Abstract
In this study, we investigated the conjugation of theophylline with different compounds of natural origin hoping to construct new hybrids with dual activity against cholinergic and inflammatory pathways as potential agents for the treatment of Alzheimer's disease (AD). Out of 28 tested hybrids, two hybrids, acefylline–eugenol 6d and acefylline–isatin 19, were able to inhibit acetylcholinesterase (AChE) at low micromolar concentration displaying IC50 values of 1.8 and 3.3 μM, respectively, when compared to the galantamine standard AChE inhibitor. Moreover, the prepared hybrids exhibited a significant anti-inflammatory effect against lipopolysaccharide induced inflammation in RAW 264.7 and reduced nitric oxide (NO), tumor necrosis alpha (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6) levels in a dose dependent manner. These hybrids demonstrated significant reductions in nitric oxide (NO), tumor necrosis alpha (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6) levels in RAW 264.7 cells induced by lipopolysaccharide (LPS). The findings of this study were further explained in light of network pharmacology analysis which suggested that AChE and nitric oxide synthase were the main targets of the most active compounds. Molecular docking studies revealed their ability to bind to the heme binding site of nitric oxide synthase 3 (NOS-3) and effectively occupy the active site of AChE, interacting with both the peripheral aromatic subsite and catalytic triad. Finally, the compounds demonstrated stability in simulated gastric and intestinal environments, suggesting potential absorption into the bloodstream without significant hydrolysis. These findings highlight the possible therapeutic potential of acefylline–eugenol 6d and acefylline–isatin 19 hybrids in targeting multiple pathological mechanisms involved in AD, offering promising avenues for further development as potential treatments for this devastating disease.
In this study, conjugation of theophylline with different compounds of natural origin led to new hybrids with dual activity against cholinergic and inflammatory pathways as potential agents for management of Alzheimer's disease (AD).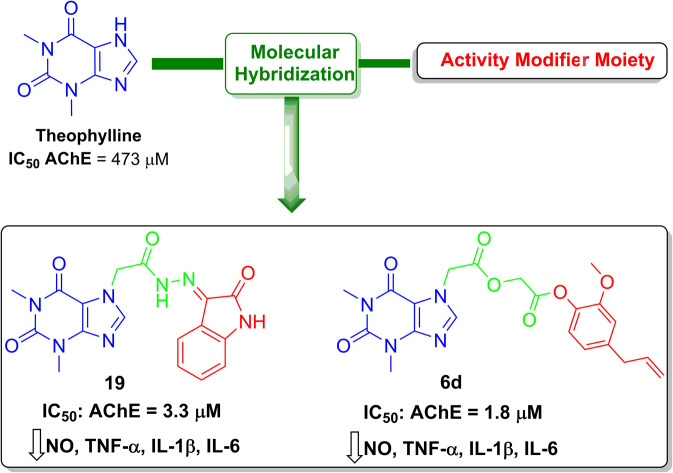
1. Introduction
Alzheimer's disease (AD) is a neurodegenerative disorder characterized by progressive cognitive decline, memory loss, and behavioral changes. The number of individuals affected by AD is alarmingly high, with an estimated 50 million people worldwide suffering from the disease. Hence, AD is placing a significant burden on individuals, families, and healthcare systems worldwide. Understanding the underlying mechanisms and developing effective therapeutic strategies for AD is of paramount importance which would eventually lead to developing innovative therapeutic strategies that target different pathways involved in AD pathology.1,2
Current therapeutic strategies for management of AD are based on the acetylcholinesterase (AChE) hypothesis which proposes that the decline in cholinergic neurotransmission contributes to the cognitive decline observed in AD as reduced levels of acetylcholine have been observed in AD patients, so Acetylcholinesterase, an enzyme responsible for acetylcholine breakdown, appears to be a promising target. As a result, cholinesterase inhibitors have been developed as a symptomatic treatment for AD, aiming to increase acetylcholine levels. However, these drugs provide only modest benefits and do not alter the underlying disease progression.3,4
Another significant theory in AD research is the amyloid hypothesis. Amyloid-beta (Aβ) peptides, derived from the amyloid precursor protein (APP), aggregate and form amyloid plaques, a hallmark of AD pathology which is linked to neurodegeneration in AD patients. This hypothesis has guided the development of therapeutic approaches targeting Aβ production or clearance.5 However, clinical trials targeting Aβ have thus far yielded disappointing results, indicating the complexity of AD pathogenesis and the need for alternative therapeutic strategies.6
Recently, the role of inflammation in AD has gained significant attention. Chronic activation of the immune cells of the central nervous system such as microglia and astrocytes, induce the release of pro-inflammatory molecules, including cytokines and reactive oxygen species. These inflammatory mediators contribute to neuronal dysfunction and promote neurodegeneration in AD.7 Targeting inflammation represents a promising approach for AD treatment, as it addresses a common pathway involved in disease progression.8
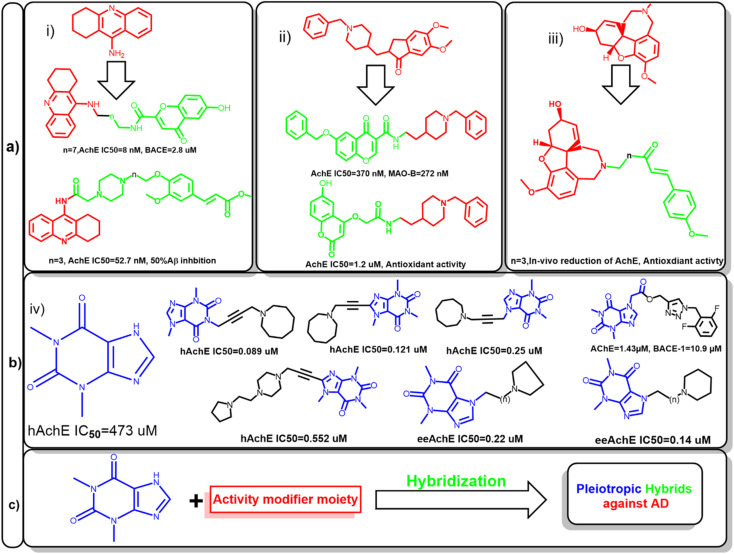
Hence, several attempts were made to modify the activity of clinically relevant agents such as tacrine, donepezil, and galantamine. This led to the development of hybrids with multiple activities against other targets involved in AD. For example, the conjugation of tacrine with ferulic acid give the produced hybrid the ability to inhibit Aβ aggregation, also beta secretase inhibition was feasible after the conjugation with chromone moiety. In case of donepezil, molecular hybridization of the benzyl piperidine moiety with chromone and coumarin scaffolds enabled these compounds to inhibit monoamineoxidase-1 (MAO-1) and increased its antioxidant activity, respectively. Also, galantamine-curcumin hybrids showed significant reduction in acetylcholinesterase and promoted the antioxidant mechanisms in vivo (Fig. 1a).9–12
Fig. 1. (a) Representative examples for previously reported inhibitor against acetylcholinesterase (AChE), (i) tacrine, (ii) donepezil, (iii) galantamine and their corresponding hybrids to modify their activity to be active against other enzymes involved in Alzheimer's disease such as monoamine oxidase (MAO) or betas secretase (BACE). (b) Theophylline (iv) has low affinity to AChE; new hybrids of theophylline had enhanced activity by linking with various moiety against human acetylcholinesterase (hAChE) or electric eel acetylcholinesterase (eeAchE). (c) Proposed strategy for anti-Alzheimer's agent based on xanthine hybrids, the counterpart of the conjugate would determine the activity of the prepared hybrids.
Amidst the search for effective AD therapeutics, natural products have emerged as a valuable source of potential compounds since they offer a vast array of chemical diversity and biological activities. In the context of AD, natural products have exhibited neuroprotective, anti-amyloidogenic, and anti-inflammatory properties, positioning them as promising candidates for drug development. Xanthine alkaloids, such as caffeine, was linked to protection against several cardiovascular and neurodegenerative diseases.13–16
Since it can cross blood–brain barrier, it would be a privileged scaffold for targeting neurodegenerative disease. Several caffeine derivatives could be accessible through chemical modification of theophylline. Acefylline, which is theophylline-7-acetic acid was extensively studied to develop new bioactive compounds. Theophylline and its derivatives have been utilized to obtain hybrid compounds combining different pharmacophores or functional groups which was proven to improve the activity of the parent drugs (Fig. 1b).17–19
Therefore, this study aims at preparing hybrid compounds based on theophylline and other known natural products to fabricate new compounds with dual activity targeting different pathways in AD as shown in Fig. 1c. In silico approaches such as network pharmacology and molecular docking would be used to identify their potential mechanism of action. Also, the stability of the most active hybrids would be assessed.
2. Results and discussion
2.1. Chemistry
Firstly, acefylline 2 (7-acetic acid derivative of theophylline) was prepared to enhance the synthetic accessibility of theophylline in further reactions. Appearance of new signals in 13C NMR (CMR) at δ 47 and 168 ppm and in 1HNMR (PMR) at δ 5–5.5 ppm as well as melting point are among compiled evidences of the success of formation of, acefylline 2.
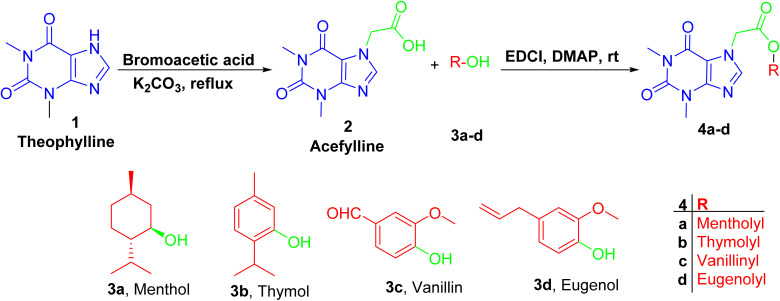
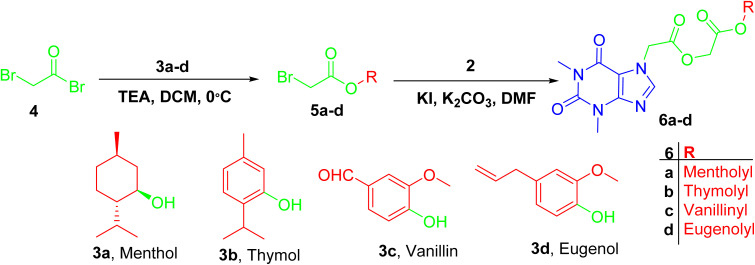
Acefylline hybrids were formed by reactions with different natural products bearing either alcohol OH (menthol) or phenolic OH (thymol, vanillin, eugenol), through Steglich esterification20 as in hybrids 4a–d (Scheme 1) or alkylation by bromoacetate derivatives 5a–d to result in hybrids 6a–d (Scheme 2).
Scheme 1. Preparation of theophylline-7-acetic acid (acefylline) followed by Steglich esterification, using EDCI as coupling agent in presence of menthol, thymol, eugenol or vanillin to give the corresponding ester.
Scheme 2. Preparation of ester derivative 6a–d was facilitated by reaction of bromoacetyl bromide with respective phenol or alcohol followed by alkylation of acefylline with acetyl derivatives.
The prepared hybrids showed characteristic spectroscopic profile including the presence of 2 signals at ranges of δ 3.2–3.4 ppm in PMR and δ 27–29 ppm in CMR assigned for methyl functionality of xanthine ring. In addition, one signal was shown at δ 5 ppm in PMR and δ 47 ppm in CMR assigned for the methylene carbon of acetic acid linker. Also, a downfield proton at ranges of δ 7.6–8 ppm in 1H NMR and a signal at δ 144 ppm in 13C NMR were observed and assigned for proton and carbon 4 of the purine ring, while the two carbonyl groups of the xanthine ring were found at δ 154 and 151 ppm.21
For hybrids 4a–d, the spectroscopic data showed consistent pattern with the reported parent compounds except the decrease in the chemical shift value of carboxylic acid of acefylline from δ 169 to 168–165 ppm confirming the formation of ester. Additionally, in case of menthol hybrid 4a, the chemical shift value of proton 1′ of menthol increased from δ 3.4 to 4.7 ppm as shown in Tables S1–S4.†
While in case of hybrids containing acetyl spacer 6a–d, acefylline 2 was reacted with bromoacetate derivatives of the corresponding alcohol or phenol. Hence, an additional signal were observed around δ 4.6 ppm (s, 2H) in PMR and δ 61 and 166 ppm in CMR corresponding to the acetyl linker as recorded in Tables S5–S8.†
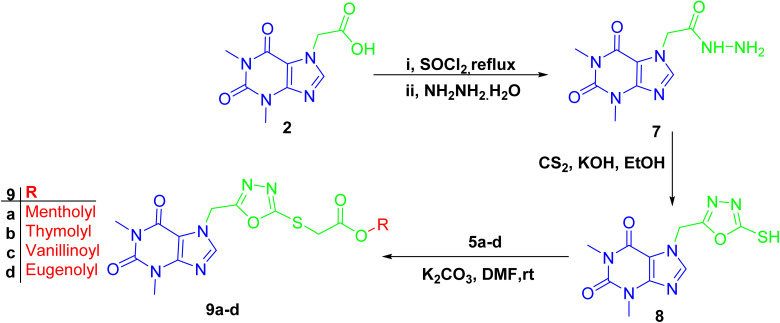
The formation of 1,3,4-oxadiazole-containing hybrids 9a–d was facile through the reaction of the hydrazide derivative of acefylline with carbon disulfide in basic medium followed by alkylation with bromoacetate derivatives of the corresponding alcohol or phenol 5a–d (Scheme 3). Thus, these derivatives exhibited similar PMR behavior as their corresponding ester derivatives 6a–d except for a new signal was shown around δ 162 ppm in CMR indicating the formation of the oxadiazole ring and the chemical shift value of the methylene linker showed significant decrease from δ 60 and 47 ppm to around δ 40 and 34 ppm, respectively, as demonstrated in Tables S9–S12.†
Scheme 3. Preparation of 1,3,4-oxadiazole-containing acefylline hybrids 9a–d.
