Abstract
Background
Quantitation of human herpesvirus-6 (HHV-6) DNA in clinical specimens is important for the diagnosis and management of HHV-6-associated infection and reactivation in immunocompromised patients, particularly transplant recipients.
Methods
The analytical performance of the Altona RealStar ASR HHV-6 qPCR on the semi-automated AltoStar AM16 system was assessed using HHV-6 reference material in plasma and cerebral spinal fluid (CSF). Qualitative and quantitative agreement was determined using 123 clinical EDTA plasma specimens tested using a laboratory-developed HHV-6 qPCR.
Results
The 95% Lower Limit of Detection was 20 IU/mL [95% confidence interval (CI): 10 to 29] in plasma and 78 IU/mL (95% CI: 55 to 146) in CSF. The assay was linear from 7.0 to 2.0 log10 IU/mL in both matrices. Overall agreement of the RealStar ASR HHV-6 qPCR on the AltoStar AM16 with a laboratory-developed test was 95.9% (95% CI: 90.8 to 98.7). Passing-Bablok analysis of specimens quantifiable by both methods and at levels >1000 copies/mL revealed a regression line of Y = 1.00*X–0.20, with neither systematic (95% CI Y-intercept: −0.66 to 0.26) nor proportional (95% CI slope: 0.89 to 1.10) bias compared to the reference.
Conclusions
The RealStar ASR HHV-6 qPCR on the AltoStar AM16 provides accurate quantitation for clinical monitoring of HHV-6 in immunocompromised hosts.
1. BACKGROUND
Human herpesvirus-6 (HHV-6) is a ubiquitous, enveloped, double-stranded DNA virus comprised of two species HHV-6A and HHV-6B in the genus Roseolovirus, which also includes human herpesvirus-7 [1,2]. HHV-6 is the causative agent of roseola infantum, a self-limited childhood rash illness characterized by three to five days of high fever that resolves abruptly, followed by a maculopapular rash that arises on the neck and trunk and spreads to the face and extremities. Following primary infection, HHV-6 establishes latency where it is capable of reactivation, and can also undergo chromosomal integration, seen in up to 1% of individuals [2].
HHV-6 reactivation may result in opportunistic disease, primarily in individuals with impaired cellular immunity, such as hematopoietic stem cell transplant (HSCT) recipients. HHV-6 reactivation can manifest non-specifically, presenting with fever and rash or myelosuppression; though subacute limbic encephalitis and delayed engraftment are also well-recognized HHV-6- associated complications in HSCT [2,3]. Detection of HHV-6 DNA in clinical specimens via quantitative PCR (qPCR) plays a key role in the diagnosis of HHV-6. Furthermore, quantitative testing may be used to inform clinical management of the immunocompromised host, including reduction of immunosuppression or initiation of antiviral therapy [3,4].
In this study we evaluate the Altona Diagnostics RealStar analyte specific reagent (ASR) HHV-6 qPCR on the semi-automated AltoStar AM16 system using HHV-6 reference panels in both plasma and cerebral spinal fluid (CSF), as well as clinical plasma specimens.
2. Materials and methods
Ethics Statement. This study was conducted with Stanford Institutional Review Board approval (protocol 46794). Individual consent was waived.
Human Herpesvirus-6 qPCR on the AltoStar AM16. The AltoStar AM16 (Altona Diagnostics) automates nucleic acid extraction and PCR set-up [5]. For each specimen, 500 μL was extracted using the AltoStar Purification Kit 1.5 and the purified nucleic acids eluted in 80 μL. Each extraction included a negative control (defibrinated human plasma; SeraCare Life Sciences), and HHV-6 low positive and high positive controls (Exact Diagnostics). The instrument also adds internal control nucleic acids to the lysed primary sample. Each reaction consisted of 18 μL general PCR reagent, 1 μL primer mix 1 μL probe mix, and 10 μL eluate. The RealStar ASR HHV-6 qPCR evaluated in this study detects and quantitates both HHV-6A and HHV-6B, but does not distinguish between types [6]. The manufacturer reports no cross-reactivity with HHV-7. Thermal cycling was performed on a Bio-Rad CFX96 real-time PCR instrument using the following conditions: 95 °C for 10 min, and then 45 cycles of 95 °C for 15 s, and 58 °C for 1 min. Each PCR plate contained a no-template control (nuclease free water; New England Biolabs) and a 4-point standard curve in copies/mL provided by Altona. Fluorescence was collected in the Altona_FAM (HHV-6) and Altona_JOE (Internal Control) channels, and thresholds were set experimentally based on background signal at 1000 and 500 relative fluorescence units (RFU), respectively.
