Abstract
Due to poor T cell infiltration, tumors evade immune surveillance. Increased CD8+ T cell infiltration in breast cancer suggests a satisfactory response to immunotherapy. COPS6 has been identified as an oncogene, but its role in regulating antitumor immune responses has not been defined. In this study, we investigated the impact of COPS6 on tumor immune evasion in vivo. Tumor transplantation models were established in C57BL/6 J mice and BALB/c nude mice. Flow cytometry was conducted to identify the role of COPS6 on tumor-infiltrating CD8+ T cells. By analyzing the TCGA and GTEx cohort, we found that COPS6 expression was significantly up-regulated in a variety of cancers. In human osteosarcoma cell line U2OS and non-small cell lung cancer cell line H1299, we showed that p53 negatively regulated COPS6 promoter activity. In human breast cancer MCF-7 cells, COPS6 overexpression stimulated p-AKT expression as well as the proliferation and malignant transformation of tumor cells, whereas knockdown of COPS6 caused opposite effects. Knockdown of COPS6 also significantly suppressed the growth of mouse mammary cancer EMT6 xenografts in BALB/c nude mice. Bioinformatics analysis suggested that COPS6 was a mediator of IL-6 production in the tumor microenvironment and a negative regulator of CD8+ T cell tumor infiltration in breast cancer. In C57BL6 mice bearing EMT6 xenografts, COPS6 knockdown in the EMT6 cells increased the number of tumor-infiltrating CD8+ T cells, while knockdown of IL-6 in COPS6KD EMT6 cells diminished tumor infiltrating CD8+ T cells. We conclude that COPS6 promotes breast cancer progression by reducing CD8+ T cell infiltration and function via the regulation of IL-6 secretion. This study clarifies the role of p53/COPS6/IL-6/CD8+ tumor infiltrating lymphocytes signaling in breast cancer progression and immune evasion, opening a new path for development of COPS6-targeting therapies to enhance tumor immunogenicity and treat immunologically “cold” breast cancer.
Keywords: immunotherapy, breast cancer, tumor immune evasion, COPS6, p53, IL-6
Introduction
Breast cancer is the most common malignant tumor worldwide, which is a serious public health problem threatening women’s health [1]. Approximately 2.3 million new breast cancer cases (11.7%) were estimated by GLOBOCAN in 2020, which has become the most common cancer [1]. After primary tumor therapy, 20%–30% of breast cancer patients are prone to show metastatic disease [2], 5%–10% of patients have metastases at diagnosis [3]. Despite considerable progress in treatment of breast cancer, the 5-year overall survival rate of patients with metastasis decreased significantly to 25% [4], compared to >90% of patients without distant metastasis [5]. In particular, recurrent and advanced triple-negative breast cancer (TNBC) patients have poor response to follow-up treatment, with median survival only about 18 months [6]. Therefore, exploring the potential mechanism and therapeutic targets of breast cancer is of great significance for the clinical treatment of breast cancer patients.
Constitutive photomorphogenesis 9 (COP9) signalosome (CSN) is an evolutionarily conserved protein complex consisting of nine subunits (CSN1–CSN8 and recently discovered CSNAP) in eukaryotes [7, 8], which was initially identified as a repressor of constitutive photomorphogenesis (COP) in Arabidopsis [8]. COPS6 was responsible for maintaining the structural integrity and function of the CSN complex in a MPN domain-dependent manner [9, 10]. CSN signalosome was involved in protein degradation, DNA repair, cell cycle control, signal transduction, transcriptional activation, and tumorigenesis [8, 9]. Analogously, COPS6 has been well characterized by coordinating cullin-RING ligase (CRL)-mediated ubiquitination activity to regulate protein degradation [9, 11]. COPS6 prevented the Lys-364-linked autoubiquitination and degradation of MDM2, subsequently enhancing p53 degradation in human cancers [12]. COPS6 promoted the Lys-214 and 217-linked autoubiquitination of TRIM21 and contributed to its ubiquitin-mediated degradation in colorectal cancer [13]. COPS6-mediated CHIP selfubiquitination maintained EGFR stability by thwarting ubiquitination and degradation in glioblastoma [14]. The above studies supported the critical roles of CNS6 in the ubiquitination and degradation of proteins, especially autoubiquitination and degradation of several E3 ligases.
In addition, accumulating evidence indicated the important roles of COPS6 in carcinogenesis. And ectopic expression or amplification of COPS6 was found in multiple human cancers, such as glioblastoma [14], colorectal cancer [13], breast cancer [12], and thyroid cancer [12], melanoma [15], hepatocellular carcinoma [16], and pancreatic cancer [17]. COPS6 potentiated the tumorigenesis in xenograft models of A549 cells by the degradation of p53 via inhibiting the autoubiquitination and degradation of MDM2 [12]. Forced expression of COPS6 facilitated the cell proliferation, migration, and invasion of hepatocellular carcinoma cells [16]. COPS6 interacted with E3 ubiquitin ligase UBR5 and decelerated UBR5 protein turnover, which expedited CDK9 ubiquitination and degradation, resulting in melanoma cell proliferation, G1 progression, migration, and invasion [15]. COPS6-involved autoubiquitination of TRIM21 initiated colorectal cancer stemness by elevating ALDH1A1 expression via OCT1 stabilization [13]. These results suggested that COPS6 plays an important role in the carcinogenesis and tumor progression by targeting various E3 ligases. However, the involvement of COPS6 in antitumor immune response remains largely unknown.
As the seventh hallmark of cancer, tumor immune microenvironment (TIME) is mainly composed of infiltrating immune cells, chemokines, and cytokines [18]. Tumor infiltrating lymphocytes (TILs) have an important role in the occurrence and progression of various solid tumors, especially the loss of CD8+ TILs is closely related to immune inactivation and evasion. The immunotherapy are emerging to improve the treatment effect for various cancers [19–21]. Sahar et al. found that the amplification of CD8+ TILs population was positively associated with tumor grades and likely conferred a prolonged survival rate in samples obtained from 1334 breast cancer patients, which may serve as an independent prognostic factor and promising therapeutic target for breast cancer patients [19, 22].
Here, we aimed to explore the potential mechanism of COPS6 in tumor immune evasion. We found that COPS6 was increased in multiple cancers and tumor suppressor gene p53 negatively regulated COPS6 expression. As an oncogene in various cancer [12–17], COPS6 promoted breast cancer progression by regulating tumor microenvironment via controlling CD8+ TILs infiltration. Therefore, our findings provided a novel mechanistic insight into the role of COPS6 in tumor immune evasion of breast cancer.
Materials and methods
Cell culture
Human umbilical vein endothelial cell line HUVEC, human osteosarcoma cell line U2OS, human non-small cell lung cancer cell line H1299, human breast cancer cell line MCF-7, human cervical cancer cell line Hela, mouse mammary cancer cell line EMT6, human cervical cancer cell line HeLa, human prostate cancer cell line PC-3, human kidney cancer cell line 786-O, and human hepatocellular carcinoma cell line HepG2 were obtained from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). MCF-7, Hela, and HepG2 cell lines were cultured in Dulbecco’s Modified Eagle’s medium (DMEM, Gibco, Shanghai, China). HUVEC, U2OS, EMT6, H1299, PC-3, and 786-O cell lines were cultured in RPMI-1640 medium (Gibco, Shanghai, China). Both culture medium were supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, USA) and the cells were grown in a humidified atmosphere containing 5% CO2.
Gene transfection and silencing
siRNAs were designed and synthesized by GenePharma (Shanghai, China). U2OS and MCF-7 cells were transfected with the small interfering RNA (siRNA) specific for COPS6 (5′-CCGUGGAAGAGAAGAUUAUTT-3′), siRNA specific for p53 (5′-GAAGAAAATTTCCGCAAAA-3′), and siRNA specific for IL-6 (5′-CTGGATTCAATGAGGAGACTT-3′) using siLentFect™ Lipid Reagent (Bio-Rad, Hercules, CA, USA) following the manufacturer’s protocol. The plasmids of pcDNA3.1-control, pcDNA3.1-COPS6, pcDNA3.1-p53, pcDNA3.1-ubiquitin were transiently transfected into cells using the X-tremeGENE HP DNA Transfection Reagent (Roche, Indianapolis, IN, USA). After 24 h of transfection, cells were harvested for the following experiments.
ActD treatment
U2OS cells (wild-type p53) were treated with the transcriptional inhibitor ActD at a final concentration of 0, 1, 5, and 10 nM. H1299 (p53-null) and U2OS (wild-type p53) cells were treated with ActD (5 nM). After 24 h, the cells were harvested and the protein levels were analyzed by immunoblotting.
Western blot analysis
Cellular extracts of 1 × 106 cells were homogenized in the RIPA lysis buffer in the presence of proteinase inhibitor cocktail, and the protein levels were measured by bicinchoninic acid protein assay kit. Then, 100 μg proteins were applied onto 10% SDS polyacrylamide gel (SDS-PAGE) for electrophoresis and then transferred onto nitrocellulose membrane. After blocking for 2 h, members were incubated overnight at 4 °C with the specific primary antibodies (COPS6, 1:2000 dilution, Enzo Life Sciences, Switzerland; p53, 1:1000 dilution, Santa Cruz Biotechnology, USA; p21, 1:1000 dilution, Santa Cruz Biotechnology, USA; Akt, 1:1000 dilution, Santa Cruz Biotechnology, USA; p-Akt473, 1:1000 dilution, Santa Cruz Biotechnology, USA; Bax, 1:1000 dilution, Santa Cruz Biotechnology, USA; Bcl-2, 1:1000 dilution, Santa Cruz Biotechnology, USA; pre-caspase3, 1:1000 dilution, Santa Cruz Biotechnology, USA; β-actin, 1:2000 dilution, Zhong-shan Biotechnology, China). After washed, the secondary antibody was added to incubate at room temperature for 2 h prior to ECL (Tanon, Shanghai, China) fluorescence imaging.
Co-immunoprecipitation
Cell lysates containing protease/phosphatase inhibitors cocktail (Sigma, St. Louis, MO, USA.) were incubated on a rocker with indicated antibodies at 4 °C overnight. Then cell lysates were immunoprecipitated by Protein A/G beads (Santa Cruz, CA, USA). Beads were centrifuged at a low speed for 10 min and supernatant was discarded. Dried beads were mixed with 1× loading buffer and boiled for 5 min. Lysate samples were loaded onto gels as performed before.
