Abstract
A practical approach to detect and identify ceftazidime-hydrolyzing extended-spectrum mutants of OXA-10 β-lactamase is presented. Large numbers of bacteria were screened by colony hybridization, a 720-bp part of blaOXA was amplified by PCR from the hybridization-positive isolates, and the products were digested by PvuII and HaeIII.
Pseudomonas aeruginosa is a virulent pathogen that is notably resistant to many antibiotics, including extended-spectrum β-lactam antibiotics. The mechanism of resistance is usually due not to extended-spectrum β-lactamases, but to hyperproduction of class C chromosomal β-lactamase or porin deficiency (1, 5).
Recently, a novel class D extended-spectrum enzyme, OXA-11, a mutant of OXA-10 (PSE-2), was found in a P. aeruginosa strain (4). Soon afterward, OXA-14 and OXA-16, other OXA-10 ceftazidime-hydrolyzing extended-spectrum derivates (OCHDs), were discovered (2, 3). These enzymes were derived from OXA-10 by single (OXA-14) or double (OXA-11 and OXA-16) base mutations. Their isoelectric points were found to be very close to each other (6.1 for OXA-10 and OXA-17, 6.2 for OXA-14 and OXA-16, and 6.4 for OXA-11).
OCHDs confer high-level ceftazidime resistance in P. aeruginosa strains. Although currently posing few problems, dissemination of the gene, blaOXA-11, or other OCHDs warrants interest and needs to be monitored. Simple methods for screening extended-spectrum β-lactamases, such as double-disk synergy, cannot be used for epidemiological purposes in this particular instance for two reasons. First, OXA-10-derived enzymes are poorly inhibited by the β-lactamase inhibitors, so synergy cannot be detected; second, P. aeruginosa strains produce chromosomal enzyme, which interferes with these synergy tests. On the other hand, isoelectric focusing cannot distinguish OXA-10 or OXA-17 from OCHDs either. Hence, studies aimed at determining the molecular basis of ceftazidime resistance or the prevalence and molecular epidemiology of OCHDs among P. aeruginosa strains require the use of sophisticated methods. Currently, cloning and sequence determination are being used to distinguish OCHDs from OXA-10. However, these methods are cumbersome and expensive and therefore poorly suited for epidemiologic studies.
We present herein a practical approach to detect OXA-10 enzymes and identify the OCHDs. We applied this approach to clinical isolates obtained from three hospitals.
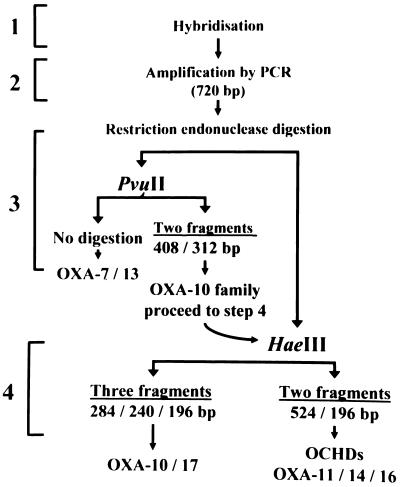
The procedure is shown in Fig. 1. Briefly, the isolates giving a positive hybridization signal were subjected to PCR. If a 720-bp amplification product cleaved to two fragments (408 and 312 bp) with PvuII and to three fragments (284, 240, and 196 bp) with HaeIII, it was assumed to be blaOXA-10 or blaOXA-17, but if the 720-bp amplification product cleaved to two fragments (524 and 196 bp) with HaeIII, it was assumed to be one of the OCHDs (Fig. 2). The presence of an OXA-10-related enzyme was further confirmed by isoelectric focusing.
FIG. 1.
Procedure for identifying OCHDs.
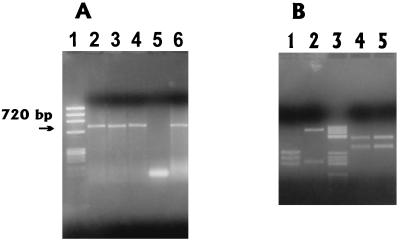
FIG. 2.
Amplification of 720-bp product (A) and restriction fragment patterns (B) obtained with HaeIII and PvuII. Lanes in panel A are as follows: 1, φX174/HaeIII marker; 2 to 4, OXA-positive isolates; 5, OXA-negative isolate; 6, positive control. Lanes in panel B are as follows: 1, HaeIII restriction pattern of blaOXA-10 or blaOXA-17; 2, HaeIII restriction pattern of OCHDs (blaOXA-11, blaOXA-14, or blaOXA-16); 3, pBR322/HaeIII marker; 4 and 5, PvuII restriction patterns of blaOXA-10 or blaOXA-17 and OCHDs, respectively.
