Abstract
Activating transcription factor 5 (ATF5) is necessary for the development of various tissues, particularly under stress. Dysfunctions of ATF5 have been shown to be involved in many diseases. The exact function of ATF5 is tissue-specific, and its role in erythropoiesis is still unknown. We here employed the loss of function strategy to investigate the role of ATF5 in murine erythropoiesis. We found that knockdown of Atf5 impaired the proliferation of fetal liver erythroid progenitors. Furthermore, erythroid differentiation was inhibited by ATF5 deficiency. Our study suggests that ATF5 may be a potential therapeutic target for treating blood diseases with ineffective erythropoiesis.
Keywords: activating transcription factor 5, erythropoiesis, fetal liver, proliferation, differentiation
As a member of the ATF/cAMP response-element binding protein (CREB) family, activating transcription factor 5 (ATF5) serves as an anti-apoptotic transcription factor and regulates cell proliferation and differentiation during the development of many tissues (Sears & Angelastro, 2017). Dysregulated ATF5 has been reported to be involved in the progression of diabetes and a variety of cancers, such as leukaemia, lymphoma, glioma, lung and breast cancers (Sears & Angelastro, 2017). However, the diverse roles of ATF5 are tissue-specific, as demonstrated by the promotion of hepatic differentiation (Du et al, 2014) and olfactory bulb development (Wang et al, 2012) but inhibition of osteogenesis (Leong et al, 2012) and other types of neurogenesis (Angelastro et al, 2005).
Atf5 is a stress-responsive gene that responds to various stresses, such as endoplasmic reticulum stress, arsenite exposure and proteasome inhibition (Zhou et al, 2008). Translation of Atf5 mRNA can be specifically upregulated by the phosphorylated eukaryotic initiation factor 2 alpha (eIF2αP) via upstream open reading frames (uORFs) under stress (Zhou et al, 2008). We recently reported that Atf5 expression was induced in basophilic erythroblasts, which are CD71high−Ter119+ early haemoglobinised erythroblasts having basophilic cytoplasm appearance, of fetal liver (FLs) under iron deficiency and furthermore, the induction of Atf5 expression by iron deficiency depended on haem-regulated eIF2α kinase (HRI) (Zhang et al, 2019). HRI is a major regulator during terminal erythropoiesis through haem- and eIF2 α P-mediated translational regulation under various stresses, including iron deficiency, arsenite exposure, heat shock and osmotic stress (Chen, 2014; Zhang et al, 2018). Although the functions of ATF5 in many types of tissue development have been characterized, especially in neurogenesis, the role of ATF5 in erythropoiesis is still unknown. In this study, we investigated the function of ATF5 in the regulation of proliferation and differentiation of murine FL erythroid progenitors and demonstrated the essential role of ATF5 in erythropoiesis.
Methods and materials
Animals and progenitor enrichment
C57BL/6J mice were maintained at the animal facility of Massachusetts Institute of Technology (MIT; Cambridge, MA), and all experiments using mice of 8–12 weeks old were carried out with protocols approved by the Committee on Animal Care (CAC). FLs from embryonic day 135 (E135) samples were used to enrich erythroid progenitors by magnetic sorting using EasySep Magnet (Stem Cell Technologies, Vancouver, Canada) as previously described (Thom et al, 2014; Zhang et al, 2019).
Knockdown of Atf5 expression
Knockdown of Atf5 gene expression was performed using recombinant retroviruses containing shRNA-expressing murine stem cell retroviral vector, MSCV-pgkGFP-U3-U6P-Bbs, a kind gift from the laboratory of Dr. Harvey F. Lodish, MIT. Five shRNA oligonucleotides targeting different regions of Atf5 mRNA were obtained from the Genetic Perturbation Platform of Broad Institute (Table SI). Preparations and infections of recombinant retroviruses were performed as previously described (Zhang et al, 2019). Briefly, erythroid progenitors (Lin−CD71−Ter119−) were infected with five Atf5 shRNAs and a Green Fluorescent Protein (GFP) control recombinant retroviruses by centrifuging at 700 g for 15 h at room temperature in the presence of 8 μg/ml polybrene (Sigma-Aldrich, Saint Louis, MO, USA).