Immunofluorescence
Cells were fixed in paraformaldehyde for 15 min and then treated with 0.5% Triton X-100 for 20 min. Goat serum (ZSGB-BIO, Beijing, China) was used for blocking for 30 min. The cells were incubated with the anti-COPS6 primary antibody (COPS6, 1:200 dilution, Enzo Life Sciences, Switzerland) overnight at 4 °C and subsequently with the secondary antibody conjugated with CoraLite488 (1:100, Proteintech, catalog: SA00013-2) after being washed thrice in PBST (PBS with 0.1% Triton X-100). Then the nucleus was stained by the DNA dye DAPI (Biosharp, China). Cells were visualized by a fluorescence microscope.
CCK8 assay
MCF-7 cells were transfected with indicated plasmids. After 6 h, cells were seeded into 96-well culture plates (Corning Incorporated, Corning, NY, USA) at a density of 4 × 103 cells. Cell proliferation was evaluated by the colorimetric water-soluble tetrazolium salt (CCK8, Vicmed, China) at time points 1 d, 2 d, 3 d and 4 d according to the manufacturer’s instructions. To test the cell viability, 100 μL serum-free culture medium and 10 μL CCK-8 solution were added into each well, then incubated at 37 °C for 2 h. The absorbance of individual well was measured at 450 nm using a Multiskan Spectrum 1500 (Thermo Labsystems, MA, USA).
EdU incorporation immunofluorescence
EdU incorporation assays were performed with EdU Imaging Kits (APExBIO Biotechnology, Houston, TX, USA) according to the manufacturer’s instructions. MCF-7 cells were transfected with indicated plasmid or siRNA and seeded on coverslips in 24-well plates. After 24 h or 48 h of the indicated treatment, each well was incubated with 200 μL of 50 μM EdU medium for 2 h. Then cells were fixed with 4% paraformaldehyde and treated with Triton X-100. Cells were stained with EdU solution and Hoechst 33342. After washing with PBS, the EdU positive cells were observed by fluorescence microscopy.
Colony formation assay
MCF-7 cells were seeded in six-well plates at a density of 500 cells per well and incubated at 37 °C in 5% CO2. Medium was changed every 3 days over 10 days of foci formation. At the end of the period, the cells were fixed in 4% formalin for 20 min and stained with 1% crystal violet for 10 min. Foci were then counted and photographed.
Transwell invasion and migration assay
To examine the invasion ability of MCF-7 cells, transwell chambers (BD Bioscience, San Jose, CA, USA) containing 8 μm pores were coated with Matrigel in the upper chamber. Cells (2.5 × 104) of serum free medium were plated in the upper chamber, the lower chamber was filled with DMEM medium supplemented with 20% FBS. For migration assay, 2.5 × 104 cells of serum free medium without Matrigel were plated in the upper chamber. After incubating 12 h for migration and 24 h for invasion, cells that migrated or invaded through the membrane were stained with crystal violet, and calculated with light microscope.
Scratch wound assay
The transfected MCF-7 cells were seeded in 6-well plates (Corning Incorporated, NY, USA). Migration was evaluated by scratching a confluent layer of MCF-7 cells using a sterile 200 μL pipette tip. We washed the well with fresh medium to remove cellular debris, and the cells were incubated at 37 °C. Images were obtained at 0, 24, 48, and 72 h.
Apoptosis assay
The Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA) was utilized to analyze cell apoptosis. MCF-7 cells were seeded onto six-well plates and transfected with COPS6 siRNA or plasmid. After 48 h or 24 h, cells were washed twice with ice-cold PBS, then incubated with 200 μL 1× binding buffer containing 5 μL Annexin V-FITC, and 300 μL 1× binding buffer containing 5 μL Propidium Iodide (PI) for 5 min avoiding light at room temperature. After incubation, cells were visualized by a fluorescence microscope.
RNA isolation and real time quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Ambion; Invitrogen, CA, USA) according to the manufacturer’s instructions. And reverse transcription of the total cellular RNA was carried out using a reverse transcripton kit (Takara, Tokyo, Japan). For qRT-PCR analysis, cDNA was amplified using SYBR Green Realtime PCR Master Mix (Takara, Tokyo, Japan). qRT-PCR reactions were performed in triplicates on ABI StepOne Plus (Carlsbad, CA, USA) following the instructions. Primers used in this study are listed as follows: COPS6-forward: 5′-TGTTTCCGTCGCTCTCCATC-3′, COPS6-reverse: 5′-CCTCGATATTTCGGCCCTCC-3′, p53-forward: 5′-ACCTATGGAAACTACTTCCTGAAA-3′, p53-reverse: 5′-CTGGCATTCTGGGAGCTTCA-3′, IL-6-forward: 5′-CCACCGGGAACGAAAGAGAA-3′, IL-6-reverse: 5′-GAGAAGGCAACTGGACCGAA-3′, GAPDH-forward: 5′-GAAGGTGAAGGTCGGAGTC-3′, GAPDH-reverse: 5′-GAAGATGGTGATGGGATTTC-3′. The levels of COPS6, p53 and IL-6 were normalized by GAPDH level. The relative expression was calculated using the relative quantification equation (RQ) = 2−ΔΔCt.
Chromatin immunoprecipitation (ChIP) assay
The EZ-ChIP kit (Upstate, NY, USA) was used according to the manufacturer’s protocol. The cells are treated with ultrasound to disrupt chromatin DNA fragments to 200–500 bp. DNA fragments were co-incubated with anti-p53 or normal rabbit IgG at 4 °C overnight. The immune complex was conjugated with 40 μL Protein A/G beads, and the DNA was purified by a DNA cleanup spin column. The presence of specific DNA sequences in the eluted sample was then measured by PCR.
Promoter cloning and generation of dual-luciferase reporter assay
A − 2000/+200 bp human COPS6 promoter was cloned and inserted in the pGL3 basic vector (Promega, USA). Further, deletion generated −500/+200 bp and −400/+200 bp reporter plasmids. Mutant COPS6 reporter plasmids were generated based on −500/+200 bp plasmid. MCF-7 cells were co-transfected with luciferase reporter plasmids and p53 plasmid. After transfection for 24 h, cells were harvested. The relative activity of firefly luciferase was assessed by the Dual-Luciferase Reporter Assay System (Promega) after 48 h of transfection and normalized to that of Renilla luciferase.
Enzyme-linked immunosorbent assay (ELISA)
MCF-7 cells were seeded in 24-well culture plates and transfected with COPS6 plasmid or siRNA. After 24 h or 48 h transfection, supernatants were harvested and measured at 450 nm using a microplate reader and Human IL-6 ELISA Kit (Elabscience Biotechnology Co., Ltd) according to manufacturer’s instructions. Ctrl, COPS6KD and COPS6KD + IL-6KD breast cancer cells were injected to C57BL/6 J to establish transplanted tumor models. After 15 days, the tumors were collected and grinded for ELISA analysis. Then, the supernatants were harvested and measured by mouse Granzyme B, Perforin1, TNF-α, IFN-γ ELISA Kit (Elabscience Biotechnology Co., Ltd) according to manufacturer’s instructions.
Evaluation of immunostaining
Tissue specimens consisted of 30 breast cancer tissues were collected from the Affiliated Hospital of Xuzhou Medical University. Patient tissue samples were obtained with informed consent, under the protocol approved by the Ethical Review Board of the Affiliated Hospital of Xuzhou Medical University. The evaluation of COPS6, p53, IL-6 and CD8a staining was blindly and independently examined by two pathologists and a consensus was reached for each core. The percentage of positive stained cells was scored into 4 categories: 1 (0–25%), 2 (26%–50%), 3 (51%–75%), and 4 (76%–100%). Staining intensity was scored 0 to 3 (0 = negative; 1 = weak; 2 = moderate; 3 = strong). The level of staining was evaluated by immunoreactive score (IRS), which was calculated by multiplying the scores of the percentage of positive cells and staining intensity. Based on the IRS, the staining pattern was defined as low (IRS: 0–6) and high (IRS: 8–12).
Tumorigenicity in vivo
The BALB/c mice (6 weeks, SPF level, 20–22 g) and C57BL/6 mice (6 weeks, 20–22 g) were purchased from Nanjing Jicui Yaokang Bioscience (Nanjing, China) and maintained in specific, pathogenfree facilities. The stable cell lines were generated by control shRNA, COPS6-shRNA (5′-GCGAAATATCGAAGTGATGAA-3′) and IL-6-shRNA (5′-GACATGTAACAAGAGTAACAT-3′) lentiviral particles. After 48 h of infection, cells stably expressing the relevant construct were selected with 2 μg/mL puromycin (Vicmed, Xuzhou, China) for 15 days. Stable EMT6 cells transfected with control shRNA, COPS6-shRNA, COPS6-shRNA+IL-6-shRNA were concentrated to 2 × 106/100 μL PBS and subcutaneously injected into the flanks of C57BL/6 mice (n = 5). Stable EMT6 cells transfected with control shRNA or COPS6-shRNA were subcutaneously injected into the flanks of BALB/c nude mice (n = 5). Tumor volumes were measured using a digital vernier caliper at specified times (5, 10, 15 days). The tumor volume was calculated as the formula length × width2 × 0.5. After 15 days, the tumors were removed for data analysis. The animal experiments involved in this study were approved by the Experimental Animal Committee of Xuzhou Medical University and the approval number is “20200048”.
Isolation and culture of CD8+ T cells from peripheral blood
Blood was obtained from healthy volunteers and written informed consent was obtained from all volunteers. Human peripheral blood mononuclear cells (hPBMCs) were separated by the Ficoll buffer (Yuanye Bio-Technology Co, Shanghai, China) via density centrifugation according to the manufacture’s instruction. After that, the CD8+ T cells were isolated from the hPBMCs by using a flow cytometer (BD, Franklin Lakes, NJ, USA) based on the protocol. CD8+ T cells were cultured in RPMI-1640 medium (Gibco, Shanghai, China) supplemented with 2 mM glutamine (Corning, Manassas, VA, USA), 10% heat-inactivated fetal bovine serum (Gibco, Gaithersburg, MD, USA), 100 U/mL penicillin and 100 μg/mL streptomycin, at 37 °C in 5% CO2.
