Abstract
What determines large-scale anatomy? DNA does not directly specify geometrical arrangements of tissues and organs, and a process of encoding and decoding for morphogenesis is required. Moreover, many species can regenerate and remodel their structure despite drastic injury. The ability to obtain the correct target morphology from a diversity of initial conditions reveals that the morphogenetic code implements a rich system of pattern-homeostatic processes. Here, we describe an important mechanism by which cellular networks implement pattern regulation and plasticity: bioelectricity. All cells, not only nerves and muscles, produce and sense electrical signals; in vivo, these processes form bioelectric circuits that harness individual cell behaviors toward specific anatomical endpoints. We review emerging progress in reading and re-writing anatomical information encoded in bioelectrical states, and discuss the approaches to this problem from the perspectives of information theory, dynamical systems, and computational neuroscience. Cracking the bioelectric code will enable much-improved control over biological patterning, advancing basic evolutionary developmental biology as well as enabling numerous applications in regenerative medicine and synthetic bioengineering.
Keywords: Bioelectricity, Ion channels, Regeneration, Morphogenesis, Embryogenesis, Patterning, Primitive cognition, Dynamical system theory, Bayesian inference
1. Introduction
1.1. To be explained: adaptive pattern regulation
It has been recognized since ancient times that the egg of a given species gives rise to an individual with the appropriate anatomy of that species (Fig. 1A). How does this occur? What is responsible for the remarkable multi-scale complexity of metazoan organisms, from the distribution of cell types among tissues to the topological shape and arrangement of the body organs, and the geometric layout of the entire bodyplan? It is widely believed that the answer lies within the genome, but it is not that simple; DNA simply encodes specific proteins – there is no direct encoding of anatomical structure. Thus, it is clear from first principles that pattern control involves a code: the encoding of anatomical positions and structures within the egg or other cell type, and the progressive decoding of this information as cells implement invariant morphogenesis (Fig. 1B). It should be noted that the current understanding of these codes is in its infancy and many fundamental questions remain to be addressed. Despite the progress of genetics and molecular genomics, we are not yet able to predict the anatomical structure of an organism from its genomic sequence (other than by comparing it to genomes whose anatomy we already know), nor in general do we know how to encode instructions to cells to induce them to develop anatomical structures to a desired functional specification.
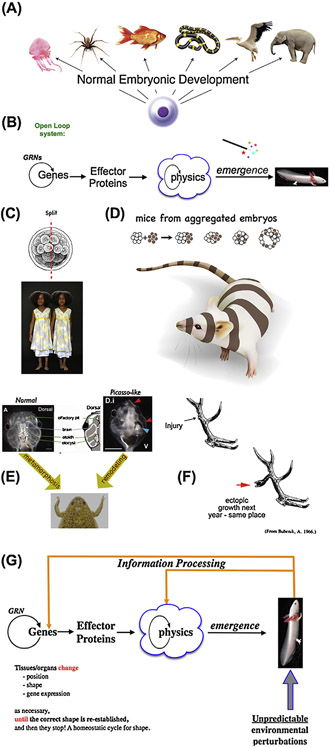
Fig. 1.
pattern regulation as a closed-loop homeostatic process.
(A) An egg will give rise to a species-specific anatomical construct. However, DNA does not directly encode geometrical layout of tissues and organs, requiring a process of decoding the genomic information into spatial configuration. (B) This process is usually described as a feed-forward system where the activity of gene-regulatory networks within cells result in the expression of effector proteins that, via structural properties and physical forces, result in the emergence of complex shape. In this view, there is no master plan for pattern – only a bottom-up emergent process driven by self-organization and parallel activity of large numbers of agents (cells); this class of models is difficult to apply to a number of biological phenomena. Some species, including many mammals, utilize regulative development which can adjust to radical deformations to the normal developmental sequence. (C) Their embryos can be divided in half, giving rise to perfectly normal monozygotic twins each of which has regenerated the missing cell mass. (D) Their embryos can also be combined, giving rise to a normal (but slightly larger) embryo in which no parts are duplicated. (E) The ability to achieve a specific target morphology despite different starting configurations (flexible morphogenesis) is clearly revealed in the Xenopus tadpole. The normal deformations of the face from that of atadpoleto that of a frog do not break down when tadpole faces are produced with organs (eyes, nostrils, etc.) in aberrant locations: rather than performing a hardwired set of movements (which would create an abnormal frog face if the components start out from incorrect locations), it orchestrates a set of appropriately altered deformations that cease when the correct frog face is produced. This kind of pattern-homeostatic process must store a set-point that serves as a stop condition; however, as with most types of memory, it can we specifically modified by experience. In the phenomenon of trophic memory (F), damage created at a specific point on the branched structure of deer antlers is recalled as ectopic branch points in subsequent years’ antler regeneration. This reveals the ability of cells at the scalp to remember the spatial location of specific damage events and alter cell behavior to adjust morphogenesis appropriately – a pattern memory that stretches across months of time and considerable spatial distance. (G) These kinds of capabilities suggest that patterning is fundamentally a homeostatic process – a closed-loop control system which employs feedback to minimize the error (distance) between a current shape and the stored target morphology. Although these kinds of decision-making models are commonplace in engineering, they are only recently beginning to be employed in biology(Barkai and Ben-Zvi, 2009; Pezzulo and Levin, 2016). Panels A,C were created by Justin Guay of Peregrine Creative. Panel C contains a photo by Oudeschool via Wikimedia Commons. Panels E,F are reprinted with permission from (Vandenberg et al., 2012) and (Bubenik and Pavlansky, 1965) respectively.
Indeed, the mystery is revealed to be even deeper than that of embryogenesis, in which the same initial starting condition (the egg) develops into the appropriate target morphology of a given species. Many types of animals exhibit extensive capacity for regeneration (Birnbaum and Alvarado, 2008) or remodeling (Farinella-Ferruzza, 1956); these organisms can restore complex body organs or appendages after dramatic morphological changes such as amputation. For example, planarian flatworms can rebuild any missing part of their body (including the head) (Lobo et al., 2012; Salo et al., 2009), while axolotls can regenerate eyes, limbs, tails, jaws, ovaries, and portions of the brain (Maden, 2008). Such examples reveal that living systems exhibit highly adaptive and context-sensitive pattern homeostasis. Individual cell behaviors are directed towards the maintenance and repair of a specific anatomical configuration. When the correct target morphology is achieved, large-scale remodeling and growth ceases.
1.2. Why do we need a code? representing homeostatic goal states in tissue properties
The current paradigm recognizes that different types of codes participate in pattern control. Examples include gradients of gene products that dictate positional information via chemical signals, such as HOX codes (Bondos, 2006), and epigenetic codes (Broccoli et al., 2015) that regulate transcriptional cascades via chromatin modification. However, the processes underlying embryogenesis are largely thought of as a ‘feed-forward’ system: the progressive unrolling of the genome in each cell results in specific cellular events which, integrated over large numbers of cellular agents over space and time, results in the emergence of a complex and highly organized forms. The mainstream consensus is that there is no overall encoding of the target morphology: the process is controlled by local events, and the resulting complex pattern is the result of emergence and self-organization.
And yet, many of the examples of complex pattern regulation are challenging to explain as an open-loop, purely-emergent process (Fig. 1C,D). For example, embryos of many species can be cut in half or deformed at early stages and yet, can still achieve the morphology of a normal organism (e.g., monozygotic twins from embryo splitting). The ability to achieve the exact same end result from different starting configurations (e.g., a planarian or salamander limb cut at different positions) is highlighted especially starkly by the process of metamorphosis. Becoming a frog requires the tadpole to rearrange its face – the various craniofacial organs move to new positions during metamorphosis. This is normally a stereotypical process, but it was recently discovered that if “Picasso” tadpoles are created (where the eyes, nostrils, and other structures are in aberrant positions), the animals will still turn into largely normal frogs (Vandenberg et al., 2012): the organs move in new ways, but still achieve normal frog face target morphology (Fig. 1E). This means that genetics does not specify hardwired movements of the organs, but rather contribute to the function of a plastic system that enables diverse responses to abnormal starting states so that an invariant (and thus encoded) outcomes result.
