Abstract
Ferredoxins are a family of iron–sulfur (Fe-S) cluster proteins that serve as essential electron donors in numerous cellular processes that are conserved through evolution. The promiscuous nature of ferredoxins as electron donors enables them to participate in many metabolic processes including steroid, heme, vitamin D, and Fe-S cluster biosynthesis in different organisms. However, the unique natural function(s) of each of the two human ferredoxins (FDX1 and FDX2) are still poorly characterized. We recently reported that FDX1 is both a crucial regulator of copper ionophore-induced cell death and serves as an upstream regulator of cellular protein lipoylation, a mitochondrial lipid-based post-translational modification naturally occurring on four mitochondrial enzymes that are crucial for TCA cycle function. Here we show that FDX1 directly regulates protein lipoylation by binding the lipoyl synthase (LIAS) enzyme promoting its functional binding to the lipoyl carrier protein GCSH and not through indirect regulation of cellular Fe-S cluster biosynthesis. Metabolite profiling revealed that the predominant cellular metabolic outcome of FDX1 loss of function is manifested through the regulation of the four lipoylation-dependent enzymes ultimately resulting in loss of cellular respiration and sensitivity to mild glucose starvation. Transcriptional profiling established that FDX1 loss-of-function results in the induction of both compensatory metabolism-related genes and the integrated stress response, consistent with our findings that FDX1 loss-of-function is conditionally lethal. Together, our findings establish that FDX1 directly engages with LIAS, promoting its role in cellular protein lipoylation, a process essential in maintaining cell viability under low glucose conditions.
Keywords: ferredoxin, lipoylation, LIAS, TCA cycle, mitochondria metabolism, Fe-S cluster biosythesis, cuproptosis, FDX2, Cancer metabolism, branchedchain amino acid metabolism, pyruvate dehydrogenase, OGDH, GCSH, GDF15, LIPT1
Ferredoxins (FDXs) are small iron-sulfur cluster-containing proteins that mediate electron shuttling among multiple metabolic pathways. FDXs generally act as electron donors and are essential in a wide array of fundamental reactions including iron–sulfur cluster and steroid hormone biosynthesis and other cytochrome-P450-associated reactions (1). Humans have two ferredoxins, FDX1 and FDX2. In human cells, FDX1 was shown to regulate steroid hormone synthesis by donating electrons to various cytochrome P450 proteins such as CYP11A1 (2, 3, 4). This function is executed only in steroidogenic tissues, explaining the particularly high mRNA levels of FDX1 in human adrenal glands (5, 6). FDX2 on the other hand was shown to be a central component of cellular iron–sulfur cluster biosynthesis (7, 8, 9, 10, 11). Many of the early studies on human ferredoxin function were conducted using bacteria or yeast reconstitution models or in vitro functional assays. As these experimental approaches did not necessarily explore the natural function of FDX1 within the human cell, they produced incomplete and sometimes contradicting findings on the contribution of human ferredoxins and FDX1, particularly in Fe-S cluster (9, 10, 12, 13) and heme biosynthesis (14, 15) and more recently in the regulation of protein lipoylation (13, 16). As such, the predominant natural role(s) of FDX1 in non-steroidogenic human cells remains poorly characterized.
We recently reported FDX1 as a crucial regulator of copper-dependent cell death (cuproptosis) (16, 17). Copper ionophores, such as elesclomol, bind and shuttle Cu (II) to mitochondria where the Cu (II) can be reduced to Cu(I) by FDX1, promoting the cytotoxic effect of copper (17, 18). Copper then induces the destabilization of Fe-S cluster proteins and can directly bind lipoylated proteins, promoting their aggregation and ultimately leading to cell death (16). Interestingly, we revealed that FDX1 loss-of-function in cancer cells also results in a lack of protein lipoylation and that FDX1 protein levels are highly correlated with overall levels of lipoylated proteins across hundreds of different cancer types (16). These results establish that FDX1 is an upstream regulator of protein lipoylation in human cells, a finding that was recently corroborated by showing that FDX1 can initiate LIAS-mediated lipoylation in vitro (13).
Protein lipoylation is a lipid post-translational modification that occurs on conserved lysine residues of only four key mitochondrial metabolic complexes. Lipoylation is essential for the enzymatic function of the lipoylated complexes which include the pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGDH), and branched-chain α-ketoacid dehydrogenase (BCKDH) complexes, and the glycine decarboxylase complex (GDC), also referred to as the glycine cleavage system. This lipoylation process is evolutionarily conserved from bacteria to humans and has been mostly characterized in bacteria and yeast systems and less so in human cells (19, 20, 21, 22). In humans, lipoic acid is synthesized de novo in a multistep process. First, an 8-carbon fatty acid (octanoic acid) synthesized in the mitochondria covalently attached to an acyl-carrier protein (ACP) is transferred to a conserved lysine residue within the Glycine Cleavage System Protein H (GCSH) protein by lipoyl (octanoyl) transferase 2 (LIPT2). Second, two sulfurs are added at C-6 and C-8 positions by the lipoyl synthase (LIAS) enzyme, a radical SAM enzyme (23, 24, 25) that uses SAM to initiate the synthesis of lipoic acid on GCSH. In this reaction, the sulfur atoms are provided by the auxiliary Fe-S cluster of LIAS, which requires Fe-S cluster reconstitution on LIAS after each catalytic cycle of lipoic acid synthesis (26, 27, 28). Therefore, any perturbation in cellular Fe-S cluster biosynthesis will result in the reduced ability of the cell to replenish the auxiliary Fe-S cluster of LIAS after each cycle of lipoic acid synthesis, and ultimately reduced levels of protein lipoylation. Given the contradicting findings on the role of FDX1 in Fe-S cluster biosynthesis and our observation that FDX1 regulates protein lipoylation (16), we set out to determine the predominant role of FDX1 in human cells and characterize the transcriptional and metabolic outcome following FDX1 loss-of-function.
Results
FDX1 regulation of lipoylation is not mediated by Fe-S cluster availability
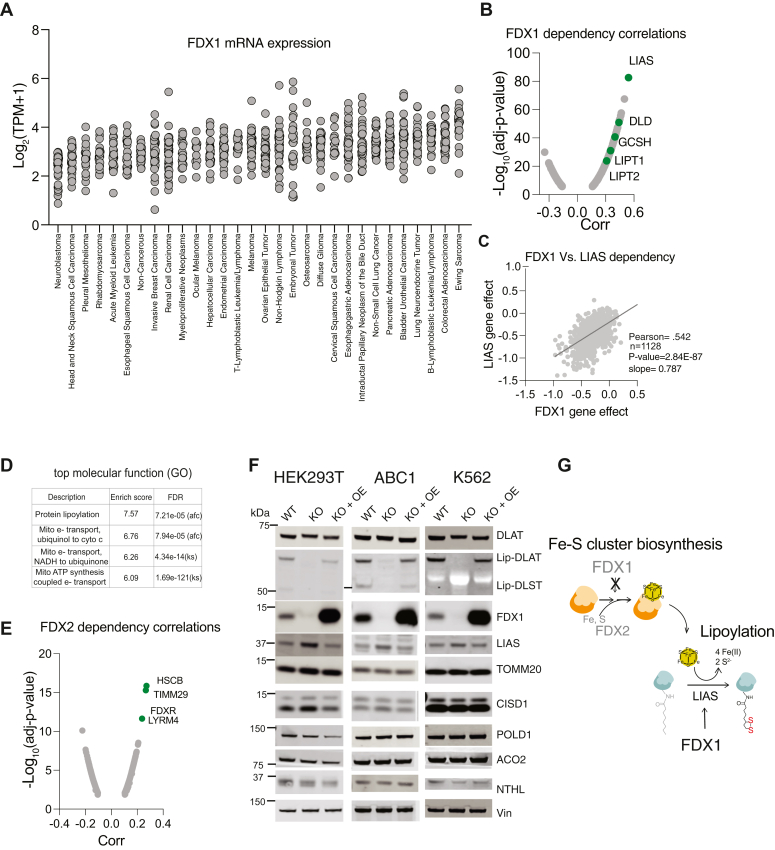
In steroidogenic tissues, FDX1 plays a unique role in the process of steroid hormone biosynthesis by contributing electrons to P450 complexes such as CYP11A1 (6). However, while most cells previously characterized for gene expression as part of the Cancer Cell Line Encyclopedia show no detectable CYP11A1 mRNA levels (Fig. S1A), FDX1 is uniformly expressed across all cell lines from different lineages (Fig. 1A). This suggests that in most human cells FDX1 has a function beyond steroid hormone biosynthesis. To better understand the function of FDX1 in these cancer cells we used the Cancer Dependency Map (www.depmap.org) resource that measures the viability effect of genome-wide CRISPR/Cas9 loss-of-function in each individual cell line across more than a thousand different cell lines. Using this resource, gene function can be inferred by correlating the effect on the viability of a specific gene loss-of-function (dependency) to the dependencies of all other genes across all examined cell lines. Gene dependencies with similar viability profiles across hundreds of cancer cell lines are expected to fall within the same pathway. This approach has been successfully applied by numerous groups to reveal unknown functions of genes of interest (29, 30, 31, 32). Using this approach, the top correlate of FDX1 dependency is LIAS, the only gene known to synthesize the lipoyl moiety in the mitochondria (Fig. 1B). This correlation between the essentiality of FDX1 and LIAS is highly significant (Fig. 1C), and the reciprocal analysis shows that FDX1 is also within the top correlates of LIAS loss-of-function (Fig. S1B). Other enzymes in the lipoylation pathway (GCSH, LIPT1, DLD, and LIPT2) also scored high, and analysis of enriched functional categories in the top 1000 genes correlating with FDX1 dependency revealed that protein lipoylation is the most enriched category (Fig. 1D and Table S1). These results were unique to FDX1, as a similar analysis for its paralog gene FDX2 did not result in any gene in the lipoylation pathway scoring in the top 1000 co-dependent genes (Fig. 1E and Table S1). Moreover, the top correlates for FDX2 were genes in the Fe-S cluster biosynthesis pathways such as HSCB and LYRM4 and the ferredoxin reductase (FDXR) enzyme. This genetic-perturbation data analysis approach first establishes FDX1 loss-of-function as a selective dependency, with most cells showing only a slight growth impairment. Second, it shows that FDX1 and FDX2 differ in their function; the predominant predicted role of FDX1 is associated with the cellular protein lipoylation pathway, whereas the role of FDX2 is shown to be involved in Fe-S cluster biosynthesis (8).
Figure 1.
