Abstract
A reverse transcription-PCR and hybridization-enzyme immunoassay (RT-PCR-EIA) has been developed to identify the major agents of bronchiolitis in infants: respiratory syncytial viruses A and B (RSVA and RSVB) and parainfluenzavirus 3 (PIV3). Two primer sets (P1-P2 and P1-P3) were selected in a conserved region of the polymerase L gene. In infected cell cultures, this method detected RSVA (n = 14), RSVB (n = 13), and PIV3 (n = 13), with the exclusion of PIV1 (n = 4), PIV2 (n = 3), measles virus (n = 6), mumps virus (n = 4), influenza A virus (n = 11), and influenza B virus (n = 4). The differentiation of the amplicons by restriction fragment length polymorphism (RFLP) showed a PvuII site for PIV3 strains and an AvaII site for RSV strains, with RSVA distinguished from RSVB by BglII. The hybridization-EIA, using three internal probes specific for each virus, correlated with the immunofluorescence assay (IFA) and RFLP results. Clinical aspirates from 261 infants hospitalized with bronchiolitis were tested by IFA, viral isolation technique (VIT), and RT-PCR-EIA. RT-PCR-EIA detected RSV sequences in 103 samples (39.4%), and IFA-VIT detected RSV sequences in 109 cases (41.7%). A few samples (2.6%) were IFA-VIT positive but PCR negative, and one sample was RT-PCR-EIA positive only. RT-PCR-EIA detected PIV3 sequences in 14 of the 15 IFA-VIT-positive isolates. The two methods showed very good correlation (96.9%), but RT-PCR-EIA was clearly more efficient in typing, leaving 5% non-A, non-B isolates, while IFA failed to resolve 23% of the isolates. The two methods contradicted each other for <5% of the isolates.
The two subgroups of respiratory syncytial virus (RSVA and RSVB) and parainfluenzavirus 3 (PIV3) are the agents most commonly involved in bronchiolitis and pneumonia in infants (4, 6, 13, 22). Early and differential diagnosis of RSV and PIV3 infections is necessary for monitoring of the infected infants, for prevention of nosocomial spread, and, in some cases, to guide the choice of a possible adapted antivirus therapy (10). An immunofluorescence assay (IFA) of viral antigens in nasal aspirates is widely used for the diagnosis of RSV and PIV infections because it is more rapid than and at least as sensitive as virus isolation technique (VIT) (5, 8). Reverse transcription-PCR (RT-PCR) has been developed recently for the detection of RSV and PIV3 sequences in nasal aspirates of children (2, 3, 9, 12, 14, 15, 19). However, each procedure detects only a single virus. The aim of this study was to define a molecular method likely to detect and identify RSV and PIV3 in a single nucleic amplification assay. Such an RT-PCR followed by a hybridization-enzyme immunoassay (RT-PCR-EIA) was developed and tested for its sensitivity toward prototype strains and wild isolates adapted to cell cultures of RSV and PIV3, its specificity toward PIV1 and PIV2, influenza viruses A and B, and mumps and measles viruses, and its use for the detection of viral sequences in nasal aspirates of hospitalized infants.
MATERIALS AND METHODS
Virus strains and respiratory specimens.
Prototype strains of RSVA (Long) and RSVB (9320) were obtained from the American Type Culture Collection (ATCC VR-26 and ATCC VR-955, respectively). PIV2 and PIV3 strains were obtained from Diagnostics Pasteur (Marnes, France), and Sendai virus and Newcastle disease virus (NDV) were obtained from P. Lebon (St. Vincent de Paul Hospital, Paris, France). Fifty-one human wild isolates of RSVA (n = 14), RSVB (n = 13), PIV3 (n = 13), PIV1 (n = 4), PIV2 (n = 3), influenza virus A (H3N2 [n = 9], H1N1 [n = 2]), influenza virus B (n = 4), and mumps (n = 3) and measles (n = 6) viruses adapted to cell cultures were obtained from our local collection. Nasal aspirates (n = 261) of children hospitalized in the pediatric units of the University Hospital of Caen were each collected from October 1995 to March 1996 in a solution containing 5 ml of viral transport medium (Eagle minimal essential medium), 5 mg of bovine albumin per ml, 4.76 mg of HEPES per ml, 1,500 U of penicillin, and 500 μg of gentamicin.
Isolation and identification of viruses by IFA and VIT.