Analysis of cell proliferation and differentiation
Infected cells were cultured in expansion medium at an initial cell density of 05 × 106 cells/ml for three days as previously described (Zhang et al, 2019), and collected daily for analyses. Proliferation of infected cells, which were GFP+, was determination by counting the nucleated cells with crystal violet stain using haemocytometers and then multiplying the percentage of GFP+ cells from flow cytometry analysis. Erythroid differentiation was performed as previously described (Zhang et al, 2018, 2019). Briefly, cells were labelled with anti-CD71 and anti-Ter119 antibodies (BioLegend, San Diego, CA, USA). 4’,6-diamidino-2-phenylindole (DAPI; Roche, Basel, Switzerland) was used to exclude dead cells. Infected GFP+ live cells were gated for erythroid differentiation analysis by flow cytometry using FACS LSR II (BD Biosciences, San Jose, CA, USA), and data were analysed with FlowJo (Tree Star, OR, USA). Erythroid differentiation was also analysed by cell morphology on cytospin slides stained with May-Grunwald/Giemsa staining (Sigma-Aldrich).
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Gene expression was performed by RT-qPCR as previously described (Zhang et al, 2018). Primers for Atf5 were forward 5′ to 3′-GGGTCATTTTAGCTCTGTGAGAGAA and reverse 5′ to 3′-ATTTGTGCCCATAACCCCTAGA. Primers for Gapdh were forward 5′ to 3′- ATGGTGAAGGTCGGTGTGAA and reverse 5′ to 3′- GAGTGGAGTCATACTGGAAC. Gapdh was used as internal control.
Statistical analysis
Independent t test (two-tailed) was used to analyse the experimental data. Data are presented as mean ± standard error (SE). *P < 005 was considered statistically significant. **P < 001; ***P < 0001.
Results and discussion