T cell migration assay
These isolated CD8+ T cells were stained with anti-human CD8α APC-Cy7 antibody (0.5 μg/1 × 106 cells) for 15 min at room temperature. The stained CD8+ T cells were adjusted to 5 × 106/mL. The CD8+ T cells (100 μL) were added into the Transwell insert (5.0 μm pore size) to allow the access to the lower compartment containing the supernatant culture medium from tumor cells under different treatment conditions. MCF-7 cells were cultured in DMEM medium and transfected with COPS6 plasmid for 24 h or COPS6 siRNA for 48 h, then the supernatant was transferred to the lower compartment for the migration assay. After 24 h co-incubation, the insert was carefully removed. The migrated CD8+ T cells were accumulated from the lower compartment and counted by flow cytometry with counting beads.
T cell proliferation assay
The isolated CD8+ T cells (5 × 104 cells) were plated into the lower compartment of Transwell chambers. The supernatant culture medium from MCF-7 cells transfected with COPS6 plasmid for 24 h or COPS6 siRNA for 48 h were plated in the upper chamber. After incubation for 24 h, cell proliferation was evaluated by CCK8 kit (CCK8, Vicmed, China) according to the manufacturer’s protocol. Briefly, the cells were incubated with the CCK-8 reaction solution for 2 h at 37 °C. The absorbance of individual wells was measured at the wavelength of 450 nm using a Multiskan Spectrum 1500 (Thermo Labsystems, USA).
Tumor T cells infiltrating and function detection
To detect tumor-infltrating lymphocytes, the tumor tissue was digested in a specific medium consisting of 100 mL of DMEM supplemented with 2 mL of FBS, 0.1 g of collagenase I (C8140, Solarbio, China), 50 μL of hyaluronidase (H8030, Solarbio, China), and DNase I (D8071, Solarbio, China) for 30 min at 37 °C. Lymphocytes were isolated by Ficoll gradient centrifugation at 800 × g for 25 min. For the detection of CD8+ T cells, cells were stained with respective antibodies (CD8a, Thermo Fisher, 46-0081-82; CD45, Thermo Fisher, 11-0451-82) for 30 min at 4 °C. Tumor tissue-infiltrating leukocytes were stained with fluorochrome-conjugated antibodies against CD3 (Thermo Fisher, 47-0031-82), CD8a, PD1 (Thermo Fisher, 11-9985-82) or TIM3 (Thermo Fisher, MA5-17957) to identify the exhausted T cells. All stained cells were measured using FACS Calibur (BD Biosciences, USA). The data were analyzed using the FlowJo software (Treestar Inc., Ashland, OR, USA).
Cancer single-cell expression map
To map the cell type-specific COPS6 expression landscape of cancer cells and tumor microenvironment in breast cancer, we utilized a public database cancer single-cell expression map (https://ngdc.cncb.ac.cn/cancerscem/index) which dedicates to collecting, analyzing, visualizing single-cell RNA-Seq data of human cancers. Multi-level analyses were performed to deeply explore the tumor microenvironment of different types of human cancers and a comprehensive online analysis platform was equipped in the database.
TNMplot: differential gene expression analysis
The pan-cancer analysis page displays the expression range for COPS6 and IL-6 gene across all tissues in all available normal and tumor RNA Seq data (RNA-seq from GTEx: 11,215 normal, RNA-seq from TCGA: 730 normal, 9886 tumor and 394 metastasis, RNA-seq from TARGET: 12 normal, 1180 tumor and 1 metastasis). The Gene signature analysis page calculated the means of COPS6 and IL-6 gene expression in breast invasive carcinoma across each patient one by one and provided a summary plot using RNA-Seq based data. Spearman correlation method was used to compare the correlation of IL-6 and CD8A, CD8B, GZMA, GZMB, PRF1 in breast invasive carcinoma based on RNA-Seq data.
Linkedomics database
LinkedOmics (http://www.linkedomics.org/login.php) is a publicly available database that includes multi-omics data from all 32 TCGA Cancer types and 10 Clinical Proteomics Tumor Analysis Consortium (CPTAC) cancer cohorts. The gene set enrichment analysis (GSEA) and GO analysis (biological process) of COPS6, IL-6 were generated from LinkedOmics.
Human Protein Atlas (HPA) database analysis
The HPA database (https://www.proteinatlas.org/) contains expression data for 17,058 proteins in cell lines, normal human tissues and tumor tissues. We used the HPA database to evaluate immunohistochemical staining of COPS6 in breast cancer. A specificity classification is used to predict the enrichment of COPS6 gene in each constituent cell type within breast tissue. The Subcellular section of the HPA provides high-resolution insights into the expression and spatiotemporal distribution of proteins encoded by 13,041 genes (65% of the human protein-coding genes). For COPS6 and IL-6, the subcellular distribution of the proteins has been investigated by immunofluorescence and confocal microscopy in U2OS and MCF-7 cell lines.
Survival analysis
Kaplan–Meier plotter (https://kmplot.com/analysis/) was used to assess the correlation between expression of genes (more than 30,000 samples from 21 tumor types) and survival rates. Sources of the database include GEO, EGA, and TCGA. Survival curves were calculated using the Kaplan–Meier method to plot survival curves, and differences between groups were compared using the log-rank test. P-values less than 0.05 were considered as a significant difference.
Analysis of immune cell infiltration
The correlation between COPS6 and immune infiltrates in breast invasive carcinoma was analyzed using a web server Tumor IMmune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/). TIMER web server is a comprehensive resource for systematical analysis of immune infiltrates across diverse cancer types. The abundances of six immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, Neutrophils, Macrophages, and Dendritic cells) are estimated by TIMER algorithm. TIMER was also utilized to explore correlations between COPS6 and CD8A, CD8B, GZMA, GZMB, IL-6, PRF1. The bioinformatics website TISIDB (http://cis.hku.hk/TISIDB/) is a web portal for tumor and immune system interaction, which integrates multiple heterogeneous data types. TISIDB was utilized to analyze the relationship between COPS6, IL-6, and lymphocytes.
Transcript expression and survival analysis using the GEPIA web tool
The online database Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) was used to analyze the RNA sequencing expression data related to our project based on The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) project. In this study, the tumor and normal tissue datasets were used to identify the expression pattern of COPS6. GEPIA generates box plots with jitter for comparing COPS6 expression in various cancer types. GEPIA2021 (http://gepia2021.cancer-pku.cn/) is a standalone extension with multiple deconvolution-based analysis for GEPIA. They deconvolute each sample tool in TCGA/GTEx with the bioinformatics tools CIBERSORT, EPIC, and quanTIseq. Survival analysis was performed to separate the samples into two groups according to the proportion of CD8+ T cell type. Then we plot the Kaplan–Meier curve of each group with the survival data. The statistical difference between the curves can be measured by the log-rank test.
Statistical analysis
Statistical analyses were performed using Statistical Package for Social Sciences Version 21.0 (SPSS 21.0) and Graphpad Prism 7.0. All values are expressed as means ± SD. Statistical significance of Student’s t-test was presented for two-group comparisons. The analysis of variance (ANOVA) test was used to compare mean values among three or more groups. Kaplan–Meier survival analyses were used to estimate the overall survival and recurrence free (RFS) survival, and the log-rank test was used to assess the differences. The correlation analysis was determined by Pearson’s test or Spearman’s test. A P < 0.05 was considered statistically significant.
Results
COPS6 was highly expressed in various cancers
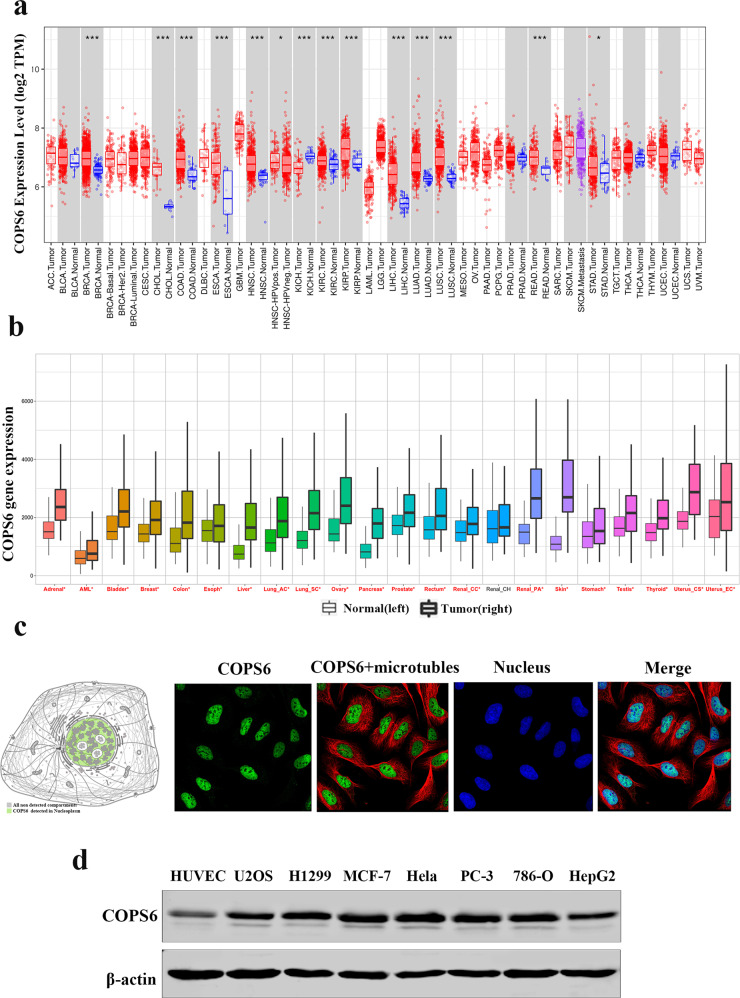
To determine the expression pattern of COPS6 in tumor and normal tissues, we evaluated COPS6 mRNA levels across different tumor types by analyzing the TCGA and GTEx cohort. The results demonstrated that COPS6 expression was significantly higher in breast carcinoma (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head-neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), rectal adenocarcinoma (READ), and stomach adenocarcinoma (STAD) (Fig. 1a, b, Supplementary Fig. S1a, b). Subcellular section of the HPA database showed that COPS6 expression was localized in the nucleus (Fig. 1c). Western blot analysis was performed to examine the COPS6 expression in normal human umbilical vein endothelial cell line HUVEC and seven human cancer cell lines. Results showed that the expression of COPS6 in cancer cell lines was higher than that in normal human cells (Fig. 1d). These data indicated that COPS6 was highly expressed in various cancers.
