Abstract
The Sonic Hedgehog (Hh) signal transduction pathway plays a critical role in many developmental processes and, when deregulated, may contribute to several cancers, including basal cell carcinoma, medulloblastoma, colorectal, prostate, and pancreatic cancer. In recent years, several Hh inhibitors have been developed, mainly acting on the Smo receptor. However, drug resistance due to Smo mutations or non-canonical Hh pathway activation highlights the need to identify further mechanisms of Hh pathway modulation. Among these, deacetylation of the Hh transcription factor Gli1 by the histone deacetylase HDAC1 increases Hh activity. On the other end, the KCASH family of oncosuppressors binds HDAC1, leading to its ubiquitination and subsequent proteasomal degradation, leaving Gli1 acetylated and not active.
It was recently demonstrated that the potassium channel containing protein KCTD15 is able to interact with KCASH2 protein and stabilize it, enhancing its effect on HDAC1 and Hh pathway.
KCTD15 and KCTD1 proteins share a high homology and are clustered in a specific KCTD subfamily. We characterize here KCTD1 role on the Hh pathway. Therefore, we demonstrated KCTD1 interaction with KCASH1 and KCASH2 proteins, and its role in their stabilization by reducing their ubiquitination and proteasome-mediated degradation. Consequently, KCTD1 expression reduces HDAC1 protein levels and Hh/Gli1 activity, inhibiting Hh dependent cell proliferation in Hh tumour cells. Furthermore, analysis of expression data on publicly available databases indicates that KCTD1 expression is reduced in Hh dependent MB samples, compared to normal cerebella, suggesting that KCTD1 may represent a new putative target for therapeutic approaches against Hh-dependent tumour.
Keywords: KCTD1, Hedgehog, KCASH1, KCTD11, KCASH2, KCTD21, KCASH3, KCTD6, HDAC1, Medulloblastoma, Cancer, Ubiquitination, Protein-stability
Introduction
The Sonic Hedgehog (Hh) signal transduction pathway plays a critical role in developmental processes and embryogenesis. In adult tissues, the Hh pathway is critical for the maintenance of stem cells, homeostasis, and tissue regeneration, but when deregulated may lead to cancerogenesis [1].
The activation of the Hh pathway occurs following the interaction between the Hh ligand and the membrane receptor Patched 1 (Ptch1). This binding interrupts the inhibitory action of Ptch1 on the Smoothened (Smo) coreceptor, allowing the release of the transcription factors Gli1, Gli2, and Gli3. Then, they migrate to the nucleus where promote the expression of a several genes involved in cell proliferation, survival, angiogenesis, and invasion [1,2] highlighting the pathway's key function in tumorigenesis processes. Indeed, alterations in Hh signaling components and non-canonical hyperactivation of Gli1 are present in many tumour types including basal cell carcinoma, medulloblastoma (MB), colorectal, prostate, pancreatic cancer [3]. Furthermore, several evidence suggested an emerging role of Hh signaling in immune crosstalk and modulation. Indeed, Hh regulates immunosuppressive mechanisms such as enhanced regulatory T-cell formation and production of immunosuppressive cytokines [4].
In recent years, some molecules have been identified to inhibit the Hh pathway, mainly acting on Smo receptor which have been approved by the FDA for therapeutic applications. However, resistance to the drug and relapse are often present, due to further Smo mutations inducing resistance or to the presence of non-canonical Hh pathway activation, independent of the binding of Smo antagonists [5]. Therefore, it is necessary to identify mechanisms of Hh pathway modulation downstream of the Smo receptor, and preferentially at the level of the Gli transcription factors. Indeed, the Gli activity is controlled also by the post-translation modification such as phosphorylation, ubiquitylation and acetylation[6]. Gli1 acetylation is a mechanism that prevents its migration into the nucleus and its transcriptional activity, while its deacetylation by the histone deacetylase HDAC1 increases its activity [7]. In turn, HDAC1 protein levels are modulated by the KCASH family of proteins (KCASH1KCTD11, KCASH2KCTD21 and KCASH3KCTD6). KCASH proteins have been identified as part of a Cullin E3 ubiquitin ligase complex which binds the histone deacetylase HDAC1, leading to its ubiquitination and subsequent proteasomal degradation [8].
Therefore, modulation of the KCASH family may represent a novel therapeutic strategy. This could be achieved by different approaches: among them the identification of transcriptional modulators that may be activated to drive the expression of KCASH genes [9], or the identification of protein stability or activity modulators, which in turn could be overexpressed or mimicked by small molecules.
Interestingly, it was recently demonstrated that the potassium channel containing protein KCTD15 can interact with KCASH2 through its BTB/POZ domain, enhancing its stability and playing an inhibitory effect on the Hh pathway-driven tumour cell proliferation [10].
However, analysis of publicly available expression databases did not highlight a strong inverse correlation between KCTD15 and Gli1 expression. This observation can be explained by the fact that KCTD15 is not a direct modulator of the Hedgehog pathway, but acts through the modulation of KCASH2, adding complexity and background noise to the analysis. At the same time, we hypothesize that other players, yet uncovered, may be involved in this regulatory mechanism.
KCTD1 encodes a KCTD protein which belongs to the same KCTD subfamily as KCTD15 [11]. These proteins share a homology of 81.4% in the BTB/POZ protein-protein interactor domain and 79.8% overall [12]. KCTD1 and KCTD15 have been suggested to have common characteristics, such as the inability to bind Cul3 and therefore to assemble an E3 ubiquitin ligase that is able to ubiquitinate and lead to proteasomal degradation its targets [13].
Given KCTD1 and KCTD15 similarities, we evaluated KCTD1 expression in normal and Hh-dependent tumour samples and characterized KCTD1 activity on KCASH proteins and on the Hh pathway, identifying a novel promising target for therapeutic approaches against Hh-dependent tumour.
KCTD1 inhibits the Hedgehog signalling reducing Gli1 activity
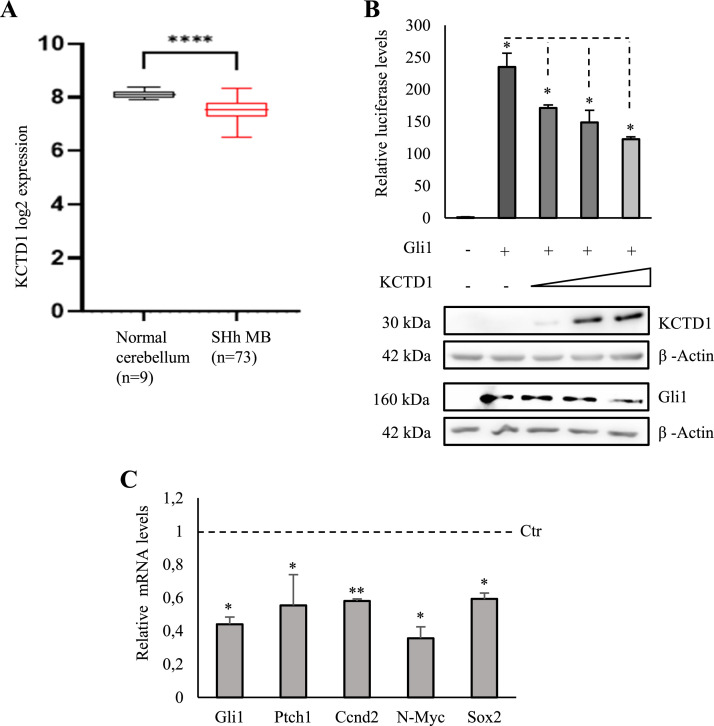
The relevance of KCTD1 in Hh dependent tumorigenesis was evaluated by the analysis of KCTD1 mRNA expression data from public databases. Indeed, these showed a significant reduction of KCTD1 expression in MB samples of the Hh group (n = 73) [14] compared to control cerebellar samples (****p < 0,0001; n=9; Fig. 1a) [15]. The downregulation of KCTD1 was further confirmed by analysis of a larger group of MB [16] (Fig. S1a; **p < 0,01).
Fig. 1.
KCTD1 suppresses the Hedgehog signalling (A) KCTD1 is downregulated in SHh-dependent MB samples. Expression levels of KCTD1 in normal cerebellum dataset and SHh-dependent MB analysed on R2 platform, as indicated in main text (****p<0.0001). (B) Upper panel: KCTD1 expression decreases Gli1-responsive element driven luciferase activity. Luciferase assays were performed on lysates from HEK293T cells transfected with 12×-GliRE-Luc alone or with Gli1, KCTD1 and pRL-TK Renilla (as a normalizer). Data are shown as mean ratios normalized to Renilla luciferase activity. Lower panel: KCTD1 and Gli1 protein levels were analysed by WB using Gli1 and KCTD1 antibodies. β-Actin protein was used as a normalizer. (C) Transcriptional activity of Gli1, Ptch1, Ccnd2, N-Myc, Sox2 is downregulated in KCTD1 transfected HEK293T cells. mRNA was assayed through RT-qPCR, normalized to HPRT, TBP and β-Actin, and represented as fold induction on the CTR. *p< 0.05; **p<0.01. Data represented mean of three independent experiments ± standard deviation (SD).