While this kind of pattern memory is clearly stable, it is not readonly – it can be rewritten (Lobo et al., 2014). Modifications made to the shape (Yamaguchi, 1977) and size (Bryant et al., 2017) of limbs in crustacea and amphibians respectively are “learned” by the system, resulting in permanent changes to the target morphology (the pattern towards which regeneration builds) upon future rounds of regeneration. A most impressive example (Fig. 1F) is that of trophic memory in deer, in which some species shed and re-grow a consistent branching pattern of antlers (bone and innervation) each year (Bubenik and Pavlansky, 1965). It was observed that damage made to one point in the branched structure resulted in ectopic branches being produced at the same point in subsequent years of growth. This means that the growth plate in the scalp somehow ‘remembers’ the location of damage for months, as the whole antler rack falls off and is regenerated, and then triggers the cell behaviors needed to form an ectopic branch in just the right place. This type of spatial memory in remaining scalp cells (recalling events that occurred at significant distance in space and time) is especially difficult to reconcile with typical “molecular pathway” arrow models (or gene-regulatory networks) and strongly suggests a spatial encoding system.
Taken together, these examples strongly suggest a ‘closed-loop’ (feed-back based) pattern homeostatic process (Fig. 1G). Systems guided by pure emergence are notoriously difficult to control and study – knowing which low-level rule to perturb experimentally and how to alter it, in order to reach a desired large-scale outcome in a recurrent process is an extremely difficult inverse problem (imagine trying to determine how to modify a function such as z = z2 + c if one wants to add an extra geometric feature to its resulting fractal image). We have argued elsewhere (Lobo et al., 2014) that the highly robust regenerative capacity of living organisms suggests that evolution has found an easier way; moreover, scientists can capitalize on aspects of top-down control to achieve progress in regenerative medicine. If modular, representational information (i.e., the encoding of large-scale structure) exists, then re-writing the code to allow the cells to “build to spec” might enable much more efficient control of growth and form compared to approaches that by micromanage individual cell behaviors.
Much of the recent progress in biology has come from exploring the extent of complex outcomes that can result in the absence of a master plan (Davidson et al., 2010; Davies and Cachat, 2016; Deglincerti et al., 2016; Halley et al., 2012; Ishimatsu et al., 2010; Raspopovic et al., 2014). In its flight from vitalism and teleology, modern biology has preferred models of emergence and de-centralized control. However, explicitly represented goal states no longer need to be anathema to biology. Over the last 50+ years, cybernetics, control systems theory, and computer science have revealed frameworks for rigorous means of implementing mechanisms that store complex states and pursue them as homeostatic setpoints (thermostats and self-driving vehicles are examples). Many fields, from cognitive neuroscience to engineering routinely utilize goal-seeking and error-minimizing homeostatic control loops to understand and create complex adaptive functionality (Pezzulo and Levin, 2016). Here, we argue that it is time to consider the possibility that the known emergent features of cell behavior are augmented by a complementary set of top-down controls in which at least some aspects of target anatomical states are encoded within tissue properties (Pezzulo and Levin, 2015). If tissues must somehow remember at least some aspect of a target state in their physico-chemical properties, then encoding and decoding is necessary, since living tissues are storing information about a future (counterfactual) state in their current structure – this is the quintessential context of a code.
What mechanisms could underlie such a morphogenetic code? Based on the observed examples of stable yet re-writeable anatomical structure, it would have to be a system that supported long-term but labile memory, with capability to sense/measure large-scale spatio-temporal signals. It would also have to have holographic properties (storing information about the whole in individual pieces) and be able to harness individual cell activities toward group-level goals. While many architectures could in principle support this kind of control system, two well-studied examples do all of the above: cognition in the living brain, and engineered artificial information-processing devices (computers). What these systems have in common is their reliance on electrical signaling. We next consider the role of bioelectrical events in encoding and decoding anatomical structure, keeping in mind that bioelectrics is perhaps but a single component of the rich morphogenetic code that regulates the shape of life at all scales.
2. Developmental bioelectricity: an encoding medium for pattern control
2.1. Evolutionary origins of bioelectricity, neural and non-neural
Brains use electrical networks to implement decision-making and memory, and to harness the triggering of specific cell behaviors (muscle contractions, gland secretions, and changes in cell proliferation and gene expression) to large-scale goals that are represented in cognitive constructs. It is becoming increasingly appreciated that communication via electrical processes is not unique to nervous systems, but in fact evolved continuously from far more evolutionarily-ancient properties that cells possessed long before nerves and brains evolved (Keijzer et al., 2013; Liebeskind et al., 2011, 2015; Mathews, 1903). The major ion channel families are present near the origin of multicellularity, and show significant expansion with the complexification of the metazoan bodyplan (Liebeskind et al., 2015; Moran et al., 2015). The same is true of neurotransmitter signaling (Buznikov et al., 2001; Levin et al., 2006; Roshchina, 2016; Venter et al., 1988), revealing that modern neural networks are an optimized form of a much more primitive cell type that utilized these same pathways to handle its physiological, behavioral, and structural decision-making.
While the somatic bioelectric dynamics in these early forms are still unknown, a large literature on electrophysiology in aneural systems from plants (Toko et al., 1990; Zimmermann et al., 2009), to fungi (Chang and Minc, 2014; Zheng et al., 2015), to unicellular algae (Goodwin and Pateromichelakis, 1979; Novák and Bentrup, 1972), to bacteria (Humphries et al., 2017; Kralj et al., 2011; Prindle et al., 2015) reveals that evolution discovered the computational value of physiological networks very early on. Recognizing the need for a medium that supports complex decision-making, classical biologists investigated developmental bioelectrics at the very dawn of mechanistic investigations into the control of body patterning (Morgan, 1904). More recently, with studies of ion channel expression and function, as well as work on pre-neural neurotransmitter roles (Buznikov et al., 1996; Levin et al., 2006; Sullivan and Levin, 2016), the parallels between brain and body in terms of electrical dynamics are becoming ever clearer (Fig. 2A). Indeed, the contribution of neural signals to patterning, seen in older work on neural determination of head-tail polarity in regeneration (Fig. 2B), as well as in more recent data on the spinal cord’s role in guiding tail regeneration (Fig. 2C) and on the brain’s role in muscle and peripheral nerve patterning (Herrera-Rincon et al., in press), helps blur the boundary between electrical activity in the brain and patterning processes in the body.
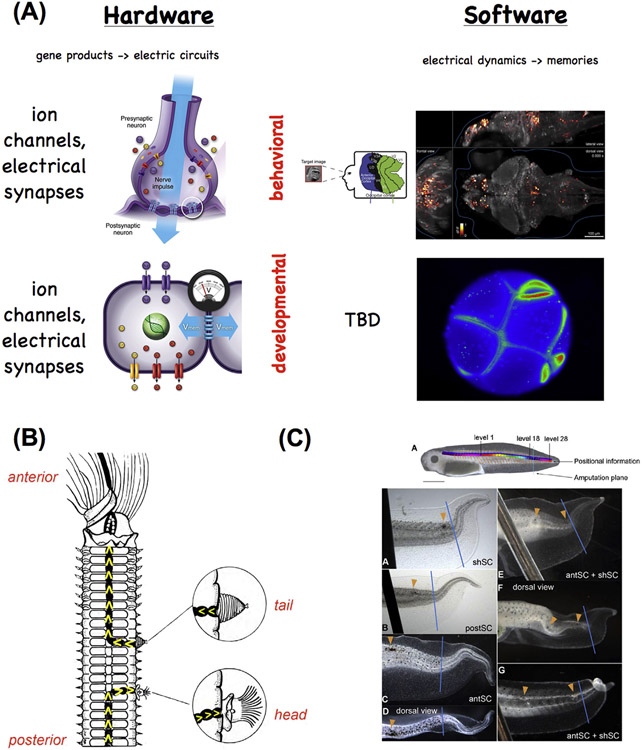
Fig. 2.
Mechanisms and functionality conserved between brain function and pattern regulation.
