Abstract
Background
The effects of prenatal antibiotic exposure on respiratory morbidity in infancy and the involved mechanisms are still poorly understood. We aimed to examine whether prenatal antibiotic exposure in the third trimester is associated with nasal microbiome and respiratory morbidity in infancy and at school age, and whether this association with respiratory morbidity is mediated by the nasal microbiome.
Methods
We performed 16S ribosomal RNA gene sequencing (regions V3–V4) on nasal swabs obtained from 296 healthy term infants from the prospective Basel–Bern birth cohort (BILD) at age 4–6 weeks. Information about antibiotic exposure was derived from birth records and standardised interviews. Respiratory symptoms were assessed by weekly telephone interviews in the first year of life and a clinical visit at age 6 years. Structural equation modelling was used to test direct and indirect associations accounting for known risk factors.
Results
α-Diversity indices were lower in infants with antibiotic exposure compared to nonexposed infants (e.g. Shannon index p-value 0.006). Prenatal antibiotic exposure was also associated with a higher risk of any, as well as severe, respiratory symptoms in the first year of life (risk ratio 1.38, 95% CI 1.03–1.84; adjusted p-value (padj)=0.032 and risk ratio 1.75, 95% CI 1.02–2.97; padj=0.041, respectively), but not with wheeze or atopy in childhood. However, we found no indirect mediating effect of nasal microbiome explaining these clinical symptoms.
Conclusion
Prenatal antibiotic exposure was associated with lower diversity of nasal microbiome in infancy and, independently of microbiome, with respiratory morbidity in infancy, but not with symptoms later in life.
Tweetable abstract
Antibiotic exposure in the third trimester of pregnancy influences postnatal nasal microbiome and infant respiratory morbidity independently. These findings add to our understanding of the effect of prenatal antibiotic use on infants’ respiratory health. https://bit.ly/42Ji1Gn
Introduction
Multiple factors have been identified as contributing to respiratory symptoms in infancy in a complex, interacting manner. These include exposure to viral infections [1], genetics [2] and environmental factors (e.g. breastfeeding [3], exposure to antibiotics [4], air pollution [5] and tobacco exposure [6]). Evidence emerging in recent years also suggests an important role of the nasal microbiome in the development of respiratory morbidity. Specifically, a dominance of genera Moraxella/Streptococcus/Haemophilus and an underrepresentation of Corynebacterium have been associated with a higher number of respiratory infections in the first year of life [7–9]. Even beyond infancy, early-life microbiome has been linked to the development of subsequent asthma at school age [10, 11]. However, some studies reported no effect [12] or found an effect on childhood asthma only in the presence of early allergic sensitisation [13].
Microbiome composition is influenced by multiple environmental factors such as breastfeeding [14] or seasonal change [15]. Additionally, postnatal antibiotics can alter the profile [16, 17]. Moreover, a previous study showed that postnatal antibiotic exposure increased the risk of asthma in childhood and that this effect was partially mediated by the microbiome [17]. Despite 20–37% of pregnant women receiving at least one antibiotic prescription during pregnancy [18, 19], the impact of prenatal maternal antibiotic use is not well studied. There are contradicting reports on whether prenatal antibiotic exposure affects subsequent infant respiratory morbidity [19–23] and it remains unclear whether this influences the neonatal nasal microbiome. Furthermore, the mediating effect of the microbiome on respiratory morbidity caused by prenatal antibiotics has only been shown in a gut microbiome mouse study [24].
To address this knowledge gap, we aimed to examine, firstly, whether the nasal microbiome at age 4–6 weeks in healthy term infants is influenced by antibiotic exposure in the third trimester of pregnancy; secondly, whether antibiotic exposure during the third trimester of pregnancy is associated with respiratory morbidity in the first year of life (symptoms, severe symptoms) as well as in early childhood (wheeze between 2 and 6 years) and the presence of atopy at 6 years. Finally, we examined the mediating effect of the nasal microbiome on the association between antibiotic exposure in the third trimester and subsequent respiratory morbidity.
Material and methods
Study population and study sample
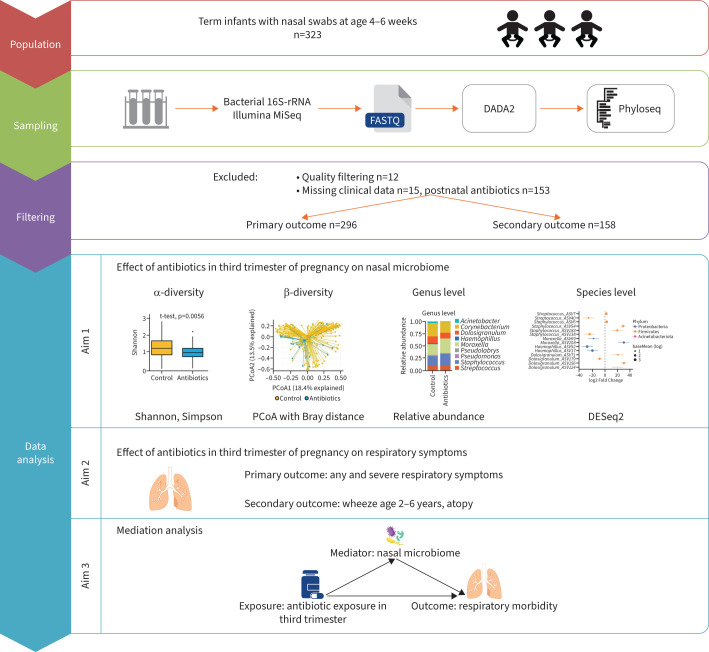
This study includes a subset of term-born infants with nasal swabs from the ongoing Basel–Bern Infant Lung Development (BILD) birth cohort [25]. Infants were recruited from 2010 to 2020, before or shortly after birth in the regions of Basel and Bern, Switzerland. Assessment in the first year of life comprised information from perinatal records, an interview and a nasal swab at age 4–6 weeks during a visit to the study centre, and weekly telephone interviews. At the age of 6 years, children visited the study centre for a follow-up appointment including an interview conducted by a study physician and a skin-prick test. Inclusion criteria for the original cohort (designed to investigate the effect of environmental factors on respiratory health) included healthy, middle-European, white infants. Exclusion criteria were need for neonatal respiratory support for >3 days postnatally, severe diseases of infant or mother and maternal drug abuse. For this study additional inclusion criteria were applied: term birth (≥37 gestational weeks), no postnatal antibiotic exposure of infants before swab and complete follow-up for respiratory symptoms. This resulted in 296 infants with symptom data in the first year of life and 158 infants with complete follow-up data at age 6 years (figure 1).
