the phylum Basidiomycota were identified in this study. Among these, 11 ascomycetous yeast species were reported as endophytic yeasts: Candida metapsilosis [48], C. orthopsilosis [49], C. parapsilosis [45],[50], C. tropicalis [48], Meyerozyma caribbica [51], M. carpophila [52], C. pseudointermedia [48], Kodamaea ohmeri [48], Pichia kluyveri [53], Pi. kudriavzevii [49], and Hanseniaspora opuntiae [53]. Among the basidiomycetous yeast, 11 species of the 15 known species were reported as endophytic yeasts: Naganishia liquefaciens, Kwoniella heveanensis, Papiliotrema aspenensis, P. laurentii, P. rajasthanensis, Rhodosporidiobolus ruineniae [48], Rhodotorula mucilaginosa [54], R. paludigena, R. taiwanensis, Moesziomyces antarcticus, and Pseudozyma churashimaensis [48].
Many yeast species were isolated from either plants, plant materials, or plant associated sources including fruits, exudates of plants, peat moss, phylloplane of plants, flowers, decomposed plants, fermented tea leaves (Miang), cacti, bark of trees, and cotton. These species include C. albicans [55], C. jaroonii [56],[57], C. palmioleophila [58], Lodderomyces elongisporus [59], Metschnikowia koreensis [57],[59]–[61], M. saccharicola [62], Cyberlindnera fabianii [57],[59], Cy. jadinii [63], Cyberflaneur subsufficiens [64], Starmera stellimalicola [65],[66], C. ethanolica [67], C. pseudolambica [57], Pichia manshurica [47], P. occidentalis [68], Kluyveromyces marxianus [47], K. starmeri [69], C. nonsorbophila [70], Diutina rugosa [57], Wickerhamiella martinezcruziae [71], Papiliotrema ruineniae [72], Rhodosporidiobolus fluvialis [73], and Rhodotorula diobovata [74].
Some yeast species obtained in the present study have not been previously reported as yeasts isolated from plants or plant materials, viz. Debaryomyces singareniensis, Ogataea thermomethanolica, Crinitomyces flavificans, Sporopachydermia lactativora, Wickerhamiella infanticola, and Apiotrichum loubieri. De. singareniensis was isolated from coal mine soil [75]. Ogataea thermomethanolica and A. loubieri were isolated from soil [76],[77]. Crinitomyces flavificans was isolated from food waste and river water [78]. Sporopachydermia lactativora was isolated from Antarctic seawater, buffalo feces, and has been reported as a contaminant yeast in wine production [77],[79],[80] while Wickerhamiella infanticola was isolated from insects in a vineyard, an infant ear, and fish intestines [81]–[83].
Fifteen yeast strains obtained during this study were identified to five genera including Candida sp. Group 1, Candida sp. Group 2, Candida sp. Group 3, Candida sp. Group 4, Starmerella sp., Zygoascus sp., Papiliotrema sp., and Rhodotorula sp. (Table S3). The plant species and environment are two factors that influence plant associated yeast diversity and community. In this study, duckweed habitats were aquatic. Therefore pH, salinity, temperature, and organic matter content or even toxic contaminants of water may affect yeast quantity and species composition. Monapathi et al. [84] found that yeasts in fresh water consist of Candida, Clavispora, Cyberlindnera, Cryptococcus, Debaryomyces, Hanseniaspora, Kluyveromyces, Metschnikowia, Meyerozyma, Pichia, Rhodotorula, Saccharomyces, Torulaspora, Trichosporon, and Yarrowia. Rich yeasts species reflect inputs from terrestrial sources such as soil and plant debris and anthropogenic activities. Since duckweed is an aquatic plant found on water surfaces, yeast species associated with duckweed may come from a terrestrial source to colonize duckweed. Generally, ascomycetous yeasts are most likely found in areas that tend to be rich in organic carbon, while basidiomycetous yeasts most likely use a broader range of carbon compounds at lower concentrations [85]. Therefore, the plant species may affect the occurrence of yeasts. This may be a consequence of the nutrient composition of each plant. In this case, the yeast community may be affected by the nutrient composition of duckweed.
It is notable that many duckweed associated yeasts isolated in this work have been reported as human and opportunistic pathogens, viz. Candida albicans [86], C. metapsilosis, C. orthopsilosis, C. parapsilosis [87],[88], C. palmioleophila [89], C. tropicalis [90], Lodderomyces elongisporus [91], and Kodamaea ohmeri [92]. These species may have come from either humans or human activities around duckweed since these samples were collected near urban areas. Glushakova et al. [93] reported the presence of pathogenic and opportunistic yeast species, C. albicans, C. glabrata, and C. parapsilosis, on the pollen of wind pollinated plants in the urban environment while these yeasts were not present on plant pollen in a forest. The same research group reported the presence of an opportunistic species, C. parapsilosis, from the internal tissue of apple and pear fruits during the entire period of fruit formation and the development is due to anthropogenic impacts in a city [45].