For IFA, 2 ml of fresh nasal aspirates in transport medium was used. The cells were separated by centrifugation, washed in phosphate-buffered saline (PBS), deposited on microscope slides, and fixed in acetone. The test used fluorescein isothiocyanate-conjugated RSV and PIV3 monoclonal antibody reagents (IMAGEN; Dako Diagnostics, Ely, United Kingdom). Monoclonal antibodies kindly supplied by E. N. Norrby (Karolinska Institute, Stockholm, Sweden) were used to identify RSVA and RSVB as previously described (7). All incubations were carried out at 37°C for 30 min, and slides were washed two times in PBS. For VIT, cultures of MRC-5 human embryonic lung fibroblasts in 25-cm2 flasks and NCI-H292 human lung mucoepidermoid cells in 48-well tissue culture plates were inoculated with, respectively, 0.2 and 0.1 ml of the resuspended specimens. In MRC-5 cultures exhibiting typical RSV cytopathic effects, infected cells were harvested, pelleted by centrifugation, deposited on microscope slides, and fixed in acetone, and the virus was identified by IFA. MRC-5 cells were kept 4 weeks before a culture was considered negative. NCI-H292 cells were incubated for 5 to 6 days, harvested by trypsination, and stained with RSV and PIV3 monoclonal antibodies. Blind passages were not routinely done.
Nucleic acid extraction.
For the prototype strains and wild virus isolates, 100 μl of the stored specimens was digested by adding a solution of 2 μl of proteinase K (10 mg/ml), 10 μl of 10% sodium dodecyl sulfate, and 1.2 μl of 1 M Tris-HCl (pH 7.6), and the samples were incubated at 56°C for 1 h. Nucleic acids were extracted with phenol-chloroform (1:1) and with chloroform-isoamylalcohol (24:1). Total nucleic acids were precipited with 66% ethanol and 0.3 M sodium acetate, pH 5.6, at −20°C overnight, collected by centrifugation, washed with 70% ethanol, and resuspended in 20 μl of H2O containing 20 U of RNase inhibitor (Boehringer Mannheim Biochemica). For clinical samples, 500 μl of nasal aspirate in transport medium stored at −80°C was extracted by the RNAzole B method (Bioprobe, Montreuil sous Bois, France).
RT-PCR.
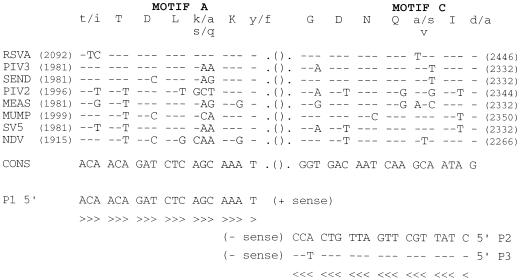
A unique (+) sense primer, P1 (5′-ACA ACA GAT CTC AGC AAA T-3′), and two alternative (−) sense primers, P2 (5′-CTA TTG CTT GAT TGT CAC C-3′) and P3 (5′-CTA TTG CTT GAT TGT CTC C-3′), were defined in the most conserved region of the genome, i.e., the functional motifs A and C of the L polymerase gene (16) (see Fig. 1). Oligonucleotides used as primers were synthesized by the Unit of Organic Chemistry, Institut Pasteur, Paris, France. For the prototype strains and wild viral isolates, the P1-P2 and P1-P3 primer sets were checked separately for RT-PCR. The two primers of each set were present for both the RT and PCR steps. For clinical sample extracts, the three primers, P1, P2, and P3, were used simultaneously. cDNA synthesis was performed for 60 min at 42°C in a 25-μl reaction volume in RT buffer (Gibco BRL, Gaithersburg, Md.) containing 5 μl of nucleic acids extract, 200 U of Moloney murine leukemia virus reverse transcriptase (Gibco BRL), 20 U of RNase inhibitor (RNasin, Promega Charbonnières, France), 40 pmol of each primer, 1 mM (each) dATP, dCTP, dGTP, dTTP, and 10 mM dithiothreitol (GIBCO BRL). For the PCR step, the reaction volume was increased to 100 μl with PCR buffer (Perkin-Elmer Cetus), 1.5 mM MgCl2, and 2.5 U of Taq polymerase (Perkin-Elmer Cetus). Amplification was performed by using an Omnigene thermocycler (Hybaid) with 30 cycles of heat denaturation at 94°C for 30 s, primer annealing at 45°C for 1.5 min, and primer extension at 72°C for 1.5 min. The PCR product was analyzed by electrophoresis in a 1.2% agarose gel. The separated fragments were stained by ethidium bromide and visualized under UV light.
FIG. 1.
Sequences deduced from the alignment of the functional motifs A and C in the L gene of RSVA, PIV3, Sendai virus (SEND), PIV2, measles virus (MEAS), mumps virus (MUMP), simian virus (SV5), and NDV. The upper line shows the amino acid sequences of the motifs (uppercase letters, conserved residues; lowercase letters, variable residues). The corresponding nucleotide sequences are aligned with reference to a consensus (CONS). Their positions are given with reference to the initiation codon of each L protein (1ATG). The structure of P3 is given with reference to P2. Dashes indicate conserved nucleotides.