FIGURE 1.
Simplified graphic overview of the workflow of the data collection and analysis. More detailed information about analysis methods can be found in the material and methods section. PCoA: principal coordinate analysis.
The Ethics Committee of Northwestern and Central Switzerland (Basel, Switzerland) and the Bernese Cantonal Ethics Research Committee (Bern, Switzerland) approved the study protocol and written informed consent was obtained at enrolment and again at follow-up.
Antibiotic exposure
At study visits, mothers were asked about antibiotic exposure during each trimester of pregnancy by trained study nurses using standardised questionnaires. We considered the third trimester as the main exposure because this period is closest to birth and the risk of infant adverse outcomes is higher for antibiotic exposure in late pregnancy [21, 26]. Infants were classified as exposed if mothers reported antibiotic use during the third trimester. Infants without exposure to antibiotics in the third trimester were considered the control (nonexposed) group. Additionally, information on infection type requiring antibiotic treatment (e.g. respiratory, urinary tract) during each trimester was obtained at interview. Birth records provided data on intrapartum antibiotic prophylaxis (IAP).
Microbiome sampling and processing
Anterior nasal swabs were taken from infants at age 4–6 weeks at the study centre. Following internal protocols, bilateral nostril swabs were taken by trained study nurses using two flexible, sterile swabs (FLOQSwabs 516CS01; Copan, Brescia, Italy) and were then placed together in a tube with 3 mL medium (UTM-RTTM in Screw-Cap Tube; Copan). In the laboratory, the medium with nasal secretion was pipetted into micro-screw tubes (Sarstedt, Nürnbrecht, Germany) and kept frozen at −80°C until further processing.
Isolation of bacterial DNA from nasal swabs, amplification of the bacterial 16S-ribosomal (r)RNA gene, and sequencing were performed by the external company Eurofins Genomics (Ebersberg, Germany) before construction of the phylogenetic library.
The NucleoSpin Food Kit (Macherey-Nagel, Düren, Germany) and protocols from Macherey-Nagel were used for DNA extraction. DNA-extracts amplification of the variable V3 and V4 regions of the bacterial 16S-rRNA gene was performed in 25 PCR cycles with the primer pair 357F/800R. Detailed descriptions of 16S-rRNA amplification can be found in [27]. 34 samples with a PCR product concentration <0.2 ng·μL−1 had to be excluded due to quality control. In addition, the included negative extraction controls showed insufficient amplification, not allowing subsequent sequencing, and were therefore excluded. Equimolar amplicon pools on the MiSeq (300PE) Platform (Illumina, San Diego, CA, USA) were used for next-generation sequencing. Detailed descriptions are available in the supplementary materials.
The raw sequencing reads were processed using DADA2 (version 1.18.0). The forward and reverse reads were trimmed to the length of 280 bp and 200 bp, respectively; otherwise, default parameters were used. Silva database version 138.1 was used for assigning taxonomy. The 16S amplicon data have been submitted to the National Center for Biotechnology Information Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/bioproject), under accession number PRJNA944094.
All microbiome analyses were done in R (version 4.2.0) using the phyloseq package. The raw amplicon sequence variants (ASV) table included a total of 8844 taxa. For the analysis, we removed ASVs with relative abundance <0.1% in all samples or with no taxa annotation at the phylum level, resulting in 136 ASVs. Details about the programmes can be found in the supplementary material.
Clinical outcomes
Primary outcomes
The information on the presence and severity of respiratory symptoms (defined as presence of wheeze, cough or breathing difficulties) during the first year of life was assessed weekly by study nurses during structured telephone interviews. The standardised symptom score was used to define the severity of symptoms, with score 0 indicating no symptoms, up to scores 3–4 indicating severe symptoms, as described in [3]. The primary outcomes were 1) weeks with any respiratory symptoms and 2) weeks with severe respiratory symptoms (defined as symptom score ≥3).
Secondary outcome
At the age of 6 years, respiratory symptoms were assessed according to an adapted International Study of Asthma and Allergies in Childhood questionnaire [25] by a study physician during the follow-up appointment. Outcome measures were 1) presence of wheeze between 2 and 6 years and 2) child atopy at 6 years (defined as presence of bronchial asthma, parent-reported rhinitis or atopic dermatitis, as described previously [28]).