(A) The hardware of the brain consists of ion channels that regulate electrical activity and highly tunable synapses that propagate electrical and neurotransmitter-mediated signals across a network. This hardware supports a wide range of electrical dynamics that implement memory and goal-seeking behavior (and are not directly encoded by the genome, and can be modified by experience, although they have default modes of inborn activity corresponding to instincts). Other (non-neural) cell types have exactly the same ion channel, electrical synapse (gap junction), and neurotransmitter machinery. They likewise support a kind of control software, implemented in time-varying electrochemical dynamics across tissues, which underlies patterning decisions. In both cases, techniques from the field of “neural decoding” can be used to extract embedded semantics (cognitive content in the case of the brain, anatomical prepatterns in the case of tissues) from bioelectrical state readings. The computational analogy is not meant to suggest that tissues (or the brain) operate specifically via the Von Neumann architecture used by today’s computers. Interestingly, the CNS provides important input into pattern regulation. When a nerve cord is cut and deviated to the side wall of worms (B), the direction of the nerve end specifies whether a tail or head anatomical structure is produced (Kiortsis and Moraitou, 1965). In tadpole tail regeneration (C), laser-induced damage created within the spinal cord produces distinct changes to the shape of the regenerating tail depending on the number and location of the pinpoint holes (Mondia et al., 2011). Left panels of (A) drawn by Jeremy Guay. Top right panel of (A) is reproduced with permission from (Naselaris et al., 2009). Bottom right panel of (A) is a frame from a time-lapse movie produced by Dany S. Adams. Panel B is modified after (Kiortsis and Moraitou, 1965). Panel C is reproduced with permission from (Mondia et al., 2011).
2.2. A brief history of bioelectrics research
Since then, the field of non-neural bioelectricity has developed in several phases. Classic work by Lund (Lund, 1947), Burr (Burr, 1944; Burr and Northrop, 1935a), and later Marsh and Beams (Marsh and Beams, 1952) used measurements of electric fields and application of physiological-strength electric gradients to investigate the link between endogenous bioelectric properties across tissues and outcomes in embryogenesis, regeneration, and cancer. A subsequent “golden age” of bioelectricity was driven by pioneers such as Jaffe (Bentrup et al., 1967; Bentrup and Jaffe, 1968; Borgens et al., 1977a, 1977b; Jaffe, 1979;Jaffe and Nuccitelli, 1977), Cone (Cone, 1970, 1971; Cone and Tongier, 1971), Nuccitelli (Kline et al., 1983; Nuccitelli, 1983; Nuccitelli, 1986, 1987; Nuccitelli, 1988; Nuccitelli and Erickson, 1983; Nuccitelli et al., 1986), Borgens (Borgens et al., 1989; Borgens, 1984, 1986; Borgens et al., 1984), Robinson (Hotary and Robinson, 1990,1991, 1992; Nuccitelli et al., 1986), and McCaig (Cao et al., 2014; Kucerova et al., 2011; Martin-Granados and McCaig, 2014; McCaig, 1986; McCaig et al., 2005; McCaig and Zhao, 1997; Yao et al., 2011); these pioneers used modern tools, such as the vibrating probe, to characterize ion fluxes in vivo and showed a range of functional phenotypes that revealed the instructive nature of bioelectric signals for determining cell behavior and growth patterns in various model systems. The last two decades have seen the development of molecular tools for probing bioelectricity (Adams and Levin, 2012, 2013; Gao et al., 2015; Nakajima et al., 2015; Oviedo et al., 2008; Reid and Zhao, 2014; Yamashita, 2013; Yamashita et al., 2013; Zhao et al., 2006), which have enabled significant advances in the mechanistic understanding of the roles of endogenous bioelectricity and the means by which they interface to transcriptional machinery.
2.3. A basic introduction to developmental bioelectricity
Bioelectric circuits consist of ion channel proteins, which passively segregate charges across the membrane, and ion pumps, which use energy to transport ions against concentration gradients. Ion translocators in the plasma membrane set the resting potential of each cell (measured in millivolts mV). In addition to local voltage potentials, bioelectric events can be propagated across long distances by ephaptic field effects (Qiu et al., 2015), transepithelial electric fields (Cortese et al., 2014), tunneling nanotubes (Wang et al., 2010), transfer of ion channels via exosomes (Wahlgren et al., 2012), and gap junctional connectivity implemented by Connexin and Innexin proteins (Mathews and Levin, 2016). Whereas ion channels and pumps exchange ions with the outside milieu, gap junctions (GJs) are channels that enable direct cell-to-cell transfer of electrical signals (and other small molecules), from the cytoplasm of one cell to its neighbor. Cells connect to each other via such electrical synapses (Palacios-Prado and Bukauskas, 2009), which facilitate the formation of bioelectrical networks that help shape the distribution of resting potential levels (Vmem) within large groups of cells, and form isopotential cell fields in vivo demarcated by GJ isolation zones (Mathews and Levin, 2016). “Vmem gradients” are defined as patterned spatial differences among the Vmem values of cells across anatomical distances (Fig. 3A). “Bioelectrical signals” are defined as a temporal change in such a pattern, which can trigger downstream patterning cascades.
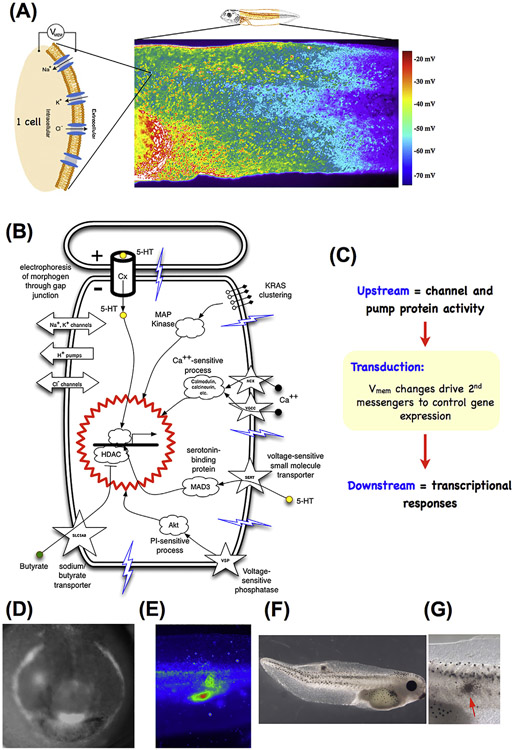
Fig. 3.
Developmental bioelectricity.
(A) Individual cells express ion channels and pumps in their membrane, which establish cell resting potentials. Voltage-sensitive fluorescent dyes can be used to view the spatio-temporal patterns of these potentials in vivo, such as the flank of a tadpole seen here (image provided by Douglas J. Blackiston). (B) Changes in voltage are transduced into second messenger cascades and downstream transcriptional responses by a variety of mechanisms including voltage-gated calcium channels, voltage-powered transporters of serotonin and butyrate, voltage-sensitive phosphatases, and electrophoresis through gap junctions. (C) Thus, activity of ion channels and pumps are transduced into changes of gene expression (which may include other ion channels, thus forming a feedback cycle). Spatial patterns of voltage and their signaling consequences serve as prepatterns for normal morphogenesis, such as the prepatterns of the tadpole face (D, reproduced from (Vandenberg et al., 2011)), or disease states such as tumors induced in tadpoles by expression of human oncogenes, detected by their bioelectric disruption (E) before they become morphologically obvious(F, close-up in G). Panel E-G reproduced with permission from (Chernet and Levin, 2013; Lobikin et al., 2012).
Cells respond to their own electrical states as well as those of their neighbors, via a range of transduction mechanisms (Fig. 3B,C; reviewed in (Levin et al., 2017)) that convert bioelectrical properties into transcriptional responses via second-messenger cascades. These include the familiar calcium pathway (Deisseroth et al., 2004; Sasaki et al., 2000), and the movement of neurotransmitter molecules under voltage-gated or electrophoretic forces (Fukumoto et al., 2005; Fukumoto and Levin, 2003; Levin et al., 2006), precisely the same strategy used in the central nervous system. One example is the electrophoretically-controlled movement of serotonin across the early left-right axis of the developing frog embryo (Fukumoto et al., 2005) – a second-messenger signal that triggers asymmetric gene expression by differentially binding an intracellular receptor and chromatin modification machinery (Carneiro et al., 2011). Another example is the bioelectrically-powered movement of butyrate in and out of cells, which targets the same chromatin modifier (histone deacetylase) to regulate tumorigenesis (Chernet et al., 2015).