Fig. 1. The expression level of COPS6 in various types of tumor and normal tissues.
a The expression level of COPS6 in different types of tumor tissues and normal tissues in TIMER database. b Pan-cancer analysis of COPS6 expression; data were derived from the TCGA and GTEx cohort. c Localization of COPS6 expression obtained from HPA database. d Western blot analysis was used to evaluate the COPS6 expression in various cancer cell lines and normal human umbilical vein endothelial cell line HUVEC.
p53 downregulated COPS6 expression at the transcriptional level
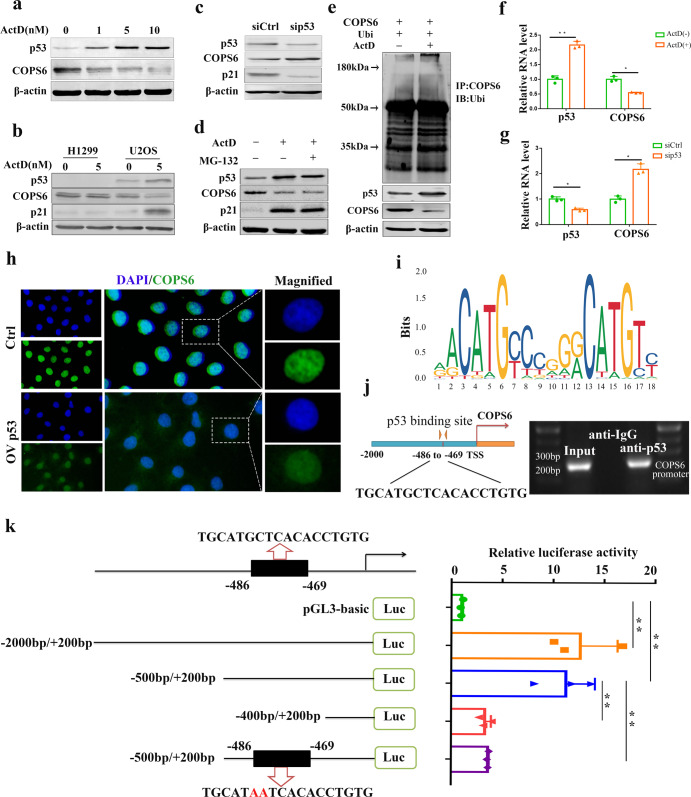
To investigate whether COPS6 protein expression was regulated by p53, we first treated U2OS (wild-type p53) cells with the transcriptional inhibitor ActD (0, 1, 5, 10 nM) for 24 h and then performed Western blot assay. Results showed that p53 expression was increased and COPS6 expression was decreased in a dose-dependent manner (Fig. 2a). Then H1299 (p53-null) and U2OS (wild-type p53) cells were treated with ActD (5 nM) for 24 h. As shown in Fig. 2b, COPS6 expression was higher in p53-negative-expressing H1299 cells than in p53-positive-expressing U2OS cells. Meanwhile, ActD had no effect on COPS6 expression in H1299 (p53-null) cells. In addition, knockdown of p53 increased the protein level of COPS6 (Fig. 2c). The decrease in protein level of COPS6 was not reversed by the proteasome inhibitor MG-132, and ActD was insufficient to modulate its expression administered by MG-132 (Fig. 2d). Furthermore, there was no significant alternation of the ubiquitination of COPS6 following ActD exposure (Fig. 2e). These data indicated that p53 was ineffectual in COPS6 protein turnover. Results from qPCR showed that cells with ActD treatment resulted in an increase of p53 mRNA concomitant with a decrease of COPS6 mRNA (Fig. 2f, g). The expression of COPS6, which was detected by immunofuorescence got the same results (Fig. 2h). p53 has been well-characterized as a transcription factor involved in tumor suppression [23]. Therefore, we conjectured that p53 might regulate COPS6 expression at the level of transcription. Potential binding “motif” sequence located in the COPS6 promoter was predicted by JASPAR and a potential p53 response element located at −486/−469 nt (TGCATGCTCACACCTGTG) of promoter region of COPS6 was also identified (Fig. 2i, j). ChIP-PCR analysis showed that p53 occupied the promoter region of COPS6 (Fig. 2j, right panel). To determine the minimum sequence required for activity, a series of five reporters constructs with progressively larger deletions from the 5′ end of the promoter were generated. Luciferase assay revealed an increased promoter activity of the pGL3–2000/+200 bp as compared to the control group in MCF-7 cells. However, when the promoter sequence was deleted to position –400 bp, the promoter activity with pGL3–400/+200 bp decreased significantly compared with the pGL3 − 500/+200, this result demonstrated that positive regulatory elements were located in the −500/−400 promoter region of COPS6 in MCF-7 cells. In addition, further mutation of this binding site effectively blocked the transcription of COPS6 (Fig. 2k). Collectively, these results suggested the existence of binding site for p53 in the promoter region of COPS6 gene.
Fig. 2. p53 negatively regulates COPS6 promoter activity.
a ActD induced an elevation in p53 level and a decrease in COPS6 level in a dose-dependent manner. b p53, COPS6 and p21 protein expression in H1299 and U2OS cells after treatment with ActD. c Knockdown of p53 upregulated the expression of COPS6. d U2OS cells were treated with ActD (5 nM) or proteasome inhibitor MG132 (50 μg/mL). Cell lysates were immunoblotted with the indicated antibodies. e U2OS cells were transfected with control COPS6, Ubi and treated with ActD 5 nM or not. The cell lysates were immunoprecipitated with anti-COPS6 and immunoblotted with anti-ubiquitin antibody. Equal amounts of the cell lysates were analyzed by immunoblotting with the indicated antibodies. f ActD induced p53 up-regulation and COPS6 down-regulation in mRNA level. g Knockdown of p53 upregulated the mRNA expression of COPS6. h The expression of COPS6 was detected by immunofuorescence. i DNA motif sequence logo of p53 was generated by JASPAR. j A schematic of the potential p53 binding sites on the promoter of COPS6. Identification of the p53 binding sequences in COPS6 promoters by ChIP-PCR. k Dual-luciferase reporter assay of COPS6 and p53. Data in f, g and k are expressed as means ± standard errors of the means of three independent experiments, *P < 0.05, **P < 0.01.
COPS6 correlates with an immunosuppressive tumor microenvironment in breast cancer
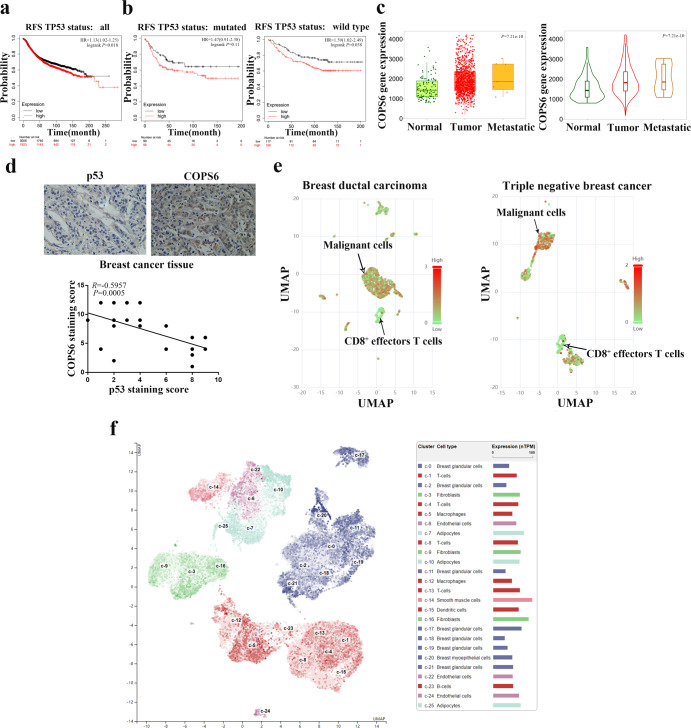
Survival analysis derived from Kaplan–Meier Plotter database revealed that higher COPS6 expression predicted poorer RFS survival in breast cancer patients with p53 wild type or mutated tumors and in all randomized patients (Fig. 3a, b). Using the online database, we compared the COPS6 expression in breast cancer and normal tissues. Through TNMplot analysis, we found that COPS6 was overexpressed in breast cancer tissues compared with normal tissues while further elevated in metastatic breast cancer (Fig. 3c). And immunohistochemical staining also showed that the expression of COPS6 was negatively related with p53 expression in breast cancer (Fig. 3d).
Fig. 3. Highly expressed COPS6 predicts a poor prognosis in breast cancer and correlates with an immunosuppressive tumor microenvironment.
a, b The prognostic value of COPS6 in breast cancer patients with p53 wild type or mutated tumors and in all randomized patients; the RFS survival was analyzed in Kaplan–Meier Plotter Database. c TNMplot database data concerning the expression pattern of COPS6 in breast cancer. d Correlation between COPS6 and p53 expression in breast tumor tissues. e Single-cell RNA-Seq data of breast dutal carcinoma in situ and triple negative breast cancer indicated the expression of COPS6 in malignant cells and CD8+ T cells. f A specificity classification was used to predict the enrichment of COPS6 gene in each constituent cell type of breast tissue.