Genetic evidence supporting the role of FDX1 in regulating protein lipoylation.A, FDX1 mRNA expression levels in cancer cell lines from the CCLE categorized by lineage. B, the CRISPR/Cas9 FDX1 loss-of-function viability outcome in 1128 cell lines correlated with the loss of function of all other tested genes. Plotted are the correlation values and the p-value for the 1000 most significant correlations. In green are genes associated with the protein lipoylation pathway. C, the correlation of LIAS and FDX1 CRISPR/Cas9 loss of function across 1128 cell lines. D, top molecular function (GO) enriched categories in the 1000 genes that have their loss-of-function most correlated with FDX1 loss-of-function. E, the CRISPR/Cas9 FDX2 loss-of-function viability outcome in 1128 cell lines correlated to the loss of function of all other tested genes. Plotted are the correlation values and the p-value for the 1000 most significant correlations. In green are genes associated with Fe-S cluster biosynthesis. F, indicated proteins were analyzed using immunoblot assays of HEK293T, K562 and ABC1 WT cells or cells with CRISPR/Cas9 induced FDX1 loss of function (KO) alone or with reconstituted FDX1 (KO+OE). G, schematic representing the regulation of Fe-S cluster biosynthesis that is upstream of protein lipoylation. FDX1 regulates lipoylation not by affecting Fe-S cluster biosynthesis. The data used to generate plots (A–D) are provided as Table S1.
To experimentally characterize the role of FDX1 we used CRISPR/Cas9 to induce FDX1 loss-of-function (we will term FDX1 KO) and controlled for off-target effects by overexpressing FDX1 in the FDX1 loss-of-function cells (KO+OE). These experiments were conducted in ABC1 (lung), HEK293 T (kidney), and K562 (myeloid) cell lines to ensure the representation of multiple cell lineages in our study. In all three cell lines, protein lipoylation was completely abolished upon FDX1 KO and completely restored upon FDX1 re-constitution (KO+OE) (Fig. 1F). This is not due to changes in total protein levels of the lipoylated proteins (as shown for DLAT). Cellular protein lipoylation is regulated by the Fe-S cluster enzyme LIAS (33, 34), which uses the sulfurs from its auxiliary Fe-S cluster to facilitate the double sulfur insertion to the sixth and eighth carbons of the octanoyl precursor of lipoic acid (26). This dependency of LIAS on Fe-S clusters for function has been shown in genetic models where perturbations to Fe-S cluster biosynthesis result in reduced levels of LIAS and consequently reduced levels of protein lipoylation (35, 36). However, in the FDX1 KO cells, LIAS levels were not reduced, and even an increase in LIAS level was observed in FDX1 KO cells as compared to WT and OE cells (Fig. 1F). This suggests that FDX1 does not regulate LIAS activity by reducing Fe-S cluster availability but rather by an upstream process that inhibits the function of LIAS, preserving its auxiliary Fe-S cluster and resulting in its stabilization (Fig. 1G). We further confirmed this model by showing that FDX1 protein levels had no effect on the protein level of other Fe-S cluster-containing proteins such as CISD1, POLD1, ACO2, and NTHL (Fig. 1F).
FDX1 regulates cell respiration and the ability to proliferate in low-glucose conditions
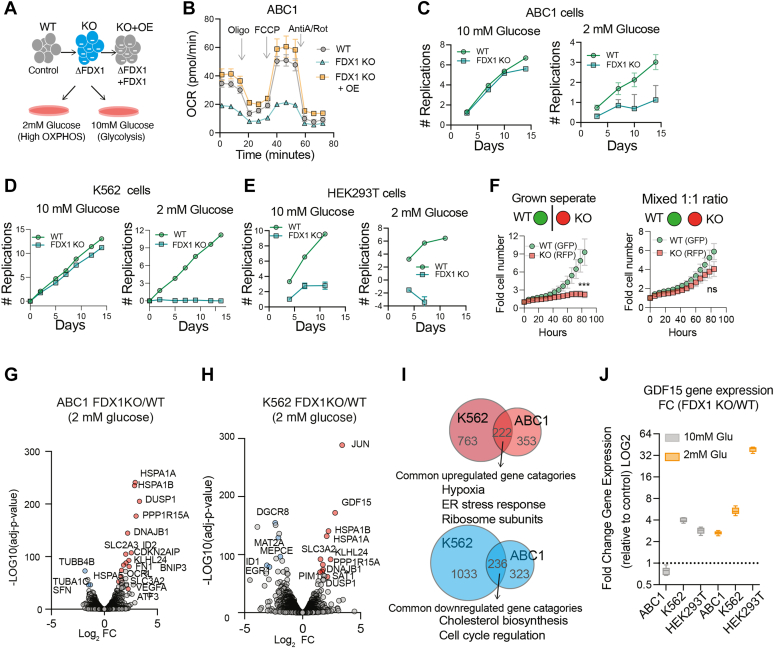
Lipoylation regulates the activity of key carbon entry points into the TCA cycle by directly regulating the activity of α-ketoacid dehydrogenases, a family of multi-subunit enzyme complexes that include the PDHC, oxoglutarate dehydrogenase complex (OGDC), and BCKDC (34). Consistent with the observed effect on protein lipoylation, FDX1 KO cells exhibit low oxygen consumption that is completely restored upon FDX1 reconstitution (Figs. 2, A and B and S2A). Despite the profound effect on cellular respiration K562 and ABC1 FDX1 KO cells exhibited only a mild growth impairment when cultured in 10 mM glucose conditions (normal media concentration) (Fig. 2, C and D). However, when glucose levels were reduced to 2 mM (low physiological level), FDX1 KO cells exhibited a strong growth defect consistent with FDX1 being essential for mitochondrial metabolism (Figs. 2, C–E and S2). A slightly different picture emerged when analyzing the HEK293T cells. In HEK293T, FDX1 KO cells showed impaired growth kinetics even when cultured in 10 mM glucose-containing media (Fig. 2E). We suspected that this FDX1 growth defect in HEK293T cells (in 10 mM glucose conditions) is at least partially due to non-cell autonomous mechanisms. To test this, we fluorescently labeled the WT and FDX1 KO HEK293T cells with either H2B-GFP or H2B-RFP that enables cell counting, and then cultured the cells either separately, or mixed in a 1:1 ratio. When cultured separately HEK293T FDX1 KO cells showed a significant growth defect as compared to WT HEK293T cells. However, when FDX1 KO cells were grown as a 1:1 mixture with WT cells, no significant proliferation differences were observed (Fig. 2F), suggesting that in HEK293T cells, deletion of FDX1 may also have non-cell autonomous growth effects. Mixing HEK293T FDX1 KO with WT cells could not rescue the growth defects of FDX1 KO when cells were grown in 2 mM glucose (Fig. S2, B and C). Together, our data show that FDX1 is an essential upstream regulator of protein lipoylation and a metabolic conditional lethality, essential in cell states where cells rely on mitochondrial energy metabolism to sustain proliferation.
Figure 2.
FDX1 loss-of-function induces the integrated stress response and inhibits proliferation and respiration in low glucose conditions.A, schematic of the experimental conditions. B, the oxygen consumption rate (OCR) was detected in ABC1 WT, FDX1 KO, and FDX1 KO cells with FDX1 expression reconstituted (KO + OE) and after the addition of oligomycin (ATP-linked), the uncoupler FCCP (maximal), or the electron transport inhibitor antimycin A/rotenone (baseline) (mean ± SD, of at least four biological replicates). C–E, proliferation of WT (green circles) or FDX1 KO (blue squares) ABC1 (C), K562 (D), and HEK293T (E) cells growing in either media containing 10 mM or 2 mM glucose. Viability was measured over a period of 14 days and represented as the number of replications (n = 3). F, the relative cell number of HEK293T WT overexpressing H2B-GFP and FDX1 KO overexpressing H2B-RFP was measured when grown either separately (left) or mixed at a 1:1 ratio (right). (n = 7). G and H, volcano plots relating to LOG2 fold-change (x-axis) and -LOG10(p-values) (y-axis) for gene expression changes observed in FDX1 KO as compared to WT in ABC1 (G) and K562 (H) cells growing in media containing 2 mM glucose. I, Venn diagrams representing the analysis presented in Table S2 showing the genes upregulated (red, top) and downregulated (blue, bottom) in FDX1 KO versus WT cells growing in 2 mM glucose conditions (genes with adjusted p-value <0.0001) and a representation of the enriched GO molecular processes gene categories of the overlapping genes (Full list in Table S2). J, relative gene expression of GDF15 in ABC1, K562, or HEK293T cells growing in either 10 mM or 2 mM glucose-containing media. (n = 3).
The transcriptional changes associated with FDX1 loss of function
To enable better characterization of the FDX1 KO cell state, we performed global gene expression analysis using a bulk RNA-seq approach. We profiled both WT and FDX1 KO cells growing in either 10 mM glucose (where FDX1 is non-essential) and 2 mM glucose (where FDX1 is essential) conditions. This analysis was done in both ABC1 and K562 cells that showed similar growth phenotypes. Principal component analysis of the gene expression in the different conditions revealed a similar trend between the different cell lines. The first principal component (PC1; explaining 42.2% and 43.7% of the variance in K562 and ABC1 cells, respectively) was largely driven by the deleterious effect of FDX1 KO under low glucose conditions (2 mM). The second principal component (PC2; explaining 32.3% and 24.3% of variance in K562 and ABC1 cells respectively) was associated with FDX1 KO in normal growth conditions where the perturbation results in minimal inhibition of proliferation rate (Fig. S2, D and E). We analyzed the genes that showed significant change following FDX1 KO (adjusted p-value <0.0001) to determine which gene changes are consistent between the different conditions and what gene categories are enriched (using GO Biological processes (37)). When comparing the significant gene changes when cells were growing in 10 mM glucose conditions, no significant categories were enriched in the upregulated gene signature, whereas genes associated with cell cycle and metabolism were commonly downregulated (Fig. S2, F and G and Table S2). On the individual gene level, FDX1 KO results in dysregulation of some metabolic genes. In ABC1 cells, there is an induction of the PDK1 (Pyruvate Dehydrogenase Kinase 1) gene alongside LDHA (lactate dehydrogenase), suggesting a suppression of the dysfunctional PDH complex and funneling of the pyruvate to the production of lactate by LDHA (Fig. S2G). There is also dysregulation of lipid metabolism genes such as SCD (downregulation) and lysophosphatidylcholine acyltransferase 1 (LPCAT1; upregulation). The induction of LPCAT1, an enzyme that can convert lysophosphatidylcholine (LPC) to phosphatidylcholine (PC), suggests lipid homeostasis dysregulation related to PC. This is further supported by the metabolomics data below that show impairment in de novo PC synthesis by the Kennedy pathway. In K562 cells, there is an increase in genes related to serine and glycine metabolism such as phosphoglycerate dehydrogenase and methylenetetrahydrofolate dehydrogenase and TCA cycle-related genes including ASS1 (argininosuccinate synthase 1) and IDH1 (isocitrate dehydrogenase 1) that all might be induced to compensate for the inactivated lipoylated complexes (Fig. S2F).