In the present study, the most prevalent species was the basidiomycetous yeast Papiliotrema laurentii, as evident by its highest relative of frequency (21.8%) and frequency of occurrence (25%). Pa. laurentii was reported as an endophytic yeast on corn leaf tissue [48]. This yeast was also found in plant materials such as tree bark and decaying fruits [94], decaying tree samples in India [95], decaying organic material collected from both primary and secondary peat swamp forests in Thailand [96], and soil [97]. This information implies that Pa. laurentii is commonly found in plant material samples such as duckweed. Moreover, Pa. laurentii has been reported to produce polyamines and promote root growth of the medicinal plant, Agathosma betulina (Berg.) Pillans [98], and to accumulate lipid for biofuel production [97],[99].
Duckweed associated yeast diversity was investigated in this study using a culture-dependent methodology, which may not have isolated all of yeast species present. However, this methodology for isolating yeasts is well adopted and reported [48],[100],[101] to assure isolation of as many yeasts as possible. The species richness estimators, Chao1, Jack1, and Bootstrap (Figure 5), showed fewer observed species than expected, indicating that some yeast species may not have been recovered. These results are in accordance with the species composition of the yeast communities associated with plants inferred by previously reported culture-dependent and culture-independent approaches [48],[73],[102],[103]. Culture-independent methods, such as using next generation sequencing (NGS) combined with a culture-dependent approach to archive complete information, may be used to study yeast diversity on duckweed. However, culture-dependent methods are beneficial in terms of yielding pure culture microorganisms that are useful bioresources for basic studies and valuable applications.
Moreover, yeasts strain obtained from natural habitats have been used to produce biochemical products. Some of the yeast species obtained from this study have been reported to have industrial biotechnological potential. For example, Metschnikowia koreensis was reported as carbonyl reductase producer [104]. Cyberlindnera subsufficiens has capabilities to produce ethanol, IAA, and extracellular enzymes [96],[105]. M. saccharicola was reported to produce a toxin that is lethal to yeasts that are pathogenic to crabs [62],[106]. Cy. jadinii was used as a source of single cell proteins [107]. Candida ethanolica has been reported as an antagonistic yeast against bacterial wilt disease of tomato plants [108]. Endophytic yeasts were reported as IAA producers, including Hanseniaspora uvarum, Meyerozyma caribbica, Rhodosporidiobolus fluvialis, and Rhodotorula mucilaginosa. Candida and Kluyveromyces species have been reported to produce alkaline proteases that inhibit phytopathogenic fungi [109],[110]. In the present study, IAA and siderophore production, as well as phosphate solubilization, were also observed in yeasts obtained (Table 2). This result is accordance with the report of Nutaratat et al. [25], which showed that epiphytic and endophytic yeasts isolated from rice and sugar cane leaves in Thailand produced plant growth promoting factors such as IAA and siderophores, as well as phosphate and zinc solubilization, 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity, and antagonistic activity against fungal rice pathogens. Furthermore, many plants associated with yeasts were reported to possess plant growth promoting capabilities. For example, endophytic yeasts from agriculturally grown fruits produced IAA [100], while endophytic yeasts from strawberry (Fragaria × ananassa) leaves and wheat (Triticum aestivum) seeds were positively identified for phosphate solubilization, siderophore production, proteolytic activity, and ammonia production [111]. The diversity of ascomycetous and basidiomycetous yeast species from duckweed may lead to potential and valuable applications.
Pichia has been reported as a plant growth promoting yeast [112]. The study of phosphate solubilizing fungi isolated and characterized from Teff (Eragrostis teff) rhizosphere soil, collected from the North Shewa and Gojam, Ethiopia, reported that P. norvegensis had phosphate solubilizing capability [113]. Pichia sp. CC1 also showed an ability to promote lettuce growth by increasing the availability of phosphorus in the soil [114]. However, there has been no report of P. kluyveri with phosphate solubilization capability.
5. Conclusions
A culture-dependent method was used to study the diversity of yeasts on duckweed (Lemnaceae). Yeast identification was based on the D1/D2 region of the large subunit (LSU) rRNA gene sequence analysis. The results revealed that of the yeasts associated with duckweed, 55.2% were ascomycetous and 44.8% were basidiomycetous. The basidiomycetous yeast Papiliotrema laurentii was identified as the most prevalent species, with a relative of frequency and frequency of occurrence of 21.8% and 25%, respectively. High values of diversity indices were shown in this study, as indicated by the Shannon-Wiener index (H′), Shannon equitability index (EH), and Simpson diversity index (1-D) values of 3.48, 0.86, and 0.96, respectively. The present study revealed that the yeast community on duckweed shows high diversity and evenness of species. The study of plant growth promoting traits of yeasts from duckweed revealed that 178 yeast strains produced IAA in the range of 0.08–688.93 mg/L. One hundred and six strains showed phosphate solubilization activity in range of 0.32–2.13 solubilization efficiency (SE) units and 173 yeast strains produced siderophores in range of 0.94–2.55 siderophore activity units (AU). This work indicates that duckweed is a potential resource to obtain plant growth promoting yeasts.
Acknowledgments
This research is funded by Kasetsart University through the Graduate School Fellowship Program to Napapohn Kajadpai. We also acknowledge a support from Science and Technology Research Partnership for Sustainable Development (SATREPS), JICA. We would also like to thank UGSAS-GU via the “Microbiology Laboratory Station for IC-GU12” at Kasetsart University and International SciKU Branding (ISB), Faculty of Science, Kasetsart University. We would like to acknowledge Assoc. Prof. Dr. Kannika Duangmal, Assist. Prof. Dr. Chanita Boonmak, Miss Sirikarn Kammanee, Miss Varunya Sakpuntoon and Miss Yuparat Saimee for providing some duckweed samples.
