Abstract
Candida inconspicua was recovered from three patients with hematological malignancies. Two patients had intravenous-catheter-associated fungemia, whereas the third had fungal hepatitis. The three cases of infection occurred over a period of 1 month in patients staying in adjacent single rooms. In vitro susceptibility testing of fungal strains showed all isolates to be resistant to fluconazole, with MICs greater than 32 μg/ml. All of the strains had identical DNA restriction profiles and randomly amplified polymorphic DNA fingerprints. These data suggest a nosocomially acquired infection emanating from a common source within the hospital environment.
Fungal infections are an increasing cause of morbidity and mortality in patients with hematological malignancies (4). Underlying malignancy, neutropenia, cytotoxic chemotherapy, advanced age, parenteral nutrition, prolonged broad-spectrum-antibiotic treatment, and use of central venous catheters (CVC) are known risk factors for these infections (7). The organisms most often responsible are Candida spp., particularly Candida albicans. Other Candida species, such as C. tropicalis, C. krusei, C. parapsilosis, C. glabrata, and C. guilliermondii, are less frequently isolated but are not uncommon, whereas C. inconspicua infections have not yet been reported for cancer patients (11). C. inconspicua has been reported for human immunodeficiency virus-infected patients with either oral or esophageal infections, for women with vaginal infections, and for patients with diabetes mellitus (2, 6). We report three cases of infection due to C. inconspicua occurring as a cluster in patients with malignancies. To our knowledge, this is the first report of C. inconspicua infections occurring as a nosocomial outbreak.
MATERIALS AND METHODS
Patient reports.
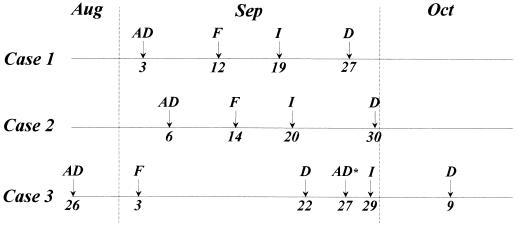
Three cases of C. inconspicua infection were identified in September 1995 in the Department of Haematology and Oncology at Pescara General Hospital, Pescara, Italy, a 900-bed teaching hospital. The patients received treatment in single adjacent rooms with restricted entry and mandatory use of sterile masks, gloves, overshoes, and dressing gowns. All patients received a cooked-food diet. Figure 1 shows a comparison of the three cases of C. inconspicua infection.
FIG. 1.
Cronology of the outbreak. AD, date of admission; F, date of onset of fever; I, date of diagnosis of C. inconspicua infection; D, date of discharge; AD*, date of readmission with fever.
Patient 1.
A 52-year-old man with acute myeloblastic leukemia (M6 FAB [where FAB stands for French-American-British classification]) in relapse was treated with cytarabine, mitoxantrone, and etoposide. Prior to intensive chemotherapy, a CVC was positioned. Prophylactic antimicrobial treatment consisted of ciprofloxacin and nystatin. During the aplastic phase, the patient developed fever (>38.5°C) and erythema of the skin at the site of CVC exit. Empiric therapy with imipenem (3 g/day) and vancomycin (30 mg/kg of body weight/day) was instituted. After 7 days of antibiotic therapy, the patient showed no response and empiric intravenous amphotericin B (AmB) (0.7 mg/kg/day) was administered. The patient improved rapidly. Three bottles of two sets of blood cultures (Vital Biomerieux Italia, Rome, Italy) done 1 day prior to the antifungal treatment grew microorganisms identified as C. inconspicua. A new CVC was placed; the removed CVC tip was cultured by a semiquantitative technique. Twenty-three CFU of yeastlike strains was isolated and subsequently identified as C. inconspicua. This high level of CVC tip colonization, in the absence of other potential sources of infection as determined through a careful clinical and laboratory evaluation, was strongly suggestive of CVC-associated candidemia (10). Cultures of blood repeatedly collected during antifungal therapy were sterile.
Patient 2.
A 56-year-old woman with acute myeloblastic leukemia (M4 FAB) in relapse was treated with cytarabine, daunorubicin, and etoposide. Prior to intensive chemotherapy, a CVC was positioned. Prophylactic antimicrobial treatment consisted of ciprofloxacin and nystatin. On day 3 of granulocytopenia during the aplastic phase, the patient developed a fever of >38.5°C unresponsive to empiric amikacin (15 mg/kg/day) and ceftazidime (6 g/day). The patient showed erythema, warmth, and tenderness over the area of the CVC exit. On day 7, a yeastlike organism was isolated from blood cultures and identified as C. inconspicua. The CVC was removed, and samples from the CVC tip cultured by a semiquantitative technique yielded 28 CFU of C. inconspicua, suggesting CVC-associated candidemia (10). Intravenous AmB (0.7 mg/kg/day) was administered and the patient improved rapidly. Cultures of blood repeatedly collected during antifungal therapy were all sterile.