Tumor microenvironment has been well-established in breast cancer aggressiveness and radiochemotherapy resistance [24, 25]. CD8+ TILs elicits significantly anti-tumor responses, and the abundance of CD8+ TILs signifies a better clinical outcome of breast cancer patients [19, 22]. To investigate whether COPS6 correlates with tumor microenvironment in breast cancer, the expression pattern of COPS6 in TILs was explored by excavating the online database Cancer single-cell expression map. It was demonstrated that COPS6 expression increased in breast cancer cells while decreased in CD8+ T cells by visualizing single-cell RNAseq data via UMAP (Fig. 3e). Moreover, a visualization of cluster cell type in HPA database indicated that COPS6 expression was relatively higher in T cells when compared to other non-tumor cell cluster in breast tissues (Fig. 3f, Supplementary Fig. S2a). These results demonstrated that tumor-promoting effects of COPS6 might be dependent on impairing CD8+ TILs.
COPS6 promotes breast cancer progression
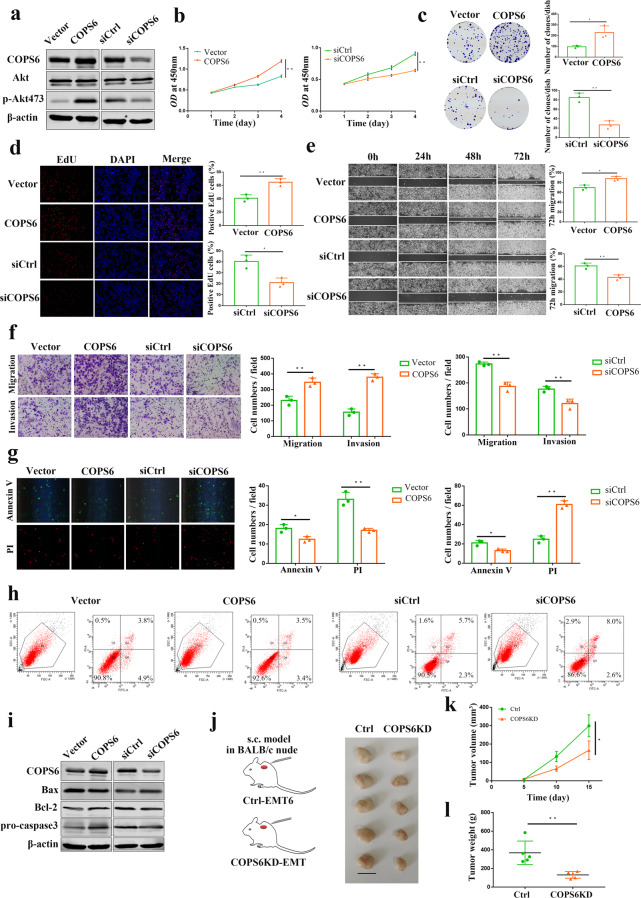
Next, we investigated the effects of COPS6 on invasive and proliferative phenotypes in breast cancer cells. Enforced expression of COPS6 stimulated p-AKT expression in breast cancer cells, knockdown of COPS6 had opposite effect (Fig. 4a), which suggested that COPS6 may activate the AKT pathway closely related to the proliferation and malignant transformation of tumor cells. CCK-8 assay was performed to test the cell proliferation. Results showed that COPS6 promoted the proliferation of breast cancer cells, COPS6-silencing reinforced cell motility (Fig. 4b). Overexpression of COPS6 potentiated the colony formation efficiency of breast cancer cells, reduction of COPS6 repressed colony formation (Fig. 4c). Breast cancer cells expressing COPS6 elevated the EdU-positive breast cancer cells, suppressing COPS6 decreased the EdU-positive cells (Fig. 4d). Wound healing and transwell assays were conducted to assess the migration and invasion ability of breast cancer cells. In vitro wound healing was detected in breast cancer cells expressing COPS6, and its loss blunted wound healing (Fig. 4e). Augment of the expression of COPS6 enhanced the migration and invasion of breast cancer cells, blockade of COPS6 retarded the migration and invasion (Fig. 4f). Furthermore, transfection of breast cancer cells with COPS6 vector impeded decreased apoptosis, silenced COPS6 expression induced apoptosis, as determined by apoptosis assay via a fluorescence microscope and flow cytometry (Fig. 4g, h). Consistent with the results from Western blot, increased Bcl-2 and pro-caspase-3 expressions were accompanied by a decrease in Bax in breast cancer cells after overexpressing COPS6, opposite patterns were observed after treatment with siCOPS6 (Fig. 4i). It is widely recognized apoptosis-regulating genes serve as a critical role in tumor growth, including the induction of pro-apoptotic Bax and anti-apoptotic Bcl-2 and pro-caspase-3 [26]. To further evaluate the oncogenic effects of COPS6 in vivo, subcutaneous inoculated tumors were established through COPS6-depleted EMT6 cells. Tumor volume increased slowly and tumor weight decreased obviously in tumors injected with anti-COPS6 expressing EMT6 cells (Fig. 4j, k). Removal of COPS6 inhibited malignant breast tumor growth in mouse transplanted tumor models (Fig. 4l). Collectively, COPS6, as oncogenic drivers, promoted breast cancer progression.
Fig. 4. COPS6 promoted breast cancer progression.
a Western blot analysis showing the expression of Akt and p-Akt473 in MCF-7 cells after COPS6 overexpression or siCOPS6 treatment. b Growth curves of MCF-7 cells treated with COPS6 overexpression or siCOPS6. c Colony formation assays in MCF-7 cells treated with COPS6 overexpression or siCOPS6. d EdU assays in MCF-7 cells treated with COPS6 overexpression or siCOPS6. e Wound healing assay in MCF-7 cells treated with COPS6 overexpression or siCOPS6. Images were captured 0, 24, 48, 72 h after wound was created. The percentage of migration was assigned as 100% when complete fusion occurred, and 0% at t = 0 h. f Transwell migration and invasion assay of MCF-7 cells treated with COPS6 overexpression or siCOPS6. g Prophase apoptotic cells were recognized by binding with FITC on the membrane (cell membrane displays green); anaphase apoptotic cells were recognized by binding with PI on the nuclei (nuclei displays red). h The cell apoptosis was detected by flow cytometry. i Western blot analysis of the expression of Bax, Bcl-2 and pro-caspase3 in MCF-7 cells after COPS6 overexpression or siCOPS6 treatment. j Tumors extracted from mice models. Scale bar, 1 cm (n = 5 mice per group). k, l Tumor growth curves and weight of the tumors for indicated groups (n = 5 mice per group). Data are expressed as means ± standard errors of the means of three independent experiments, *P < 0.05, **P < 0.01.
COPS6 inhibits CD8+ T cell infiltration and IL-6 production in breast cancer
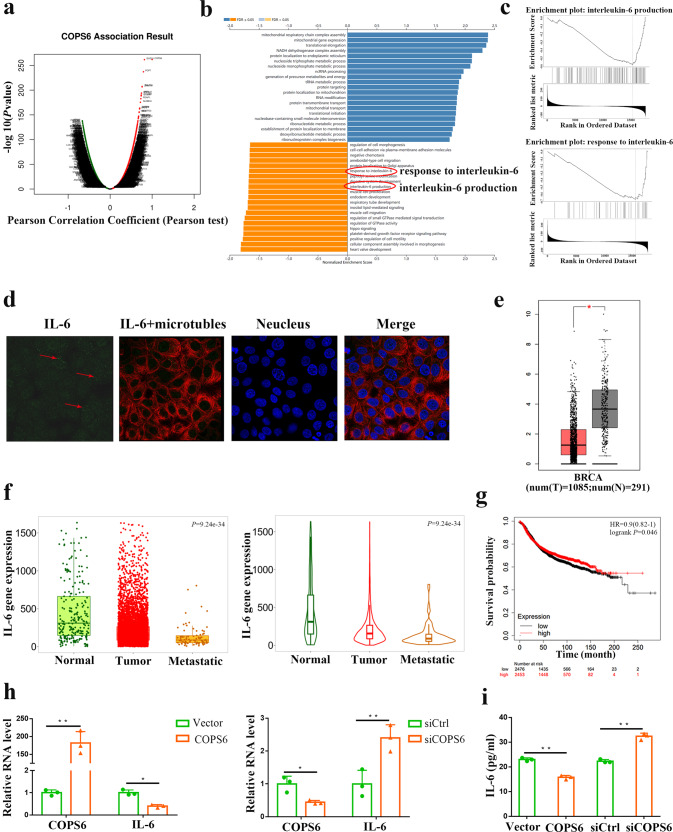
Subsequently, bioinformatics analysis was employed to address the detailed mechanism of COPS6 driving breast cancer progression. A volcanic map and differential genes heatmaps were generated to visualize the differential expression genes in breast cancer after COPS6 amplification by the online database of Linkedomics (Fig. 5a, Supplementary Fig. S3a, b). The possible biological progression was assessed by GO biological process analysis according to the COPS6-involved differential genes (Fig. 5b). It is noteworthy that COPS6 was negatively correlated the response to (interleukin 6, IL-6) and IL-6 production pathways (Fig. 5b). More specifically, gene-set enrichment analysis (GSEA) indicated the significant enrichments of response to IL-6 and IL-6 production pathways, which are involved in the amplification of COPS6 in breast cancer (Fig. 5c). IL-6 characterized as the multi-functional, multi-effect cytokine and the core inflammatory factor in the tumor microenvironment plays an important role in the tumor progression by mainly activating JAK/STAT3, Ras/MAPK, PI3K-PKB/Akt signaling pathways [27–29]. Increased serum IL-6 levels and ectopic expression of IL-6 receptor are closely related to the occurrence and development of various human tumors [29, 30]. Subcellular localization of IL-6 protein in both the cytoplasm and nucleus was visualized by immunofluorescence staining (Fig. 5d).
Fig. 5. COPS6 inhibits the IL-6 production in breast cancer.
a A volcanic map indicated the differential expression genes of COPS6 in breast cancer. b, c GO biological process analysis and GESA of COPS6-involved differential genes. d Subcellular localization of IL-6 protein obtained from HPA database. e Expression pattern of IL-6 in breast cancer tissues and corresponding normal tissues. f TNMplot database data concerning the expression pattern of IL-6 in breast cancer. g The prognostic value of IL-6 in breast cancer; the overall survival were analyzed in Kaplan–Meier Plotter Database (https://kmplot.com/analysis/). h The mRNA expression of IL-6 after overexpression or knockdown of COPS6. i The release of IL-6 in different cultured groups measured by ELISA. *P < 0.05, **P < 0.01.
