Abstract
Purpose
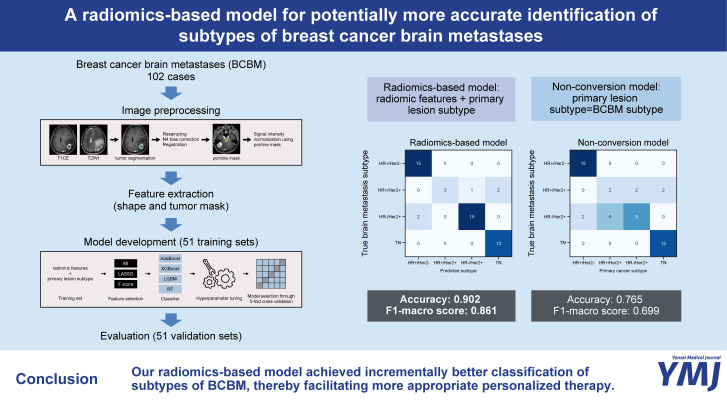
Breast cancer brain metastases (BCBM) may involve subtypes that differ from the primary breast cancer lesion. This study aimed to develop a radiomics-based model that utilizes preoperative brain MRI for multiclass classification of BCBM subtypes and to investigate whether the model offers better prediction accuracy than the assumption that primary lesions and their BCBMs would be of the same subtype (non-conversion model) in an external validation set.
Materials and Methods
The training and external validation sets each comprised 51 cases (102 cases total). Four machine learning classifiers combined with three feature selection methods were trained on radiomic features and primary lesion subtypes for prediction of the following four subtypes: 1) hormone receptor (HR)+/human epidermal growth factor receptor 2 (HER2)-, 2) HR+/HER2+, 3) HR-/HER2+, and 4) triple-negative. After training, the performance of the radiomics-based model was compared to that of the non-conversion model in an external validation set using accuracy and F1-macro scores.
Results
The rate of discrepant subtypes between primary lesions and their respective BCBMs were 25.5% (n=13 of 51) in the training set and 23.5% (n=12 of 51) in the external validation set. In the external validation set, the accuracy and F1-macro score of the radiomics-based model were significantly higher than those of the non-conversion model (0.902 vs. 0.765, p=0.004; 0.861 vs. 0.699, p=0.002).
Conclusion
Our radiomics-based model represents an incremental advance in the classification of BCBM subtypes, thereby facilitating a more appropriate personalized therapy.
Keywords: Breast cancer, brain metastasis, radiomics, magnetic resonance imaging
Graphical Abstract
INTRODUCTION
Breast cancer is the second most common cancer that metastasizes to the central nervous system after lung cancer, and the incidence of breast cancer brain metastases (BCBM) is increasing with advances in systemic therapy and neuroimaging.1,2 Breast cancer is clinically classified according to estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) subtypes, and subtype identification is a key factor to prolonging survival and maintaining quality of life in the treatment of metastatic breast cancer.3 For example, hormone receptor (HR)-positive breast cancer can be treated using endocrine therapy. For patients with HER2-overexpressing breast cancer, HER2-targeted therapy, such as trastuzumab, is now routinely administered primarily in combination with adjuvant chemotherapy.4 Meanwhile, for triple-negative breast cancer (TNBC), which is known to be the most malignant breast cancer subtype with a poor prognosis, standardized TNBC treatment regimens are lacking.5
Discrepancies in subtypes between primary breast cancer and metastatic breast cancer are increasingly being reported, with overall discordant rates up to 40.3%.6,7,8,9,10,11,12,13,14,15,16 Such variation can have a significant impact on patient treatment strategies, responses to treatment, and ultimately, prognosis.10,12,17,18,19,20 While a unanimous consensus is currently lacking in treatment guidelines regarding the biological features that should guide treatment decision-making in cases of subtype discrepancy, identifying the subtypes of metastatic breast cancer holds clinical significance. For example, in patients with discordant HER2 status, where it was negative at baseline but positive in the metastatic setting, the use of a HER2-targeted therapy should be considered regardless of timing. Additionally, for patients with metastatic TNBC, irrespective of the subtype in the primary breast cancer, it is advisable to consider the use of chemotherapy with or without immune checkpoint inhibitor therapy or targeted therapies like antibody-drug conjugates.21 In this context, the American Society of Clinical Oncology and the Joint European Association of Neuro-Oncology–European Society for Medical Oncology guidelines recommend evaluating the receptor expression status of distant metastases of breast cancer through biopsy whenever possible.21,22 However, obtaining BCBM material from all patients through surgery or biopsy may not be practical or feasible depending on the condition of the patient or the location of the lesion. In addition, considering the risk of surgery and possible bias in obtaining samples due to the intrinsic heterogeneity of BCBM lesions, it should be considered that these procedures cannot always give accurate results.7,9,13 These issues highlight the need for innovative approaches to identifying biomarkers for metastatic cancer.