2.4. What bioelectric signals do: instructive influence over morphogenesis
Bioelectric properties control cell behaviors (Cao et al., 2013; Pan and Borgens, 2012; Tai et al., 2009; Yamashita, 2013), often acting as a switch between the organized collective behavior of a patterned tissue and tumorigenesis (Chernet and Levin, 2013, 2014; Lobikin et al., 2012; Oviedo and Beane, 2009). For example, transmembrane voltage levels control the proliferation of a wide range of cell types (Blackiston et al., 2009), and Vmem regulates differentiation in a range of stem/progenitor and IPS cells (Sundelacruz et al., 2008, 2009). Proliferation (MacFarlane and Sontheimer, 2000; Ouadid-Ahidouch and Ahidouch, 2008, 2013; Ouadid-Ahidouch et al., 2016; Putney and Barber, 2003; Valenzuela et al., 2000) and cell shape (Blackiston et al., 2011; Morokuma et al., 2008) are known to be regulated by voltage properties, which encode important parameters at the level of individual cells. However, rich encoding properties for bioelectricity are revealed when network (multicellular)-level roles are examined.
Classic investigations of bioelectricity utilized applied electric fields via electrodes (Lee et al., 1993; McCaig et al., 2005), which showed that numerous cell types read electric field lines to decode positional information (Shi and Borgens, 1995) and direction (Gruler and Nuccitelli, 1991; Mycielska and Djamgoz, 2004) information for morphogenesis and migration respectively. New techniques include voltage-sensitive fluorescent dyes to read out Vmem information in vivo, and the misexpression of ligand- or light-gated ion channels and pumps (Adams et al., 2014; Adams et al., 2013) to write desired voltage changes into tissues in a spatially- and temporally-controlled manner. Computational tools that provide physiology-level modeling platforms for understanding bioelectric dynamics (Pietak and Levin, 2016) have also been important and quantitative theory for inferring information processing aspects of bioelectric state change (Tononi et al., 1998; Tononi and Sporns, 2003; Tononi et al., 1994). All of these developments have proceeded in parallel with similar efforts in neural decoding (Nishimoto et al., 2011; Quian Quiroga and Panzeri, 2009) and inception of false memories via bioelectric editing (Liu et al., 2014; Ramirez et al., 2013) in the field of neuroscience.
The information content of bioelectrical cell states is thus an active area of investigation (Moore et al., 2017). One property likely mediated by these gradients is that of positional information, as long-range bioelectrical gradients are an ideal modality for coordinating spatial configuration with functional decision-making at the cell level. Future work will determine how electrically-mediated positional signaling integrates with the molecular-genetic systems of positional memory, such as the HOX code in the skin and muscle (Kragl et al., 2009; Kragl et al., 2008; McCusker and Gardiner, 2013, 2014; Rinn et al., 2006; Witchley et al., 2013). Importantly however, recent work has begun mechanistically linking bioelectric activity in cells with canonical pathways such as BMP (Dahal et al., 2017; Dahal et al., 2012) and Hedgehog (Belgacem and Borodinsky, 2015; Swapna and Borodinsky, 2012).
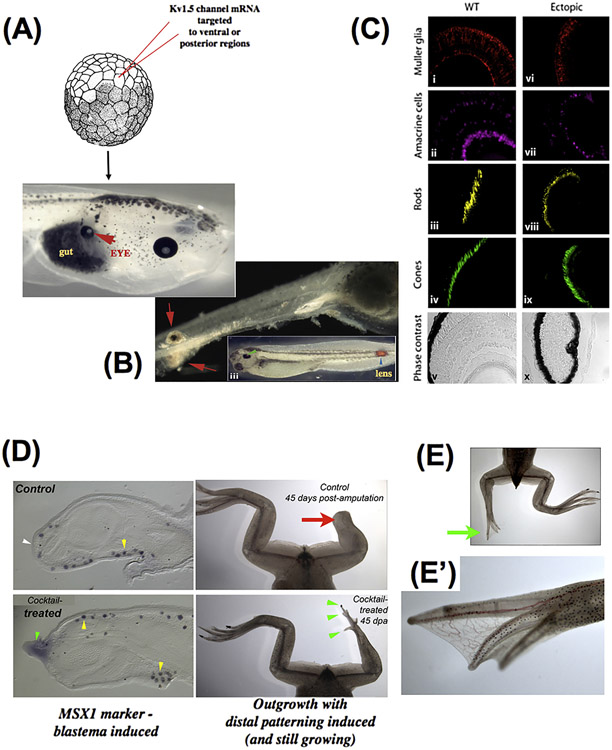
Using a range of new strategies for functionally investigating the importance of bioelectric states, recent work has revealed that voltage gradients instruct much more than cell-level behaviors. Specific bioelectric prepatterns (Fig. 3D) have been found in the face (Adams et al., 2016a; Vandenberg et al., 2011) and brain (Pai et al., 2015a,b), which appear to encode locations of specific anatomical features. When these prepatterns are artificially altered (Fig. 3E-G), predictable changes in downstream gene expression and anatomy result. Moreover, alterations of bioelectrical pattern (by introducing new ion channels, or using drugs to target endogenous channels) can induce regeneration of entire appendages in tadpoles (Adams et al., 2007), change the size of body structures in planaria and zebrafish (Beane et al., 2013; Perathoner et al., 2014), and induce reversal or randomization of the whole body axes, regulating left-right asymmetry in chick and frog (Adams et al., 2006; Levin et al., 2002) and anterior-posterior anatomical polarity in planarian regeneration (Beane et al., 2011; Durant et al., 2017).
All of this work clearly demonstrates that bioelectricity is an important input into pattern regulation, operating in embryogenesis, regeneration, and cancer suppression (Levin, 2011a, 2014; Tseng and Levin, 2013). But does it implement a code? Or is it just another set of mechanisms, like the biochemical gradients and mechanical forces that also participate in pattern regulation? Since these codes must be physically embodied, we must next decide what features constitute a code, and demarcate a set of events as best described as a coding/decoding system as opposed to a purely mechanical mechanism. A thorough philosophical discussion of principled criteria for treating systems from mechanical vs. information-processing perspectives has been presented pre viously (Dennett, 1987; Rosen, 1985).
3. Cracking the bioelectric code
3.1. Why is bioelectric signaling an example of a code?
We use several key features to define the existence of a “code”, in distinction to a purely mechanistic set of consecutive physical states: when does some property “encode” information, rather than merely causing downstream events? This is a complex philosophical issue related to an extensive literature on causation and semiotics (Baslow, 2011; Hoffmeyer, 2000; Pattee, 1982, 2001), and it is likely that the distinction is a continuum without a sharp definitive distinction. Here we attempt to only give a few practical guidelines, to illustrate what is meant by a code in the context of biological patterning.
The first feature focuses on the fact that a signal’s encoded meaning is not inherently tied to the physical property of said signal, but rather it is arbitrary and defined by (evolutionary) convention – amply illustrated by the extensive use of symbolic codes in human technology. A signal encodes some state if its presence triggers that state to occur because of how the system interprets that signal, not because it’s a physical event that forces the outcome. An electric field may force the movement of a charged particle – this is not an example of a code; a pattern of electric events may be interpreted by cells and cue them to differentiate – this is a code in the sense that those electrical events do not in themselves require any link to differentiation, and evolution could have wired the system so that those same events instead signaled the need for programmed cell death. As another example, the HOX code is a code because a combination of HOX transcripts causes specific structures (e.g., wings vs. legs) to form in an organism, but none of those proteins actually have any wing- or leg-like properties or activities – it is the interpretation of this arbitrary signal by cell groups that gives it meaning and a functional context. Thus, the decoding process is key. For example, a heat pulse that denatures proteins and induces cell death is not an encoded signal because its effects are physical and direct, not requiring an evolved system to interpret it. The bioelectric code is a code in this sense because the voltage properties do not themselves imply any particular kind of patterned structure – it is the act of decoding of voltage-mediated signals by cell groups that interprets the code in constructing and remodeling anatomy.
