Abstract
Background
Early identification of dementia is crucial for prompt intervention for high-risk individuals in the general population. External validation studies on prognostic models for dementia have highlighted the need for updated models. The use of machine learning in dementia prediction is in its infancy and may improve predictive performance. The current study aimed to explore the difference in performance of machine learning algorithms compared to traditional statistical techniques, such as logistic and Cox regression, for prediction of all-cause dementia. Our secondary aim was to assess the feasibility of only using clinically accessible predictors rather than MRI predictors.
Methods
Data are from 4,793 participants in the population-based AGES-Reykjavik Study without dementia or mild cognitive impairment at baseline (mean age: 76 years, % female: 59%). Cognitive, biometric, and MRI assessments (total: 59 variables) were collected at baseline, with follow-up of incident dementia diagnoses for a maximum of 12 years. Machine learning algorithms included elastic net regression, random forest, support vector machine, and elastic net Cox regression. Traditional statistical methods for comparison were logistic and Cox regression. Model 1 was fit using all variables and model 2 was after feature selection using the Boruta package. A third model explored performance when leaving out neuroimaging markers (clinically accessible model). Ten-fold cross-validation, repeated ten times, was implemented during training. Upsampling was used to account for imbalanced data. Tuning parameters were optimized for recalibration automatically using the caret package in R.
Results
19% of participants developed all-cause dementia. Machine learning algorithms were comparable in performance to logistic regression in all three models. However, a slight added performance was observed in the elastic net Cox regression in the third model (c = 0.78, 95% CI: 0.78–0.78) compared to the traditional Cox regression (c = 0.75, 95% CI: 0.74–0.77).
Conclusions
Supervised machine learning only showed added benefit when using survival techniques. Removing MRI markers did not significantly worsen our model’s performance. Further, we presented the use of a nomogram using machine learning methods, showing transportability for the use of machine learning models in clinical practice. External validation is needed to assess the use of this model in other populations. Identifying high-risk individuals will amplify prevention efforts and selection for clinical trials.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12911-023-02244-x.
Keywords: Dementia, Machine learning, Prediction model
Introduction
Dementia is characterized by debilitating cognitive impairment that increases the risk of mortality [1], while quality of life decreases for both the patient and his or her caregivers. Currently, 50 million people in the world have dementia, which is expected to triple by 2050 [2]. While much research has been done on the risk factors for dementia, no effective treatment is available [3]. Further, by the time of diagnosis, the brain has already substantially declined in function [4]. Thus, early classification is crucial for prompt intervention and better outcomes for high-risk individuals. Many prognostic models for incident dementia have been developed using ‘traditional’ statistical techniques, such as logistic or Cox regression [5–8]. However, external validation of these models showed poor calibration and performance [9, 10], highlighting the need for updated models for prognostication of dementia. The recent increased application of machine learning for disease prediction offers the possibility to improve dementia prognostic models. Machine learning can aid in unraveling complex relationships between predictors, taking into account nonlinear relationships and interactions, while additionally using that information to increase a model’s predictive performance [11].
Research thus far using machine learning for dementia prediction is in its infancy and current models primarily focus on magnetic resonance imaging (MRI) for prediction (please see these recent reviews for an overview [12–14]). Some studies have explored demographic factors [15, 16] and plasma proteomic data [17–19], but no studies have yet also explored some commonly assessed biomarkers (e.g., glucose, cholesterol, blood pressure) along with demographic and lifestyle information in dementia prediction using machine learning classifiers [12]. A recent review also highlighted the need for the development of new prognostic models for dementia that focus on clinical variables over imaging variables [12]. An emphasis on predictors that are more clinically accessible than MRI is crucial for the potential future use of prognostic models for dementia in clinical practice. Focusing on accessible predictors will allow for wider generalizability of the assessment of high-risk individuals for dementia into the general population. It follows the order and flow of the diagnostic process, by focusing first on cheaper, less invasive, and potentially more accessible predictors in a general practice setting, the starting point for a patient, as opposed to in a memory clinic.
Previous studies using machine learning methods have mostly used the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort for algorithm testing [12], with relatively limited sample sizes (i.e., less than 1,000 participants). Discrimination has focused on differentiating mild cognitive impairment [15] from Alzheimer’s disease [12], the leading cause of dementia. Further, most studies that implemented machine learning methods did not take class imbalance into account [12], which focuses on negative predictive value over positive predictive value and introduces possible bias. As previous studies have also focused on cohorts that have more cases than controls, the possible generalizability of the prognostic model decreases [14]. Therefore, there is a current gap in developing a dementia risk model using machine learning for the general population, using a large sample size.
Our research questions were the following: (1) What is the added performance of machine learning algorithms (i.e., elastic net regression, random forest, support vector machine) for dementia prognosis compared to traditional statistical techniques (e.g., logistic and Cox regression) in a large, population-based cohort from Reykjavik, Iceland of almost 5,000 individuals without dementia or mild cognitive impairment (average age: 76 years, 69% female, 29% with college/university level education)? (2) What is the difference in performance when focusing only on clinically accessible predictors? (3) What is the difference in performance when assessing women and men separately?
Methods
This study was reported following the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) Statement [20].
Study sample
Data originated from the Age, Gene/Environment Susceptibility (AGES)-Reykjavik Study, a community-based cohort study of individuals 65 years or older living in the Reykjavik area. More details are provided elsewhere [21]. In brief, participants from the AGES-Reykjavik Study stem from the Reykjavik study, initiated in 1967 by the Icelandic Heart Association. Between 2002 and 2006, 5,764 individuals randomly selected from survivors of the Reykjavik Study were included. Baseline cognitive, biometric, and MRI assessments were done at the Reykjavik research center. Individuals with dementia or mild cognitive impairment at baseline were excluded from the current analysis, leaving 4,793 individuals in the analytical sample. Cognitive, biometric, and MRI assessments were done at baseline between 2002 and 2006, with follow-up of incident dementia diagnoses for a maximum of 12 years. Written informed consent was obtained from all participants. The Icelandic National Bioethics Committee (VSN: 00–0063), the Icelandic Data Protection Authority, and the Institutional Review Board for the National Institute on Aging, NIH approved this study.