Radiomics is an emerging field in medical imaging that extracts high-dimensional features from images to reveal potential biological characteristics of tumors.23 Several previous studies have proposed radiomics-based models to predict the subtype of primary breast cancer. However, their usefulness has yet to be extensively investigated in BCBM.24,25,26,27
Therefore, the goal of this study was to develop a radiomics-based model for multiclass classification of the subtypes of BCBM using preoperative brain MRI and to investigate whether radiomic features could improve the accuracy of classification over the presumption that BCBM subtypes would be identical to those of the primary breast cancer lesion.
MATERIALS AND METHODS
Study population
This retrospective study was approved by the Gangnam Severance Hospital Institutional Review Board (no. 3-2022-0371), and the requirement for obtaining informed consent was waived.
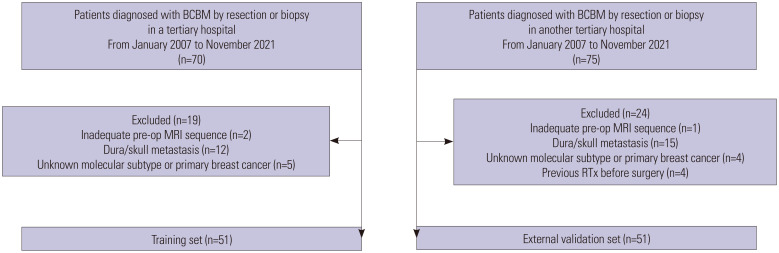
Patients with brain metastases from primary breast cancer, confirmed by surgical resection or biopsy between January 2007 and November 2021, were identified at two different tertiary hospitals. Patients from one hospital were assigned to the training set, whereas patients from the other hospital were designated as the external validation set. The inclusion criteria were as follows: 1) patients with brain parenchymal metastases from breast cancer confirmed by surgical resection or biopsy and 2) patients who underwent preoperative brain MRI including both T2-weighted imaging (T2WI) and 3D gradient echo contrast-enhanced T1-weighted imaging (T1CE) sequences. The exclusion criteria were as follows: 1) absence of any T2WI or T1CE sequence in preoperative MRI (n=2 in the training set, n=1 in the external validation set); 2) dura or skull metastasis that was surgically resected (n=12 in the training set, n=15 in the external validation set); 3) unknown molecular subtype of primary breast cancer (n=5 in the training set, n=4 in the external validation set); and 4) history of previous radiation therapy on the resected lesion before surgery (n=4 in the external validation set). Consequently, 51 and 51 patients (102 patients in total) constituted the training and external validation sets, respectively (Fig. 1).
Fig. 1. Flowchart of the study population for the training and external validation sets. BCBM, breast cancer brain metastases; RTx, radiation therapy.
Pathologic examination
Pathological diagnosis and subtype were determined based on postoperative tissue samples according to the American Society of Clinical Oncology/College of American Pathologists guidelines.28 For ER and PR, greater than 1% expression indicated positivity, and HR-positive was defined as either ER- or PR-positive. For HER2, a score of 3+ on immunohistochemistry indicated positivity. In cases of equivocal expression of HER2 (2+), 2+ on fluorescence in situ hybridization or HER2 amplification on next-generation sequencing indicated positivity. Finally, subtypes were classified into four classes for further analysis: 1) HR-positive and HER2-negative (HR+/HER2-), 2) HR-positive and HER2-positive (HR+/HER2+), 3) HR-negative and HER2-positive (HR-/HER2+), and 4) triple-negative (TN).
Image acquisition
Magnetic resonance (MR) scans of the training set were acquired using 3-T scanners (MAGNETOM Vida, Siemens Healthineers, Erlangen, Germany; Discovery MR750, GE Healthcare, Chicago, IL, USA) with an eight-channel head coil. MR scans of the external validation set were performed using 3-T MR scanners (Achieva or Ingenia; Philips Medical Systems, Amsterdam, Netherlands) with an eight-channel head coil. Imaging protocols in both institutions included T2WI and T1CE after intravenous administration of a gadolinium-based contrast agent (ProHance, 0.2 mL/kg; Bracco, Milan, Italy). The imaging parameters for the training set were as follows: 1) T2WI: repetition time/echo time (TR/TE), 4000–6000/100 ms; section thickness, 5 mm; field of view (FOV), 230 mm; matrix, 352×352–512×325. 2) T1CE: TR/TE, 8.2/3.2 ms; section thickness, 1 mm; FOV, 230 mm; matrix, 256×256 or 224×224. The imaging parameters for the external validation set were as follows: 1) T2WI: TR/TE, 4000–9000/80–120 ms; section thickness, 5 mm; FOV, 220–240 mm; matrix, 256×256. 2) T1CE: TR/TE, 6.3–8.3/3.1–4 ms; section thickness, 1 mm; FOV, 240 mm; matrix, 192×192.
