Abstract
Humans are significantly impacting riverine systems worldwide, prompting us to investigate the effects of water pollution on the gut microbiome of Cyprinus carpio (common carp). Using 16S rRNA gene sequencing, we compared the gut microbiomes of common carp from two sites along river Yamuna with different pollution levels. Water pollution significantly altered the fish gut microbiome structure and microbial composition. Proteobacteria dominated in both sampling sites, while Bacteroidota prevailed in polluted water samples, indicating sewage and fecal contamination. Less polluted samples exhibited Verrucomicrobiae and Planctomycetes, negatively correlated with pollution levels. The polluted site had higher prevalence of potentially pathogenic and heavy metal-resistant bacteria, as well as microbial communities associated with wastewater treatment systems. Functional prediction highlighted the significant role of the gut microbiome in digestion and metabolism, with active enzymes for breaking down various organic substances. Biosynthetic pathways for leucine, valine, and isoleucine were present in both sites, known to be involved fish immunity. The host maintained a stable and diverse bacterial consortium, while microbial diversity became more specialized due to human activities, adapting to anthropogenic stress and selection pressures.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03747-0.
Keywords: Microbiome, Common carp, Anthropogenic activities, 16S rRNA gene
Introduction
Microorganisms form indispensable and multifaceted mutual associations with hosts, and these microbial communities are jointly known as microbiome (Ursell et al. 2012). Explicitly, the gut microbiome is of great interest as the intestine is a multifunctional organ system and harbors greater microbial diversity contrasted to other organs (Colston and Jackson 2016; Degregori et al. 2021). Microbial communities in the gastrointestinal tract (GI) of the host may influence development, growth, ecology, metabolism, reproduction, immune system, and evolution (Pérez et al. 2010). In spite of growing curiosity in vertebrate’s gut microbiome, understanding of microbial diversity composition and evolutionary dynamics of fishes is comparatively limited (Ghanbari et al. 2015; Tarnecki et al. 2019). With a diversity of 34,600 species (FishBase 2021), presence of broad ecological varieties, and characteristic microbial profile within their GI tracts, fish can represent a relevant model organism to examine the association of microbial communities with their hosts (Nelson et al. 2016). Interaction of several factors such as quality of surrounding water, diet, host genetics, developmental stage, immune status, and other host-specific pressures influences gut microbiota in fishes (Llewellyn et al. 2014; Khurana et al. 2021). Intraspecific variation in the gut microbiome can provide an understanding of the adaptive potential of species challenged with environmental stressors (Des Roches et al. 2018; Walter et al. 2019; Xue et al. 2006).
While variation in physicochemical parameters of surrounding water may induce natural responses in the host, human-induced disturbances can have more drastic consequences and these effects can be further exaggerated in fragile ecosystems like rivers. Owing to the industrial revolution and anthropogenic activities, Yamuna has become one of the most polluted rivers in the world (CPCB 2006; Sharma et al. 2017, 2020). As the river flows through distinct regions of Delhi, various drains and untreated wastewater severely deteriorate its water quality (Dhillon et al. 2013; Said and Hussain 2019). The fish diversity of the river Yamuna from Dakpathar to Allahabad constitutes 143 fish species (10 orders, 29 families, and 73 genera) with Cyprinidae being the most abundant family followed by Schilbeidae, Bagridae, and Sisoridae (Sharma et al. 2017; Koushlesh et al. 2021). The rise in pollution load has led to declined fish catches, a remarkable increase in invasive fish populations, and shifts in the fish species composition (Sharma et al. 2017). Due to these dynamics, this river is facing a hard battle against pollution and can be used for understanding the influence of anthropogenic activities on the microflora that are sentinels of water quality. Pollution also has been studied to have profound effects on species’ ecology as well as on their gut microbiomes (Degregori et al. 2021; Xia et al. 2014; Silbiger et al. 2018).
In spite of these facts, the impact of deteriorated water quality of river Yamuna on taxonomic and functional aspects of habitant fish’s gut microbiome remains unexplored. The common carp (Cyprinus carpio Linnaeus, 1758) is a widespread freshwater fish that is included in the world’s 100 worst invasive species (Courtenay Walter and Welcomme 1989; Dwivedi et al. 2016; GISD 2021). C. carpio serves as a keystone ecosystem engineer, as its feeding method involves agitating sediments at the bottom of the water body, thus causing uprooting macrophytes and modification of habitats for native fish and other aquatic species (FAO 2009). The microbial composition residing in the gut of C. carpio has formerly been explored by cultivation-based approaches (Sugita et al. 1990; Namba et al. 2007; Tsuchiya et al. 2008) but culture-independent methods such as 16S rRNA gene screening serve as a more reliable and detailed strategy to evaluate microbial community composition along with the functional potential in the GI tract of this fish (van Kessel et al. 2011; Eichmiller et al. 2016; Kakade et al. 2020). Being an aquatic bioindicator (Yeşilbudak and Erdem 2014), screening the intraspecies microbial diversity would lead the way for understanding the issues that resident fishes are facing in the polluted river Yamuna.
In this backdrop, our goal was to perform a comparative study of the microbial diversity and functional attributes of river Yamuna water and gut microbiome samples of C. carpio by using Illumina HiSeq 2500 platform for high-throughput sequencing of 16S rRNA gene. Samples were collected from two different geographical locations (~ 120 km away) of river Yamuna (i) anthropogenically impacted and highly polluted Wazirabad barrage, Delhi (site-α) (Bharti et al. 2022), and (ii) a comparatively less polluted site in Dabarkipar, Karnal, Haryana (site-β), to understand whether a core microbial consortia can be defined or are host host-specific selective pressures overwhelmed by the environmental conditions, in particular water pollution. Also, capturing the patterns of variation across the intraspecific gut microbial community composition would shed light on the consequences of changes in the environment.
Methodology
Collection of samples and amplicon sequencing
Water and fish samples were collected from the highly polluted site at (1) Wazirabad barrage, Delhi (site-α) (28° 40′ 5.53′′ N, 77° 15′ 0.35′′ E) (Supplementary1), (2) and a relatively less polluted site Dabarkipar, Karnal, Haryana (site-β); (29.6556º N, 77.1198º E), river Yamuna, India. Twenty samples of common carp from each site were collected by cast net of different mesh sizes and were brought to the laboratory. Length–weight (45.12 ± 3.21 cm; 620 ± 32.9 g (site-α) and 50.13 ± 2.11 cm; 700 ± 13.8 g (site-β)) estimation of all sampled fishes was performed. Physicochemical parameters of water (pH, temperature, total dissolved solids (TDS), dissolved oxygen (DO), electrical conductivity (EC), and chemical oxygen demand (COD)) were recorded on the spot by Orion 5-star Portable Multimeter (Thermo Fisher Scientific Inc. [NYSE: TMO], MA, USA) from both the sampling sites (Supplementary S1). Estimation of heavy metal traces of iron (Fe), zinc (Zn), cadmium (Cd), lead (Pb), nickel (Ni), and chromium (Cr) in the water samples was done by employing Atomic Absorption Spectroscopy (Sensa AAS Dual, GBC, Australia) with EPA’s acid digestion procedure (EPA, 3050B). Aseptic dissection of 20 fish samples was performed followed by removal of gastrointestinal tract (GIT) and storage in sterile cryotubes at – 80 °C. Water samples were filtered using 0.22 µm filter (MF-Millipore TM), and filters were carried on with DNA extraction. Power Soil® DNA isolation kit (MO BIO) was used to extract metagenomic DNA from water sample (5L) and gut contents (700 mg). Amplicon sequencing was performed using the Illumina HiSeq 2500 sequencing platform (Novogene Co., Ltd., China) by PCR (341 F and 805R barcoded fusion primers) from V3–V4 region of 16S rRNA genes.
Data curation, OTU clustering and taxonomic analysis
Low-quality bases, adaptor, barcodes, primer sequences were removed employing cut adapt in QIIME 2.2019.7 (Bolyen et al. 2019), and paired-end reads were merged using FLASH (v1.2.7) (Magoč and Steven 2011). Further, downstream analysis was performed with the processed reads (> 70% bases; Phred score ≥ 30). QIIME 2.2019.7 was employed for data analysis using de novo approach. Identification of operational taxonomy units (OTUs) (clustered at 97% similarity), taxonomic affiliation using a naïve Bayesian classifier (Wang et al. 2007), and SILVA 138 database (updated 2014) (Quast et al. 2013) using UCHIME algorithm (Edgar 2013) were performed. Phylum hierarchy was visualized using Circos v 0.63.10.