Detection and typing of amplified products by restriction fragment length polymorphism (RFLP) and hybridation-EIA.
The PCR products (5 μl) were subjected to restriction endonucleases under conditions specified by the manufacturer (Bethesa Research Laboratories) and analyzed by electrophoresis on a 1.2% agarose gel containing ethidium bromide. Three restriction enzymes, AvaII, BglII, and PvuII, which are highly conserved within each virus group but display differentiating capacities between groups, were selected to identify RSVA, RSVB, and PIV3. Specific genomic (minus sense) probes internal to the amplified products were selected from the multiple alignment by using both the sequences arising from this study and others available from the DNA databases (GenBank): RSVA probe, 5′-2289-TACATTGTTAGGATCTACAGTATGATC TCCTATATAGGGG-2250-3′; RSVB probe, 5′-AACTTCATTAAGATTGACAACATGA TCCTTTATGAAAGGA-2250-3′; PIV3 probe, 5′-2095-GAGGGTGTAACCAATTAAA CAATTTATTTAATCCAAATA-2056-3′). Their positions are given with reference to the initiation codon of each L protein (1ATG), except for the RSVB L gene, which is not available in the database. The probes were synthesized and 5′ biotinylated at the Unit of Organic Chemistry, Institut Pasteur. A microtiter plate was coated with single-stranded DNA probes with streptavidin-biotin bond. The PCR products were hybridized and then revealed by an antibody detecting double-stranded DNA in an EIA (GEN-ETI-K DEIA; Sorin) performed as recommended by the manufacturer. The optimal concentrations of the RSVA, RSVB, and PIV3 probes were set at 0.1, 1, and 0.5 ng/μl, respectively.
RESULTS
Sensitivity and specificity of RT-PCR for prototype strains and wild isolates.
Previous comparisons between the deduced amino acid sequences of L proteins from nonsegmented negative-stranded RNA viruses exhibited six blocks of strong conservation separated by variable links. Block III, which corresponds to the catalytic domain for polymerization, contained 4 to 5 motifs highly conserved among the RNA-dependent polymerases (RNA polymerases and reverse transcriptases) (16, 17, 20, 21). We first aligned the conserved block IIIs of the paramyxoviruses available in the database: RSVA, PIV3, Sendai, PIV2, measles, mumps, simian virus 5, and NDV. The primers were then designed to specifically detect RSV and PIV3 in the conserved motifs A and C (Fig. 1). Each primer was designed in a consensus way to allow acceptable complementarity to the sequences. A unique consensus (+) sense primer (P1) was generated with only two consecutive mismatches compared to the sequences of RSVA and PIV3 and at least three mismatches compared to the other sequences. Two (−) sense primers, P2 and P3, were necessary to take into consideration the sequence variability between PIV3 and RSVA. They differ by a single nucleotide in position 17 of the primer (i.e., −3 from the first nucleotide incorporated). P2 contains one mismatch with RSVA and two with PIV3. Inversely, P3 contains one mismatch with PIV3 and two with RSVA. Both primers were more divergent for the other viruses, except for the murine Sendai virus (one mismatch with P2 and two with P3, like RSVA). According to the database, both primer sets were expected to amplify L gene DNA segments of 355 bp for RSVA or RSVB and 352 bp for PIV3 and Sendai virus.
The specificities of the P1-P2 and P1-P3 primer sets were established on prototype strains of RSVA, RSVB, PIV3, and Sendai virus, as well as on a panel of 70 clinical isolates of para- and orthomyxoviruses identified by IFA. On ethidium bromide agarose gel electrophoresis, RSVA and Sendai virus gave the expected amplicons with both the P1-P2 and P1-P3 primer sets. For RSVB, the expected amplicon was observed only with the P1-P2 primer set (Table 1). One RSVB isolate (no. 9401891) was amplified with both the P1-P2 and P1-P3 primer sets. For PIV3, the prototype strain and six isolates (46%) reacted with the P1-P2 primer set, while all of them reacted with the P1-P3 primer set. Such a result is in accordance with the better complementarity of the P3 primer to the PIV3 sequence (Fig. 1). Finally, no specific amplication was obtained with any of the primer sets for other paramyxoviruses (PIV1 [n = 4], PIV2 [n = 3], measles virus [n = 6], and mumps virus [n = 3]), orthomyxoviruses (influenza virus A [H3N2] [n = 9], influenza virus A [H1N1] [n = 2], and influenza virus B [n = 4]), or uninfected cell extracts used as controls (data not shown).
TABLE 1.