Image processing
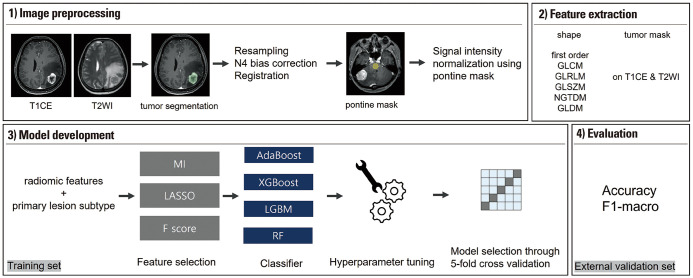
A schematic of the pipeline is shown in Fig. 2. Semi-automated segmentation, including signal intensity thresholding and region growing, was performed using 3D slicer software (version 4.11.0; https://www.slicer.org/) on T1CE images by a neuroradiologist with 4 years of experience in neuro-oncological imaging. In cases where there were multiple metastatic lesions, only the resected lesion was segmented and used for further processing in each patient. Additionally, if there were multiple lesions resected, we specifically segmented the largest one.
Fig. 2. Study pipeline. T2WI, T2-weighted imaging; T1CE, 3D gradient echo contrast-enhanced T1-weighted imaging; GLCM, gray-level co-occurrence matrix; GLRLM, gray-level run length matrix; GLSZM, gray-level size zone matrix; NGTDM, neighboring gray tone difference matrices; GLDM, gray-level dependence matrix; MI, mutual information; LASSO, least absolute shrinkage and selection operator; AdaBoost, Adaptive Boosting; XGBoost, Extreme Gradient Boosting; LGBM, LightGBM: RF, Random Forest.
Next, resampling of T1CE, T2WI, and tumor segmentation masks into a uniform voxel size of 1×1×1 mm3, N4 bias-correction, and co-registration of T2WI to T1CE were sequentially performed using Advanced Normalization Tools (https://stnava.github.io/ANTs).29 Finally, the image signal intensities were normalized as follows: First, pontine masks were manually drawn on the central portion of the pons across seven consecutive slices (mean volume of the masks: 1224 mm3) on the preprocessed T1CE images. Second, mean and standard deviation values of the signal intensity of the pontine masks were calculated for the preprocessed T1CE and T2WI images, respectively. Third, Z-score normalization of the segmented tumor lesion was performed using the mean and standard deviation values of the pontine masks on the respective preprocessed T1CE and T2WI images for each patient.
Radiomic feature extraction
Radiomic features were extracted using the PyRadiomics Python package (version 3.0.1; https://pyradiomics.readthedocs.io/).30 Eighteen first-order features and 75 texture features, including 24 gray-level co-occurrence matrices, 16 gray-level size zone matrices, 16 gray-level run length matrices, 5 neighboring gray tone difference matrices, and 14 gray-level dependence matrices, were extracted from the tumor masks on the preprocessed T1CE and T2WI images, with a bin number of 32 to discretize intensities. In addition, 14 shape features were extracted from the tumor masks. Consequently, 200 radiomic features were extracted for each examination. The radiomic features followed the standard sets of the Image Biomarker Standardization Initiative.31
Model development
Radiomics-based machine learning models for multiclass classification were developed using Python 3 with the Scikit-learn library (version 1.2.0; https://scikit-learn.org/). For radiomics-based models, the original subtype of primary breast cancer was added as a feature along with the radiomic features. In the non-conversion model, the BCBM subtype was presumed to be identical to that of the primary breast cancer.
Before development of the radiomics-based model, all feature values were scaled using the ‘RobustScaler’ function of the Scikit-learn library. Next, feature reduction was performed using three different methods: 1) univariate selection based on F score, using the ‘f_classif’ function of the Scikit-learn library; 2) univariate selection based on mutual information, using the ‘mutual_info_classif’ function of the Scikit-learn library; and 3) least absolute shrinkage and selection operator (LASSO), using the ‘LassoCV’ function of the Scikit-learn library. The following four machine learning classifiers were used in combination with the three feature selection methods: 1) Adaptive Boosting (AdaBoost), 2) Extreme Gradient Boosting (XGBoost), 3) LightGBM (LGBM), and 4) Random Forest. For each combination of feature selection and classification method, hyperparameters, such as learning rate, number of estimators, maximum depth, or gamma, were optimized through stratified 5-fold cross-validation in the training set, and the model with the highest accuracy was chosen as the best performing model.
Model performance evaluation and explanation
The best performing model in the 5-fold cross-validation in the training set was selected as the radiomics-based model, and its diagnostic performance was compared with that of the non-conversion model in the external validation set using accuracy and F1-macro score as diagnostic metrics. Accuracy was compared using the generalized estimating equation (GEE), and F1-macro score was compared using bootstrapping with 1000 repetitions. Additionally, to compare the diagnostic performance of the two models for each subtype, the correct classification rate was analyzed using GEE. Statistical analyses were performed using SAS (version 9.4, SAS Institute Inc., Cary, NC, USA). p<0.05 was considered statistically significant.