Analytical Evaluation. Custom panels comprised of HHV-6B were manufactured by Exact Diagnostics. The lower limit of detection (LLOD) panels consisted of EDTA plasma or CSF at 1000, 750, 500, 250, 125, 75, and 10 IU/mL. For EDTA plasma, 24 replicates were tested at each level; 8 were tested per day on 3 separate days. For CSF, 16 replicates were tested at each level; 8 were tested per day on 2 separate days. The linearity panels consisted of EDTA plasma or CSF at 10-fold dilutions from 7.0 to 2.0 log10 IU/mL. Four replicates were tested at each of the 7.0, and 6.0 log10 IU/mL, whereas 8 replicates were tested at each of the remaining levels. The linearity panels were tested on a single day.
Clinical specimens. This study utilized 123 EDTA plasma specimens submitted to the Stanford Health Care Clinical Virology Laboratory between September 2016 and April 2021 for quantitative HHV-6 testing. All specimens were stored at −80 °C before testing.
Reference HHV-6 qPCR. Total nucleic acids were purified using 400 μL of EDTA plasma using the EZ1 Virus Mini Kit version 2.0 on the EZ1 Advanced XL instrument (Qiagen). The extracts were eluted in 60 μL of buffer AVE. HHV-6 DNA was quantitated using a laboratory-developed qPCR assay targeting a conserved region of the HHV-6 U66 gene, as previously described [7]. This qPCR detects both HHV-6A and HHV-6B but does not distinguish between them. Each reaction was performed using the QuantiFast Pathogen + IC kit on the Rotor-Gene Q instrument (Qiagen), and contained 10 μL of eluate, with a final reaction volume of 25 μL. Primers (HHV6Q_FWD: GAACACGTGGGTCAGATAGTTGAT; HHV6Q_REV: CATCGCCGTCACCAAACTT) and hydrolysis probe (HHV6Q Probe: FAM-CACGATTGGCTAAAGC-MGB-NFQ) were added at final concentrations of 400 nM and 200 nM, respectively. Cycling conditions were hold at 95 °C for 5 min, then 45 cycles of 95 °C for 15 s, then at 60 °C for 30 s. Detection was performed in the green (HHV-6) and yellow (internal control) channels; the threshold was set at 0.1 for both channels. The assay was calibrated using the Exact Diagnostics HHV-6 Verification Panel (Bio-Rad) and is linear from 2.7 to 6.7 log10 copies/mL.
Typing of Clinical Samples via Sequencing. Archived nucleic acid eluates from HHV-6 positive clinical samples extracted for reference qPCR testing with cycle threshold (Ct) values < 32 cycles were selected for sequence typing [8]. The U95 (HHV6–U95–F1: TAATATGATCAATCCCATCAAAC; HHV6–U95-R2: ATCCATTCATCGCCGGAAGCCGT) and U86 (HHV6–U86–F1: AACTCTTACAAAAAACATCATGAC; HHV6–U86-R1: CCTTCTTCAGAGCTACTGGAAT) genes were amplified using LongAmp PCR reagents (New England Biolabs) on the Veriti 96-well Fast thermal cycler (Thermo Fisher Scientific). Each separate, gene-specific reaction contained 12.5 μL master mix, 5 μL of eluate, and 400 nM of each primer in a final reaction volume of 25 μL. Cycling conditions were as follows: hold at 94 °C for 2 min, then 45 cycles of 95 °C for 15 s, 55 °C for 30 s, and 68 °C for 1 min, and final hold at 68 °C for 10 min. Amplicons of the correct size (U95: 342 bases; U86: 290 bases) were visualized by gel electrophoresis on 1% agarose plus ethidium bromide. In reactions containing extra bands, the band of the expected size was gel purified using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific). Amplicons were submitted for Sanger sequencing at Elim Biopharm (Hayward, California) using the same primers used for amplification. The HHV-6 type was determined by comparing the FASTA files to reference HHV-6A (NC_001664.4) and HHV-6B (NC_000898.1) sequences using the National Center for Biotechnology Information (NCBI) nucleotide Basic Local Alignment Search Tool (BLAST). The HHV-6 type was called only for samples in which both gene targets were amplified and >200 bases aligned to at least one of the reference sequences in both the forward and reverse directions. The Exact Diagnostics HHV-6B plasma reference material at 5.0 log10 IU/mL was used as control.