Analysis of RSVA, RSVB, PIV3, and Sendai virus strains by IFA, RT-PCR, RFLP, and hybridization-EIA
| Virusa and specimen | RT-PCR result with product of indicated expected size (bp)
|
RFLP result for fragment with indicated endpoints (bp)
|
Hybridization-EIA result (index value)b
|
|||||
|---|---|---|---|---|---|---|---|---|
| P1-P2 | P1-P3 | AvaII | BglII | PvuII | RSVA | RSVB | PIV3 | |
| RSVA | 355 | 355 | 263/92 | 177/172 | ||||
| Long | + | + | + | + | − | 10 | 0.6 | 0.2 |
| 9345839 | + | + | + | − | − | 8.4 | 0.2 | 0.1 |
| 9343012 | + | + | + | − | − | 9.5 | 0.1 | 0.4 |
| 9346219 | + | + | + | − | − | 10.5 | 0.4 | 0.2 |
| 9344506 | + | + | + | − | − | 1.5 | 0.1 | 0.2 |
| 9400961 | + | + | + | − | − | 10.9 | 0.1 | 0.2 |
| 9404584 | + | + | + | − | − | 10 | 0.2 | 0.1 |
| 9402928 | + | + | + | + | − | 9.6 | 0.3 | 0.1 |
| 9401927 | + | + | + | + | − | 8 | 0.2 | 0.1 |
| 9344539 | + | + | + | − | − | 11.3 | 0.2 | 0.2 |
| 9400220 | + | + | + | − | − | 1.3 | 0.2 | 0.3 |
| 9400932 | + | + | + | − | − | 2.6 | 0.03 | 0.2 |
| 9401887 | + | + | + | − | − | 8.3 | 0.04 | 0.3 |
| 9400972 | + | + | + | − | − | 6.7 | 0.2 | 0.3 |
| 283/66 | ||||||||
| 9400504 | + | + | + | + | − | 0.2 | 5.3 | 0.1 |
| RSVB | ||||||||
| 9320 | + | − | + | + | − | 0.3 | 10.3 | 0.3 |
| 9400906 | + | − | + | + | − | 0.2 | 11.7 | 0.1 |
| 9342557 | + | − | + | + | − | 0.3 | 11.4 | 0.2 |
| 9346188 | + | − | + | + | − | 0.4 | 12.9 | 0.1 |
| 9346000 | + | − | + | + | − | 0.3 | 7.7 | 0.2 |
| 9401761 | + | − | + | + | − | 0.7 | 5.5 | 0.1 |
| 9346417 | + | − | + | + | − | 0.2 | 4.3 | 0.2 |
| 9401990 | + | − | + | + | − | 0.4 | 9.2 | 0.1 |
| 9402975 | + | − | + | + | − | 0.5 | 10.5 | 0.2 |
| 9400215 | + | − | + | + | − | 0.2 | 8.6 | 0.2 |
| 9344256 | + | − | + | + | − | 0.7 | 8.8 | 0.1 |
| 9401023 | + | − | + | + | − | 0.3 | 9.3 | 0.2 |
| 9338709 | + | − | + | + | − | 0.2 | 8.5 | 0.2 |
| 9401891 | + | + | + | − | − | 2.0 | 2.4 | 0.2 |
| PIV3 | 352 | 352 | 295/57 | |||||
| Pasteur | + | + | − | − | + | 0.5 | 0.3 | 12.3 |
| 9441219 | + | + | − | − | + | 0.2 | 0.1 | 11.1 |
| 9439318 | + | + | − | − | + | 0.1 | 0.3 | 11.4 |
| 9439389 | + | + | − | − | + | 0.4 | 0.2 | 13.0 |
| 9426479 | − | + | − | − | + | 0.2 | 0.4 | 10.3 |
| 9426019 | + | + | − | − | + | 0.2 | 0.4 | 5.8 |
| 9426354 | − | + | − | − | + | 0.2 | 0.2 | 9.4 |
| 9431422 | − | + | − | − | + | 0.2 | 0.2 | 10.5 |
| 9504839 | + | + | − | − | + | 0.3 | 0.3 | 8.4 |
| 9511637 | − | + | − | − | + | 0.2 | 0.2 | 6.4 |
| 9436203 | − | + | − | − | + | ND | ND | ND |
| 9509919 | + | + | − | − | + | ND | ND | ND |
| 9438367 | − | + | − | − | + | ND | ND | ND |
| 9419101 | − | + | − | − | + | ND | ND | ND |
| Sendai virus | + | + | − | − | + | 0.5 | 0.20 | 0.25 |
Determined by IFA.
Hybridization is positive if the index value is >1 (positive hybridization is indicated by boldface type). Index value = sample OD/cut-off OD. ND, not done.