Statistical analysis
Alpha diversity metrics analysis was done using the rarefaction QIIME (2.2019.7) process with default parameters (Bolyen et al. 2019). For comparison of alpha diversity across water and gut microbiome samples, Shannon, Simpson, ACE, Chao1, and Good-coverage indices were calculated and visualized in R software (V.2.15.3) (R-core-team 2013). The significant differences in alpha diversity index between samples were checked with the Kruskal–Wallis test (p < 0.05). We compared beta diversity across the samples using the UniFrac distance metrics (Lozupone and Knight 2005). Consequently, Bray–Curtis distance-based principal coordinate analysis (PCoA) plot was generated to visualize the microbiome samples’ beta diversity pattern. Likewise, NMDS (non-metric multidimensional scaling) was performed for visualization of variation in microbial community composition between samples. The PCoA and NMDS dimensionality reduction maps were generated in R software.
To perform OTU network-based analysis, network maps were constructed using QIIME and visualized using Cytoscape (v. 3.0.1) (Shannon et al. 2003). OTU table (97% sequence similarity) was converted to the Cytoscape format. The edges connecting nodes representing fish and water samples (circles) to species-level OTUs in a particular sample are colored according to the host–habitat type (edge-weighted spring embedded model in Cytoscape v. 3.0.1).
Functional prediction
We employed PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2) for prediction of the functional repertoire of microbial consortia in water and gut samples from site-α and site-β (Douglas et al. 2020). KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways and enzymes were deciphered.
Results
Physicochemical analysis of water samples
Physicochemical characteristics from both sampling sites are provided in Supplementary S1. The pH values pointed out neutral to alkaline nature of the examined water samples. Low DO (1.8 ± 0.8 mg/L) whereas elevated BOD (102.4 ± 1.4 mg/L), COD (266.4 ± 3.10 mg/L) and TDS (1050.7 ± 3.34 mg/L) values were observed in site-α water sample (Bharti et al. 2022). In site-β, DO, BOD, and COD values were 4.8 ± 0.7, 19.5 ± 1.8, and 103 ± 2.0, respectively. Also, in site-α, among the tested heavy metals, Fe (2.89 ± 0.07 mg/L), Cr (1.06 ± 0.04 mg/L), and Zn (2.25 ± 0.07 mg/L) were beyond their permissible limit, i.e., 2 mg/L, 0.05 mg/L, 0.01 mg/L (WHO 2011), respectively. In site-β, all the physicochemical parameters were within the permissible range for water samples. Overall, based on the above parameters, it could be inferred that these two sampling sites are physicochemically different.
Sequencing and diversity analysis
Illumina paired-end platform was used for amplicon sequencing to produce 250 bp paired-end raw reads (30,44,701) followed by merging to obtain clean tags. The SILVA database was used to quantify the chimeric sequences in clean tags, and these sequences were removed to obtain 1,346,070 effective tags which were then used for subsequent analysis (Table 1). After clustering at 97% sequence similarity, a total of 1318 OTUs were identified. Statistical indices of alpha diversity at clustering threshold 97% (number of reads chosen for normalization: cutoff = 19,448) are depicted in Table 1.
Table 1.
Sequencing statistics and alpha diversity indices of samples collected from polluted and less polluted sites of river Yamuna
| Sample | Effective tags | Average length (nt) | Q30 | GC % | Alpha diversity indices | ||||
|---|---|---|---|---|---|---|---|---|---|
| Shannon | Simpson | ACE | Chao1 | Goods coverage | |||||
| Cyprinus carpio (α-C1) | 93,304 | 455 | 96.9 | 54.3 | 2.7 | 0.7 | 166.4 | 158.2 | 0.9 |
| Cyprinus carpio (α-C2) | 459,351 | 457 | 98.2 | 55.0 | 3.3 | 0.8 | 153 | 132.1 | 0.9 |
| Water (α-W1) | 25,118 | 443 | 97.1 | 50.6 | 4.9 | 0.9 | 485.3 | 473 | 0.9 |
| Water (α-W2) | 512,921 | 455 | 98.4 | 50.9 | 5.4 | 0.9 | 565.7 | 592 | 0.9 |
| Cyprinus carpio (β-C1) | 36,820 | 468 | 99.7 | 55.4 | 2.3 | 0.7 | 44 | 44 | 1.0 |
| Cyprinus carpio (β-C2) | 31,655 | 469 | 99.6 | 55.9 | 2.9 | 0.7 | 39 | 41 | 1.0 |
| Water (β-W1) | 92,580 | 426 | 95.7 | 52.9 | 3.3 | 0.5 | 745 | 693 | 0.9 |
| Water (β-W2) | 94,321 | 427 | 95.9 | 52.8 | 3.5 | 0.6 | 512 | 473 | 0.9 |
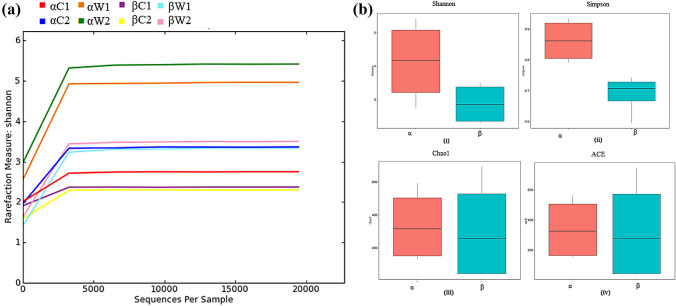
The rarefaction curve showed that each sample flattened with high sequence numbers, thus indicating reasonable sequencing depth (Fig. 1a). Alpha diversity calculated as Shannon and Simpson’s indices showed fluctuations in the diversity across the gut and water samples from site-α and site-β (Table 1; Fig. 1b). Shannon values were in range from 2.3 to 5.4 (minimum in gut samples from site-β and highest in water samples from site-α). Collectively, all samples from site-α showed higher diversity as compared to site-β (Fig. 1b). Simpson’s index (equitability) values were in range 0.5–0.9: maximum in site-α water samples and minimum in site-β water samples. Chao1 and ACE indices reflected the species richness and showed that fish gut from site-β had a lower microbial community richness compared to the fish gut samples from site-α (Table 1). From both the sites, water samples possessed greater microbial community richness as compared to the gut samples. Good’s coverage values (≥ 0.9) signify that good sequencing coverage was accomplished. As Kruskal–Wallis test showed p > 0.05, and thus, no statistically significant relationship was found between the standard deviations of the alpha diversity values (Shannon/Simpson).
Fig. 1.
Comparative rarefaction curves and alpha diversity of gut and water samples based on Shannon, Simpson, Chao 1, ACE indices. a Rarefaction curve indicated the sequencing depth of eight samples; C. carpio gut samples (αC1, αC2) and water samples (αW1, αW2) from polluted site-α; as well as C. carpio gut samples (βC1, βC2)and water samples (βW1, βW2) from the less polluted site-β; b Boxplots illustrated deviations in alpha diversity between the polluted (site-α) and less polluted (site-β). These deviations were assessed using four indices: (i) Shannon index, (ii) Simpson index, (iii) chao1, (iv) ACE index. Significance testing was conducted using the Kruskal–Wallis test (p > 0.05), revealing a relationship between the standard deviations of the alpha diversity values
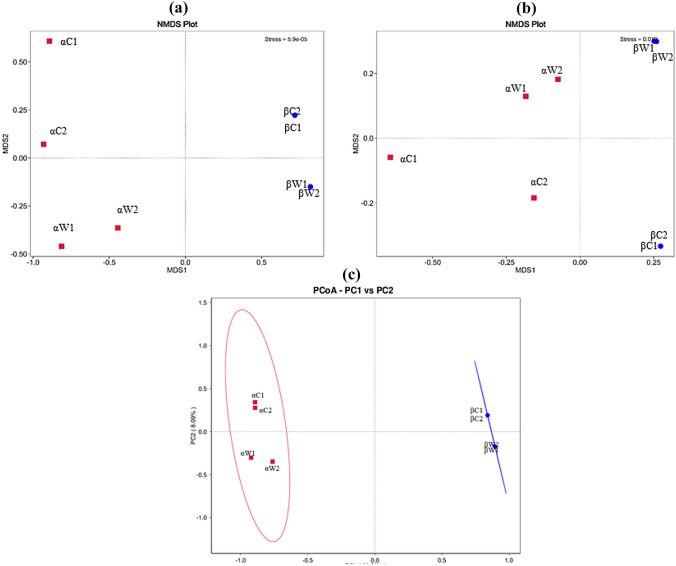
Non-metric multidimensional (NMDS) analysis was employed using weighted and unweighted UniFrac distance matrix (stress values: 5.9e–05; 0.019) and showed that gut and water samples from site-α clustered together, whereas samples from site-β tended to form a separate group indicating differences in bacterial composition between the two sites (Fig. 2a, b). The PCoA clustering employing weighted UniFrac distances showed concordant results where microbiome samples of gut and water from site-α and site-β separated along PC1 with 90.52% of total variation, whereas along PC2, variation was 8.09% and overall total variation was 98.61% (Fig. 2c). The ordinated beta diversity analysis supports the hypothesis that water and gut microbiomes from site-α differ from the microbiomes from site-β and implies that different environmental conditions and water quality has a huge influence on the bacterial community composition of the samples. The results implied the presence of different community dynamics in these geographically and physicochemically heterogenic ecosystems.