Patient 3.
On 26 August 1995 a 42-year-old female with centroblastic follicular non-Hodgkin’s lymphoma (stage III), splenectomized for massive extravascular bleeding and fibrosis, was hospitalized to undergo autologous peripheral hematopoietic stem cell transplantation. The patient was prophylactically treated with ciprofloxacin and oral fluconazole. During the aplastic phase, the patient developed a fever of >38.5°C. All blood cultures were negative. Imipenem (3 g/day) and vancomycin (30 mg/kg/day) were administered. Intravenous AmB was empirically added after 7 days of uneffective antibiotic treatment. Despite the AmB treatment, an intermittent high fever continued for another 7 days. Apyrexia was achieved concomitantly with bone marrow recovery. At day 27, after autologous transplantation, intravenous antifungal treatment was discontinued and the patient was discharged. Five days later, the patient was readmitted with a fever of >38.5°C, abdominal pain, vomiting, and diarrhea but without any apparent clinical signs of candidiasis. The neutrophil count was 7.1 × 109/liter, but tests showed abnormal liver function: alkaline phosphatase = 610 U/ml (n = 120); gamma glutamyl transpeptidase = 589 U/ml (n = 60), glutamic-oxaloacetic transaminase = 168 U/ml (n < 35), glutamic pyruvic transaminase = 220 U/ml (n < 40), and C-reactive protein = 89 mg/liter (n < 10). Blood cultures were negative. An abdominal ultrasonography (US) revealed multiple hypoechoic lesions in the liver, and the computerized tomography scan showed multiple low-density areas. A US-guided fine-needle liver biopsy was performed. Histopathologic examination using the periodic acid-Schiff stain reaction showed numerous fungal organisms in the necrotic areas (Fig. 2), subsequently identified as C. inconspicua. A treatment with intravenous AmB (0.8 mg/kg) was started. After 10 days of therapy, the patient became afebrile and was discharged. The patient continued antifungal therapy at home on alternate days. During the treatment, surveillance by abdominal US and computerized tomography showed progressive regression of the liver lesions. Three months later, these lesions had disappeared.
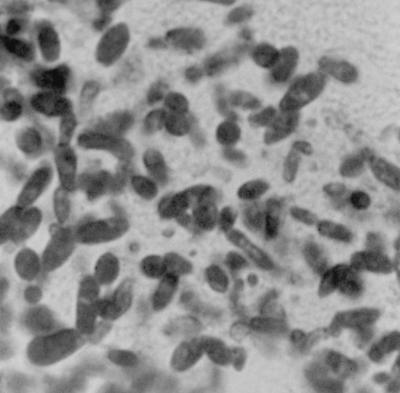
FIG. 2.
Budding yeast with elongated blastoconidia consistent with the morphology of C. inconspicua in necrotic liver tissue stained with the periodic acid-Schiff stain. Magnification, ×1,000.
Surveillance cultures.
A routine surveillance program for infection control is carried out on all patients admitted to the Department of Haemotology and Oncology and includes quantitative aerobic, anaerobic, and fungal culture of oral swabs, stools, and urine performed upon admission and twice a week thereafter. Environmental cultures and cultures of samples from health-care workers are performed only occasionally and are not included in this routine surveillance program.
Specimen collection and processing.
Blood samples (10 ml from the CVC and 10 ml from a peripheral vein) were inoculated into aerobic and anaerobic bottles (Vital Biomerieux). These were incubated at 37°C and read twice daily with an automatic detector (Vital Biomerieux). For the bottles indicated as positive, an aliquot of the blood-broth mixture was removed for Gram staining. Based on the Gram stain results, the samples were subcultured onto appropriate media. When yeastlike cells were observed, the samples were subcultured onto Sabouraud glucose agar (SGA) plates (Difco Laboratories, Detroit, Mich.) and into Sabouraud dextrose broth (SDB) (Difco).
In the case of catheter removal, the CVC tip was rolled onto an SGA (Difco) plate, and after 48 h of incubation at 37°C the colonies were counted and the organisms were identified. Catheter-related sepsis was defined as a count of ≥15 CFU per plate with no other definite source of infection (3, 10).