Nosocomial P. aeruginosa isolates were obtained from three university hospitals located in distinct regions of Turkey. Susceptibilities were determined on Mueller-Hinton agar (Oxoid, Unipath Ltd., Basingstoke, United Kingdom) plates according to the recommendations of the National Committee for Clinical Laboratory Standards. Antibiotic disks were obtained from Oxoid.
PCR was designed to amplify a 720-bp fragment of the OXA-10, -17, -11, -14, and -16 genes with the sense primer OPR1 (5′-GTCTTTCGAGTACGGCATTA-3′) at position 35 and the antisense primer OPR2 (5′-ATTTTCTTAGCGGCAACTTAC-3′) at position 755 of blaOXA-10. Total bacterial DNAs for PCR tests were obtained simply by boiling a dense suspension of bacteria in 100 μl of TE buffer (10 mM Tris-acetate, 1 mM EDTA [pH 8]). Amplification was achieved with 35 cycles as follows: 1 min at 95°C, 1 min at 54°C, and 3 min at 72°C. The OXA-10 probe was obtained by labelling the PCR product with the digoxigenin labelling and detection kit (Boehringer-Mannheim, Mannheim, Germany) as described by the manufacturer. Colony hybridization was achieved on a positively charged nylon membrane (Boehringer-Mannheim). The details of the colony hybridization (7) and isoelectric focusing procedures (6) were described elsewhere.
Amplified 720-bp products were precipitated by sodium-acetate (1.10 volume, 3 M [pH 5.5])-cold ethanol (2 volumes) for 30 min at −70°C and centrifuged for 10 min at 12,000 × g. The pellets were washed once with pure ethanol and once with 70% ethanol, air dried, and resuspended in 50 μl of TE buffer. Purified products were digested with 10 U of PvuII or HaeIII (MBI, Fermentas, Lithuania) for 3 h at 37°C in 20-μl volumes.
The DNA sequences of the PCR products obtained by amplification with the primers OPR1 and OPR2 were determined by an automated cycle sequencing system (MWG-BIOTECH; Biotechnologie GmbH, Ebersberg, Germany). Products were analyzed on a LI-COR 4200 automated sequencer.
Recently, PER-1, an extended-spectrum β-lactamase conferring high-level ceftazidime resistance, was found to be highly prevalent among P. aeruginosa strains in Turkey (8). In the present study, we looked for the presence of this enzyme to explain the source of ceftazidime resistance in the isolates giving the OXA-10 restriction pattern (Table 1). PER-1 was searched for by colony hybridization with a specific probe, and the presence of the enzyme was confirmed by isoelectric focusing. The details of these methods were described previously (7).
TABLE 1.
Susceptibility testing of hybridization-positive isolates
| Isolate no. | Restriction pattern | Presence of PER-1 | Zone diam (mm) for indicated diska:
|
|||||
|---|---|---|---|---|---|---|---|---|
| CFOX | ATM | CPM | CTAZ | CTAX | AMOX-CLAV | |||
| Ps-162b | OCHDs | Negative | 0 | 0 | 0 | 0 | 9 | 0 |
| Ps-119 | OXA-10 type | Negative | 0 | 15 | 15 | 15 | 9 | 0 |
| Ps-83 | OXA-10 type | Negative | 0 | 16 | 15 | 12 | 13 | 0 |
| Ps-80 | OXA-10 type | Negative | 0 | 20 | 15 | 25 | 20 | 0 |
| Ps-79 | OXA-10 type | Negative | 0 | 20 | 15 | 10 | 15 | 0 |
| Ps-145 | OXA-10 type | Positive | 0 | 0 | 0 | 0 | 0 | 0 |
| Ps-94 | OXA-10 type | Positive | 0 | 0 | 0 | 0 | 0 | 0 |
| Ps-67 | OXA-10 type | Positive | 0 | 0 | 0 | 0 | 0 | 0 |
| Ps-AT56 | OXA-10 type | Positive | 0 | 0 | 11 | 0 | 0 | 0 |
| Ps-1 | OXA-10 type | Positive | 0 | 0 | 9 | 0 | 0 | 0 |
| Ps-158 | OXA-10 type | Positive | 0 | 0 | 0 | 0 | 0 | 0 |
| Ps-29 | OXA-10 type | Positive | 0 | 0 | 10 | 0 | 0 | 0 |
| Ps-T3349 | OXA-10 type | Positive | 0 | 0 | 10 | 0 | 0 | 0 |
Abbreviations: AMOX-CLAV, amoxicillin-clavulanate; ATM, aztreonam; CPM, cefepime; CFOX, cefoxitin; CTAX, cefotaxime; CTAZ, ceftazidime.
Confirmed OXA-14 producer.