Dementia assessment
Details regarding the procedure for dementia ascertainment can be found elsewhere [22–24]. In brief, a three-step procedure based on international guidelines [21] was used. First, all participants underwent neuropsychological testing of cognition using the Mini-Mental State Examination (MMSE) and the Digit Symbol Substitution Test [23], with the next step in those who screened positive undergoing further neuropsychological examination. In the third step, in those who screened positive on the neuropsychological examinations, further proxy and diagnostic assessments were performed regarding the Activities of Daily Living (ADL), as well as social and cognitive functioning. Then, a multidisciplinary panel including a neurologist, geriatrician, neuroradiologist, and neuropsychologist performed a consensus diagnosis that included exam measures and brain MRI [24]. Additional dementia cases were also obtained through medical and nursing home records as well as in death certificates. Dementia cases obtained through nursing homes were collected following a standardized protocol in Icelandic nursing homes [25]. The current study focused on all-cause dementia only.
Demographics
Age (continuous), sex (dichotomous), education (categorical; categorized as primary school, secondary school, college, or university), and current marital status (married/living together, widowed, divorced, single) were collected by questionnaire at baseline.
Clinical variables
A wide range of clinical variables were used, including metabolic, lipid, and inflammatory levels, as well as medical diagnoses (more information in Supplementary Info 1).
Medication use
Medication use was treated as dichotomous (yes/no) for benzodiazepines, beta-adrenergic blockers, glucocorticoids, psycholeptics, or anti-depressants.
Lifestyle variables
We included the following continuous variables: alcohol consumption, mental leisure activity (days per month), social leisure activity (days per month), number of close friends, and number of living close relatives. The categorical variables we included are as follows: smoking status (current, former, never), physical activity within the last 12 months (never, rarely, occasionally, moderate, high), difficulty in walking 2 km (very easy, somewhat easy, not that easy), difficulty in walking 500 m (very easy, somewhat easy, not that easy), and how often fish is consumed as the main meal (never, less than once a week, 1–2 times a week, 3–4 times a week, 5–6 times a week, daily, more than once a day).
Cognitive assessment
The raw total score of the test of global cognitive function, the MMSE, was the only variable used to assess cognition.
Neuroimaging variables
MR images were collected using 1.5T brain MRI (Signa TwinSpeed; General Electric Medical Systems). For more information on the MRI protocol, refer to [26–28]. Log-transformed white matter lesion volume and hippocampal volume, as well as the ratio of gray matter/intracranial volume (to account for correlation), and the number of cerebral microbleeds were entered as continuous predictors. The presence of infarcts (yes/no) was entered as a dichotomous variable.
Statistical analyses
All analyses were performed in R (v 4.0.3). Before beginning the analyses, data were split into a two-thirds (proportion: 0.66) training set and a one-third test set, ensuring for balanced incident dementia cases in the train/test sets by using the split_df() function in R.
Sample size calculations
We performed a post-hoc sample size calculation using pmsampsize package in R to calculate the number of events/cases required using logistic regression as best-case-scenario [29]. If all predictors are included, the required sample size is at least 1,691, which is less than the current sample of 4,793.
Missing data
Half of the individuals (55%) had at least one missing value on predictors (max: 27% missing on ability to walk 2 km or 500 m). There were no missing values on the outcome (i.e., dementia). Missing data were handled with multiple imputation using the mice package in R separately in the training and test sets using ten imputed datasets. The predictor matrix for the training set was used for imputation in the test set. All predictors as well as the outcome were used in the imputation process. A random imputed dataset from a total of ten was selected for further analyses for both the training and test sets as pooling methods for machine learning prognostic models have yet to be validated. See Supplementary Table 1 for an overview of predictors and outcome in both training and test sets.
Model building
The caret package in R [30] was used for all prediction models, i.e. elastic net regression, random forest, support vector machine, and logistic regression. To take time-to-event and censoring into account, we also performed a regular Cox regression using the glmnet package [31] and elastic net Cox regression using the hdnom package [32] in R. For the support vector machine classifier, a radial kernel was used to allow for nonlinear separations of the data. Hyperparameter tuning was performed automatically by caret. Pseudocode can be found in Supplementary Code 1. The models were first fitted with all features (model 1). Then, models were fit after feature selection using the Boruta package in R [33] for more parsimonious models (model 2). In short, Boruta uses a random forest classifier and applies mean decrease accuracy to evaluate each feature’s importance based on 99 iterations. Tentative features were not included. Lastly, to evaluate a clinically accessible model (i.e., one that does not include MRI features), models were fit only with features selected from Boruta that were not MRI (model 3). Tuning parameters were optimized for recalibration and varied across all three models (Supplementary Table 2).
Internal validation
Using cross-validation, more variability is introduced into the training of each classifier. Ten-fold cross-validation, repeated ten times, for a total of 100 times, was used in training each machine learning algorithm. The training data are divided into ten folds, with the given classifier trained on nine folds, using the tenth for testing. This is repeated until each of the ten folds is held back for testing. The performance metrics are then averaged across all repetitions. Further, upsampling was performed to handle imbalanced data and was implemented during cross-validation. This is done by resampling with replacement our class with incident dementia (i.e., the minority class) to be the same size as those who do not develop dementia (i.e., the majority class). If models failed to converge with upsampling, downsampling was used, which deletes samples from the majority class (i.e., those who do not develop dementia). Additionally, we tested different thresholds for classification other than 0.5, ranging from 0.10 to 0.90 by steps of 0.02.
Performance metrics
The following performance measures were used to assess the models: area under the receiver operating characteristic (ROC) curve (AUC), sensitivity, specificity, positive predictive value, and negative predictive value. The model with the highest AUC was then used for the test set. For the survival models, the c-statistic was used. C-statistics and AUC values are comparable to assess performance. The MLeval package in R was used to calculate 95% confidence intervals. Bootstrapping using the hdnom package was done to calculate 95% confidence intervals in the elastic net Cox regression models. The hdnom package was used to create calibration plots for the elastic net Cox regression as well as to create a clinically relevant nomogram.
Sensitivity analysis
To assess if the prognostic model has similar performance in men and women, the trained model in both sexes was tested on men and women separately.
Results
During an average of 9 ± 3 years of follow-up, 892 (n = 750 from nursing homes) individuals developed dementia. Mean (SD) age at baseline for all participants was 76 [6] years and 59% were female. Demographic and clinical information for the full study sample on all predictor variables and the outcome are shown in Table 1.