We also investigated which features contributed the most to the decision of the radiomics-based model by Shapley additive explanations (SHAP) value analysis, which is a method originally based on cooperative game theory and used to offer a high level of interpretability of machine learning models by assigning an importance value to each feature for a particular prediction, using the “shap” Python library.32
RESULTS
The overall patient demographics of the training and external validation sets are presented in Table 1. The median size of brain metastatic lesions on which the radiomics study was performed in the training set was found to be significantly smaller than that of the external validation set [8106.4 (interquartile range: 5254.2–15226.6) mm3 vs. 14786.8 (9901.2–26713.6) mm3, p<0.001]. Deep gray matter lesions were included only in the training set, while cerebellar lesions were more common in the external validation set. There were no other significant differences between the two groups. In both the training and external validation sets, all patients underwent surgical resection, and no instances of biopsy were recorded. Approximately 25% of patients exhibited subtype conversion in both the training and external validation sets. The respective conversion rates in the training and external validation sets were as follows: in the training set, 1) HR+/HER2- to TN (n=1), 2) HR+/HER2+ to HR+/HER2- (n=2), 3) HR+/HER2+ to HR-/HER2+ (n=5), 4) HR+/HER2+ to TN (n=1), 5) HR-/HER2+ to HR+/HER2+ (n=3), and 6) TN to HR-/HER2+ (n=1); in the external validation set, 1) HR+/HER2- to HR-/HER2+ (n=2), 2) HR+/HER2+ to HR-/HER2+ (n=6), 3) HR-/HER2+ to HR+/HER2+ (n=2), and 4) TN to HR+/HER2+ (n=2).
Table 1. Demographics and Clinical Characteristics of the Patients.
| Training set (n=51) | External validation set (n=51) | p value | ||
|---|---|---|---|---|
| Age (yr) | 49.8±9.1 | 51.8±10.3 | 0.306 | |
| Interval between MRI and operation (day) | 6.2± 6.0 | 6.3±5.6 | 0.959 | |
| Size of brain lesion (mm3)* | 8106.4 (5254.2–15226.6) | 14786.8 (9901.2–26713.6) | <0.001 | |
| Location | 0.033 | |||
| Cerebellum | 15 (29.4) | 23 (45.1) | ||
| Deep gray matter | 5 (9.8) | 0 (0.0) | ||
| Cerebral lobes | 31 (60.8) | 28 (54.9) | ||
| Molecular subtype of brain lesion | 0.573 | |||
| HR+/HER2- | 9 (17.6) | 15 (29.4) | ||
| HR+/HER2+ | 7 (13.7) | 6 (11.8) | ||
| HR-/HER2+ | 19 (37.3) | 17 (33.3) | ||
| TN | 16 (31.4) | 13 (25.5) | ||
| Molecular subtype of primary lesion | 0.174 | |||
| HR+/HER2- | 8 (15.7) | 17 (33.3) | ||
| HR+/HER2+ | 12 (23.5) | 8 (15.7) | ||
| HR-/HER2+ | 16 (31.4) | 11 (21.6) | ||
| TN | 15 (29.4) | 15 (29.4) | ||
| Molecular subtype conversion | 13 (25.5) | 12 (23.5) | >0.999 | |
HR, hormone receptor; HER2, human epidermal growth factor receptor 2; TN, triple negative.
*The size of the brain metastatic lesions on which the radiomics study was performed is reported using the median value and interquartile range after undergoing a normality test. A comparison between the two median values was performed using the Mann-Whitney test.
The best performance was achieved when LASSO was used as a feature selection method for all four classifiers, with the following four features selected: “glszm_LargeAreaLowGrayLevelEmphasis” on the T1CE image, “gldm_LargeDependenceLowGrayLevelEmphasis” on the T2WI image, “firstorder_Energy” on the T2WI image, and the original subtype of the primary breast cancer. After hyperparameter tuning, the best accuracies of the four classifiers on the 5-fold cross-validation in the training set were as follows: AdaBoost, 0.651±0.194; XGBoost, 0.805±0.084; LGBM, 0.784±0.148; and Random Forest, 0.725±0.116. Therefore, a combination of the LASSO selection method and XGBoost was selected as the radiomics-based model.
In the training set, the accuracy and F1-macro score of the non-conversion model were 74.5% [95% confidence interval (CI): 62.5–86.5] and 72.3% (95% CI: 60.0–84.6), respectively. In contrast, the radiomics-based model achieved an accuracy of 90.2% (95% CI: 82.0–98.4) and an F1-macro score of 88.5% (95% CI: 79.8–97.3).
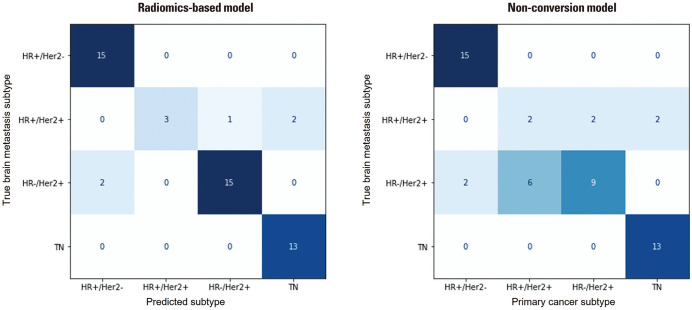
In the external validation set, the accuracy of the radiomics-based model was significantly higher than that of the non-conversion model [90.2% (95% CI: 82.0–98.4) vs. 76.5% (95% CI: 64.8–88.1), p=0.004]. The radiomics-based model also showed significantly higher F1-macro score than the non-conversion model [86.1% (95% CI: 74.7–97.3) vs. 69.9% (95% CI: 59.6–82.4), p=0.002] (Table 2). The radiomics-based model successfully corrected seven of the 12 subtype conversion cases in the external validation set: six cases were HR-/HER2+ subtype converted from HR+/HER2+, and the other case was HR+/HER2+ subtype converted from HR-/HER2+. In analysis of correct classification rates for each subtype, there was a significant difference in the prediction performance for the HR-/HER2+ subtype between the two models (p=0.002) (Fig. 3 and Supplementary Table 1, only online).