Fig. 2.
The microbial community differences among gut samples of fish and water samples were visualized using non-metric multidimensional plot (NMDS) and principal coordinate analysis (PCoA). The stress values for the NMDS plot were 5.9e–05; 0.019 indicating a good representation of the data. The dots on the plot were color-coded, with red representing samples from polluted site (site-α) and blue representing samples from less polluted site (site-β). The pairwise distances between these samples were determined using both a weighted and b unweighted UniFrac algorithms; c Principal coordinate analysis (PCoA) was performed using the weighted UniFrac algorithm. In the NMDS plot and PCoA, the gut and water samples from the polluted site (site-α) clustered together, while the samples from less polluted site (site-β) formed a separate group. This clustering indicates significant differences in bacterial composition between the two sites, and this analysis was conducted using R (v-3.7) software
Taxonomic profiles
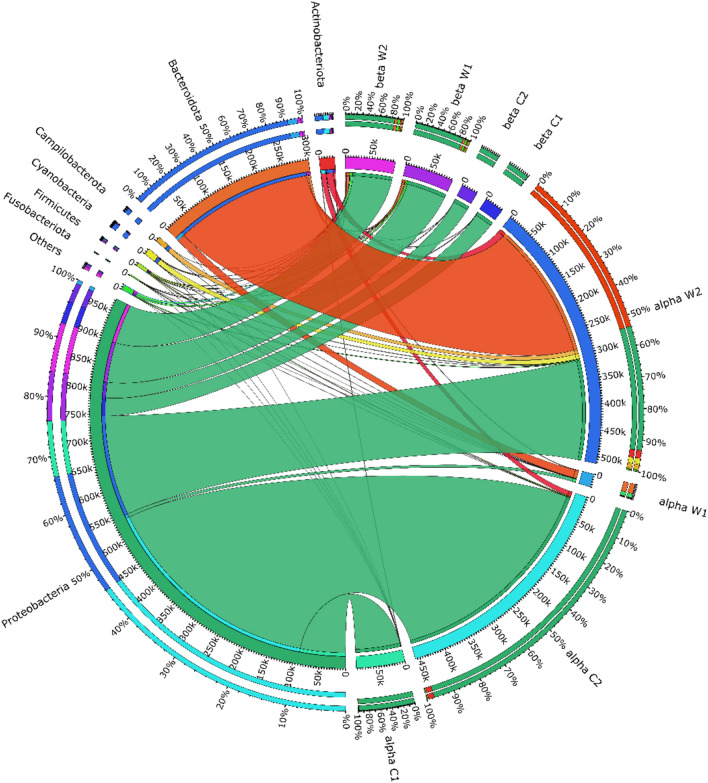
Multiple differences were observed in the relative abundance proportions of various taxa from phylum to genus level while comparing site-α and site-β samples. Proteobacteria was omnipresent in all the examined samples. In the area designed as site-α, Proteobacteria (97.7%) and Actinobacteria (1.2%) were detected in the gut samples (Bharti et al. 2022), while in site-β, we observed the dominance of Proteobacteria (98.5%), Fusobacteria (1.04%) and Firmicutes (0.3%) in the fish gut (Fig. 3). Water samples pointed out a remarkable difference showing Bacteroidota (60.5%) as a major phylum followed by Proteobacteria (31.1%), Actinobacteria (3.6%), and Cyanobacteria (2.3%) in site-α, whereas Proteobacteria (85.8%), Planctomycetes (12.03%), Bacteroidota (4.4%), Verrucomicrobiota (3.7%), Actinobacteria (2.5%), and Firmicutes (1.8%) were enriched in the water samples from site-β.
Fig. 3.
Microbial community’s abundance at the phylum level was assessed using Circos (v0.63-10). The analysis involved mapping eight samples based on their abundances, which were depicted in the outer circle, while the inner circle represents the synteny with the respective phyla. In the gut samples from site-α, Proteobacteria (97.7%) and Actinobacteria (1.2%) were enriched, whereas site-β exhibited dominance of Proteobacteria (98.5%), Fusobacteria (1.04%), and Firmicutes (0.3%). In the water samples, Bacteroides (60.5%); Proteobacteria (31.1%), Actinobacteria (3.6%), and Cyanobacteria (2.3%) in site-α. For site-β, the water samples were enriched in Proteobacteria (85.8%), Planctomycetes (12.03%), Bacteroidota (4.4%), and Verrucomicrobiota (3.7%)
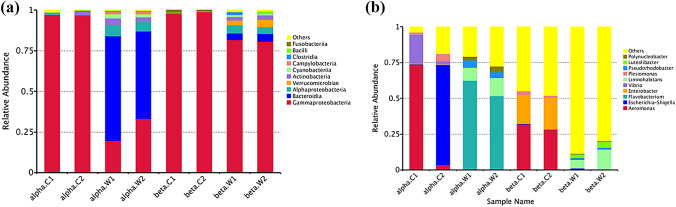
At the class rank level, Gammaproteobacteria was highly frequent in all the samples. In common carp from site-α, besides Gammaproteobacteria (97.4%), Actinobacteria (1.27%) was also found. In site-β, Gammaproteobacteria, (98.5%), Fusobacteria (1.04%), and Bacilli (0.2%) dominated the fish gut. Bacteroidia (60.5%) was highest followed by Gammaproteobacteria (24.2%), Alphaproteobacteria (6.9%), and Actinobacteria (2.3%) in polluted water, while in the less polluted water samples, Gammaproteobacteria (81.1%), Alphaproteobacteria (4.6%), Bacteroidia (4.4%), Verrucomicrobia (3.7%), and Actinobacteria (2.4%) were enriched (Fig. 4).
Fig. 4.
A multi-stacked histogram plot was generated to illustrate the relative abundances of microbial diversity up to the class and genus level. a The plot depicted the distribution of the most abundant class distribution in gut samples, where Gammaproteobacteria accounted for 97.4% in site-α and 98.5% in site-β. Water samples in site-α exhibited the highest abundance of Bacteroidia (60.5%), followed by Gammaproteobacteria (24.2%) and Alphaproteobacteria (6.9%). In site-β water samples, Gammaproteobacteria (81.1%), Alphaproteobacteria (4.6%), and Bacteroidia (4.4%) were the enriched classes; b Multi-stacked histogram plot representing relative abundance at genus level. In the fish gut of site-α, the most abundant genera were Aeromonas (36.6%), Escherichia-Shigella (35.9%), and Vibrio (6.1%). In site-β, the dominant genera in the fish gut were Aeromonas (31.9%), Enterobacter (20.1%), and Plesiomonas (2.6%). Water samples from site-α showed an enrichment of Flavobacterium (58.6%), Limnohabitans (10.1%), and Pseudorhodobacter (4.6%). On the other hand, water samples from site-β comprised Limnohabitans (5.9%) Luteolibacter (2.5%), and Flavobacterium (1.1%)
At genus level, Aeromonas (36.6%), Escherichia-Shigella (35.9%) and, Vibrio (6.1%) were abundantly found in the fish gut in site-α, whereas in site-β, Aeromonas (31.9%), Enterobacter (20.1%), Plesiomonas (2.6%), Cetobacterium (1.3%), and Kosakonia (0.8%) harbored the fish gut. In water samples from site-α, Flavobacterium (58.6%), Limnohabitans (10.1%), Pseudorhodobacter (4.6%), and Polynucleobacter (2.5%) were enriched, while Limnohabitans (5.9%), Luteolibacter (2.5%), Flavobacterium (1.1%), Pseudorhodobacter (0.9), Rhodobacter (0.6%), Novosphingobium (0.5%), Dechloromonas (0.03), Dechlorobacter (0.01%), Azoarcus (0.01%), Candidatus “Nitrospira” (0.01%), and Sulphuritalea (0.01%) were detected in water samples from site-β (Fig. 4). The results showed that the microbial composition was different in site-α and site-β samples.