Table 1.
Characteristics of the predictors in the study sample (n = 4793)
| Mean (SD) or n (%) | % missing per variable | |
|---|---|---|
| Demographics | ||
| Age (years)* + | 76 (6) | 0% |
| Sex (female)* + | 2822 (59%) | 0% |
| Education (college + university) | 1392 (29%) | 6% |
| Neuroimaging variables | ||
| Log-transformed white matter lesion volume (ml)* | 13.5 (2.5) | 18% |
| Hippocampal volume (ml)* | 5.6 (0.7) | 17% |
| Number of microbleeds* | 0.3 (1.6) | 17% |
| Presence of infarcts | 1491 (31%) | 16% |
| Gray matter volume (ml)* | 676 (63) | 18% |
| Intracranial volume (ml)* | 1501 (148) | 18% |
| Clinical variables | ||
| Abdominal circumference (cm) | 101 (12) | 1% |
| Carotid intima-media thickness test (CIMT) | 1 (0.1) | 10% |
| High-density lipoprotein (mmol/L) | 1.6 (0.5) | < 1% |
| Low-density lipoprotein (mmol/L) | 3.5 (1.0) | < 1% |
| Triglycerides (mmol/L) | 1.2 (0.7) | < 1% |
| Fasting glucose (mmol/L) | 5.8 (1.2) | < 1% |
| B-hemoglobin A1c (g/dl) | 0.5 (0.1) | 8% |
| High-sensitive c-reactive protein (mg/L) | 3.8 (6.7) | < 1% |
| Systolic blood pressure (mmHg) | 142 (21) | 1% |
| Diastolic blood pressure (mmHg) | 74 (10) | 1% |
| Hypertension | 3855 (80%) | 1% |
| Coronary artery disease | 842 (18%) | 0% |
| Diabetes mellitus | 591 (12%) | 0% |
| Metabolic syndrome | 1499 (31%) | 1% |
| Stroke/blood clot in the brain | 297 (6%) | 2% |
| History of cancer | 753 (16%) | 1% |
| Experienced a head trauma or lost consciousness | 416 (9%) | 5% |
| Subjective memory decline*+ | 1431 (30%) | 3% |
| Often forget the names of a friend | 1522 (32%) | 5% |
| Often forget where items are*+ | 2083 (44%) | 5% |
| Difficulty finding the right words | 1517 (32%) | 5% |
| Difficulty finding the way to familiar places*+ | 385 (8%) | 5% |
| Inability in managing money*+ | 132 (3%) | 4% |
| Inability in dressing oneself*+ | 29 (1%) | 6% |
| Intermit claudication in legs | 227 (5%) | 5% |
| Insomnia | 1438 (30%) | 3% |
| Poor health status | 276 (6%) | 1% |
| ADL score, full dependence on all items*+ | 52 (1%) | 6% |
| Morning salivary cortisol (nmol/L) | 19.8 (13.3) | 9% |
| Evening salivary cortisol (nmol/L) | 3.8 (6.6) | 9% |
| GDS-15 sum score*+ | 2.3 (2.1) | 6% |
| All anxiety questions ‘yes’ | 40 (1%) | 1% |
| Diagnosis of current GAD, social phobia, panic disorder, or agoraphobia | 98 (2%) | 5% |
| Current/past diagnosis of major depressive disorder | 248 (5%) | 5% |
| Medication use | ||
| Benzodiazepines | 396 (8%) | 0% |
| Beta-adrenergic blockers | 1660 (35%) | 0% |
| Glucocorticoids | 171 (4%) | 0% |
| Psycholeptics | 818 (17%) | 0% |
| Anti-depressants | 662 (14%) | 0% |
| Lifestyle variables | ||
| Current smoker, % | 582 (12%) | 4% |
| Alcohol consumption (g/week) | 16 (33) | 4% |
| Moderate/high physical activity | 1509 (31%) | 7% |
| Mental leisure activity (days per month) | 7 (6) | 6% |
| Social leisure activity (days per month) | 4 (4) | 6% |
| Single marital status, % | 288 (6%) | 6% |
| Number of close friends | 4 (4) | 6% |
| Not that easy to walk 2 km*+ | 960 (20%) | 27% |
| Not that easy to walk 500 m*+ | 233 (5%) | 27% |
| Number of living close relatives | 7 (4) | 6% |
| Never fish consumption, % | 26 (1%) | 6% |
| Cognitive assessment | ||
| MMSE total score*+ | 27 (3) | 1% |
| Outcome | ||
| Incident dementia | 892 (19%) | 0% |
| Follow-up time (years) | 9 (3) | 0% |
* marks variables entered in model 2. + marks variables entered in model 3. GAD = generalized anxiety disorder. GDS-15 = Geriatric Depression Scale-15. CVLT = California Verbal Learning Test
Model performance
Logistic regression (AUC = 0.73, 95% CI: 0.71–0.75) had a similar AUC to the elastic net regression (AUC = 0.74, 95% CI: 0.72–0.76) and random forest classifiers (AUC = 0.74, 95% CI: 0.72–0.76) in model 1 (i.e., the full model), as well as in the model after feature selection and after removal of neuroimaging variables (Table 2). Support vector machine showed lower performance compared to all other machine learning classifiers and the logistic regression. Both logistic regression and the elastic net regression had the same performance in model 3 without neuroimaging variables (AUC = 0.71, 95% CI: 0.68–0.74) (Table 2).
Table 2.