Next, we verified whether KCTD1 expression affects Gli1-dependent transcription: indeed, the expression of exogenous KCTD1 significantly decreased the transcriptional activity of a Gli1-responsive luciferase reporter (Fig. 1b). In agreement with these results, HEK293T cells overexpressing KCTD1 presented a reduced expression of the endogenous Hh target Gli1, which is the most reliable readout of Hh activity. Furthermore, other known Hh targets where modulated: PTCH1, the Hh transmembrane receptor, responsible for a negative feedback loop; cyclin D1, which is involved the progression of cell proliferation; N-Myc, important regulator of the early stages of cell cycle progression and Sox2, known as a master transcriptional regulator of stemness (Fig. 1c) [17], [18], [19], [20], [21]. Notably, KCTD1 overexpression did not affect significantly Gli2 and Gli3 protein levels (Fig. S1b, c).
KCTD1 modulates KCASH1 and KCASH2 protein stability
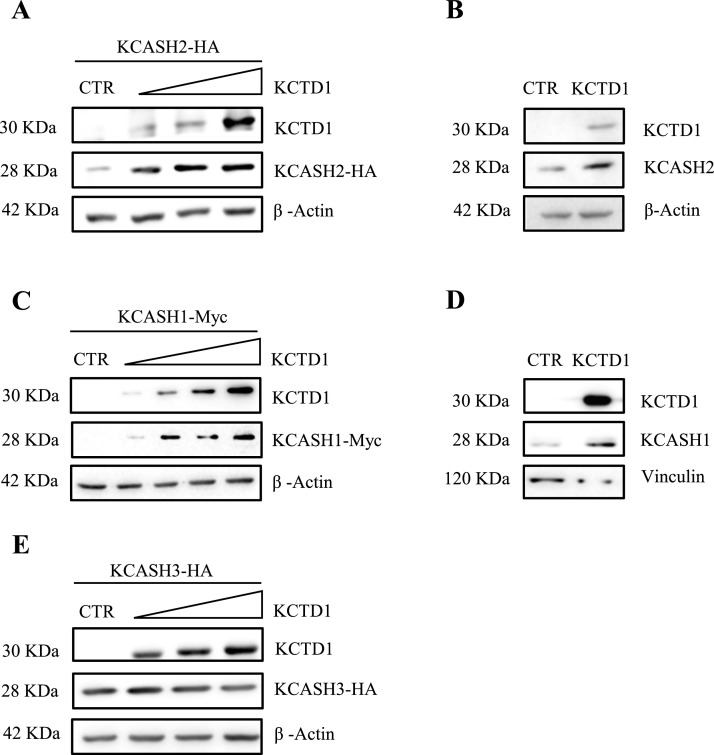
The ability of KCTD1 to suppress the activity of Gli1, and its homology with KCTD15 prompted us to investigate whether KCTD1 can inhibit the Hh signalling by controlling the stability of KCASH2 [10]. KCTD1 overexpression in HEK293T cells promotes a dose-dependent increase in KCASH2 protein levels, modulating both exogenous and endogenous protein (Fig. 2a, b).
Fig. 2.
KCTD1 expression increases KCASH2 and KCASH1 protein levels but does not modulate KCASH3. (A) HEK293T cells were transfected with HA-tagged KCASH2 and different amounts of KCTD1. After 24 h were lysed and analysed by WB using anti-HA and -KCTD1 antibodies. β-Actin protein was used as a normalizer. (B) HEK293T were transfected with empty vector or KCTD1 expressing vector; 24 h later, cells were lysed and endogenous KCASH2 proteins was analysed by WB. β-Actin was used as loading control. (C) HEK293T cells were transfected with Myc-tagged KCASH1 and different amounts of KCTD1, then lysed and analysed by WB using anti-Myc and -KCTD1 antibodies. β-Actin protein is shown as a normalizer. (D) HEK293T were transfected with empty vector or KCTD1 expressing vector; 24 h later, cells were lysed and endogenous KCASH1 was analysed by WB. Vinculin was used as loading control. (E) HEK293T cells were transfected with HA-tagged KCASH3 and different amounts of KCTD1, 24 later lysed and analysed by WB using anti-HA and -KCTD1 antibodies. β-Actin protein was used as a normalizer.
Since KCASH2 shares several structural and functional features with its paralogues, KCASH1 and KCASH3, we evaluated whether KCTD1 overexpression can induce stabilization of the other KCASH family members.
Exogenous expression of KCTD1 increases both exogenous and endogenous KCASH1 protein levels (Fig. 2c, d), while it does not affect KCASH3 protein levels (Fig. 2e).
Analysis of protein-protein interaction between KCTD1 and KCASH proteins
To evaluate biochemically the interaction between KCASH family members and KCTD1, we expressed and purified the different proteins and performed microscale thermophoresis [22].
The expression and purification for all the proteins was carried out as already described elsewhere [23], [24], [25] except for KCASH2. KCASH2 protein was purified by a two-step procedure consisting of a GST- affinity chromatography, a proteolytic digestion in the presence of thrombin followed by a further GST-affinity chromatography, and finally a gel filtration. The final yield of KCASH2 was of 2 mg/L.
To get insight into the structural integrity of KCASH2, the protein conformation was investigated by far-UV Circular dichroism (CD) spectroscopy. The spectrum revealed not only that the protein is correctly folded, but also resembles the one typical of an alpha-βeta-structured protein as expected from three-dimensional model derived from Alphafold predictions (Fig. S2a) [26].
The thermal stability of KCASH2 was also explored by far-UV CD spectroscopy following CD signal at 220 nm. Although a detailed thermodynamic characterization was hampered by protein precipitation upon unfolding, this study indicates that the domain is stable up to 63 °C (Fig. S2b). Comparing with KCASH1 [23] which displays a Tm of 45 °C, KCASH2 shows an 18 °C higher Tm indicating that KCASH2 is significantly more stable.
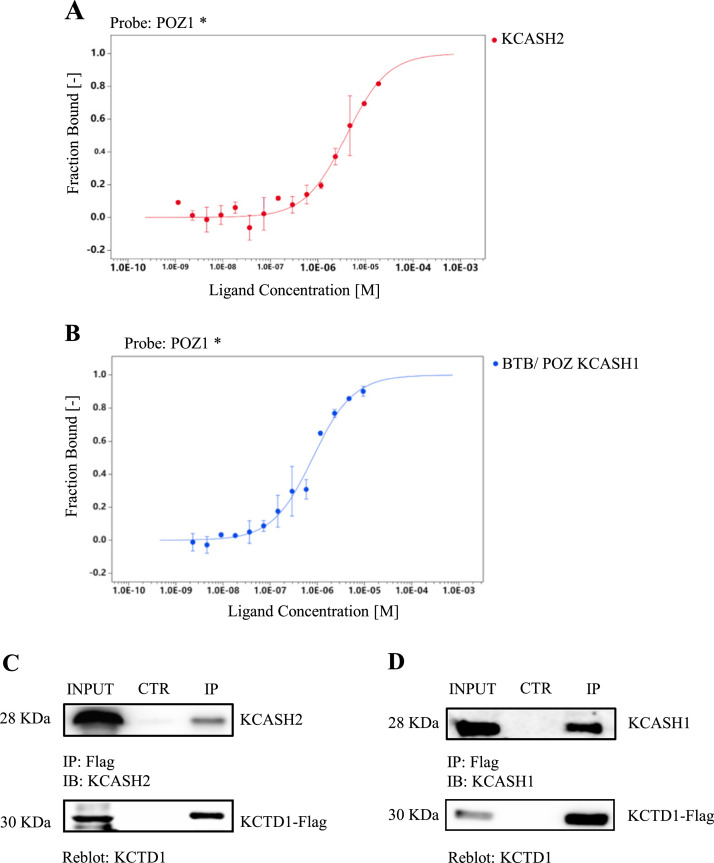
MST analyses showed that the C-terminal BTB/POZ containing fragment of KCTD1 (BTB/POZ1) binds KCASH2 with a good affinity (KD 3.9 ± 0.3 μM; Fig. 3a) comparable with the affinity observed for the BTB/POZ domain of KCTD15 (BTB/POZ15) binding to KCASH2 (KD of 6,6 μM ± 1,3 μM; Fig. S3a-b). The data observed for the interaction between the BTB/POZ15 and KCASH2 where consistent with the previously demonstrated ability to co-immunoprecipitate of the two full length proteins [10]. Interestingly, analysis of the affinity between BTB/POZ15 and BTB/POZ-KCASH1 seems to suggest a good affinity also between these two proteins (Fig. S3c). It must be noted that in previous work the interaction between the two full length proteins, expressed in cell cultures, could not be demonstrated [10], suggesting that the C-terminal regions of the two proteins may negatively affect their interaction, potentially through conformational changes or post-translational modifications.