Statistical analysis
Within-sample (α-diversity) and between-sample diversity (β-diversity) were calculated using the phyloseq package. The difference in α-diversity metrics (Shannon, Simpson) and most abundant genera between infants with and without exposure to antibiotics during the third trimester were first evaluated using the t-test and the Mann–Whitney test, respectively. Then, a multivariable linear regression model with adjustment (according to [29]) for birth mode (Caesarean/vaginal delivery), older siblings (yes/no), season of swab collection (winter/spring/summer/autumn) and study centre (Basel/Bern) was used to compare α-diversity metrics between groups. Associations of antibiotic exposure with the most abundant genera were analysed by similarly adjusted multiple linear regression using log2-transformed count (Streptococcus and Corynebacterium) and negative binominal regression (Staphylococcus, Moraxella, Dolosigranulum) to account for the sparse, overdispersed count. To compare β-diversity between groups, principal coordinate analysis (PCoA) with the Bray–Curtis distance based on ASV count and permutational multivariate ANOVA (PERMANOVA) [30] with the same adjustments as for α-diversity metrics was used. Next, the DESeq2 method [31], also with the same adjustments, was applied to investigate the difference in the abundance of ASVs between the infants with and without antibiotic exposure during the third trimester.
Respiratory symptom scores with a large number of zeroes (i.e. weeks without respiratory symptoms) had an overdispersed Poisson distribution. Thus, associations of respiratory symptoms in the first year of life with antibiotics exposure and the microbiome were analysed using multivariable negative binomial regression analysis adjusting for sex, birth mode, weeks of any breastfeeding, season of swab collection, maternal atopy (defined as bronchial asthma, allergic rhinitis or atopic dermatitis; yes/no), maternal smoking during pregnancy (yes/no), presence of older siblings, childcare (yes/no) and study centre, according to previously established regression models [3, 32]. To assess the relationship of antibiotics exposure in the third trimester and wheeze and atopy development in childhood, logistic regression was used with adjustment for the same covariates. All regression analyses were performed in Stata 16 (StataCorp, College Station, TX, USA).
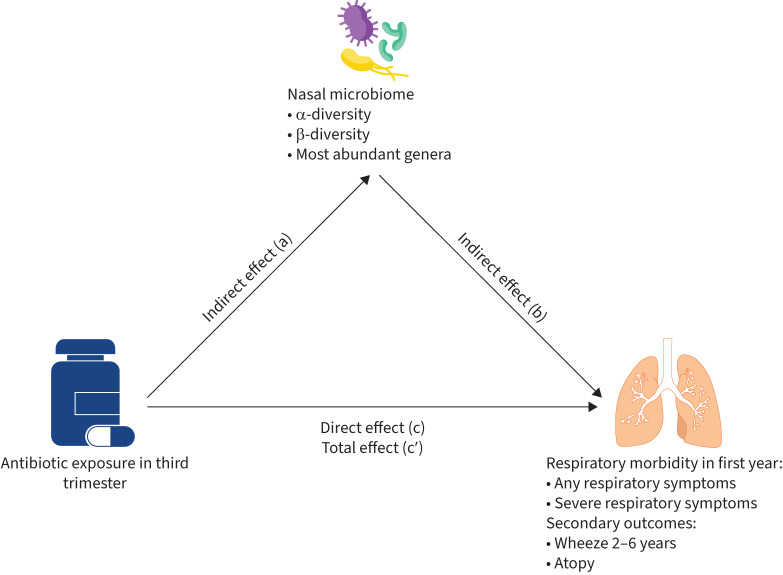
Mediation analysis was performed with generalised structural equation modelling in Stata 16, as done in [17, 33]. Figure 2 shows a simplified visualisation of the mediation statistical framework.
FIGURE 2.
Pathway of mediation effect of antibiotics exposure in third trimester on respiratory morbidity/atopy. c: direct effect of antibiotics exposure in third trimester on respiratory morbidity/atopy after controlling for microbiome; a×b: indirect effect of antibiotics exposure in third trimester on respiratory morbidity/atopy; c′: total effect of antibiotics exposure in third trimester on respiratory morbidity/atopy (c′=c+a×b).
In sensitivity analyses, we repeated all analyses with additional adjustment for IAP and performed a subgroup analysis after excluding infants exposed to IAP from the control group. IAP was not included in the primary analysis because it had no effect on the microbiome or respiratory morbidity. Furthermore, we repeated the analysis after restriction of the study sample to infants of nonatopic mothers. The same method was applied for mediation analysis.
p-values were adjusted for Benjamini–Hochberg false discovery rate (padj-value): p<0.05 and padj<0.05 were considered as significant.
Results
Descriptive statistics
We included a total of 296 term-born infants with analysed nasal swabs, complete data for symptoms in the first year of life, and no postnatal antibiotics before swab. Secondary outcomes at 6 years of age include 158 children. Demographic and clinical characteristics of included infants are outlined in table 1 and, for the sample at 6 years of age, supplementary table S1. 34 (11%) children were exposed to antibiotics in the third trimester. There was no difference in clinical characteristics between exposed and control groups.
TABLE 1.