Footnotes
Funding information: This work was financially supported by Kasetsart University Research and Development Institute (KURDI) under the project FF(KU) 4.64 (Utilization of duckweeds and associated microbes for wastewater treatment and bioplastic development).
Conflict of interest: All authors declare that there are no conflicts of interest in this paper.
Author contributions: N.K. performed the data curation, formal analysis, investigation, methodology and writing-original draft. J.A. performed formal analysis. P.K. revised the manuscript and N.S. provided conceptualization, funding acquisition, resources, supervision, review and editing. All authors have read and agreed to the published version of the manuscript.
References
- 1.Wang W, Wu Y, Yan Y, et al. DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol. 2010;10:1–11. doi: 10.1186/1471-2229-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landolt E, Kandeler R. Biosystematic investigations in the family of duckweeds (Lemnaceae), Vol. 4: the family of Lemnaceae-a monographic study, Vol. 2 (phytochemistry, physiology, application, bibliography) Veroeffentlichungen des Geobotanischen Instituts der ETH, Stiftung Ruebel (Switzerland) 1987 doi: 10.1007/BF00037640. [DOI] [Google Scholar]
- 3.Tippery NP, Les DH. Tiny plants with enormous potential: phylogeny and evolution of Duckweeds. In: Cao X.H., Fourounjian P., Wang W., editors. The Duckweed Genomes. Cham: Springer International Publishing; 2020. pp. 19–38. [DOI] [Google Scholar]
- 4.Sońta M, Rekiel A, Batorska M. Use of duckweed (Lemna L.) in sustainable livestock production and aquaculture–a review. Ann Anim Sci. 2019;19:257–271. doi: 10.2478/aoas-2018-0048. [DOI] [Google Scholar]
- 5.Bhanthumnavin K, Mcgarry MG. Wolffia arrhiza as a possible source of inexpensive protein. Nature. 1971;232:495–495. doi: 10.1038/232495a0. [DOI] [PubMed] [Google Scholar]
- 6.Appenroth KJ, Sree KS, Böhm V, et al. Nutritional value of duckweeds (Lemnaceae) as human food. Food Chem. 2017;217:266–273. doi: 10.1016/j.foodchem.2016.08.116. [DOI] [PubMed] [Google Scholar]
- 7.Edelman M, Colt M. Nutrient value of leaf vs. seed. Front Chem. 2016;4:32. doi: 10.3389/fchem.2016.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appenroth KJ, Sree KS, Bog M, et al. Nutritional value of the duckweed species of the genus Wolffia (Lemnaceae) as human food. Front Chem. 2018;6:483. doi: 10.3389/fchem.2018.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acosta K, Appenroth KJ, Borisjuk L, et al. Return of the Lemnaceae: Duckweed as a model plant system in the genomics and postgenomics era. Plant Cell. 2021;33:3207–3234. doi: 10.1093/plcell/koab189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yinglin W, Yuanyuan Y, Qian X, et al. Research advances on application of duckweed in bioremediation of polluted water. Ecotoxicol Environ Saf. 2022;74–85 doi: 10.7524/AJE.1673-5897.20210831002. [DOI] [Google Scholar]
- 11.Cedergreen N, Madsen TV. Nitrogen uptake by the floating macrophyte Lemna minor. New Phytol. 2002;155:285–292. doi: 10.1046/j.1469-8137.2002.00463.x. [DOI] [PubMed] [Google Scholar]
- 12.Yan A, Wang Y, Tan SN, et al. Phytoremediation: a promising approach for revegetation of heavy metal-polluted land. Front Plant Sci. 2020;11:359. doi: 10.3389/fpls.2020.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegler P, Sree K, Appenroth KJ. Duckweeds for water remediation and toxicity testing. Toxicol Environ Chem. 2016;98:1127–1154. doi: 10.1080/02772248.2015.1094701. [DOI] [Google Scholar]
- 14.Zhou H, Liu X, Chen X, et al. Characteristics of removal of waste-water marking pharmaceuticals with typical hydrophytes in the urban rivers. Sci Total Environ. 2018;636:1291–1302. doi: 10.1016/j.scitotenv.2018.04.384. [DOI] [PubMed] [Google Scholar]
- 15.Ekperusi AO, Nwachukwu EO, Sikoki FD. Assessing and modelling the efficacy of Lemna paucicostata for the phytoremediation of petroleum hydrocarbons in crude oil-contaminated wetlands. Sci Rep. 2020;10:1–9. doi: 10.1038/s41598-020-65389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osama R, Awad HM, Ibrahim MG, et al. Mechanistic and economic assessment of polyester wastewater treatment via baffled duckweed pond. J Water Process Eng. 2020;35:101179. doi: 10.1016/j.jwpe.2020.101179. [DOI] [Google Scholar]
- 17.Cheng JJ, Stomp AM. Growing duckweed to recover nutrients from wastewaters and for production of fuel ethanol and animal feed. Clean-Soil Air Water. 2009;37:17–26. doi: 10.1002/clen.200800210. [DOI] [Google Scholar]