For compounds containing 1,2,3-triazole linker 15a–c, they were successfully obtained through copper catalyzed azide alkyne cycloaddition reaction of the propargylated derivative of acefylline 14 with respective azides 10a-b and 13 that prepared from bromoacetylated derivatives 5a-b and 4-chloro-coumarin 12, respectively, as demonstrated in Scheme 4. The reaction was confirmed by the appearance of additional signals around δ 8 (1H) and 5.2 (2H) ppm corresponding to the triazole ring and the methylene bridge between the xanthine moiety and the triazole ring, respectively. Moreover, three new signals were observed in CMR corresponding to the two olefinic carbons in the triazole ring and the methylene bridge around δ 142, 122, and 60 ppm, respectively.
Scheme 4. Preparation of acefylline triazole hybrids 15a–c.
The other data showed no significant change in the NMR profile in comparison to that found in the parent compounds when compared to spectroscopic data in literature22 except for 4-hydroxycoumarin derivative 13 where carbon 4′ showed significant upfield shift from δ 162 to 154 ppm due to the substitution of hydroxyl group with azide group as recorded in Tables S13–S15.†
Finally, hydrazone linker-tethered derivatives 18a–l and 19 were furnished through coupling of hydrazide 7 with various aromatic aldehydes, 16a–l, and isatin 17, respectively as shown in Scheme 5. The disappearance of aldehydic proton around δ 9–10 ppm and the appearance of two new signals at δ 11 and 8 ppm corresponding to the hydrazone linker protons. Furthermore, the decrease in chemical shift of aldehydic carbon from δ 190 to 140 ppm were among the spectroscopic features to confirm the formation of acefylline-hydrazone hybrids 18a–l. Lastly, the methylene group of acefylline showed significant decrease in chemical shift from around δ 60 to 47 ppm. For acefylline–isatin hybrid 19, the hydrazone linker was observed at δ 11.36 and 143.21 ppm as explained in Tables S16–S28.†
Scheme 5. Preparation of acefylline hydrazone hybrids 18a–j and 19 through the reaction of hydrazide with corresponding aldehydes or ketone.
2.2. Biological evaluation
2.2.1. Network pharmacology
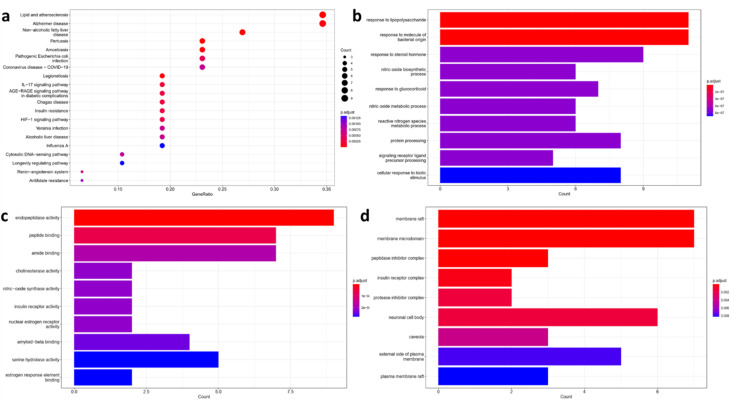
The chemical structure of the hybrids was submitted to network pharmacology analysis to identify potential mechanisms of theophylline hybrids for management of AD. Target fishing showed that there are potential 332 molecular targets for acefylline conjugates. Among them 25 target was shown to be correlated with molecular targets associated with atherosclerosis and AD pathogenesis as revealed by Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Fig. 2a), inducible nitric oxide synthase (iNOS), endoplasmic nitric oxide synthase (eNOS), several cytokines such as TNF-α, IL-1β and IL-6 and Glycogen Synthase Kinas-3 (GSK-3) were found to be the most relevant to the top five KEGG pathways-gene network. This is in agree with previous reports highlighting that atherosclerosis patients were found to be more susceptible to AD risk, as both disease involve the activation of inflammatory cascades and dysregulation of inflammation resolving pathway. Moreover, atherosclerosis causes reduced blood flow to the brain which participate in accumulation of amyloid plaques and neurofibrillary tangles.23–25 Remarkably, caffeine which is a methylated derivative of theophylline has been studied thoroughly for its role in management of metabolic syndrome and atherosclerosis by decreasing cholesterol levels.26 Also, chronic administration caffeine to of Alzheimer's transgenic mice was reported to decrease the levels of insoluble brain Aβ leading to restore of their cognitive performance.27
Fig. 2. Enrichment analysis results of Alzheimer's disease-related targets in relation with acefylline hybrids significantly enriched terms in, (a) KEGG, (b) BP, (c) MF, and (d) CC.
Also, gene enrichment reveals other genes involved in biological processes (BP), molecular functions (MF) and cellular component (CC) associated with the predicted targets. The top five enriched terms in BP was related to response to stimuli that trigger or suppress inflammation such as LPS, NO or glucocorticoid respectively (Fig. 2b). For MF, endopeptidase activity, peptide binding, amide binding, choline esterase activity and nitric oxide activity were among the top five enriched terms where targets such as nitric oxide synthase, beta secretase, acetylcholinesterase and butyl cholinesterase appeared as the most significant ones as depicted in Fig. 2c. Finally, the top enriched terms of CC were a reflection to what have been found in KEGG analysis where membrane rafts and membrane microdomain were found to be the most significant term with P ≥ 0.002 (Fig. 2d). Interestingly, lipid rafts were shown to play important role in the development of AD through regulation of Aβ deposition and AChE signaling.28
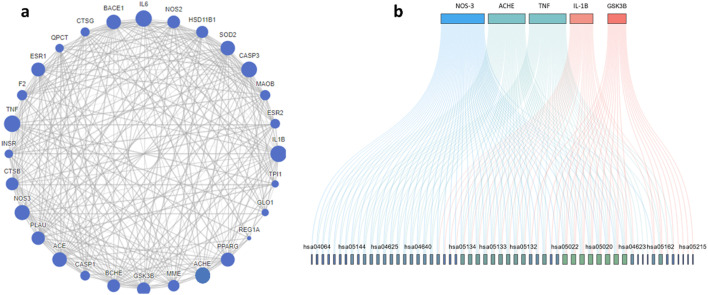
Protein–protein interaction (PPI) of the enriched targets was also studied, where the constructed network showed eNOS, AChE, TNF, IL-1β and GSK-3β as the most prominent targets. Still eNOS and AChE were the most prominent target with 47/68 and 40/68 interactions respectively as shown in Fig. 3a and b. AChE targeting is one of the clinical pillars for management of AD while NOS is still under investigation, nevertheless the correlation between overexpression NOS and AD is well established,29 also NOS inhibitors have shown neuroprotective activity in experimental animal.30
Fig. 3. (a) Protein–protein interactions of targets predicted to be relevant to acefylline hybrid activity. (b) Top five targets included in KEGG pathways where the greater the degree of overlap of genes, the more the same pathways involved. Lines with different colors indicate different gene-pathway relationships.
In summary, network pharmacology suggests that the ability of the prepared hybrids to modulate pathways correlated to AD through regulation of inflammatory response and cholinesterase interactions with lipid rafts which would consequently reduce Aβ precipitation and plaque formation.
2.3. In vitro anti-inflammatory assay of acefylline-hybrids
2.3.1. LPS-induced inflammation in RAW 264.7 cell line
Since network pharmacology revealed that the prepared hybrids might possess anti-inflammatory effect through modulation of NO production, LPS induced inflammation in RAW 264.7 cell line was used as a primary screening to determine the most active hybrids. Initially, the cytotoxicity of the compounds were assessed at 25 μM using MTT assay. The preliminary results of this screening revealed that all tested acefylline hybrids do not halt the proliferation of RAW 264.7 cell line (Table 1).
Cytotoxic effect of acefylline hybrid on RAW 264.7 using MTT assay at 50 μMa.
| Code | Viability (%) |
|---|---|
| 2 | 85 ± 1.7 |
| 4a | 91.4 ± 0.1 |
| 4b | 94 ± 0.2 |
| 4c | 90 ± 1 |
| 4d | 93 ± 1 |
| 6a | 88 ± 0.8 |
| 6b | 89 ± 1 |
| 6c | 90 ± 1.1 |
| 6d | 92 ± 1 |
| 9a | 90 ± 1.1 |
| 9b | 93 ± 1 |
| 9c | 92 ± 1 |
| 9d | 98 ± 0.6 |
| 15a | 86 ± 1.3 |
| 15b | 97 ± 0.6 |
| 15c | 88 ± 0.8 |
| 18a | 94 ± 0.2 |
| 18b | 87 ± 0.8 |
| 18c | 88 ± 0.8 |
| 18d | 90 ± 1 |
| 18e | 92 ± 1 |
| 18f | 91 ± 1 |
| 18g | 92 ± 1 |
| 18h | 93 ± 1 |
| 18i | 104.7 ± 1.2 |
| 18j | 89 ± 1 |
| 18k | 94.7 ± 1.2 |
| 18l | 93 ± 1 |
| 19 | 89.5 ± 1.1 |
Data represent the mean and SD of % viability based on two independent experiment.
2.3.2. NO level in LPS induced RAW 264.7
Cytotoxicity assay results suggested that evaluating the anti-inflammatory of acefylline hybrids at concentration lower than 25 μM would not affect RAW 264.7 viability. So, all the compounds were tested for their ability to inhibit NO production in LPS induced RAW 264.7 in comparison to L-NG-nitro arginine methyl ester (l-NAME) as standard at 250 μM. All of the prepared hybrids was able to reduce NO level in comparison to the parent compound acefylline. Still, only ten hybrids were able to achieve better inhibition of NO than the standard L-NG-nitro arginine methyl ester (l-NAME), among them there were 3 hybrids 4d, 6d and 9d containing eugenol conjugates through direct esterification, acetyl linker or oxadiazole linker, respectively. Three hydrazones which are based on benzaldehyde derivatives (18h–18l) and isatin 19, two vanillin conjugates through oxadiazole and hydrazone linker (9c, 18j), thymol conjugate 9b linked by oxadiazole spacer and finally coumarin conjugate tethered with triazole ring (15c). The % inhibition of all the hybrids are presented in Table 2.
Inhibitory effects of acefylline hybrids at 25 μM on NO production in RAW264.7 stimulated by 10 μg ml−1 of LPS.
| No. | Code | % inhibition |
|---|---|---|
| 1 | 2 | 58.7 ± 1.82 |
| 2 | 4a | 66.4 ± 0.79 |
| 3 | 4b | 70.7 ± 0.87 |
| 4 | 4c | 65.4 ± 0.84 |
| 5 | 4d | 86.8 ± 0.87 |
| 6 | 6a | 79.3 ± 0.85 |
| 7 | 6b | 74 ± 0.87 |
| 8 | 6c | 68.6 ± 0.92 |
| 9 | 6d | 88 ± 1 |
| 10 | 9a | 69.6 ± 0.92 |
| 11 | 9b | 92.8 ± 0.9 |
| 12 | 9c | 86.4 ± 0.86 |
| 13 | 9d | 93.2 ± 0.87 |
| 14 | 15a | 61.1 ± 0.9 |
| 15 | 15b | 68.6 ± 0.92 |
| 16 | 15c | 85 ± 0.88 |
| 17 | 18a | 65.4 ± 0.84 |
| 18 | 18b | 71.8 ± 0.9 |
| 19 | 18c | 75 ± 0.88 |
| 20 | 18d | 70.7 ± 0.87 |
| 21 | 18e | 71.8 ± 0.9 |
| 22 | 18f | 63.2 ± 0.93 |
| 23 | 18g | 66.4 ± 0.79 |
| 24 | 18h | 96.6 ± 0.87 |
| 25 | 18i | 63.2 ± 0.93 |
| 26 | 18j | 92.2 ± 0.86 |
| 27 | 18k | 65.4 ± 0.84 |
| 28 | 18l | 85.4 ± 0.87 |
| 29 | 19 | 90 ± 0.88 |
| 30 | l-NAMEa | 84.8 ± 0.9 |
l-NAME was used at 250 μM, data represent the mean and SD of % inhibition based on two independent experiment.
2.3.3. Effect of acefylline hybrids on inhibition of AChE
As the second axis of the proposed mechanism of action of the prepared hybrids involve the inhibition of acetylcholinesterase (AChE). Compounds that emerged as the best inhibitor of NO production was tested at 25 μM as demonstrated in Table 3. Five of them namely 6d, 19, 9c, 15c and 18j showed inhibition more than 75%. All the prepared hybrid showed improved activity over the parent drug acefylline which only achieved 6.5% inhibition to AChE at 25 μM.
Inhibitory effects of selected acefylline hybrids at 25 μM on acetylcholinesterase activitya.
| No. | Compound | Inhibition (%) |
|---|---|---|
| 1 | Acefylline | 6.5 ± 1.5 |
| 2 | 4d | 61.39 ± 3.79 |
| 3 | 6d | 97 ± 3.2 |
| 4 | 9b | 61.07 ± 1.79 |
| 5 | 9c | 88.2 ± 3.7 |
| 6 | 9d | 41.33 ± 3.57 |
| 7 | 15c | 85 ± 2.8 |
| 8 | 18h | 73.6 ± 3 |
| 9 | 18j | 78 ± 3.9 |
| 10 | 18l | 55.42 ± 2.07 |
| 11 | 19 | 92.5 ± 2.4 |
Data represent the mean and SD of % inhibition based on two independent experiment.
It is worthy to mention that acefylline–eugenol conjugates through acetyl linker had more advantage over other conjugation strategies such as oxadiazole and direct esterification where the first achieved 97% inhibitory effect, while the later achieved only 61% and 41% inhibition against AChE. For acefylline–vanillin conjugates, hybrids containing oxadiazole linker achieved better inhibition than that observed in case of the hydrazone spacer.