Population characteristics in the first year of life
| Total | No antibiotics in third trimester | Antibiotics in third trimester | p-value # | |
| Infants | 296 | 262 | 34 | |
| Male | 155 (52.4) | 139 (53.1) | 16 (47.1) | 0.510 |
| Gestational age, weeks | 39.7±1.1 | 39.8±1.1 | 39.7±1.1 | 0.619 |
| Duration of any breastfeeding, weeks | 33.1±15.7 | 33.1±15.5 | 33.5±16.9 | 0.886 |
| Birth mode: Caesarean section | 85.0 (28.7) | 72 (27.5) | 13.0 (38.2) | 0.192 |
| Postnatal age at swab, days | 36.0±6.7 | 36.0±6.7 | 35.5±6.5 | 0.665 |
| Season of swab collection | 0.260 | |||
| Winter | 67 (22.6) | 62 (23.7) | 5 (14.7) | |
| Spring | 75 (25.3) | 62 (23.7) | 13 (38.2) | |
| Summer | 77 (26.0) | 68 (26.0) | 9 (26.5) | |
| Autumn | 77 (26.0) | 70 (26.7) | 7 (20.6) | |
| Study centre: Basel | 172 (58.1) | 145 (55.3) | 27 (79.4) | 0.007 |
| Maternal atopy ¶ | 91 (30.8) | 85 (32.6) | 6 (17.6) | 0.076 |
| Intrapartum antibiotic prophylaxis | 78 (26.4) | 68 (26.0) | 10 (29.4) | 0.667 |
| Maternal smoking during pregnancy | 12 (4.1) | 10 (3.8) | 2 (5.9) | 0.635 |
| Presence of older siblings | 173 (58.4) | 152 (58.0) | 21 (61.8) | 0.676 |
| Attendance in childcare | 162 (54.7) | 142 (54.2) | 20 (58.8) | 0.610 |
| Maternal infections in third trimester requiring antibiotic treatment | ||||
| Urinary tract/gynaecological infection | 16 (5.4) | 16 (47.1) | ||
| Respiratory infection | 8 (2.7) | 8 (23.5) | ||
| ENT infection+ | 4 (1.4) | 4 (11.8) | ||
| Other infection§ | 2 (0.7) | 2 (5.9) | ||
| Unknown | 4 (1.4) | 4 (11.8) | ||
| Symptoms in first year, weeks | 6.2±5.0 | 6.0±4.8 | 7.2±6.1 | 0.282 |
| Severe symptoms in first year, weeks | 0.7±1.2 | 0.6±1.1 | 1.2±1.7 | 0.030 |
Data are presented as n, n (%) or mean±sd. unless otherwise stated. ENT: ear, nose and throat; #: p-values were obtained using t-test, Mann–Whitney test, Pearson Chi-squared test and Fisher exact test, as appropriate; ¶: defined as bronchial asthma, allergic rhinitis or atopic dermatitis; +: defined as infection of ears, neck, throat, e.g. otitis media; §: defined as infection not classified above, e.g. gastrointestinal infection.
Association of antibiotic exposure with microbiome
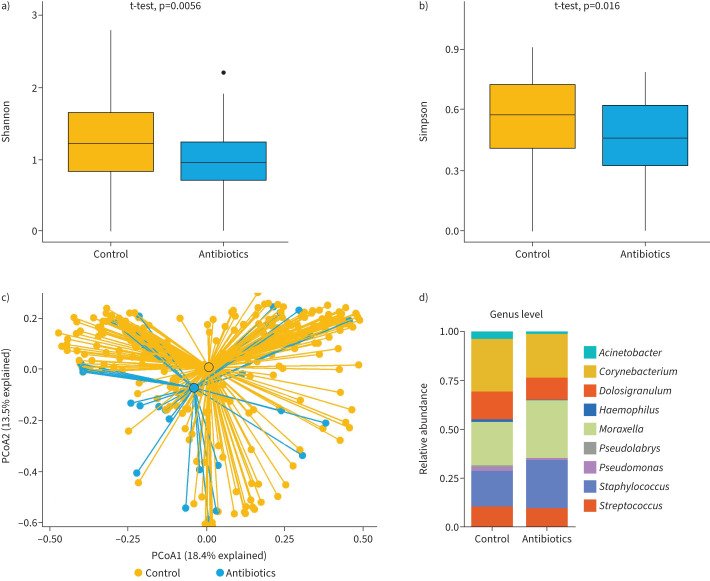
α-diversity indices (Shannon and Simpson) were lower in the group of infants with antibiotic exposure in the third trimester compared to control group in both unadjusted and adjusted analysis (figure 3a, b and table 2). PCoA of nasal microbiome showed no difference between infants with and those without prenatal antibiotic exposure (figure 3c; PERMANOVA p-value=0.120).
FIGURE 3.
Comparison of different microbiome metrics between infants with and without antibiotic exposure in third trimester: α-diversity represented by a) Shannon and b) Simpson (p-values were obtained using t-test); c) β-diversity: principal coordinate analysis (PcoA) based on Bray distance permutational multivariate ANOVA p=0.120; d) relative abundance of the most frequent genera.
TABLE 2.
Adjusted associations of antibiotic exposure in the third trimester with microbiome
| Coefficient (95% CI) | p-value | padj-value # | |
| α-Diversity | |||
| Shannon | −0.26 (−0.47– −0.06) | 0.012 | 0.012 |
| Simpson | −0.11 (−0.19– −0.03) | 0.010 | 0.012 |
| β-Diversity | |||
| PCoA1 | −0.04 (−0.14–0.05) | 0.391 | 0.391 |
| PCoA2 | −0.08 (−0.15–0.00) | 0.041 | 0.082 |
| Genus | |||
| Staphylococcus | −0.05 (−0.88–0.79) | 0.908 | 0.908 |
| Streptococcus | −0.73 (−1.93–0.47) | 0.231 | 0.593 |
| Moraxella | 0.64 (−0.95–2.24) | 0.430 | 0.717 |
| Dolosigranulum | −0.19 (−1.39–1.01) | 0.752 | 0.908 |
| Corynebacterium | −1.76 (−3.14– −0.38) | 0.013 | 0.070 |
Linear regression was used for α-/β-diversity and for log2-transformed count of Streptococcus and Coryebacterium. Negative binominal regression was used for Staphylococcus, Moraxella and Dolosigranulum. Associations were adjusted for birth mode, presence of older siblings, season of swab collection and study centre. Bold type represents statistical significance. PCoA: principal component analysis. #: adjusted p-value (padj-value) was obtained using Benjamini–Hochberg correction for multiple comparisons within group.