- 18.Cui W, Cheng J. Growing duckweed for biofuel production: a review. Plant Biol. 2015;17:16–23. doi: 10.1111/plb.12216. [DOI] [PubMed] [Google Scholar]
- 19.Guo L, Jin Y, Xiao Y, et al. Energy-efficient and environmentally friendly production of starch-rich duckweed biomass using nitrogen-limited cultivation. J Clean Prod. 2020;251:119726. doi: 10.1016/j.jclepro.2019.119726. [DOI] [Google Scholar]
- 20.Ma Y, Zhu M, Yu C, et al. Large-scale screening and characterisation of Lemna aequinoctialis and Spirodela polyrhiza strains for starch production. Plant Biol. 2018;20:357–364. doi: 10.1111/plb.12679. [DOI] [PubMed] [Google Scholar]
- 21.Sowinski EE, Gilbert S, Lam E, et al. Linkage structure of cell-wall polysaccharides from three duckweed species. Carbohydr Polym. 2019;223:115119. doi: 10.1016/j.carbpol.2019.115119. [DOI] [PubMed] [Google Scholar]
- 22.Pagliuso D, Grandis A, Lam E, et al. High saccharification, low lignin, and high sustainability potential make duckweeds adequate as bioenergy feedstocks. Bioenergy Res. 2021;14:1082–1092. doi: 10.1007/s12155-020-10211-x. [DOI] [Google Scholar]
- 23.Ren H, Jiang N, Wang T, et al. Enhanced biogas production in the duckweed anaerobic digestion process. J Energy Resour Technol. 2018;140 doi: 10.1115/1.4039782. [DOI] [Google Scholar]
- 24.Kogel KH, Franken P, Hückelhoven R. Endophyte or parasite-what decides? Curr Opin Plant Biol. 2006;9:358–363. doi: 10.1016/j.pbi.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Nutaratat P, Srisuk N, Arunrattiyakorn P, et al. Plant growth-promoting traits of epiphytic and endophytic yeasts isolated from rice and sugar cane leaves in Thailand. Fungal Biol. 2014;118:683–694. doi: 10.1016/j.funbio.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Joubert PM, Doty SL. Endophytic yeasts: Biology, ecology and applications. In: Pirttilä A., Frank A., editors. Endophytes of forest trees. Cham: Springer; 2018. pp. 3–14. [DOI] [Google Scholar]
- 27.Saimee Y, Duangmal K. Streptomyces spirodelae sp. nov., isolated from duckweed. Int J Syst Evol Microbiol. 2021;71:005106. doi: 10.1099/ijsem.0.005106. [DOI] [PubMed] [Google Scholar]
- 28.Ishizawa H, Kuroda M, Inoue D, et al. Community dynamics of duckweed-associated bacteria upon inoculation of plant growth-promoting bacteria. FEMS Microbiol Ecol. 2020;96:fiaa101. doi: 10.1093/femsec/fiaa101. [DOI] [PubMed] [Google Scholar]
- 29.Zuki NAAM, Yahya H, Ariffin N, et al. The classification of duckweed and its bacterial community: A review. Malaysian J Sci. 2022;8:14–26. doi: 10.33102/2022238. [DOI] [Google Scholar]
- 30.Kittiwongwattana C, Vuttipongchaikij S. Biodiversity of endophytic bacteria isolated from duckweed (Landoltia punctata) and their IAA production. Sci Technol Asia. 2015:1–11. [Google Scholar]
- 31.Bog M, Appenroth KJ, Sree KS. Key to the determination of taxa of Lemnaceae: An update. Nordic J Botany. 2020;38 doi: 10.1111/njb.02658. [DOI] [Google Scholar]
- 32.Ruiz-Barba JL, Maldonado-Barragán A, Jiménez Díaz R. Small-scale total DNA extraction from bacteria and yeast for PCR applications. Anal Biochem. 2005;347 doi: 10.1016/j.ab.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 33.Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/A:1001761008817. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vu D, Groenewald M, Szöke S, et al. DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud Mycol. 2016;85:91–105. doi: 10.1016/j.simyco.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon CE. A mathematical theory of communication. Mob Comput Commun Rev. 2001;5:3–55. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 37.Spellerberg IF, Fedor PJ. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’Index. Glob Ecol Biogeogr. 2003;12:177–179. doi: 10.1046/j.1466-822X.2003.00015.x. [DOI] [Google Scholar]
- 38.Simpson EH. Measurement of diversity. Nature. 1949;163:688–688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 39.Colwell RK, Chao A, Gotelli NJ, et al. Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. Plant Ecol. 2012;5:3–21. doi: 10.1093/jpe/rtr044. [DOI] [Google Scholar]
- 40.Hammer Ø, Harper DA, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9. [Google Scholar]
- 41.Nutaratat P, Amsri W, Srisuk N, et al. Indole-3-acetic acid production by newly isolated red yeast Rhodosporidium paludigenum. J Gen Appl Microbiol. 2015;61:1–9. doi: 10.2323/jgam.61.1. [DOI] [PubMed] [Google Scholar]