Upon data analysis, we found a repertoire of potentially pathogenic microbes, viz. Aeromonas veronii (0.3% C; 0.1% W), Escherichia coli (0.3% C; 0.01% W), Vibrio cholerae (0.9% C; 0.01% W), Acinetobacter junii (0.03% C; 0.1% W), Streptococcus iniae (0.2% C; 0.1% W), Shewanella putrefaciens (0.03% C; 0.8% W), Pseudomonas aeruginosa (0.08% C), and Staphylococcus aureus (0.03% C) in gut and water samples from site-α. These potentially pathogenic genera were also detected from site-β but with very low abundance (Table 2). We calculated the pathogenic OTUs from both the sites, and as a result, 68 pathogenic OTUs from site-α and 25 OTUs from site-β were detected.
Table 2.
Comparative description of microbial diversity along with their biotechnological potential from polluted and less polluted sites of river Yamuna, India
| Characteristics | Species | Gut microbiome samples from polluted site | Gut microbiome samples from less polluted site | Water microbiome samples from polluted site | Water microbiome samples from less polluted site | References |
|---|---|---|---|---|---|---|
| Pathogens | Aeromonas sp. | + | + | + | + | Kim et al. (2021) |
| Escherichia sp. | + | + | + | + | Del Rio‐Rodriguez et al. (1997) | |
| Vibrio cholerae | + | ND | + | + | Senderovich et al. (2010) | |
| Acinetobacter junii | + | + | + | + | Malick et al. (2020) | |
| Streptococcus iniae | + | ND | + | ND | Agnew and Barnes (2007) | |
| Shewanella putrefaciens | + | ND | + | ND | Allameh et al. (2014) | |
| Pseudomonas aeruginosa | + | ND | ND | ND | Thomas et al. (2014) | |
| Staphylococcus aureus | + | ND | ND | ND | Agnew and Barnes (2007) | |
| Flavobacterium sp. | ND | ND | + | + | Cai et al. (2013) | |
| Probiotic potential | Bacillus velezensis | + | ND | ND | ND | Khalid et al. (2021) |
| Lactobacillus plantarum | + | ND | ND | ND | Cebeci and Gürakan (2003) | |
| Enterococcus faecalis | + | ND | ND | ND | Allameh et al. (2014) | |
| Bifidobacterium longum | + | ND | ND | ND | Vijayaram and Kannan (2018) | |
| Lactococcus lactis | + | ND | ND | ND | Arriba et al. (2021) | |
| Leuconostoc falkenbergense | + | ND | ND | ND | Arriba et al. (2021) | |
| Cetobacterium somerae | ND | + | ND | ND | Kim et al. (2021) | |
| Blautia glucerasea | ND | + | ND | ND | Uyar GÖ and Yildiran (2019) | |
| Bioremediation | Leucobacter chromiireducens | + | ND | + | ND | Tahri et al. (2016) |
| Pseudomonas aeruginosa | + | ND | ND | ND | Jaber and Al-Mayahi (2020) | |
| Pseudomonas fluorescens | + | ND | ND | ND | Sharma et al. (2006) | |
| Methanothrix harundinacea | + | ND | ND | ND | Wagner et al. (2002) | |
| Dechloromonas agitata | ND | ND | + | ND | Wagner et al. (2002) | |
| Nitrospira defluvii | ND | ND | + | + | Silyn-Roberts and Lewis (2001) | |
| Thauera humireducens | ND | ND | + | ND | Wagner et al. (2002) | |
| Zoogloea ramigera | ND | ND | + | ND | Cydzik-Kwiatkowska and Zielińska (2016) | |
| Zoogloea oryzae | ND | ND | ND | + | Cydzik-Kwiatkowska and Zielińska (2016) |
An interesting observation from site-α host microbiome was the presence of potentially probiotic microbes such as Bacillus velezensis (0.5%), Lactobacillus plantarum (0.7%), Enterococcus faecalis (0.3%), Bifidobacterium longum (0.8%), Lactococcus lactis (0.3%), and Leuconostoc falkenbergense (0.1%), while site-β gut microbiome showed the presence of Cetobacterium somerae (1.3%), and Blautia glucerasea (0.01%) were found.
Heavy metal and organic matter tolerants
Physicochemical parameters of water samples from both sites revealed that heavy metals Cr, Fe, and Zn were exceeding the permissible limit in site-α as compared to site-β, and therefore, we attempted to explore heavy metal microbial tolerants. Leucobacter chromiireducens (1.05% C; 0.08% W), Pseudomonas aeruginosa (0.08%), and Pseudomonas fluorescens (0.2%) were found in gut samples from site-α, and opportunistic pathogen Pseudomonas aeruginosa (0.0005%) was detected from water samples from site-β. Furthermore, microbial communities involved in organic matter decomposition were also found which is in line with the high pollution load in river Yamuna. In site-α, Methanothrix harundinacea (0.25%) was detected in gut samples and Dechloromonas agitata (0.65%), Candidatus “Nitrospira defluvii” (0.5%), Thauera humireducens (0.12%), and Zoogloea ramigera (0.5%) were found in water samples. In water samples from site-β, only Zoogloea oryzae (0.02%), Candidatus “Nitrospira defluvii” (0.01%) were detected (Tables 2, 3). From site-α gut samples, a novel bacterium strain MB25T, designated as Sporosarcina cyprini sp. nov., was isolated (Bharti et al. 2022). This bacterium demonstrated moderate tolerance to Cr+6 (20 mgL−1) and Cd+2 (20 mgL−1) which is line with the heavy metal pollution status of site-α.
Table 3.
Variation in microbial composition in fish gut and water samples from polluted and less polluted site
| Species | Polluted site (%) | Less polluted site (%) |
|---|---|---|
| Gut samples | ||
| Aeromonas sp. | 36.60 | 31.9 |
| Vibrio cholerae | 6.0 | 0.06 |
| Plesiomonas shigelloides | 3.0 | 2.0 |
| Cetobacterium sp. | 0.09 | 1.04 |
| Pseudomonas aeruginosa | 0.08 | 0.005 |
| Bifidobacterium sp. | 0.06 | 0.005 |
| Enterobacter sp. | 0.36 | 20.1 |
| Water samples | ||
| Flavobacterium sasangense | 6.27 | 0.01 |
| Pseudorhodobacter sp. | 4.5 | 1.2 |
| Curvibacter sp. | 1.3 | 1.7 |
| Acidovorax sp. | 0.96 | 0.09 |
| Hydrogenophaga sp. | 0.6 | 0.2 |
| Polynucleobacter cosmopolitanus | 0.6 | 0.2 |
| Nitrospira defluvii | 0.5 | 0.01 |
| Acinetobacter junii | 0.15 | 0.03 |
| Shewanella sp. | 0.06 | 0.005 |
| Arcobacter cryaerophilus | 0.04 | 0.01 |
| Zoogloea sp. | 0.04 | 0.02 |
| Caulobacter sp. | 0.03 | 0.005 |
| Luteolibacter sp. | 0.01 | 0.03 |
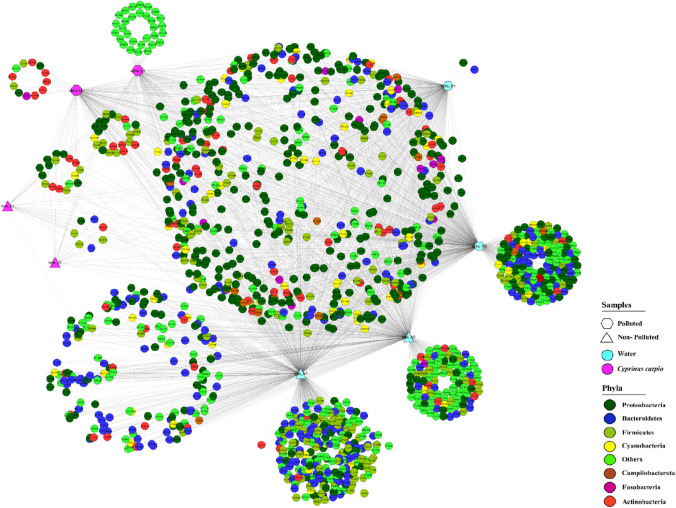
Also, OTU network-based approach was employed to examine core and unique gut microbial communities clusters with the habitats together at the phyla level (Fig. 5). In the results, an expanded node represented a host–habitat sample and the OTUs were interconnected to the host and habitat in which they were residing. In accordance with the specific microbial compositional differences observed in the OTU network-based analysis, the OTU nodes of the host within the same habitat were more likely to connect with each other rather than to those from different habitats (Fig. 5). Here, we calculated unique OTUs in all the samples alpha-C1 (n = 30); alpha-C2 (n = 12); alpha-W1 (n = 2); alpha-W2 (n = 132); and beta-W1 (n = 228); beta-W2 (n = 95), while in the case of beta-C1, C2 samples, all the OTUs were sharing homology with different samples. The shared OTUs among the gut samples from both the sites were n = 106, whereas among water microbiome samples were n = 96. Shared OTUs between all the samples from site alpha were 320 and from all the samples from site beta were 250. The results suggested that microbial diversity in fish from polluted site was higher than that from less polluted site clearly evident with the results of alpha diversity. Also, less number of shared OTUs between gut samples of both the sites suggested environment-driven variation was minimal while intraspecific microbiota was perpetuated.