A closely-related property of codes is that the physical implementation is not as important as the information content. This is also true of the bioelectric code because it has been repeatedly found that cells can respond not to individual channel proteins or individual ion species, but to resting potential – voltage, which is an aggregate property and can be encoded by a range of chloride, sodium, and potassium concentrations. There are exceptions to this (e.g., ion channels that operate as binding proteins for other partners, and calcium, which function in extremely small quantities as a unique chemical signal rather than setting Vmem), but overall the same outcome can be achieved by triggering appropriate Vmem states regardless of which ion translocator or ion is used. For example, tail regeneration in tadpoles can be rescued after shutdown of the native V-ATPase proton pump by misexpression of a completely different pump from yeast that has no sequence or structural homology (Adams et al., 2007). Likewise, eye patterning can be induced by a range of channels that set Vmem to a specific level (Pai et al., 2012), while metastatic behavior in normal melanocytes can be induced or rescued using chloride, sodium, or potassium as long as the appropriate Vmem signal is provided. The bioelectric code signals by virtue of its physiological state, with no 1:1 correspondence to any specific gene product.
Aside from philosophical debates on the precise definition of a “code” (Barbieri, 2008; Pattee, 1982, 2001), the value of a metaphor lies in its explanatory power – its ability to drive new research, uncover new phenomena, and enable capabilities that competing paradigms did not facilitate. One example of a conceptual gap left by today’s paradigm concerns the most important aspect of regeneration: the stop condition. How do regenerative processes know when to stop – when the correct target morphology has been achieved? This question can be put into sharp focus with the following thought experiment (Fig. 4). Suppose neoblasts (adult stem cells) from two planaria with distinct head shapes are combined into one body, and that body is decapitated. What head shape will result? Would it be an average between the two, a dominant shape, or a continuously metamorphosing head (as neither population of neoblasts is ever entirely happy with the current head shape)? The striking fact is that, despite numerous high-resolution analyses of stem cell function and gene-regulatory circuits in planaria (Aboobaker, 2011; Shibata et al., 2010; Vogg et al., 2014), there are no models in this field that can make a prediction on this very basic question. The understanding of regeneration and the link of known facts to the key question of how it knows what to build and when to stop exhibits a profound conceptual gap not filled by existing models, coding or otherwise.
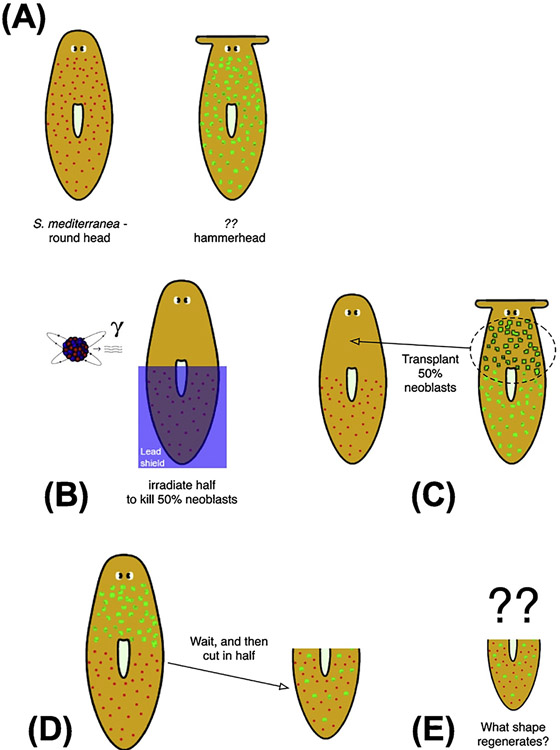
Fig. 4.
How to determine what pattern cells will build to?
This schematic describes a thought experiment to focus attention on the stop condition for regeneration: how do cells decide what pattern constitutes ‘correct, finished’ repair so that they can stop growth and remodeling? (A) Different species of planaria have (and regenerate) different shapes of heads, for example round or flat. (B) If half of the stem cells (neoblasts) of one species are destroyed (by irradiation), and some neoblasts from another species are transplanted (C), we can amputate the resulting worm (D) and ask: what head shape will it regenerate? Perhaps it will be an in-between (averaged) shape, or perhaps one of the sets of neoblasts is dominant, or perhaps the head will undergo continuous and unceasing deformation as neither set of neoblasts is ever satisfied with the current shape of the head. It is important to note that none of the excellent molecular-genetic work in this field has given rise to a model which can make a prediction (or constrain) the outcome of this kind of question. This thought experiment illustrates the fact that questions of control theory, representation (encoding), and algorithmic control over regeneration have been so far largely left out of mechanistic work in pattern control.
The hypothesis that bioelectric signaling is truly implementing a code makes a number of unique predictions. What has been the benefit of treating bioelectrics as a computational layer? Over the last 20 years, we have used concepts from computer and cognitive science to model bioelectrics as an endogenous computational medium that processes information which must be encoded and decoded by cell groups. By suggesting the first applications of neurotransmitter, ion channel, and electrical synapse-modifying methods to pattern regulation, this perspective has given rise to a variety of unique new findings.
3.2. Testing the predictions of a code-based view of pattern regulation
The first prediction was that bioelectric properties would encode patterns at levels above the cellular level – large-scale features of anatomy that cannot be defined at the single cell state (Fig. 5). Experiments designed to target regions of living frog embryos with specific ion channels and thereby alter local voltage maps in a whole area during development revealed that this strategy can indeed be used to induce whole eye formation in the gut or other regions outside the head (Pai et al., 2012). Thus, it is possible to rewrite the morphogenetic fate of not just single cells, but tissues in a modular fashion – specifying the message “build an eye here” without needing to directly micromanage each cell’s differentiation and placement into the incredibly complex structure of a whole eye. A similar phenomenon was observed during the reprogramming of a planarian blastema from tail to a complete head (Beane et al., 2011), and in the induction of complete tail regeneration (Tseng et al., 2010). In each of these cases, the information content provided by the exogenous channel was very small – certainly not enough to directly specify the structure of the newly-formed organ. This is one sure sign of a code, in which a simple message can trigger far more complex responses after it is decoded by the recipient. This is a clear example in which such questions bear on practical issues of biomedicine: this type of “subroutine call” effect, where a simple master trigger causes a complex, self-limiting morphogenetic response, may allow regenerative medicine to provide repair of organs long before we understand everything needed to directly build a complex structure from scratch. It should be noted also that in the case of the eye, molecular-genetic master regulators, like Pax6, cannot initiate eye formation outside the anterior neural field. Thus, the bioelectric data not only reveal extensions to overly-limiting views of tissue competence that emerged from biochemical studies, but enable a novel capability not previously possible to achieve.
Fig. 5.
Bioelectric modification of large-scale pattern.
Counter to early predictions that voltage outside the nervous system was a housekeeping parameter, it has been observed that specific alteration of resting potential patterns in vivo by misexpression of new ion channels can give rise to coherent, modular changes in anatomical arrangement. In Xenopus laevis, when specific ion channel mRNA is injected into embryonic blastomeres, regions of the animal – even those outside the anterior neural field – can be induced to form a complete eye. Eye structures can be induced inside the gut (A), tail, or spinal cord (B); in some cases, these eyes possess all the correct tissue layers of a normal eye (C), revealing master-level regulator control of organ formation (triggered by a simple manipulation, not micromanagement of individual cell fates and positions). These data also reveal the ability of bioelectric signals to overcome traditional limits on tissue competence (as, for example, the master eye gene Pax6 cannot produce eyes outside the head, and it was thought that gut endoderm was not competent to form eyes). (D) Drug cocktails using blockers and activators of endogenous channels can be used to trigger regenerative response; shownhere is a monensin ionophore cocktail inducing the expression of the MSX1 gene and subsequent regeneration of the leg (including distal elements such as toes and toenails, E, close-up in E’) in a post-metamorphic frog that normally does not regenerate legs. Images in A-C are reproduced with permission from (Pai et al., 2012). Images in D-E’ are the work of Aisun Tseng.
A second prediction was that the bioelectric code should allow a rich layer of control that can override genome-default outcomes. One example of this is recent work showing that, contrary to the view of cancer as exclusively an irrevocable phenotype caused by clonal expansion of genetically mutated founder cells, metastatic disease can be triggered by disruption of bioelectric signaling (Blackiston et al., 2011; Lobikin et al., 2015; Morokuma et al., 2008), while oncogene-induced tumors can be suppressed by enforcing appropriate bioelectric state (Chernet et al., 2016; Chernet and Levin, 2013, 2014) despite the fact that the oncogene is still strongly expressed. Similarly, it was shown that brain defects induced by a mutated Notch gene can be rescued, resulting in normal brain structure, gene expression, and behavior (IQ), by artificially inducing a brain-specific bioelectric pre-patterning during Xenopus development (Pai et al., 2015b). All of these examples illustrate the dissociation of genetic state from outcome (predictions based on transcriptomic, proteomic, or genomic analyses of each of those cases would have been incorrect), and highlight the essential nature of bioelectrically-encoded information in health and disease.