Relative abundance comparisons at genus level did not reveal significant differences between infants with and without antibiotic exposure in the third trimester (figure 3d). In adjusted analysis, a decreased relative abundance of Corynebacteria was found in infants exposed to antibiotics in the third trimester (coefficient (β) −1.76, 95% CI −3.15–−0.36; p=0.014; table 2). However, after adjustment for multiple comparisons, this association was no longer significant (padj=0.070).
Differential abundance analysis identified 14 bacterial ASVs that were differentially abundant in infants with and without exposure to antibiotics in third trimester (supplementary figure S1), e.g. Streptococcus mitis group (ASV7) demonstrated a lower expression in infants with third-trimester antibiotic exposure.
Association of antibiotic exposure with clinical outcomes (direct effect)
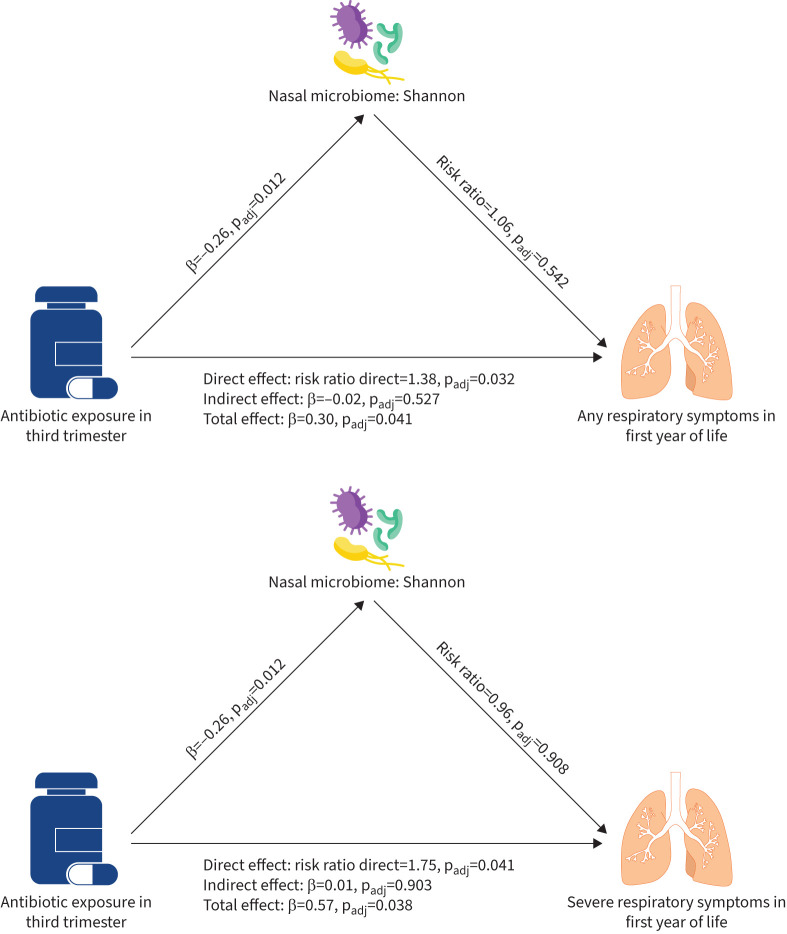
Exposure to antibiotics in the third trimester was associated with increased risk of any respiratory symptoms (risk ratio 1.38, 95% CI 1.03–1.84; padj=0.032) and severe respiratory symptoms (risk ratio 1.75, 95% CI 1.02–2.97; padj=0.041; figure 4) in the first year of life. In contrast, there was no association of antibiotic exposure with wheeze between 2 and 6 years and atopy at 6 years (supplementary table S3).
FIGURE 4.
Structural equation modelling of antibiotic exposure in the third trimester and respiratory morbidity in the first year of life mediated by Shannon index for a) any respiratory symptoms as outcome, b) severe respiratory symptoms as outcome. Linear regression (adjusted for birth mode, presence of older siblings, season of swab collection and study centre) was used to test an association between antibiotic exposure in third trimester and α-diversity indices. Negative binominal regression (adjusted for sex, birth mode, weeks of any breastfeeding, season of swab collection, maternal atopy, maternal smoking during pregnancy, presence of older siblings, childcare and study centre) was used to assess the associations with any/severe respiratory symptoms. Adjusted p-value (padj-value) was obtained using Benjamini–Hochberg correction for multiple comparisons within group (e.g. direct effect α-diversity measures).
Mediation analysis
The indirect effect represents the indirect route of antibiotics exposure, which first affects the respiratory microbiome and then may lead to increased susceptibility to respiratory infection. As mediators, we included α-diversity metrics, the most abundant genera and the first two principal components for β-diversity. No mediating effect was found for all microbiome measures (figure 3, supplementary table S2), since there was no significant association between microbiome measures and subsequent respiratory morbidity (supplementary table S7). The total effect showing the overall association between antibiotic exposure and respiratory morbidity was significant for respiratory and severe symptoms due to the significant direct effect.
Sensitivity analysis
Sensitivity analysis was completed with additional adjustment for IAP, in the subgroup after excluding infants exposed to IAP from the control group and in a subgroup which included only infants of nonatopic mothers. However, results did not change substantially (supplementary tables S4–S6).