Summary of cross-validated prediction models on trained data (n = 3473)
| Model | AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|
| Model 1 | |||||
| Logistic regression |
0.73 [0.71–0.75] |
64 [60–68] |
70 [68–71] |
32 [30–35] |
89 [88–91] |
| Elastic net |
0.74 [0.72–0.76] |
68 [64–71] |
69 [67–71] |
33 [31–36] |
90 [89–92] |
| Random forest |
0.74 [0.72–0.76] |
6 [4–8] |
99 [99–99] |
60 [47–71] |
82 [81–83] |
| SVM |
0.65 [0.62–0.68] |
49 [45–53] |
73 [71–74] |
29 [27–32] |
86 [85–88] |
| Model 2 | |||||
| Logistic regression |
0.74 [0.72–0.76] |
67 [63–70] |
70 [68–72] |
34 [31–36] |
90 [89–91] |
| Elastic net |
0.74 [0.72–0.76] |
67 [63–70] |
69 [67–71] |
33 [30–36] |
90 [89–91] |
| Random forest |
0.74 [0.72–0.76] |
47 [43–51] |
84 [82–85] |
40 [36–44] |
88 [86–89] |
| SVM |
0.73 [0.71–0.75] |
72 [69–76] |
63 [61–65] |
31 [28–33] |
91 [89–92] |
| Model 3 | |||||
| Logistic regression |
0.71 [0.68–0.74] |
64 [60–68] |
68 [66–70] |
31 [29–34] |
89 [88–91] |
| Elastic net |
0.71 [0.68–0.74] |
64 [60–67] |
67 [65–69] |
31 [28–33] |
89 [88–90] |
| Random forest |
0.71 [0.68–0.74] |
55 [51–59] |
75 [73–77] |
34 [31–37] |
88 [87–89] |
| SVM |
0.70 [0.67–0.73] |
69 [65–73] |
61 [59–63] |
29 [27–31] |
90 [88–91] |
AUC = area under the ROC curve. SVM = support vector machine
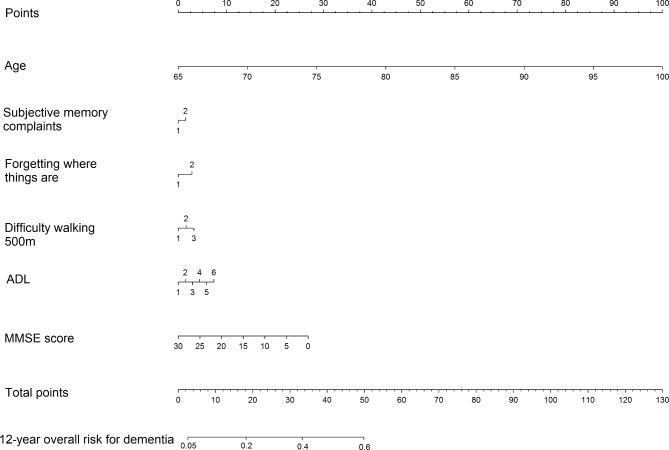
When taking time-to-event into account with the elastic net Cox model, the c-statistic was high (c = 0.80, 95% CI: 0.79–0.80) in model 1 and higher than the traditional Cox model (c = 0.78, 95% CI: 0.77–0.79). The same c-statistics and confidence intervals were seen in model 2. Performance slightly lowered in model 3, but the elastic net Cox regression still showed higher c-statistics (c = 0.78, 95% CI: 0.78–0.78, model 3) compared to the traditional Cox model (c = 0.75, 95% CI: 0.74–0.77). The results of the elastic net Cox regression for model 3 are presented as a nomogram in Fig. 1 for 12-year overall risk. To predict the patient’s risk for dementia, one can draw a vertical line to the top given each variable to get the number of points per that variable. The points from each variable are then summed and the total number of points is used to give a patient’s overall 12-year risk.
Fig. 1.
Predictive nomogram for 12-year overall risk for incident dementia in the elastic net Cox regression for model 3. To predict the patient’s risk for dementia, one can draw a vertical line to the top given each variable to get the number of points per that variable. The points from each variable are then summed and the total number of points represents a patient’s overall 12-year risk
When testing different thresholds, all classifiers demonstrated optimal sensitivity and specificity at 0.50.
Regarding resampling, up-sampling was used for all models except for all support vector machine models. Down-sampling was used instead for model convergence.
Feature selection
For feature selection, Boruta ranked the following variables as most important: age, hippocampal volume, log-transformed white matter lesion volume, gray matter/intracranial volume ratio, MMSE score, difficulty finding the way to familiar places, difficulty in dressing oneself, subjective memory decline, the ADL score, forgetting where items are, number of microbleeds, the sum score of the Geriatric Depression Scale-15, how difficult it is to walk 500 m, sex, inability to manage money, and how difficult it is to walk 2 km (Supplementary Fig. 1). These variables were then used as the predictors in the parsimonious model (model 2), and then the MRI variables were removed for the clinically accessible model (model 3).
Variable importance slightly differed per algorithm in model 3. The least amount of variables used were in the elastic net regression (Supplementary Fig. 2). As there is no built-in variable importance for support vector machine, the AUC is shown instead on the x-axis.
Internal validation
As the elastic net model performed the best regarding AUC, sensitivity, and specificity, it was chosen as the classifier to be used on the test data. The AUC was the same for both models 1 and 2 (AUC = 0.73; 95% CI: 0.70–0.76) and slightly decreased in model 3 when MRI variables were removed (AUC = 0.72; 95% CI: 0.69–0.75) (Table 3). Sensitivity was the same in all models (Sensitivity = 61%; 95% CI: 56–66%), and specificity was highest in model 2 (Specificity = 71%; 95% CI: 69–74%) (Table 3). For the elastic net Cox model, c-statistics were comparable for all three models (model 3: c = 0.77; 95% CI: 0.77–0.78).
Table 3.