Fig. 5.
An OTU network-based analysis was conducted to explore the shared and unique microbial communities present in fish gut and water samples from polluted and less polluted sites. The analysis involved connecting nodes representing fish and water samples (circles) to species-level OTUs, with color-coded edges indicating the host–habitat type. The edge-weighted spring embedded model in Cytoscape (v-3.0.1) was utilized for this purpose. Notably, OTU nodes belonging to the same habitat displayed a higher likelihood of being interconnected. In the gut samples, 106 shared OTUs were found to be shared, while in the water samples, 96 OTUs were shared. Furthermore, there were 320 shared OTUs among all the samples from site-α (polluted site) and 250 shared OTUs among all the samples from site-β (less polluted site). Within the gut samples, Proteobacteria exhibited the highest abundance, with 268,744 and 33,741 OTUs in site-α and site-β, respectively. In contrast, Bacteroidota (OTUs = 145,888) and Proteobacteria (OTUs = 80,173) were dominant taxa in water samples from site-α and site-β, respectively
Functional prediction
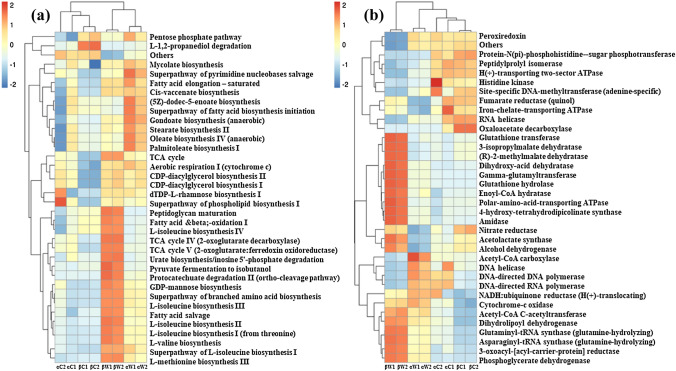
To understand variations in functional repertoire of the intestinal and surrounding water microflora as a consequence of anthropogenic activities and pollution, metagenomes’ functional potential between the two groups were predicted by PICRUSt2. About 422 pathways were enriched among which 35 were found to be completed as shown in the heatmap (Supplementary S2, Fig. 6a). A maximum number of predicted functions were allocated to metabolic pathways in which biosynthesis pathways for L-isoleucine, cis-vaccenate, stearate, and palmitoleate, bifidobacterium shunt, and ectoine were present in the fish gut from site-α and menaquinol pathways were present in gut samples from both sites. Pathways for aerobic respiration, L-isoleucine biosynthesis, L-isoleucine biosynthesis, L-valine biosynthesis, and fatty acid and beta-oxidation were enriched in water samples from both sites.
Fig. 6.
Abundances of UniRef90 families and predicted metabolic pathways were represented using PICRUSt2 (Douglas et al. 2020). a Functional pathway prediction, b predicted enzymes of microbial communities in microbiome samples. A total of 422 pathways were enriched, with 35 complete pathways depicted in the heatmap. Gut samples from site-α exhibited pathways for L-isoleucine, cis-vaccenate, stearate, and menaquinone and stress protectant ectoine biosynthetic pathway. In the water samples from both sites, enriched pathways included L-isoleucine biosynthesis, L-isoleucine biosynthesis, L-valine biosynthesis involved in immunological function; The fish gut samples from both sites displayed the presence of cellulose, lysophospholipase, lysozyme, and chitinase enzymes, involved in the digestion of organic matter. The samples from site-β exhibited a higher predominance of nitrate reductase. In the water sample from site-α, the presence of lignin-degrading enzymes, such as peroxiredoxin and glutathione peroxidase, indicated the prevalence of debris and organic matter at the polluted site
PICRUSt2 also showed the incidence of enzymes involved in digestion and metabolism in the gut samples (Supplementary S3, Fig. 6b). Enzymes active against cellulose, hemicellulose, lignin, debris, and organic matter (cellulase (EC 3.2.1.4), lysophospholipase (EC 3.1.1.5), lysozyme (EC 3.2.1.17), chitinase (EC 3.2.1.14)) were found to be active in fish gut from both the sites as the principal nutritional constituent of this invasive fish includes phytoplankton, plant debris, and detritus. Commonly, peptidylprolyl isomerase (EC 5.2.1.8) and fumarate reductase (EC 1.3.5.4) were enriched in all gut samples. Nitrate reductase (EC 1.7.99.4) was more predominant in site-β samples and was also detected in gut samples from site-α. The presence of lignin-degrading enzymes such as peroxiredoxin (EC 1.11.1.15) and glutathione peroxidase (EC 1.11.1.9) in the water samples from site-α indicated the prevalence of debris and organic matter at the region; besides this enzyme chitinase (EC 3.2.1.14) was also present in the site-α samples.
Discussion
Microbial community structure of carps along with the function potential has been examined to be strongly influenced by the environment (Kakade et al. 2020; Jing et al. 2021). In present study, water contamination’s influence on the microbiome dynamics of common carp was well attained by contrasting metagenomics analysis at two geographically unrelated and physicochemically different sites of river Yamuna. Low concentration of DO and elevated levels of BOD are pointers of worsening quality of the river water and are consistent with the anthropogenic activities and pollution at site-α of river Yamuna. In addition, high species diversity was observed in samples from polluted site as contamination can supplement surplus bacteria load and hence can lead to inflated alpha diversity (Minich et al. 2019). Beta diversity analysis revealed water contamination enlarged the differences in the composition of microbiome within the same species as the microbiome samples from two sites formed distinct clusters.
Taxonomic profiling showed that among all the classified sequence reads, phylum Proteobacteria predominated the fish gut irrespective of the site and could be signified as the core microbiome. The similar trend of pre-eminence of Proteobacteria in the fish gut has earlier been supported by several research studies (Talwar et al. 2018; Liu et al. 2016; Tyagi et al. 2019; Johny et al. 2021). The occurrence of alike bacterial phylum irrespective of the geographical site specifies its role in vital functions such as digestion, nutrient absorption, and generation of the immune response. At genus level, Aeromonas and Plesiomonas, members of Gammaproteobacteria, were ubiquitous in all examined gut samples. Numerous studies have suggested that Aeromonas is a predominant genus in the gut of fishes feeding on detritus of plant origin and in omnivorous freshwater fishes as cellulose degrading species (Khurana et al. 2020, 2021). Likewise, Plesiomonas has been previously found in the gut microbiome of several fish species (Zhang et al. 2020, 2021). Cetobacterium somerae, a member of phylum Fusobacteria, deserves special mention as its presence might reflect combination of a fermentative metabolism and vitamin production in the GI tract. Several studies have demonstrated the role of Cetobacterium in many freshwater fish to satisfy their vitamin B12 dietary needs and can prevent the growth of harmful pathogens (Khurana et al. 2021; Sugita et al. 1990; Kim et al. 2021). The fact that carps do not have a dietary vitamin B12 requirement is well explained by the incidence of Cetobacterium in their gut (van Kessel et al. 2011). Another member with similar role is Enterobacter that has been earlier reported to be involved in production of cellulose, amylase, and protease (Ray et al. 2012) stipulating its role in digestion.
In the water samples, from the site with better water quality, Proteobacteria was the most abundant that is consistent with the past studies of river Yamuna (Mittal et al. 2019; Rajeev et al. 2021). Another phylum within our amplicon sequences from the less polluted water samples was the Verrucomicrobiae that has been negatively correlated with pollution levels (Berg et al. 2012) and is involved in aerobic/obligate anaerobic fermentative metabolism. Additionally, occurrence of Planctomycetes, a bioindicator phylum, in site-β water samples and its absence in site-α indicate decent water quality in site-β, since its richness seems to decrease with increased pollution (Chen et al. 2019). Contrastingly, in the polluted site, a remarkable difference was observed with Bacteroidota being the predominant phyla followed by Proteobacteria that is distinct from previous metagenomics studies on River Yamuna (Mittal et al. 2019; Rajeev et al. 2021). This could be due to the fact that the phylum Bacteroidota is influenced with physicochemical parameters such as temperature and TDS and seasonal variation TDS (Kaevska et al. 2016; Zhang et al. 2012) and its prevalence in a polluted water sample indicates poor water quality, fecal contamination, and organic pollution (Tani et al. 2002; Ahmed et al. 2016). Also, the incidence of members of Actinobacteria in all samples from polluted site may suggest the increased susceptibility of the fish toward pathogens as Actinobacteria are known as antibiotic factories and are able to produce secondary metabolites against pathogenic microbes (Jami et al. 2015).