Table 2. Diagnostic Performance of the Radiomics-Based and Non-Conversion Models in the Training and External Validation Sets.
| Model | Training set | External validation set | ||||
|---|---|---|---|---|---|---|
| Accuracy | F1-macro score | Accuracy | p value | F1-macro score | p value | |
| Radiomics-based model | 0.902 (0.820–0.984) | 0.885 (0.798–0.973) | 0.902 (0.820–0.984) | 0.004 | 0.861 (0.747–0.973) | 0.002 |
| Non-conversion model | 0.745 (0.625–0.865) | 0.723 (0.600–0.846) | 0.765 (0.648–0.881) | 0.699 (0.596–0.824) | ||
Parentheses indicate 95% confidence intervals.
Fig. 3. Confusion matrices for the radiomics-based and non-conversion models in the external validation set. HR, hormone receptor; HER2, human epidermal growth factor receptor 2; TN, triple negative.
According to SHAP value analysis of the radiomics-based model on the external validation set, the top two most contributing features for the multiclass classification were primary lesion subtype and the ‘firstorder_energy’ feature on T2WI. A variance importance plot, summary plots, and waterfall plots with representative cases of the external validation set are shown in Supplementary Figs. 1 and 2 (only online).
DISCUSSION
In this study, we developed a radiomics-based model for multiclass classification of BCBM subtypes that incorporates both radiomic features and information on the subtype of primary breast lesions. We showed that the radiomics-based model significantly improved subtype prediction over the presumption that the subtype of BCBM is identical to that of the primary breast cancer lesion, with an accuracy of 0.902 (95% CI, 0.820–0.984) and an F1-marco score of 0.861 in an external validation set.
Approximately 25% of patients exhibited subtype conversion in both the training and external validation sets. Among the subtype conversion cases, the most frequent conversion was from HR+/HER2+ to HR-/HER2+ in both the training set (5/13) and the external validation set (6/12). Notably, for both HR and HER, positive to negative conversion was more frequently observed than negative to positive conversion in both the training and external validation sets: for HR, n=7 vs. n=3 in the training set and n=8 vs. n=4 in the external validation set; for HER, n=3 vs. n=1 in the training set and n=4 vs. n=0 in the external validation set. This tendency is consistent with previous research on subtype discordance. According to a recent meta-analysis study, including 39 studies assessing receptor conversion between primary and metastatic breast cancer, positive to negative receptor conversions occurred more frequently than from negative to positive.16
Radiomics, which is used for medical image analysis by converting medical images into quantitative data, can improve the accuracy of diagnosis, evaluate prognosis and response, and ultimately lead to better clinical decisions.33 Many radiomics research models have investigated breast cancer receptor expression and molecular subtypes. Leithner, et al.26 applied a radiomics model to differentiate receptor status and molecular subtypes (luminal A, luminal B, TN) of breast cancer and reported that radiomics-based models were useful for the assessment of breast cancer subtypes with >80% accuracy.34 In addition, a radiomics approach has been reported to have predictive value not only for distinguishing TNBC from other breast cancer subtypes, but also for distinguishing different TNBC molecular subtypes, as well as prognostic value in patients with TNBC.35,36 However, radiomics studies for predicting subtypes in breast cancer metastases are limited, with, to the best of our knowledge, only one study published to date. Luo, et al.37 constructed machine learning-based radiomic signatures and predicted the receptor status of BCBM on preoperative brain MRI with accuracies of 78.3%, 82.6%, and 82.6% for ER, PR, and HER2, respectively, in a test set.
We developed a radiomics-based machine-learning model capable of multiclass classification of BCBM. Because our model was not restricted to binary classification, it could be more relevant and useful in clinical practice, given that breast cancer subtypes are classified into multiple subtypes. In addition, the diagnostic performance of our model was validated using an external validation set and measured using F1-macro score, which is an appropriate metric for multiclass classification.38 Our model successfully corrected seven of the 12 subtype conversion cases in the external validation set. In particular, all six BCBM cases with the HR-/HER2+ subtype, which were converted from the HR+/HER2+ subtype, were accurately identified using our model. Interestingly, all conversion cases (n=12) in the external validation showed HR status conversion, and all corrected predictions of the conversion cases (n=7) resulted from the correction of HR status, whereas the radiomics-based model did not improve the prediction of HER2 status over the non-conversion model. The small number of HER2 receptor conversions (n=4) that occurred in the external validation set may be a contributing factor. In addition, we cannot rule out that HER2 receptor status has little or no effect on imaging features. Further studies with larger sample sizes are warranted to address this issue.