In case of isatin hydrazone, it achieved the 2nd best inhibitory activity, also the coumarin triazole hybrid inhibit AChE by 85%. So, the ability of these 5 hybrids to inhibit the enzyme at different doses was studied and their IC50 was calculated. Compounds 6d and 19 demonstrated the best IC50 = 1.8 and 3.3 μM respectively which was comparable to the standard AChE inhibitor galantamine as shown in Table 4 and Fig. S1†. While other hybrids showed relatively higher IC50 but still stronger by thirty-fold stronger inhibitory effect compared to the parent drug theophylline.
IC50 inhibition data of acetylcholinesterase (AChE) activity with acefylline based hybrids and reference AChE inhibitor galantamine.
| No. | Activity modifying moiety | Linker | Compound | IC50 (μM) |
|---|---|---|---|---|
| 1 | Eugenol | Acetyl | 6d | 1.8 ± 0.3 |
| 2 | Vanillin | Oxadiazole | 9c | 5.55 ± 0.33 |
| 3 | 4-Hydroxy coumarin | Triazole | 15c | 9.8 ± 0.7 |
| 4 | Vanillin | Hydrazone | 18j | 14.6 ± 0.5 |
| 5 | Isatin | Hydrazone | 19 | 3.3 ± 0.2 |
| 6 | — | — | Galantamine | 1.3 ± 0.25 |
2.3.4. Effect of acefylline hybrids on cytokine levels in RAW 264.7 induced by LPS
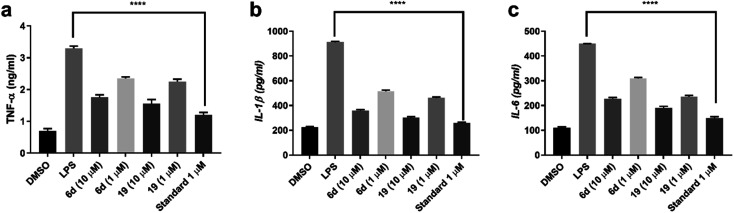
Since compounds 6d and 19 showed significant NO and AChE inhibitory activity, their ability to reduce induced cytokine levels by LPS in RAW 264.7 was assessed. At dose 10 μM compound 6d and 19 reduced TNF-α by 49% and 53% respectively in comparison to LPS treated cells while in case of IL-1β it was downregulated by 61% and 66.7% respectively. Also, IL-6 levels was decreased by 49.3% and 57.7% respectively. This observed decrease in the cytokines level was also still applicable at 1 μM where compound 6d and 19 lowered TNF-α by 28.78% and 31.8% respectively in comparison to LPS treated cells. On the other hand, IL-1β was reduced by 43.4% and 49.4% respectively. Finally, IL-6 levels was lowered by 31.3% and 47.5% respectively in comparison to cells induced by LPS as shown in Fig. 4a–c.
Fig. 4. Effect of compounds 6d and 19 on (a) TNF-α, (b) IL-1β, and (c) IL-6 levels in RAW 264.7 treated with 1 μg ml−1 LPS in comparison to dexamethasone as standard. ****Significantly different from control group at p < 0.0001.
The biological evaluation of acefylline hybrids indicated their dual activity as anti-inflammatory and acetylcholinesterase inhibitor as suggested by network pharmacology. Thymol, eugenol, coumarin and aldehydes conjugates with acefylline through acetyl, oxadiazole, triazole and hydrazone linker were observed as the most actives. Two compounds 6d which is acefylline–eugenol conjugate through acetyl linker and 19 which is acefylline conjugate through hydrazone linker achieved the best activity among the prepared hybrids.
While theophylline itself is understudied as anti-inflammatory agent it was reported to suppress TNF production at 20 μM in LPS stimulated RAW264.7 (ref. 31) On the other hand, caffeine which is the methylated analogue of theophylline is well studied. For example, caffeine and its analogue pentoxifylline were previously reported to possess anti-inflammatory activity in RAW264.7 induced by LPS but at relatively high dose 100–800 μM and 0.5–8 mM respectively.32,33 Also, caffeine attenuated neuroinflammation induced by LPS in BV2 microglial cells, where pro-inflammatory such as TNF-α and iNOS through prevention of phosphorylation of AKT and ERK leading to suppress of the downstream inflammatory cascade.34
Remarkably, the neuroprotective effect of caffeine against LPS induced neurodegeneration in mice through activation of nuclear factor erythroid 2-related factor 2 (Nrf2) and reduction of NO, TNF-α and oxidative stress.35 Moreover, it decreased number of activated microglia in hippocampus of experimental animals subjected to inflammation associated with LPS administration or aging.36
Furthermore, caffeine was found to protect against cognitive impairment in AD transgenic mice through decreasing Aβ levels and β-secretase.37 Despite caffeine is a weak inhibitor to AChE, it was found to have indirect inhibitory effect on AChE through inactivation of protein kinase A, which is correlated with enhancing the activity of AChE to hydrolyze acetylcholine.38–40 In this context, chemical derivatization of theophylline and caffeine led to the development of novel acetylcholinesterase inhibitors41–43 which support our proposal to choose xanthine alkaloid to design hybrids for management of AD.
For the counterpart moiety of hybrid 6d, eugenol is known for its ability to exert substantial anti-inflammatory and antioxidant activity in vitro and in vivo which is attributed to the modulation of several and overlapping pathways.44 So, numerous studies investigated its application in management of AD. For instance, eugenol at 0.01 mg kg−1 was able to reduce plaques induced in rats injected by Aβ which was reflected by improvement in memory of the treated rat.45 Also, it mitigated the cholinergic dysfunction in hippocampus induced by scopolamine through decreasing in AChE activity.
It also protected against neurotoxicity by decreasing glutamate levels, boosting antioxidant mechanisms through increasing SOD and catalase activity and decreasing mitochondrial reactive oxygen species (ROS) and NO production.46 In combination with psilocybin, it was able to protect against inflammation induced in mice brain by LPS where several cytokines levels such as TNF, IL-1β, IL-6 were reduced.47 This was also reproducible in other experimental model based on aluminum chloride induced neurotoxicity.48
Furthermore, eugenol was evaluated as potential pleotropic agent against AD. While it showed remarkable protection against hydrogen peroxide and Aβ toxicity to PC12 cell line, it showed weak inhibition to AChE and β-secretase.49 Still chemical derivatization has shown to improve the activity to micromolar range.50,51 Taking these studies in consideration the conjugation of acefylline and eugenol hybrid through acetyl linker could be an intuitive approach to target different pathways involved in AD. Also, since hybrid compound could be considered as prodrug, acetylcholinesterase itself would help to release its original parent compounds once it reached the action site leading to extended efficacy.52
In case of compound 19, isatin was linked to acefylline. Isatin is a remarkable molecule with plethora of activities. In the last decade, it was thoroughly investigated for its ability to modulate molecular targets associated with AD since it is one of the endogenous neuroprotector molecules.53,54 So, isatin itself was recognized as a ligand for multiple binding sites of Aβ preventing its interaction with several target and maintain it in monomer state allowing their physiological degradation.55
In Aβ-induced Human Neuroblastoma (SK-N-SH Cells), isatin thiosemicarbazone increased the survival rate from 20% to approximately 100% which was explained by their inhibitory effect against GSK-3β which is one of the molecular targets predicted by our network pharmacology analysis.56 Moreover, several isatin derivatives were designed as AChE inhibitors; interestingly, isatin–tacrine and isatin–triazole hybrids had pronounced ability to prevent Aβ aggregation.57–59
Regarding the contribution of isatin in resolving inflammation, isatin, its derivatives were able to reduce the levels of COX-2, TNF and iNOS in LPS challenged RAW 264.7 at relatively high dose (100 μM)60 but when conjugated with Nalidixic acid through amino acid linkers, the prepared hybrids were able to inhibit iNOS by 80% at 10 μM.61 Hence, the observed acetylcholinesterase and anti-inflammatory effect of compound 19 could be justified in the light of these studies.
2.4. In silico analysis
2.4.1. Molecular dockings
Since NOS-3 and AChE emerged as relevant targets for the prepared hybrids, we utilized molecular docking to obtain insights about binding mode in the active site of these targets. CB-Dock2 server was employed to perform Template based docking where the active sites were determined based on grid box around the co-crystallized ligand. Redocking of the co-crystallized ligand produce predicted poses with RMSD less than 1.5 indicating the validity of the software (Fig. S2†). Binding energy for the 2 hybrids was calculated. Compound 6d and 19 showed excellent binding comparable to the co-crystallized ligand (Table 5) and post docking analysis was done to identify important interactions of each compound with the active site.
Blind docking results of acefylline based hybrids in binding site eNOS, ACHE.
| Target | Docking size (X, Y, Z) | Center box (X, Y, Z) | Vina score of 19 | Vina score of 6d | Vina score standarda |
|---|---|---|---|---|---|
| eNOS | 10, 11.5, 53 | 22, 22, 22 | −9.6 | −9.3 | RMSD = 0.87b |
| −9.7 | |||||
| ACHE | −14, −43.8, 27.6 | 27, 27, 27 | −10.3 | −10.9 | −12.2 |
| RMSD = 0.385c |
Co-crystallized ligand.
W69, PDB:6AV7.
E20, PDB:4EY7.
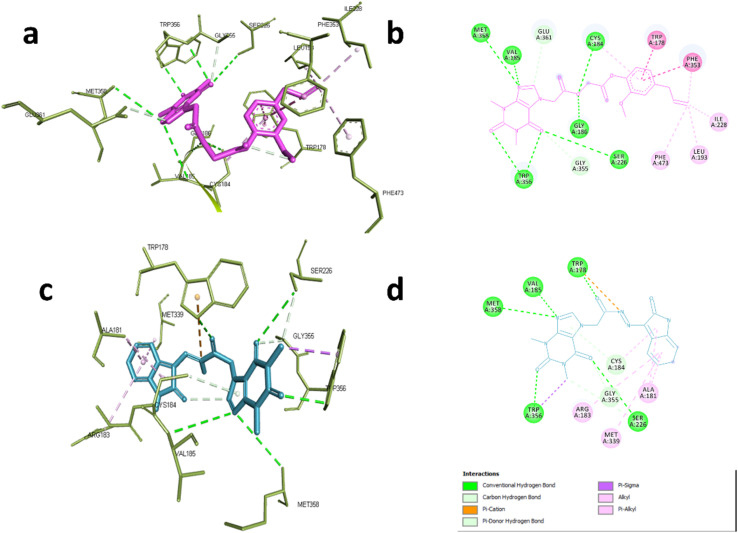
In case of NOS-3, the co-crystallized ligand was found to be near the heme-porphyrin catalytic site, and it interacted with Trp356, Met358, Glu361 through hydrogen bonding. On the other hand, hydrophobic interactions with Pro334, Val 336 Met339, and Phe353 were also identified.62 Compound 6d showed similar interaction profile where it was able to form hydrogen bonds with Cys184, Val185, Gly186, Ser226, TRP356,Met358 and Glu361 through acefylline moiety while other hydrophobic interactions such as TRP178, Leu193, Ile228, Phe353 and Phe473 were exerted through the eugenyl moiety as shown in Fig. 5a and b. This was also the case of compound 19 where the acefylline scaffold formed hydrogen bond with TRP178, Val185, Ser226, TRP356, Met 358 while isatin moiety formed hydrophobic interactions with Ala181, Arg183, Cys184, Met339 and Gly355 as shown in Fig. 5c and d.
Fig. 5. Interactions with eNOS complex (PDB:6AV7). (a) 3D and (b) 2D interactions of 6d (magenta). (c) 3D and (d) 2D interactions of 19 (cyan).
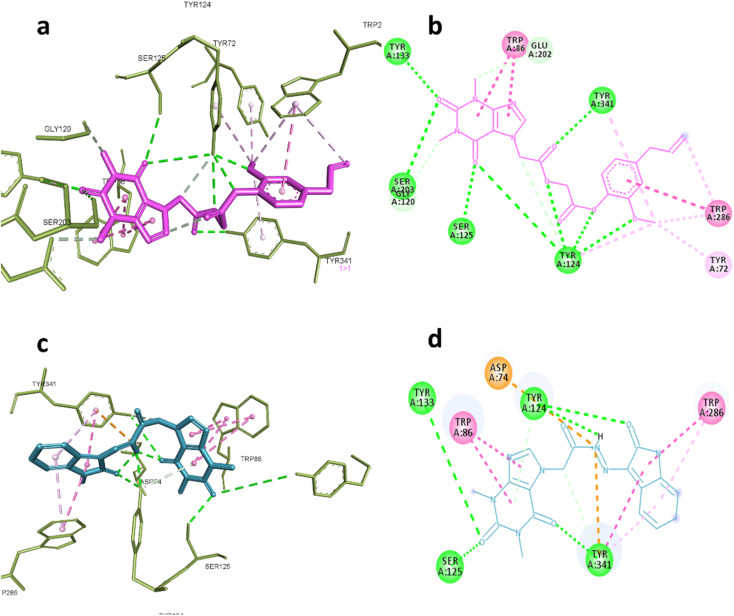
AChE was one of the predicted targets for the prepared hybrids and compounds 6d and 19 also showed good inhibitory activity against this enzyme experimentally. Post docking analysis gave insights about their binding mode in comparison to known inhibitors. The co-crystallized ligand donepezil mainly exert its remarkable inhibitory activity through hydrophobic interactions with Trp86, Trp286, Tyr337, Phe338, Tyr341and one hydrogen bond with Phe295 while galantamine the standard inhibitor inhibitory effect depends on blocking the catalytic active site by interaction with Glu202, Ser203, Tyr337 through hydrogen bonding and Trp86, Gly122 through hydrophobic interactions.