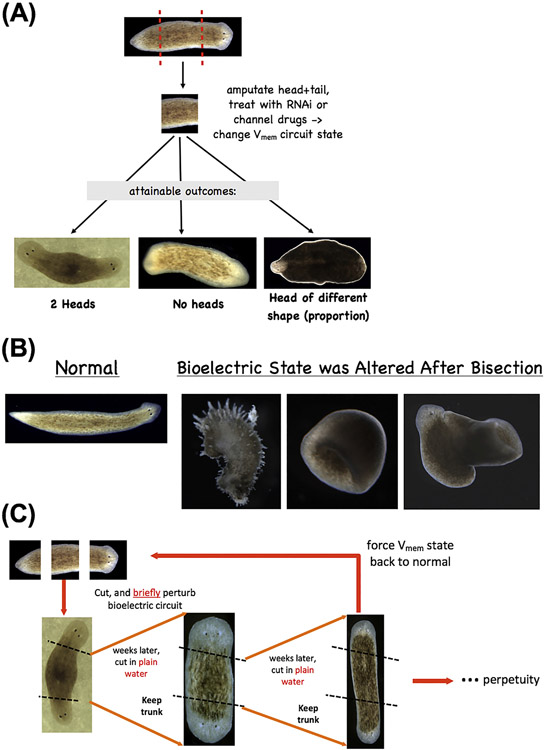
A third prediction is that it should be possible to coax significantly different anatomies from the same wild-type genome, using simple perturbations that result in coherent changes in large-scale structure without requiring extensive tweaking of underlying mechanisms (Fig. 6A,B). A recent example of this is the observation (Emmons-Bell et al., 2015) that temporary reduction of the bioelectric connectivity in planarian tissues during regeneration could induce a piece of Girardia dorotocephala planarian to regenerate heads appropriate to two other species of planaria. Fragments of Girardia flatworms, treated with a gap junction uncoupler, regenerated head shapes, brain morphology, and stem cell distributions appropriate to two other extant species of planaria and completely different from their genome-default morphologies. Despite a wild-type genomic sequence, bodies can produce anatomical structures quantitatively similar to those of species 150 million years distant. The evolutionary prevalence of morphological change via changes in the bioelectric layer remains to be studied. However, the fact that other species’-specific anatomies can emerge from the same genome is likely to be an important part of understanding the relationship between genotype and anatomical phenotype during evolution for the following reason. Consider planaria; their most frequent mode of reproduction is fission followed by regeneration (Neuhof et al., 2016); thus, they escape Weissmann’s Barrier – without an obligate sperm and egg stage, somatic mutations propagate into the next generation (as long as they are not lethal to the cell). Planaria have been subject to millions of years of somatic mutations and yet their morphology is robust – they regenerate a correct planarian anatomy every single time, without developing cancer or aging. Moreover, planaria are the only model system in which no genetic patterning mutant lines are available. Every other model (C. elegans, zebrafish, chick, mouse, frog, etc.) offers genetic mutants with altered morphology. With only one exception (produced by a bioelectric perturbation described below), flatworm lines are always normal, perfect planaria.
Fig. 6.
Re-writing form by targeting bioelectric circuits.
(A) Altering the electrical properties of tissue (by targeting ion channels, pumps, and gap junctional connectivity) in planarian fragments can be used to alter the head-tail polarity (producing double- or no-headed worms) or change the size of head structures. Moreover, such manipulations (B) can give rise to drastically altered forms which even depart from the normal flat anatomy of planaria, all with a wild-type genomic sequence. Most importantly (C), such changes can be permanent (Oviedo et al., 2010): double-headed planaria produced in this way continue to regenerate as double-headed when the ectopic heads are amputated, in plain water, revealing that brief (48 h) targeting of the bioelectric network to induce a different circuit state (with depolarized regions at both ends) permanently re-species the target morphology to which each piece of this worm would regenerate upon damage. The worms can be set back to normal by treatment with pump-blocking reagents that restore the normal bioelectric encoded pattern (Durant et al., 2017). Panels in (A) are Reproduced with permission from (Nogi and Levin, 2005) and (Beane et al., 2013). Panels in B are reproduced with permission from (Sullivan et al., 2016).
How is it possible that with a highly variable genome (Benya et al., 2017; Nishimura et al., 2015; Peiris et al., 2016), as befits a system in which somatic mutations are heritable, planarian regenerative and developmental processes exhibit the highest fidelity of any other model species? The inability of any existing data or models to answer this question (or indeed, to address the more general question of how much mutagenesis we would expect a genome to experience before anatomical change results), highlights how much remains to be learned about morphogenetic codes beyond the genetic code. All of the above examples reveal the importance of information encoded in physiological networks as a mediator of the morphogenetic code between the genome and the anatomy. Bioelectric signaling is a new type of epigenetic process, with dynamics quite distinct from that of chromatin-modifying effects as it is fundamentally distributed (multi-cellular scale) and encodes patterning, not only metabolic and differentiation state information.
Finally, a most fundamental aspect of the bioelectric code model is the implication that it should be possible to edit the encoded target state to which cells build, as an alternative to revising the local rules followed by each cell. If target morphology is indeed encoded as a kind of memory for anatomical patterning, then it should be possible to re-write it. to A key property of memory is its lability. Can pattern be permanently altered without genomic editing? A set of studies over the last few years (Oviedo et al., 2010) has established a model of trophic memory more tractable than deer antlers and crab claws: the planarian (Fig. 6C). Altering the bioelectric network of regenerating planaria, using a pharmacological reagent that leaves tissues within 24 h, produces animals that are indistinguishable from normal by anatomy, histology, key marker gene expression, or stem cell distribution; however, if amputated in plain water, a proportion of these animals give rise to double-headed worms. This stochastic state, in which the pattern to which they will regenerate has been de-coupled from their current pattern (a unique outcome in regeneration), persists indefinitely – it is stable, with no more perturbation. The only thing different about such “cryptic” animals is that their endogenous bioelectric states differ from true wild-type worms – a fact that can be decoded (read out) by human observers using voltage-reporting dyes, and by cells themselves during regeneration. Moreover, double-head worms produced by this process remain double-headed in perpetuity (Oviedo et al., 2010), despite a normal genomic sequence, revealing that target morphology can be re-written by bioelectric circuit change. Every piece of that worm acquires a different (i.e., double-headed) encoded goal state to which it will regenerate, and this process is stable to the worm’s normal mode of reproduction (fission), revealing a novel epigenetic pathway that may be important for evolutionary plasticity. Most crucially, these manipulations reveal the existence of an encoded pattern memory, because they re-write it, altering what the tissue will do in the future (latency – a key aspect of the definition of memory).
3.3. Major knowledge gaps
The idea of electrical properties of tissue being important, even instructive, aspects of patterning has been around for a long time; prescient classical workers like H. S. Burr long ago suspected that bioelectric prepatterns encoded anatomical states (Burr and Bullock, 1941; Burr and Northrop, 1935b; Burr and Sinnott, 1944). However, modern formulations of the bioelectric code concept that cohere with the advances in information sciences, computational neuroscience, and molecular evolutionary genetics comprise an emerging field. Emerging tools are coming on-line to begin to interrogate the encoding process and structure, and the mechanisms by which this code is read and could be re-written.
One major knowledge gap is not knowing precisely what is being encoded. Models that appear appropriate to specific cases include: individual cell states (cancer, mitotic control), paint-by-numbers direct prepatterns for gene expression domains (craniofacial patterning), positional or size information (neural tube, tail size), or selection of discrete patterning subroutines at the organ (limb, tail) and axial polarity (left-right and head-tail decisions). It is essential to extend the existing tools to enable the writing of arbitrary bioelectric state to determine which aspects of pattern control can be written and whether the encoding of these messages are via spatial patterns of voltage, temporal fluctuations, or both. The ability to read a bioelectric pattern and predict the resulting anatomy, and conversely, to induce a bioelectric pattern that will drive morphogenesis of a desired form, is the long-term goal of the research program focused on cracking the bioelectric code.