To characterize the function of ATF5 in murine erythropoiesis, we used shRNAs to knockdown the expression of Atf5 mRNA in the erythroid progenitors of Lin−CD71−Ter119− from E135 FLs. We prepared five shRNA constructs specifically targeting different regions of Atf5 mRNA, and produced the respective recombinant retroviruses. Among the five shRNAs, two, A1 and A5, reached more than 80% knockdown efficiency, as compared to the non-targeting GFP control shRNA (Ctrl) (Fig 1A). The efficiency of infections of erythroid progenitors by recombinant retroviruses were around 50%, as shown by percentages of GFP+ cells (Fig 1B). Infected progenitors were cultured in expansion phase for three days. Knockdown of Atf5 reduced the proliferation capacity of erythroid progenitors by about 50% on day 3 for both A1 and A5 as compared to Ctrl (Fig 1C). Furthermore, the differentiation of erythroid progenitors was also impaired by knockdown of Atf5 on both days 2 and 3, especially on day 3, as shown by the increased percentage of erythroid progenitors at stage (S1) but decreased percentage at the more matured stages (S2 and S3) as compared to Ctrl (Fig 2A,B). The inhibition of erythroid differentiation of erythroid progenitors by knockdown of Atf5 was further confirmed by cell morphology demonstrated by the larger and earlier erythroid progenitors upon knockdown of Atf5 compared to Ctrl (Fig 2C). We did not observe promoted proliferation and inhibited differentiation in A3 and A4 as compared to Ctrl. Given that Atf5 knockdown cells were not proliferating in differentiating medium, no sufficient cells were available for differentiation and enucleation assays in differentiation phase.
Fig 1.
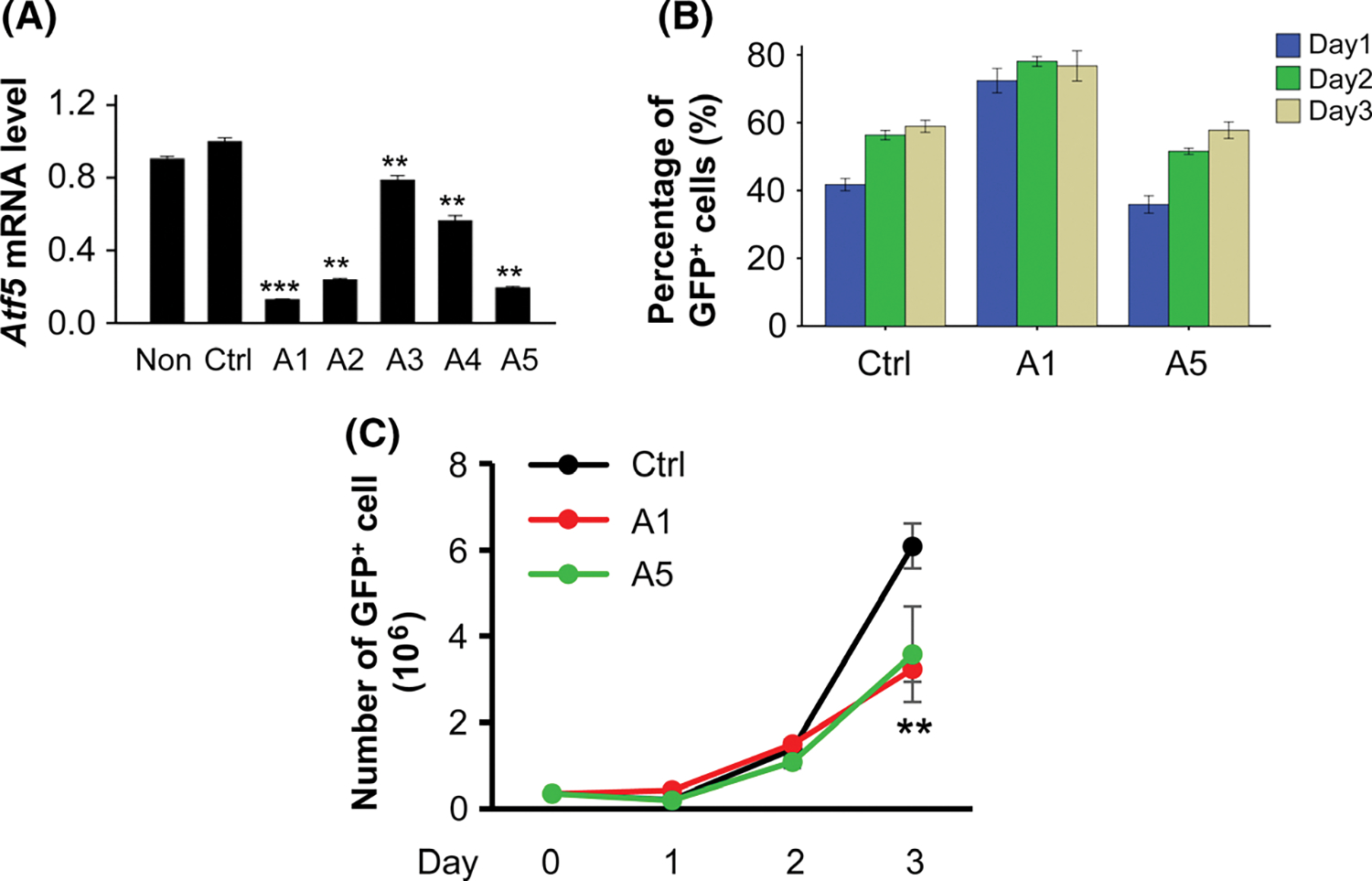
Proliferation of FL erythroid progenitors upon Atf5 knockdown by shRNAs. (A) Knockdown efficiency of Atf5 by shRNAs in the purified erythroid progenitors of fetal livers. Expression level in the non-targeting GFP control shRNA (Ctrl) was defined as 1 (n = 3). (B) Percentages and (C) daily numbers of GFP+ cells after retrovirus infection (n = 3). Non: uninfected; Ctrl: infected with retrovirus expressing non-targeting control shRNA; A1–A5: infected with retroviruses expressing five different shRNAs targeting Atf5 mRNA. Data are presented as mean ± SE. **P < 001; ***P < 0001 as compared to Ctrl.
Fig 2.