These interactions were reproducible by compound 6d where it formed hydrogen bonds with Ser203, Gly120, Tyr124, Ser125, Tyr133 and Tyr 341, also hydrophobic interactions with Tyr72, Trp86, Trp286 indicating that the compound 6d occupy the full active site gorge-channel which is similar to the binding mode of donepezil rather than galantamine. Compound 19 was also able to interact with Try124, Tyr133, Ser125 and Try341 through hydrogen bonding and Trp86 and Trp286 through hydrophobic interactions as demonstrated in Fig. 6a–d.
Fig. 6. Interactions with AChE (PDB:4EY7). (a) 3D and (b) 2D interactions of 6d (magenta). (c) 3D and (d) 2D interactions of 19 (cyan).
Nevertheless, the length of linker may explain the difference in the inhibitory activity of compound 6d and 19 where the acetyl linker in case of the first allowed the xanthine moiety to reach the catalytic triad and interact with Ser203, which is important to achieve good inhibitory activity.63 Overall, the analysis of binding mode of 6d and 19 showed their ability to bind to both of catalytic site and peripheral aromatic site (PAS) suggesting that their ability to neutralize the pathological role of AChE through preventing acetyl choline hydrolysis and the aggregation of Aβ by disrupting their interaction with PAS of AChE.64
2.5. Preliminary stability studies
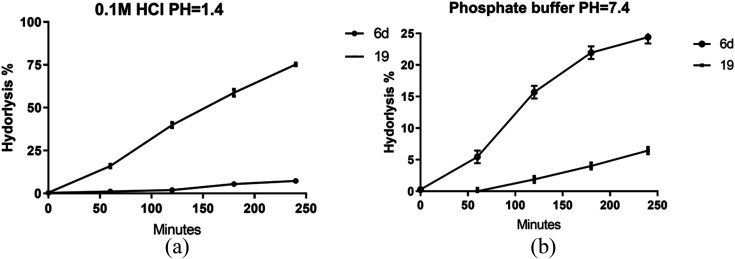
Given the enhanced bioactivity observed in compounds 6d and 19 relative to other hybrid derivatives, our investigation centered on evaluating their stability under simulated gastric and intestinal conditions. Compound 6d demonstrated exceptional stability when exposed to 0.1 M HCl, exhibiting a half-life (t1/2) of 35 hours, while compound 19 exhibited a significantly shorter t1/2 of 1.8 hours, as shown in Fig. 7a.
Fig. 7. Stability of prepared hybrids at different buffers (a) pH = 1.4, (b) phosphate buffer pH = 7.4. Data presented as mean values ±SD of three independent experiments.
However, a contrasting pattern emerged when the pH was adjusted to 7.4. Notably, due to its ester composition, compound 6d displayed increased susceptibility to hydrolysis, resulting in a t1/2 of 11.3 hours. Conversely, compound 19 displayed a prolonged t1/2 of 76 hours, as depicted in Fig. 7b. These findings are supported by prior investigations, emphasizing the diminished stability of hydrazones within acidic pH in comparison with basic or neutral environments. In contrast, esters assume a prodrug role in relatively alkaline pH levels, facilitating sustained release of the active substrate.
Importantly, both compounds exhibited substantive resistance to acid hydrolysis.65,66 Based on the data we gathered, it can be inferred that both compounds are not greatly affected by gastric or intestinal fluids. This suggests that they have the ability to be absorbed into the body without undergoing substantial breakdown.
3. Conclusion
This study investigated the conjugation of theophylline with different compounds of natural origin hoping to construct new hybrids with dual activity against cholinergic and inflammatory pathways as potential agents for treatment of Alzheimer's disease (AD). In this study, a series of 28 theophylline-based hybrids were synthesized and investigated against acetylcholinesterase (AChE) and inflammation. Out of 28 tested hybrids; two hybrids, acefylline–eugenol 6d and acefylline–isatin 19 were able to inhibit acetylcholinesterase (AChE) at low micromolar concentration when compared to “galantamine” a known standard AChE inhibitor. Moreover, the prepared hybrids possessed significant anti-inflammatory effect against lipopolysaccharide induced inflammation in RAW 264.7 and reduced nitric oxide (NO), tumor necrosis alpha (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6) levels in dose dependent manner. The findings of this study were further explained in the light of network pharmacology analysis which suggested that AChE and nitric oxide synthase are the main targets of the most active compounds. Our molecular docking studies supported the obtained results. Finally, the two promising hybrids proved to be stable when studied in medium simulating gastric and intestinal environment suggesting their possible mode of absorption without substantial breakdown.
Particularly, the hybrids of theophylline with either eugenol or isatin revealed very promising and unleashing their therapeutic potential for managing complex health disorders, e.g., Alzheimer's disease and other neurodegenerative disorders. However, further investigation of the kinetic properties of these hybrids is necessary before translating these findings into preclinical and clinical practice.
4. Material and methods
4.1. Chemistry
4.1.1. General
Melting points were measured with a FALC melting point apparatus and were uncorrected. The NMR spectra were recorded by Bruker spectrometer at 400 MHz 13C NMR spectra were run at 101 MHz in deuterated dimethyl sulfoxide (DMSO-d6) or chloroform (CDCl3). Chemical shifts (δ ppm) were reported relative to the solvent DMSO-d6 or CDCl3. All coupling constant (J) values were given in hertz. Mass spectrometry were carried out using mass spectrometer: thermo scientific ISQLT single quadrupole (USA). Microanalyses were carried out using PerkinElmer 240 elemental analyzer for elements C, H, N, and S results were obtained within the accepted limits. Optical rotation was measured with a PolyScience polarimeter model SR-6. Unless otherwise noted, all solvents and reagents were commercially available and used without further purification.
4.1.2. Synthesis of acefylline (2)
A solution of bromoacetic acid (14 g, 0.1 mol) in water (250 ml) containing potassium bicarbonate (0.5 g, 0.5 mol) was added drop-wisely, with stirring, to another solution of theophylline (1) (18 g, 0.1 mol) and potassium bicarbonate (0.5 g, 0.5 mol) in water (250 ml). The mixture was refluxed for 8 h then cooled and filtered. The filtrate was acidified with dilute hydrochloric acid to reach pH value of 2. The formed precipitate was collected by filtration and recrystallized from methanol to give white needles (m.p. 267–290 °C)67 (Rf = 0.2 DCM : methanol 7 : 3).
4.1.3. Synthesis of acefylline hybrid ester (4a–d)
In a 25 ml round bottom flask, a mixture of acefylline 2 (90 mg, 0.4 mmol), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDCI) (0.86 g, 0.45 mmol), and 4-dimethylaminopyridine (DMAP) (0.05 g, 0.04 mM) in dry methylene chloride (25 ml) was stirred for 10 minutes in an ice salt bath. After that, a chosen hydroxyl-containing compound 2a–d (0.45 mmol) was added in a dropwise manner with constant stirring over a period of 1 h, maintaining the temperature constant, then the reaction mixture was stirred further 48 h at room temperature until the reaction was completed. The reaction mixture was filtered and evaporated then the crude product was purified using column chromatography using dichloromethane: ethyl acetate gradient elution from 100% to 70% dichloromethane to give compounds (4a–d).
4.1.3.1. 2-Isopropyl-5-methylcyclohexyl-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetate (4a)
Yield 75% as semisolid, [α]25D = −52.5 (c 15 g l−1, EtOH). 1H NMR (400 MHz, CDCl3) δ ppm: 7.59 (s, 1H), 5.01 (s, 2H), 4.71 (td, J = 10.9, 4.4 Hz, 1H), 3.53 (s, 3H), 3.31 (s, 3H), 1.96 (d, J = 11.5 Hz, 1H), 1.78 (qt, J = 7.2, 3.2 Hz, 1H), 1.61 (dt, J = 12.1, 2.8 Hz, 2H), 1.36 (dtt, J = 38.3, 11.3, 3.0 Hz, 2H), 1.05–0.89 (m, 2H), 0.83 (t, J = 7.0 Hz, 7H), 0.68 (d, J = 6.9 Hz, 3H); 13C NMR (101 MHz, CDCl3) δ ppm: 166.60, 155.16, 151.69, 148.29, 141.79, 107.20, 76.88, 47.62, 46.90, 40.62, 34.02, 31.40, 29.91, 27.93, 26.19, 23.27, 21.95, 20.76, 16.23; displayed a molecular ion at m/z 376.46 calculated for the molecular formula anal. calcd for C19H28N4O4, C, 60.62; H, 7.5; N, 14.88; found C, 60.71; H, 7.27; N, 14.95.
4.1.3.2. 2-Isopropyl-5-methylphenyl-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetate (4b)
Yield 65%, 1H NMR (400 MHz, CDCl3) δ ppm: 7.64 (d, J = 6.2 Hz, 1H), 7.12 (dd, J = 8.0, 2.2 Hz, 1H), 7.00–6.93 (m, 1H), 6.80 (d, J = 2.2 Hz, 1H), 5.28 (d, J = 2.3 Hz, 2H), 3.53 (d, J = 2.2 Hz, 3H), 3.33 (d, J = 2.2 Hz, 3H), 2.98–2.86 (m, 1H), 2.22 (s, 3H), 1.09 (dd, J = 6.9, 2.2 Hz, 6H); 13C NMR (101 MHz, CDCl3) δ ppm: 165.80, 155.19, 151.57, 148.36, 147.18, 141.61, 136.79, 136.70, 127.70, 126.54, 122.14, 107.12, 47.44, 29.84, 27.87, 26.86, 23.00, 20.73; displayed a molecular ion at m/z 370.41 calculated for the molecular formula anal. calcd for C19H22N4O4, C, 61.61; H, 5.99; N, 15.13; found C, 61.64; H, 5.42; N, 15.26.
4.1.3.3. 4-Formyl-2-methoxyphenyl-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetate (4c)
Yield 25%, 1H NMR (400 MHz, CDCl3) δ ppm: 9.84 (s, 1H), 7.85 (s, 1H), 7.48–7.41 (m, 2H), 7.06 (d, J = 8.5 Hz, 1H), 3.98 (d, J = 5.1 Hz, 2H), 3.68 (s, 3H), 3.51 (d, J = 5.9 Hz, 6H); 13C NMR (101 MHz, CDCl3) δ ppm: 191.11, 168.81, 156.26, 151.67, 148.99, 140.23, 127.71, 114.55, 107.03, 56.26, 50.97, 30.40, 28.63; displayed a molecular ion at m/z 372.34 calculated for the molecular formula anal. calcd for C17H16N4O6, C, 54.84; H, 4.33; N, 15.05; found C, 55.11; H, 4.89; N, 14.54.
4.1.3.4. 4-Allyl-2-methoxyphenyl-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetate (4d)
Yield 75%, 1H NMR (400 MHz, CDCl3) δ ppm: 7.63 (s, 1H), 6.96 (d, J = 7.8 Hz, 1H), 6.69 (d, J = 10.4 Hz, 2H), 5.91–5.81 (m, 1H), 5.30 (s, 2H), 5.06–4.98 (m, 2H), 3.74 (s, 3H), 3.52 (s, 3H), 3.33 (s, 3H), 3.29 (d, J = 6.9 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ ppm: 165.49, 155.29, 151.70, 150.41, 148.50, 141.99, 139.76, 137.27, 136.85, 122.27, 120.78, 116.35, 112.74, 107.16, 55.85, 47.14, 40.05, 29.86, 27.96; displayed a molecular ion at m/z 384.39 calculated for the molecular formula anal. calcd for C19H20N5O5, C, 77.49; H, 10.22% found C, 77.93; H, 10.03% C, 59.37; H, 5.24; N, 14.58; found C, 59.89; H, 4.78; N, 14.26.
4.1.4. Synthesis of bromo acetyl derivatives (5a–d)
Acetyl derivatives were prepared adopting previously reported method with slight modifications.68 In a 100 ml round bottom flask, a mixture of a chosen hydroxyl-containing compound 3a–d [menthol (a), thymol (b), vanillin (c), or eugenol (d)] (10 mmol) and triethylamine (1.01 g, 10 mmol) in DCM (25 ml) was cooled in an ice salt mixture to −10 °C. To this reaction mixture, bromoacetyl bromide (2.02 g, 10 mmol) in methylene chloride (25 ml) was added in a dropwise manner with constant stirring over a period of 1 h, maintaining the temperature constant, then the reaction mixture was stirred for further 5 h until the reaction is completed. The reaction mixture was filtered and evaporated then the crude product was purified using column chromatography using petroleum ether: DCM gradient elution from 100% to 80% petroleum ether to give compounds (5a–d).
4.1.5. Synthesis of acefylline hybrid acetyl linker derivative (6a–d)
In a 25 ml round flask, a mixture of bromoacetate derivative 5a–d (0.25 mmol), acefylline 2 (50 mg, 0.2 mmol), potassium carbonate (55 mg, 0.3 mmol) and potassium iodide (0.69 m, 0.02 mmol) in DMF (10 ml) was stirred for 48 hours in room temperature. The reaction mixture was poured into crushed ice with stirring and extracted with ethyl acetate (3 × 25 ml). The organic layer was dried over anhydrous sodium sulphate, filtered and the solvent was removed under reduced pressure to obtain semisolid residue, which was chromatographed on silica gel column using methylene chloride: ethyl acetate gradient elution from 100% to 70% methylene as eluent to give compounds (6a–d).