Another key area of current investigation is the ontogenic origin of the code: where do bioelectric prepatterns come from? In regeneration, and in clonal reproduction (e.g., in planarian reproductive fission), the patterns can be driven by remaining tissue – they serve as pattern memories that instruct new growth, as recently shown in flatworms (Durant et al., 2017). What about organisms that develop from a single fertilized egg cell? In this case, there are two (not mutually exclusive) ways for bioelectric patterns to arise. One is via self-organization and symmetry breaking driven by amplification and feedback loops in the electric circuit. This has been well-studied for biochemical signals (Raspopovic et al., 2014; Schiffmann, 2005; Watanabe and Kondo, 2015), and likewise occurs in electric networks (neural and non-neural) (de Roos et al., 1997; McNamara et al., 2016; Mustard and Levin, 2014; Pietak and Levin, 2016). The implication is that while every multi-cellular electric circuit has a default bioelectric dynamic, this dynamic can be changed by experience (environmental or experimental perturbation) and thus diverge from the genome-default. This means that bioelectric circuits are analogous to Turing-type self-organization in the sense that they emerge from symmetry-breaking generic processes, but have additional capabilities because tunable synapses (voltage-gated channels and gap junctions) enable the stable patterns to change based on its physiological history.
A second possible origin of macroscopic bioelectric patterns in development should be mentioned, although it is at present purely hypothetical. It is possible that at least some aspects of large-scale organismic bioelectric prepatterns are projected upon (represented in) subcellular domains on the surface of the egg cell. As the cells proliferate during embryonic cleavage stages, these domains would be partitioned into differential bioelectric properties among distinct cells (and of course become modified beyond simple expansion by the complex feedback loops in electrically-interacting cells). It is known that cells bear many different domains on their surface, as small as a few microns in size, each of which can support distinct Vmem even though adjacent (Adams and Levin, 2013; Kline et al., 1981; Martens et al., 2004; O’Connell et al., 2006; O’Connell and Tamkun, 2005). While the functional relevance of these domains is completely unknown, it is conceivable that even the bioelectric distributions on the surface of a single cell encode important information. If true, such a phenomenon on the surface of an egg cell provides a potential mechanism for passing complex patterns across generations without the need for the pattern encoding to start de novo in each generation (Neuhof et al., 2016). Evidence for such a conjecture can be sought for in future work using optogenetics and lipid raft targeting mechanisms in large eggs such as Xenopus, although it has already begun to be addressed as a computational medium in neuroscience (Wallace, 2007).
4. Unification: neural vs. non-neural bioelectric codes
How related are developmental bioelectric codes to the more familiar electrical signaling that encodes information in the nervous system? The mechanisms that implement the bioelectric code are not new, exotic inventions in the evolutionary process. These are ancient, primitive, highly-conserved properties that evolved from basic physiological functions in pre-metazoa which had to use ion flux to drive adaptive behavior, patterning, and metabolism all in one cell. It is therefore no coincidence that evolution speed-optimized these functions in organisms with brains, using nerves to drive the behavior of muscles (and thus rapidly move the organism through 3D space) while other cell types continue to talk to each other using slower bioelectric signals (moving the body configuration through morphospace (Stone, 1997)). We recently tested another prediction of this view at the level of evolutionary molecular biology, by asking how much overlap there was between genes involved in patterning and those involved in memory and cognition. We extracted all the genes from the REGene database that are implicated in regeneration (Zhao et al., 2016) and subjected these to a subnetwork enrichment analysis in Pathway Studio (Elsevier) as described in Pai et al. (2016) to determine what biological processes were related to this unique list. Of the more than ~2000 transcripts collected in “model species” (e.g. chimpanzee, frog, chicken, zebrafish, fruitfly, and others) from REGene, 1309 unique transcripts were mapped successfully into Pathway Studio using gene Name + Alias (Resnet 11.0 is a mammal-dominated database and not all transcripts collected from the model species in REGene could be mapped). The term “Regeneration” was ranked as the top biological process related to the transcripts extracted from the REGene database (Supplemental Data 1). There were a number of additional processes related to the gene set used for “regeneration”, notably learning/memory (enrichment P < 0.0001). Of the 700 transcripts related to learning/memory in the Resnet database, 177 of these overlapped with the original list from REGene, thus ~25% of transcripts implicated in regeneration are also involved in the process of learning/memory based upon the two databases (Supplemental Data 1 contains these transcripts). Of the genes that mapped successfully into Pathway Studio, there was 30% overlap between the two categories. The analysis also identified significant overlap between genes involved in learning/memory and those involved in development and cancer. Given the ancient origin of bioelectricity and the molecular conservation of mechanisms and algorithms by which the brain and body compute, how much overlap might there be between the developmental bioelectric code and the neural bioelectric code that underlies cognition?
We suggest that the bioelectric code is a linking nexus between cognitive neuroscience, developmental biology, and the field of primitive cognition. It is likely that many tissues in vivo are a kind of excitable medium, which supports morphological computation via a range of physiological signals, including bioelectric ones. If so, they are a remarkably useful proof-of-principle model for the field of unconventional computation – living tissues erase the boundary between the computational unit and the body it drives; they process information about the shape changes to make to their own structure. A computational architecture that can robustly reason about its own shape and change that shape ‘on the fly’ is the dream of today’s robotics and morphological computation communities (Doursat and Sanchez, 2014; Doursat et al., 2012, 2013; Fernandez et al., 2012). Indeed, recent studies have shown that brains retain information despite drastic remodeling – this is seen in the work (Blackiston et al., 2008) on persistence of memory during the transition from caterpillar to butterfly (in which the brain is largely destroyed and recreated), and in recent work (Shomrat and Levin, 2013) confirming McConnell’s early studies showing that when trained planaria are decapitated, their tails grow new heads and remember their original learning (Corning, 1966,1967). The interface of information encoding in brain and body, especially during contexts such as regeneration of the CNS in which both episodic and somatic memories must play a role, reveals the lack of a sharp dividing line between neuroscience and pattern formation.
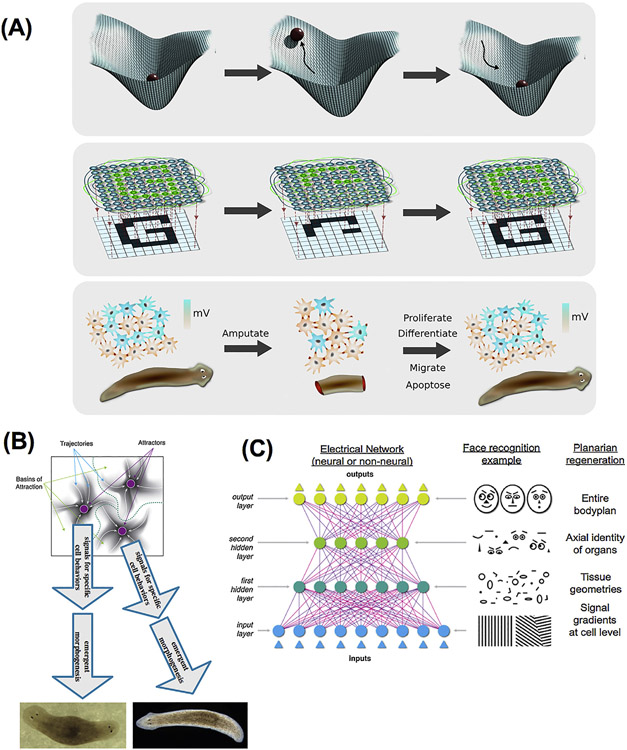
Information encoding in living tissue is the fundamental umbrella under which regenerative biology, neuroscience, robotics, etc. all operate. It is likely that conceptual tools of computational neuroscience will help crack the patterning bioelectric code, for example by determining how spatial patterns can be represented as memories within a (non)neural network (Fig. 7). It is just as likely that progress on decoding the simpler (?) encoding of patterns in non-neural contexts will help efforts to develop neural decodings (i.e., to extract first-person cognitive content from brain scans). Integrated cases (such as storage of learned memories in the bodies of planaria and their imprinting on the nascent brain) are expected not only to enrich our understanding of the material embodiments of mind, but to also drive transformative applications in memory technology. Research in regenerative biology is already driving extensions of computer science, such as the investigations of the stability properties of artificial neural networks under topology change (Hammelman et al., 2016); this has only begun to be explored, and is not only useful as a model for traumatic brain injury repair and computational psychiatry (Adams et al., 2016b) but also as an extension of the artificial neural network (ANN) paradigm and connectionist theory in general to non-neural substrates.