The sensitivity of the RT-PCR test was examined by analyzing serial 10-fold dilutions of RSVA and RSVB prototype strains with virus titers of 103 and 104 50% tissue culture infective doses (TCID50)/ml in MRC-5-infected cells, respectively. Amplified products were detected on an agarose gel at 10−1 and 10−2 dilutions for RSVA and RSVB, respectively, and after hybridization at 10−2 and 10−3 dilutions for RSVA and RSVB, respectively (data not shown). Thus, the minimal amount of viral RNA detected by RT-PCR-EIA was approximately 10 TCID50.
Identification and typing of viruses by RFLP.
As shown in Table 1, AvaII cleaved exclusively the 355-bp RSV amplicons (endpoints, bp 263 and 92), and PvuII cleaved the 352-bp PIV3 amplicons (bp 295 and 57). Thus, RFLP data allowed simple distinctions between RSV and PIV amplicons on the basis of the mutually exclusive presence of the AvaII and PvuII restriction sites, respectively. Moreover, further analysis of the fragment length obtained by BglII digestion showed that the bp 283 and 66 couple was typical of RSVB isolates, while only 21% of the RSVA isolates were cleaved into a bp 177 and 172 couple, indicating greater genetic heterogeneity. Discordant typing between IFA and RFLP was observed for two RSV strains (7.5%). For RSV “A” 9400504, this discordance was complete, since the BglII pattern (bp 283 and 66) was clearly of RSV “B” type. For RSV “B” 9401891, the situation was more ambiguous, since the absence of cleavage by BglII is not a strict signature of the RSV “A” type. There were no BglII sites in the PIV3 amplicons.
Typing of viruses by hybridization-EIA.
The sequences of the amplicons of several representative isolates were determined (not shown) and compared to their counterparts in the database. Then, oligonucleotides probes were searched in regions typical for each virus group. The RSVA and RSVB probes were chosen for positions 159 to 200 of the amplicon, where the two sequences are the most divergent (40%). The PIV3 probe was chosen for positions 77 to 118 of the amplicon, in one area rather divergent from the RSV sequences (about 55%) and including, notably, a unique 3-nucleotide gap. The RSVA, RSVB, and PIV3 probes were tested for typing in a hybridization-EIA test on the prototype strains and viral isolates (Table 1). The results were considered positive when the index value (sample optical density [OD]/cut-off OD [negative control OD + 0.150]) was >1. For each of the three probes, hybridization was positive with the homologous prototype strains with high index values (>10) and systematically negative (index value, <1) with heterologous prototype strains, indicating the absence of cross-hybridization. In addition, 97.5% of the tested amplicons from the clinical isolates were exclusively positive with the homologous probe. Notably, this included isolate no. 9400504, which was then determined to be definitively of the RSVB subgroup according to molecular data, in contradiction to the antigenic data. However, isolate no. 9401891, RSVB by IFA, conserved its ambiguous nature by showing cross-hybridization with both the RSVA probe (index value, 2) and the RSVB probe (index value, 2.4).
Detection of viruses in clinical samples by RT-PCR-EIA.
RT-PCR-EIA, using the three primers (P1, P2, and P3) simultaneously, and hybridization with the RSVA, RSVB, and PIV3 probes were compared to the routine techniques (IFA and VIT) on 261 nasal aspirates (Table 2). RT-PCR-EIA detected RSV sequences in 103 samples (39.4%), and IFA and VIT detected RSV sequences in 109 cases (41.7%). Seven samples (2.6%) were IFA and VIT positive but PCR negative, and only one sample (0.4%) was RT-PCR-EIA positive only. The concordance between IFA-VIT and RT-PCR-EIA was 96.9% (253/261) for the detection of RSV-infected samples. For RSV typing, the abilities of the techniques were 77.1% (84/109) for IFA and 94.2% (97/103) for RT-PCR-EIA, with 22.9% and 5.8% undetermined results, respectively. All of the 63 RSVA-positive samples, but only 17 of the 21 RSVB-positive samples, were identified by IFA or RT-PCR-EIA. Four of the RSVB-positive samples gave positive signals with the RSVA probe. For 25 RSV infections, the subgroup, A or B, could not be defined by using monoclonal antibodies, and half of them were identified as RSVA by RT-PCR-EIA. RT-PCR-EIA was clearly more efficient in typing, leaving 5% non-A, non-B isolates, while IFA failed to resolve 23% of the isolates. The two methods contradicted each other for <5% of the isolates. Finally, the concordance between IFA and RT-PCR-EIA was 84.3% (86/102) for typing RSV in infected samples. Fifteen of the 261 samples (5.7%) were PIV3 positive by IFA-VIT, and RT-PCR-EIA detected PIV3 sequences in 14 of them, confirming the slight advantage in sensitivity of IFA-VIT observed for RSV infection.