4.1.5.1. 2-Isopropyl-5-methylcyclohexyl-2-(2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetoxy)acetate (6a)
Yield 60%, [α]25D = −67.5(c 15 g l−1, EtOH). 1H NMR (400 MHz, CDCl3) δ ppm: 7.63 (s, 1H), 5.18 (s, 2H), 4.70 (td, J = 11.0, 4.5 Hz, 1H), 4.64 (s, 2H), 3.54 (s, 3H), 3.32 (s, 3H), 1.95–1.86 (m, 1H), 1.79–1.67 (m, 1H), 1.66–1.57 (m, 2H), 1.47–1.26 (m, 2H), 1.06–0.90 (m, 2H), 0.82 (dd, J = 8.7, 6.7 Hz, 7H), 0.67 (d, J = 7.0 Hz, 3H); 13C NMR (101 MHz, CDCl3) δ ppm: 166.68, 166.57, 155.29, 151.64, 148.43, 141.95, 107.06, 76.21, 61.92, 47.09, 46.89, 40.67, 34.05, 31.39, 29.94, 27.96, 26.26, 23.38, 21.96, 20.69, 16.27; displayed a molecular ion at m/z 434.49 calculated for the molecular formula anal. calcd for C21H30N4O6 C, 58.05; H, 6.96; N, 12.9; found C, 58.25; H, 6.96; N, 12.6%.
4.1.5.2. 2-(2-Isopropyl-5-methylphenoxy)-2-oxoethyl-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetate (6b)
Yield 45%, 1H NMR (400 MHz, CDCl3) δ ppm: 7.60 (s, 1H), 7.13 (d, J = 7.9 Hz, 1H), 6.98 (dd, J = 8.0, 1.8 Hz, 1H), 6.77–6.70 (m, 1H), 5.19 (s, 2H), 4.94 (s, 2H), 3.53 (s, 3H), 3.30 (s, 3H), 2.85 (dt, J = 13.7, 6.8 Hz, 1H), 2.23 (s, 3H), 1.09 (d, J = 6.9 Hz, 6H); 13C NMR (101 MHz, CDCl3) δ ppm: 166.73, 165.92, 155.30, 151.61, 148.44, 147.04, 141.89, 136.89, 136.87, 127.77, 126.69, 122.25, 107.05, 61.71, 47.09, 29.94, 27.93, 27.06, 23.02, 20.79; displayed a molecular ion at 428.45 m/z calculated for the molecular formula anal. calcd for C21H24N4O6 C, 58.87; H, 5.65; N, 13.08; found C, 58.42; H, 6.13; N, 13.
4.1.5.3. 2-(4-Formyl-2-methoxyphenoxy)-2-oxoethyl-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetate (6c)
Yield 25%, 1H NMR (400 MHz, CDCl3) δ ppm: 9.88 (s, 1H), 7.61 (d, J = 1.8 Hz, 1H), 7.42 (dt, J = 9.7, 1.6 Hz, 2H), 7.18 (dd, J = 7.9, 3.0 Hz, 1H), 5.25–5.15 (m, 2H), 4.97 (d, J = 17.3 Hz, 2H), 3.83 (d, J = 3.9 Hz, 3H), 3.53 (s, 3H), 3.30 (d, J = 2.1 Hz, 3H); 13C NMR (101 MHz, CDCl3) δ ppm: 190.95, 166.61, 164.67, 155.32, 151.62, 148.55, 148.51, 143.78, 141.94, 135.66, 124.66, 123.17, 110.94, 107.06, 61.36, 56.20, 47.07, 29.91, 27.94; displayed a molecular ion at 430.37 m/z calculated for the molecular formula anal. calcd for C19H18N4O8 C, 53.03; H, 4.22; N, 13.02; found C, 53.07; H, 3.63; N, 13.59.
4.1.5.4. 2-(4-Allyl-2-methoxyphenoxy)-2-oxoethyl-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetate (6d)
Yield 45%, 1H NMR (400 MHz, CDCl3) δ ppm: 7.59 (s, 1H), 6.89 (d, J = 8.0 Hz, 1H), 6.74–6.66 (m, 2H), 5.93–5.82 (m, 1H), 5.18 (s, 2H), 5.08–4.98 (m, 2H), 4.95 (s, 2H), 3.73 (s, 3H), 3.53 (s, 3H), 3.36–3.27 (m, 2H), 3.30 (s, 3H); 13C NMR (101 MHz, CDCl3) δ ppm: 166.61, 165.38, 155.31, 151.63, 150.50, 148.47, 141.95, 139.74, 137.04, 136.85, 122.19, 120.73, 116.35, 112.79, 107.06, 61.55, 55.86, 47.03, 40.07, 29.92, 27.94; displayed a molecular ion at 442.43 m/z calculated for the molecular formula anal. calcd for C21H22N4O7, C, 57.01; H, 5.01; N, 12.66; found C, 56.86; H, 4.64; N, 13.15.
4.1.6. Synthesis of acefylline hybrid oxadiazole linker (9a–d)
A solution of acefylline 2 (2 g, 8 mM) was refluxed in thionyl chloride 25 ml for 6 hours and the reaction mixture was evaporated under pressure, then 25 ml isopropanol was added and stirred for 0.5 h. Before adding 20 ml of hydrazine hydrate and the reaction mixture was refluxed for 3 hours to give the corresponding hydrazide 7. The reaction was left to cool down, filtered under vacuum, and washed with isopropanol to give off white powder (m.p. 276–280 °C).
A mixture of hydrazide 7 (1.26 g, 5 mM) and potassium hydroxide (0.28, 5 mM) was formed by dissolving them in absolute ethanol (50 ml), carbon disulfide (0.82 ml) was added, and reaction mixture was refluxed for 6 hours. Completion of reaction was monitored through TLC, then the reaction mixture was cooled, and excess chilled water was added, acidified with dilute hydrochloric acid (10%) and the precipitate of 1,3,4-oxadiazole derivative of acefylline 8 was formed which were filtered off under vacuum to give off white powder (m.p. 252–253 °C) (reported m.p. 254 °C).69
In a 25 ml round flask, a mixture of bromoacetate 5a–d (0.2 mM), 1,3,4-oxadiazole derivative 8 (60 mg, 0.2 mM), and potassium carbonate (50 mg, 0.2 mM) in DMF (5 ml) was stirred for 24 hours in room temperature. The reaction mixture was poured into crushed ice with stirring and extracted with ethyl acetate (3 × 25 ml). The organic layer was dried over anhydrous sodium sulphate, filtered and the solvent was removed under reduced pressure to obtain semisolid residue, which was chromatographed on silica gel column using methylene chloride: ethyl acetate gradient elution from 100% to 60% methylene chloride as eluent to give compounds (9a–d).
4.1.6.1. 2-Isopropyl-5-methylcyclohexyl-2-((5-((1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)methyl)-1,3,4-oxadiazol-2-yl)thio)acetate (9a)
Yield 75%, [α]25D = −46 (c 15 g l−1, EtOH). 1H NMR (400 MHz, CDCl3) δ ppm: 7.68 (s, 1H), 5.73 (s, 2H), 4.67 (td, J = 10.9, 4.4 Hz, 1H), 3.97 (s, 2H), 3.54 (s, 3H), 3.33 (s, 3H), 1.97–1.87 (m, 1H), 1.77 (ddd, J = 11.3, 8.4, 5.7 Hz, 2H), 1.66–1.56 (m, 2H), 1.38–1.21 (m, 1H), 1.04–0.90 (m, 2H), 0.82 (dd, J = 9.0, 6.7 Hz, 7H), 0.66 (d, J = 6.9 Hz, 3H); 13C NMR (101 MHz, CDCl3) δ ppm: 166.68, 165.19, 162.15, 155.19, 151.56, 148.66, 141.35, 106.70, 76.93, 46.89, 40.58, 40.38, 34.64, 34.05, 31.39, 29.92, 28.04, 26.18, 23.30, 21.97, 20.73, 16.21; displayed a molecular ion at 490.58 m/z calculated for the molecular formula anal. calcd for C22H30N6O5S, C, 53.86; H, 6.16; N, 17.13; S, 6.54; found C, 54.09; H, 5.62; N, 17.42; S, 6.63.
4.1.6.2. 2-Isopropyl-5-methylphenyl-2-((5-((1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)methyl)-1,3,4-oxadiazol-2-yl)thio)acetate (9b)
Yield 55%, 1H NMR (400 MHz, CDCl3) δ ppm: 7.68 (s, 1H), 7.12 (d, J = 7.9 Hz, 1H), 7.01–6.94 (m, 1H), 6.78–6.73 (m, 1H), 5.73 (s, 2H), 4.24 (s, 2H), 3.53 (s, 3H), 3.33 (s, 3H), 2.89 (p, J = 6.9 Hz, 1H), 2.23 (s, 3H), 1.08 (d, J = 6.9 Hz, 6H); 13C NMR (101 MHz, CDCl3) δ ppm: 166.10, 164.89, 162.40, 155.18, 151.54, 148.61, 147.58, 141.33, 136.84, 136.82, 127.73, 126.60, 122.23, 106.69, 40.43, 34.26, 29.95, 28.04, 26.98, 23.05, 20.83; displayed a molecular ion at 484.53 m/z calculated for the molecular formula anal. calcd for C22H24N6O5S, C, 54.54; H, 4.99; N, 17.35; S, 6.62; found C, 54.56; H, 5.4; N, 17.43; S, 7.04.
4.1.6.3. 4-Formyl-2-methoxyphenyl-2-((5-((1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)methyl)-1,3,4-oxadiazol-2-yl)thio)acetate (9c)
Yield 15%, 1H NMR (400 MHz, CDCl3) δ ppm: 9.87 (s, 1H), 7.69 (s, 1H), 7.40 (d, J = 6.9 Hz, 2H), 7.23–7.15 (m, 1H), 5.73 (s, 2H), 4.25 (s, 2H), 3.79 (s, 3H), 3.50 (s, 3H), 3.31 (s, 3H); 13C NMR (101 MHz, CDCl3) δ ppm: 190.97, 164.99, 164.62, 162.53, 155.17, 151.60, 151.51, 148.69, 144.26, 141.44, 135.60, 124.68, 123.15, 110.89, 106.68, 56.18, 40.41, 33.88, 29.89, 28.02; displayed a molecular ion at 486.46 m/z calculated for the molecular formula anal. calcd for C20H18N6O7S, C, 49.38; H, 8.23; N, 3.69; S, 4.22; found C, 48.82; H, 7.64; N, 3.76; S, 4.35.
4.1.6.4. 4-Allyl-2-methoxyphenyl-2-((5-((1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)methyl)-1,3,4-oxadiazol-2-yl)thio)acetate (9d)
Yield 25%, 1H NMR (400 MHz, CDCl3) δ ppm: 7.68 (d, J = 1.9 Hz, 1H), 6.91–6.84 (m, 1H), 6.68 (t, J = 4.6 Hz, 2H), 5.94–5.79 (m, 1H), 5.73 (d, J = 2.1 Hz, 2H), 5.07–4.98 (m, 2H), 4.26–4.19 (m, 2H), 3.69 (s, 3H), 3.58–3.47 (m, 3H), 3.35–3.27 (m, 5H); 13C NMR (101 MHz, CDCl3) δ ppm: 165.66, 164.81, 162.35, 155.18, 151.53, 150.46, 148.68, 141.40, 139.68, 137.54, 136.84, 122.11, 120.69, 116.36, 112.74, 106.69, 55.85, 40.38, 40.07, 34.00, 29.87, 28.02; displayed a molecular ion at 498.51 m/z calculated for the molecular formula anal. calcd for C22H22N6O6S. C, 53.01; H, 4.45; N, 16.86; S, 6.43; found C, 52.65; H, 4.47; N, 17.01; S, 6.82.
4.1.7. Synthesis of azide derivative (10a-b and 13)
In order to perform this reaction, bromoacetate derivatives 5a-b and 4-hydroxy coumarin 11 was transformed to the corresponding azide. Compounds, 5a-b (2 mmol) were stirred with sodium azide (3 mmol) in solution of ethanol/water (1 : 1) for 3 h, then the reaction mixture was evaporated under vacuum and extracted with ethyl acetate (3 × 25 ml). The organic layer was dried over anhydrous sodium sulphate, filtered and the solvent was evaporated in vacu to obtain oily liquid residues of 10a-b which were used without further purification.
On the other hands, 11 (1.3 mg, 8 mmol) was charged in phosphorus oxychloride (20 ml), then, triethylamine (1.5 ml, 12 mmol) was added drop wisely, and the mixture was refluxed for 4 h. Then the reaction mixture was distilled to give yellowish brown residue which give white needles upon crystallization from petroleum ether to give 4-chloro coumarin 12, with m.p. range of 85–87 °C (reported m.p. 84–86 °C (ref. 70)).
To a solution of 12 (540 mg, 3 mmol) in dry acetone, sodium azide (292.2 mg, 4.5 mmol) was added and stirred in room temperature for 5 h. The reaction mixture was evaporated in vacu and 50 ml water was added then extracted with ethyl acetate (3 × 25 ml). The organic layer was dried over anhydrous sodium sulphate, filtered and the solvent was evaporated in-vacu to obtain white needles of 13 with m.p. range of 160–163 °C (reported m.p. 165–167 °C (ref. 71)).
4.1.8. Synthesis of acefylline hybrid with 1,2,3 triazole linker (15a–c)
Acefylline 2 (2 g, 0.7 mmol) was stirred with potassium carbonate (1.7 g, 1 mmol) in DMF for 20 minutes then a solution of propargyl bromide (1.19 ml, 1.5 mmol) in DMF was added and the reaction mixture was stirred in room temperature for 8 h. After that, the reaction was terminated based on TLC analysis by pouring it into crushed ice with stirring and extracted with ethyl acetate (3 × 25 ml). The organic layer was dried over anhydrous sodium sulphate, filtered and the solvent was removed under reduced pressure to obtain white needles which were recrystallized from methanol to give 14 (m.p. 170–171 °C, reported m.p. 168–170 °C (ref. 72)) which was used without further purification.
Copper sulphate pentahydrate (108.4 mg, 1.2 equivalent) and sodium ascorbate (86.12 mg, 1.2 equivalent) in water was added to a stirred solution of 14 in DMF, then an azide derivative (10a-b or 13) (100 mg, 1.1 equivalent) was added to the reaction mixture and stirred in room temperature for 48 h. The reaction mixture was poured into crushed ice with stirring and extracted with ethyl acetate (3 × 25 ml). The organic layer was dried over anhydrous sodium sulphate, filtered and the solvent was removed under reduced pressure to obtain semisolid residue, which was chromatographed on silica gel column using methylene chloride: ethyl acetate gradient elution from 100% to 60% methylene chloride as eluent to give compounds (15a–c).