We additionally attempted to explain the results of our model by applying SHAP value analysis, which assigns an importance value to each feature for a particular prediction.32 According to the SHAP value analysis of the external validation set, the subtype of the primary breast cancer lesion contributed the most to subtype prediction, which was to be expected given the conversion rate of 23.5%. The most contributing radiomic feature was the “firstorder_energy” feature on T2WI, which is a measure of the magnitude of voxel gray values in an image. Interestingly, for HR+/HER2+ subtype prediction, the largest contributing factor was not the primary lesion subtype, but the “firstorder_energy” feature on T2WI. This may be attributed to the fact that half of the conversion cases occurred in the HR+/HER2+ primary lesion subtype and that one-third of the conversion cases changed to the HR+/HER2+ subtype in BCBM. Despite attempts to interpret the results using the SHAP value analysis, however, the nature of multiclass classification in this study’s design hindered an intuitive interpretation. To solve the current “black box” problem of radiomics-based models and hence make this approach more applicable in clinical practice, additional research should be undertaken to better interpret the relationship between radiomics-based predictors and outcomes.
Notably, our model showed comparable performance in both the training and external validation sets. We postulate that the signal intensity normalization method using the mean and standard deviation values of signal intensity of the pons area was robust for signal normalization while preserving the signal characteristics of the tumor lesions. Owing to the multiplicity of BCBM with accompanying peritumoral edema, commonly used signal normalization methods, such as the Nyul or WhiteStripe methods, were inapplicable in this study.39,40 Since the pons is relatively less affected by brain metastases, it may serve as a reliable reference for brain parenchymal signal intensity in radiomics studies of brain metastases.
Our study had some limitations. As this was a retrospective study, there may be selection bias. However, we minimized selection bias by consecutively including all patients who underwent surgery or biopsy during the defined time period in the dataset. Second, the sample size may not have been sufficiently large. The number of available cases with complete pathological information, including subtype, for both primary cancer and BCBM was restricted. Third, all corrected predictions of the conversion cases resulted from the correction of HR status. The limited number of HER2 receptor conversions that occurred in the external validation set (n=4) may be a contributing factor. In addition, while our study primarily focused on the development and validation of the radiomics-based model, it is important to note that the implementation of the model in real-world clinical settings remains unexplored. To address these important aspects, we are planning to conduct a prospective study with a larger sample size. This future study aims to thoroughly investigate the model’s performance and evaluate its impact on clinical outcomes, thereby facilitating its practical utilization by clinicians. Finally, we conducted semi-automated segmentation of BCBM lesions, which might have introduced a certain level of bias. However, in this study, it was necessary to manually verify that only resected tumor lesions were segmented; hence, algorithms recently developed for automatic segmentation of brain tumors were not universally applicable.
In conclusion, our radiomics-based model has the potential for more accurately identifying subtypes of BCBR than the assumption that the primary lesion and its metastatic brain lesions are of the same subtype, thereby facilitating a more appropriate personalized therapy for metastatic breast cancer.
ACKNOWLEDGEMENTS
The authors acknowledge the statistical support of the Biostatistics Collaboration Unit, Medical Research Support Services of Yonsei University College of Medicine.
This study was supported by Bracco Imaging Korea (3-2022-0371 to B.J.); This study was partially supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI21C1161 to S.S.A.).
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Bio Joo.
- Data curation: Mina Park, Sung Jun Ahn, Yae Won Park, and Sung Soo Ahn.
- Formal analysis: Bio Joo.
- Funding acquisition: Bio Joo and Sung Soo Ahn.
- Investigation: Seonghyeon Cho and Bio Joo.
- Methodology: Bio Joo.
- Resources: Bio Joo and Sung Soo Ahn.
- Supervision: Sang Hyun Suh and Seung-Koo Lee.
- Validation: Bio Joo.
- Visualization: Bio Joo.
- Writing—original draft: Seonghyeon Cho.
- Writing—review & editing: Bio Joo.
- Approval of final manuscript: all authors.
DATA AVAILABILITY STATEMENT
Radiomic feature data are available upon reasonable request to the corresponding author. Image data is the property of each institution.
SUPPLEMENTARY MATERIALS
Correct Classification Rate for Each Subtype in the External Validation Set
Variance importance plot (A) and summary plots for each prediction (B) in the external validation set.