- 42.Louden BC, Haarmann D, Lynne AM. Use of blue agar CAS assay for siderophore detection. J Microbiol Biol Educ. 2011;12:51–53. doi: 10.1128/jmbe.v12i1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fomina MA, Alexander IJ, Colpaert JV, et al. Solubilization of toxic metal minerals and metal tolerance of mycorrhizal fungi. Soil Biol Biochem. 2005;37:851–866. doi: 10.1016/j.soilbio.2004.10.013. [DOI] [Google Scholar]
- 44.Bibi F, Ali Z. Measurement of diversity indices of avian communities at Taunsa Barrage Wildlife Sanctuary, Pakistan. J Anim Plant Sci. 2013;23:469–474. [Google Scholar]
- 45.Glushakova A, Kachalkin A. Endophytic yeasts in Malus domestica and Pyrus communis fruits under anthropogenic impact. Microbiology. 2017;86:128–135. doi: 10.1134/S0026261716060102. [DOI] [PubMed] [Google Scholar]
- 46.Isaeva O, Glushakova A, Garbuz S, et al. Endophytic yeast fungi in plant storage tissues. Biol Bull Rev. 2010;37:26–34. doi: 10.1134/S1062359010010048. [DOI] [PubMed] [Google Scholar]
- 47.Limtong S, Koowadjanakul N. Yeasts from phylloplane and their capability to produce indole-3-acetic acid. World J Microbiol. 2012;28:3323–3335. doi: 10.1007/s11274-012-1144-9. [DOI] [PubMed] [Google Scholar]
- 48.Khunnamwong P, Jindamorakot S, Limtong S. Endophytic yeast diversity in leaf tissue of rice, corn and sugarcane cultivated in Thailand assessed by a culture-dependent approach. Fungal Biol. 2018;122:785–799. doi: 10.1016/j.funbio.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 49.Matos ÍTSR, de Souza VA, D'Angelo GdR, et al. Yeasts with fermentative potential associated with fruits of camu-camu (Myrciaria dubia, Kunth) from North of Brazilian Amazon. Sci World J. 2021;2021 doi: 10.1155/2021/9929059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaz AB, Mota RC, Bomfim MRQ, et al. Antimicrobial activity of endophytic fungi associated with Orchidaceae in Brazil. Can J Microbiol. 2009;55:1381–1391. doi: 10.1139/W09-101. [DOI] [PubMed] [Google Scholar]
- 51.Islam F, Salam MA, Rahman MA, et al. Plant endophytic yeasts Pichia fermentans and Meyerozyma caribbica improve growth, biochemical composition, haematological parameters and morphology of internal organs of premature Barbonymus gonionotus. Aquac Res. 2021;19:100575. doi: 10.1016/j.aqrep.2020.100575. [DOI] [Google Scholar]
- 52.de Lima Targino HM, Silva VSL, Escobar IEC, et al. Maize-associated Meyerozyma from the Brazilian semiarid region are effective plant growth-promoting yeasts. Rhizosphere. 2022:100538. doi: 10.1016/j.rhisph.2022.100538. [DOI] [Google Scholar]
- 53.Ling L, Li Z, Jiao Z, et al. Identification of novel endophytic yeast strains from tangerine peel. Curr Microbiol. 2019;76:1066–1072. doi: 10.1007/s00284-019-01721-9. [DOI] [PubMed] [Google Scholar]
- 54.Bura R, Vajzovic A, Doty SL. Novel endophytic yeast Rhodotorula mucilaginosa strain PTD3 I: production of xylitol and ethanol. J Ind Microbiol Biotechnol. 2012;39:1003–1011. doi: 10.1007/s10295-012-1109-x. [DOI] [PubMed] [Google Scholar]
- 55.Ramirez-Castrillon M, Usman LM, Silva-Bedoya LM, et al. Dominant yeasts associated to mango (Mangifera indica) and rose apple (Syzygium malaccense) fruit pulps investigated by culture-based methods. An Acad Bras Cienc. 2019;91 doi: 10.1590/0001-3765201920190052. [DOI] [PubMed] [Google Scholar]
- 56.Imanishi Y, Jindamorakot S, Mikata K, et al. Two new ascomycetous anamorphic yeast species related to Candida friedrichii—Candida jaroonii sp. nov., and Candida songkhlaensis sp. nov.—isolated in Thailand. Antonie van Leeuwenhoek. 2008;94:267–276. doi: 10.1007/s10482-008-9242-2. [DOI] [PubMed] [Google Scholar]
- 57.Limtong S, Kaewwichian R. The diversity of culturable yeasts in the phylloplane of rice in Thailand. Ann Microbiol. 2015;65:667–675. doi: 10.1007/s13213-014-0905-0. [DOI] [PubMed] [Google Scholar]
- 58.Kachalkin AV, Yurkov AM. Yeast communities in Sphagnum phyllosphere along the temperature-moisture ecocline in the boreal forest-swamp ecosystem and description of Candida sphagnicola sp. nov. Antonie van Leeuwenhoek. 2012;102:29–43. doi: 10.1007/s10482-012-9710-6. [DOI] [PubMed] [Google Scholar]
- 59.Limtong S, Kaewwichian R, Yongmanitchai W, et al. Diversity of culturable yeasts in phylloplane of sugarcane in Thailand and their capability to produce indole-3-acetic acid. World J Microbiol. 2014;30:1785–1796. doi: 10.1007/s11274-014-1602-7. [DOI] [PubMed] [Google Scholar]