Discussion
To the best of our knowledge, our study is the first to show the effect of antibiotic exposure during the third trimester on postnatal nasal microbiome. Moreover, we identified that prenatal antibiotic exposure increased the risk of developing any respiratory symptoms in the first year of life. For severe symptoms the effect was even stronger, but no effect persisted at 2–6 years. We did not find an association between nasal microbiome and early-life respiratory outcomes, either in α-diversity, or in the abundance of genera. Furthermore, by using mediation analysis, our findings suggest that the nasal microbiome is not a mediator of the association of prenatal antibiotics with respiratory symptoms in the first year of life and later childhood.
In our study, infants exposed to antibiotics in the third trimester showed a lower α-diversity of nasal microbiome. However, β-diversity was not different between infants exposed to antibiotics and the control (nonexposed group) infants. Concerning abundance at genus level, Corynebacterium tended to be decreased in infants exposed to antibiotics. These findings are in line with other studies reporting similar effects of postnatal antibiotic treatment [16, 33]. Moreover, an underrepresentation of Corynebacterium was identified as a risk profile for respiratory morbidity in infancy [8]. A previous study has already demonstrated that an altered vaginal microbiome changes the nasal and gut microbiome of the 1-week-old child [34]. Stokholm et al. [35] reported that antibiotic use during pregnancy altered the mother's vaginal microbiome; thus, we can explain the found changes in infants’ nasal microbiome by considering the transmission of the mother's altered vaginal (and other body habitats) microbiome.
Additionally, we could show an association between antibiotic exposure in the third trimester and an increased risk of any respiratory and severe respiratory symptoms in the first year of life. These findings are comparable with previous studies [20, 21, 23]. In addition, we addressed the hypothesis raised by meta-analysis [19, 22], that these results might be confounded by maternal atopy. Therefore, we accounted for this effect by adjusting for maternal atopy in regression analyses. Furthermore, in sensitivity analysis the observed effects remained stable after exclusion of children with atopic mothers. However, there is no effect of prenatal antibiotics on wheeze and atopy in childhood. Even though this is plausible (as events in pregnancy are likely to become less important over time), results for the secondary outcomes need to be interpreted cautiously since power is low (>40% of the children have not reached the age for follow-up (6 years)).
Our data, in line with others, demonstrates no effect of α-diversity metrics on subsequent respiratory morbidity [8, 36]. In contrast with other studies, the abundance of genera was not associated with number and severity of respiratory symptoms [7–9, 37]. This difference in findings between studies might be related to heterogeneous definition of the outcome (respiratory morbidity in the first year of life) and different methodological approaches [7–9, 37]. Furthermore, the effect of prenatal antibiotics on respiratory symptoms was not mediated by the nasal microbiome. Our findings suggest that there might be other mechanisms linking prenatal antibiotics with respiratory morbidity. In utero infection per se may affect fetal lung development via exposure to inflammatory cytokines leading to modification of alveolarisation and subsequent respiratory morbidity [38, 39].
We found no evidence that IAP may affect nasal microbiome or respiratory morbidity. A meta-analysis had reported controversial results of different studies performed with gut microbiome, noting that most of them were conducted by inclusion of only vaginally born or breastfed infants [40]. In our cohort, additional adjustment for IAP and exclusion of infants with IAP exposure in the control group did not change our main findings.
One of the main strengths of the current study is the prospective weekly assessment of respiratory symptoms and possible risk factors (breastfeeding, childcare) by study nurses in a homogeneous, healthy population. Our study also has limitations. The sample size of exposed children was, especially for secondary outcomes, rather small. Therefore, one could not discriminate between different types of antibiotics and infections. In addition, there were no maternal microbiome samples to test a possible causal effect of antibiotics on the infant microbiome via altered maternal microbiome. Finally, our cohort includes healthy infants from a white, middle-European population, which may not be generalisable to other populations and ethnicities.
In conclusion, our findings from a prospective birth cohort of healthy term infants suggest that prenatal antibiotic exposure in the third trimester can influence the nasal microbiome at age 4–6 weeks. Moreover, although infants with exposure to prenatal antibiotics experience more weeks with any and severe respiratory symptoms in first year of life, the nasal microbiome is not a mediator between prenatal antibiotics and subsequent respiratory morbidity. No effect can be seen after the first year.
These results contribute to a better understanding of the pathway of adverse effects of prenatal antibiotic treatment on susceptibility to respiratory infection in infancy and could henceforward lead to a more thoughtful administration of antibiotics to pregnant women. Future studies with larger numbers of exposed infants are critical for understanding the mechanisms underlying infants’ respiratory health and nasal microbiome associated with duration, spectrum, and indication of prenatal antibiotic treatment and for further investigation of the effects on respiratory morbidity beyond the first year of life.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00225-2023.SUPPLEMENT (518.9KB, pdf)
Acknowledgements
The authors thank all the families who participated in this study and the BILD study group and the BILD cohort team (University of Basel Children's Hospital, Basel, Switzerland; Division of Respiratory Medicine, Department of Pediatrics, Inselspital, Bern University Hospital, University of Bern, Switzerland) for their assistance with recruiting and data collection. Furthermore we would like to thank Fiona Beck (University Children’s Hospital Basel (UKBB), University of Basel, Basel) for manuscript editing.
Provenance: Submitted article, peer reviewed.