Summary of the elastic net models on test data (n = 1870), as well as stratified by sex
| AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|
| Model 1 |
0.73 [0.70–0.76] |
61 [55–66] |
71 [69–73] |
33 [29–37] |
89 [87–91] |
| Women |
0.74 [0.70–0.78] |
60 [53–67] |
70 [67–73] |
35 [30–40] |
87 [84–89] |
| Men |
0.73 [0.67–0.79] |
64 [55–73] |
71 [67–75] |
29 [23–35] |
92 [89–94] |
| Model 2 |
0.73 [0.70–0.76] |
61 [56–66] |
71 [69–74] |
33 [29–37] |
89 [87–91] |
| Women |
0.73 [0.69–0.77] |
59 [52–66] |
71 [67–74] |
35 [30–40] |
87 [84–89] |
| Men |
0.73 [0.67–0.79] |
63 [54–72] |
72 [68–76] |
29 [24–36] |
92 [89–94] |
| Model 3 |
0.72 [0.69–0.75] |
61 [56–66] |
69 [66–71] |
31 [28–35] |
89 [86–90] |
| Women |
0.71 [0.67–0.75] |
59 [52–65] |
69 [66–72] |
33 [29–38] |
86 [84–89] |
| Men |
0.72 [0.66–0.78] |
66 [57–75] |
67 [63–71] |
27 [22–33] |
92 [89–94] |
AUC = area under the ROC curve; PPV = positive predictive value; NPV = negative predictive value
Calibration
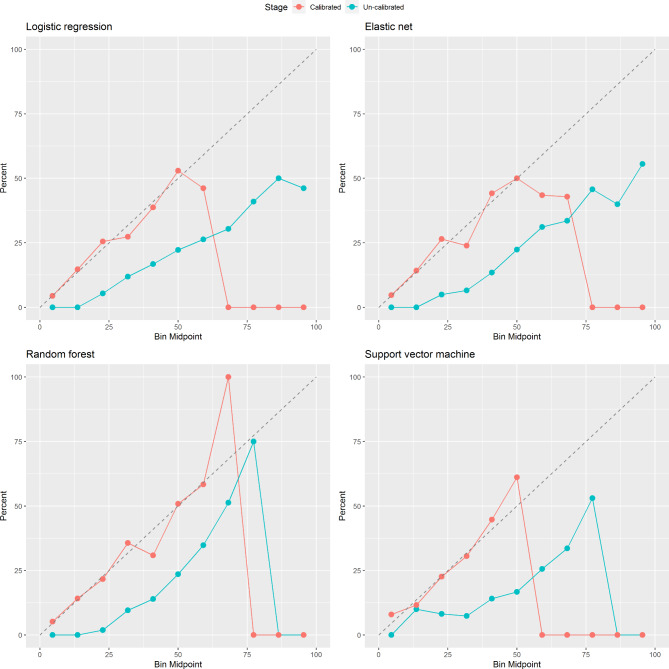
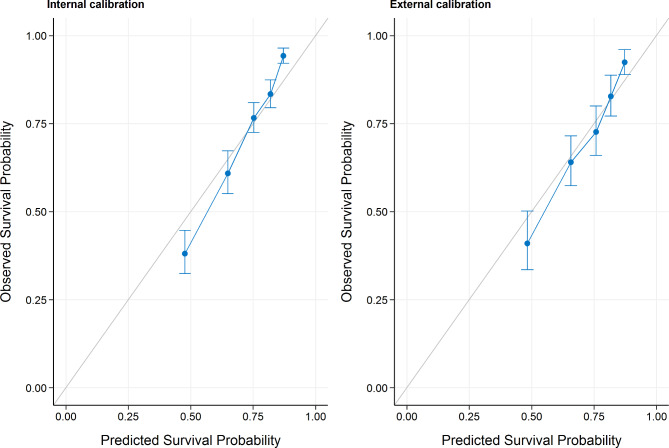
Calibration was assessed for all models. All models showed overfitting, which was resolved after re-calibration (Fig. 2). Re-calibration was performed by training a logistic regression using the uncalibrated probabilities as a predictor. In the elastic net Cox regression, calibration was optimal in both our training (internal calibration) and testing sets (external calibration) (Fig. 3).
Fig. 2.
Calibration plots for logistic regression, elastic net regression, random forest, and support vector machine in model 3 (clinically accessible model) both before and after recalibration. Performance above the diagonal represents under-forecasting and performance below the diagonal represent over-forecasting. There were no individuals in the bins after 77
Fig. 3.
Calibration plots for the elastic net Cox regression in both the training set (internal calibration) and in the test set (‘external’ calibration). Performance above the diagonal represents under-forecasting and performance below the diagonal represents over-forecasting
Sex stratification
Models were also tested on women only and men only to assess possible differences in predictive accuracy when stratified by sex. Across all models using elastic net regression, men and women had similar AUCs. Sensitivity was slightly higher in men, whereas specificity was slightly higher in women (Table 3). However, confidence intervals overlapped. In the elastic net Cox regression model, men (c = 0.86, 95% CI: 0.85–0.87, model 3) had higher c-statistics than women (c = 0.73, 95% CI: 0.72–0.74, model 3) in all three models.
Discussion
The current study aimed to explore the difference in performance between machine learning algorithms and traditional statistical methods for a prognostic model for dementia. We further aimed to assess the feasibility of only using clinically accessible predictors compared to including structural brain MRI, as well as exploring model performance when stratifying by sex. Machine learning only showed benefit over traditional statistical methods when using survival methods. When removing imaging variables from the prediction model, AUC and c-statistic values slightly lowered but remained high. Models performed similarly in men and women in the elastic net regression; however, in the elastic net Cox regression, men had higher c-statistics compared to women.
The current study explored the difference in performance when using machine learning methods compared to traditional statistical techniques. Previous prediction models using machine learning yielded high performance accuracy when using only MRI variables [34], yet systematic reviews have highlighted the lack of exploration on other, more clinically accessible variables for dementia prediction [12, 35]. Machine learning showed added benefit only when using survival techniques, as our elastic net Cox regression outperformed the regular Cox regression. A recent comparative study on various machine learning survival models and Cox regression for dementia prediction also found similar accuracy across techniques [36], which is also in line with previous studies assessing possible performance differences between conventional regression techniques and machine learning [37, 38]. Further, a study predicting two-year incident dementia also found similar performance across traditional techniques (i.e., logistic regression) and machine learning algorithms, with a slight added benefit of machine learning models regarding positive predictive value [39]. The current study found a slight advantage over elastic net regression, which was also found in a simulation study [38]. To note, elastic net reduces the risk of overfitting by penalizing the estimates. This also increases comprehensibility of the prognostic model by decreasing the number of required variables. We were also able to build a nomogram from our elastic net Cox regression, highlighting the feasibility and explainability of using machine learning in clinical settings [40]. This study highlights the importance of censoring in risk prediction as well as the use of algorithms that can capture interactions and high-dimensional relationships within predictors, such as with machine learning [41]. Further, when removing neuroimaging markers, the performance of all models, including those using traditional statistical techniques, lowered, but remained high overall.