Furthermore, the prevalence of fish disease-relevant pathogens (A. veronii, E. coli, V. cholera, A. junii S. putrefaciens, P. aeruginosa, S. iniae, S. aureus) in polluted site indicated that deteriorating surroundings facilitated the invasion of pathogenic bacteria in fish gut eco-environment. Moreover, in the GI tract of fishes from polluted environment, probiotic members (B. velezensis, L. plantarum, E. faecalis, L. lactis, and L. falkenbergense) could be serving as a defense mechanism against above-mentioned fish pathogens by the production of active metabolites and bacteriocins (Meidong et al. 2017; Khurana et al. 2021). Hence, the incidence of both potentially pathogenic and probiotic strains in fish gut from the polluted region suggests symbiotic relationship and reflects the affinity of the host for the microflora that contributes to the maintenance of immune function in stressed environments.
Chromium degrading L. chromiireducens and P. aeruginosa could be correlated with the high concentration of chromium in the polluted site (Joshi-Tope and Francis 1995; Austin and Allen-Austin 1985; Zhu et al. 2008; Bakiyaraj et al. 2014; Jaber and Al-Mayahi 2020). Past studies have explained the metabolic strategies underlying the possible mechanisms of bacterial resistance to heavy metals, involving direct ion export or reduction to a lower toxic/soluble (Silver and Phung 1996). Nitrospira species are anaerobic bacteria, involved in nitrogen cycling, and thus, their presence could offer new solutions for the removal of nitrogen from polluted water (Silyn-Roberts and Lewis 2001). A denitrifying member Zoogloea sp. was enriched in polluted site as compared to the other site. It is known for its role in poly-B-hydroxybutyrate production, metal biosorption, and bioremediation and is generally enriched in organically polluted waters (Wagner et al. 2002; Saǧ and Kutsal 2000).
Functional prediction emphasized the significance of gut microbial community as contributors to the digestion and metabolism. Biosynthetic pathway for leucine, valine, isoleucine found in gut samples of both sites has been reported to have a role in immunity in fishes by enhancing lymphocyte proliferation in response to environmental pollutants (Tarnecki et al. 2019). Incidence of menaquinone pathway suggests role of gut microbiome in vitamin K2 production as fish receive its menaquinones from gut microflora, and it is essential for posttranslational modification of certain proteins required for blood coagulation (Krossøy et al. 2011). In gut samples from polluted site, ectoine biosynthesis constructed via an evolutionarily conserved biosynthetic pathway and is a stress protectant (Richter et al. 2019), while Bifidobacterium shunt (Bif Shunt) pathway has a unique feature for energy harvesting and its presence depicts an evolutionary adaptation confers a fitness advantage during natural colonization (Sanders et al. 2018). Thus, this study reflects that anthropogenic stress can modulate the fish gut microbiomes by influencing not only the microbial diversity, but also their potential function as well. On the one side, the host retains the stable multifunctional bacterial consortia, while on the other side, microbial diversity becomes specialized due to anthropogenic stress and selection pressure.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary File S1: Physicochemical properties of water collected from sampling site. Supplementary file1 (XLSX 11 KB)
Supplementary File S2: Relative abundance of different pathways in gut samples and water samples. Supplementary file2 (XLSX 63 KB)
Supplementary File S3: Relative abundance of different enzymes in gut samples and water samples. Supplementary file3 (XLSX 260 KB)
Acknowledgements
This work was supported by funds from the Institute of Eminence (IoE) and the Indian Council of Agricultural Research—National Bureau of Agriculturally Important Microorganisms funded project (ICAR-NBAIM) [grant number NBAIM/AMAAS/2017-20/GF/1a/512]. MB and SN thank the Council of Scientific and Industrial Research (CSIR) for providing doctoral fellowships.
Author contributions
RKN proposed the idea. MB wrote the manuscript. MB and SN performed the analysis. RKN and SN critically reviewed the manuscript and improved it. All authors read and approved the final manuscript.
Funding
National Bureau of Agriculturally Important Microorganisms, NBAIM/AMAAS/2017-20/GF/1a/512, Ram Krishan Negi
Data availability
The raw sequencing read data have been deposited in the National Centre for Biotechnology Information (NCBI) under the project Accession Number PRJNA809116.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
No special permission was required for this study.
Contributor Information
Meghali Bharti, Email: meghalibharti25@gmail.com.
Shekhar Nagar, Email: snagar@db.du.ac.in.
Ram Krishan Negi, Email: negigurukul@gmail.com.
References
- Agnew W, Barnes AC. Streptococcus iniae: an aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. Vet Microbiol. 2007;122:1–15. doi: 10.1016/j.vetmic.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Ahmed W, Hughes B, Harwood VJ. Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water. 2016;8:231. doi: 10.3390/w8060231. [DOI] [Google Scholar]
- Allameh KS, Ringø E, Yusoff MF, et al. Properties of Enterococcus faecalis, a new probiotic bacterium isolated from the intestine of snakehead fish (Channa striatus Bloch) Afr J Microbiol Res. 2014;8:2215–2222. doi: 10.5897/AJMR2013.5830. [DOI] [Google Scholar]
- Arriba LMG, Alcántara HAM, Mohedano ML. Lactic Acid bacteria isolated from fermented doughs in Spain produce dextrans and riboflavin. Foods. 2021;10:2004. doi: 10.3390/foods10092004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin B, Allen-Austin D. A review: bacterial pathogens of fish. J Appl Bacteriol. 1985;58:483–506. doi: 10.1111/j.1365-2672.1985.tb01490.x. [DOI] [PubMed] [Google Scholar]
- Bakiyaraj R, Baskaran L, Chidambaram A. Bioremediation of chromium by Bacillus subtilis and Pseudomonas aeruginosa. Int J Curr Microbiol App Sci. 2014;3:715–719. [Google Scholar]
- Berg J, Brandt KK, Al-Soud WA, Holm PE, Hansen LH, Sørensen SJ, et al. Selection for Cu-tolerant bacterial communities with altered composition, but unaltered richness, via long-term Cu exposure. Appl Environ Microbiol. 2012;78:7438–7446. doi: 10.1128/AEM.01071-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti M, Nagar S, Khurana H, et al. Metagenomic insights to understand the role of polluted river Yamuna in shaping the gut microbial communities of two invasive fish species. Arch Microbiol. 2022;204:1–12. doi: 10.1007/s00203-022-03127-x. [DOI] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, De La Fuente L, Arias RC. Biofilm formation by the fish pathogen Flavobacterium columnare: development and parameters affecting surface attachment. Appl Environ Microbiol. 2013;79:5633–5642. doi: 10.1128/AEM.01192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebeci A, Gürakan C. Properties of potential probiotic Lactobacillus plantarum strains. Food Microbiol. 2003;20:511–518. doi: 10.1016/S0740-0020(02)00174-0. [DOI] [Google Scholar]
- Chen J, McIlroy SE, Archana A, Baker DM, Panagiotou G. A pollution gradient contributes to the taxonomic, functional, and resistome diversity of microbial communities in marine sediments. Microbiome. 2019;7:1–12. doi: 10.1186/s40168-019-0714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colston TJ, Jackson CR. Microbiome evolution along divergent branches of the vertebrate tree of life: what is known and unknown. Mol Ecol. 2016;25:3776–3800. doi: 10.1111/mec.13730. [DOI] [PubMed] [Google Scholar]
- Courtenay Walter R, Welcomme RL (1989) International Introductions of Inland Aquatic Species". Copeia 520
- CPCB (2006) Assessment and development of river basin series: ADSORBS/41/2006-07. Water quality status of Yamuna River (1999–2005). Central Pollution Control Board, Ministry of Environment & Forests, New Delhi