Following analyzing the clinical significance of IL-6 expression through the GEPIA2 web server, it showed that IL-6 was significantly increased in breast cancer tissues, compared with normal tissues (Fig. 5e). Accordingly, IL-6 was increased in breast cancer tissues compared with normal tissues while further elevated in metastasis breast cancer tissues through TNMplot analysis (Fig. 5f). Increased IL-6 led to dismal prognosis for breast cancer patients GC patients by searching the Kaplan–Meier plotter database (Fig. 5g). Furthermore, qPCR showed that COPS6 significantly decreased IL-6 mRNA, and COPS6 knockdown induced its expression in breast cancer cells (Fig. 5h). IL-6 has been considered as a key cytokine in the link between inflammation and cancer, and has received a lot of attention as the indicators of progression and prognosis of multiple human cancers [29–31]. IL-6 production collected from culture supernatants following COPS6 upregulation and downregulation in breast cancer cells was quantitatively determined by ELISA assays. Overexpression of COPS6 impaired secretion of IL-6, while COPS6 depletion induced secretion (Fig. 5i). In general, these data support a major role of IL-6 in COPS6-induced breast cancer progression.
IL-6 positively regulates CD8+ T cells in breast cancer
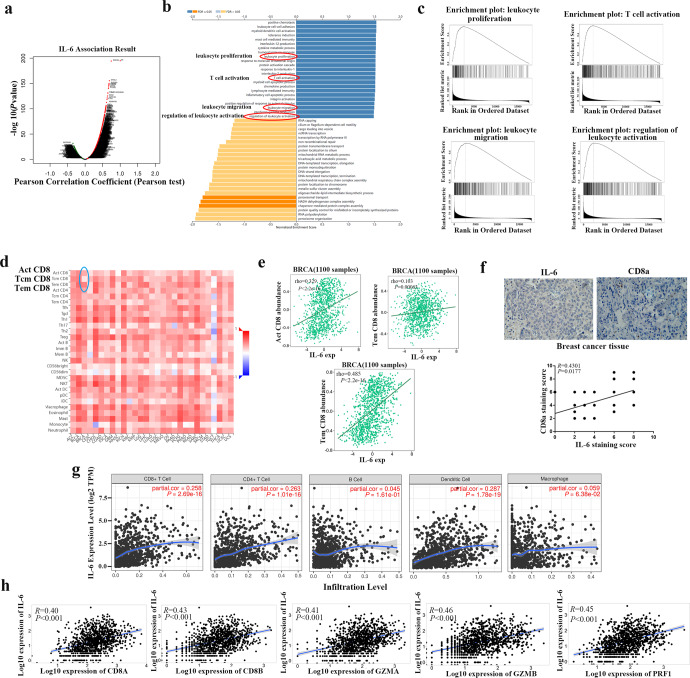
The distribution of differential genes following IL-6 amplification was visualized and shown in Fig. 6a and Supplementary Fig. S4a. By analyzing TCGA cohort, we demonstrated that IL-6 expression was significantly lower in various tumors (Supplementary Fig. S4b, c). GO biological process analysis indicated that IL-6 was involved in immune regulation, including leukocyte proliferation, T cell activation, leukocyte migration, and regulation of leukocyte activation (Fig. 6b). Subsequent GSEA analysis also supported enrichments of gene sets associated with leukocyte proliferation, T cell activation, leukocyte migration, and regulation of leukocyte activation in response to IL-6 (Fig. 6c). Considering the negative regulation of CD8+ TILs and IL-6 secretion by COPS6, we next assessed the mechanisms whereby IL-6 affects the immune cell infiltration in breast cancer. The correlations of immune cell infiltrations and IL-6 were systematically screened in various cancers through TISIDB database, which revealed positive correlations between IL-6 expression and CD8 T-cell subsets (Act, Tcm, and Tem CD8) in BRCA (Fig. 6d). And positive correlations between IL-6 expression and Act/Tem CD8 T-cell subsets in breast cancer were also predicted from TISIDB (Fig. 6e). Furthermore, immunohistochemical staining also showed that the expression of IL-6 was positively related with CD8a in breast cancer (Fig. 6f). The relationships between IL-6 expression and immune infiltration abundance in tumor microenvironment were analyzed in breast cancer based on TIMER database, which revealed that IL-6 showed significant correlation with infiltrating levels of CD4+ T cells, CD8+ T cells, B cells, macrophages, and dendritic cells (Fig. 6g). TNMplot database was also enrolled to visualize the positive correlations between IL-6 expression and highly connected CD8+ T genes, including CD8A/B, GZMA/B, and PRF1 in breast cancer (Fig. 6h). These data highlighted that IL-6 promoted breast cancer progression by reprogramming tumor microenvironment via inducing immune cell infiltration.
Fig. 6. IL-6 positively regulates CD8+ T cells in breast cancer.
a A volcanic map indicated the differential expression genes of IL-6 in breast cancer. b, c GO biological process analysis and GESA of IL-6-involved differential genes. d, e Bioinformatics analysis of the correlation between IL-6 and immune effector cells using TISIDB database. f Correlation between IL-6 and CD8a expression in breast tumor tissues. g Correlation between IL-6 expression and CD8+ T cell, CD4+ T cell, B cell, dentritic cell, and macrophage infiltration in breast cancer. h Correlation between IL-6 expression and highly connected CD8+ T genes, including CD8A/B, GZMA/B, and PRF1 in breast cancer.
COPS6 inhibits CD8+ T cell infiltration via regulating IL-6 production in breast cancer
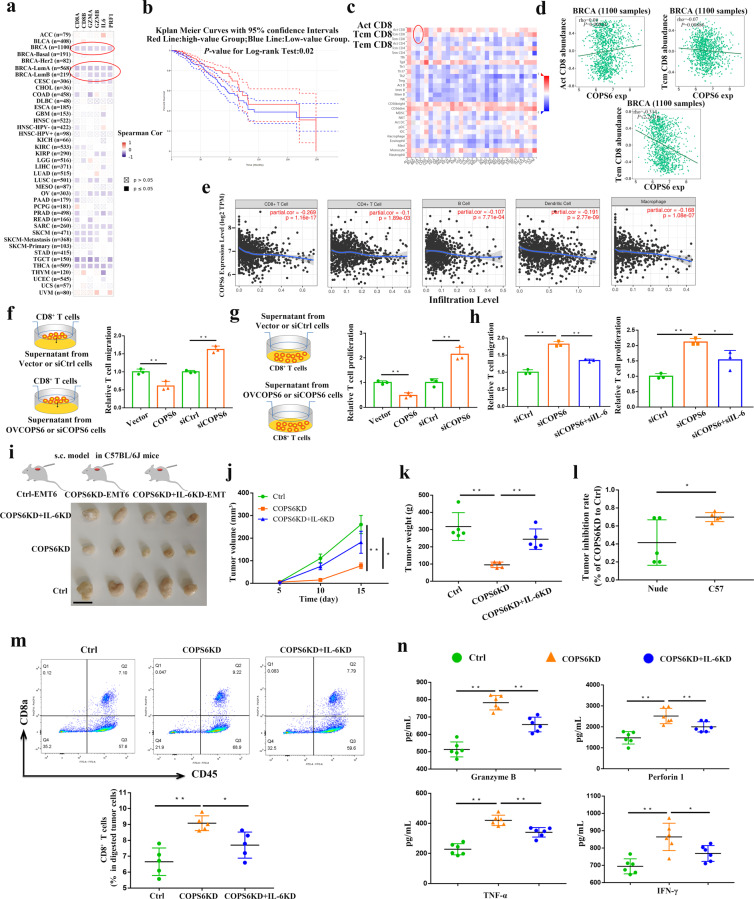
TIMER web server was applied to assess the effects of COPS6 on the functional indicators of CD8+ T cells in cancers. The inhibitory effects of COPS6 on the functional indicators CD8A/B, GZMA/B, PRF1 and IL-6 expression were obviously observed in breast cancer patients including LumB or LumB subtypes (Fig. 7a). Breast cancer patients with abundant CD8+ T cells predicted improved overall survival by GEPIA web tool (Fig. 7b). The correlations of immune cell infiltrations and COPS6 were also systematically screened in various cancers by TISIDB (Fig. 7c), which displayed a negative correlation between COPS6 and Tem CD8 T-cell subset rather than Act and Tcm subsets (Fig. 7c). Negative correlation between COPS6 and Tem CD8 T-cell subset rather than Act and Tcm subsets was also observed by TISIDB (Fig. 7d). TIMER database analysis displayed that COPS6 showed a negative correlation with infiltrating levels of CD8+ T cells in breast cancer patients (Fig. 7e), whereas there was no compelling association of COPS6 with CD4+ T cells, B cells, macrophages, and dendritic cells (Fig. 7e). Noteworthy, IL-6 positively regulated the infiltration of CD8+ T cells and tumor microenvironment as described above (Fig. 6d, e), so we could conclude that COPS6 impaired CD8+ TILs and induced tumor immune evasion by regulating IL-6 production. Finally, we investigated the potential of COPS6 gain/loss-of-function on the migration and proliferation of CD8+ T cells by the supernatant of breast cancer cells with COPS6 upregulation or downregulation. Transwell assays showed that COPS6 expression inhibited the migration of CD8+ T cells, and silenced COPS6 expression contributes to cell migration (Fig. 7f). COPS6 also suppressed the proliferation of CD8+ T cells, COPS6 deficiency induced the proliferation CD8+ T cells (Fig. 7g). However, knockdown of IL-6 in COPS6KD cells could rescue the enhancement of T cell migration and proliferation (Fig. 7h). To further identify the role of COPS6 in tumor immune regulation, mouse-derived Ctrl, COPS6KD and COPS6KD + IL-6KD breast cancer cells have been injected to C57 BL/6 J to establish transplanted tumor models. Results showed that knockdown of IL-6 increased COPS6KD-EMT6 tumor growth in C57BL/6 J mice (Fig. 7i–k). Knockdown of COPS6 in EMT6 cells resulted in a tumor inhibition rate of about 70% when the cells were inoculated in C57BL/6 J mice. However, depletion of COPS6 in EMT6 cells only caused about 41% tumor inhibition rate in BALB/c-nude mice (Fig. 7l), indicating a potential role of COPS6 in the regulation of antitumor immune responses. To analyze the number of immune cells in tumor tissues, we performed flow cytometry and found that COPS6 knockdown in the EMT6 cells increased the number of tumor-infiltrating CD8+ T cells, while knockdown of IL-6 in COPS6KD EMT6 cells decreased tumor infiltrating CD8+ T cells in C57BL6 mice (Fig. 7m). In addition, PD1+ and TIM3+ CD8+ T cells in COPS6KD tumors had significantly lower amount than Ctrl tumors, while knockdown of IL-6 rescued COPS6KD induced lower amount of PD1+ and TIM3+ CD8+ T cells in C57BL6 mice (Supplementary Fig. S5a, b). Furthermore, COPS6KD tumors had significantly higher amounts of TNF-α, IFN-γ, Granzyme B and perforin1 than Ctrl tumors, while knockdown of IL-6 rescued COPS6KD induced high soluble factors in C57BL6 mice (Fig. 7n). Thus, COPS6 promoted breast cancer progression by reducing CD8+ T cell infiltration and function via the regulation of IL-6 secretion (Fig. 8).