The most important variables for prediction in our final elastic net Cox regression included age, subjective memory complaints, and MMSE score. Subjective memory decline has been shown to be present years before mild cognitive impairment and later dementia [42], highlighting its possible use in early prediction. Further, variables such as ‘forgetting where things are’ or ‘difficulty dressing oneself’ were also present in our final model, which are items similar to those being used to create a telephonic interview for dementia prediction [43]. Functional limitations were also found in previous studies to be highly predictive of later developing dementia [44, 45]. Previous studies have explored the use of neuropsychological assessments for prognostic models of dementia [9, 46], however the current study only used the MMSE and still showed high performance. To note, the variables with most predictive power in our model were used in the three-step procedure to diagnose dementia during follow-up at the clinic, i.e., the MMSE and the ADL score, which may have induced overfitting into our model. However, our study focused on the feasibility of using machine learning methods for dementia prediction.
One recent study using population-based data from the UK Biobank also explored the use of machine learning for dementia prediction, with five and ten-year predictions [47]. However, one of the top predictors was APOE e4 genotype, making this model less clinically accessible due to the need for genotyping. APOE e4 genotype was also used in some previous prediction models, focusing on individuals already at risk (i.e., those with amnestic mild cognitive impairment) [48], and it is also included in the well-known Disease State Index (DSI) model [49]. The current study focused on the feasibility of using clinically accessible variables; therefore, we aimed to assess if performance can remain high for prediction even without genotyping.
While performing sex-stratified validation of prediction models is still quite novel and explorative, our study found differences in the elastic net Cox regression when testing our prediction model in women and men separately. As sex differences in dementia have been highlighted previously with the push for sex-based prognostic models [50, 51], future studies should further explore the possible benefit of creating sex-stratified prognostic models.
Strengths of the current study include using multiple imputation to address missing data and cross-validation to increase variability in training of the prediction models. We additionally address differences between novel machine learning classifiers, classical logistic and Cox regression, and using a survival-based machine learning method (i.e., the elastic net Cox regression). The current study also had a large sample size from a well-phenotyped, community-based population. We also report calibration, which has been highlighted as lacking in previous prognostic studies [37, 52]. Further, tuning of the machine learning classifiers was done for recalibration. We also were able to extract a clinically relevant nomogram from our elastic net Cox regression that makes our machine learning methods translatable to clinical practice. Lastly, we performed resampling and threshold adjustment which further helps address imbalanced classification.
The current study also had limitations. The models presented first need to be externally validated to assess its transportability to other populations. Further, the ascertainment of dementia was done with a three-step procedure that consisted of the ADL and MMSE, which were also used as predictors. Further, the AGES-Reykjavik cohort is predominantly White; therefore, it is crucial for the validation of this model in marginally underrepresented populations. Further, development of prognostic models in systemically minoritized groups should also be prioritized for future research. Lastly, we did not assess different time-windows for our survival models as we solely aimed to assess the comparability of techniques. Future studies should assess which models suit best for shorter- or longer-term prediction of dementia.
Our results showed that prediction models developed using supervised machine learning classifiers are feasible and add to the model’s performance, only when using survival methods. We also exemplify ways to implement machine learning in a classical point-based method using a nomogram. Additionally, model performance remained high after the removal of MRI variables. As dementia becomes a leading problem in developing countries, focusing on clinically accessible variables for the prognostication of dementia is crucial.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The AGES-Reykjavik study was funded by the Icelandic Heart Association, National Institute of Aging contracts (N01-AG-12100 and HHSN271201200022C), the Intramural Program at National Institute of Aging, and Althingi (the Icelandic Parliament). This study was supported by a grant from Alzheimer Nederland (WE.03-2017-06).
Authors’ contributions
ELT, CA, and MIG played a role in the design of the work. ELT conducted analyses. LJL and VG played substantial roles in collecting the data. All authors revised the manuscript.
Funding
The AGES-Reykjavik study was funded by the Icelandic Heart Association, National Institute of Aging contracts (N01-AG-12100 and HHSN271201200022C), the Intramural Program at National Institute of Aging, and Althingi (the Icelandic Parliament). This study was supported by grants from Alzheimer Nederland (WE.03-2017-06, PI Geerlings) and (WE.03.2021-09, PI Geerlings).
Data Availability
Data from the AGES-Reykjavik study are available through collaboration (AGES_data_request@hjarta.is) under a data usage agreement with the IHA.
Declarations
Ethics approval and consent to participate
Written informed consent was given from all participants. The Icelandic National Bioethics Committee (VSN: 00–063), the Icelandic Data Protection Authority, and the Institutional Review Board for the National Institute on Aging, NIH approved this study. All the steps/ methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Taudorf L, Nørgaard A, Brodaty H, Laursen TM, Waldemar G. Dementia increases mortality beyond effects of comorbid conditions: a national registry-based cohort study. Eur J Neurol. 2021;28(7):2174–84. doi: 10.1111/ene.14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livingston G, Huntley J, Sommerlad A, Ames D, Ballard C, Banerjee S, et al. Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. Lancet. 2020;396(10248):413–46. doi: 10.1016/S0140-6736(20)30367-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tisher A, Salardini A. A comprehensive update on treatment of Dementia. Semin Neurol. 2019;39(2):167–78. doi: 10.1055/s-0039-1683408. [DOI] [PubMed] [Google Scholar]
- 4.Ewers M, Sperling RA, Klunk WE, Weiner MW, Hampel H. Neuroimaging markers for the prediction and early diagnosis of Alzheimer’s disease dementia. Trends Neurosci. 2011;34(8):430–42. doi: 10.1016/j.tins.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes DE, Beiser AS, Lee A, Langa KM, Koyama A, Preis SR, et al. Development and validation of a brief dementia screening indicator for primary care. Alzheimers Dement. 2014;10(6):656–65e1. doi: 10.1016/j.jalz.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kivipelto M, Ngandu T, Laatikainen T, Winblad B, Soininen H, Tuomilehto J. Risk score for the prediction of dementia risk in 20 years among middle aged people: a longitudinal, population-based study. Lancet Neurol. 2006;5(9):735–41. doi: 10.1016/S1474-4422(06)70537-3. [DOI] [PubMed] [Google Scholar]