Statistical analysis. Lower limit of detection (LLOD) was calculated in R using probit regression. Precision was determined using a custom R script implementing the formula described in Chesher, 2008 [9]. Linearity was assessed using ordinary least squares regression in Prism version 6.0g (GraphPad, La Jolla, CA). Prism was also used to generate correlation and Bland-Altman plots. Positive percent agreement (PPA) and negative percent agreement (NPA) were calculated using MedCalc Diagnostic test evaluation calculator (https://www.medcalc.org/calc/diagnostic_test.php, Version 20.027). Kappa statistics were determined using www.graphpad.com/quickcalcs/kappa2/. Passing-Bablok regression analyses were performed using XLSTAT (2022.4.1.1368).
3. RESULTS
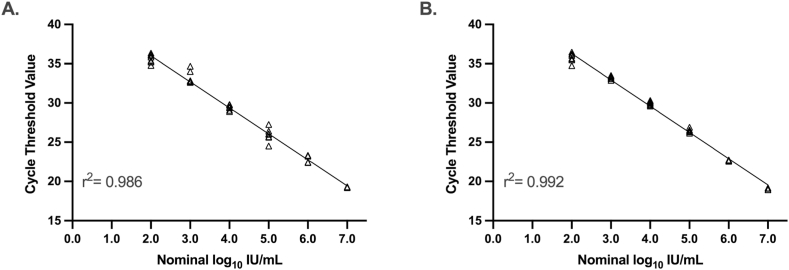
Analytical Performance of the RealStar ASR HHV-6 qPCR on the AltoStar AM16. The 95% lower limit of detection was 20 IU/mL (95% CI 10 to 29) for EDTA plasma and 78 IU/mL (95% CI 55 to 146) for CSF. Table 1, Table 2 describe assay imprecision at low levels of HHV-6 DNA for EDTA plasma and CSF, respectively. Next, linearity was assessed by plotting the observed Ct values against the nominal values and performing ordinary least squares regression (Fig. 1A and B). The Altona HHV-6 qPCR is linear from 7.0 to 2.0 log10 IU/mL in both EDTA plasma and CSF.
Table 1.
Imprecision of the Altona HHV-6 qPCR at low levels of HHV-6 DNA in EDTA plasma.
| Nominal Concentration (IU/mL) | Nominal Concentration (log10 IU/mL) | Observed Mean (log10 copies/mL) | SD Within-Run | % CV Within-Run | SD Between-Run | % CV Between-Run | SD Total | % CV Total |
|---|---|---|---|---|---|---|---|---|
| 1000 | 3.00 | 2.83 | 0.09 | 3.22 | 0.08 | 2.72 | 0.11 | 4.06 |
| 750 | 2.88 | 2.68 | 0.08 | 3.00 | 0.14 | 5.05 | 0.16 | 5.78 |
| 500 | 2.70 | 2.51 | 0.12 | 4.81 | 0.10 | 4.03 | 0.15 | 6.04 |
| 250 | 2.40 | 2.20 | 0.18 | 7.95 | 0.12 | 5.39 | 0.20 | 9.18 |
| 125 | 2.10 | 1.85 | 0.20 | 10.63 | 0.09 | 4.74 | 0.20 | 11.02 |
IU, International Unit; SD, standard deviation; CV, coefficient of variation.
Table 2.
Imprecision of the Altona HHV-6 qPCR at low levels of HHV-6 DNA in Cerebral Spinal Fluid.
| Nominal Concentration (IU/mL) | Nominal Concentration (log10 IU/mL)) | Observed Mean (log10 copies/mL) | SD Within-Run | % CV Within-Run | SD Between-Run | % CV Between-Run | SD Total | % CV Total |
|---|---|---|---|---|---|---|---|---|
| 1000 | 3.00 | 2.77 | 0.08 | 2.77 | 0.12 | 4.41 | 0.14 | 5.12 |
| 750 | 2.88 | 2.60 | 0.09 | 3.48 | 0.12 | 4.51 | 0.14 | 5.56 |
| 500 | 2.70 | 2.32 | 0.11 | 4.60 | 0.13 | 5.64 | 0.16 | 7.10 |
| 250 | 2.40 | 2.00 | 0.18 | 9.01 | 0.31 | 15.62 | 0.35 | 17.75 |
| 125 | 2.10 | 1.55 | 0.27 | 17.48 | 0.24 | 15.35 | 0.35 | 22.43 |
IU, International Unit; SD, standard deviation; CV, coefficient of variation.