The liver biopsy sample taken from patient 3 was homogenized in sterile saline in a biological safety cabinet. Portions of the tissue homogenate were cultured for aerobic and anaerobic bacteria by conventional methods (5). Moreover, aliquots of the specimens were directly plated onto SGA (Difco) and inoculated into SDB (Difco).
Strain identification.
The yeastlike isolates were identified according to their morphological characteristics and biochemical profiles. The morphological features of all organisms were examined on a medium for germ tube production (medium for blastesis; Diagnostic Pasteur, Marnes-La-Croquette, France) and on cornmeal agar (Unipath S.p.A., Garbagnate Milanese, Milan, Italy) slide cultures. Growth was performed on a chemically defined vitamin-free medium (vitamin-free yeast base; Difco), by incubating preparations at 30°C for 7 days. Biochemical tests were performed by using ID32 C strips with an ATB (automation test bactériologique) reader (API system; BioMerieux). Antifungal susceptibility testing for AmB and fluconazole was performed according to the National Committee for Clinical Laboratory Standards guidelines (12).
DNA typing. (i) Restriction fragment length polymorphism (RFLP).
Isolates were grown overnight in 10 ml of YPD broth (1% yeast extract, 2% peptone, 2% dextrose) (Oxoid, Unipath S.p.A.). The culture was spun down, and the DNA was extracted for restriction as described by Scherer and Stevens (14). DNA was resuspended in a final volume of 50 μl, and 15 μl was digested with EcoRI for 6 h by using buffer supplied by the manufacturer (Boehringer Mannheim GmbH, Mannheim, Germany). Fragments were separated by electrophoresis through 0.9% agarose in TBE buffer (0.089 M Tris, 0.089 M boric acid, 2 mM Na2EDTA), stained with ethidium bromide, and photographed with a Polaroid type 557 camera.
(ii) Randomly amplified polymorphic DNA (RAPD).
PCR DNA was further purified with a Geneclean II kit (Bio 101 Inc., La Jolla, Calif.) to remove inhibitors present in the samples. Two decanucleotide primers (Perkin-Elmer S.p.A., Milan, Italy) were used.
Primer sequences were as follows: for CI12, 5′-ACGGTACCAG-3′, and for CI25, 5′-GAACAGCTGG-3′. PCRs were performed with 5 U of AmpliTaq DNA polymerase (Stoffel fragment) by using supplied buffer: dCTP, dGTP, dATP, and dTTP, each at 200 μM; 5 mM MgCl2; 200 pmol of primer; and 2 μl of DNA. All reagents were supplied by Perkin-Elmer.
The RAPD assay was carried out with 50-μl volumes in a GeneAmp PCR System 2400 (Perkin-Elmer S.p.A.) programmed as follows: 94°C for 3 min, followed by 45 cycles of 94°C for 1 min, 34°C for 1 min, and 72°C for 1 min. The final extension step was prolonged to 7 min at 72°C.
The resulting DNA fragments were separated through a 1.5% agarose gel and visualized as described above.
C. inconspicua strains.
Five strains of C. inconspicua were isolated from our three patients. Patients 1 and 2 had two isolates each, from blood culture and the CVC tip, respectively. The fifth strain was isolated from a liver biopsy sample from patient 3. An American Type Culture Collection (ATCC) human strain of C. inconspicua (ATCC 16783) was used as a reference strain. No epidemiologically unrelated strains were available to be used as controls, since there were no previous isolates of C. inconspicua from either surveillance cultures or clinical samples.
RESULTS
C. inconspicua was never isolated from surveillance cultures from patients admitted to the Department, including those in this study, or from environmental samples and cultures from health-care workers.
The fungal agents of these three infective episodes showed identical phenotypic characteristics and were identified as C. inconspicua (8). The cells were ovoid on SDB after 72 h at 25°C, measuring 1.5 by 3 μm to 3.0 by 5 μm. On SGA, the streak cultures were greyish white and smooth. The yeasts did not form true hyphae or pseudohyphae. Only a very primitive pseudomycelium consisting of branched chains of ovoid cells was observed in the cornmeal agar. Direct readings from the API system showed an ID32 C profile of 0200010005 (good identification for C. inconspicua). The organism did not grow in the presence of cycloheximide. Strain ATCC 16783 yielded an identical code with the API profile. Neither our isolates nor the ATCC reference strain grew on vitamin-free medium. All C. inconspicua isolates were resistant to fluconazole (MICs, >32 μg/ml) and susceptible to AmB (MICs, 0.004 μg/ml).