- 7.Stephan BCM, Gaughan DM, Edland S, Gudnason V, Launer LJ, White LR. Mid- and later-life risk factors for predicting neuropathological brain changes associated with Alzheimer’s and vascular dementia: The Honolulu Asia Aging Study and the Age, Gene/Environment Susceptibility-Reykjavik Study. Alzheimers Dement. 2022. [DOI] [PMC free article] [PubMed]
- 8.Tang EY, Harrison SL, Errington L, Gordon MF, Visser PJ, Novak G, et al. Current developments in Dementia Risk Prediction Modelling: an updated systematic review. PLoS ONE. 2015;10(9):e0136181. doi: 10.1371/journal.pone.0136181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vonk JMJ, Greving JP, Gudnason V, Launer LJ, Geerlings MI. Dementia risk in the general population: large-scale external validation of prediction models in the AGES-Reykjavik study. Eur J Epidemiol. 2021;36(10):1025–41. doi: 10.1007/s10654-021-00785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Licher S, Yilmaz P, Leening MJG, Wolters FJ, Vernooij MW, Stephan BCM, et al. External validation of four dementia prediction models for use in the general community-dwelling population: a comparative analysis from the Rotterdam Study. Eur J Epidemiol. 2018;33(7):645–55. doi: 10.1007/s10654-018-0403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang D, Frederick DA, Lledo EE, Rosenfield N, Berardi V, Linstead E, et al. Examining the utility of nonlinear machine learning approaches versus linear regression for predicting body image outcomes: the U.S. Body Project I. Body Image. 2022;41:32–45. doi: 10.1016/j.bodyim.2022.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Javeed A, Dallora AL, Berglund JS, Ali A, Ali L, Anderberg P. Machine learning for Dementia Prediction: a systematic review and future research directions. J Med Syst. 2023;47(1):17. doi: 10.1007/s10916-023-01906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolcet-Negre MM, Imaz Aguayo L, García-de-Eulate R, Martí-Andrés G, Fernández-Matarrubia M, Domínguez P, et al. Predicting Conversion from Subjective Cognitive decline to mild cognitive impairment and Alzheimer’s Disease Dementia using Ensemble Machine Learning. J Alzheimers Dis. 2023;93(1):125–40. doi: 10.3233/JAD-221002. [DOI] [PubMed] [Google Scholar]
- 14.Goerdten J, Čukić I, Danso SO, Carrière I, Muniz-Terrera G. Statistical methods for dementia risk prediction and recommendations for future work: a systematic review. Alzheimers Dement (N Y) 2019;5:563–9. doi: 10.1016/j.trci.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gómez-Ramírez J, Ávila-Villanueva M, Fernández-Blázquez M. Selecting the most important self-assessed features for predicting conversion to mild cognitive impairment with random forest and permutation-based methods. Sci Rep. 2020;10(1):20630. doi: 10.1038/s41598-020-77296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grassi M, Rouleaux N, Caldirola D, Loewenstein D, Schruers K, Perna G, et al. A Novel ensemble-based machine learning algorithm to predict the Conversion from mild cognitive impairment to Alzheimer’s Disease using Socio-Demographic characteristics, clinical information, and neuropsychological measures. Front Neurol. 2019;10:756. doi: 10.3389/fneur.2019.00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kivisäkk P, Magdamo C, Trombetta BA, Noori A, Kuo YKE, Chibnik LB, et al. Plasma biomarkers for prognosis of cognitive decline in patients with mild cognitive impairment. Brain Commun. 2022;4(4):fcac155. doi: 10.1093/braincomms/fcac155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casanova R, Varma S, Simpson B, Kim M, An Y, Saldana S, et al. Blood metabolite markers of preclinical Alzheimer’s disease in two longitudinally followed cohorts of older individuals. Alzheimers Dement. 2016;12(7):815–22. doi: 10.1016/j.jalz.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiandaca MS, Zhong X, Cheema AK, Orquiza MH, Chidambaram S, Tan MT, et al. Plasma 24-metabolite Panel predicts preclinical transition to clinical stages of Alzheimer’s Disease. Front Neurol. 2015;6:237. doi: 10.3389/fneur.2015.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. The TRIPOD Group. Circulation. 2015;131(2):211–9. doi: 10.1161/CIRCULATIONAHA.114.014508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris TB, Launer LJ, Eiriksdottir G, Kjartansson O, Jonsson PV, Sigurdsson G, et al. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165(9):1076–87. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sigurdsson S, Aspelund T, Kjartansson O, Gudmundsson EF, Jonsdottir MK, Eiriksdottir G, et al. Incidence of Brain Infarcts, Cognitive Change, and risk of Dementia in the General Population: the AGES-Reykjavik Study (Age Gene/Environment Susceptibility-Reykjavik Study) Stroke. 2017;48(9):2353–60. doi: 10.1161/STROKEAHA.117.017357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saczynski JS, Sigurdsson S, Jonsdottir MK, Eiriksdottir G, Jonsson PV, Garcia ME, et al. Cerebral infarcts and cognitive performance: importance of location and number of infarcts. Stroke. 2009;40(3):677–82. doi: 10.1161/STROKEAHA.108.530212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu C, Cotch MF, Sigurdsson S, Jonsson PV, Jonsdottir MK, Sveinbjrnsdottir S, et al. Cerebral microbleeds, retinopathy, and dementia: the AGES-Reykjavik Study. Neurology. 2010;75(24):2221–8. doi: 10.1212/WNL.0b013e3182020349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jørgensen LM, el Kholy K, Damkjaer K, Deis A, Schroll M. [“RAI”--an international system for assessment of nursing home residents] Ugeskr Laeger. 1997;159(43):6371–6. [PubMed] [Google Scholar]
- 26.Sveinbjornsdottir S, Sigurdsson S, Aspelund T, Kjartansson O, Eiriksdottir G, Valtysdottir B, et al. Cerebral microbleeds in the population based AGES-Reykjavik study: prevalence and location. J Neurol Neurosurg Psychiatry. 2008;79(9):1002–6. doi: 10.1136/jnnp.2007.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scher AI, Gudmundsson LS, Sigurdsson S, Ghambaryan A, Aspelund T, Eiriksdottir G, et al. Migraine headache in middle age and late-life brain infarcts. JAMA. 2009;301(24):2563–70. doi: 10.1001/jama.2009.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sigurdsson S, Aspelund T, Forsberg L, Fredriksson J, Kjartansson O, Oskarsdottir B, et al. Brain tissue volumes in the general population of the elderly: the AGES-Reykjavik study. NeuroImage. 2012;59(4):3862–70. doi: 10.1016/j.neuroimage.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riley RD, Snell KI, Ensor J, Burke DL, Harrell FE, Jr, Moons KG, et al. Minimum sample size for developing a multivariable prediction model: PART II - binary and time-to-event outcomes. Stat Med. 2019;38(7):1276–96. doi: 10.1002/sim.7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn M. Building Predictive Models in R using the caret Package. J Stat Softw. 2008;28(5):1–26. doi: 10.18637/jss.v028.i05. [DOI] [Google Scholar]
- 31.Friedman JH, Hastie T, Tibshirani R. Regularization Paths for generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33(1):1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao N, Xu Q-S, Li M-Z. hdnom: Building Nomograms for Penalized Cox Models with High-Dimensional Survival Data. bioRxiv. 2016:065524.