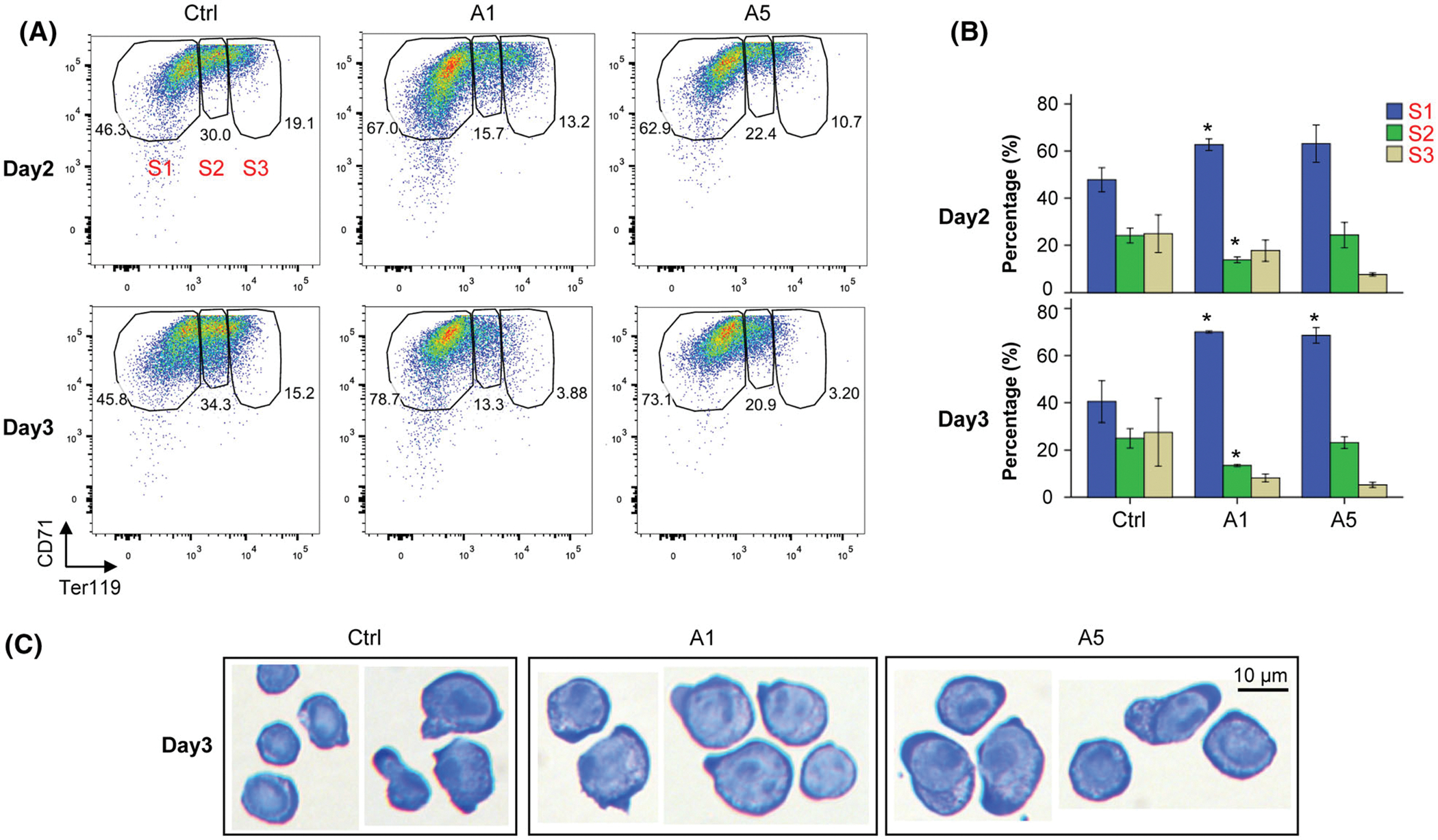
Differentiation of FL erythroid progenitors upon Atf5 knockdown. (A) Representative flow cytometric plots of cell differentiation and (B) the percentages of cells at different erythroid differentiation stages for the GFP+ cells on days 2 (upper panels) and 3 (bottom panels) (n = 3). Data are presented as mean ± SE. *P < 005 as compared to Ctrl. (C) Representative images of cell morphology on day 3. Scale bar = 10 μm. Ctrl: infected with retrovirus expressing non-targeting shRNA; A1 and A5: infected with retroviruses expressing A1 and A5 shRNAs targeting Atf5 mRNA. Data are presented as mean ± SE. *P < 005 as compared to Ctrl.
ATF5 was shown to be significantly upregulated in mature hepatocytes and function as a maturation factor in lineage-reprogrammed human embryonic fibroblasts (Du et al, 2014). In osteogenic differentiation, ATF5 was reported to be a suppressor (Leong et al, 2012). The roles of ATF5 are complicated in the development of neural tissues. ATF5 was shown to be required for the survival and differentiation of olfactory sensory neurons (Wang et al, 2012). However, ATF5 inhibits the differentiation of neural progenitors into astrocytes (Angelastro et al, 2005). The role of ATF5 in erythropoiesis as revealed in this study is similar to its functions in hepatic development and, especially, in olfactory bulb development.