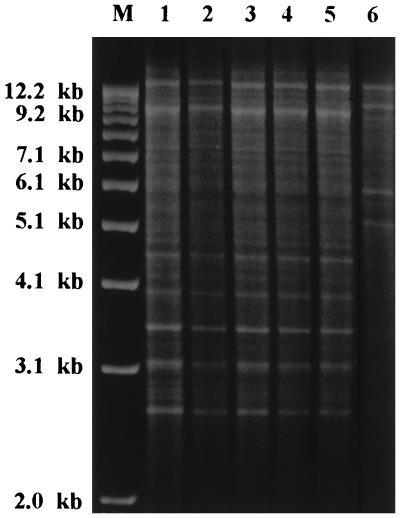
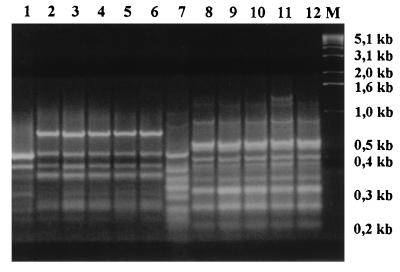
We performed DNA typing on both the clinical isolates of C. inconspicua and the ATCC reference strain. All typing systems gave unequivocal results. All the clinical isolates had identical DNA restriction profiles after digestion with EcoRI (Fig. 3). When primer CI12 was used, the RAPD patterns of our isolates were also indistinguishable. Primer CI25 revealed just a slight difference in the profile of the CVC tip isolate from patient 2, due to the presence of a supplementary band of 1.2 kb (Fig. 4, lane 11). The ATCC control strain was clearly distinguishable from the other strains when typed by either RFLP or RAPD.
FIG. 3.
Restriction endonuclease pattern (EcoRI digests of whole chromosomal DNA) of clinical isolates and an ATCC reference strain of C. inconspicua. Lanes: M, size markers (1-kb DNA ladder; GIBCO BRL, Gaithersburg, Md.); 1 and 2, blood and catheter tip isolates from patient 1; 3 and 4, blood and catheter tip isolates from patient 2; 5, hepatic isolate from patient 3; 6, ATCC 16783 (human isolate).
FIG. 4.
RAPD fingerprints of clinical isolates and an ATCC reference strain of C. inconspicua obtained with primers CI12 (lanes 1 to 6) and CI25 (lanes 7 to 12). Lanes: 1, ATCC 16783 (human isolate); 2 and 3, blood and catheter tip isolates from patient 1; 4 and 5, blood and catheter tip isolates from patient 2; 6, hepatic isolate from patient 3; 7, ATCC 16783; 8 and 9, blood and catheter tip isolates from patient 1; 10 and 11, blood and catheter tip isolates from patient 2; 12, hepatic isolate from patient 3; M, size markers (1-kb DNA ladder).
DISCUSSION
This paper describes a small cluster of infections due to C. inconspicua in patients with malignancies. Several pieces of evidence are confirmatory of the epidemic nature of this cluster. The three cases of infection occurred over a period of 1 month and involved patients in adjacent single rooms. No previous isolate of C. inconspicua from clinical, surveillance, or environmental samples has been reported at our Department. No more C. inconspicua infections were detected after that episode. All our C. inconspicua isolates are genetically indistinguishable by both REA and RAPD analysis. These identical profiles are not attributable to a lack of discriminative power of our molecular typing systems, given their capability to differentiate our isolates from the ATCC reference strain, which was used specifically to validate our typing systems. The slightly different RAPD profile generated with primer CI25 for the CVC tip isolate from patient 2 is likely to be expression of a microevolutionary change within the infecting clonal population (9, 13).
It is difficult to answer the question of whether these patients became infected from a common source within the hospital environment or whether the strain was brought to the hospital by the index patient (case 1), from whom it subsequently spread into the hospital environment. The latter hypothesis seems unlikely, since these patients were screened for oral, fecal, and urinary yeast carriage on admission, in the context of a global infection control program, and were negative for C. inconspicua. The alternative explanation may be considered more reliable, but since this outbreak was only retrospectively identified and investigated, no screening of the ward was carried out at the time of the outbreak to detect a potential source of the strain within the hospital environment.
Our patients have survived despite having disseminated infections, suggesting a low virulence for the epidemic strain of C. inconspicua.