- 60.Hong SG, Chun J, Oh HW, et al. Metschnikowia koreensis sp. nov., a novel yeast species isolated from flowers in Korea. Int J Syst Evol Microbiol. 2001;51:1927–1931. doi: 10.1099/00207713-51-5-1927. [DOI] [PubMed] [Google Scholar]
- 61.Camargo FP, Araujo ACV, de Moraes EM, et al. A comparison between cactophilic yeast communities isolated from Cereus hildmannianus and Praecereus euchlorus necrotic cladodes. Fungal Biol. 2016;120:1175–1183. doi: 10.1016/j.funbio.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 62.Kaewwichian R, Yongmanitchai W, Kawasaki H, et al. Metschnikowia saccharicola sp. nov. and Metschnikowia lopburiensis sp. nov., two novel yeast species isolated from phylloplane in Thailand. Antonie van Leeuwenhoek. 2012;102:743–751. doi: 10.1007/s10482-012-9774-3. [DOI] [PubMed] [Google Scholar]
- 63.Sibirny A. Non-conventional yeasts: from basic research to application: Springer. 2019. [DOI]
- 64.Lopes MR, Lara CA, Moura ME, et al. Characterisation of the diversity and physiology of cellobiose-fermenting yeasts isolated from rotting wood in Brazilian ecosystems. Fungal Biol. 2018;122:668–676. doi: 10.1016/j.funbio.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki M, Nakase T, Komagata K. Candida stellimalicola, a new species of anamorphic yeast isolated from star apple in Thailand. J Gen Appl Microbiol. 1994;40:115–121. doi: 10.2323/JGAM.40.115. [DOI] [Google Scholar]
- 66.da Cunha T, Ferraz LP, Wehr PP, et al. Antifungal activity and action mechanisms of yeasts isolates from citrus against Penicillium italicum. Int J Food Microbiol. 2018;276:20–27. doi: 10.1016/j.ijfoodmicro.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 67.Kanpiengjai A, Chui-Chai N, Chaikaew S, et al. Distribution of tannin-'tolerant yeasts isolated from Miang, a traditional fermented tea leaf (Camellia sinensis var. assamica) in northern Thailand. Int J Food Microbiol. 2016;238:121–131. doi: 10.1016/j.ijfoodmicro.2016.08.044. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, Shang Y, Chen J, et al. Diversity of non-Saccharomyces yeasts of grape berry surfaces from representative Cabernet Sauvignon vineyards in Henan Province, China. FEMS Microbiol Lett. 2021;368:fnab142. doi: 10.1093/femsle/fnab142. [DOI] [PubMed] [Google Scholar]
- 69.Freitas LF, Batista TM, Santos AR, et al. Yeast communities associated with cacti in Brazil and the description of Kluyveromyces starmeri sp. nov. based on phylogenomic analyses. Yeast. 2020;37:625–637. doi: 10.1002/yea.3528. [DOI] [PubMed] [Google Scholar]
- 70.Nakase T, Jindamorakot S, Am-In S, et al. Candida nonsorbophila sp. nov., a new ascomycetous yeast species isolated in Thailand. FEMS Yeast Res. 2009;9:663–667. doi: 10.1111/j.1567-1364.2009.00512.x. [DOI] [PubMed] [Google Scholar]
- 71.Maciel NO, Santos AR, Felix CR, et al. Wickerhamiella martinezcruziae fa, sp. nov., a yeast species isolated from tropical habitats. Int J Syst Evol Microbiol. 2021;71:005092. doi: 10.1099/ijsem.0.005092. [DOI] [PubMed] [Google Scholar]
- 72.Li AH, Yuan FX, Groenewald M, et al. Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud Mycol. 2020;96:17–140. doi: 10.1016/j.simyco.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Into P, Pontes A, Sampaio JP, et al. Yeast diversity associated with the phylloplane of corn plants cultivated in Thailand. Microorganisms. 2020;8:80. doi: 10.3390/microorganisms8010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osman ME, Abdel-Razik AB, Zaki KI, et al. Isolation, molecular identification of lipid-producing Rhodotorula diobovata: optimization of lipid accumulation for biodiesel production. J Genet Eng Biotechnol. 2022;20:1–15. doi: 10.1186/s43141-022-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saluja P, Prasad GS. Debaryomyces singareniensis sp. nov., a novel yeast species isolated from a coal mine soil in India. FEMS Yeast Res. 2007;7:482–488. doi: 10.1111/j.1567-1364.2006.00182.x. [DOI] [PubMed] [Google Scholar]
- 76.Limtong S, Srisuk N, Yongmanitchai W, et al. Pichia thermomethanolica sp. nov., a novel thermotolerant, methylotrophic yeast isolated in Thailand. Int J Syst Evol Microbiol. 2005;55:2225–2229. doi: 10.1099/ijs.0.63712-0. [DOI] [PubMed] [Google Scholar]