TABLE 2.
Comparison of IFA-VIT, RT-PCR, and hybridization-EIA for detection of RSV in 261 nasal aspirates of hospitalized infants
| IFA-VIT result | No. of specimens
|
|||
|---|---|---|---|---|
| RT-PCR positive
|
RT-PCR and hybridizationEIA negative | |||
| Hybridization-EIA positive
|
Hybridization-EIA negative | |||
| Probe A | Probe B | |||
| Positive | ||||
| RSVA | 63 | 0 | 0 | 0 |
| RSVB | 4 | 17 | 0 | 0 |
| RSV (nontypeable) | 12 | 0 | 6 | 7 |
| Negative | 1 | 0 | 0 | 151 |
DISCUSSION
Antigen detection with commercially available monoclonal antibodies remains today the most commonly used method for the detection of the Paramyxoviridae (RSVA, RSVB, and PIV3) associated with bronchiolitis in hospitalized infants (4, 6, 22). The IFA is rapid but less cost effective than the reference technique, VIT. Alternative molecular methods have been developed for detection of RSV (3, 9, 14, 15, 19) and PIV3 (12) sequences in cell culture extracts or nasal aspirates. The N gene has been the focus for most RT-PCR methods because it is one of the most highly transcribed, which could account for better sensitivity, and one of the most conserved, which accounts for greater specificity. Indeed, 96% similarity is observed between the N genes of RSVA and RSVB (11). The primers defined by Cane and Pringle (2) for RSV were used by Cubie et al. (3) as outer primers in a nested PCR and by ourselves in an RT-PCR in which we have defined an internal sequence used as a probe in a nonisotopic hybridization assay (9). Milaan et al. (14) tried to define target sequences for amplification and hybridization in flanking regions of the N gene, close to the end of the proximal 1B gene, and others tried more distal genomic regions like the F and G genes (15, 19). However, all studies found better sensitivity with the N gene (15, 18). For PIV3, an RT-PCR assay has been reported by Karron et al. (12) using primers in a highly conserved region of the hemagglutin-neuraminidase gene.
The weakness of these molecular procedures is that detection is limited to one type of virus. The purpose of this study was to assess a single broad and sensitive assay able to detect and identify the RSVA, RSVB, and PIV3 genomes and transcripts. Because of the great genetic divergence between the targeted viruses, appropriate primers were available only in the most stable genomic region, i.e., catalytic domain III of the L polymerase (1, 16, 17, 21). The possible disadvantage of the L gene could be the poor level of transcription limiting the sensitivity of the assay. However, Sacramento et al. (18) previously demonstrated on rabies virus that RT-PCR sensitivity when amplifying the cDNA to the N gene was somehow similar to that of the cDNA to the N mRNA transcript. This suggested that the mRNAs were more sensitive to cytoplasmic nucleases than (− or +) genomic RNA which remains protected by the N protein within the ribonucleoprotein structure. According to this hypothesis, the disadvantage in sensitivity of targeting the L rather than N gene for diagnosis is reduced.
The L primer sets have been based on the functional motifs A (XTDLXKX) and C (GDNQXIX) within block III of the paramyxovirus L proteins available in the data banks (16, 17) (Fig. 1). To avoid decreasing the efficiency of amplification with the sequence length, they were separated by a short distance (352 to 355 nucleotides). To increase the chance of amplification with genetically divergent isolates, the complementarity between the last 3′ nucleotides of each primer and the template has been optimized by carefully taking into account the flexibility of the genetic code. The P1 (+) primer ends with four very conserved residues: a strictly invariant K codon and the common T nucleotide of a variable Y/F position. The P2 and P3 (−) primers end with the longest continuous stretch of invariant amino acids, GDNQ, between L proteins (17). However, they differ by a single mutation in order to take into account the natural “wobble” position of the G codon.