- Cydzik-Kwiatkowska A, Zielińska M. Bacterial communities in full-scale wastewater treatment systems. World J Microbiol Biotechnol. 2016;32:66. doi: 10.1007/s11274-016-2012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degregori S, Casey JM, Barber PH. Nutrient pollution alters the gut microbiome of a territorial reef fish. Mar Pollut Bull. 2021;169:112522. doi: 10.1016/j.marpolbul.2021.112522. [DOI] [PubMed] [Google Scholar]
- Del Rio-Rodriguez RE, Inglis V, Millar SD. Survival of Escherichia coli in the intestine of fish. Aquac Res. 1997;28:257–264. doi: 10.1111/j.1365-2109.1997.tb01041.x. [DOI] [Google Scholar]
- Des Roches S, Post DM, Turley NE, et al. The ecological importance of intraspecific variation. Nat Ecol Evol. 2018;2:57–64. doi: 10.1038/s41559-017-0402-5. [DOI] [PubMed] [Google Scholar]
- Dhillon MK, George MP, Mishra S. Water quality of River Yamuna-Delhi stretch. Int J Environ Sci. 2013;3:1416. doi: 10.6088/ijes.2013030500012. [DOI] [Google Scholar]
- Douglas GM, Maffei VJ, Zaneveld J, et al. PICRUSt2: an improved and customizable approach for metagenome inference. Bio Rxiv. 2020 doi: 10.1101/672295. [DOI] [Google Scholar]
- Dwivedi AC, Mayank P, Tiwari A. The River as transformed by human activities: the rise of the invader potential of Cyprinus carpio and Oreochromis niloticus from the Yamuna River, India. J Earth Sci Clim. 2016;7:361. [Google Scholar]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Eichmiller JJ, Hamilton MJ, Staley C. Environment shapes the fecal microbiome of invasive carp species. Microbiome. 2016;4:1–13. doi: 10.1186/s40168-016-0190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2009) Cyprinus carpio. In Cultured aquatic species fact sheets. Text by Peteri, A. Edited and compiled by Valerio Crespi and Michael New. CD-ROM (multilingual)
- FishBase (2021) Retrieved April 12, 2021, from https://www.fishbasese/search.php?c_code=356#country
- Ghanbari M, Kneifel W, Domig KJ. A new view of the fish gut microbiome: advances from next-generation sequencing. Aquac. 2015;448:464–475. doi: 10.1016/j.aquaculture.2015.06.033. [DOI] [Google Scholar]
- Global Invasive Species Database (2021) Downloaded from http://www.iucngisd.org/gisd/100_worst.php on 09-02-2021
- Jaber SM, Al-Mayahi FSA. Screening and characterization of Pseudomonas aeruginosa resistant for heavy metal from surface sediment of Euphrates River, Iraq. Biochem Cell Arch. 2020;20:5203–5210. [Google Scholar]
- Jami M, Ghanbari M, Kneifel W, et al. Phylogenetic diversity and biological activity of culturable Actinobacteria isolated from freshwater fish gut microbiota. Microbiol Res. 2015;175:6–15. doi: 10.1016/j.micres.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Jing X, Su S, Zhang C, et al. Dynamic changes in microbial community structure in farming pond water and their effect on the intestinal microbial community profile in juvenile common carp (Cyprinus carpio L.) Genomics. 2021;113:2547–2560. doi: 10.1016/j.ygeno.2021.05.024. [DOI] [PubMed] [Google Scholar]
- Johny TK, Puthusseri RM, Bhat SG. Metagenomic landscape of taxonomy, metabolic potential and resistome of Sardinella longiceps gut microbiome. Arch Microbiol. 2021;27:87. doi: 10.1007/s00203-021-02675-y. [DOI] [PubMed] [Google Scholar]
- Joshi-Tope G, Francis AJ. Mechanisms of biodegradation of metal-citrate complexes by Pseudomonas fluorescens. J Bacteriol. 1995;177:1989–1993. doi: 10.1128/jb.177.8.1989-1993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaevska M, Videnska P, Sedlar K, Slana I. Seasonal changes in microbial community composition in river water studied using 454-pyrosequencing. Springerplus. 2016;5:1–8. doi: 10.1186/s40064-016-2043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakade A, Salama ES, Pengya F, et al. Long-term exposure of high concentration heavy metals induced toxicity, fatality, and gut microbial dysbiosis in common carp, Cyprinus carpio. Environ Pollut. 2020;266:115293. doi: 10.1016/j.envpol.2020.115293. [DOI] [PubMed] [Google Scholar]
- Khalid F, Khalid A, Fu Y. Potential of Bacillus velezensis as a probiotic in animal feed: a review. J Microbiol. 2021;59:627–633. doi: 10.1007/s12275-021-1161-1. [DOI] [PubMed] [Google Scholar]
- Khurana H, Sharma M, Verma H, Lopes BS, Lal R, Negi RK. Genomic insights into the phylogeny of Bacillus strains and elucidation of their secondary metabolic potential. Genomics. 2020;112:3191–3200. doi: 10.1016/j.ygeno.2020.06.005. [DOI] [PubMed] [Google Scholar]
- Khurana H, Sharma M, Bharti M, Singh DN, Negi RK. Gut milieu shapes the bacterial communities of invasive silver carp. Genomics. 2021;113:815–826. doi: 10.1016/j.ygeno.2021.01.013. [DOI] [PubMed] [Google Scholar]
- Kim PS, Shin NR, Lee JB, et al. Host habitat is the major determinant of the gut microbiome of fish. Microbiome. 2021;9:1–16. doi: 10.1186/s40168-021-01113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushlesh SM, Sajina AM, Roshith CM. Ichthyofaunal diversity of the major Indian rivers: a review. J Inland Fish Soc India. 2021;53:22–35. doi: 10.47780/jifsi.53.1&2.2021.115769. [DOI] [Google Scholar]
- Krossøy C, Waagbø R, Ørnsrud R. Vitamin K in fish nutrition. Aquac Nutr. 2011;17:585–594. doi: 10.1111/j.1365-2095.2011.00904.x. [DOI] [Google Scholar]
- Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Guo X, Gooneratne R, et al. The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci Rep. 2016;6:24340. doi: 10.1038/srep24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn MS, Boutin S, Hoseinifar SH, Derome N. Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front Microbiol. 2014;5:207. doi: 10.3389/fmicb.2014.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, Steven LS. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;2:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malick RC, Bera AK, Chowdhury H, et al. Identification and pathogenicity study of emerging fish pathogens Acinetobacter junii and Acinetobacter pittii recovered from a disease outbreak in Labeo catla (Hamilton, 1822) and Hypophthalmichthys molitrix (Valenciennes, 1844) of freshwater wetland in West Bengal, India. Aqua Res. 2020;51:2410–2420. doi: 10.1111/are.14584. [DOI] [Google Scholar]
- Meidong R, Doolgindachbaporn S, Sakai K, et al. Isolation and selection of lactic acid bacteria from Thai indigenous fermented foods for use as probiotics in tilapia fish Oreochromis niloticus. Aquacult Aquar Conserv Legis. 2017;10:455–463. [Google Scholar]
- Minich JJ, Sanders JG, Amir A, et al. Quantifying and understanding well-to-well contamination in microbiome research. Msystems. 2019;4:e00186–e219. doi: 10.1128/mSystems.00186-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal P, Prasoodanan PKV, Dhakan DB, Kumar S, Sharma VK. Metagenome of a polluted river reveals a reservoir of metabolic and antibiotic resistance genes. Environ Microbiome. 2019;14:1–12. doi: 10.1186/s40793-019-0345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba A, Mano N, Hirose H. Phylogenetic analysis of intestinal bacteria and their adhesive capability in relation to the intestinal mucus of carp. J Appl Microbiol. 2007;102:1307–1317. doi: 10.1111/j.1365-2672.2006.03204.x. [DOI] [PubMed] [Google Scholar]
- Nelson JS, Grande TC, Wilson MV. Fishes of the world. Wiley; 2016. [Google Scholar]
- Pérez T, Balcázar JL, Ruiz-Zarzuela I, et al. Host–microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010;3:355–360. doi: 10.1038/mi.2010.12. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeev AC, Sahu N, Arvind K, Deori M, Grace T, Dev SA, et al. Exploring prevalence of potential pathogens and fecal indicators in geographically distinct river systems through comparative metagenomics. Environ Pollut. 2021;282:117003. doi: 10.1016/j.envpol.2021.117003. [DOI] [PubMed] [Google Scholar]
- Ray AK, Ghosh K, Ringø E. Enzyme-producing bacteria isolated from fish gut: a review. Aquac Nutr. 2012;18:465–492. doi: 10.1111/j.1365-2095.2012.00943.x. [DOI] [Google Scholar]
- R-Core-Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