Fig. 1.
Linear regression of Altona HHV-6 qPCR cycle threshold values using 10-fold dilutions of HHV-6B in EDTA plasma (A) and cerebral spinal fluid (B).
Evaluation of the RealStar ASR HHV-6 qPCR on the AltoStar AM16 Using Clinical Plasma Samples. A total of 123 EDTA plasma samples were tested using the original laboratory-developed HHV-6 qPCR results as reference. Overall agreement was 90.2% [111/123; 95% confidence interval (CI): 83.6%–94.9%], positive percent agreement was 85.3% (58/68; 95% CI: 74.6%–92.7%) and negative percent agreement was 96.4% (53/55; 95% CI: 87.5%–99.6%). The kappa coefficient was 0.81 (95% CI: 0.70 to 0.91).
The low positive percent agreement raised concern for viral DNA degradation during storage; all discrepant and concordant positive samples were re-extracted and tested using the reference HHV-6 qPCR. Of the 10 reference detected/Altona not detected discrepant samples, 6 samples were not detected by the reference qPCR upon re-testing. Of the 2 reference not detected/Altona detected discrepant samples, 1 sample was detected by the reference qPCR upon retesting. All five of the reproducibly discrepant samples had HHV-6 DNA levels below the assays’ lower limit of quantitation (Reference <2.7 log10 copies/mL; Altona <2.0 log10 copies/mL). When accounting for the results of re-testing, the overall agreement was 95.9% (118/123; 95% CI: 90.8%–98.7%), positive percent agreement was 93.7% (59/63; 95% CI: 84.5%–98.2%), and negative percent agreement was 98.3% (59/60; 95% CI: 91.1%–100.0%) (Table 3). The kappa coefficient was 0.92 (95% CI: 0.85 to 0.99).
Table 3.
Qualitative agreement of Altona HHV-6 with Reference HHV-6 qPCR Testing.
| Reference |
Positive Percent Agreement (95% CI) | Negative Percent Agreement (95% CI) | |||
|---|---|---|---|---|---|
| Detected | Not Detected | ||||
| Altona | Detected | 59 | 1 | 93.7% (84.5–98.2) | 98.3% (91.1–100.0) |
| Not Detected | 4 | 59 | |||
CI, confidence interval.
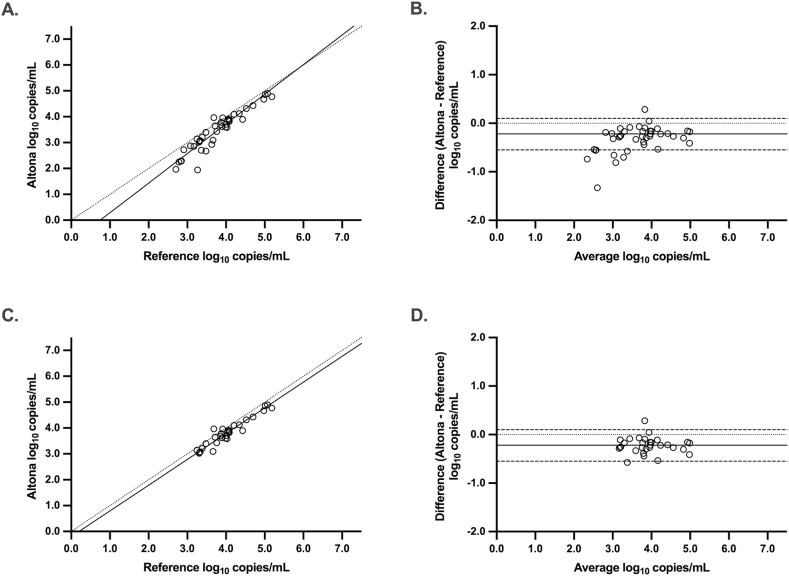
Passing-Bablok regression was performed using the values obtained from the re-extracted samples to determine the quantitative agreement between the Altona HHV-6 qPCR and the reference (Fig. 2A). This analysis resulted in a regression line of Y = 1.15*X – 0.86. The 95% confidence interval of the slope (1.03–1.29) denotes a positive proportional bias, whereas the 95% confidence interval of the Y-intercept (−1.42 to −0.37) shows a negative systematic bias. Bland-Altman analysis of this data (Fig. 2B) showed a bias of −0.32 log10 copies/mL (Altona – Reference HHV-6 qPCR) and 95% limits of agreement of −0.86 to 0.21.