Fig. 3.
KCTD1 can bind both KCASH2 and KCASH1 proteins. (A) MST fluorescence signal (fraction bound) of BTB/POZ1 plotted against increasing concentration of KCASH2. (B) MST fluorescence signal (fraction bound) of BTB/POZ1 plotted against increasing concentration of BTB/POZ11. (C) Co-IP assays were performed on total lysates from HEK293T cells transfected with expression vector encoding for Flag-tagged KCTD1 and immunoprecipitated (IP) with anti-Flag agarose beads. IP samples and a fraction of the total lysate were separated on SDS-PAGE gels. Blots were immunoblotted (IB) with anti-KCASH2 antibody and reblotted with anti-KCTD1 antibody. (D) Co-IP assays were performed on total lysates from HEK293T cells transfected with expression vectors encoding Flag-tagged KCTD1 and the immunoprecipitated (IP) with anti-Flag agarose beads. IP samples and a fraction of the total lysate were separated on SDS-PAGE gels. Blots were immunoblotted (IB) with anti-KCASH1 antibody and reblotted with anti-KCTD1 antibody.
As expected, BTB/POZ1 binds also KCASH1 with an affinity (KD 0.8 ± 0.1 μM) comparable to KCASH2 (Fig. 3b and S4a-b).
Interestingly, BTB/POZ1 can interact with the BTB/POZ domain of KCTD15 (KD: ± 0,9 μM ± 0,05 μM); and this interaction is confirmed with the same affinity when using the full length KCTD1 (flKCTD1), confirming that in this context the interactions are driven mainly by the BTB/POZ domains (Fig. S5a, c).
In vitro co-immunoprecipitation assay (Co-IP), following the overexpression of Flag-tagged KCTD1 and HA-tagged KCASH2, Myc-tagged KCASH1 in HEK293T cells, confirmed that KCTD1 interacts with these proteins (Fig. 3c, d).
KCTD1 increases KCASH1 and KCASH2 half-life by inhibiting their proteasome-dependent degradation
We hypothesized that KCTD1 increases KCASH2 and KCASH1 stability by inhibiting their degradation.
To verify this hypothesis, HEK293T cells were transfected with a control or a KCTD1 expressing plasmid, then treated with cycloheximide (CHX), an inhibitor of protein synthesis.
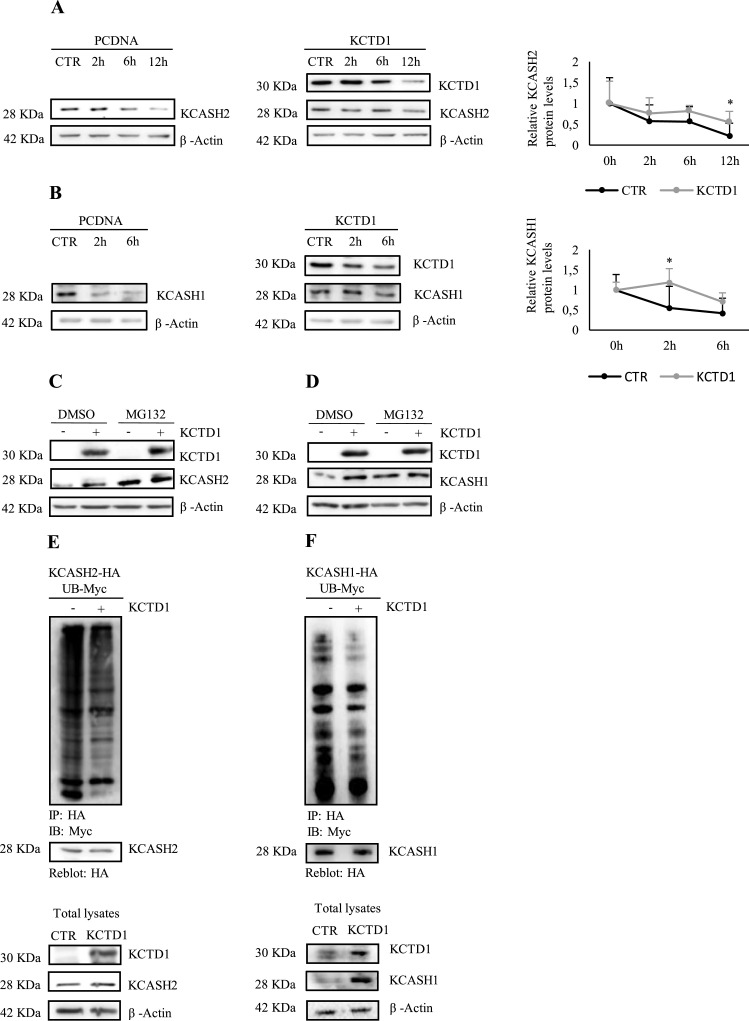
In the control transfected cells, KCASH2 exhibited a half-life between 6 and 12 hours, while KCASH1 displays a half-life shorter than 2 hours (Fig. 4a, b). This result is coherent with the higher protein stability of KCASH2 shown in Fig. S2.
Fig. 4.
KCTD1 modulates KCASH1 and KCASH2 half-life by inhibiting their proteasome-dependent degradation. (A-B) HEK293T were transfected with empty vector or KCTD1 expressing vector; then, 24 h later, were treated with CHX (200 µg/ml) for several hours as reported in figure. Protein lysates were analysed by WB, using KCTD1, KCASH2, KCASH1 antibodies. β-Actin protein was used as a normalizer. Data represented mean of three independent experiments ± standard deviation (SD) *p<0,05. (C-D) Cell treatments with the proteasome inhibitor MG132 increases the levels of KCASH1 and KCASH2 proteins and prevents their degradation. HEK293T were transfected with empty vector or KCTD1 expressing vector; 24 h later, they were treated with MG132 (5 μM) for 16 h. Protein lysates were analysed by WB, using KCTD1, KCASH2 and KCASH1 antibodies. β-Actin protein was used as loading control. (E-F) Cell lysates from HEK293T cells, transfected with the indicated plasmids, were immunoprecipitated with anti-HA agarose beads and immunoblotted with anti-Myc to detect conjugated Myc-Ub. The blot was re-probed with an anti-HA antibody to monitor immunoprecipitation efficiency. Total protein lysates were analysed by WB, using KCTD1, KCASH2 and KCASH1 antibodies. β-Actin protein was used as loading control.
In KCTD1 overexpressing cells, KCASH2 and KCASH1 half-life was significantly extended (Fig. 4a, b) compared to control cells, suggesting that KCTD1 interferes with KCASH protein turn over. Of note, KCTD1 expression has a relatively stronger effect on KCASH1 stability, allowing it to reach a half-life similar to KCASH2 protein.
We hypothesized that KCASH proteins degradation may be mediated by the proteasome, similarly to other KCTD family members, such as KCTD10 [27].
As shown in Fig. 4c and d, in untreated cells the exogenous expression of KCTD1 results in increased levels of KCASH2 and KCASH1 (see lane 1 versus lane 2) comparable to levels that can be seen after treatment with the proteasome inhibitor MG132 (lanes 3 and lane 4). Interestingly, following MG132 treatment, the expression of KCTD1 does not further raise KCASH levels, indicating that KCTD1 mechanisms of action interferes with proteasome mediated degradation.
Coherently with this hypothesis, we observed that KCTD1 can decrease the ubiquitination of KCASH2 and KCASH1 which is known to be fundamental for protein targeting to the proteasome [28] (Fig. 4e, f).