When cells are shifted to low glucose conditions (2 mM), where FDX1 is essential, a more significant overlap in gene categories is observed between the cell lines. FDX1 KO resulted in the suppression of lipid metabolism and cell cycle–related genes and the induction of stress-related gene signatures that include hypoxia-induced genes (NOP53, PDK1, BACH1, BNIP3L, BNIP3, CDKN1B, and more), ER stress-related genes (USP25, PPP1R15A, HSPA1A, DNAJB9 and more), and integrated stress response (ISR) genes such as the metabokine GDF15 that is responsive to mitochondrial dysfunction (38, 39) (Fig. S2, G–I). We validated some of the RNA-seq results (Fig. S2, H and I) and specifically show that GDF15 mRNA levels are highly induced in FDX1 KO cells growing in low glucose media (Fig. 2J). These data suggest that tolerance to FDX1 KO between different cell lines, as detected by the induction of the ISR or cell proliferation inhibition, differs between cell lines and depends on the natural metabolic wiring of each cell type and environmental metabolite availability.
The effect of FDX1 loss of function on cellular metabolism
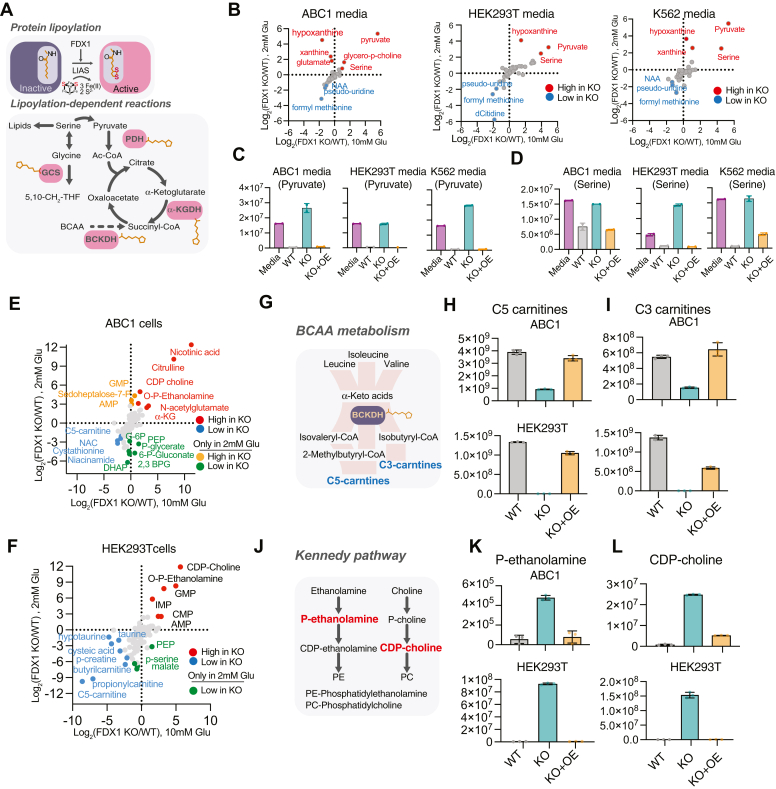
FDX1 is essential for cells that rely on mitochondrial metabolism. To gain further insight into the direct effect of FDX1 on cellular metabolite levels, we profiled the metabolic consequences of FDX1 KO in three different cell lines cultured in both normal and low glucose conditions. We hypothesized that the major metabolic changes would result from dysfunctional lipoylated complexes (Fig. 3A) and that these will reflect the aberrant mitochondrial metabolism in FDX1-KO cells providing the etiology for the conditional essentiality of this gene in low glucose. We tested our hypothesis by conducting both intracellular and media metabolite profiling of WT, FDX1 KO, and FDX1 KO with reconstituted FDX1 (KO+OE) cells. These experiments were conducted in ABC1 (lung), HEK293T (kidney), and K562 (myeloid) cell lines to enable the discrimination between universal effects of FDX1 compared to lineage or cell state-specific ones in our analysis (Table S3). FDX1 KO-induced changes in media metabolites were similar across all three cell lines with higher levels of pyruvate, serine, and glycine and lower levels of formyl-methionine in media of FDX1 KO cells (Fig. 3B). One likely interpretation is that high levels of pyruvate in FDX1 KO cells (Fig. 3, B and C) result from the malfunction of the non-lipoylated PDH complex. High levels of serine observed in media of FDX1 KO cells (Fig. 3, B and D) could result from an impaired glycine cleavage system (GCS) that will result in the accumulation of serine by conversion from accumulated glycine by the mitochondrial SHMT2 enzyme (40). Moreover, dysfunctional GCS will inhibit the synthesis of 10-formyl THF which is the formyl donor for formyl-methionine (41, 42) (Fig. 3A) explaining the observed increase in uptake of formyl-methionine by FDX1 KO cells.
Figure 3.
FDX1 loss-of-function results in metabolite signature consistent with protein lipoylation deficiency.A, schematic of the metabolic processes where lipoylated proteins regulate key TCA cycle (PDH, α-KGDH, and BCKDH) and one-carbon metabolism (GCSH) entry points. B, LOG2 fold change of metabolites in the media of either ABC1, HEK293T, or K562 FDX1 KO as compared to WT cells grown in media containing either 2 mM (y-axis) or 10 mM (x-axis) glucose. C and D, changes in media pyruvate (C) or serine (D) levels in WT, FDX1 KO, or FDX1 KO cells with reconstituted FDX1 (KO+OE) HEK293T, ABC1 and K562 cells. E and F, intracellular metabolite level changes between FDX1 KO and WT ABC1 (E), and HEK293T (F) cells (presented as LOG2 fold change of KO/WT) grown in media containing either 2 mM (y-axis) or 10 mM (x-axis) glucose. G–L, changes in intracellular levels of C5- (H), C3- (I) carnitines, phospho-ethanolamine (K), and CDP-choline (L) in either WT, FDX1 KO, or FDX1 KO cells with reconstituted FDX1 (KO + OE) HEK293T and ABC1 cells growing in 2 mM glucose conditions (n = 3). (G) Schematic of branched chain amino acids (BCAA) metabolism and (J) Schematic of the Kennedy Pathway.
FDX1 KO-induced changes in intracellular metabolites were more variable between the different examined cell lines. However, there were specific metabolites that showed a significant change across all three cell lines. First, there was a consistent decrease in both C3- and C5-carnitine levels (Fig. 3, H and I) in FDX1 KO cells across all three models. C3- and C5-carnitines are downstream products of branched-chain amino acid (BCAA) metabolism that is mediated by the lipoylated BCKDH complex; the decreased levels in FDX1 KO cells suggest the BCKDH complex is inhibited in these cells (Fig. 3G). When cells were grown in low glucose conditions, these metabolic changes were further enhanced and a stronger decrease in C3- and C5- carnitines in FDX1-KO cells is observed (Figs. 3, E–I and S3, A–C), implying a failed attempt to channel BCAA as an alternative fuel for the TCA cycle (43). Second, there was a dramatic increase in two intermediates (CDP-choline and phospho-ethanolamine) of the Kennedy pathway (Figs. 3, K and L and S3, D and E), which is responsible for the de novo synthesis of phosphatidylethanolamine and phosphatidylcholine (Fig. 3J). It is yet unclear if this is due to impairment in lipid metabolism that could be related to the observed dysregulation of lipid metabolism genes such as OCRL, LPCAT1, and SCD (Fig. 2) or as a cellular response to impaired mitochondrial respiration given that phosphoethanolamine directly regulates mitochondrial respiration (44, 45). Third, reduced levels of phosphoenolpyruvate (PEP) and other glycolytic intermediates such as 2,3-bisphosphoglyceric acid (2,3-BPG), and glucose-6-phosphate (G-6-P) were observed in FDX1 KO cells only when cultured in low glucose, likely reflecting altered metabolic flux (Fig. 3, E and F). Finally, there were some effects that seemed to be cell line specific. In ABC1 cells, there is an observed increase in both α-ketoglutarate and citrulline (Fig. 3E). While the increase in α-KG is expected when DLST (a component of the α-KG dehydrogenase complex) is not lipoylated, the increase in citrulline could be due to altered proline and arginine metabolism that was previously reported in LIPT1 KO models (46).