On clinical and prototype strains, both sets of primers were reactive with RSVA, whereas P1-P2 and P1-P3 seemed to be more specific for RSVB and PIV3, respectively. Thus, the three primers, P1, P2, and P3, were mixed in the same RT-PCR mixture for the simultaneous diagnosis of any RSVA, RSVB, or PIV3 infection in nasal aspirates, with the exclusion of other para- and orthomyxoviruses. However, this method does not differentiate between these viruses. Two typing tools for distinction between RSVA, RSVB, and PIV3 have been evaluated with clinical strains. RFLP analysis of the amplicons by three restriction enzymes, AvaII, PvuII, and BglII, has shown a PvuII site, typical of PIV3 isolates, and an AvaII site, typical of RSV isolates. Further distinction between RSVA and RSVB can be performed by BglII, which cleaves systematically RSVB and only 15% of RSVA at a different site. The differential hybridization assay by an EIA using the three oligonucleotide probes chosen to be specific for each virus and different from one virus to the other exhibited a 100% correlation with IFA for PIV3, being positive with the homologous probe (index value, >1) and systematically negative (index value, <1) with heterologous probe. This indicates the absence of cross-hybridization. However, for RSVA and RSVB strains, the correlation was only 92.5%, since there were two discrepancies among the 27 isolates between the results of IFA and RT-PCR-EIA. One result could be explained by the simultaneous presence of the two viruses, RSVA and RSVB, in a specimen (94001891) identified as RSVB by IFA but which hybridized with the two probes and had no conclusive RFLP pattern. The other result (with specimen no. 9400504) is a clear contradiction between the antigenic and the genetic methods of typing: it was identified as RSVA by IFA but hybridized exclusively with the RSVB probe and showed unambiguously the RSVB RFLP pattern, while the amplicon exhibited the RSVB sequence (not shown).
Among the 261 nasal aspirates, the tendencies observed with the clinical strains were confirmed and enlarged. The IFA-VIT and RT-PCR-EIA tests gave concordant results for 96.9% of the samples for RSV infection and 93.3% for PIV3 infection. This indicates that the RT-PCR-EIA method described here is an alternative diagnosis tool. However, 1 of 14 IFA-VIT samples positive for PIV3 was negative by PCR-EIA. On the other hand, 7 of 109 IFA-VIT samples positive for RSV were negative by PCR amplification, and in 6 additional cases the amplified products could not be hybridized by EIA. Taken together, these data suggest that IFA-VIT is slightly more sensitive than RT-PCR-EIA for diagnosis (41.7% positive samples versus 39.4%, respectively). In contrast, the RT-PCR-EIA method is clearly more efficient than IFA in differentiating RSVA and RSVB samples. It failed to type only 3.7 to 5.8% of the RSV-positive clinical isolates or aspirates, while IFA failed for 23%. It is of note that the two methods are in complete disagreement (RSVA versus RSVB) for 3.7 to 4.7% of the samples without any consistent explanation to date.
Different hypotheses could be proposed to explain the slightly better sensitivity of IFA-VIT over PCR-EIA. The genetic diversity of several RSV isolates could impair the correct hybridization of the primer and/or the probes, despite their selection in the most stable genomic region of the L polymerase (16, 17, 21). The fact that seven RT-PCR-EIA RSV-negative samples were in the group of those impossible to type by IFA is an important argument for their divergent nature. Alternatively, technical impairments could be involved, such as the presence of inhibitors of the enzymatic steps in nasal aspirates or the loss and/or degradation of RNA during storage (−80°C) or the purification procedure. Despite this slight difference, the sensitivity of the RT-PCR-EIA described here is comparable to that of IFA-VIT. Similar observations were made for two earlier PCR assays in the absence of hybridization of the amplified products (3, 15). However, with the use of hybridization, an increased sensitivity over IFA-VIT was reported (9, 14). The reasons for such differences are unclear and still under investigation.
In conclusion, we have developed a molecular method allowing detection in a single step and further typing of paramyxoviruses RSVA, RSVB, and PIV3, which are responsible for lower respiratory tract infections. This method promises to be an efficient and competitive alternative to antigenic detection or to VIT if its use is not constrained by economic and convenience aspects.
ACKNOWLEDGMENTS
We thank all members of the Lyssavirus Laboratory, Institut Pasteur, Paris, France, for moral support and expert technical advice.
C. Bahloul was a recipient of a fellowship from the Tunisian government, and H. Badrane was a recipient of a fellowship from the “Cooperation Franco-Marocaine” and from the “Fondation Merieux.”