Fig. 2.
Passing-Bablok regression (A, C) and Bland-Altman analysis (B, D) comparing HHV-6 DNA levels (log10 copies/mL) between the Altona and reference qPCR methods in EDTA plasma. Panels A and B set the quantifiable ranges at 2.0 log10 copies/mL for Altona and 2.7 log10 copies/mL for the reference. Panels C and D set the quantifiable ranges at 3.0 log10 copies/mL for both methods. Passing-Bablok: Solid line, regression line; dotted line, line of identity. Bland-Altman: Solid line, mean difference; dotted line, zero difference; dashed lines; 95% confidence intervals.
Quantitative agreement improved if the low end of the quantifiable range was set to 3.0 log10 copies/mL for both methods. Passing-Bablok regression of this data set resulted in a regression line of Y = 1.00*X – 0.20 (Fig. 2C). The 95% confidence interval of the slope (0.89–1.10) included one and the 95% confidence interval of the Y-intercept (−0.66 to 0.26) included zero, indicating neither proportional, nor systematic bias. Bland-Altman analysis revealed a bias of −0.22 log10 copies/mL (Altona – Reference HHV-6 qPCR) and 95% limits of agreement of −0.55 to 0.10.
A subset of the clinical samples was typed via sequencing of the U95 and U86 genes. Adequate sequence was obtained for 68.2% (15/22) of the selected samples. All sequenced samples were HHV-6B.
4. Discussion
In this study we evaluated the RealStar ASR HHV-6 qPCR on the semi-automated AltoStar AM16 using reference material and clinical specimens. Analytical performance of EDTA plasma and CSF met requirements for clinical use. In addition, qualitative agreement was observed when the Altona HHV-6 qPCR was compared with a laboratory developed reference qPCR using clinical EDTA plasma specimens. Finally, neither systematic nor proportional bias was observed when the lower limit of quantitation was set to 1000 copies/mL.
Strategies for the diagnosis of HHV-6-associated diseases after transplantation may include symptom-based testing, as well as pre-emptive approaches in high-risk recipients [2,3,[10], [11], [12]]. Once a positive result is obtained, however, thresholds for the reduction of immunosuppression or initiation of antiviral therapy may vary widely based on the source of the hematopoietic stem cells (e.g., unrelated cord blood transplant), clinical context (e.g., whether the patient is symptomatic versus asymptomatic), as well as institution-specific protocols [10,[13], [14], [15], [16], [17]]. The challenge in setting a clinical threshold is compounded, at least in part, due to the lack of harmonization among different HHV-6 qPCR assays [18,19]. Nevertheless, after immunosuppression has been adjusted or antiviral therapy initiated, monitoring patients for the clearance of HHV-6 DNA in blood, and if serially collected, CSF, is generally accepted [3,11,12].
This study has several strengths, including the number of replicates used in the analytical evaluation and the number of clinical samples used in comparing qPCR methods. In addition, the AltoStar AM16 system automates nucleic acid extraction and PCR set-up. Up to 4 qPCR assays can be set-up and performed from a single nucleic acid eluate, and because HHV-6 and the other RealStar transplant virus ASRs, such as adenovirus and BK virus, share common cycling parameters, this combination of semi-automation and compatible qPCR reagents allows efficient virologic testing post-transplantation.
Limitations include the absence of HHV-6A reference materials in the analytical evaluation and the sequence confirmation of only HHV-6B in a subset of the clinical specimens used in the qPCR method comparison. These shortcomings, however, are unlikely to impact our overall conclusions, as HHV-6B causes the majority of HHV-6-associated disease in transplant recipients [4]. Should additional data more strongly implicate HHV-6A in post-transplant sequelae or HHV-6 DNA monitoring becomes more commonly utilized in other patient populations, further verification of both the analytical and clinical performance of this assay for HHV-6A may be warranted.