FDX1 binds LIAS in vitro and in cells
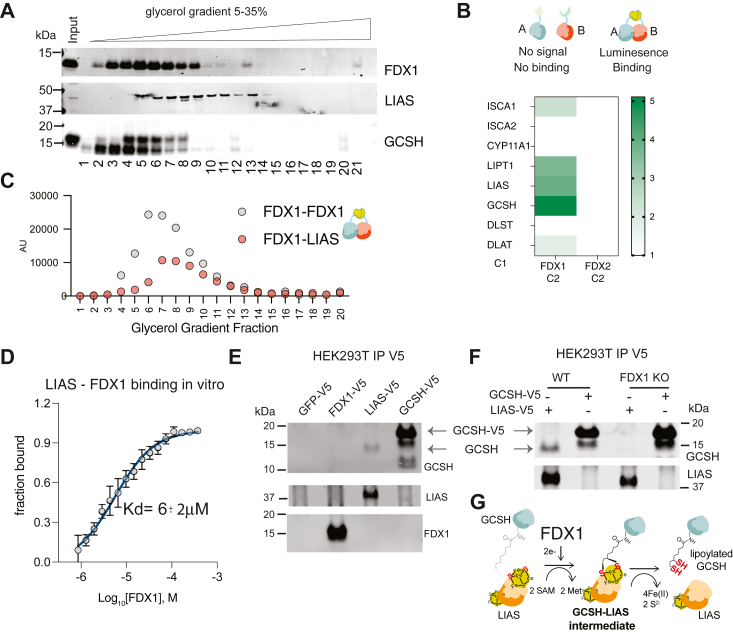
Given the essential role of FDX1 in regulating cellular protein lipoylation, we next determined if indeed FDX1 directly engages with the lipoylation machinery in the cells. To do so we implemented several orthogonal approaches. First, using a glycerol gradient sedimentation assay we show that endogenous FDX1, LIAS, and GCSH co-migrate (Fig. 4A), suggesting they might form a functional complex in the cells. We then implemented a Nano Luciferase (NanoLuc)-based complementation assay to explore the intracellular protein-protein interactions of FDX1 (47, 48). In this assay, two complementary fragments of NanoLuc (C1 and C2) are fused to two different proteins (A and B) of interest. The strength of the protein-protein interaction in cells can be quantified based on the intensity of luminescence (Fig. S4A) (47). One complementing fragment of Nanoluc was fused to the C-terminus of FDX1 (C2) and the other fragment (C1) was fused to other proteins of interest. Using this approach, we show that FDX1 forms a strong interaction with itself but not with FDX2, which was used as a control (Fig. S4B), suggesting FDX1 functions as a multi-subunit complex (dimer or oligomer) in cells. Next, we used this assay to establish the binding of FDX1 with specific proteins in the lipoylation pathway. Strong binding of FDX1 was observed with the lipoyl synthase (LIAS), the lipoylated enzyme GCSH and the lipoyl transferase (LIPT1). We did not observe significant binding with the lipoylated proteins DLAT and DLST or the other suggested FDX1 interacting proteins (ISCA2, CYP11A1). The binding of FDX1 to the lipoylating enzymes was specific for FDX1 and was not observed for FDX2 suggesting FDX1 could be an integral part of the lipoylation complex in cells (Fig. 4B). Further glycerol gradient sedimentation assays on cells overexpressing either FDX1 or FDX1 with LIAS with complementary NanoLuc tags (C1 and C2) showed that the measured peak of luminescence corresponds to the fractions where the endogenous proteins co-migrate (Fig. 4C), suggesting the complementation assay measurements reflect the endogenous state. Third, we used a purified in-vitro experimental approach using a Microscale Thermophoresis assay (MST) that enables the characterization of binding without the need for immobilization of the proteins and can ensure the detection of weak or transient interactions (49). This MST binding approach revealed that purified FDX1 and LIAS enzymes (Fig. S4, C–F) bind in vitro with a calculated Kd in the 6 μM range (Fig. 4D), establishing direct binding of FDX1 and LIAS. Finally, we undertook an immunoprecipitation approach to better characterize the interaction of FDX1 with LIAS and other lipoylation machinery proteins. We were not able to show direct protein-protein interaction of FDX1-LIAS when immunoprecipitating either FDX1 or LIAS. This is either due to the transient nature of the interaction or the limitations of the antibodies used. As such, we decided to determine if we can detect the binding of GCSH with LIAS in cells as in vitro studies showed that in the initial step of insertion of two sulfur atoms at C6 and C8 by of an (N6-octanoyl)-lysyl residue on the carrier peptide by LIAS resulted in the formation of a covalent cross-link bond between LIAS and the lipoyl carrier protein (36, 50, 51). The binding of GSCH-LIAS reflects an active process of lipoylation. Immunoprecipitation of V5 tagged overexpressed LIAS resulted in the co-immunoprecipitation of the endogenous GCSH protein (Figs. 4E and S4G) establishing this interaction can be detected in human cells. We did not detect this interaction when immunoprecipitating overexpressed V5 tagged GCSH, suggesting that either the c-terminal tagging of GCSH disrupts this binding or the LIAS antibody is not sensitive enough for the detection of the co-immunoprecipitated LIAS (or both). In FDX1 KO cells, this binding of overexpressed LIAS to endogenous GCSH is completely abolished (Figs. 4F and S4H), suggesting FDX1 is essential for the formation of the LIAS-GCSH bond and lipoylation activation (Fig. 4G). Together, these findings establish a model whereby FDX1 directly binds LIAS and serves as the electron donor for the SAM-dependent LIAS-mediated lipoylation reaction as recently shown in vitro (13) (Fig. 4G).
Figure 4.
FDX1 directly binds LIAS in cells and in vitro.A, HEK293T cell lysates were subjected to a 5 to 30% glycerol gradient and FDX1, LIAS, and GCSH protein sedimentation was analyzed using immunoblot analysis. B, FDX1 or FDX2 used as a control tagged with fragment 1 of the complementation Nano-luciferase (C1) were overexpressed in the presence of indicated proteins tagged with fragment 2 of the complementation Nano-luciferase (C2). Luminescence was measured 24 h after transfection and presented as the calculated NLR score (n = 3). C, FDX1-C2 was overexpressed in HEK293T cells together with either FDX1-C1 or LIAS-C1 and lysates were analyzed for luminescence following sedimentation in 5 to 30% glycerol gradient. D, MST results from titration of FDX1 to Red-NHS dye-labeled LIAS are shown, averaged raw data (n = 3, open circles), the calculated binding curve fit from which the Kd was extracted (solid blue line), and the standard deviations (vertical black error bars). The fraction bound of labeled LIAS is plotted against the concentration of titrated FDX1 ligand and a Kd of 6 ± 2 μM was determined. E, overexpressed V5-tagged GFP, FDX1, LIAS, and GCSH in HEK293T cells were immunoprecipitated using V5 antibody, and the co-immunoprecipitated GCSH, LIAS, and FDX1 proteins were analyzed by immunoblotting. F, overexpressed, V5-tagged LIAS or GCSH used as a control were immunoprecipitated using V5 antibody, and the co-immunoprecipitated GCSH and LIAS were analyzed by immunoblotting. G, schematic representing the mechanism of FDX1 regulation of protein lipoylation based on both previous findings (13, 16, 51, 53) and the data provided in this work.
Discussion
In this work, we explored the role and mechanism by which FDX1 regulates protein lipoylation in human cells. We were motivated by our recent finding that showed FDX1 is an upstream regulator of protein lipoylation in many cancer cell types (16). The goal of this study was first to establish if FDX1-regulation of protein lipoylation is direct or indirect by inducing global Fe-S cluster biosynthesis defects. The second goal was to characterize the essentiality of FDX1-regulated lipoylation in cells growing in different glucose conditions. And the third goal was to map the transcriptional and metabolic networks that are most affected by disruption of this poorly characterized pathway in human cells.
Our findings revealed that depletion of FDX1 results in a cell state with no detectable lipoylated proteins, minimal respiration, and growth defects that are strongly affected by glucose availability in the media. FDX1-regulated protein lipoylation defects are not due to reduced Fe-S cluster biosynthesis as previous findings suggested (8, 9, 11). In fact, we saw no effect at all on the protein level of any of the tested Fe-S cluster proteins in cells with FDX1 loss-of-function. On the contrary, FDX1 loss of function resulted in the stabilization of the LIAS protein, as expected if FDX1 is an upstream regulator of LIAS activity. Inactive LIAS will not breakdown its auxiliary Fe-S cluster that is used as a substrate in the processes of lipoylation and as such will result in the stabilization of the enzyme consistent with recent reports (13, 52). A recent report showed in vitro that reduced FDX1 (by the action of NADPH and FDXR) could efficiently replace dithionite as an electron source to initiate the SAM-dependent activity of LIAS (13). In the work above we provide additional evidence that refines the role of FDX1 in human cells. First, we establish the binding of FDX1 with LIAS both in vitro and in cells. Second, using in-cell complementation and co-sedimentation assays, we show that FDX1 engages not only with LIAS but also with the lipoyl carrier enzyme GCSH and lipoyl transferase LIPT1, suggesting these could all be part of an active lipoylation complex. Lastly, it was previously established in vitro that during lipoyl synthesis by LIAS, a covalent interaction of LIAS (lipoyl synthase) with the lipoyl carrier peptide (which would be GCSH in cells) is transiently formed (36, 51, 53). Building on these findings, we show above that such binding between LIAS-GCSH can be detected in cells and is completely dependent on the presence of FDX1. In FDX1 KO cells, LIAS-GCSH interaction is no longer detected, suggesting FDX1 is essential for LIAS activation in cells. Together, our initial observation that FDX1 regulates lipoylation in multiple cancers (16), the recently published mechanistic studies on FDX1 regulation of lipoylation in vitro (13) and the outcome of this regulation in hypoxic conditions (52) and the work described above, provide compelling evidence on a new and important role of FDX1 in directly regulating LIAS mediated protein lipoylation in human cells. Future work will determine if this is a newly acquired function of ferredoxins in evolution.
We further assessed the metabolic and transcriptional outcome of FDX1 KO with the goal of establishing a robust FDX1 metabolite and transcriptional profile. Using three distinct cell line models, this approach was only partially successful suggesting that the cellular response to lipoylation inhibition is influenced by the basal metabolic cell state and differs between cell lines. Despite this, some very consistent changes in metabolite profiling were revealed across all examined cell lines. FDX1 KO cells exhibited reduced utilization of pyruvate, reduced accumulation of BCAA breakdown products and in some cases accumulation of α-KG (ABC1 cells). These results are consistent with lipoylated protein complex function inhibition and reproduce previous metabolite assessment in models where lipoylated complexes were perturbed (42, 46). Interestingly, a very strong and reproducible accumulation of the Kennedy pathway intermediates (CDP-choline and phospho-ethanolamine) was observed in all tested FDX1 KO cell lines. Phospho-ethanolamine has been shown to directly regulate mitochondrial respiration (44, 45); it remains to be determined if dysregulation of these metabolites is due to direct regulation by FDX1 or compensatory mechanisms that involve the rewiring of lipid metabolism, as described in LIPT1 mutant cells (46).
The transcriptional profiling was less conclusive and did not result in a clear FDX1-dependent gene signature. Overall, in the stressed conditions, there is an induction of cell metabolism, hypoxia, ER-stress, and the ISR related genes, consistent with previous reports that indicated that the predominant gene signature activated by mitochondrial dysfunction is related to the ISR (54). GDF15, a metabokine that is responsive to mitochondrial dysfunction (38, 39), is highly induced in FDX1 KO cells in particular when growing in low glucose media (Fig. 2J), possibly explaining some of the non-cell autonomous effects of FDX1 KO we observed. The genes that are downregulated following FDX1 KO are largely related to either cell cycle gene categories or lipids, specifically cholesterol metabolism categories possibly contributing to the observed accumulation of the Kennedy pathway intermediates and consistent with previous analysis of LIPT1 mutant cells (46) that showed global lipid biosynthesis defects.
It was surprising to reveal that despite the complete loss of protein lipoylation in FDX1 KO cells, only a mild fitness defect was observed when cells were grown in high glucose conditions (in two of the three cell lines examined). Moreover, across hundreds of cancer cell lines, FDX1 loss of function is most correlated with LIAS loss of function and shows only a modest cell growth inhibition effect in most cell lines tested (tested in normal media conditions). Only in a small subset of cancer cell lines the loss of FDX1 or LIAS is essential and results in complete growth inhibition (depmap.org). The sensitivity of these cell lines to FDX1 and LIAS KO in normal media is most likely driven by the increased dependency of these cells on the function of the lipoylated α-KGDH complex. This is reflected by the high correlation of OGDH and DLST KO effects on viability with those of LIAS KO (Fig. S1B) and the previous findings showing specific sensitivity of cancer cells to perturbation of the α-KGDH complex (55, 56). The dependency on other lipoylated complexes is most likely masked by the high glucose levels and could be flushed out by growing cells in lower glucose conditions.