- 77.Lorliam W, Akaracharanya A, Suzuki M, et al. Diversity and fermentation products of xylose-utilizing yeasts isolated from buffalo feces in Thailand. Microbes Environ. 2013;28:354–360. doi: 10.1264/jsme2.me13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sakpuntoon V, Péter G, Groenewald M, et al. Description of Crinitomyces reliqui gen. nov., sp. nov. and reassignment of Trichosporiella flavificans and Candida ghanaensis to the genus Crinitomyces. J Fungi. 2022;8:224. doi: 10.3390/jof8030224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodrigues de Miranda L. A new genus: Sporopachydermia. Antonie van Leeuwenhoek. 1978;44:439–450. doi: 10.1007/BF00394320. [DOI] [PubMed] [Google Scholar]
- 80.Perpetuini G, Rossetti AP, Battistelli N, et al. Adhesion properties, biofilm forming potential, and susceptibility to disinfectants of contaminant wine yeasts. Microorganisms. 2021;9:654. doi: 10.3390/microorganisms9030654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camilo S, Chandra M, Branco P, et al. Wine microbial consortium: Seasonal sources and vectors linking vineyard and winery environments. Ferment. 2022;8:324. doi: 10.3390/fermentation8070324. [DOI] [Google Scholar]
- 82.Kurtzman CP. New anamorphic yeast species: Candida infanticola sp. nov., Candida polysorbophila sp. nov., Candida transvaalensis sp. nov. and Trigonopsis californica sp. nov. Antonie Van Leeuwenhoek. 2007;92:221–231. doi: 10.1007/s10482-007-9150-x. [DOI] [PubMed] [Google Scholar]
- 83.Kolochani MK, Mousavi SM, Zamani I, et al. Isolation and identification of lactic acid bacteria and yeasts with probiotic ability from the intestine of gilthead seabream. Iran Vet J. 2022;18:74–86. doi: 10.22055/IVJ.2022.353171.2482. [DOI] [Google Scholar]
- 84.Monapathi ME, Bezuidenhout CC, James Rhode OH. Aquatic yeasts: diversity, characteristics and potential health implications. J Water Health. 2020;18:91–105. doi: 10.2166/wh.2020.270. [DOI] [PubMed] [Google Scholar]
- 85.Suh SO, Blackwell M, Kurtzman CP, et al. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia. 2006;98:1006–1017. doi: 10.3852/mycologia.98.6.1006. [DOI] [PubMed] [Google Scholar]
- 86.Kim J, Sudbery P. Candida albicans, a major human fungal pathogen. J Microbiol. 2011;49:171–177. doi: 10.1007/s12275-011-1064-7. [DOI] [PubMed] [Google Scholar]
- 87.Brilhante RSN, Sales JA, da Silva MLQ, et al. Antifungal susceptibility and virulence of Candida parapsilosis species complex: an overview of their pathogenic potential. J Med Microbiol. 2018;67:903–914. doi: 10.1099/jmm.0.000756. [DOI] [PubMed] [Google Scholar]
- 88.Bertini A, De Bernardis F, Hensgens LA, et al. Comparison of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis adhesive properties and pathogenicity. Int J Med Microbiol. 2013;303:98–103. doi: 10.1016/j.ijmm.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 89.Jensen RH, Arendrup MC. Candida palmioleophila: characterization of a previously overlooked pathogen and its unique susceptibility profile in comparison with five related species. J Clin Microbiol. 2011;49:549–556. doi: 10.1128/JCM.02071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ann Chai LY, Denning DW, Warn P. Candida tropicalis in human disease. Crit Rev Microbiol. 2010;36:282–298. doi: 10.3109/1040841X.2010.489506. [DOI] [PubMed] [Google Scholar]
- 91.Al-Obaid K, Ahmad S, Joseph L, et al. Lodderomyces elongisporus: a bloodstream pathogen of greater clinical significance. New Microbes New Infect. 2018;26:20–24. doi: 10.1016/j.nmni.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ioannou P, Papakitsou I. Kodamaea ohmeri infections in humans: A systematic review. Mycoses. 2020;63:636–643. doi: 10.1111/myc.13094. [DOI] [PubMed] [Google Scholar]
- 93.Glushakova A, Kachalkin A, Zheltikova T, et al. Yeasts associated with wind-pollinated plants—leading pollen allergens in Central Russia. Microbiology. 2015;84:722–725. doi: 10.1134/S0026261715050082. [DOI] [PubMed] [Google Scholar]
- 94.Yalçın HT, Fındık B, Terzi Y, et al. Isolation and molecular identification of industrially important enzyme producer yeasts from tree barks and fruits. Arch Microbiol. 2021;203:1079–1088. doi: 10.1007/s00203-020-02104-6. [DOI] [PubMed] [Google Scholar]
- 95.Prakash A, Randhawa HS, Khan ZU, et al. Environmental distribution of Cryptococcus species and some other yeast-like fungi in India. Mycoses. 2018;61:305–313. doi: 10.1111/myc.12741. [DOI] [PubMed] [Google Scholar]
- 96.Boonmak C, Khunnamwong P, Limtong S. Yeast communities of primary and secondary peat swamp forests in southern Thailand. Antonie Van Leeuwenhoek. 2020;113:55–69. doi: 10.1007/s10482-019-01317-0. [DOI] [PubMed] [Google Scholar]