This study is also limited by the lack of calibration to the 1st World Health Organization (WHO) International Standard for HHV-6B DNA in the quantitative comparison of qPCR methods [20]. Though the Altona and laboratory-developed qPCRs used in this study were calibrated using two different assay-specific standards, quantitative agreement was observed over a clinically relevant range of HHV-6 DNA levels. Future studies are required to ensure that harmonization to the WHO international standard improves quantitative agreement and that the international standard is commutable among available HHV-6 nucleic acid amplification tests.
FUNDING
Altona Diagnostics funded this work and provided the RealStar reagents and AltoStar AM16 instrument used in this study.
Author statement
Jordan Mah: Data curation, Formal analysis, Writing - original draft, Writing - review & editing; ChunHong Huang: Data curation, Investigation; Validation, Writing - review & editing; Malaya K. Sahoo: Data curation, Formal analysis, Investigation, Validation, Writing - review & editing; Benjamin A. Pinsky: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing - original draft, Writing - review & editing.
Declaration of competing interest
Benjamin A. Pinsky performed contracted research for Altona Diagnostics and Hologic, Inc. He also received travel support from Altona. The other authors report no declarations of interest.
Data availability
Data will be made available on request.
References
- 1.De Bolle L., Naesens L., De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin. Microbiol. Rev. 2005;18(1):217–245. doi: 10.1128/CMR.18.1.217-245.2005. PubMed PMID: 15653828; PubMed Central PMCID: PMC544175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agut H., Bonnafous P., Gautheret-Dejean A. Laboratory and clinical aspects of human herpesvirus 6 infections. Clin. Microbiol. Rev. 2015;28(2):313–335. doi: 10.1128/CMR.00122-14. PubMed PMID: 25762531; PubMed Central PMCID: PMC4402955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward K.N., Hill J.A., Hubacek P., de la Camara R., Crocchiolo R., Einsele H., et al. Guidelines from the 2017 European Conference on Infections in Leukaemia for management of HHV-6 infection in patients with hematologic malignancies and after hematopoietic stem cell transplantation. Haematologica. 2019;104(11):2155–2163. doi: 10.3324/haematol.2019.223073. Epub 20190829. PubMed PMID: 31467131; PubMed Central PMCID: PMC6821622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agut H., Bonnafous P., Gautheret-Dejean A. Human herpesviruses 6A, 6B, and 7. Microbiol. Spectr. 2016;4(3) doi: 10.1128/microbiolspec.DMIH2-0007-2015. PubMed PMID: 27337451. [DOI] [PubMed] [Google Scholar]
- 5.Mah J., Huang C.H., Sahoo M.K., Pinsky B.A. Evaluation of a semiautomated system for the quantitation of human adenovirus DNA from clinical samples. Microbiol. Spectr. 2023;11(2) doi: 10.1128/spectrum.05010-22. Epub 20230227. PubMed PMID: 36847504; PubMed Central PMCID: PMC10100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altona Diagnostics. RealStar ASR HHV-6. 2023. [cited 2023 May 6, 2023]. Available from: https://altona-diagnostics.com/en/products/reagents/realstar-real-time-pcr-reagents/realstar-asr-hhv-6.html.