The disruption of FDX1-regulated lipoylation has both cell autonomous and non-autonomous effects. In HEK293T cells, FDX1 KO resulted in impaired growth in the presence of 10 mM glucose (unlike K562 and ABC1 cells). However, no growth kinetic differences were observed between WT and FDX1 KO cells when they were grown mixed together (1:1 ratio) (Fig. 2F). This suggests that beyond the cell-intrinsic effects, FDX1 KO may effect cell growth by other cell non-autonomous mechanisms such as secretion of cell growth inhibitory factors (such as GDF15) or by toxic alteration of metabolite availability. Interestingly, in a parallel study, it was established that proliferation defects observed in some lipoylation null cancer cells, when growing in high glucose conditions, can be rescued if cultured in hypoxic conditions (52). Hypoxia attenuated FDX1 KO-induced inhibition of cell proliferation and completely blocked the induction of GDF15 and other mitochondrial stress genes (52). Together these findings indicate that the dependency of cells on FDX1 and lipoylation pathways is contingent on both the intrinsic metabolic wiring of the cell and nutrient availability involving both cell autonomous and non-autonomous mechanisms. Revealing the specific genetic and cellular states that might affect this dependency will establish if FDX1-regulated protein lipoylation is a targetable vulnerability in a subset of cancers.
Experimental procedures
Correlation analysis
CRISPR FDX1, FDX2, and LIAS dependency scores and FDX1 and CYP11A1 gene expression across 1128 cell lines from the Broad Institute’s public 22Q4 dependency map were correlated with other CRISPR dependency scores from the Broad Institute’s 22Q4 Dependency Map/CCLE release. Pearson’s correlations were performed in R using the cor.test function. p values were then corrected for false discovery using the Benjamini-Hochberg method of the p.adjust function in R and q-values were -log10-normalized. The top 1000 correlating genes were plotted using GraphPad Prism v8.3.0. Some of the results were used for gene enrichment analysis using STRING (57) version 11.5.
Cell lines and culturing conditions
HEK293T, K562, and ABC1 cells used in this study were grown in RPMI media supplemented with 10% FBS and PS (for consistency between cell line experiments). For the glucose experiments, cells were grown in RPMI without glucose (ThermoFisher Scientific) media that was supplemented with 10% dialyzed fetal bovine serum and 1 mM pyruvate, and 2 mM fresh glutamine and indicated concentrations of glucose (2 mM–10 mM). Uridine was not added to the media as it showed no effect on the growth of any of the examined cell lines.
Generation of FDX1 loss of function cell lines
The general strategy for creating the FDX1 loss-of-function was to nucleofect a ribonucleoprotein (RNP) complex of CRISPR-Cas9 with the FDX1 targeting gRNA and then do single cell cloning to ensure maximal depletion of FDX1 gene expression. We could not grow our single-cell clones of ABC1 cells, so for these cells we undertook an additional step before the nucleofection step where ABC1 cells were pre-transduced with the previously validated gRNA targeting FDX1 (16) cloned into BRD016 guide only lentivector. Cells were selected for 1 week to ensure optimal gRNA expression and then subjected to the nucleofection protocol.
Generation of FDX1 KO using nucleofection
The ribonucleoprotein (RNP) used in FDX1 KO by nucleofection was formed using a 1:1 ratio of Alt-R CRISPR-Cas9 tracrRNA (100 μM) and Alt-R CRISPR-Cas9 predesigned crRNA (100 μM) (Hs.Cas9.FDX1.1.AD IDT guide) (Integrated DNA Technologies). The mixture was incubated at 95 °C for 5 min and then brought to room temperature to form the gRNA duplex. The gRNA duplex (50 μM) was then combined with Alt-R Cas9 enzyme (61 μM) (Integrated DNA Technologies) at a ratio of 3:2 and incubated at room temperature for 10 to 20 min to form the RNP that was used for the nucleofection described below.
Nucleofection of ABC1, HEK293T, and K562 cells was conducted according to the manufacturer’s protocol (Lonza). 500,000 cells per sample were pelleted by centrifugation at 4 °C using a tabletop centrifuge at 3000 rpm for 5 min. Cell culture media supernatant was aspirated and discarded, and the cells were resuspended in 30uL of a mixture containing 20 μl Lonza SF cell line solution, 5 μl FDX1 Cas9 RNP, 1 μl electroporation enhancer, and 4 μl PBS (Lonza Biosciences). Each cell suspension was added to individual wells of an electroporation cuvette (Lonza Biosciences), and then nucleofected using the Lonza 4D system program A549 pulse code CM-130. Cells were then transferred from the cuvette into pre-warmed media in a 6-well or 12-well culture plate and allowed to grow to 80% confluence. The HEK 293T and K562 nucleofected cells were further single-cell plated to generate FDX1 KO single-cell clones. ABC1 FDX1 KO cells could not be single cell cloned and the bulk nucleofected population was validated for high-efficiency KO and used in subsequent experiments.
Generation of the FDX1 reconstituted cell lines (KO+OE)
FDX1 was further introduced to the FDX1 loss-of-function cell line clones selected to be used in the study. FDX1 was cloned into the pMT025 backbone vector (Addgene plasmid # 158579). Viral particles were produced in HEK293T cells and FDX1 loss-of-function cells (ABC1, HEK293T and K562) were infected and selected with puromycin to ensure homogenous FDX1 gene expression.
Immunoblot analysis
Cells were lysed using RIPA lysis buffer (Sigma-Aldrich) with protease inhibitor cocktail tablets (Sigma-Aldrich) and then incubated on ice for 30 min to homogenize. Samples were subjected to centrifugation for 10 min at 10,000 RPM to pellet cell debris. The supernatant was collected for quantification and the cell debris pellet was discarded. Protein was quantified using the bicinchoninic acid (BCA) method with a bovine-specific albumin standard curve for normalization. Each protein extract was boiled in 1× LDS sample buffer with a 1:10 TCEP solution reducing agent and then size fractionated via pre-cast SDS-PAGE Bis-Tris 4 to 12% gels (Thermo Fisher Scientific), and transferred onto nitrocellulose membranes with the iBlot-2 system (Thermo Fisher Scientific). Each membrane was then incubated at room temperature for 1 h in LICOR Odyssey blocking buffer and then incubated at 4 °C with the respective antibody for 1 h overnight. The membranes were then washed four times with 1× TBS with 0.1% Tween20 (TBST) and incubated with fluorophore-specific IRDye secondary antibodies (LI-COR). The membranes were then washed again four times with TBST before being imaged on a LI-COR Odyssey machine. Antibodies used in this study include: FDX1 (abcam ab108257), Lipoic acid (Millipore, cat# 437695 and abcam ab58724), LIAS (Proteintech 11577-1-AP), DLAT (Cell signaling 12362S and Thermo), DLST (cell signaling 5556S), Vinculin (abcam ab130007), CISD1 (Thermo Scientific 16006-1-AP), NTHL (Invitrogen PA316563), Actin (Cell signaling 8H10D10), ACO-2 (abcam ab129069), TOMM20 (abcam ab186735), POLD1 (abcam ab186407), and GCSH (Proteintech 16726-1-AP).
RNA extraction
RNA was extracted from cells using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Library preparation with polyA selection and illumina sequencing
Library preparation and sequencing were conducted by Azenta life sciences according to the following protocol. RNA samples were quantified using a Qubit 2.0 Fluorometer (ThermoFisher Scientific) and RNA integrity was checked using TapeStation (Agilent Technologies). RNA sequencing libraries were prepared using the NEBNext Ultra II RNA Library Prep for Illumina using the manufacturer’s instructions (New England Biolabs). Briefly, mRNAs were initially enriched with Oligod(T) beads. Enriched mRNAs were fragmented for 15 min at 94 °C. First-strand and second-strand cDNA were subsequently synthesized. cDNA fragments were end-repaired and adenylated at 3′ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by PCR with limited cycles. The sequencing libraries were validated on the Agilent TapeStation (Agilent Technologies), and quantified using a Qubit 2.0 Fluorometer (ThermoFisher Scientific) as well as by quantitative PCR (KAPA Biosystems).
The sequencing libraries were multiplexed and clustered onto a flowcell. After clustering, the flowcell was loaded on the Illumina instrument according to the manufacturer’s instructions. The samples were sequenced using a 2 × 150 bp paired-end (PE) configuration. Image analysis and base calling were conducted by the control software. Raw sequence data (.bcl files) generated from the sequencer were converted into fastq files and de-multiplexed using Illumina's bcl2fastq 2.20 software. One mismatch was allowed for index sequence identification. After investigating the quality of the raw data, sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the reference genome available on ENSEMBL using the STAR aligner v.2.5.2b. BAM files were generated during this step. Unique gene hit counts were calculated by using feature Counts from the Subread package v.1.5.2. Only unique reads that fell within exon regions were counted.
RNA-seq analysis
The exploratory analysis used TPM-normalized counts of RNAseq data. The data were first filtered to only those genes that had counts greater than two (TPM > 2) in all conditions, and subsequently log-transformed. Each cell line was then analyzed individually. We centered and scaled the log (TPM + 1) data such that each gene’s expression is centered at 0 with a standard deviation of 1. Using the cleaned and transformed data, we conducted Principal Component Analysis (shown in Fig. S2), which revealed clear separations between samples. We then applied DESeq2 for each of our hypotheses. Our hypotheses were (1) H1-Given wild type, low glucose versus high glucose, (2) H2- Given low glucose, FDX1 KO versus wild type, (3) H3-Given high glucose, FDX1 KO versus wild type, and (4) H4- Given FDX1 KO, low glucose versus high glucose. Lastly, using the results from DESeq2, we determined the genes with an adjusted p-value <0.0001 that are either upregulated or downregulated in both cell lines examined (ABC1 and K562) in the different growth conditions (2 mM and 10 mM glucose). Gene categories enriched in these gene sets were analyzed using Geneontology (GO) (58) specifically looking for gene categories enriched using GO molecular processes (37).
cDNA synthesis and RT-qPCR
Extracted RNA was used to make cDNA using a Bio-Rad iScript cDNA synthesis kit (Catalog #: 1708891) following the manufacturer's protocol. 500 ng of RNA was used as input. cDNA was diluted 1:10 prior to qPCR. qPCR was done in triplicate, using Applied Biosystems Power SYBR Green PCR Master Mix (Catalog #: 43-676-59). 10 μl of diluted cDNA was used per replicate, and 15 μl of Primer/SYBR Green Master Mix (12.5 μl 2× SYBR green master mix, 0.25 μl of 10 mM forward & reverse primer, and 2.25 of ddH2O per sample). See Primer sequences in table below. RT-qPCR was done using a ThermoFisher QStudio 6 FLX real-time PCR system using the following protocol: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Ct values were normalized to each sample's β-Actin transcript levels, then compared to WT samples to obtain ddCt values, which were used to quantify fold change in gene expression from WT cells to FDX1 KO cells.