Author contributions: C. Rüttimann, O. Gorlanova and U. Frey designed the study. C. Rüttimann, A. Nissen-Kratzert, N. Künstle, A. Gisler, O. Gorlanova, S. Yammine and R. Steinberg assisted in collection of the clinical and metadata. M. Hilty and N. Mostacci were responsible for sample amplicon sequencing and bioinformatics analyses. C. Rüttimann and O. Gorlanova performed the data analysis. C. Rüttimann and O. Gorlanova wrote the main manuscript with input from the coauthors. C. Rüttimann, O. Gorlanova, U. Frey and M. Hilty contributed to the statistical interpretation of results. P. Latzin and U. Frey are the principal investigators of the BILD cohort. All authors read and approved the final manuscript.
Support statement: The samples used in this study are part of the Basel–Bern Infant Lung Development birth cohort study, which is funded by the Swiss National Science Foundation (grant 320030_204717). Funding information for this article has been deposited with the Crossref Funder Registry.
Conflict of interest: C. Rüttimann is partially funded by the Freie Akademische Gesellschaft Basel. The other authors declare no conflict of interest.
References
- 1.Feldman AS, He Y, Moore ML, et al. . Toward primary prevention of asthma. Reviewing the evidence for early-life respiratory viral infections as modifiable risk factors to prevent childhood asthma. Am J Respir Crit Care Med 2015; 191: 34–44. doi: 10.1164/rccm.201405-0901PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang HHF, Teo SM, Sly PD, et al. . The intersect of genetics, environment, and microbiota in asthma – perspectives and challenges. J Allergy Clin Immunol 2021; 147: 781–793. doi: 10.1016/j.jaci.2020.08.026 [DOI] [PubMed] [Google Scholar]
- 3.Gorlanova O, Thalmann S, Proietti E, et al. . Effects of breastfeeding on respiratory symptoms in infancy. J Pediatr 2016; 174: 111–117. doi: 10.1016/j.jpeds.2016.03.041 [DOI] [PubMed] [Google Scholar]
- 4.Donovan BM, Abreo A, Ding T, et al. . Dose, timing, and type of infant antibiotic use and the risk of childhood asthma. Clin Infect Dis 2020; 70: 1658–1665. doi: 10.1093/cid/ciz448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern G, Latzin P, Röösli M, et al. . A prospective study of the impact of air pollution on respiratory symptoms and infections in infants. Am J Respir Crit Care Med 2013; 187: 1341–1348. doi: 10.1164/rccm.201211-2008OC [DOI] [PubMed] [Google Scholar]
- 6.Goksör E, Amark M, Alm B, et al. . The impact of pre- and post-natal smoke exposure on future asthma and bronchial hyper-responsiveness. Acta Paediatr 2007; 96: 1030-1035. doi: 10.1111/j.1651-2227.2007.00296.x [DOI] [PubMed] [Google Scholar]
- 7.Neumann RP, Hilty M, Xu B, et al. . Nasal microbiota and symptom persistence in acute respiratory tract infections in infants. ERJ Open Res 2018; 4: 00066-2018. doi: 10.1183/23120541.00066-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch A, de Steenhuijsen Piters WAA, van Houten MA, et al. . Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am J Respir Crit Care Med 2017; 196: 1582–1590. doi: 10.1164/rccm.201703-0554OC [DOI] [PubMed] [Google Scholar]
- 9.Toivonen L, Hasegawa K, Waris M, et al. . Early nasal microbiota and acute respiratory infections during the first years of life. Thorax 2019; 74: 592–599. doi: 10.1136/thoraxjnl-2018-212629 [DOI] [PubMed] [Google Scholar]
- 10.Bisgaard H, Hermansen MN, Buchvald F, et al. . Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med 2007; 357: 1487–1495. doi: 10.1056/NEJMoa052632 [DOI] [PubMed] [Google Scholar]
- 11.Tang HHF, Lang A, Teo SM, et al. . Developmental patterns in the nasopharyngeal microbiome during infancy are associated with asthma risk. J Allergy Clin Immunol 2021; 147: 1683–1691. doi: 10.1016/j.jaci.2020.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Meel ER, Jaddoe VWV, Looman KIM, et al. . Airway bacterial carriage and childhood respiratory health: a population-based prospective cohort study. Pediatr Allergy Immunol 2020; 31: 774–782. doi: 10.1111/pai.13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teo SM, Tang HHF, Mok D, et al. . Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe 2018; 24: 341–352. doi: 10.1016/j.chom.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, DeWan AT. Microbiome links breast-feeding and asthma protection: a cohort study helps elucidate this underlying player. J Allergy Clin Immunol 2022; 150: 587–588. doi: 10.1016/j.jaci.2022.07.017 [DOI] [PubMed] [Google Scholar]
- 15.McCauley KE, Flynn K, Calatroni A, et al. . Seasonal airway microbiome and transcriptome interactions promote childhood asthma exacerbations. J Allergy Clin Immunol 2022; 150: 204–213. doi: 10.1016/j.jaci.2022.01.020 [DOI] [PubMed] [Google Scholar]
- 16.Teo SM, Mok D, Pham K, et al. . The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015; 17: 704–715. doi: 10.1016/j.chom.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toivonen L, Schuez-Havupalo L, Karppinen S, et al. . Antibiotic treatments during infancy, changes in nasal microbiota, and asthma development: population-based cohort study. Clin Infect Dis 2021; 72: 1546–1554. doi: 10.1093/cid/ciaa262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jonge L, Bos HJ, van Langen IM, et al. . Antibiotics prescribed before, during and after pregnancy in the Netherlands: a drug utilization study. Pharmacoepidemiol Drug Saf 2014; 23: 60–68. doi: 10.1002/pds.3492 [DOI] [PubMed] [Google Scholar]