KCTD1 reduces HDAC1 protein levels through KCASH1 and KCASH2
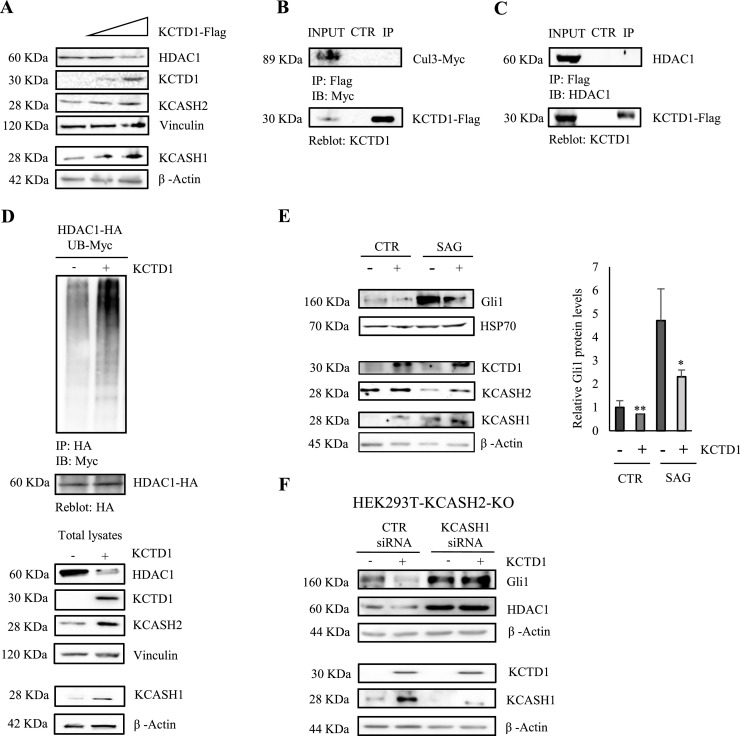
Since KCASH oncosuppressors are known to regulate the Hh pathway through promotion of HDAC1 degradation, we wondered whether KCTD1 overexpression affects the modulation of HDAC1. For this purpose, HEK293T cells were transfected with increasing amounts of the vector encoding Flag- tagged KCTD1. The overexpression of KCTD1 leads to an increase in KCASH1 and KCASH2 and a decrease in HDAC1 protein levels (Fig. 5a).
Fig. 5.
KCTD1 reduces Gli1 transcriptional activity and HDAC1 protein levels but does not interact with Cul3 and HDAC1. (A) KCTD1 expression reduces HDAC1 protein levels. HEK293T cells were transfected with different amounts of KCTD1; after 24 h, cells were lysed and endogenous HDAC1, KCASH1 and KCASH2 protein levels were analysed by WB. Vinculin and β-Actin were used as loading controls. KCTD1 is not able to interact with Cul3 (B) and HDAC1 (C). Co-IP assays were performed from HEK293T cells transfected with expression vector Flag-tagged KCTD1, Myc-tagged Cul3 (B); from HEK293T cells transfected with expression vector Flag-tagged KCTD1 (C). Total lysates were immunoprecipitated (IP) with anti-Flag agarose beads and IP samples and a fraction of the total lysate were separated on SDS-PAGE gels. Blots were immunoblotted (IB) with anti-HDAC1 antibody, anti-Cul3 antibody, and reblotted with anti-KCTD1 antibody. (D) On the top, the KCTD1 presence promotes HDAC1 ubiquitination. Lysates from HEK293T cells, transfected with the indicated plasmids, were immunoprecipitated with anti-HA agarose beads and immunoblotted with anti-Myc to detect conjugated Myc-Ub. The blot was re-probed with an anti-HA antibody to monitor immunoprecipitation efficiency. On the bottom, HDAC1, KCTD1, KCASH2 and KCASH1 total protein levels. Vinculin and β-Actin protein were used as loading controls. (E) KCTD1 is not able to decrease Hedgehog activity in absence of KCASH1 and KCASH2. HEK293T KCASH2-KO cells were transfected with scrambled siRNA (siCTR) or with KCASH1 siRNA (siKCASH1) and 48 h later protein lysates were analysed by WB, using HDAC1, KCTD1, KCASH1 and Gli1 antibodies. β-Actin protein was used as a normalizer. (F) KCTD1 reduces Gli1 activity in cells treated with Smo agonist SAG. HEK293T were transfected with empty vector or KCTD1 expressing vector, pre-starved in serum-free medium for 24 h and then treated with 200nM SAG for 48 h. Protein lysates were analysed by WB, using KCTD1, Gli1, KCASH2 and KCASH1 antibodies. β-Actin protein was used as loading control.
Although KCTD1 shares a considerable degree of similarity with the KCASH protein family in the BTB/POZ domain, it was observed that KCTD1 is not able to interact with Cul3 [13]. We confirmed these data by Co-IP in HEK293T cells expressing Flag-tagged KCTD1 (Fig. 5b). We next performed Co-IP experiments to verify if KCTD1 has a direct interaction with HDAC1. As expected, KCTD1, as KCTD15, does not co-immunoprecipitate with HDAC1, indicating that KCTD1 acts on HDAC1 indirectly, through the modulation of KCASH1 and KCASH2 (Fig. 5c).
Next, we verified if the expression of KCTD1 promotes HDAC1 ubiquitination. For this purpose, we performed ubiquitination assays on cell lysates from HEK293T cells transfected with the HA-tagged HDAC1 and Myc-tagged Ubiquitin, in presence or absence of Flag-tagged KCTD1. As expected, the presence of KCTD1 promotes the ubiquitination of HDAC1 (Fig. 5d) which will lead to its proteasomal degradation. The effect of KCTD1 on Gli1 was confirmed not only at the basal level of Hh activity, but also after the stimulation of the Hedgehog pathway with the Smo agonist SAG [29], which increased significantly Gli1 expression. Also in this condition, KCTD1 overexpression was able to increase KCASH1 and KCASH2 protein levels and turn off the Hh pathway significantly (Fig. 5e).
Based on our observations, the ability of KCTD1 to induce HDAC1 degradation and inhibit Hh activity (and therefore Gli1 expression) seems to relay on the presence of KCASH2 and KCASH1. Indeed, when we silenced KCASH1 expression in KCASH2 KO cells (generated using Crispr/Cas9; Fig. S6), generating a double knock-out, the effects of KCTD1 overexpression on Gli1 and HDAC1 protein levels were abolished (Fig. 5f).
KCTD1 expression inhibits Hedgehog dependent cell proliferation in medulloblastoma cells
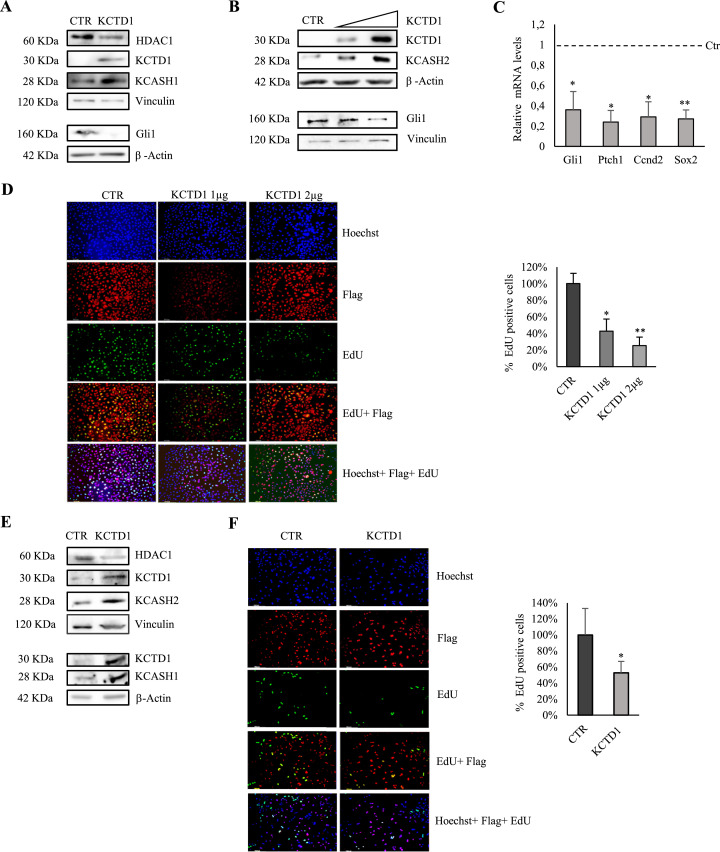
To evaluate the relevance of KCTD1 in the Hh signaling in a tumoral context, we used a Hh dependent MB cell line (DAOY). Overexpression of KCTD1 led to an increase of endogenous KCASH1 and KCASH2 protein levels and a concomitant reduction in Hh activity, measured by monitoring Gli1 protein levels (Fig. 6a, b). Similarly, KCTD1 expression reduced the expression of Hh target genes, such as Gli1, Ptch1, Cyclin D2 and Sox2. [17,21,30] (Fig. 6c). Furthermore, overexpression of KCTD1 in DAOY cells reduced their proliferation, as monitored by EdU incorporation assay (Fig. 6d).
Fig. 6.