4.1.8.1. 2-Isopropyl-5-methylcyclohexyl-2-(4-((2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetoxy)methyl)-1H-1,2,3-triazol-1-yl)acetate (15a)
Yield 25%,[α]25D = −56.7(c 15 g l−1, EtOH). 1H NMR (400 MHz, CDCl3) δ ppm: 7.79 (s, 1H), 7.57 (s, 1H), 5.32 (s, 2H), 5.16–5.03 (m, 4H), 4.72 (td, J = 10.9, 4.4 Hz, 1H), 3.54 (s, 3H), 3.31 (d, J = 1.1 Hz, 3H), 2.00–1.89 (m, 1H), 1.72 (pd, J = 6.9, 2.5 Hz, 1H), 1.67–1.57 (m, 2H), 1.46–1.38 (m, 1H), 1.37–1.29 (m, 1H), 1.06–0.89 (m, 2H), 0.84 (t, J = 7.0 Hz, 7H), 0.68 (d, J = 6.9 Hz, 3H); 13C NMR (101 MHz, CDCl3) δ ppm: 167.02, 165.65, 155.30, 151.62, 148.52, 141.85, 125.75, 107.19, 77.13, 59.11, 51.10, 47.41, 46.84, 40.65, 33.96, 31.38, 29.92, 27.93, 26.30, 23.29, 21.94, 20.73, 16.26; displayed a molecular at 515.57 m/z calculated for the molecular formula anal. calcd for C24H33N7O6, C, 55.91; H, 6.45; N, 19.02; found C, 55.54; H, 6.9; N, 18.81.
4.1.8.2. (1-(2-(2-Isopropyl-5-methylphenoxy)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)methyl-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetate (15b)
Yield 15%, 1H NMR (400 MHz, CDCl3) δ ppm: 7.93 (s, 1H), 7.65 (s, 1H), 7.22 (d, J = 7.9 Hz, 1H), 7.08 (d, J = 7.9 Hz, 1H), 6.86 (s, 1H), 5.48 (s, 2H), 5.41 (s, 2H), 5.12 (s, 2H), 3.61 (s, 3H), 3.37 (s, 3H), 2.90 (p, J = 6.9 Hz, 1H), 2.32 (s, 3H), 1.17 (d, J = 6.8 Hz, 6H); 13C NMR (101 MHz, CDCl3) δ ppm: 167.04, 164.95, 155.26, 151.58, 148.37, 147.06, 142.39, 141.88, 137.00, 136.62, 127.99, 126.75, 125.76, 122.10, 107.11, 59.06, 53.49, 50.95, 30.00, 27.95, 27.18, 23.02, 20.82; displayed a molecular ion at 509.52 m/z calculated for the molecular formula anal. calcd for C24H27N7O6, C, 56.58; H, 5.34; N, 19.24; found C, 56.45; H, 5.58; N, 19.25.
4.1.8.3. (1-(2-Oxo-2H-chromen-4-yl)-1H-1,2,3-triazol-4-yl)methyl-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetate (15c)
Yield 10%, 1H NMR (400 MHz, DMSO-d6) δ ppm: 8.86 (s, 1H), 8.09 (s, 1H), 7.85–7.75 (m, 2H), 7.60 (d, J = 8.4 Hz, 1H), 7.45 (t, J = 7.8 Hz, 1H), 6.96 (s, 1H), 5.42 (s, 2H), 5.27 (s, 2H), 3.44 (d, J = 6.8 Hz, 3H), 3.16 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.05, 159.92, 154.98, 154.15, 151.48, 148.55, 146.30, 143.70, 142.72, 134.04, 127.35, 125.90, 125.52, 117.70, 114.78, 111.28, 106.84, 58.41, 47.62, 30.00, 27.95; displayed a molecular ion at 463.41 m/z calculated for the molecular formula anal. calcd for C24H27N7O6, C, 54.43; H, 3.7; N, 21.16; found C, 54.09; H, 3.63; N, 21.46.
4.1.9. Synthesis of acefylline hybrid with hydrazone linker (18a–l, 19)
Aldehydes 16a–l or isatin 17 (0.1 mmol) were added to a solution of 7 (25 mg, 0.1 mmol) in absolute ethanol and catalytic amount of glacial acetic acid was further added, then the mixture was refluxed for 1–2 h. Upon completion of the reaction, it was filtered while hot, the precipitate was washed with hot ethanol under vacuum to obtain (18a–l) and 19.
4.1.9.1. E-N′-Benzylidene-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetohydrazide (18a)
White powder (yield 65%); m.p. 269–270 °C (reported 272–274 °C (ref. 73)); 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.80 (s, 1H), 8.12–8.05 (m, 2H), 7.75 (d, J = 7.0 Hz, 2H), 7.47 (d, J = 6.3 Hz, 3H), 5.57 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.43, 154.95, 151.51, 148.34, 144.79, 144.13, 134.31, 130.61, 129.35, 127.41, 107.19, 47.87, 29.95, 27.91; displayed a molecular ion at 340.34 m/z calculated for the molecular formula anal. calcd for C16H16N6O3, C, 56.47; H, 4.74; N, 24.69; found C, 56.91; H, 4.33; N, 25.07.
4.1.9.2. (E)-2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N′-(4-hydroxybenzylidene)acetohydrazide (18b)
White powder (yield 80%); m.p. 235–238 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.58 (s, 1H), 9.98 (s, 1H), 8.06 (s, 1H), 7.95 (s, 1H), 7.56 (dd, J = 9.6, 2.9 Hz, 2H), 6.87–6.79 (m, 2H), 5.51 (s, 2H), 3.46 (s, 3H), 3.21 (d, J = 1.7 Hz, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.02, 159.87, 154.92, 151.50, 148.30, 145.04, 144.12, 129.14, 125.33, 116.18, 107.18, 47.83, 29.93, 27.89; displayed a molecular ion at 356.34 m/z calculated for the molecular formula anal. calcd for C16H16N6O4, C, 53.93; H, 4.53; N, 23.58; found C, 53.91; H, 4.28; N, 24.03.
4.1.9.3. E-N′-(2,5-Dimethoxybenzylidene)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetohydrazide (18c)
White powder (yield 75%); m.p. 275–280 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.74 (s, 1H), 8.36 (s, 1H), 8.07 (d, J = 6.5 Hz, 1H), 7.40 (d, J = 3.0 Hz, 1H), 7.10–6.97 (m, 2H), 5.55 (s, 2H), 3.82 (s, 3H), 3.76 (s, 3H), 3.46 (s, 3H), 3.20 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.35, 154.90, 153.71, 152.64, 151.48, 148.30, 144.06, 140.16, 122.85, 117.76, 113.75, 109.96, 107.18, 56.66, 55.95, 47.91, 29.93, 27.89; displayed a molecular ion at 400.40 m/z calculated for the molecular formula anal. calcd for C18H20N6O5, C, 54; H, 5.04; N, 20.99; found C, 54.03; H, 5.31; N, 20.6.
4.1.9.4. E-N′-(3,4-Dimethoxybenzylidene)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetohydrazide (18d)
White powder (yield 65%); m.p. 280–282 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.68 (s, 1H), 8.08 (d, J = 3.3 Hz, 1H), 7.98 (s, 1H), 7.36 (d, J = 1.9 Hz, 1H), 7.26–7.19 (m, 1H), 7.03 (d, J = 8.3 Hz, 1H), 5.55 (s, 2H), 3.82 (d, J = 4.7 Hz, 6H), 3.46 (d, J = 3.7 Hz, 3H), 3.21 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.19, 154.95, 151.53, 151.22, 149.56, 148.36, 144.89, 144.12, 127.08, 121.89, 112.06, 109.06, 107.23, 56.07, 56.00, 47.92, 29.95, 27.90; displayed a molecular ion at 356.34 m/z calculated for the molecular formula anal. calcd for C18H20N6O5, C, 54; H, 5.04; N, 20.99; found C, 53.8; H, 5.64; N, 21.25.
4.1.9.5. (E)-2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N′-(3,4,5-trimethoxybenzylidene)acetohydrazide (18e)
White powder (yield 75%); m.p. > 300 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.82 (d, J = 12.5 Hz, 1H), 8.09 (s, 1H), 7.98 (s, 1H), 7.05 (d, J = 13.7 Hz, 2H), 5.57 (s, 2H), 3.85 (s, 6H), 3.71 (s, 3H), 3.47 (s, 3H), 3.21 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.42, 154.94, 153.67, 151.52, 148.36, 144.64, 144.11, 139.63, 129.84, 107.22, 104.85, 104.66, 60.61, 56.43, 47.98, 29.97, 27.91; displayed a molecular ion at 430.42 m/z calculated for the molecular formula anal. calcd for C19H22N6O6, C, 53.02; H, 5.15; N, 19.53; found C, 53.26; H, 4.57; N, 19.
4.1.9.6. (E)-2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N′-(3-methoxybenzylidene)acetohydrazide (18f)
White powder (yield 75%); m.p. 255–257 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.82 (s, 1H), 8.15 (s, 1H), 8.10 (s, 1H), 7.98 (s, 1H), 7.17–7.00 (m, 3H), 5.58 (s, 2H), 3.79 (d, J = 53.2 Hz, 6H), 3.22 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.42, 154.94, 153.67, 151.52, 148.36, 144.64, 144.11, 139.63, 129.84, 107.22, 104.85, 104.66, 56.43, 47.98, 29.97, 27.91; displayed a molecular at 370.37 m/z calculated for the molecular formula anal. calcd for C17H18N6O4, 55.13; H, 4.9; N, 22.69; found C, 55.33; H, 4.95; N, 23.1.
4.1.9.7. (E)-2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N′-(2-hydroxybenzylidene)acetohydrazide (18g)
White powder (yield 45%); m.p. 222–225 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.71 (s, 1H), 10.11 (s, 1H), 8.37 (s, 1H), 8.07 (s, 1H), 7.77 (dd, J = 7.9, 1.7 Hz, 1H), 7.35–7.23 (m, 1H), 6.96–6.84 (m, 2H), 5.54 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.11, 156.92, 154.95, 151.51, 148.32, 144.13, 141.92, 131.86, 129.48, 126.47, 119.89 (d, J = 3.6 Hz), 116.66, 107.19, 47.84, 29.95, 27.91; displayed a molecular ion in its electron impact mass spectrum at 356.34 m/z calculated for the molecular formula anal. calcd for C16H16N6O4, 55.93; H, 4.53; N, 23.58; found C, 55.4; H, 4.47; N, 24.06.
4.1.9.8. (E)-N′-(Benzo[d][1,3]dioxol-5-ylmethylene)-2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetohydrazide (18h)
Yield 25%, white powder, m.p. > 300 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.68 (s, 1H), 8.07 (s, 1H), 7.96 (s, 1H), 7.37 (s, 1H), 7.17 (d, J = 8.1 Hz, 1H), 7.00 (d, J = 8.0 Hz, 1H), 6.10 (s, 2H), 5.54 (s, 2H), 3.47 (s, 3H), 3.21 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.28, 154.93, 151.51, 149.54, 148.49, 148.33, 144.45, 144.10, 128.79, 123.75, 108.92, 107.21, 105.50, 102.05, 47.91, 29.95, 27.91; displayed a molecular ion at 384.35 m/z calculated for the molecular formula anal. calcd for C17H16N6O5, 53.12; H, 4.2; N, 21.87; found C, 53.7; H, 4.28; N, 21.33.
4.1.9.9. (E)-2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N′-(3-hydroxy-4-methoxybenzylidene)acetohydrazide (18i)
White powder (yield 75%); m.p. 265–268 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.61 (s, 1H), 9.30 (s, 1H), 8.07 (s, 1H), 7.92 (s, 1H), 7.24 (t, J = 3.0 Hz, 1H), 7.11–7.04 (m, 1H), 6.98 (dd, J = 8.4, 4.9 Hz, 1H), 5.53 (s, 2H), 3.82 (d, J = 5.2 Hz, 3H), 3.47 (s, 3H), 3.21 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.04, 154.96, 151.52, 150.26, 148.33, 147.30, 145.05, 144.18, 127.15, 120.65, 112.61, 112.30, 107.18, 56.07, 47.78, 29.95, 27.91; displayed a molecular ion at 386.37 m/z calculated for the molecular formula anal. calcd for C17H18N6O5, C, 52.85; H, 4.7; N, 21.75; found C, 52.92; H, 5.06; N, 21.95.
4.1.9.10. (E)-2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N′-(4-hydroxy-3-methoxybenzylidene)acetohydrazide (18j)
White powder (yield 65%); m.p. 274–276 °C (reported 278–280 °C (ref. 74)); 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.61 (s, 1H), 9.58 (s, 1H), 8.08 (d, J = 2.2 Hz, 1H), 7.94 (s, 1H), 7.35–7.26 (m, 1H), 7.11 (td, J = 8.9, 1.9 Hz, 1H), 6.84 (dd, J = 8.1, 3.4 Hz, 1H), 5.54 (s, 2H), 3.84 (s, 3H), 3.47 (s, 3H), 3.21 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.08, 154.94, 151.52, 149.39, 148.47, 148.34, 145.17, 144.13, 125.75, 121.96, 115.98, 109.87, 107.22, 56.05, 47.93, 29.96, 27.91; displayed a molecular ion at 386.37 m/z calculated for the molecular formula anal. calcd for C17H18N6O5, C, 52.85; H, 4.7; N, 21.75; found C, 53.13; H, 4.19; N, 21.46.