Primers used:
| GAPDH_Fw | GGTGTGAACCATGAGAAGTATGA |
| GAPDH_Rv | GAGTCCTTCCACGATACCAAAG |
| β-actin_Fw | GGATCAGCAAGCAGGAGTATG |
| β-actin_Rv | AGAAAGGGTGTAACGCAACTAA |
| HPRT_Fw | CGAGATGTGATGAAGGAGATGG |
| HPRT_Rv | TTGATGTAATCCAGCAGGTCAG |
| GDF15_Fw | CTACAATCCCATGGTGCTCAT |
| GDF15_Rv | TCATATGCAGTGGCAGTCTTT |
| PDK1_Fw | AAGGATGCAAGGACCAGAAG |
| PDK1_Rv | GGTTCCAGATGCCAGTTATCA |
| LDHA_Fw | AGATTCCAGTGTGCCTGTATG |
| LDHA_Rv | ACCTCTTTCCACTGTTCCTTATC |
| PHGDH_Fw | GATCTGGCCTCTCTGTGATTT |
| PHGDH_Rv | GGGCAAAGGTGTTGTCATTC |
| HSPA1A_Fw | CCACCATTGAGGAGGTAGATTAG |
| HSPA1A_Rv | CAGGAAATTGAGAACTGACAAACA |
| HSPA1B_Fw | CTACCATTGAGGAGGTGGATTAG |
| HSPA1B_Rv | CAAAGAAGTGAAGCAGCAAAGA |
| LPCAT1_Fw | TGAAGGGAGGAGAGAAGATAGG |
| LPCAT1_Rv | GCTCTCGTCGAACAGTGAAA |
| DNAJB1_Fw | CTGTCTTCTCTTTGGCCATCTC |
| DNAJB1_Rv | CTGCTGGAACGAGAGGTATTG |
| SLC2A3_Fw | AGGATGCAGGTGTTCAAGAG |
| SLC2A3_Rv | GCCCTTTCCACCAGAAATAGA |
Metabolite profiling by mass spectrometry for detection of polar metabolites
Cells from the culture were washed with ice-cold 0.9% NaCl briefly and extracted in buffer (80% Methanol, 25 mM Ammonium Acetate, and 2.5 mM Na-Ascorbate prepared in LC-MS water, supplemented with isotopically labeled amino acid standards [Cambridge Isotope Laboratories, MSK-A2-1.2], aminopterin, and reduced glutathione standard [Cambridge Isotope Laboratories, CNLM-6245-10]) and transferred into 1.5 ml tubes. Samples were vortexed for 10 s, then centrifuged for 10 min at 18,000g to pellet cell debris. The supernatant was dried on ice using a liquid nitrogen dryer. Metabolites were reconstituted in 15 μl water supplemented with QReSS [Cambridge Isotope Laboratories, MSK-QRESS-KIT] and 1 μl was injected into a ZIC-pHILIC 150 × 2.1 mm (5 μm particle size) column (EMD Millipore) operated on a Vanquish Flex UHPLC System (Thermo Fisher Scientific). Chromatographic separation was achieved using the following conditions: buffer A was acetonitrile; buffer B was 20 mM ammonium carbonate, 0.1% ammonium hydroxide in water. Gradient conditions were: linear gradient from 20% to 80% B; 20 to 20.5 min: from 80% to 20% B; 20.5 to 28 min: hold at 20% B at 150 ml/min flow rate. The column oven and autosampler tray were held at 25 °C and 4 °C, respectively. MS data acquisition was performed using a QExactive benchtop orbitrap mass spectrometer equipped with an Ion Max source and a HESI II probe (Thermo Fisher Scientific) and was performed in positive and negative ionization mode in a range of m/z = 70 to 1000, with the resolution set at 70,000, the AGC target at 1 × 106, and the maximum injection time (Max IT) at 20 ms.
Data analysis for metabolomics
Relative quantification of polar metabolites was performed with TraceFinder 5.1 (Thermo Fisher Scientific) using a 7 ppm mass tolerance and referencing an in-house library of chemical standards. Pooled samples and fractional dilutions were prepared as quality controls and injected at the beginning and end of each run. In addition, pooled samples were interspersed throughout the run to control for technical drift in signal quality as well as to serve to assess the coefficient of variability (CV) for each metabolite. Data from TraceFinder was further consolidated and normalized with an in-house R script: (https://github.com/FrozenGas/KanarekLabTraceFinderRScripts/blob/main/MS_data_script_v2.4_20221018.R). Briefly, this script performs normalization and quality control steps: (1) extracts and combines the peak areas from TraceFinder output.csvs; (2) calculates and normalizes to an averaged factor from all mean-centered chromatographic peak areas of isotopically labeled amino acid internal standards within each sample; (3) filters out low-quality metabolites based on user inputted cut-offs calculated from pool reinjections and pool dilutions; (4) calculates and normalizes for biological material amounts based on the total integrated peak area values of high-confidence metabolites. In this study, the linear correlation between the dilution factor and the peak area cut-offs are set to RSQ >0.95 and the CV <30%. Finally, data were Log transformed and Pareto scaled within the MetaboAnalyst-based statistical analysis platform (PMID:25897128) to generate principal component analysis, PLSDA, and heatmaps. In Table S3 missing data were replaced with limit of detection value (1/10 of minimum positive value of each variable).
Complementation assay
The vectors used for the Nano-Luc complementation assay, was created by inserting the FDX1 and other genes of interest into N2H plasmids (47) (pDEST-C1 or -C2 kindly received from Yves Jacob, https://www.addgene.org/Marc_Vidal/) using the Gateway LR reaction. For the complementation assay 100 ng of total DNA (1:1 ratio of plasmids) was transfected into 293T cells in 96-well plates using TansIT(R)-LT1 (Mirus) transfection reagent according to the manufacturer’s protocol. Luminescence was measured 24 to 36 h after transfection using Nano-glo (Promega) and viability (that was not affected) was measured using cell-titer glo (Promega) to ensure there were no viability differences (>10%) between the samples. All experiments were conducted with at least three biological replicates of each condition. In Figure 4, the interaction is presented as the normalized luminescence ratio (NLR). The NLR score represents the raw luminescence value of the tested pair (A–B) divided by the sum of the luminescence value from the two individual proteins (A-alone + B-alone).
For the glycerol gradient, cells growing in 6-wells were transfected with FDX1-C2 and either FDX1-C1 or LIAS-C1 (2.5 μg total plasmid at 1:1 ratio). 36 h after transfection cells were lysed and subjected to glycerol gradient as described below.
Glycerol gradient
The 5 to 30% glycerol gradient was generated using the gradient master mixer (BioComp Instruments) with buffers containing 20 mM HEPES-KOH (pH 7.9), 150 mM NaCl, 1.5 mM MgCl2 and either 5% or 30% glycerol. HEK293T cells were lysed in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1% NP-40, 1X Halt protease, and phosphatase inhibitors). Cell lysates were spun at 20,000g for 10 min at 4 °C, and supernatant was loaded onto the glycerol gradient. Gradients were spun at 45,000 RPM for 16 h at 4 °C in an SW55Ti rotor using an ultracentrifuge (Beckman). After centrifugation, 250 μl fractions were collected off the top of the glycerol gradient. For total protein lysates, NuPage sample buffer was added for a final concentration of 1× sample buffer, and 16 μl of sample was run on a 4 to 12% Bis-Tris gel following a routine immunoblot procedure.
Mitochondrial isolation
Mitochondria were isolated using ThermoFisher Scientific Mitochondria Isolation Kit for Cultured Cells (Catalog #: 89874) following the manufacturer's protocols using the dounce homogenization method. Using 20 million cells as input, cells were resuspended in kit buffer A and homogenized/lysed using a dounce tissue grinder. Cell lysate with intact mitochondria was centrifuged at 700g for 10 min, washing the cell lysate and removing cellular debris. The cytosolic and mitochondrial supernatant fractions were transferred to new tubes and centrifuged at 4000g for 12 min. The cytosolic supernatant fraction was removed and discarded leaving the isolated mitochondrial pellet remaining. The mitochondrial cell pellet was washed using kit buffer C and pelleted again by centrifuging at 5000g for 5 min. The supernatant was discarded, and the pure mitochondrial pellet was lysed for 30 min on ice using a lysis buffer mix (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1× Thermo Scientific Halt Protease inhibitor, and 1% NP40). The protein lysate was now centrifuged at 5000g for 15 min, then quantified using the ThermoFisher Scientific Pierce BCA assay, and used as input for Immunoprecipitation assay.
Immunoprecipitation assay
One milligram of isolated mitochondria lysate was incubated with 2 μg of V5 monoclonal antibody (Invitrogen, Catalog #: R960-25) overnight at 4 °C at 10 RPM. 30 μl of Protein G Dynabeads (Invitrogen, Catalog #: 10003D) per sample were pre-washed with wash buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 1× Thermo Scientific Halt Protease inhibitor). Washed Dynabeads were added to protein lysates and incubated at 4 °C at 10 RPM for 2 h. Using a magnetic tube rack, the supernatant fraction was removed, and beads with immunoprecipitated protein were washed 3× using the same wash buffer recipe as before, then eluted using 2× NuPage LDS sample buffer (Invitrogen, Catalog #: NP0007). Elution was boiled at 95 °C for 10 min and transferred to new tubes, cooled, then run on gels following the routine immunoblot protocol previously described.
Viability measurements using H2GFP/RFP
HEK293T WT and FDX1 KO cells were transduced with H2B-GFP or H2B-RFP, respectively. For the viability assays, cells were counted and plated at indicated ratios and viability was monitored by counting of the number of GFP or RFP positive cells over time using Incucyte (standard protocols).
Recombinant LIAS and FDX1 proteins over-production in E. coli and purification
Genes encoding LIAS (aa 28–372) and FDX1 (aa 61–184) lacking their respective mitochondrial transit peptides were custom synthesized at ThermoFisher Scientific after codon optimization for recombinant protein overexpression in Escherichia coli. The LIAS gene was subcloned into a modified pSUMO vector (LifeSensors Inc), (pDWSUMO), while the FDX1 gene was subcloned into pET28a vector. Both genes were cloned using NdeI and XhoI restriction sites of the appropriate vector. After gene sequence verification at the Penn State Genomics Core Facility (University Park, PA), the plasmids were separately used to transform E. coli BL21 (DE3) cells carrying pDB1282, the plasmid which harbors the isc operon from A. vinelandii (50). As cloned, LIAS was over-expressed as a fusion with a ULP1 protease cleavable N-terminal SUMO tag that also carried a hexa-histidine tag at its N-terminus, while FDX1 was overproduced with an N-terminal hexa-histidine tag. During purification, the SUMO tag was cleaved from the fusion protein affording pure LIAS with a Gly-His appendage at the N-terminus. Both proteins were overexpressed, purified and their respective iron-sulfur clusters reconstituted following procedures as previously published for LIAS with no further modifications (26).