ATF5 is involved in the integrated stress responses (ISR) through regulating endoplasmic reticulum unfolded protein response (UPR) and cytosolic heat shock response (Sears & Angelastro, 2017), similar to the role of ATF4, which is regulated by HRI in iron-deficient erythropoiesis (Chen, 2014). HRI-mediated ISR is necessary for effective erythropoiesis in iron deficiency (Zhang et al, 2018). The dependence of Atf5 expression on HRI supports the involvement of ATF5 in HRI-activated ISR under iron deficiency (Zhang et al, 2019). HRI-deficient mice develop ineffective erythropoiesis during iron deficiency (Han et al, 2001). Furthermore, HRI deficiency exacerbates severity of beta-thalassaemia in murine models (Han et al, 2005). Our study demonstrates that ATF5 is important for the proliferation and differentiation of late erythroid colony-forming units. Thus, Atf5 may be a new therapeutic target for the treatment of diseases with ineffective erythropoiesis, e.g. thalassaemia. Furthermore, ATF5 was shown to be a regulator of mammalian mitochondrial URP (UPRmt) and is required for maintaining mitochondrial function and the recovery from mitochondrial stress (Fiorese et al, 2016). Recently, we identified the role of HRI in the translational regulation of both cytoplasmic and mitochondrial UPR (Zhang et al, 2019). The HRI-ATF4 axis is necessary to induce the expression of downstream genes for UPRmt, redox homeostasis and metabolic reprogramming to maintain mitochondrial respiration and erythroid differentiation (Zhang et al, 2019). Atf5 mRNA is a downstream target of ATF4 (Zhou et al, 2008), indicating the association between HRI and ATF5 in regulating UPRmt under stress.
Conclusion
This study investigated, for the first time, the function of ATF5 in erythropoiesis and demonstrated the requirement of ATF5 in the proliferation and differentiation of erythroid progenitors. Our data may provide a new therapeutic target for the treatment of diseases with ineffective erythropoiesis, such as thalassaemia.
Supplementary Material
Table SI. DNA sequences of shRNA oligonucleotides.
Acknowledgements
This work was supported by National Institute of Health Grant RO1 DK087984.
Footnotes
Competing interests
All authors declare no competing interests.
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- Angelastro JM, Mason JL, Ignatova TN, Kukekov VG, Stengren GB, Goldman JE & Greene LA (2005) Downregulation of activating transcription factor 5 is required for differentiation of neural progenitor cells into astrocytes. Journal of Neuroscience, 25, 3889–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ (2014) Translational control by hemeregulated eIF2alpha kinase during erythropoiesis. Current Opinion in Hematology, 21, 172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Wang J, Jia J, Song N, Xiang C, Xu J, Hou Z, Su X, Liu B, Jiang T, Zhao D, Sun Y, Shu J, Guo Q, Yin M, Sun D, Lu S, Shi Y & Deng H (2014) Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell, 14, 394–403. [DOI] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin Y-F, Rosin N, Pellegrino MW & Haynes CM (2016) The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Current Biology, 26, 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, Fleming M, Leboulch P, Orkin SH & Chen JJ (2001) Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO Journal, 20, 6909–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han AP, Fleming MD & Chen JJ (2005) Heme-regulated eIF2alpha kinase modifies the phenotypic severity of murine models of erythropoietic protoporphyria and beta-thalassemia. Journal of Clinical Investigation, 115, 1562–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong DT, Abraham MC, Gupta A, Lim T-C, Chew FT & Hutmacher DW (2012) ATF5, a possible regulator of osteogenic differentiation in human adipose-derived stem cells. Journal of Cellular Biochemistry, 113, 2744–2753. [DOI] [PubMed] [Google Scholar]
- Sears TK & Angelastro JM (2017) The transcription factor ATF5: role in cellular differentiation, stress responses, and cancer. Oncotarget, 8, 84595–84609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom CS, Traxler EA, Khandros E, Nickas JM, Zhou OY, Lazarus JE, Silva AP, Prabhu D, Yao Y, Aribeana C, Fuchs SY, Mackay JP, Holzbaur EL & Weiss MJ (2014) Trim58 degrades Dynein and regulates terminal erythropoiesis. Developmental Cell, 30, 688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-Z, Ou J, Zhu LJ & Green MR (2012) Transcription factor ATF5 is required for terminal differentiation and survival of olfactory sensory neurons. Proceedings of the National Academy of Sciences of the United States of America, 109, 18589–18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Macias-Garcia A, Velazquez J, Paltrinieri E, Kaufman RJ & Chen J-J (2018) HRI coordinates translation by eIF2αP and mTORC1 to mitigate ineffective erythropoiesis in mice during iron deficiency. Blood, 131, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Macias-Garcia A, Ulirsch JC, Velazquez J, Butty VL, Levine SS, Sankaran VG & Chen J-J (2019) HRI coordinates translation necessary for protein homeostasis and mitochondrial function in erythropoiesis. eLife, 8, e46976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA & Wek RC (2008) Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. Journal of Biological Chemistry, 283, 7064–7073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table SI. DNA sequences of shRNA oligonucleotides.