- 7.Yang C.H., Sahoo M.K., Fitzpatrick M., Lau A.H., Pinsky B.A., Martinez O.M. Evaluating for human herpesvirus 6 in the liver explants of children with liver failure of unknown etiology. J. Infect. Dis. 2019;220(3):361–369. doi: 10.1093/infdis/jiy644. Epub 2018/11/13. PubMed PMID: 30418598. [DOI] [PubMed] [Google Scholar]
- 8.Isegawa Y., Mukai T., Nakano K., Kagawa M., Chen J., Mori Y., et al. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 1999;73(10):8053–8063. doi: 10.1128/JVI.73.10.8053-8063.1999. PubMed PMID: 10482554; PubMed Central PMCID: PMC112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesher D. Evaluating assay precision. Clin. Biochem. Rev. 2008;29(Suppl 1):S23–S26. Epub 2008/10/15. PubMed PMID: 18852851; PubMed Central PMCID: PMC2556577. [PMC free article] [PubMed] [Google Scholar]
- 10.Hill J.A. Human herpesvirus 6 in transplant recipients: an update on diagnostic and treatment strategies. Curr. Opin. Infect. Dis. 2019;32(6):584–590. doi: 10.1097/QCO.0000000000000592. PubMed PMID: 31567413; PubMed Central PMCID: PMC7141773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogata M., Uchida N., Fukuda T., Ikegame K., Kamimura T., Onizuka M., et al. Clinical practice recommendations for the diagnosis and management of human herpesvirus-6B encephalitis after allogeneic hematopoietic stem cell transplantation: the Japan Society for Hematopoietic Cell Transplantation. Bone Marrow Transplant. 2020;55(6):1004–1013. doi: 10.1038/s41409-019-0752-5. Epub 20191119. PubMed PMID: 31745253. [DOI] [PubMed] [Google Scholar]
- 12.Olson A.L., Politikos I., Brunstein C., Milano F., Barker J., Hill J.A., et al. Guidelines for infection prophylaxis, monitoring and therapy in cord blood transplantation. Transplant Cell Ther. 2021;27(5):359–362. doi: 10.1016/j.jtct.2021.01.024. PubMed PMID: 33965172. [DOI] [PubMed] [Google Scholar]
- 13.Ogata M., Kikuchi H., Satou T., Kawano R., Ikewaki J., Kohno K., et al. Human herpesvirus 6 DNA in plasma after allogeneic stem cell transplantation: incidence and clinical significance. J. Infect. Dis. 2006;193(1):68–79. doi: 10.1086/498531. Epub 20051130. PubMed PMID: 16323134. [DOI] [PubMed] [Google Scholar]
- 14.Ogata M., Satou T., Kawano R., Takakura S., Goto K., Ikewaki J., et al. Correlations of HHV-6 viral load and plasma IL-6 concentration with HHV-6 encephalitis in allogeneic stem cell transplant recipients. Bone Marrow Transplant. 2010;45(1):129–136. doi: 10.1038/bmt.2009.116. Epub 20090525. PubMed PMID: 19465942. [DOI] [PubMed] [Google Scholar]
- 15.Dulery R., Salleron J., Dewilde A., Rossignol J., Boyle E.M., Gay J., et al. Early human herpesvirus type 6 reactivation after allogeneic stem cell transplantation: a large-scale clinical study. Biol. Blood Marrow Transplant. 2012;18(7):1080–1089. doi: 10.1016/j.bbmt.2011.12.579. Epub 20111230. PubMed PMID: 22212513. [DOI] [PubMed] [Google Scholar]
- 16.Hill J.A., Koo S., Guzman Suarez B.B., Ho V.T., Cutler C., Koreth J., et al. Cord-blood hematopoietic stem cell transplant confers an increased risk for human herpesvirus-6-associated acute limbic encephalitis: a cohort analysis. Biol. Blood Marrow Transplant. 2012;18(11):1638–1648. doi: 10.1016/j.bbmt.2012.04.016. Epub 20120504. PubMed PMID: 22564265; PubMed Central PMCID: PMC3816521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogata M., Satou T., Kadota J., Saito N., Yoshida T., Okumura H., et al. Human herpesvirus 6 (HHV-6) reactivation and HHV-6 encephalitis after allogeneic hematopoietic cell transplantation: a multicenter, prospective study. Clin. Infect. Dis. 2013;57(5):671–681. doi: 10.1093/cid/cit358. Epub 20130530. PubMed PMID: 23723198. [DOI] [PubMed] [Google Scholar]
- 18.Flamand L., Gravel A., Boutolleau D., Alvarez-Lafuente R., Jacobson S., Malnati M.S., et al. Multicenter comparison of PCR assays for detection of human herpesvirus 6 DNA in serum. J. Clin. Microbiol. 2008;46(8):2700–2706. doi: 10.1128/JCM.00370-08. Epub 20080611. PubMed PMID: 18550745; PubMed Central PMCID: PMC2519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Pagter P.J., Schuurman R., de Vos N.M., Mackay W., van Loon A.M. Multicenter external quality assessment of molecular methods for detection of human herpesvirus 6. J. Clin. Microbiol. 2010;48(7):2536–2540. doi: 10.1128/JCM.01145-09. Epub 20100210. PubMed PMID: 20147642; PubMed Central PMCID: PMC2897485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization Collaborative study to establish the 1st WHO international standard for human herpes virus 6B (HHV-6B) DNA for nucleic acid amplification technique (NAT)-based assays. 2017. https://www.who.int/publications/m/item/WHO-BS-2017.2321 [cited 2023 February 15, 2023]. Available from:
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.