- 97.Vieira NM, Ventorim RZ, de Moura Ferreira MA, et al. Insights into oleaginous phenotype of the yeast Papiliotrema laurentii. Fungal Genet Biol. 2020;144:103456. doi: 10.1016/j.fgb.2020.103456. [DOI] [PubMed] [Google Scholar]
- 98.Cloete KJ, Valentine AJ, Stander MA, et al. Evidence of symbiosis between the soil yeast Cryptococcus laurentii and a sclerophyllous medicinal shrub, Agathosma betulina (Berg.) Pillans. Microb Ecol. 2009;57:624–632. doi: 10.1007/s00248-008-9457-9. [DOI] [PubMed] [Google Scholar]
- 99.Wang G, Liu L, Liang W. Single cell oil production from hydrolysates of inulin by a newly isolated yeast Papiliotrema laurentii AM113 for biodiesel making. Biotechnol Appl Biochem. 2018;184:168–181. doi: 10.1007/s12010-017-2538-9. [DOI] [PubMed] [Google Scholar]
- 100.Kachalkin A, Glushakova A, Streletskii R. Diversity of endophytic yeasts from agricultural fruits positive for phytohormone iaa production. BioTech (Basel) 2022;11:38. doi: 10.3390/biotech11030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gai CS, Lacava PT, Maccheroni Jr W, et al. Diversity of endophytic yeasts from sweet orange and their localization by scanning electron microscopy. J Basic Microbiol. 2009;49:441–451. doi: 10.1002/yea.3528. [DOI] [PubMed] [Google Scholar]
- 102.Kaewkrajay C, Putchakarn S, Limtong S. Cultivable yeasts associated with marine sponges in the Gulf of Thailand, South China Sea. Antonie van Leeuwenhoek. 2021;114:253–274. doi: 10.1007/s10482-021-01518-6. [DOI] [PubMed] [Google Scholar]
- 103.Srisuk N, Nutaratat P, Surussawadee J, et al. Yeast communities in sugarcane phylloplane. Microbiology. 2019;88:353–369. doi: 10.1134/S0026261719030135. [DOI] [Google Scholar]
- 104.Singh A, Chisti Y, Banerjee UC. Production of carbonyl reductase by Metschnikowia koreensis. Bioresour Technol. 2011;102:10679–10685. doi: 10.1016/j.biortech.2011.09.032. [DOI] [PubMed] [Google Scholar]
- 105.Jaiboon K, Lertwattanasakul N, Limtong P, et al. Yeasts from peat in a tropical peat swamp forest in Thailand and their ability to produce ethanol, indole-3-acetic acid and extracellular enzymes. Mycol Prog. 2016;15:755–770. doi: 10.1007/s11557-016-1205-9. [DOI] [Google Scholar]
- 106.Tan C, Wang L, Xue Y, et al. Purification and molecular characterization of a Metschnikowia saccharicola killer toxin lethal to a crab pathogenic yeast. FEMS Microbiol Lett. 2018;365 doi: 10.1093/femsle/fny038. [DOI] [PubMed] [Google Scholar]
- 107.Sousa-Silva M, Vieira D, Soares P, et al. Expanding the knowledge on the skillful yeast Cyberlindnera jadinii. J Fungi. 2021;7:36. doi: 10.3390/jof7010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nguyen MT, Ranamukhaarachchi SL, Hannaway DB. Efficacy of antagonist strains of Bacillus megaterium, Enterobacter cloacae, Pichia guilliermondii and Candida ethanolica against bacterial wilt disease of tomato. J Phytol. 2011;3 [Google Scholar]
- 109.Agrawal T, Kotasthane AS. Chitinolytic assay of indigenous Trichoderma isolates collected from different geographical locations of Chhattisgarh in Central India. Springerplus. 2012;1:1–10. doi: 10.1186/2193-1801-1-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vernekar JV, Ghatge MS, Deshpande VV. Alkaline protease inhibitor: a novel class of antifungal proteins against phytopathogenic fungi. Biochem Biophys Res Commun. 1999;262:702–707. doi: 10.1006/bbrc.1999.1269. [DOI] [PubMed] [Google Scholar]
- 111.Petkova M, Petrova S, Spasova-Apostolova V, et al. Tobacco plant growth-promoting and antifungal activities of three endophytic yeast strains. Plants. 2022;11:751. doi: 10.3390/plants11060751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nimsi KA, Manjusha K, Kathiresan K, et al. Plant growth-promoting yeasts (PGPY), the latest entrant for use in sustainable agriculture: a review. J Appl Microbiol. 2022;134 doi: 10.1093/jambio/lxac088. [DOI] [PubMed] [Google Scholar]
- 113.Gizaw B, Tsegay Z, Tefera G, et al. Phosphate solubilizing fungi isolated and characterized from teff rhizosphere soil collected from north showa and gojam, Ethiopia. J Fertil Pestic. 2017;8 doi: 10.4172/2471-2728.1000180. [DOI] [Google Scholar]
- 114.Nakayan P, Shen FT, Hung MH, et al. Effectiveness of Pichia sp. CC1 in decreasing chemical fertilization requirements of garden lettuce in pot experiments. J Food Ag-Ind. 2009 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.