4.1.9.11. (E)-2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N′-((1E,2E)-3-Phenylallylidene)acetohydrazide (18k) C18H18N6O3
White powder (yield 45%); m.p. 240–242 °C (reported m.p. 237–239 °C (ref. 75)); 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.68 (s, 1H), 8.06 (s, 1H), 7.87 (d, J = 12.3 Hz, 1H), 7.65–7.61 (m, 3H), 7.45–7.38 (m, 2H), 7.24 (d, J = 28.9 Hz, 1H), 7.05 (dd, J = 29.9, 12.6 Hz, 1H), 5.45 (s, 2H), 3.46 (s, 3H), 3.21 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.08, 154.94, 151.50, 149.82, 148.32, 147.40, 144.18, 139.93, 136.23, 129.33, 127.62, 125.25, 107.13, 47.72, 29.95, 27.91; displayed a molecular ion at 366.38 m/z calculated for the molecular formula anal. calcd for C18H18N6O3, C, 59.01; H, 4.95; N, 22.94; found C, 58.9; H, 5.04; N, 22.38.
4.1.9.12. (E)-2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N′-(4-methoxybenzylidene)acetohydrazide (18l)
White powder (yield 50%); m.p. 245–248 °C (reported m.p. 248–250 °C (ref. 73)); 1H NMR (400 MHz, DMSO-d6) δ ppm: 11.7 (s, 1H), 8.1 (s, 1H), 8.0 (s, 1H), 7.7 (dd, J = 7.2, 5.1 Hz, 2H), 7.0 (dd, J = 9.0, 3.1 Hz, 2H), 5.5 (s, 2H), 3.8 (s, 3H), 3.5 (s, 3H), 3.2 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 168.1, 161.3, 154.9, 151.5, 148.3, 147.7, 144.6, 144.1, 129.0, 126.9, 114.8, 107.2, 55.8, 47.8, 29.9, 27.9; displayed a molecular ion at 370.37 m/z calculated for the molecular formula anal. calcd for C17H18N6O4, C, 55.13; H, 4.9; N, 22.69; found C, 55.34; H, 4.95; N, 23.06.
4.1.9.13. (E)-2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)-N′-(2-oxoindolin-3-ylidene)acetohydrazide (19)
Yellow powder (yield 85%); m.p. 240–243 °C; 1H NMR (400 MHz, DMSO-d6) δ ppm: 12.72 (s, 1H), 11.36 (s, 1H), 8.16–8.10 (m, 1H), 7.53 (dd, J = 55.5, 29.1 Hz, 2H), 7.03 (dd, J = 54.1, 28.9 Hz, 2H), 5.73 (s, 2H), 3.57 (s, 3H), 3.21 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ ppm: 162.83, 154.97, 151.50, 148.44, 144.07, 143.21, 132.48, 123.17, 121.32, 119.89, 111.78, 107.05, 47.44, 29.98, 27.92; displayed a molecular ion in its electron impact mass spectrum at 381.35 m/z calculated for the molecular formula anal. calcd for C17H15N7O4, 53.54; H, 3.96; N, 25.71; found C, 53.39; H, 3.98; N, 25.85.
4.2. Biological evaluation
4.2.1. Network pharmacology
To identify potential molecular targets of compound 6d and 19 their 3d chemical structures were submitted to PharmMapper server (https://www.lilab-ecust.cn/pharmmapper/)76 and Swiss target prediction server.77 The predicted targets were merged, duplicates were removed and saved in CSV file. Molecular targets associated with Alzheimer's disease (AD) were retrieved from input 2.0 server, an interface for network pharmacology.78 Then, Venny 2.1.0 program (https://bioinfogp.cnb.csic.es/tools/venny/) was utilized to determine common targets between the predicted and AD related targets.
The identified targets were submitted to enrichment analysis module integrated in INPUT 2.0 server where KEGG pathways analysis was preformed and most relevant BP, MF, and CC were identified to establish pathways-genes network using the most significantly enriched terms. Then protein–protein interaction network was constructed using confidence score = 0.7 as cut off and finally gene distribution analysis was performed to identify targets that are significantly correlated with pathways involved in AD.
4.2.2. Cell culture
RAW 264.7 was obtained from VACSERA, Egypt. The cell lines were maintained at 37 °C and 5% CO2 and were cultured in DMEM containing 10% Fetal Bovine Serum and 1% pen-strep antibiotics until reaching 80% confluence. For cytotoxicity studies the cells were seeded in 96 well plates for cytokine estimation the cells were seeded in 6 well plates. Cytotoxic activity of the prepared compounds against and RAW 264.7 cell lines was evaluated at 50 μM after 24 hours using MTT assay. For all coming studies, the compounds were used at subtoxic dose to the cells to avoid any cytotoxic effect.
4.2.3. Estimation of nitric oxide reduction in RAW 264.7 cell line stimulated by LPS
In this experiment, cells were seeded in six well plate at concentration of 1 × 106 cells per ml and treated with prepared hybrids at 10 μM or l-NAME as standard inhibitor for NO production at dose 250 μM for one hour. Following this treatment, the cells were stimulated with LPS 1 μg ml−1 LPS (Sigma-Aldrich). They were then incubated for 24 hours. After the incubation period, the supernatant was collected and used to determine the level of nitric oxide using Griess reagent (Bio-vision, USA) according to manufacture instructions then, the absorbance of the mixture was determined at 540 nm using a microplate reader. The obtained values were then compared with the values obtained from sodium nitrite standards that were assessed simultaneously. By doing so, the nitrite concentration in the media of the treated cells was calculated.
4.2.4. Acetylcholinesterase inhibition assay
Elmann's method was used to perform the assay with slight modifications.63 In brief, a 96-well plate was used for the assay. Initially, 10 μL of the Ellman's reagent (0.4 mM in 100 mM tris buffer, pH 7.5) was added to each well. This was followed by the addition of 20 μL of the enzyme solution (acetylcholine esterase at a final concentration of 0.02 U ml−1 in 50 mM tris buffer containing 0.1% bovine serum albumin and pH was adjusted to 7.5). Subsequently, 20 μL of the sample or standard solution was added to each well, followed by 140 μL of buffer. The mixture was allowed to stand at room temperature for 15 minutes. Afterward, 10 μL of the substrate (0.4 mM acetylcholine iodide) in buffer was immediately added to all wells. The plate was then incubated in a dark chamber at room temperature for 20 minutes and the absorbance of the developed color was measured at 412 nm data were represented as means ± SD and IC50 for compounds achieved more than 75% inhibition at 25 μM was calculated using sigmoidal curve.
4.2.5. Estimation of cytokine reduction in LPS induced inflammation in RAW 264.7
Compounds 6d and 19 have been selected to investigate their ability to alleviate LPS cytokine production. For this purposes RAW 264.7 cells were cultured at 1 × 106 confluence and cells were divided to control cells (DMSO only), cells treated with 1 or 10 μM of the tested compounds or 1 μM of dexamethasone as standard agent for 24 h. and cells induced by LPS. After that, the cells were stimulated with LPS 1 μg ml−1 (Sigma-Aldrich) except the control cells. Cells supernatant was collected, centrifuged at 4 °C at 1000 × g for 20 minutes and clear supernatant was collected and the levels of cytokines such as TNF-α, IL-1β and IL-6 using ELISA kits according to the manufacturer's instructions (FineTest®, China).
4.3. Statistical analysis
Group comparison was conducted using GraphPad Prism version 6.01 (GraphPad Software, San Diego, CA, USA). The data were expressed as mean ± SEM and analyzed using one-way analysis of variance (one-way ANOVA). A p-value of ≤ 0.05 was considered statistically significant.
4.4. In silico investigation of potential mechanism of action of acefylline hybrid
4.4.1. Molecular docking
NOS-3 and AChE were shown to be the most significant potential targets for the prepared hybrids, molecular docking was used to shed light on their binding mode. Briefly, 3D chemical structure of compound 6d and 19 were uploaded to ligand preparation module in CB-Dock2 online server while The PDB file of NOS-3:16v7 and AChE:4EY7 were retrieved from protein data bank and submitted to the docking interface provided by CB-Dock2. Template based docking was used to study the ability of the compounds to interact with different detected binding cavities then the software rank the poses according to their score.79 Finally, the visualization of the best pose was carried out using discovery studio visualizer to study their interaction with the binding site.80
4.5. Chemical stability studies
4.5.1. HPLC analysis
Dionex UltiMate 3000RS HPLC system was used for all chromatographic runs. Injection volume was (20 μL) and reversed phase C18 column with dimensions of 150 × 4.6 mm × 5 μm (Inertsil, Thermofisher) was used for all chromatographic studies. The chromatographic method was based on previously reported method with slight modification81 where the mobile phase consisted of a mixture of acetonitrile and 0.1 formic acid in water using a ratio of 10 : 90 (v/v%), and the elution process was carried out at a flow rate of 1 ml min−1. To establish the calibration curve, the peak areas of acefylline hybrids 6d and 19 were plotted against the corresponding standard concentrations of acefylline hybrid in methanol solutions, ranging from 12.5 to 150 μM.
4.5.2. In vitro hydrolysis
The hydrolysis kinetics of the prodrugs were examined in aqueous buffer solutions at pH 1.2 simulating gastric environment and pH 7.4 for simulation of intestinal and systemic circulation environment. Solutions containing 75 μM of the hybrid compounds were prepared in 50 ml of either 0.1 M HCl (pH 1.4) or phosphate buffer (pH 7.4). These solutions were placed in capped 50 ml Erlenmeyer flasks and kept at a constant temperature of 37 °C. At specific time intervals (0, 60, 120, 180, 240 minutes), samples were withdrawn from the flasks, and their % hydrolysis was determined using HPLC analysis. Reaction rate constants (Kobs) and half-life (t1/2) values were subsequently calculated.82
Data availability
The data in this manuscript would be provided by authors upon request.
Conflicts of interest
The authors declare that there is no conflict of interest, and no funding was associated with this manuscript.
Supplementary Material
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04867e
References
- Tatulian S. A. Drug Discovery Today. 2022;27:1027–1043. doi: 10.1016/j.drudis.2022.01.016. [DOI] [PubMed] [Google Scholar]
- Tahami Monfared A. A. Byrnes M. J. White L. A. Zhang Q. Neurol. Ther. 2022;11:553–569. doi: 10.1007/s40120-022-00338-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-R. Huang J.-B. Yang S.-L. Hong F.-F. Molecules. 2022;27:1816. [Google Scholar]
- Stanciu G. D. Luca A. Rusu R. N. Bild V. Beschea Chiriac S. I. Solcan C. Bild W. Ababei D. C. Biomolecules. 2020;10:40. doi: 10.3390/biom10010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani R. J. Plascencia-Villa G. Perry G. Lab. Invest. 2019;99:958–970. doi: 10.1038/s41374-019-0231-z. [DOI] [PubMed] [Google Scholar]
- Ceyzériat K. Zilli T. Millet P. Frisoni B. G. Garibotto V. Tournier B. B. Curr. Alzheimer Res. 2020;17:112–125. doi: 10.2174/1567205017666200304085513. [DOI] [PubMed] [Google Scholar]
- Walker K. A. Ficek B. N. Westbrook R. ACS Chem. Neurosci. 2019;10:3340–3342. doi: 10.1021/acschemneuro.9b00333. [DOI] [PubMed] [Google Scholar]
- Xie J. Van Hoecke L. Vandenbroucke R. E. Front. Immunol. 2022;12:796867. doi: 10.3389/fimmu.2021.796867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubley A. Erofeev A. Gorelkin P. Beloglazkina E. Majouga A. Krasnovskaya O. Int. J. Mol. Sci. 2023;24:1717. doi: 10.3390/ijms24021717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M. Kaur M. Chadha N. Silakari O. Mol. Diversity. 2016;20:271–297. doi: 10.1007/s11030-015-9628-9. [DOI] [PubMed] [Google Scholar]
- George N. Jawaid Akhtar M. Al Balushi K. A. Alam Khan S. Bioorg. Chem. 2022;127:105941. doi: 10.1016/j.bioorg.2022.105941. [DOI] [PubMed] [Google Scholar]