- 19.Momen NC, Liu X. Maternal antibiotic use during pregnancy and asthma in children: population-based cohort study and sibling design. Eur Respir J 2021; 57: 2000937. doi: 10.1183/13993003.00937-2020 [DOI] [PubMed] [Google Scholar]
- 20.Stokholm J, Sevelsted A, Bønnelykke K, et al. . Maternal propensity for infections and risk of childhood asthma: a registry-based cohort study. Lancet Respir Med 2014; 2: 631–637. doi: 10.1016/S2213-2600(14)70152-3 [DOI] [PubMed] [Google Scholar]
- 21.Zhong Y, Zhang Y, Wang Y, et al. . Maternal antibiotic exposure during pregnancy and the risk of allergic diseases in childhood: a meta-analysis. Pediatr Allergy Immunol 2021; 32: 445–456. doi: 10.1111/pai.13411 [DOI] [PubMed] [Google Scholar]
- 22.Bai L, Zhao D, Cheng Q, et al. . Trimester-specific association between antibiotics exposure during pregnancy and childhood asthma or wheeze: the role of confounding. Ann Epidemiol 2019; 30: 1–8. doi: 10.1016/j.annepidem.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 23.Goth FEM, Schmidt BJ, Green K, et al. . Neonatal FeNO, risk factors, and respiratory morbidity in infants: a cohort study. Pediatr Pulmonol 2021; 56: 3174–3182. doi: 10.1002/ppul.25585 [DOI] [PubMed] [Google Scholar]
- 24.Alhasan MM, Cait AM, Heimesaat MM, et al. . Antibiotic use during pregnancy increases offspring asthma severity in a dose-dependent manner. Allergy 2020; 75: 1979–1990. doi: 10.1111/all.14234 [DOI] [PubMed] [Google Scholar]
- 25.Fuchs O, Latzin P, Kuehni CE, et al. . Cohort profile: the Bern infant lung development cohort. Int J Epidemiol 2012; 41: 366–376. doi: 10.1093/ije/dyq239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uldbjerg CS, Miller JE, Burgner D, et al. . Antibiotic exposure during pregnancy and childhood asthma: a national birth cohort study investigating timing of exposure and mode of delivery. Arch Dis Child 2021; 106: 888–894. doi: 10.1136/archdischild-2020-319659 [DOI] [PubMed] [Google Scholar]
- 27.Hilty M, Qi W, Brugger SD, et al. . Nasopharyngeal microbiota in infants with acute otitis media. J Infect Dis 2012; 205: 1048–1055. doi: 10.1093/infdis/jis024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorlanova O, Appenzeller R, Mahmoud YS, et al. . Effect of breastfeeding duration on lung function, respiratory symptoms and allergic diseases in school-age children. Pediatr Pulmonol 2020; 55: 1448–1455. doi: 10.1002/ppul.24733 [DOI] [PubMed] [Google Scholar]
- 29.Korten I, Mika M, Klenja S, et al. . Interactions of respiratory viruses and the nasal microbiota during the first year of life in healthy infants. mSphere 2016; 1: e00312-16. doi: 10.1128/mSphere.00312-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol 2001; 26: 32–46. Doi: 10.1111/j.1442-9993.2001.01070.pp.x [DOI] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. doi: 10.1186/gb-2014-15-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Usemann J, Xu B, Delgado-Eckert E, et al. . Dynamics of respiratory symptoms during infancy and associations with wheezing at school age. ERJ Open Res 2018; 4: 00037-2018. doi: 10.1183/23120541.00037-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patrick DM, Sbihi H, Dai DLY, et al. . Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: evidence from population-based and prospective cohort studies. Lancet Respir Med 2020; 8: 1094–1105. doi: 10.1016/S2213-2600(20)30052-7 [DOI] [PubMed] [Google Scholar]
- 34.Rasmussen MA, Thorsen J, Dominguez-Bello MG, et al. . Ecological succession in the vaginal microbiota during pregnancy and birth. ISME J 2020; 14: 2325–2335. doi: 10.1038/s41396-020-0686-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stokholm J, Schjørring S, Eskildsen CE, et al. . Antibiotic use during pregnancy alters the commensal vaginal microbiota. Clin Microbiol Infect 2014; 20: 629–635. doi: 10.1111/1469-0691.12411 [DOI] [PubMed] [Google Scholar]
- 36.Korten I, Ramsey K, Mika M, et al. . Nasal microbiota and respiratory tract infections: the role of viral detection. Am J Respir Crit Care Med 2019; 199: 919–922. doi: 10.1164/rccm.201710-2020LE [DOI] [PubMed] [Google Scholar]
- 37.de Steenhuijsen Piters WA, Heinonen S, Hasrat R, et al. . Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016; 194: 1104–1115. doi: 10.1164/rccm.201602-0220OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velten M, Heyob KM, Rogers LK, et al. . Deficits in lung alveolarization and function after systemic maternal inflammation and neonatal hyperoxia exposure. J Appl Physiol 2010; 108: 1347-1356. doi: 10.1152/japplphysiol.01392.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Algert CS, Bowen JR, Lain SL, et al. . Pregnancy exposures and risk of childhood asthma admission in a population birth cohort. Pediatr Allergy Immunol 2011; 22: 836–842. doi: 10.1111/j.1399-3038.2011.01206.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dierikx TH, Visser DH, Benninga MA, et al. . The influence of prenatal and intrapartum antibiotics on intestinal microbiota colonisation in infants: a systematic review. J Infect 2020; 81: 190–204. doi: 10.1016/j.jinf.2020.05.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00225-2023.SUPPLEMENT (518.9KB, pdf)