Fig. 7. COPS6 inhibits CD8+ T cell infiltration via IL-6 production in breast cancer.
a Correlations between COPS6 expression and CD8A/B, GZMA/B, PRF1, IL-6 gene expression in pan-cancer. b Association of CD8+ T cell infiltration level with survival of breast cancer patients. c, d Bioinformatics analysis of the correlation between COPS6 expression and immune effector cells using TISIDB database. e Correlation between COPS6 expression and CD8+ T cell, CD4+ T cell, B cell, dentritic cell, and macrophage infiltration in breast cancer. f–h Relative migration or proliferation of human CD8+ T cells co-incubated with culture medium supernatant from indicated treatment MCF-7 cells. i Tumors extracted from mice models. Scale bar, 1 cm (n = 5 mice per group). j, k Tumor growth curves and weight of the tumors for indicated groups (n = 5 mice per group). l Tumor inhibition rate for COPS6KD EMT6 cells relative to control cells after inoculation into BALB/c-nude mice or C57BL/6 J, respectively. m The effects of knocking down COPS6 and IL-6 in EMT6 tumors on CD8+ T cell infiltration were analyzed by flow cytometry. n The secretion of Granzyme B, Perforin1, TNF-α and IFN-γ in mouse tumors was analyzed by ELISA Kit. *P < 0.05, **P < 0.01.
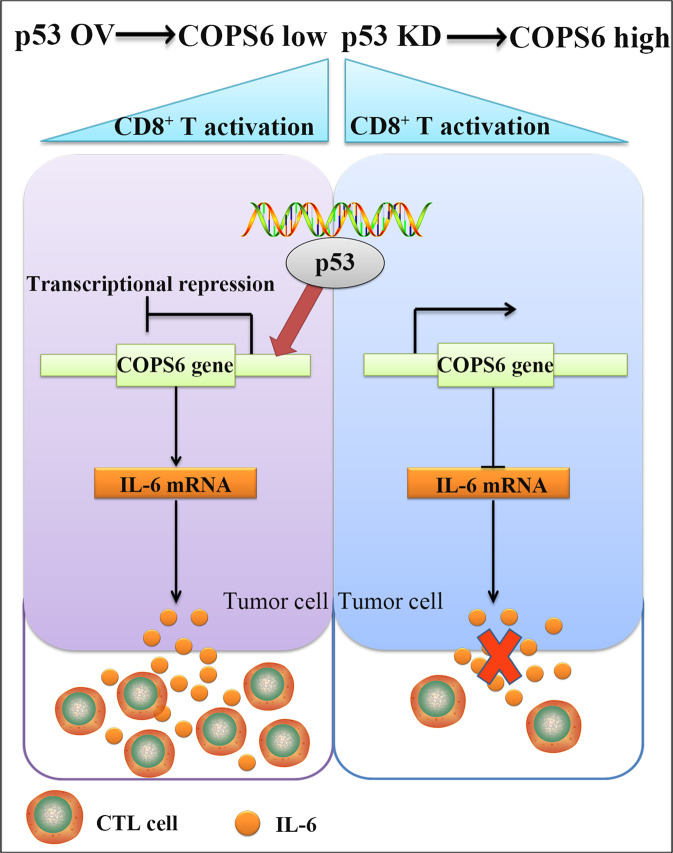
Fig. 8. Model of COPS6-mediated immunosuppressive effect in breast cancer.
COPS6, negatively regulated by p53, inhibits CD8+ T cells infiltration by repressing the production of IL-6 in breast cancer.
Discussion
COP9 signalosome regulated multiple important cellular biological processes, such as ubiquitin/proteasome system, DNA damage repair, transcription regulation, phosphorylation modification, embryonic development, physiological rhythm and tumorigenesis [8, 9, 32]. Growing evidence has emerged to highlight the roles of CSN subunits in carcinogenesis and tumor progression [32–34]. Of all the subunits, CSN5 and COPS6 were relatively well studied in tumor progression [33, 34]. For example, CSN5 depletion inhibited the proliferation of HeLa cells via elevating ROS production [35]. CSN5-mediated p14ARF degradation facilitated gastric cancer progression [36]. Analogously, COPS6 possessed oncogenic potential in multiple human cancers, and its overexpression was detected in glioblastoma [14], colorectal cancer [13], melanoma [15], hepatocellular carcinoma [16] and pancreatic cancer [17]. COPS6 promoted initiation and progression of cancer mainly by regulating the protein degradation of downstream partners, such as CDK9 [17], Snail1 [37], PD-L1 [38], p16INK4a [39], and EGFR [14]. However, the underlying molecular mechanisms by which COPS6 affect tumor immune microenvironment and tumor infiltrating lymphocytes remain undefined. In the present study, we identified the ectopic expression of COPS6 in breast cancer, and p53-mediated transcriptional activation was responsible for COPS6 expression. COPS6 served as a novel inducer of proliferation, migration and invasion of breast cancer cells. Mechanism dissection revealed that COPS6 restrained CD8+ TILs and facilitated tumor immune evasion through negative modulating IL-6 production in breast cancer. These findings provided evidence of a de novo COPS6-IL-6-CD8+ TILs signal in breast cancer progression, and possibly, antitumor immunity.
Initially, we found that COPS6 was up-regulated in various cancers by excavating the online database, and COPS6 overexpression predicted poor prognosis of breast cancer patients, which were consistent with previous studies related to the roles of COPS6 in cancers [13–15, 17, 19]. The C-terminal domain of COPS6 was essential for maintaining the integrity of CSN complex and binding to Cullin-RING E3 ubiquitin ligases [40]. It has been fully characterized that COPS6 played a critical role in ubiquitin-proteasome dependent degradation by regulating E3 ubiquitin ligases such as MDM2 [12] and COP1 [41]. Tumor suppressor gene p53 was negatively regulated by COPS6 [42–44]. COPS6 potentiated the degradation of p53 by thwarting the Lys-364-linked autoubiquitination and degradation of MDM2 in human cancers [12]. In addition, COPS6 expression strongly negatively correlated with mutant-type p53 in 92 cases of invasive breast cancer [42]. We found that p53 suppressed COPS6 expression in breast cancer and the enrichment of p53 at COPS6 promoter regions. We demonstrated that p53 occupied the promoter of COPS6 and induced its transcription inhibition in breast cancer. Therefore, it might be inferred that p53 and COPS6 were negatively correlated each other in cancers. Further studies are needed to explore the negative feedback loop between p53 and COPS6 in cancer initiation and progression.
We provided evidence that COPS6 promoted the proliferation, migration and invasion of breast cancer cells and inhibited apoptosis by the gain-of-function and loss-of-function experiments. Previous studies also identified COPS6 as an oncogenic driver in various cancers, such as hepatocellular carcinoma [16], melanoma [15], colorectal cancer [13], pancreatic cancer [17], glioblastoma [38], gastric cancer [36], endometrial cancer [43], and breast cancer [37]. However, the regulation of anti-tumor immunity by COPS6 remains elusive. Bioinformatics analysis identified the functional enrichment of response to IL-6 and IL-6 production after COPS6 ectopic expression in breast cancer. IL-6 is a cytokine that regulates inflammatory response and inhibits inflammation-related apoptosis, and plays an important role in immune escape and tumor progression [27–29]. The anti-tumor potential of IL-6 depends on blunting tumor immunity [45]. Our results demonstrated that COPS6 decreased IL-6 expression and impaired secretion of IL-6 in breast cancer. IL-6 participated in the immune regulation and progression of breast cancer by activating CD8+ TILs. The abundance of CD8+ TILs exerts anti-tumorigenic effects, which exhibits a better prognosis for breast cancer patients [19, 22]. Also as expected, COPS6 was implicated in antitumor immunity directed by tumor-infiltrating lymphocytes except for the involvement modulating tumor growth and metastasis. COPS6 amplification was associated with decreased CD8+ TILs and activation. And COPS6 effectuated the growth-promoting pro-metastatic effects on CD8+ T cells. Therefore, these results demonstrated that COPS6 in promoting breast cancer malignancy might be dependent on impairing CD8+ TILs via the regulation of IL-6 secretion.
In summary, our results provided evidence connecting COPS6, CD8+ TILs and breast cancer progression mediated by IL-6, which unveiled a previously unrecognized role for COPS6 in antitumor immunity. We confirmed that COPS6 was highly expressed in various cancers and COPS6 expression confered a poor prognosis in breast cancer. Tumor suppressor p53 was responsible for the ectopic expression of COPS6. Intriguingly, we found that COPS6 correlated with an immunosuppressive tumor microenvironment in breast cancer. Mechanism dissection revealed that COPS6 could serve as an immunosuppressive factor via impairing CD8+ TILs infiltration in breast cancer induced by secretion of IL-6, contributing to breast cancer progression. This study highlighted the de novo COPS6-IL-6-CD8+ TILs signal and provided the novel mechanistic insight into the role of COPS6 in carcinogenesis and tumor progression. Thus, our findings suggested a promising therapeutic target to perturb tumor immune escape via interruption of COPS6.