We studied 75 nosocomial P. aeruginosa isolates. A positive hybridization signal with the OXA probe was detected in 13 strains (17%) (Table 1), and a 720-bp product was successfully amplified from all of them. PvuII cleaved the products of all the strains to two fragments, typical for the OXA-10 family. HaeIII cleaved PCR products from 12 of these strains to three fragments, as expected for OXA-10 and -17, but one PCR product, from isolate Ps-162, was cleaved to two fragments, a rather typical pattern for OCHDs (Fig. 2). Sequences of two isolates, Ps-29 and Ps-162, were analyzed and confirmed to be OXA-10 and OXA-14, respectively. Isoelectric points from these enzymes were cofocused with the related controls. Double-disk diffusion tests failed to show any synergy between an amoxicillin-clavulanate disk and aztreonam, cefotaxime, ceftazidime, or cefepime. Isoelectric focusing revealed 6.0 to 6.4 enzymes in all hybridization-positive isolates.
Although OXA-10 and OXA-17 do not confer resistance to ceftazidime, eight isolates with the OXA-10-type restriction pattern were found to be highly resistant to ceftazidime (Table 1). They were producing a second enzyme, PER-1-type extended-spectrum β-lactamase, which confers high-level ceftazidime resistance in P. aeruginosa strains (8).
The G-to-A mutation at position 470 of blaOXA-10 causes the replacement of glycine by aspartate. Because this mutation is common among the OCHDs and unique to OXA-14 (Table 2), to identify this mutation as the one responsible for the extended-spectrum activity would not be incorrect. This point is the target of HaeIII, so when a mutation occurs here and the spectrum expands to hydrolyze ceftazidime, HaeIII loses its restriction site and the restriction pattern changes. Therefore, the method presented here is designed to identify OCHDs but not to differentiate OXA-17 from OXA-10.
TABLE 2.
Positions of base differences of related OXA-type β-lactamases and percentage of homology to blaOXA-10
| Enzyme | Position
|
Homology (%) | Accession no.b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 97 | 348 | 370 | 428 | 468 | 470a | 471 | 521 | |||
| OXA-10 | A | T | G | A | A | G | C | A | U37105 | |
| OXA-11 | G | A | 0.99 | Z22590 | ||||||
| OXA-14 | A | 0.99 | L38523 | |||||||
| OXA-16 | A | A | 0.99 | |||||||
| OXA-17 | G | 0.99 | ||||||||
| OXA-7 | G | G | T | T | 0.95 | X75562 | ||||
| OXA-13c | G | G | T | T | 0.96 | U59183 | ||||
With the guanine-to-adenine change at position 470, glycine replaced by aspartate, giving resistance to ceftazidime.
Accession numbers were obtained from the GENIUSnet database of the German Cancer Research Center.
Only the differences common to OXA-13 and OXA-7 are shown.
Type II endonucleases recognize and cleave DNA at a specific site. Application of this highly reproducible and specific method to a PCR-generated DNA fragment is very easy. Nevertheless, despite the specificity of the digestion step, contamination of amplicons from prior reactions can adversely affect the specificity of the PCR phase. Therefore, it is highly recommended that only PCR results from isolates with positive hybridization reactions be considered valid.
We conclude that colony hybridization is useful in screening (positive predictive value, 100%) and that restriction analysis is convenient for detecting OXA-10-type β-lactamases and identifying ceftazidime-hydrolyzing mutants.
Acknowledgments
We thank Ayhan Yücel for giving us the opportunity to study in his laboratory and Victor L. Yu for reviewing the manuscript.
REFERENCES
- 1.Chen H Y, Yuan M, Livermore D M. Mechanisms of resistance to beta-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J Med Microbiol. 1995;43:300–309. doi: 10.1099/00222615-43-4-300. [DOI] [PubMed] [Google Scholar]
- 2.Danel F, Hall L M C, Gur D, Livermore D M. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1996. OXA-16: a new OXA-10-related β-lactamase giving ceftazidime resistance in P. aeruginosa from Turkey, abstr. C27; p. 39. [Google Scholar]
- 3.Danel F, Hall L M C, Gur D, Livermore D M. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1881–1884. doi: 10.1128/aac.39.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall L M C, Livermore D M, Gur D, Akova M, Akalin H E. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:1637–1644. doi: 10.1128/aac.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock R E, Woodruff W A. Roles of porin and beta-lactamase in beta-lactam resistance of Pseudomonas aeruginosa. Rev Infect Dis. 1988;10:770–775. doi: 10.1093/clinids/10.4.770. [DOI] [PubMed] [Google Scholar]
- 6.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytic isoelectric focusing for detection and identification of b-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 7.Vahaboglu H, Dodanli S, Eroglu C, Öztürk R, Soyletir G, Yildirim I, Avkan V. Characterization of multiple-antibiotic-resistant Salmonella typhimurium strains: molecular epidemiology of PER-1-producing isolates and evidence for nosocomial plasmid exchange by a clone. J Clin Microbiol. 1996;34:2942–2946. doi: 10.1128/jcm.34.12.2942-2946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vahaboglu H, Öztürk R, Aygün G, Coşkunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik İ, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]