- 33.Kursa MB, Rudnicki WR. Feature selection with the Boruta Package. J Stat Softw. 2010;36(11):1–13. doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 34.Gupta Y, Lama RK, Kwon GR. Prediction and classification of Alzheimer’s Disease based on combined features from Apolipoprotein-E genotype, Cerebrospinal Fluid, MR, and FDG-PET imaging biomarkers. Front Comput Neurosci. 2019;13:72. doi: 10.3389/fncom.2019.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Oh I, Schindler S, Lai AM, Payne PRO, Gupta A. Machine learning for modeling the progression of Alzheimer disease dementia using clinical data: a systematic literature review. JAMIA Open. 2021;4(3):ooab052. doi: 10.1093/jamiaopen/ooab052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M, Greenberg M, Forkert ND, Chekouo T, Afriyie G, Ismail Z, et al. Dementia risk prediction in individuals with mild cognitive impairment: a comparison of Cox regression and machine learning models. BMC Med Res Methodol. 2022;22(1):284. doi: 10.1186/s12874-022-01754-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019;110:12–22. doi: 10.1016/j.jclinepi.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Austin PC, Harrell FE, Steyerberg EW. Predictive performance of machine and statistical learning methods: impact of data-generating processes on external validity in the “large N, small p” setting. Stat Methods Med Res. 2021;30(6):1465–83. doi: 10.1177/09622802211002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.James C, Ranson JM, Everson R, Llewellyn DJ. Performance of Machine Learning Algorithms for Predicting Progression to Dementia in Memory Clinic Patients. JAMA Netw Open. 2021;4(12):e2136553. doi: 10.1001/jamanetworkopen.2021.36553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin SA, Townend FJ, Barkhof F, Cole JH. Interpretable machine learning for dementia: a systematic review. Alzheimers Dement. 2023;19(5):2135–49. doi: 10.1002/alz.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Sperrin M, Ashcroft DM, van Staa TP. Consistency of variety of machine learning and statistical models in predicting clinical risks of individual patients: longitudinal cohort study using cardiovascular disease as exemplar. BMJ. 2020;371:m3919. doi: 10.1136/bmj.m3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell AJ, Beaumont H, Ferguson D, Yadegarfar M, Stubbs B. Risk of dementia and mild cognitive impairment in older people with subjective memory complaints: meta-analysis. Acta Psychiatr Scand. 2014;130(6):439–51. doi: 10.1111/acps.12336. [DOI] [PubMed] [Google Scholar]
- 43.Makino K, Lee S, Bae S, Chiba I, Harada K, Katayama O, et al. Development and validation of new screening tool for predicting dementia risk in community-dwelling older japanese adults. J Transl Med. 2021;19(1):448. doi: 10.1186/s12967-021-03121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aschwanden D, Aichele S, Ghisletta P, Terracciano A, Kliegel M, Sutin AR, et al. Predicting Cognitive Impairment and Dementia: A Machine Learning Approach. J Alzheimers Dis. 2020;75(3):717–28. doi: 10.3233/JAD-190967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cleret de Langavant L, Bayen E, Yaffe K. Unsupervised machine learning to identify high likelihood of Dementia in Population-Based surveys: Development and Validation Study. J Med Internet Res. 2018;20(7):e10493. doi: 10.2196/10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira T, Ferreira FL, Cardoso S, Silva D, de Mendonça A, Guerreiro M, et al. Neuropsychological predictors of conversion from mild cognitive impairment to Alzheimer’s disease: a feature selection ensemble combining stability and predictability. BMC Med Inform Decis Mak. 2018;18(1):137. doi: 10.1186/s12911-018-0710-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.You J, Zhang YR, Wang HF, Yang M, Feng JF, Yu JT, et al. Development of a novel dementia risk prediction model in the general population: a large, longitudinal, population-based machine-learning study. EClinicalMedicine. 2022;53:101665. doi: 10.1016/j.eclinm.2022.101665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chun MY, Park CJ, Kim J, Jeong JH, Jang H, Kim K, et al. Prediction of conversion to dementia using interpretable machine learning in patients with amnestic mild cognitive impairment. Front Aging Neurosci. 2022;14:898940. doi: 10.3389/fnagi.2022.898940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mattila J, Koikkalainen J, Virkki A, van Gils M, Lötjönen J. Design and application of a generic clinical decision support system for multiscale data. IEEE Trans Biomed Eng. 2012;59(1):234–40. doi: 10.1109/TBME.2011.2170986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferretti MT, Martinkova J, Biskup E, Benke T, Gialdini G, Nedelska Z, et al. Sex and gender differences in Alzheimer’s disease: current challenges and implications for clinical practice: position paper of the Dementia and Cognitive Disorders Panel of the European Academy of Neurology. Eur J Neurol. 2020;27(6):928–43. doi: 10.1111/ene.14174. [DOI] [PubMed] [Google Scholar]
- 51.Ren L, Liang J, Wan F, Wang Y, Dai X-j. Development of a clinical risk score Prediction Tool for 5-, 9-, and 13-Year risk of Dementia. JAMA Netw Open. 2022;5(11):e2242596–e. doi: 10.1001/jamanetworkopen.2022.42596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andaur Navarro CL, Damen JAA, Takada T, Nijman SWJ, Dhiman P, Ma J, et al. Completeness of reporting of clinical prediction models developed using supervised machine learning: a systematic review. BMC Med Res Methodol. 2022;22(1):12. doi: 10.1186/s12874-021-01469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the AGES-Reykjavik study are available through collaboration (AGES_data_request@hjarta.is) under a data usage agreement with the IHA.