Microscale thermophoresis (MST)
To further confirm the interactions between FDX1 and LIAS in vitro, their binding affinity was measured via MST. For these experiments, a Monolith NT.115 Microscale Thermophoresis instrument (NanoTemper Technologies GmbH) was used. For the binding assays, a Monolith RED-NHS second-generation protein labeling kit (NanoTemper Technologies GmbH) was used to label LIAS, and all the labeling and binding steps of the experiment were performed in an anaerobic chamber (Coy Laboratory products,). The labeling, binding, and data analysis steps were carried out following the procedures as recommended by the manufacturer of the labeling kit. The MST experiments were performed in triplicate at room temperature in MST Buffer (PBS buffer (19 mM Na2HPO4, 2 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.5 mM KCl) and with a 300 nM final concentration of the labeled LIAS protein as the target, while FDX1 was serially diluted (375–0.009 μM) as the ligand. The LIAS-FDX1 mixtures were incubated for 30 min at room temperature and then separately loaded into MST standard capillaries. The capillaries were then sealed and removed from the anaerobic chamber for binding measurements. Measurements were performed at 22 °C using medium MST power and 100% excitation power settings on the MO.control data collection software (NanoTemper Technologies GmbH). The data were analyzed and fit using the MO. Affinity Analysis software (NanoTemper Technologies GmbH) to determine the Kd of FDX1 binding to LIAS. The data were plotted using GraphPad Prism v8.3.0.
Data availability
All data supporting the findings of this study are available within this manuscript. Any further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peter Tsvetkov (ptsvetko@broadinstitute.org).
Supporting information
This article contains supporting information.
Conflict of interest
P. T. was a consultant and has received research funding from Riva Therapeutics.
Acknowledgments
We thank Joshua Dempster for help with the gene expression analysis.
Author contributions
P. T. conceptualization; M. B. D., N. R. B., P. T., B. P., N. K., D. M. W., S. J. B., and A. C. investigation; M. B. D., N. R. B., P. T., B. P., and N. K. formal analysis; D. M. W., S. J. B., P. T., and T. R. G. supervision; P. T. writing–original draft.
Funding and additional information
This work was supported by the National Cancer Institute grant 1 R35 CA242457-01 (T. R. G.) and R01CA279550 (P. T.), by Riva therapeutics (P.T., N. R. B. and M. B. D.), Research and Recruitment Funding by BCH (B. P. and N. K.), National Institutes of Health (awards GM-122595 to S. J. B.), the National Science Foundation (MCB-1716686 to S. J. B.), and the Eberly Family Distinguished Chair in Science (to S. J. B.). S. J. B. is an investigator of the Howard Hughes Medical Institute. N. K. is a Pew Biomedical Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by George M. Carman
Supporting information
Gene expression and dependency scores from CCLE.
Gene expression of HEK293T and K562 cells (WT, FDX1 KO and FDX1 KO +OE).
Metabolite analyses of HEK293T and K562 cells (WT, FDX1 KO and FDX1 KO +OE).
A, CYP11A1 mRNA expression levels in cancer cell lines from the CCLE categorized by lineage. B, the CRISPR/Cas9 LIAS loss-of-function viability outcome in 1128 cell lines correlated to the loss-of-function of all other tested genes. Plotted are the correlation values and the p-value for the 1000 most significant correlations. In green are genes associated with protein lipoylation pathway.
Supporting Figure S2: A, basal OCR was measured in HEK293T WT, FDX1 KO and FDX1 KO cells with FDX1 reconstitute (KO + OE). Mean ± SD, of at least eight biological replicates). B and C, viability of HEK293T WT and FDX1 KO overexpressing either H2B-GFP or H2B-RFP (respectively) was analyzed by cell counting when cells were grown separately (B) or as a 1:1 ratio mixture (C) in media containing 2 mM glucose. D and E, PCA of gene expression levels derived from RNA-seq in K562 (D) and ABC1 (E) WT (gray) and FDX1 KO (green) cells growing in either 2 mM (square) or 10 mM (circle) glucose conditions (n = 2 for each condition). F and G, volcano plots relating to LOG2 fold-change (x-axis) and -LOG10 (p-values) (y-axis) for gene expression changes observed in FDX1 KO as compared to WT in K562 (F) and ABC1 (G) cells growing in media containing 10 mM glucose. (H and I) The relative gene expression in FDX1 KO versus WT ABC1 (H) or K562 (I) WT and FDX1 KO cells growing in either 2 mM or 10 mM glucose media conditions. Relative gene expression is presented as a box plot of fold-change of mRNA (that was normalize din each cell ine to ACTNB) in FDX1 KO over WT cells as measured in three biological and two technical replicates.
Supporting Figure S3: A, intracellular metabolite level changes between FDX1 KO and WT K562 cells (presented as LOG2 fold change of KO/WT) grown in media containing either 2 mM (y-axis) or 10 mM (x-axis) glucose. B–E, Changes in intracellular levels of C5- (B), C3- (C) carnitines, phospho-ethanolamine (D) and CDP-choline (E) in either WT, FDX1 KO, or FDX1 KO cells with reconstituted FDX1 (KO+OE) K562 cells.
Supporting Figure S4: A, schematic describing the nano-luciferase complementation assays used and the equation used to normalize luminescence ratio (NLR) score corresponding to the raw luminescence value of the tested pair (A and B) divided by the sum of luminescence value from the two individual proteins (A-alone + B-alone). B, the oligomerization of FDX1 was measured by overexpressing FDX1 tagged with fragment 1 of the complementation Nano-luciferase (C1) together with either FDX1 tagged with fragment 2 (C2) or FDX2 tagged with fragment 2 (C2) used as a negative control. Luminescence was measured 24 h after transfection (n = 3) and presented as the calculated NLR score. G and H, the total protein input of the immunoprecipitation experiments presented in Fig. 4, E and F respectively.
References
- 1.Hall D.O., Cammack R., Rao K.K. Role for ferredoxins in the origin of life and biological evolution. Nature. 1971;233:136–138. doi: 10.1038/233136a0. [DOI] [PubMed] [Google Scholar]
- 2.Ewen K.M., Kleser M., Bernhardt R. Adrenodoxin: the archetype of vertebrate-type [2Fe-2S] cluster ferredoxins. Biochim. Biophys. Acta. 2011;1814:111–125. doi: 10.1016/j.bbapap.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Ewen K.M., Ringle M., Bernhardt R. Adrenodoxin--a versatile ferredoxin. IUBMB Life. 2012;64:506–512. doi: 10.1002/iub.1029. [DOI] [PubMed] [Google Scholar]
- 4.Grinberg A.V., Hannemann F., Schiffler B., Muller J., Heinemann U., Bernhardt R. Adrenodoxin: structure, stability, and electron transfer properties. Proteins. 2000;40:590–612. doi: 10.1002/1097-0134(20000901)40:4<590::aid-prot50>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 5.Miller W.L. Minireview: regulation of steroidogenesis by electron transfer. Endocrinology. 2005;146:2544–2550. doi: 10.1210/en.2005-0096. [DOI] [PubMed] [Google Scholar]
- 6.Miller W.L., Auchus R.J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freibert S.A., Goldberg A.V., Hacker C., Molik S., Dean P., Williams T.A., et al. Evolutionary conservation and in vitro reconstitution of microsporidian iron-sulfur cluster biosynthesis. Nat. Commun. 2017;8 doi: 10.1038/ncomms13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange H., Kaut A., Kispal G., Lill R. A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1050–1055. doi: 10.1073/pnas.97.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheftel A.D., Stehling O., Pierik A.J., Elsasser H.P., Muhlenhoff U., Webert H., et al. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11775–11780. doi: 10.1073/pnas.1004250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y., Ghosh M., Kovtunovych G., Crooks D.R., Rouault T.A. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim. Biophys. Acta. 2012;1823:484–492. doi: 10.1016/j.bbamcr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webert H., Freibert S.A., Gallo A., Heidenreich T., Linne U., Amlacher S., et al. Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin. Nat. Commun. 2014;5:5013. doi: 10.1038/ncomms6013. [DOI] [PubMed] [Google Scholar]
- 12.Cai K., Tonelli M., Frederick R.O., Markley J.L. Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase donate electrons iron-sulfur cluster biosynthesis. Biochemistry. 2017;56:487–499. doi: 10.1021/acs.biochem.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz V., Basu S., Freibert S.A., Webert H., Boss L., Muhlenhoff U., et al. Functional spectrum and specificity of mitochondrial ferredoxins FDX1 and FDX2. Nat. Chem. Biol. 2022;19:206–217. doi: 10.1038/s41589-022-01159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bareth B., Dennerlein S., Mick D.U., Nikolov M., Urlaub H., Rehling P. The heme a synthase Cox15 associates with cytochrome c oxidase assembly intermediates during Cox1 maturation. Mol. Cell. Biol. 2013;33:4128–4137. doi: 10.1128/MCB.00747-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barros M.H., Carlson C.G., Glerum D.M., Tzagoloff A. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 2001;492:133–138. doi: 10.1016/s0014-5793(01)02249-9. [DOI] [PubMed] [Google Scholar]
- 16.Tsvetkov P., Coy S., Petrova B., Dreishpoon M., Verma A., Abdusamad M., et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254–1261. doi: 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsvetkov P., Detappe A., Cai K., Keys H.R., Brune Z., Ying W., et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat. Chem. Biol. 2019;15:681–689. doi: 10.1038/s41589-019-0291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zulkifli M., Spelbring A.N., Zhang Y., Soma S., Chen S., Li L., et al. FDX1-dependent and independent mechanisms of elesclomol-mediated intracellular copper delivery. Proc. Natl. Acad. Sci. U. S. A. 2023;120 doi: 10.1073/pnas.2216722120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris T.W., Reed K.E., Cronan J.E., Jr. Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan S.W., Cronan J.E., Jr. A new metabolic link. The acyl carrier protein of lipid synthesis donates lipoic acid to the pyruvate dehydrogenase complex in Escherichia coli and mitochondria. J Biol Chem. 1997;272:17903–17906. doi: 10.1074/jbc.272.29.17903. [DOI] [PubMed] [Google Scholar]
- 21.Reed L.J. A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J Biol Chem. 2001;276:38329–38336. doi: 10.1074/jbc.R100026200. [DOI] [PubMed] [Google Scholar]
- 22.Cronan J.E., Zhao X., Jiang Y. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv Microb Physiol. 2005;50:103–146. doi: 10.1016/S0065-2911(05)50003-1. [DOI] [PubMed] [Google Scholar]
- 23.Broderick J.B., Broderick W.E., Hoffman B.M. Radical SAM enzymes: nature's choice for radical reactions. FEBS Lett. 2023;597:92–101. doi: 10.1002/1873-3468.14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broderick J.B., Duffus B.R., Duschene K.S., Shepard E.M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014;114:4229–4317. doi: 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landgraf B.J., McCarthy E.L., Booker S.J. Radical S-adenosylmethionine enzymes in human health and disease. Annu. Rev. Biochem. 2016;85:485–514. doi: 10.1146/annurev-biochem-060713-035504. [DOI] [PubMed] [Google Scholar]