- Richter AA, Mais CN, Czech L, et al. Biosynthesis of the stress-protectant and chemical chaperon ectoine: biochemistry of the transaminase EctB. Front Microbial. 2019;10:2811. doi: 10.3389/fmicb.2019.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saǧ Y, Kutsal T. Determination of the biosorption heats of heavy metal ions on Zoogloea ramigera and Rhizopus arrhizus. Biochem Eng J. 2000;6:145–151. doi: 10.1016/S1369-703X(00)00083-8. [DOI] [PubMed] [Google Scholar]
- Said S, Hussain A. Pollution mapping of Yamuna River segment passing through Delhi using high-resolution GeoEye-2 imagery. Appl Water Sci. 2019;9:1–8. doi: 10.1007/s13201-019-0923-y. [DOI] [Google Scholar]
- Sanders ME, Benson A, Lebeer S, et al. Shared mechanisms among probiotic taxa: implications for general probiotic claims. Curr Opin Biotechnol. 2018;49:207–216. doi: 10.1016/j.copbio.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Senderovich Y, Izhaki I, Halpern M. Fish as reservoirs and vectors of Vibrio cholerae. PLoS ONE. 2010;5:e8607. doi: 10.1371/journal.pone.0008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Sundaram CS, Luthra PM, et al. Role of proteins in resistance mechanism of Pseudomonas fluorescens against heavy metal induced stress with proteomics approach. J Biotechnol. 2006;126:374–382. doi: 10.1016/j.jbiotec.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Sharma AP, Das MK, Vass KK, et al. Patterns of fish diversity, community structure and ecological integrity of River Yamuna, India. Aquat Ecosyst Health Manag. 2017;20:30–42. doi: 10.1080/14634988.2017.1265879. [DOI] [Google Scholar]
- Sharma R, Singh NS, Singh DK. Impact of heavy metal contamination and seasonal variations on enzyme’s activity of Yamuna River soil in Delhi and NCR. Appl Water Sci. 2020;10:1–8. doi: 10.1007/s13201-020-1166-7. [DOI] [Google Scholar]
- Silbiger NJ, Nelson CE, Remple K, Sevilla JK, Quinlan ZA, Putnam HM, et al. Nutrient pollution disrupts key ecosystem functions on coral reefs. Proc Royal Soc B. 2018;285:20172718. doi: 10.1098/rspb.2017.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Ann Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- Silyn-Roberts G, Lewis G. In situ analysis of Nitrosomonas spp. in wastewater treatment wetland biofilms. Water Res. 2001;35:2731–2739. doi: 10.1016/S0043-1354(00)00544-3. [DOI] [PubMed] [Google Scholar]
- Sugita H, Miyajima C, Kobayashi H, et al. Distribution of microflora in the intestinal tract of Carp Cyprinus carpio. Nippon Suisan Gakk. 1990;56:1133–1138. doi: 10.2331/suisan.56.1133. [DOI] [Google Scholar]
- Tahri Joutey N, Bahafid W, Sayel H, et al. Leucobacter chromiireducens CRB2, a new strain with high Cr (VI) reduction potential isolated from tannery-contaminated soil (Fez, Morocco) Ann Microbiol. 2016;66:425–436. doi: 10.1007/s13213-015-1125-y. [DOI] [Google Scholar]
- Talwar C, Nagar S, Lal R, et al. Fish gut microbiome: current approaches and future perspectives. Indian J Microbiol. 2018;58:397–414. doi: 10.1007/s12088-018-0760-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani K, Ogawa M, Kenzaka T (2002) Distribution of fecal bacterial groups in the river and lake water in the city of Hanoi, Vietnam. Annual Report of FY 2000, The Core University Program between Japan Society for the Promotion of Science (JSPS) and National Centre for Natural Science and Technology (NCST), pp. 94–100
- Tarnecki AM, Brennan NP, Schloesser RW, Rhody NR. Shifts in the skin-associated microbiota of hatchery-reared common snook Centropomusundecimalis during acclimation to the wild. Microb Ecol. 2019;77:770–781. doi: 10.1007/s00248-018-1252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Thanigaivel S, Vijayakumar S. Pathogenecity of Pseudomonas aeruginosa in Oreochromis mossambicus and treatment using lime oil nano emulsion. Colloids Surf B: Biointerfaces. 2014;116:372–377. doi: 10.1016/j.colsurfb.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Tsuchiya C, Sakata T, Sugita H. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett Appl Microbiol. 2008;46:43–48. doi: 10.1111/j.1472-765X.2007.02258.x. [DOI] [PubMed] [Google Scholar]
- Tyagi A, Singh B, Thammegowda NKB, et al. Shotgun metagenomics offers novel insights into taxonomic compositions, metabolic pathways and antibiotic resistance genes in fish gut microbiome. Arch Microbiol. 2019;201:295–303. doi: 10.1007/s00203-018-1615-y. [DOI] [PubMed] [Google Scholar]
- Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the Human. Microbiome Nutr. 2012;70:S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyar GÖ, Yildiran H. A nutritional approach to microbiota in Parkinson’s disease. Bio Sci Microb Food H. 2019;19:002. doi: 10.12938/bmfh.19-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kessel MA, Dutilh BE, Neveling K, et al. Pyrosequencing of 16S rRNA gene amplicons to study the microbiota in the gastrointestinal tract of carp (Cyprinus carpio L.) AMB Express. 2011;1:1–9. doi: 10.1186/2191-0855-1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaram S, Kannan S. Probiotics: The marvelous factor and health benefits. Biomed Biotechnol Res J. 2018;2:1. doi: 10.4103/bbrj.bbrj_87_17. [DOI] [Google Scholar]
- Wagner M, Loy A, Nogueira R, et al. Microbial community composition and function in wastewater treatment plants. Antonie Van Leeuwenhoek. 2002;81:665–680. doi: 10.1023/A:1020586312170. [DOI] [PubMed] [Google Scholar]
- Walter JM, Andrea B, Daniela MP. Insights into the potential of the Atlantic cod gut microbiome as biomarker of oil contamination in the marine. Microorganisms. 2019;7:209. doi: 10.3390/microorganisms7070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . Guidelines for drinking-water quality. 4. Geneva: World Health Organization (WHO); 2011. [Google Scholar]
- Xia JH, Lin G, Fu GH, et al. The intestinal microbiome of fish under starvation. BMC Genom. 2014;15:1–11. doi: 10.1186/1471-2164-15-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Jia J, Yue X, et al. River contamination shapes the microbiome and antibiotic resistance in sharpbelly (Hemiculterleucisculus) Environ Pollut. 2006;268:115796. doi: 10.1016/j.envpol.2020.115796. [DOI] [PubMed] [Google Scholar]
- Yeşilbudak B, Erdem C. Cadmium accumulation in gill, liver, kidney and muscle tissues of common carp, Cyprinus carpio, and Nile tilapia, Oreochromis niloticus. Bull Environ Contam Toxicol. 2014;92:546–550. doi: 10.1007/s00128-014-1228-3. [DOI] [PubMed] [Google Scholar]
- Zhang M, Yu N, Chen L, et al. Structure and seasonal dynamics of bacterial communities in three urban rivers in China. Aquat Sci. 2012;74:113–120. doi: 10.1007/s00027-011-0201-z. [DOI] [Google Scholar]
- Zhang H, Ding Q, Wang A, Liu Y, Teame T, Ran C, et al. Effects of dietary sodium acetate on food intake, weight gain, intestinal digestive enzyme activities, energy metabolism and gut microbiota in cultured fish: Zebrafish as a model. Aquac. 2020;523:735188. doi: 10.1038/s41598-019-57238-5. [DOI] [Google Scholar]
- Zhang Y, Wen B, David MA, Gao JZ, Chen ZZ. Comparative analysis of intestinal microbiota of discus fish (Symphysodon haraldi) with different growth rates. Aquac. 2021;540:736740. doi: 10.1016/j.aquaculture.2021.736740. [DOI] [Google Scholar]
- Zhu W, Yang Z, Ma Z, et al. Reduction of high concentrations of chromate by Leucobacter sp. CRB1 isolated from Changsha, China. World J Microbiol Biotechnol. 2008;24:991–996. doi: 10.1007/s11274-007-9564-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File S1: Physicochemical properties of water collected from sampling site. Supplementary file1 (XLSX 11 KB)
Supplementary File S2: Relative abundance of different pathways in gut samples and water samples. Supplementary file2 (XLSX 63 KB)
Supplementary File S3: Relative abundance of different enzymes in gut samples and water samples. Supplementary file3 (XLSX 260 KB)
Data Availability Statement
The raw sequencing read data have been deposited in the National Centre for Biotechnology Information (NCBI) under the project Accession Number PRJNA809116.