KCTD1 overexpression inhibits Hedgehog signalling in MB cell lines, increasing KCASH1 and KCASH2 protein levels. (A-B) KCASH1 and KCASH2 protein levels are increased in MB cells expressing KCTD1 while HDAC1 and Gli1 protein are reduced. (A) DAOY were transfected with empty vector (Ctr) or KCTD1 expressing vector and 24 h later protein lysates were analysed by WB, using HDAC1, KCTD1, KCASH1 and Gli1 antibodies. β-Actin and Vinculin protein were used as a normalizer. (B) DAOY cells were transfected with different amounts of KCTD1, 24 h later, they were lysed and analysed by WB, using HDAC1, KCTD1, KCASH2 and Gli1 antibodies. β-Actin or Vinculin were used as loading controls. (C) Transcriptional activity of Gli1, Ptch1, Ccnd2, Sox2 is reduced in KCTD1 expressing HEK293T cells. The mRNA levels were evaluated by RT-qPCR, normalized to β-Actin, HPRT and β2m, and represented as % of the Ctr. (D) KCTD1 overexpression reduced EdU incorporation in DAOY cells. DAOY cells were transfected with the control plasmid PCDNA-Flag and different amounts of KCTD1-Flag for 24 h. Following 1h incubation with EdU, the cells were fixed and stained. Percentage of EdU positive cells was calculated over total transfected cells. (E) KCASH1 and KCASH2 protein levels are increased in D283-Med MB cells following expression of KCTD1 while HDAC1 protein is reduced. D283 cells were transfected with empty vector or KCTD1 expressing vector and 24 h later protein lysates were analysed by WB, using anti-HDAC1, KCTD1, KCASH1 and KCASH2 antibodies. β-Actin and Vinculin protein were used as a normalizer. (F) Proliferation of D283 cells is decreased following KCTD1 overexpression. D283 cells were transfected with the control plasmid or KCTD1-Flag for 24 h. Following 1h incubation with EdU, the cells were fixed and stained with the kit. Percentage of EdU positive cells was calculated over total transfected cells. (*p< 0.05, **p< 0.01; results are expressed as the mean ± SD of three independent experiments, Student's t-test).
The interplay between KCTD1-KCASHs-HDAC1 was confirmed also in a second human medulloblastoma cell line, the D283-Med (D283). D283, although not considered properly a Hh-dependent cell line (it is often classified as Group 3/4 MB), have been previously shown to respond to Hh inhibition [31] and to HDAC inhibition [32] providing a good enough model. Data on the D283 cells confirmed an increase of the levels of KCASH1 and KCASH2 following KCTD1 overexpression, and a consequent reduction of HDAC1 levels (Fig. 6e). At the same time, we could demonstrate a reduction of cell proliferation following overexpression of KCTD1, both with the EdU incorporation assay and the Ki67 marker of proliferation (Fig. 6f and Fig.S7). The reduction of D283 cell proliferation is coherent with a previous observation on D283 cells, following KCASH1 overexpression [33].
Discussion
Here we report the identification and characterization of KCTD1 as a novel positive modulator of the KCASH family of oncosuppressors, revealing a new function for KCTD1 as a suppressor of the Hedgehog pathway.
KCTD1 was first identified as a transcriptional repressor of AP2α [12,34,35].
It was also demonstrated that mutations of the KCTD1 gene, especially at the level of the BTB/POZ domain, are associated with the onset of SEN, a rare autosomal dominant disease, characterized by cutis aplasia of the scalp, syndactyly, anomalies of the external ears and nails; and malformations of the breast [36,37]. Although part of the phenotype of the SEN (i.e. cutis aplasia) could be linked to the potential suppression of AP2α transcriptional activity, other ectodermal abnormalities are likely a consequence of altered KCTD1 activity on other developmental pathways.
Recently, a role for KCTD1 in cancer has been suggested, proposing that KCTD1 in colon adenocarcinoma cell lines may act as a tumour suppressor, due to its ability to downregulate β-catenin, a central player in the WNT/β-catenin signalling pathway [38].
KCTD1 is part of KCTD family and is clustered in clade A together with KCTD15. These two proteins share an 80% homology [12] and both have been involved in the onset of neurodevelopmental disorders [39].
Previous data have shown that KCTD15 is involved in the Hh modulation, acting as stabilizer of the oncosuppressor KCASH2 [10]. Given the high similarity of KCTD1 and KCTD15, we investigated if KCTD1 may play an analogous role in this context.
We demonstrate here that KCTD1 can stabilize and consequently increase the KCASH2 protein levels. Interestingly, while KCTD15 modulates only KCASH2, KCTD1 can bind to and stabilize also KCASH1. Although KCTD1 is not able to interact directly with KCASH3, it has to be noted that KCASH3 does not bind directly HDAC1 but acts on it only through heterodimerization with KCASH1 [8]. Consequently, modulation of KCASH1 and KCASH2 is sufficient to affect also KCASH3 activity.
We also report that KCASH1 and KCASH2 stability is regulated by ubiquitination and proteasomal degradation and that KCTD1 binding to KCASH1 and KCASH2 interferes with their ubiquitination. This effect may be mediated by a change of KCASHs conformations which may affect their protein-protein interaction capability or may mask their ubiquitination sites.
As a consequence of KCASH1 and KCASH2 stabilization, KCTD1 overexpression leads to an increase of HDAC1 ubiquitination and proteasomal degradation, acting as a negative regulator of the Hh pathway.
The contribution of KCTD1 in the control of the Hh pathway was also confirmed in a pathological setting. Indeed, overexpression of KCTD1 in human MB cell lines increases KCASH1 and KCASH2 protein levels, reducing HDAC1 levels and suppressing Hh signaling and MB cell proliferation.
Furthermore, in silico analysis of expression data on publicly available databases indicates that KCTD1 expression is reduced in Hh dependent MB samples, compared to normal cerebella. Of note, KCTD1 expression is reduced also in WNT dependent MB samples (see Fig. S1), coherently with the recently described role for KCTD1 in modulating also the β-catenin/WNT pathway [38]. Furthermore, the observation of a downregulation of KCTD1 also in the worst prognosis Group 3 MB, which is often characterized by Myc amplification [40], hints to further not yet identified anti tumoral functions of KCTD1, including the ability of this gene to negatively modulate the transcriptional machinery [16].
According to our data and published literature, it is important to highlight that KCTD1 may act at the crossroads of different pathways involved in development and cancerogenesis. Interestingly the phenotype observed in KCTD1 mutated SEN patients does not appear to be due only to alteration of the AP2α signaling but is likely to be associated to alteration of other signalling. Indeed, athelia has been associated also to alterations in the WNT signalling [41]. Likewise, anomalies in the Hh pathway, may lead to a pattern of developmental defect that partially overlap with the SEN syndrome. In particular, the presence of alteration of keratinocyte homeostasis (leading often to Basal Cell carcinomas) and syndactyly are common features of the Gorlin-Goltz syndrome, a rare autosomal dominant disorder caused by mutations in the Hedgehog signalling pathway [42].
Similarly, the observation of altered mobility and stemness in cells lacking KCTD1 [38], may be due not only to alterations in the WNT pathway, but also in the Hh pathway, which is involved in cell stemness and metastasis [43].
The data presented here, by identifying the new Hh modulator KCTD1, contributes to a deeper understanding of the mechanism modulating KCASH proteins, and in turn their ability to suppress Gli1 activity. This information may provide the bases for new therapeutic approaches against Hh dependent tumours.
Materials and methods
Cell culture and transfection
Medulloblastoma cell line DAOY (ATCC HTB-186) was cultured in Minimum Essential Medium (Gibco-Thermo Fisher Scientific, Massachusetts, United States) supplemented with 10% heat-inactivated foetal bovine serum (FBS), 1% sodium pyruvate, 1% non-essential amino acid solution, 1% L-glutamine, and 1% penicillin/streptomycin.
Medulloblastoma cell line D283 Med (ATCC HTB-185) was cultured in Minimum Essential Medium (Gibco-Thermo Fisher Scientific, Massachusetts, United States) supplemented with 20% heat-inactivated foetal bovine serum (FBS), 1% sodium pyruvate, 1% non-essential amino acid solution, 1% L-glutamine, and 1% penicillin/streptomycin.
DAOY and D283 Med cells were transfected with Lipofectamine Plus, according to the manufacturer's instructions (Invitrogen-Thermo Fisher Scientific, California, United States).
HEK293T cells (ATCC CRL-3216) were cultured in Dulbecco Modified Eagle Medium (Gibco) supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin. HEK293T cells were transfected with Lipofectamine 2000, according to the manufacturer's instructions (Invitrogen).