- 26.Warui D.M., Sil D., Lee K.H., Neti S.S., Esakova O.A., Knox H.L., et al. In vitro demonstration of human lipoyl synthase catalytic activity in the presence of NFU1. ACS Bio. Med. Chem. Au. 2022;2:456–468. doi: 10.1021/acsbiomedchemau.2c00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saudino G., Ciofi-Baffoni S., Banci L. Protein-interaction affinity gradient drives [4Fe-4S] cluster insertion in human lipoyl synthase. J. Am. Chem. Soc. 2022;144:5713–5717. doi: 10.1021/jacs.1c13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendricks A.L., Wachnowsky C., Fries B., Fidai I., Cowan J.A. Characterization and reconstitution of human lipoyl synthase (LIAS) supports ISCA2 and ISCU as primary cluster donors and an ordered mechanism of cluster assembly. Int. J. Mol. Sci. 2021;22:1598. doi: 10.3390/ijms22041598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amici D.R., Jackson J.M., Truica M.I., Smith R.S., Abdulkadir S.A., Mendillo M.L. FIREWORKS: a bottom-up approach to integrative coessentiality network analysis. Life Sci. Alliance. 2021;4 doi: 10.26508/lsa.202000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayraktar E.C., La K., Karpman K., Unlu G., Ozerdem C., Ritter D.J., et al. Metabolic coessentiality mapping identifies C12orf49 as a regulator of SREBP processing and cholesterol metabolism. Nat. Metab. 2020;2:487–498. doi: 10.1038/s42255-020-0206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan J., Meyers R.M., Michel B.C., Mashtalir N., Sizemore A.E., Wells J.N., et al. Interrogation of mammalian protein complex structure, function, and membership using genome-scale fitness screens. Cell Syst. 2018;6:555–568.e7. doi: 10.1016/j.cels.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T., Yu H., Hughes N.W., Liu B., Kendirli A., Klein K., et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 2017;168:890–903.e15. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solmonson A., DeBerardinis R.J. Lipoic acid metabolism and mitochondrial redox regulation. J. Biol. Chem. 2018;293:7522–7530. doi: 10.1074/jbc.TM117.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowland E.A., Snowden C.K., Cristea I.M. Protein lipoylation: an evolutionarily conserved metabolic regulator of health and disease. Curr. Opin. Chem. Biol. 2018;42:76–85. doi: 10.1016/j.cbpa.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maio N., Rouault T.A. Iron-sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery. Biochim. Biophys. Acta. 2015;1853:1493–1512. doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy E.L., Booker S.J. Biochemical approaches for understanding iron-sulfur cluster regeneration in Escherichia coli lipoyl synthase during catalysis. Methods Enzymol. 2018;606:217–239. doi: 10.1016/bs.mie.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montero R., Yubero D., Villarroya J., Henares D., Jou C., Rodriguez M.A., et al. GDF-15 is elevated in children with mitochondrial diseases and is induced by mitochondrial dysfunction. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yatsuga S., Fujita Y., Ishii A., Fukumoto Y., Arahata H., Kakuma T., et al. Growth differentiation factor 15 as a useful biomarker for mitochondrial disorders. Ann. Neurol. 2015;78:814–823. doi: 10.1002/ana.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilsson R., Jain M., Madhusudhan N., Sheppard N.G., Strittmatter L., Kampf C., et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minton D.R., Nam M., McLaughlin D.J., Shin J., Bayraktar E.C., Alvarez S.W., et al. Serine catabolism by SHMT2 is required for proper mitochondrial translation initiation and maintenance of formylmethionyl-tRNAs. Mol. Cell. 2018;69:610–621.e5. doi: 10.1016/j.molcel.2018.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukha D., Fokra M., Feldman A., Sarvin B., Sarvin N., Nevo-Dinur K., et al. Glycine decarboxylase maintains mitochondrial protein lipoylation to support tumor growth. Cell Metab. 2022;34:775–782.e9. doi: 10.1016/j.cmet.2022.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newgard C.B., An J., Bain J.R., Muehlbauer M.J., Stevens R.D., Lien L.F., et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fontana D., Mauri M., Renso R., Docci M., Crespiatico I., Rost L.M., et al. ETNK1 mutations induce a mutator phenotype that can be reverted with phosphoethanolamine. Nat. Commun. 2020;11:5938. doi: 10.1038/s41467-020-19721-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gohil V.M., Zhu L., Baker C.D., Cracan V., Yaseen A., Jain M., et al. Meclizine inhibits mitochondrial respiration through direct targeting of cytosolic phosphoethanolamine metabolism. J. Biol. Chem. 2013;288:35387–35395. doi: 10.1074/jbc.M113.489237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni M., Solmonson A., Pan C., Yang C., Li D., Notzon A., et al. Functional assessment of lipoyltransferase-1 deficiency in cells, mice, and humans. Cell Rep. 2019;27:1376–1386.e6. doi: 10.1016/j.celrep.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi S.G., Olivet J., Cassonnet P., Vidalain P.O., Luck K., Lambourne L., et al. Maximizing binary interactome mapping with a minimal number of assays. Nat. Commun. 2019;10:3907. doi: 10.1038/s41467-019-11809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsvetkov P., Eisen T.J., Heinrich S.U., Brune Z., Hallacli E., Newby G.A., et al. Persistent activation of mRNA translation by transient Hsp90 inhibition. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seidel S.A., Dijkman P.M., Lea W.A., van den Bogaart G., Jerabek-Willemsen M., Lazic A., et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanz N.D., Grove T.L., Gogonea C.B., Lee K.H., Krebs C., Booker S.J. RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins. Methods Enzymol. 2012;516:125–152. doi: 10.1016/B978-0-12-394291-3.00030-7. [DOI] [PubMed] [Google Scholar]
- 51.McLaughlin M.I., Lanz N.D., Goldman P.J., Lee K.H., Booker S.J., Drennan C.L. Crystallographic snapshots of sulfur insertion by lipoyl synthase. Proc. Natl. Acad. Sci. U. S. A. 2016;113:9446–9450. doi: 10.1073/pnas.1602486113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joshi P.S., Sadre S., Guo X.A., McCoy J.G., Mootha V.K. Lipoylation is dependent on FDX1 and dispensable under hypoxia in human cells. J. Biol. Chem. 2023 doi: 10.1016/j.jbc.2023.105075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lanz N.D., Pandelia M.E., Kakar E.S., Lee K.H., Krebs C., Booker S.J. Evidence for a catalytically and kinetically competent enzyme-substrate cross-linked intermediate in catalysis by lipoyl synthase. Biochemistry. 2014;53:4557–4572. doi: 10.1021/bi500432r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mick E., Titov D.V., Skinner O.S., Sharma R., Jourdain A.A., Mootha V.K. Distinct mitochondrial defects trigger the integrated stress response depending on the metabolic state of the cell. Elife. 2020;9 doi: 10.7554/eLife.49178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen E.L., Ulanet D.B., Pirman D., Mahoney C.E., Coco J., Si Y., et al. Differential aspartate usage identifies a subset of cancer cells particularly dependent on OGDH. Cell Rep. 2016;17:876–890. doi: 10.1016/j.celrep.2016.09.052. [DOI] [PubMed] [Google Scholar]
- 56.Ilic N., Birsoy K., Aguirre A.J., Kory N., Pacold M.E., Singh S., et al. PIK3CA mutant tumors depend on oxoglutarate dehydrogenase. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E3434–E3443. doi: 10.1073/pnas.1617922114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression and dependency scores from CCLE.
Gene expression of HEK293T and K562 cells (WT, FDX1 KO and FDX1 KO +OE).
Metabolite analyses of HEK293T and K562 cells (WT, FDX1 KO and FDX1 KO +OE).
A, CYP11A1 mRNA expression levels in cancer cell lines from the CCLE categorized by lineage. B, the CRISPR/Cas9 LIAS loss-of-function viability outcome in 1128 cell lines correlated to the loss-of-function of all other tested genes. Plotted are the correlation values and the p-value for the 1000 most significant correlations. In green are genes associated with protein lipoylation pathway.
Supporting Figure S2: A, basal OCR was measured in HEK293T WT, FDX1 KO and FDX1 KO cells with FDX1 reconstitute (KO + OE). Mean ± SD, of at least eight biological replicates). B and C, viability of HEK293T WT and FDX1 KO overexpressing either H2B-GFP or H2B-RFP (respectively) was analyzed by cell counting when cells were grown separately (B) or as a 1:1 ratio mixture (C) in media containing 2 mM glucose. D and E, PCA of gene expression levels derived from RNA-seq in K562 (D) and ABC1 (E) WT (gray) and FDX1 KO (green) cells growing in either 2 mM (square) or 10 mM (circle) glucose conditions (n = 2 for each condition). F and G, volcano plots relating to LOG2 fold-change (x-axis) and -LOG10 (p-values) (y-axis) for gene expression changes observed in FDX1 KO as compared to WT in K562 (F) and ABC1 (G) cells growing in media containing 10 mM glucose. (H and I) The relative gene expression in FDX1 KO versus WT ABC1 (H) or K562 (I) WT and FDX1 KO cells growing in either 2 mM or 10 mM glucose media conditions. Relative gene expression is presented as a box plot of fold-change of mRNA (that was normalize din each cell ine to ACTNB) in FDX1 KO over WT cells as measured in three biological and two technical replicates.
Supporting Figure S3: A, intracellular metabolite level changes between FDX1 KO and WT K562 cells (presented as LOG2 fold change of KO/WT) grown in media containing either 2 mM (y-axis) or 10 mM (x-axis) glucose. B–E, Changes in intracellular levels of C5- (B), C3- (C) carnitines, phospho-ethanolamine (D) and CDP-choline (E) in either WT, FDX1 KO, or FDX1 KO cells with reconstituted FDX1 (KO+OE) K562 cells.
Supporting Figure S4: A, schematic describing the nano-luciferase complementation assays used and the equation used to normalize luminescence ratio (NLR) score corresponding to the raw luminescence value of the tested pair (A and B) divided by the sum of luminescence value from the two individual proteins (A-alone + B-alone). B, the oligomerization of FDX1 was measured by overexpressing FDX1 tagged with fragment 1 of the complementation Nano-luciferase (C1) together with either FDX1 tagged with fragment 2 (C2) or FDX2 tagged with fragment 2 (C2) used as a negative control. Luminescence was measured 24 h after transfection (n = 3) and presented as the calculated NLR score. G and H, the total protein input of the immunoprecipitation experiments presented in Fig. 4, E and F respectively.
Data Availability Statement
All data supporting the findings of this study are available within this manuscript. Any further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peter Tsvetkov (ptsvetko@broadinstitute.org).