Supplementary information
Acknowledgements
This work was supported by the Graduate Student Scientific Research Innovation Projects in Jiangsu Province (KYCX22-2944) and Xuzhou Key Research and Development Projects (KC22119).
Author contributions
DSP designed the study; WQD and ZMZ performed the Research; XJ and MJK analyzed the data; WQD, ZMZ, and XJ wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Wen-qi Du, Zhi-man Zhu, Xin Jiang
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-023-01085-8.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Liang Y, Zhang H, Song X, Yang Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin Cancer Biol. 2020;60:14–27. doi: 10.1016/j.semcancer.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Lim B, Hortobagyi GN. Current challenges of metastatic breast cancer. Cancer Metastasis Rev. 2016;35:495–514. doi: 10.1007/s10555-016-9636-y. [DOI] [PubMed] [Google Scholar]
- 4.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Prim. 2019;5:66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 5.Lucci A, Hall CS, Lodhi AK, Bhattacharyya A, Anderson AE, Xiao L, et al. Circulating tumour cells in non-metastatic breast cancer: a prospective study. Lancet Oncol. 2012;13:688–95. doi: 10.1016/S1470-2045(12)70209-7. [DOI] [PubMed] [Google Scholar]
- 6.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379:2108–21. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 7.Fuzesi-Levi MG, Fainer I, Ivanov Enchev R, Ben-Nissan G, Levin Y, Kupervaser M, et al. CSNAP, the smallest CSN subunit, modulates proteostasis through cullin-RING ubiquitin ligases. Cell Death Differ. 2020;27:984–98. doi: 10.1038/s41418-019-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubiel W, Chaithongyot S, Dubiel D, Naumann M. The COP9 Signalosome: a multi-DUB complex. Biomolecules. 2020;10:1082. doi: 10.3390/biom10071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou J, Cui H. CSN6: a promising target for cancer prevention and therapy. Histol Histopathol. 2020;35:645–52. doi: 10.14670/HH-18-206. [DOI] [PubMed] [Google Scholar]
- 10.Du W, Zhang R, Muhammad B, Pei D. Targeting the COP9 signalosome for cancer therapy. Cancer Biol Med. 2022;19:573–90. doi: 10.20892/j.issn.2095-3941.2021.0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin N, Xu D, Li J, Deng XW. COP9 signalosome: discovery, conservation, activity, and function. J Integr Plant Biol. 2020;62:90–103. doi: 10.1111/jipb.12903. [DOI] [PubMed] [Google Scholar]
- 12.Zhao R, Yeung SC, Chen J, Iwakuma T, Su CH, Chen B, et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest. 2011;121:851–65. doi: 10.1172/JCI44111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin B, Zou S, Li K, Wang H, Wei W, Zhang B, et al. CSN6-TRIM21 axis instigates cancer stemness during tumorigenesis. Br J Cancer. 2020;122:1673–85. doi: 10.1038/s41416-020-0779-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou J, Deng Q, Zhou J, Zou J, Zhang Y, Tan P, et al. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene. 2017;36:1134–44. doi: 10.1038/onc.2016.280. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Hou J, Shi S, Du J, Liu Y, Huang P, et al. CSN6 promotes melanoma proliferation and metastasis by controlling the UBR5-mediated ubiquitination and degradation of CDK9. Cell Death Dis. 2021;12:118. doi: 10.1038/s41419-021-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu M, Zhen L, Lin L, Wu K, Wang Y, Cai X. Overexpression of CSN6 promotes the epithelial-mesenchymal transition and predicts poor prognosis in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2020;44:340–8. doi: 10.1016/j.clinre.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Shi J, Guan X, Zhan F, Liu C, Li Z, Yao Y, et al. CSN6 expression is associated with pancreatic cancer progression and predicts poor prognosis. Cancer Biol Ther. 2019;20:1290–9. doi: 10.1080/15384047.2019.1632143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 19.Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer. 2016;4:59. doi: 10.1186/s40425-016-0165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang SS, Liu W, Ly D, Xu H, Qu L, Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol. 2019;16:6–18. doi: 10.1038/s41423-018-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valpione S, Mundra PA, Galvani E, Campana LG, Lorigan P, De Rosa F, et al. The T cell receptor repertoire of tumor infiltrating T cells is predictive and prognostic for cancer survival. Nat Commun. 2021;12:4098. doi: 10.1038/s41467-021-24343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29:1949–55. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 23.Xia Z, Kon N, Gu AP, Tavana O, Gu W. Deciphering the acetylation code of p53 in transcription regulation and tumor suppression. Oncogene. 2022;41:3039–50. doi: 10.1038/s41388-022-02331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehraj U, Dar AH, Wani NA, Mir MA. Tumor microenvironment promotes breast cancer chemoresistance. Cancer Chemother Pharmacol. 2021;87:147–58. doi: 10.1007/s00280-020-04222-w. [DOI] [PubMed] [Google Scholar]
- 25.Deepak KGK, Vempati R, Nagaraju GP, Dasari VR, S N, Rao DN, et al. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol Res. 2020;153:104683. doi: 10.1016/j.phrs.2020.104683. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Lu Z, Zhao X. Targeting Bcl-2 for cancer therapy. Biochim Biophys Acta Rev Cancer. 2021;1876:188569. doi: 10.1016/j.bbcan.2021.188569. [DOI] [PubMed] [Google Scholar]
- 27.Jones SA, Jenkins BJ. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat Rev Immunol. 2018;18:773–89. doi: 10.1038/s41577-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 28.Unver N, McAllister F. IL-6 family cytokines: key inflammatory mediators as biomarkers and potential therapeutic targets. Cytokine Growth Factor Rev. 2018;41:10–7. doi: 10.1016/j.cytogfr.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taher MY, Davies DM, Maher J. The role of the interleukin (IL)-6/IL-6 receptor axis in cancer. Biochem Soc Trans. 2018;46:1449–62. doi: 10.1042/BST20180136. [DOI] [PubMed] [Google Scholar]
- 30.Abaurrea A, Araujo AM, Caffarel MM. The role of the IL-6 cytokine family in epithelial-mesenchymal plasticity in cancer progression. Int J Mol Sci. 2021;22:8334. doi: 10.3390/ijms22158334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kampan NC, Xiang SD, McNally OM, Stephens AN, Quinn MA, Plebanski M. Immunotherapeutic interleukin-6 or interleukin-6 receptor blockade in cancer: challenges and opportunities. Curr Med Chem. 2018;25:4785–806. doi: 10.2174/0929867324666170712160621. [DOI] [PubMed] [Google Scholar]
- 32.Lee MH, Zhao R, Phan L, Yeung SC. Roles of COP9 signalosome in cancer. Cell Cycle. 2011;10:3057–66. doi: 10.4161/cc.10.18.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan C, Wang D, Liu G, Pan Y. Jab1/Cops5: a promising target for cancer diagnosis and therapy. Int J Clin Oncol. 2021;26:1159–69. doi: 10.1007/s10147-021-01933-9. [DOI] [PubMed] [Google Scholar]
- 34.Guo Z, Wang Y, Zhao Y, Shu Y, Liu Z, Zhou H, et al. The pivotal oncogenic role of Jab1/CSN5 and its therapeutic implications in human cancer. Gene. 2019;687:219–27. doi: 10.1016/j.gene.2018.11.061. [DOI] [PubMed] [Google Scholar]
- 35.Pandey P, Khan F, Maurya P. Targeting Jab1 using hesperidin (dietary phytocompound) for inducing apoptosis in HeLa cervical cancer cells. J Food Biochem. 2021;45:e13800. doi: 10.1111/jfbc.13800. [DOI] [PubMed] [Google Scholar]
- 36.Wang L, Du WQ, Xie M, Liu MR, Huo FC, Yang J, et al. Jab1 promotes gastric cancer tumorigenesis via non-ubiquitin proteasomal degradation of p14ARF. Gastric Cancer. 2020;23:1003–17. doi: 10.1007/s10120-020-01087-z. [DOI] [PubMed] [Google Scholar]
- 37.Mou J, Wei L, Liang J, Du W, Pei D. CSN6 promotes the cell migration of breast cancer cells by positively regulating Snail1 stability. Int J Med Sci. 2020;17:2809–18. doi: 10.7150/ijms.50206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su L, Guo W, Lou L, Nie S, Zhang Q, Liu Y, et al. EGFR-ERK pathway regulates CSN6 to contribute to PD-L1 expression in glioblastoma. Mol Carcinog. 2020;59:520–32. doi: 10.1002/mc.23176. [DOI] [PubMed] [Google Scholar]
- 39.Du W, Liu Z, Zhu W, Li T, Zhu Z, Wei L, et al. CSN6 promotes tumorigenesis of gastric cancer by ubiquitin-independent proteasomal degradation of p16(INK4a) Cancer Biol Med. 2019;16:514–29. doi: 10.20892/j.issn.2095-3941.2018.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faull SV, Lau AMC, Martens C, Ahdash Z, Hansen K, Yebenes H, et al. Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome. Nat Commun. 2019;10:3814. doi: 10.1038/s41467-019-11772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi HH, Zou S, Wu JL, Wang H, Phan L, Li K, et al. EGF relays signals to COP1 and facilitates FOXO4 degradation to promote tumorigenesis. Adv Sci (Weinh) 2020;7:2000681. doi: 10.1002/advs.202000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Tang M, Zhang L, Xu X, Qi X, Yang Y, et al. Clinical implications of CSN6 protein expression and correlation with mutant-type P53 protein in breast cancer. Jpn J Clin Oncol. 2013;43:1170–6. doi: 10.1093/jjco/hyt148. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Chen T. CUL4A regulates endometrial cancer cell proliferation, invasion and migration by interacting with CSN6. Mol Med Rep. 2021;23:23. doi: 10.1007/s00894-016-3198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue Y, Chen J, Choi HH, Phan L, Chou PC, Zhao R, et al. HER2-Akt signaling in regulating COP9 signalsome subunit 6 and p53. Cell Cycle. 2012;11:4181–90. doi: 10.4161/cc.22413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chonov DC, Ignatova MMK, Ananiev JR, Gulubova MV. IL-6 activities in the tumour microenvironment. Part 1. Open Access Maced J Med Sci. 2019;7:2391–8. doi: 10.3889/oamjms.2019.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.