Representative case in which a subtype conversion from HR+/HER2+ to HR-/HER2+ occurred. T1CE and T2WI images are shown in (A). In the waterfall plot of the case in (B), red arrows indicate feature effects driving the prediction value higher, and blue arrows indicate effects driving the prediction value lower, depicted with the magnitude of the corresponding feature’s effect. Feature 0, feature 1, feature 2, and feature 3 denote “glszm_LargeAreaLowGrayLevelEmphasis” on the T1CE image, “gldm_LargeDependenceLowGrayLevelEmphasis” on the T2WI image, “firstorder_Energy” on the T2WI image, and the primary lesion subtype, respectively. The value of 2 for feature 3 indicates the HR+/HER2+ subtype of the primary lesion. For comparison, T1CE and T2WI images of the cases with the highest SHAP values for prediction of HR+/HER2+ subtype and HR-/HER2+ subtype are depicted in (C) and (D), respectively. The predictions of the model were correct in both cases.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Achrol AS, Rennert RC, Anders C, Soffietti R, Ahluwalia MS, Nayak L, et al. Brain metastases. Nat Rev Dis Primers. 2019;5:5. doi: 10.1038/s41572-018-0055-y. [DOI] [PubMed] [Google Scholar]
- 3.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 4.Higgins MJ, Baselga J. Targeted therapies for breast cancer. J Clin Invest. 2011;121:3797–3803. doi: 10.1172/JCI57152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Zhang H, Merkher Y, Chen L, Liu N, Leonov S, et al. Recent advances in therapeutic strategies for triple-negative breast cancer. J Hematol Oncol. 2022;15:121. doi: 10.1186/s13045-022-01341-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, Qarmali M, Siegal GP, Wei S. Receptor conversion in metastatic breast cancer: analysis of 390 cases from a single institution. Mod Pathol. 2020;33:2499–2506. doi: 10.1038/s41379-020-0615-z. [DOI] [PubMed] [Google Scholar]
- 7.Yeung C, Hilton J, Clemons M, Mazzarello S, Hutton B, Haggar F, et al. Estrogen, progesterone, and HER2/neu receptor discordance between primary and metastatic breast tumours-a review. Cancer Metastasis Rev. 2016;35:427–437. doi: 10.1007/s10555-016-9631-3. [DOI] [PubMed] [Google Scholar]
- 8.Priedigkeit N, Hartmaier RJ, Chen Y, Vareslija D, Basudan A, Watters RJ, et al. Intrinsic subtype switching and acquired ERBB2/HER2 amplifications and mutations in breast cancer brain metastases. JAMA Oncol. 2017;3:666–671. doi: 10.1001/jamaoncol.2016.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timmer M, Werner JM, Röhn G, Ortmann M, Blau T, Cramer C, et al. Discordance and conversion rates of progesterone-, estrogen-, and HER2/neu-receptor status in primary breast cancer and brain metastasis mainly triggered by hormone therapy. Anticancer Res. 2017;37:4859–4865. doi: 10.21873/anticanres.11894. [DOI] [PubMed] [Google Scholar]
- 10.Pedrosa RMSM, Mustafa DA, Soffietti R, Kros JM. Breast cancer brain metastasis: molecular mechanisms and directions for treatment. Neuro Oncol. 2018;20:1439–1449. doi: 10.1093/neuonc/noy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulsbergen AFC, Claes A, Kavouridis VK, Ansaripour A, Nogarede C, Hughes ME, et al. Subtype switching in breast cancer brain metastases: a multicenter analysis. Neuro Oncol. 2020;22:1173–1181. doi: 10.1093/neuonc/noaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperduto PW, Mesko S, Li J, Cagney D, Aizer A, Lin NU, et al. Estrogen/progesterone receptor and HER2 discordance between primary tumor and brain metastases in breast cancer and its effect on treatment and survival. Neuro Oncol. 2020;22:1359–1367. doi: 10.1093/neuonc/noaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan AJ, Giannoudis A, Palmieri C. The genomic landscape of breast cancer brain metastases: a systematic review. Lancet Oncol. 2021;22:e7–e17. doi: 10.1016/S1470-2045(20)30556-8. [DOI] [PubMed] [Google Scholar]
- 14.Karagöz Özen DS, Ozturk MA, Aydin Ö, Turna ZH, Ilvan S, Özgüroglu M. Receptor expression discrepancy between primary and metastatic breast cancer lesions. Oncol Res Treat. 2014;37:622–626. doi: 10.1159/000368312. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y, Tong Y, Chen X, Shen K. Association of biomarker discrepancy and treatment decision, disease outcome in recurrent/metastatic breast cancer patients. Front Oncol. 2021;11:638619. doi: 10.3389/fonc.2021.638619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrijver WAME, Suijkerbuijk KPM, van Gils CH, van der Wall E, Moelans CB, van Diest PJ. Receptor conversion in distant breast cancer metastases: a systematic review and meta-analysis. J Natl Cancer Inst. 2018;110:568–580. doi: 10.1093/jnci/djx273. [DOI] [PubMed] [Google Scholar]
- 17.Hanley BP, Walsh SM, O’Leary DP, MacNally SP, Power C, Farrell M, et al. The significance of receptor status discordance between breast cancer primary and brain metastasis. Breast J. 2018;24:683–685. doi: 10.1111/tbj.13028. [DOI] [PubMed] [Google Scholar]
- 18.Xiao W, Li X, Yang A, Chen B, Zheng S, Zhang G, et al. Analysis of prognostic factors affecting the brain metastases free survival and survival after brain metastases in breast cancer. Front Oncol. 2020;10:431. doi: 10.3389/fonc.2020.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi ZB, Yu P, Zhang S, Wang WN, Han YQ, Ouyang QC, et al. Profile and outcome of receptor conversion in breast cancer metastases: a nation-wide multicenter epidemiological study. Int J Cancer. 2021;148:692–701. doi: 10.1002/ijc.33227. [DOI] [PubMed] [Google Scholar]