REFERENCES
- 1.Banerjee A K. Transcription and replication of rhabdoviruses. Microbiol Rev. 1987;51:66–87. doi: 10.1128/mr.51.1.66-87.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cane P A, Pringle C R. Respiratory syncytial virus heterogeneity during an epidemic: analysis by limited nucleotide sequencing (SH gene) and restriction. J Gen Virol. 1991;72:349–357. doi: 10.1099/0022-1317-72-2-349. [DOI] [PubMed] [Google Scholar]
- 3.Cubie H A, Inglis J M, Leslie E E, Edmunds A T, Totapally B. Detection of respiratory syncytial virus in acute bronchiolitis infants. J Med Virol. 1992;38:283–287. doi: 10.1002/jmv.1890380410. [DOI] [PubMed] [Google Scholar]
- 4.Downham M A, McQuillin J, Gardner P S. Diagnosis and significance of parainfluenza virus infections in children. Arch Dis Child. 1979;49:8–15. doi: 10.1136/adc.49.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freymuth F. Rapid diagnosis of respiratory syncytial virus infections in children. Lancet. 1980;ii:539–540. doi: 10.1016/s0140-6736(80)91866-8. [DOI] [PubMed] [Google Scholar]
- 6.Freymuth F, Quibriac M, Petitjean J, Daon F, Amiel M L. Les virus responsables d’infections respiratoires en pédiatrie. Bilan de 3480 aspirations nasales realisées chez l’enfant en une période de six ans. Ann Pediatr (Paris) 1987;34:493–501. [PubMed] [Google Scholar]
- 7.Freymuth F, Petitjean J, Pothier P, Norby E, Brouard J. Prevalence of respiratory syncytial virus subgroups A and B in France, 1982 to 1990. J Clin Microbiol. 1991;29:653–655. doi: 10.1128/jcm.29.3.653-655.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freymuth F, Petitjean J, Eugene G, Vabret A, Brouard J, Duhamel J F, Guillois B. Diagnostic des infections à virus respiratoire syncytial. Med Mal Infect. 1995;S23:824–829. [PubMed] [Google Scholar]
- 9.Freymuth F, Eugene G, Vabret A, Petitjean J, Gennetay E, Brouard J, Duhamel J F, Guillois B. Detection of respiratory syncytial virus by reverse transcription-PCR and hybridization with a DNA enzyme immunoassay. J Clin Microbiol. 1995;33:3352–3355. doi: 10.1128/jcm.33.12.3352-3355.1995. . (Erratum, 34:1601, 1996.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall C B, McBride J T, Gala C L, Hildreth S W, Schnabel K C. Ribavirin treatment of respiratory syncytial viral infection in infants with underlying cardiopulmonary disease. JAMA. 1985;254:3047–3051. [PubMed] [Google Scholar]
- 11.Johnson P R, Collins P L. The 1B (NS2), 1C (NS1) and N proteins of human respiratory syncytial virus (RSV) of antigenic subgroups A and B: sequence conservation and divergence within RSV genomic RNA. J Gen Virol. 1989;70:1539–1547. doi: 10.1099/0022-1317-70-6-1539. [DOI] [PubMed] [Google Scholar]
- 12.Karron R A, Froehlich J L, Bobo L, Belshe R B, Yolken R H. Rapid detection of parainfluenza virus type 3 RNA in respiratory specimens: use of reverse transcription-PCR-enzyme immunoassay. J Clin Microbiol. 1994;32:484–488. doi: 10.1128/jcm.32.2.484-488.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin A J, Gardner P S, McQuillin J. Epidemiology of respiratory viral infection among paediatric inpatients over a six year period in North East England. Lancet. 1978;ii:1035–1038. doi: 10.1016/s0140-6736(78)92351-6. [DOI] [PubMed] [Google Scholar]
- 14.Milaan A J, Sprenger M J W, Rothbarth P H, Brandeburg A H, Nasurel N, Class E C J. Detection of respiratory syncytial virus by RNA-polymerase chain reaction and differentiation of subgroups with oligonucleotide probes. J Med Virol. 1993;44:80–87. doi: 10.1002/jmv.1890440115. [DOI] [PubMed] [Google Scholar]
- 15.Paton A W, Paton J C, Lawrence A J, Goldwater P N, Harris R J. Rapid detection of respiratory syncytial virus in nasopharyngeal aspirates by reverse transcriptase and polymerase chain reaction amplification. J Clin Microbiol. 1992;30:901–904. doi: 10.1128/jcm.30.4.901-904.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding element. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poch O, Blumberg B M, Bourgueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand viruses: theoretical assignment of functional domains. J Gen Virol. 1990;71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 18.Sacramento D, Bourhy H, Tordo N. PCR technique as an alternative method for diagnosis and molecular epidemiology of rabies virus. Mol Cell Probes. 1991;6:229–240. doi: 10.1016/0890-8508(91)90045-l. [DOI] [PubMed] [Google Scholar]
- 19.Sullender W M, Sun L, Anderson L J. Analysis of respiratory syncytial virus genetic variability with amplified cDNAs. J Clin Microbiol. 1993;31:1224–1231. doi: 10.1128/jcm.31.5.1224-1231.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tordo N, Poch O. Structure of rabies virus. In: Campbell J B, Charlton K M, editors. Rabies. Boston, Mass: Kluwer Academic Publishers; 1988. pp. 25–45. [Google Scholar]
- 21.Tordo N, De Haan P, Goldbach R, Poch O. Evolution of negative-stranded RNA genomes. Semin Virol. 1992;3:341–357. [Google Scholar]
- 22.Welliver R. Natural history of parainfluenza virus infection in childhood. J Paediatr. 1982;101:180–187. doi: 10.1016/s0022-3476(82)80113-3. [DOI] [PubMed] [Google Scholar]