Mycoplasma contamination in cells cultures was routinely screened by using PCR detection kit (Applied Biological Materials, Richmond, BC, Canada).
HEK293T cells were transfected with using Lipofectamine 2000 (Invitrogen, Carlsbad,
CA, USA) according to the manufacturer's instructions. DAOY cells were transfected with Lipofectamine Plus, according to the manufacturer's instructions (Invitrogen-Thermo Fisher Scientific, California, United States).
Reagents
Cycloheximide (CHX) was provided by SERVA (Heidelberg, Germany). MG-132 was purchased from Sigma-Aldrich (Darmstadt, Germany). HEK293T were incubated with 200μg/ml CHX for 2h, 6h and 12 h; and with MG-132 (5 μM) for 16 h.
Hyperconfluent HEK293T cells pre-starved for 24 h in serum-free DMEM medium were treated with 200 nM Smoothened Agonist (SAG, Sigma-Aldrich, Steinheim, Germany) for 48 h.
Gene silencing using small-interfering RNA (siRNA)
KCTD11/KCASH1 RNA interference (siRNA) was performing using Silencer® Select Pre-designed siRNA (16708A, Ambion-Thermo Fisher Scientific). The gene silencing effects were evaluated by Western blotting.
Plasmids
The following plasmids were used: 12X Gli-Luc and pRL-TK Renilla (kind gift from R. Toftgård, Karolinska Institutet, Stockholm); pCDNA Cul3-myc (kind gift M. Pagano, New York University school of Medicine, NY); c-Flag pcDNA3 (20011; Addgene), pEGFP-N1 was obtained from Clontech (Takara, Saint-Germain-en-Laye, France). pCMV-KCTD1-Flag; pCXN-hKCASH1/KCTD11-Myc; pCXN2-hKCASH2/KCTD21-HA; pCXN2-h KCASH3/KCTD6-HA; pCDNA3.1-HDAC1-HA; pCDNA3.1-Gli2-Flag; pCDNA3.1-Gli3-Flag were generated with standard cloning techniques and verified by sequencing.
Luciferase activity assay
Dual luciferase assay reactions were prepared using Firefly Luciferase Assay Kit 2.0 (Biotium, California, USA), following manufacturer's instructions. Luciferase activity was quantified using GloMax® Discover Microplate Reader (Promega). Results are expressed as Luciferase/Renilla ratios.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cells was extracted using TRizol (Invitrogen) and RNA Clean & Concentrator™-5 (R1014, Zymo Research, California, USA). cDNA synthesis was performed using the High-Capacity cDNA reverse transcription kit (BIO-65054, Meridian Bioscience, Ohio, USA) according to the manufacturer's instructions. Quantitative real-time PCR analysis of CCND1, CCND2, N-Myc, Sox2, Ptch1 and Gli1 messenger RNA (mRNA) was performed on cDNAs employing TaqMan gene expression assay (Applied Biosystem - Thermo Fisher Scientific) and using the ViiATM7 Real-Time PCR System (Applied Biosystem) as previously described [44]. All results were normalized to the endogenous controls: TBP (4326322E), ß2M (4326319E), HPRT (4326321E) and ß-Actin (4326315E, Applied Biosystem).
Western blot and co-immunoprecipitation assay
Western blotting (WB) was performed by lysing cells in denaturing buffer SDS-urea (50 mM Tris HCl pH 7.8, 2% SDS, 10% glycerol, 10 mM Na4P2O7, 100 mM NaF, 6 M urea, 10 mM EDTA). Extracts were then sonicated, quantified, and loaded onto SDS–polyacrylamide gel. The gel was then transferred to a nitrocellulose membrane (#NBA085C001EA, PerkinElmer), which was then blocked with 5% milk in TBS-T (Tris HCl with 0.1% Tween 20) and incubated with primary antibodies overnight and HPR-conjugated secondary antibodies, diluted in 5% milk solution. Detection of the HRP signal was performed by using ECL (#K-12045-D50, Advansta).
For the Co-immunoprecipitation, the cells were lysed with buffer containing 50 mM Tris-HCl pH 7.6, 1% deoxycholic acid sodium salt, 150 mM NaCl, 1% NP40, 5 mM EDTA, 100 mM NaF, supplemented with phosphatase inhibitor and Halt Protease Inhibitor cocktail (Thermo Fisher Scientific). The lysates were incubated with Flag beads (A2220; Sigma-Aldrich Merck, Darmstadt, Germany) and HA beads (A2095; Sigma-Aldrich Merck, Darmstadt, Germany) for 2 h at 4°C . Control samples were saturated with anti-Flag peptide (F3290; Sigma-Aldrich Merck) or with HA Synthetic Peptide (26184; Thermo Fisher Scientific). Beads were washed extensively with lysis buffer, and the complexes were evaluated by WB analysis. We used the antibodies listed below: mouse monoclonal antibody against β-actin (AC-15, A5441, Sigma-Aldrich Merck), mouse monoclonal anti-Vinculin (SC-73614; Santa Cruz Biotechnology), rabbit monoclonal anti-KCTD21/KCASH2 (AB192259; Abcam, Cambridge, UK), rabbit polyclonal anti-KCTD1 (PA5-24877; Invitrogen), rabbit polyclonal anti-KCTD11/KCASH1 (PA5-41088; Invitrogen), rabbit monoclonal anti-HDAC1 (H3284; Sigma-Aldrich Merck), rabbit polyclonal anti-Gli1 (2553; cell signalling); mouse monoclonal anti-Cul3 (SC-166110; Santa Cruz Biotechnology), mouse monoclonal anti-HA (SC-7392; Santa Cruz Biotechnology), mouse monoclonal anti-Flag M2 (F3165; Sigma-Aldrich), rabbit monoclonal anti-c-Myc (C3966; Sigma-Aldrich Merck). Secondary antibody anti–mouse (SC-516102) or anti-rabbit (SC-2357), conjugated with HRP were from Santa Cruz.
Ubiquitination assay
The HEK293T cells, after 24 h from transfection, were lysed in a solution containing RIPA buffer (50 mM Tris-HCl at pH 7.6, 150 mM NaCl, 0.5% sodium deoxycholic, 5 mM EDTA, 0.1% SDS, 100 mM NaF, 2 mM Tetrasodium pyrophosphate and 1% NP-40) supplemented with protease and phosphatase inhibitors. Lysates were subjected to immunoprecipitation with HA beads (Sigma-Aldrich Merk, A2095) for 2 hours, at 4°C, with rotation. The immunoprecipitated proteins were then washed five times with the RIPA lysis buffer, resuspended in sample loading buffer, boiled for 5 min, resolved in SDS-PAGE, and then subjected to immunoblot analysis.
CRISPR/Cas9
sgRNA (single-guide RNA) specific for KCASH2 gene was designed using the informatics platform Benchling (Benchling, 2019, https://benchling.com). PAM sequence (NGG) was located before the stop sequence. The sg-RNA was cloned into the LentiCRISPRV2-puro vector (Addgene plasmid #52961). HEK293T transfected cells were selected with Puromycin and subcloned to single-cell to obtain a line KCASH2 Knock-out. Subsequently, the absence of KCASH2 was confirmed through sequencing.
Proliferation assay and Immunofluorescence
An assay kit (C10337; Invitrogen) was adopted to inquire the cell proliferation ability. D283 Med cells were transferred to coverslips pre-coated with Poly-L-Lysine (10µg/ml in ddH2O, catalogue number P6516, Sigma-Aldrich). DAOY and D283 Med cells were transfected with the indicated plasmids and incubated with EdU buffer at 37 °C for 1 h, fixed with 4% formaldehyde for 15 minutes and permeabilized with 0.1% Triton X-100 for 20 min. EdU solution was added into culture followed by the staining of nuclei with Hoechst. To identify KCTD1-Flag and PCDNA-Flag we used the Anti-Flag antibody (F7425, Sigma Aldrich); To detect the presence of Ki67 was used the Ki-67 Antibody (H-300): sc-15402 (Santa Cruz Biotechnology); the secondary antibody Alexa Fluor 594 (A-11012, Thermo Fisher Scientific). Images were acquired by a fluorescence microscope.
Production of recombinant proteins
KCTD1 FL, BTB/POZ1, BTB/POZ11 and BTB/POZ15 proteins were produced as described elsewhere [12,23,25,45].
The BTB/POZ domains of the potassium channel tetramerization domain proteins prevalently assume pentameric states [45].