- 20.Zhao W, Sun L, Dong G, Wang X, Jia Y, Tong Z. Receptor conversion impacts outcomes of different molecular subtypes of primary breast cancer. Ther Adv Med Oncol. 2021;13:17588359211012982. doi: 10.1177/17588359211012982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gennari A, André F, Barrios CH, Cortés J, de Azambuja E, DeMichele A, et al. ESMO clinical practice guideline for the diagnosis, staging and treatment of patients with metastatic breast cancer. Ann Oncol. 2021;32:1475–1495. doi: 10.1016/j.annonc.2021.09.019. [DOI] [PubMed] [Google Scholar]
- 22.Van Poznak C, Somerfield MR, Bast RC, Cristofanilli M, Goetz MP, Gonzalez-Angulo AM, et al. Use of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2015;33:2695–2704. doi: 10.1200/JCO.2015.61.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 24.Crivelli P, Ledda RE, Parascandolo N, Fara A, Soro D, Conti M. A new challenge for radiologists: radiomics in breast cancer. Biomed Res Int. 2018;2018:6120703. doi: 10.1155/2018/6120703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davey MG, Davey MS, Boland MR, Ryan ÉJ, Lowery AJ, Kerin MJ. Radiomic differentiation of breast cancer molecular subtypes using pre-operative breast imaging - A systematic review and meta-analysis. Eur J Radiol. 2021;144:109996. doi: 10.1016/j.ejrad.2021.109996. [DOI] [PubMed] [Google Scholar]
- 26.Leithner D, Horvat JV, Marino MA, Bernard-Davila B, Jochelson MS, Ochoa-Albiztegui RE, et al. Radiomic signatures with contrast-enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: initial results. Breast Cancer Res. 2019;21:106. doi: 10.1186/s13058-019-1187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Son J, Lee SE, Kim EK, Kim S. Prediction of breast cancer molecular subtypes using radiomics signatures of synthetic mammography from digital breast tomosynthesis. Sci Rep. 2020;10:21566. doi: 10.1038/s41598-020-78681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Arch Pathol Lab Med. 2010;134:e48–e72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- 29.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. [cited 2023 March 1];arXiv [Preprint] 2016 doi: 10.48550/arXiv.1612.07003. Available at: [DOI] [Google Scholar]
- 32.Lundberg S, Lee SI. A unified approach to interpreting model predictions. [cited 2023 March 1];arXiv [Preprint] 2017 doi: 10.48550/arXiv.1705.07874. Available at: [DOI] [Google Scholar]
- 33.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leithner D, Bernard-Davila B, Martinez DF, Horvat JV, Jochelson MS, Marino MA, et al. Radiomic signatures derived from diffusion-weighted imaging for the assessment of breast cancer receptor status and molecular subtypes. Mol Imaging Biol. 2020;22:453–461. doi: 10.1007/s11307-019-01383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang L, You C, Xiao Y, Wang H, Su GH, Xia BQ, et al. Radiogenomic analysis reveals tumor heterogeneity of triple-negative breast cancer. Cell Rep Med. 2022;3:100694. doi: 10.1016/j.xcrm.2022.100694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng X, Xia L, Sun S. A pre-operative MRI-based brain metastasis risk-prediction model for triple-negative breast cancer. Gland Surg. 2021;10:2715–2723. doi: 10.21037/gs-21-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo X, Xie H, Yang Y, Zhang C, Zhang Y, Li Y, et al. Radiomic signatures for predicting receptor status in breast cancer brain metastases. Front Oncol. 2022;12:878388. doi: 10.3389/fonc.2022.878388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erickson BJ, Kitamura F. Magician’s corner: 9. Performance metrics for machine learning models. Radiol Artif Intell. 2021;3:e200126. doi: 10.1148/ryai.2021200126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Med Imaging. 2000;19:143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- 40.Shinohara RT, Sweeney EM, Goldsmith J, Shiee N, Mateen FJ, Calabresi PA, et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 2014;6:9–19. doi: 10.1016/j.nicl.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correct Classification Rate for Each Subtype in the External Validation Set
Variance importance plot (A) and summary plots for each prediction (B) in the external validation set.
Representative case in which a subtype conversion from HR+/HER2+ to HR-/HER2+ occurred. T1CE and T2WI images are shown in (A). In the waterfall plot of the case in (B), red arrows indicate feature effects driving the prediction value higher, and blue arrows indicate effects driving the prediction value lower, depicted with the magnitude of the corresponding feature’s effect. Feature 0, feature 1, feature 2, and feature 3 denote “glszm_LargeAreaLowGrayLevelEmphasis” on the T1CE image, “gldm_LargeDependenceLowGrayLevelEmphasis” on the T2WI image, “firstorder_Energy” on the T2WI image, and the primary lesion subtype, respectively. The value of 2 for feature 3 indicates the HR+/HER2+ subtype of the primary lesion. For comparison, T1CE and T2WI images of the cases with the highest SHAP values for prediction of HR+/HER2+ subtype and HR-/HER2+ subtype are depicted in (C) and (D), respectively. The predictions of the model were correct in both cases.
Data Availability Statement
Radiomic feature data are available upon reasonable request to the corresponding author. Image data is the property of each institution.