- Mezeiova E. Spilovska K. Nepovimova E. Gorecki L. Soukup O. Dolezal R. Malinak D. Janockova J. Jun D. Kuca K. Korabecny J. J. Enzyme Inhib. Med. Chem. 2018;33:583–606. doi: 10.1080/14756366.2018.1443326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sari D. R. T. and Krisnamurti G. C., Proceeding International Conference on Religion, Science and Education, 2022, vol. 1, pp. 685–692 [Google Scholar]
- Maia L. De Mendonça A. Eur. J. Neurol. 2002;9:377–382. doi: 10.1046/j.1468-1331.2002.00421.x. [DOI] [PubMed] [Google Scholar]
- Londzin P. Zamora M. Kąkol B. Taborek A. Folwarczna J. Nutrients. 2021;13:537. doi: 10.3390/nu13020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel Bar F. M. Elimam D. M. Mira A. S. El-Senduny F. F. Badria F. A. Nat. Prod. Res. 2019;33:2591–2599. doi: 10.1080/14786419.2018.1462177. [DOI] [PubMed] [Google Scholar]
- Sharma M. Sharma A. Nuthakki V. K. Bhatt S. Nandi U. Bharate S. B. Drug Dev. Res. 2022;83:1803–1821. doi: 10.1002/ddr.21998. [DOI] [PubMed] [Google Scholar]
- Yousaf M. Zahoor A. F. Faiz S. Javed S. Irfan M. J. Heterocycl. Chem. 2018;55:2447–2479. [Google Scholar]
- Reshetnikov D. V. Ivanov I. D. Baev D. S. Rybalova T. V. Mozhaitsev E. S. Patrushev S. S. Vavilin V. A. Tolstikova T. G. Shults E. E. Molecules. 2022;27:8787. doi: 10.3390/molecules27248787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neises B. Steglich W. Angew Chem. Int. Ed. Engl. 1978;17:522–524. [Google Scholar]
- Zhao X. Ma H. Pan Q. Wang H. Qian X. Song P. Zou L. Mao M. Xia S. Ge G. Yang L. Drug Metab. Dispos. 2020;48:345–352. doi: 10.1124/dmd.119.089565. [DOI] [PubMed] [Google Scholar]
- Yamaji T., Saito T., Hayamizu K., Yamamoto O., Yanagisawa M., Wasada N. and Tamura T., Spectral Database for Organic Compounds, SDBS, AIST, http://sdbs.db.aist.go.jp/sdbs/cgibin/cre_index.cgi [Google Scholar]
- Xie B. Shi X. Xing Y. Tang Y. Brain Behav. 2020;10:e01601. doi: 10.1002/brb3.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredman G. Nat. Rev. Cardiol. 2019;16:259–260. doi: 10.1038/s41569-019-0182-5. [DOI] [PubMed] [Google Scholar]
- Casserly I. Topol E. J. Lancet. 2004;363:1139–1146. doi: 10.1016/S0140-6736(04)15900-X. [DOI] [PubMed] [Google Scholar]
- Alhadi I. A. Al Ansari A. M. AlSaleh A. F. F. Alabbasi A. M. A. BMC Endocr. Disord. 2023;23:34. doi: 10.1186/s12902-023-01288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C. Cirrito J. R. Lin X. Wang L. Verges D. K. Dickson A. Mamcarz M. Zhang C. Mori T. Arendash G. W. Holtzman D. M. Potter H. J. Alzheimer's Dis. 2009;17:681–697. doi: 10.3233/JAD-2009-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks D. Nalivaeva N. Turner A. Front. Physiol. 2012;3:189. doi: 10.3389/fphys.2012.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Monte S. M. Chiche J.-D. von dem Bussche A. Sanyal S. Lahousse S. A. Janssens S. P. Bloch K. D. Lab. Invest. 2003;83:287–298. doi: 10.1097/01.lab.0000056995.07053.c0. [DOI] [PubMed] [Google Scholar]
- Favié L. M. A. Cox A. R. van den Hoogen A. Nijboer C. H. A. Peeters-Scholte C. M. P. C. D. van Bel F. Egberts T. C. G. Rademaker C. M. A. Groenendaal F. Front. Neurol. 2018;9:258. doi: 10.3389/fneur.2018.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W.-F. Chen Y.-M. Lin C.-Y. Huang H.-L. Yeh H. Chang Y.-T. Huang K.-T. Lin M.-C. J. Inflammation. 2018;15:3. doi: 10.1186/s12950-018-0179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.-H. Kim K.-J. Ryu S.-J. Lee B.-Y. Chem.-Biol. Interact. 2016;248:1–7. doi: 10.1016/j.cbi.2016.01.020. [DOI] [PubMed] [Google Scholar]
- Vadiei K. Tucker S. D. Lopcz-Berestein G. Wasan K. M. Pharmacol. Toxicol. 1996;78:174–180. doi: 10.1111/j.1600-0773.1996.tb00200.x. [DOI] [PubMed] [Google Scholar]
- Kang C.-H. Jayasooriya R. G. P. T. Dilshara M. G. Choi Y. H. Jeong Y.-K. Kim N. D. Kim G.-Y. Food Chem. Toxicol. 2012;50:4270–4276. doi: 10.1016/j.fct.2012.08.041. [DOI] [PubMed] [Google Scholar]
- Badshah H. Ikram M. Ali W. Ahmad S. Hahm J. R. Kim M. O. Biomolecules. 2019;9:719. doi: 10.3390/biom9110719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothers H. M. Marchalant Y. Wenk G. L. Neurosci. Lett. 2010;480:97–100. doi: 10.1016/j.neulet.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendash G. W. Schleif W. Rezai-Zadeh K. Jackson E. K. Zacharia L. C. Cracchiolo J. R. Shippy D. Tan J. Neuroscience. 2006;142:941–952. doi: 10.1016/j.neuroscience.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Grifman M. Arbel A. Ginzberg D. Glick D. Elgavish S. Shaanan B. Soreq H. Mol. Brain Res. 1997;51:179–187. doi: 10.1016/s0169-328x(97)00246-5. [DOI] [PubMed] [Google Scholar]
- Kolahdouzan M. Hamadeh M. J. CNS Neurosci. Ther. 2017;23:272–290. doi: 10.1111/cns.12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli-Alinovi C. Ficarra S. Russo A. M. Giunta E. Barreca D. Galtieri A. Misiti F. Tellone E. Biochimie. 2016;121:52–59. doi: 10.1016/j.biochi.2015.11.022. [DOI] [PubMed] [Google Scholar]
- Fabiani C. Biscussi B. Munafó J. P. Murray A. P. Corradi J. Antollini S. S. Mol. Pharmacol. 2022;101:154–167. doi: 10.1124/molpharm.121.000415. [DOI] [PubMed] [Google Scholar]
- Reshetnikov D. V. Ivanov I. D. Baev D. S. Rybalova T. V. Mozhaitsev E. S. Patrushev S. S. Vavilin V. A. Tolstikova T. G. Shults E. E. Molecules. 2022;27:8787. doi: 10.3390/molecules27248787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscussi B. Murray A. P. Chem. Proc. 2022;12:62. [Google Scholar]
- Parween N. Jabeen A. Prasad B. J. Adv. Sci. Res. 2021;12:8–12. [Google Scholar]
- Taheri P. Yaghmaei P. Tehrani H. S. Ebrahim-Habibi A. Neurophysiology. 2019;51:114–119. [Google Scholar]
- Garabadu D. Sharma M. Neurotoxic. Res. 2019;35:848–859. doi: 10.1007/s12640-019-0008-6. [DOI] [PubMed] [Google Scholar]
- Zanikov T. Gerasymchuk M. Ghasemi Gojani E. Robinson G. I. Asghari S. Groves A. Haselhorst L. Nandakumar S. Stahl C. Cameron M. Li D. Rodriguez-Juarez R. Snelling A. Hudson D. Fiselier A. Kovalchuk O. Kovalchuk I. Molecules. 2023;28:2624. doi: 10.3390/molecules28062624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said M. M. Rabo M. M. A. Arch. Ind. Hyg. Toxicol. 2017;68:27. [Google Scholar]
- Chowdhury S. Kumar S. J. Food Biochem. 2021;45:e13571. doi: 10.1111/jfbc.13571. [DOI] [PubMed] [Google Scholar]
- Topal F. Gulcin I. Dastan A. Guney M. Int. J. Biol. Macromol. 2017;94:845–851. doi: 10.1016/j.ijbiomac.2016.10.096. [DOI] [PubMed] [Google Scholar]
- Genç Bilgiçli H. Kestane A. Taslimi P. Karabay O. Bytyqi-Damoni A. Zengin M. Gulçin İ. Bioorg. Chem. 2019;88:102931. doi: 10.1016/j.bioorg.2019.102931. [DOI] [PubMed] [Google Scholar]
- Toublet F.-X. Lecoutey C. Lalut J. Hatat B. Davis A. Since M. Corvaisier S. Freret T. Sopkova de Oliveira Santos J. Claeysen S. Boulouard M. Dallemagne P. Rochais C. Molecules. 2019;24:2786. doi: 10.3390/molecules24152786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev A. Buneeva O. Glover V. Biol.: Targets Ther. 2007;1:151–162. [PMC free article] [PubMed] [Google Scholar]
- Medvedev A., Buneeva O., Gnedenko O., Fedchenko V., Medvedeva M., Ivanov Y., Glover V. and Sandler M., in Oxidative Stress and Neuroprotection, ed. H. Parvez and P. Riederer, Springer Vienna, Vienna, 2006, 10.1007/978-3-211-33328-0_11, pp. 97–103 [DOI] [PubMed] [Google Scholar]
- Medvedev A. E. Buneeva O. A. Kopylov A. T. Gnedenko O. V. Medvedeva M. V. Kozin S. A. Ivanov A. S. Zgoda V. G. Makarov A. A. Int. J. Mol. Sci. 2015;16:476–495. doi: 10.3390/ijms16010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavroidi B. Kaminari A. Matiadis D. Hadjipavlou-Litina D. Pelecanou M. Tzinia A. Sagnou M. Brain Sci. 2022;12:806. doi: 10.3390/brainsci12060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazimontazer E. Sadeghpour H. Nadri H. Sakhteman A. Tüylü Küçükkılınç T. Miri R. Edraki N. Bioorg. Chem. 2019;89:103006. doi: 10.1016/j.bioorg.2019.103006. [DOI] [PubMed] [Google Scholar]
- Bhagat K. Singh J. V. Sharma A. Kaur A. Kumar N. Gulati H. K. Singh A. Singh H. Bedi P. M. S. J. Mol. Struct. 2021;1245:131085. [Google Scholar]
- Ozgun D. O. Yamali C. Gul H. I. Taslimi P. Gulcin I. Yanik T. Supuran C. T. J. Enzyme Inhib. Med. Chem. 2016;31:1498–1501. doi: 10.3109/14756366.2016.1149479. [DOI] [PubMed] [Google Scholar]
- Matheus M. E. Violante F. d. A. Garden S. J. Pinto A. C. Fernandes P. D. Eur. J. Pharmacol. 2007;556:200–206. doi: 10.1016/j.ejphar.2006.10.057. [DOI] [PubMed] [Google Scholar]
- Naglah A. M. Ahmed A. F. Wen Z.-H. Al-Omar M. A. Amr A. E.-G. E. Kalmouch A. J. Biomol. Struct. Dyn. 2016;21:498. [Google Scholar]
- Boumezber S. Yelekçi K. J. Biomol. Struct. Dyn. 2023;41:3607–3629. doi: 10.1080/07391102.2022.2054471. [DOI] [PubMed] [Google Scholar]
- Islam M. S. Al-Majid A. M. Azam M. Verma V. P. Barakat A. Haukka M. Elgazar A. A. Mira A. Badria F. A. ACS Omega. 2021;6:31539–31556. doi: 10.1021/acsomega.1c03978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahidul Islam M. Al-Majid A. M. Azam M. Prakash Verma V. Barakat A. Haukka M. Domingo L. R. Elgazar A. A. Mira A. Badria F. A. ChemistrySelect. 2021;6:14039–14053. [Google Scholar]
- Sánchez D. A. Tonetto G. M. Ferreira M. L. Catalysts. 2023;13:473. [Google Scholar]
- Matesic L. Locke J. M. Vine K. L. Ranson M. Bremner J. B. Skropeta D. Bioorg. Med. Chem. 2011;19:1771–1778. doi: 10.1016/j.bmc.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Pirinccioglu N. Williams A. J. Chem. Soc., Perkin Trans. 2. 1998:37–40. [Google Scholar]
- Manon B. Sharma P. D. Indian J. Chem., Sect. B: Org. Chem. Incl. Med. Chem. 2009;48:1279–1287. [Google Scholar]
- Shahzadi I. Zahoor A. F. Rasul A. Rasool N. Raza Z. Faisal S. Parveen B. Kamal S. Zia-ur-Rehman M. Zahid F. M. J. Heterocycl. Chem. 2020;57:2782–2794. [Google Scholar]
- Rao M. L. Kumar A. Tetrahedron. 2014;70:6995–7005. [Google Scholar]
- Zhao J.-W. Wu Z.-H. Guo J.-W. Huang M.-J. You Y.-Z. Liu H.-M. Huang L.-H. Eur. J. Med. Chem. 2019;181:111520. doi: 10.1016/j.ejmech.2019.07.023. [DOI] [PubMed] [Google Scholar]
- Ruddarraju R. R. Murugulla A. C. Kotla R. Tirumalasetty M. C. B. Wudayagiri R. Donthabakthuni S. Maroju R. Baburao K. Parasa L. S. Eur. J. Med. Chem. 2016;123:379–396. doi: 10.1016/j.ejmech.2016.07.024. [DOI] [PubMed] [Google Scholar]
- Klosa J. Arch. Pharm. 1955;288:114–119. doi: 10.1002/ardp.19552880304. [DOI] [PubMed] [Google Scholar]
- Gopinatha V. K. Mantelingu K. Rangappa K. S. J. Chin. Chem. Soc. 2020;67:1453–1461. [Google Scholar]
- Alafeefy A. M. Alqasoumi S. I. Abdel hamid S. G. El-Tahir K. E. Mohamed M. Zain M. E. Awaad A. S. J. Enzyme Inhib. Med. Chem. 2014;29:443–448. doi: 10.3109/14756366.2013.795957. [DOI] [PubMed] [Google Scholar]
- Wang X. Shen Y. Wang S. Li S. Zhang W. Liu X. Lai L. Pei J. Li H. Nucleic Acids Res. 2017;45:W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller D. Michielin O. Zoete V. Bioinformatics. 2013;29:3073–3079. doi: 10.1093/bioinformatics/btt540. [DOI] [PubMed] [Google Scholar]
- Li X. Tang Q. Meng F. Du P. Chen W. Comput. Struct. Biotechnol. J. 2022;20:1345–1351. doi: 10.1016/j.csbj.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Senduny F. F. Elgazar A. A. Alwasify H. A. Abed A. Foda M. Abouzeid S. Lewerenz L. Selmar D. Badria F. Planta Med. 2023;89:964–978. doi: 10.1055/a-2058-3863. [DOI] [PubMed] [Google Scholar]
- Elimam D. M. Elgazar A. A. El-Senduny F. F. El-Domany R. A. Badria F. A. Eldehna W. M. J. Enzyme Inhib. Med. Chem. 2022;37:39–50. doi: 10.1080/14756366.2021.1988944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L.-N. Mao M.-Q. Zhao X.-H. Yang L. Jia H. Xia S.-Y. J. Pharm. Biomed. Anal. 2019;174:220–225. doi: 10.1016/j.jpba.2019.05.039. [DOI] [PubMed] [Google Scholar]
- Dhokchawle B. V. Tauro S. J. Bhandari A. B. Drug Res. 2016;66:46–50. doi: 10.1055/s-0035-1548908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data in this manuscript would be provided by authors upon request.