The two patients with C. inconspicua fungemia associated with CVC infections did not develop mural thrombosis and were successfully treated with a course of AmB and removal of the CVC. The patient with C. inconspicua hepatitis also had an improvement from AmB therapy. This is the first case of an hepatic localization of C. inconspicua. Mostly C. albicans and C. tropicalis have been reported as causative agents of hepatic and/or splenic infection in patients with malignancies receiving chemotherapy (1, 15).
In summary, the spectrum of Candida spp. responsible of disseminated infections continues to expand, including species that were rarely considered pathogens. C. inconspicua should be taken in proper consideration as a potential agent of nosocomial invasive infections in immunocompromised patients.
ACKNOWLEDGMENTS
We thank A. Nanetti (Istituto di Microbiologia, Università di Bologna, Bologna, Italy) and C. Farina (Istituto di Microbiologia, Ospedali Riuniti di Bergamo, Bergamo, Italy) for confirming the identity of C. inconspicua. We also thank BioMerieux for confirming the API profiles of our isolates.
This work was supported in part by the Associazione Donatori Sangue, Pescara, Italy.
REFERENCES
- 1.Akihiko, C., I. Miura, A. Ohshima, T. Nishinari, T. Nimura, H. Niitsu, and A. B. Miura. 1994. Risk factors for hepatosplenic abscesses in patients with acute leukemia receiving empiric azole treatment. Am. J. Med. Sci. 308(Suppl. 6):309–312. [DOI] [PubMed]
- 2.Aly, F. Z., C. C. Blackwell, D. A. C. MacKenzie, D. M. Weir, R. A. Elton, C. G. Cumming, J. A. Sofaer, and B. F. Clarke. 1991. Chronic atrophic oral candidiasis among patients with diabetes mellitus—role of secretor status. Epidemiol. Infect. 106(Suppl. 2):355–363. [DOI] [PMC free article] [PubMed]
- 3.Andremont A, Paulet R, Nitenberg G, Hill C. Value of semiquantitative cultures of blood drawn through catheter hubs for estimating the risk of catheter tip colonization in cancer patients. J Clin Microbiol. 1988;26:2297–2299. doi: 10.1128/jcm.26.11.2297-2299.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodey G P, Bolwar R, Fainstein V. Infectious complications in leukemic patients. Semin Hematol. 1993;19:193–226. [PubMed] [Google Scholar]
- 5.Forbes B A, Granato P A. Processing specimens for bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 265–281. [Google Scholar]
- 6.Gerhard, I., D. Ohlhorst, W. Eggert-Kruse, and B. Nebaum. 1989. Topical one-day treatment with ketoconazole: a double-blind randomized control study on vaginal candidosis. Mycoses 32(Suppl. 5):253–265. [PubMed]
- 7.Karabinis A, Hill C, Leclercq B, Tancrède C, Baume D, Andremont A. Risk factors for candidemia in cancer patients: a case-control study. J Clin Microbiol. 1988;26:429–432. doi: 10.1128/jcm.26.3.429-432.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreger-van Rij N J W, editor. The yeasts: a taxonomic study. 3rd ed. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1984. pp. 708–709. [Google Scholar]
- 9.Lockhart S R, Fritch J J, Meier A S, Schröppel K, Srikantha T, Galask R, Soll D R. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol. 1995;33:1501–1509. doi: 10.1128/jcm.33.6.1501-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maki D G, Weise C E, Sarafin H W. A semiquantitative culture method for identifying intravenous catheter-related infections. N Engl J Med. 1977;296:1305–1309. doi: 10.1056/NEJM197706092962301. [DOI] [PubMed] [Google Scholar]
- 11.Meunier, F., M. Aoun, and N. Bitar. 1992. Candidemia in immunocompromised patients. Clin. Infect. Dis. 14(Suppl. 1):120–125. [DOI] [PubMed]
- 12.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. Proposed standard NCCLS document M27-P. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1992. [Google Scholar]
- 13.Pujol C, Joly S, Lockhart S R, Noel S, Tibayrenc M, Soll D R. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J Clin Microbiol. 1997;35:2348–2358. doi: 10.1128/jcm.35.9.2348-2358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherer S, Stevens D A. Application of DNA typing methods in epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thaler M, Pastakia B, Shawker T, O’Leary T, Pizzo P A. Hepatic candidiasis in cancer patients: the evolving picture of the syndrome. Ann Intern Med. 1988;108:88–100. doi: 10.7326/0003-4819-108-1-88. [DOI] [PubMed] [Google Scholar]