The full-length sequence of KCASH2 was cloned in pGEX4T3 using the restriction sites BamHI and EcoRI. The recombinant protein GST-KCASH2 was purified by a GST-Trap FF (Cytiva, Milan, Italy). The fraction of interest was dialyzed against 50 mM Tris/HCl pH 7.5, 150 mM NaCl, 1 mM DTT at 4°C and successively digested with the specific protease (Thrombin, SIGMA Aldrich). Successively, the sample was loaded again on the GST-affinity chromatography and finally obtained pure on a gel-filtration chromatography (Superdex 200 10 × 300, Cytiva, Milan, Italy).
Circular dichroism
KCASH2 CD spectra were recorded at 20°C using a Jasco J-1500 spectropolarimeter equipped with a Peltier thermostatic cell holder. Far-UV measurements (190–260 nm) were carried out using a 0.1-cm path length cell in 10 mM Tris, 50 mM NaCl, 1 mM DTT, pH 7.4, at a protein concentration of 10 μM. Thermal denaturation was performed from 20 to 100°C with an increment of temperature of 1°C/min−1 monitoring CD signal at 220 nm. CD spectra were averaged over at least three independent scans and the baseline corrected by subtracting the buffer contribution.
Microscale thermophoresis
MST experiments were performed on a Monolith NT 115 system (Nano Temper Technologies) using 60 % LED and 20 % IR-laser power. BTB/POZ15 and BTB/POZ1were labelled with reactive dyes using N-Hydroxysuccinimide (NHS)-ester chemistry, which reacts efficiently with the primary amines of the proteins to form a stable dye-protein conjugate. For labelling, protein concentration was adjusted to 20 μM in labelling buffer (Nano Temper Technologies), while the dye concentration was adjusted to a threefold concentration of the protein (60 μM). The proteins and the fluorescent dye solution were incubated for 60 min at room temperature in the dark. A 16-point serial dilution (1:1) was prepared for KCASH2 or BTB/POZ KCASH1 (BTB/POZ11) or KCTD1FL or BTB/POZ1 at the final concentration ranged from 30 μM to 0.00183 μM. The samples were filled into Premium capillaries and the measurements were conducted at 25°C in 20 mM NaP, 200 mM NaCl, 1 mM DTT, 0.05 % Tween-20 pH 7.4 buffer. An equation implemented by the software MO-S002 MO Affinity Analysis provided by the manufacturer was used for fitting baseline-corrected fraction bound values at different peptides concentrations.
Public dataset gene expression analysis
R2-Genomics analysis and visualization platform (http://r2.amc.nl) were used for gene expression analysis as previously described [46]. Data were also analysed with the GraphPad Prism software.
Statistical analysis
The statistical significance of differences between tested groups was analysed using Student's t-test and statistical significance was set at *p < 0.05, **p < 0.01 or ***p<0.001. Results are expressed as mean ± S.D. All experiments were replicated biologically at least three times.
Data availability
All data generated or analysed during this study are included in this published article, or available in the public databases indicated in the article.
CRediT authorship contribution statement
A. Di Fiore: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. S. Bellardinelli: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. L. Pirone: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. R. Russo: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. A. Angrisani: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. G. Terriaca: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. M. Bowen: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. F. Bordin: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. Z.M. Besharat: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. G. Canettieri: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. F. Fabretti: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. S. Di Gaetano: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. L. Di Marcotullio: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. E. Pedone: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. M. Moretti: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. E. De Smaele: Conceptualization, Data curation, Funding acquisition, Methodology, Software, Validation, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Adriano Apostolico and Faranak Taj Mir from the De Smaele lab for the helpful discussion of the project.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2023.100926.
Appendix. Supplementary materials
References
- 1.Zhang Y., Beachy P.A. Cellular and molecular mechanisms of Hedgehog signalling. Nat. Rev. Mol. Cell Biol. 2023 doi: 10.1038/s41580-023-00591-1. [DOI] [PubMed] [Google Scholar]
- 2.Pandolfi S., Stecca B. Cooperative integration between Hedgehog-GLI signalling and other oncogenic pathways: implications for cancer therapy. Expert Rev. Mol. Med. 2015;17 doi: 10.1017/erm.2015.3. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigafoos A.N., Paradise B.D., Fernandez-Zapico M.E. Hedgehog/GLI Signaling Pathway: Transduction, Regulation, and Implications for Disease. Cancers (Basel) 2021;(14):13. doi: 10.3390/cancers13143410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grund-Gröschke S., Stockmaier G., Aberger F. Hedgehog/GLI signaling in tumor immunity - new therapeutic opportunities and clinical implications. Cell Commun. Signal. 2019;17(1):172. doi: 10.1186/s12964-019-0459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pietrobono S., Gagliardi S., Stecca B. Non-canonical Hedgehog signaling pathway in cancer: activation of GLI transcription factors beyond smoothened. Front. Genet. 2019;10:556. doi: 10.3389/fgene.2019.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulino A., Di Marcotullio L., Canettieri G., De Smaele E., Screpanti I. Hedgehog/Gli control by ubiquitination/acetylation interplay. Vitam. Horm. 2012;88:211–227. doi: 10.1016/B978-0-12-394622-5.00009-2. [DOI] [PubMed] [Google Scholar]
- 7.Canettieri G., Di Marcotullio L., Greco A., Coni S., Antonucci L., Infante P., Pietrosanti L., De Smaele E., Ferretti E., Miele E., et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 2010;12(2):132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 8.De Smaele E., Di Marcotullio L., Moretti M., Pelloni M., Occhione M.A., Infante P., Cucchi D., Greco A., Pietrosanti L., Todorovic J., et al. Identification and characterization of KCASH2 and KCASH3, 2 novel Cullin3 adaptors suppressing histone deacetylase and Hedgehog activity in medulloblastoma. Neoplasia. 2011;13(4):374–385. doi: 10.1593/neo.101630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angrisani A., Di Fiore A., Di Trani C.A., Fonte S., Petroni M., Lospinoso Severini L., Bordin F., Belloni L., Ferretti E., Canettieri G., et al. Specific protein 1 and p53 interplay modulates the expression of the KCTD-containing cullin3 adaptor suppressor of Hedgehog 2. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.638508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spiombi E., Angrisani A., Fonte S., De Feudis G., Fabretti F., Cucchi D., Izzo M., Infante P., Miele E., Po A., et al. KCTD15 inhibits the Hedgehog pathway in medulloblastoma cells by increasing protein levels of the oncosuppressor KCASH2. Oncogenesis. 2019;8(11):64. doi: 10.1038/s41389-019-0175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinkas D.M., Sanvitale C.E., Bufton J.C., Sorrell F.J., Solcan N., Chalk R., Doutch J., Bullock A.N. Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases. Biochem. J. 2017;474(22):3747–3761. doi: 10.1042/BCJ20170527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pirone L., Smaldone G., Spinelli R., Barberisi M., Beguinot F., Vitagliano L., Miele C., Di Gaetano S., Raciti G.A., Pedone E. KCTD1: A novel modulator of adipogenesis through the interaction with the transcription factor AP2α. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864(12) doi: 10.1016/j.bbalip.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Angrisani A., Di Fiore A., De Smaele E., Moretti M. The emerging role of the KCTD proteins in cancer. Cell Commun. Signal. 2021;19(1):56. doi: 10.1186/s12964-021-00737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kool M., Jones D.T., Jäger N., Northcott P.A., Pugh T.J., Hovestadt V., Piro R.M., Esparza L.A., Markant S.L., Remke M., et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25(3):393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth R.B., Hevezi P., Lee J., Willhite D., Lechner S.M., Foster A.C., Zlotnik A. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7(2):67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 16.Weishaupt H., Johansson P., Sundström A., Lubovac-Pilav Z., Olsson B., Nelander S., Swartling F.J. Batch-normalization of cerebellar and medulloblastoma gene expression datasets utilizing empirically defined negative control genes. Bioinformatics. 2019;35(18):3357–3364. doi: 10.1093/bioinformatics/btz066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J., Zhou X., Yang J., Sun Q., Liu Y., Li N., Zhang Z., Xu H. Circ-GLI1 promotes metastasis in melanoma through interacting with p70S6K2 to activate Hedgehog/GLI1 and Wnt/β-catenin pathways and upregulate Cyr61. Cell Death. Dis. 2020;11(7):596. doi: 10.1038/s41419-020-02799-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferretti E., De Smaele E., Di Marcotullio L., Screpanti I., Gulino A. Hedgehog checkpoints in medulloblastoma: the chromosome 17p deletion paradigm. Trends Mol. Med. 2005;11(12):537–545. doi: 10.1016/j.molmed.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Oliver T.G., Grasfeder L.L., Carroll A.L., Kaiser C., Gillingham C.L., Lin S.M., Wickramasinghe R., Scott M.P., Wechsler-Reya R.J. Transcriptional profiling of the Sonic Hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. U. S. A. 2003;100(12):7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Justilien V., Walsh M.P., Ali S.A., Thompson E.A., Murray N.R., Fields A.P. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell. 2014;25(2):139–151. doi: 10.1016/j.ccr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diao Y., Rahman M.F., Vyatkin Y., Azatyan A., St Laurent G., Kapranov P., Zaphiropoulos P.G. Identification of novel GLI1 target genes and regulatory circuits in human cancer cells. Mol. Oncol. 2018;12(10):1718–1734. doi: 10.1002/1878-0261.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senatore E., Chiuso F., Rinaldi L., Intartaglia D., Delle Donne R., Pedone E., Catalanotti B., Pirone L., Fiorillo B., Moraca F., et al. The TBC1D31/praja2 complex controls primary ciliogenesis through PKA-directed OFD1 ubiquitylation. EMBO J. 2021;40(10) doi: 10.15252/embj.2020106503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Correale S., Pirone L., Di Marcotullio L., De Smaele E., Greco A., Mazzà D., Moretti M., Alterio V., Vitagliano L., Di Gaetano S., et al. Molecular organization of the cullin E3 ligase adaptor KCTD11. Biochimie. 2011;93(4):715–724. doi: 10.1016/j.biochi.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Smaldone G., Pirone L., Capolupo A., Vitagliano L., Monti M.C., Di Gaetano S., Pedone E. The essential player in adipogenesis GRP78 is a novel KCTD15 interactor. Int. J. Biol. Macromol. 2018;115:469–475. doi: 10.1016/j.ijbiomac.2018.04.078. [DOI] [PubMed] [Google Scholar]
- 25.Smaldone G., Pirone L., Balasco N., Di Gaetano S., Pedone E.M., Vitagliano L. Cullin 3 recognition is not a universal property among KCTD proteins. PLOS One. 2015;10(5) doi: 10.1371/journal.pone.0126808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esposito L., Balasco N., Vitagliano L. Alphafold predictions provide insights into the structural features of the functional oligomers of all members of the KCTD family. Int. J. Mol. Sci. 2022;23(21) doi: 10.3390/ijms232113346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu X., Yan F., Wang F., Yang Z., Xiao L., Li L., Xiang S., Zhou J., Ding X., Zhang J. TNFAIP1 interacts with KCTD10 to promote the degradation of KCTD10 proteins and inhibit the transcriptional activities of NF-κB and AP-1. Mol. Biol. Rep. 2012;39(11):9911–9919. doi: 10.1007/s11033-012-1858-7. [DOI] [PubMed] [Google Scholar]
- 28.Rousseau A., Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018;19(11):697–712. doi: 10.1038/s41580-018-0040-z. [DOI] [PubMed] [Google Scholar]
- 29.Norris A.M., Appu A.B., Johnson C.D., Zhou L.Y., McKellar D.W., Renault M.A., Hammers D., Cosgrove B.D., Kopinke D. Hedgehog signaling via its ligand DHH acts as cell fate determinant during skeletal muscle regeneration. Nat. Commun. 2023;14(1):3766. doi: 10.1038/s41467-023-39506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenney A.M., Rowitch D.H. Sonic Hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 2000;20(23):9055–9067. doi: 10.1128/MCB.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.La Regina G., Bai R., Coluccia A., Famiglini V., Pelliccia S., Passacantilli S., Mazzoccoli C., Ruggieri V., Sisinni L., Bolognesi A., et al. New pyrrole derivatives with potent tubulin polymerization inhibiting activity as anticancer agents including Hedgehog-dependent cancer. J. Med. Chem. 2014;57(15):6531–6552. doi: 10.1021/jm500561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaeger M., Ghisleni E.C., Fratini L., Brunetto A.L., Gregianin L.J., Brunetto A.T., Schwartsmann G., de Farias C.B., Roesler R. Viability of D283 medulloblastoma cells treated with a histone deacetylase inhibitor combined with bombesin receptor antagonists. Childs Nerv. Syst. 2016;32(1):61–64. doi: 10.1007/s00381-015-2963-4. [DOI] [PubMed] [Google Scholar]
- 33.Di Marcotullio L., Ferretti E., De Smaele E., Argenti B., Mincione C., Zazzeroni F., Gallo R., Masuelli L., Napolitano M., Maroder M., et al. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc. Natl. Acad. Sci. U. S. A. 2004;101(29):10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding X., Luo C., Zhou J., Zhong Y., Hu X., Zhou F., Ren K., Gan L., He A., Zhu J., et al. The interaction of KCTD1 with transcription factor AP-2alpha inhibits its transactivation. J. Cell. Biochem. 2009;106(2):285–295. doi: 10.1002/jcb.22002. [DOI] [PubMed] [Google Scholar]
- 35.Ding X.F., Luo C., Ren K.Q., Zhang J., Zhou J.L., Hu X., Liu R.S., Wang Y., Gao X. Characterization and expression of a human KCTD1 gene containing the BTB domain, which mediates transcriptional repression and homomeric interactions. DNA Cell Biol. 2008;27(5):257–265. doi: 10.1089/dna.2007.0662. [DOI] [PubMed] [Google Scholar]
- 36.Marneros A.G., Beck A.E., Turner E.H., McMillin M.J., Edwards M.J., Field M., de Macena Sobreira N.L., Perez A.B., Fortes J.A., Lampe A.K., et al. Mutations in KCTD1 cause scalp-ear-nipple syndrome. Am. J. Hum. Genet. 2013;92(4):621–626. doi: 10.1016/j.ajhg.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smaldone G., Balasco N., Pirone L., Caruso D., Di Gaetano S., Pedone E.M., Vitagliano L. Molecular basis of the scalp-ear-nipple syndrome unraveled by the characterization of disease-causing KCTD1 mutants. Sci. Rep. 2019;9(1):10519. doi: 10.1038/s41598-019-46911-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smaldone G., Pecoraro G., Pane K., Franzese M., Ruggiero A., Vitagliano L., Salvatore M. The oncosuppressive properties of KCTD1: its role in cell growth and mobility. Biology. 2023;(3):12. doi: 10.3390/biology12030481. Basel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng X., Aouacheria A., Lionnard L., Metz K.A., Soane L., Kamiya A., Hardwick J.M. KCTD: A new gene family involved in neurodevelopmental and neuropsychiatric disorders. CNS Neurosci. Ther. 2019;25(7):887–902. doi: 10.1111/cns.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Northcott P.A., Buchhalter I., Morrissy A.S., Hovestadt V., Weischenfeldt J., Ehrenberger T., Gröbner S., Segura-Wang M., Zichner T., Rudneva V.A., et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borck G., de Vries L., Wu H.J., Smirin-Yosef P., Nürnberg G., Lagovsky I., Ishida L.H., Thierry P., Wieczorek D., Nürnberg P., et al. Homozygous truncating PTPRF mutation causes athelia. Hum. Genet. 2014;133(8):1041–1047. doi: 10.1007/s00439-014-1445-1. [DOI] [PubMed] [Google Scholar]
- 42.Fernández L.T., Ocampo-Garza S.S., Elizondo-Riojas G., Ocampo-Candiani J. Basal cell nevus syndrome: an update on clinical findings. Int. J. Dermatol. 2022;61(9):1047–1055. doi: 10.1111/ijd.15884. [DOI] [PubMed] [Google Scholar]
- 43.Sari I.N., Phi L.T.H., Jun N., Wijaya Y.T., Lee S., Kwon H.Y. Hedgehog signaling in cancer: a prospective therapeutic target for eradicating cancer stem cells. Cells. 2018;(11):7. doi: 10.3390/cells7110208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abballe L., Mastronuzzi A., Miele E., Carai A., Besharat Z.M., Moretti M., De Smaele E., Giangaspero F., Locatelli F., Ferretti E., et al. Numb isoforms deregulation in medulloblastoma and role of p66 isoform in cancer and neural stem cells. Front. Pediatr. 2018;6:315. doi: 10.3389/fped.2018.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smaldone G., Pirone L., Pedone E., Marlovits T., Vitagliano L., Ciccarelli L. The BTB domains of the potassium channel tetramerization domain proteins prevalently assume pentameric states. FEBS Lett. 2016;590(11):1663–1671. doi: 10.1002/1873-3468.12203. [DOI] [PubMed] [Google Scholar]
- 46.Di Giulio S., Colicchia V., Pastorino F., Pedretti F., Fabretti F., Nicolis di Robilant V., Ramponi V., Scafetta G., Moretti M., Licursi V., et al. A combination of PARP and CHK1 inhibitors efficiently antagonizes MYCN-driven tumors. Oncogene. 2021;40(43):6143–6152. doi: 10.1038/s41388-021-02003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article, or available in the public databases indicated in the article.