Fig. 7.
A neural network view of bioelectric circuits.
(A) One way to think about the regenerative (pattern-homeostatic) process is as a dynamical system in which the correct shape represents an attractor. Damage raises the energy of the system (here represented by the ball leaving the lowest-energy state), but it settles back to the correct state by a least-action mechanism minimizing error. The field of artificial neural networks (ANNs) and computational neuroscience, which have begun to explain the holographic storage of patterning information in networks, provides a number of conceptual frameworks (Hartwell et al., 1999) in which to understand the function of non-neural bioelectric networks as implementing a distributed, self-correcting pattern mechanism. (B) Specifically, in certain kinds of networks (Hopfield, 1982), attractors in their state space correspond to specific memories. We propose a model in which attractor states of bioelectric circuits correspond to specific anatomical layouts by controlling cell behaviors such as proliferation and differentiation. This provides a quantitative, mechanistic approach to understand how electrical signaling encodes pattern memories. Deforming the landscape, or altering cells’ interpretation of the network’s instructions, are both ways to manipulate the outcome. (C) Another important insight provided by the field of artificial neural networks is that of encoding “high level” items: middle layers of ANNs encode emergent features of the inputs; this provides a way to think about how cell signaling networks (in particular, electrical networks) could encode and decode information about parameters above the single cell level (organ size, topological arrangement, etc.). Images in panel C produced by Justin Guay of Peregrine Creative. Panel A produced by Alexis Pietak.
5. Conclusion
Most aspects of biomedicine boil down to the control of shape: birth defects, traumatic injury, aging, degenerative disease – all could potentially be resolved if we knew how to tell cells to build specific new complex structures to spec, on demand. Even cancer, the defection of cells from the body’s patterning cues, could be addressed via reprograming (normalization), as could the creation of arbitrarily-complex artificial constructs in vitro. Likewise, we cannot understand the evolutionary process until we really understand the relationship between genomes (what is targeted by mutation) and anatomy (what is tested for fitness by selection). It is consequently of the utmost importance to understand how shape is encoded in cellular properties and learn to control that shape. Ever-finer reductive drilldown into single-cell molecular events is not going to be sufficient; a complementary synthetic effort to understand the algorithms and encoded meaning (not just the mechanisms) of pattern regulation is essential.
Alongside the genetic and (chromatin) epigenetic codes operates a crucial set of controls called the bioelectric code. While the encoding of pattern outcomes to bioelectric states is only known for a small handful of examples, it is clear that profound lessons about the source and nature of information that determines anatomical pattern remain to be learned. Manipulation of long-term, large-scale anatomical structure can now be achieved by transiently re-writing the bioelectric states of living tissue. Work in tractable animal models is now beginning to be done in human cells (Golding et al., 2016; Li et al., 2016; McNamara et al., 2016; Pai et al., 2016; Sundelacruz et al., 2015), and this field has many applications in human therapeutics aimed at cellular control and the creation of novel bioengineered constructs (Kamm and Bashir, 2014).
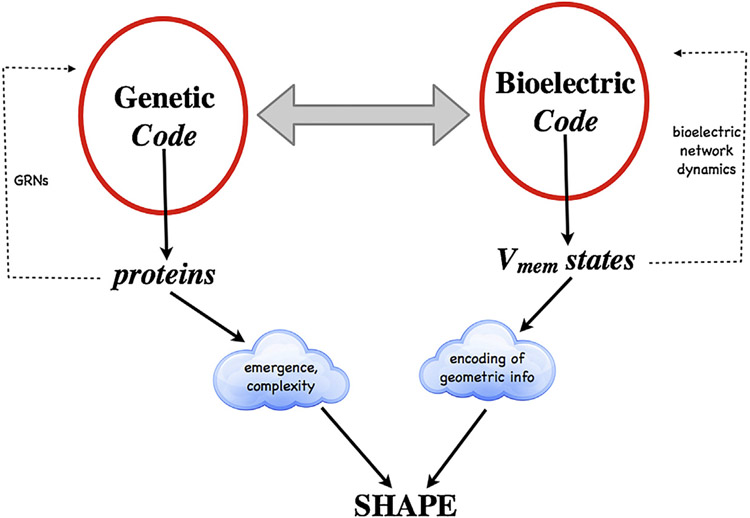
Bioelectricity is not just another pathway. It is uniquely suited for the control of complex outcomes (encoding) because voltage-sensitive ion channels and electrical synapses implement signaling elements that are sensitive to history (voltage-gated ion currents – in effect, transistors). Those elements are easily turned into memory circuits and logic gates, which in turn enable flexible, robust computational properties. Still, it is not just about bioelectricity; ionic circuits are only one layer of the morphogenetic code (Fig. 8). The main point is that biological pattern regulation is a combination of emergent features that fill in local features and top-down controls that make decisions about large-scale patterning. One direction for future work is to attempt to re-write the encoded goal state, instead of manipulating individual cells; to the extent that cells may be “universal constructors”, deforming their perception space (Bugaj et al., 2017) landscape and letting them do what they are good at (finding stable attractors within that space), might be a most efficient path to pattern control.
Fig. 8.
The relationship of the bioelectric and genetic code.
The genetic code specifies proteins via gene-regulatory networks, the activity of which results in emergent patterning events. Coupled to this (but having its own distinct dynamics and capabilities) is the bioelectric code, in which cell networks use direct, indirect, or neural-like representations of specific patterns at different scales of organization to specify instructions for modifying anatomy. Together, these codes serve as pattern memory over evolutionary timescales and provide distinct opportunities for biomedical intervention.
Major opportunities in this field includes the development of quantitative theory – conceptual models based on dynamical systems theory, least-action field-like principles, and Bayesian inference (Friston, 2013; Friston et al., 2014, 2010; Goodwin, 1985; Yoshida and Kaneko, 2009). Integrating fundamentals of bioelectrical dynamics with the increasing progress in the understanding of gene-regulatory networks and physical forces in patterning will be an essential next step. One important aspect however is scale-invariance: numerous aspects of pattern control, such as intercalation of positional information values, appear to work similarly in single cells as they do in multicellular organisms (Brandts, 1993; French et al., 1976; Jerka-Dziadosz et al., 1995). Thus, it may be possible to derive fundamental laws in which patterning outcomes can utilize diverse underlying mechanisms to implement them. If indeed information processing is ancient, one of such fundamental laws may be the drive to reduce high-level measurable such as surprise (Friston et al., 2010). Cells may have an innate capacity to learn to anticipate their environment and act in a way that maximizes reward; this has already been shown for cultured neurons in vitro, which can learn to operate a virtual flight simulator “body” (DeMarse and Dockendorf, 2005). Thus, direct training of non-neural tissues by providing positive and negative rein-forcement for patterning behavior could be a way to offload the computational complexity of patterning tasks from the bioengineer onto the living system, rewarding for desired behavior and letting the system self-organize internal processes to achieve the necessary goal.
This kind of generalized plasticity in service to specific outcomes is closely related to a key insight that drove the development of the computer science revolution – the independence of hardware and software, and the ability to run the same software on different hardware, or obtain different behavior from the same hardware by changing the software. If bioelectric dynamics running on genome-specified ion channel complements in cells can be treated as a kind of software, the next revolution in biology could be likewise driven in part by the realization that we do not have to manipulate living systems at the level of their “machine code” (affecting specific molecules), but at the level of information – re-writing the encoded goal states and thus gaining a more top-down control over growth and form with myriad applications to biomedicine (Levin, 2011b) and robust technology (Mange et al., 2000a,b; Tempesti et al., 2007, 1999, 2005).
Supplementary Material
Acknowledgements
This paper is dedicated to the memory of H. S. Burr, who was the first to suggest that bioelectric prepatterns form a code instructing future patterning events. M. L. gratefully acknowledges supported by an Allen Discovery Center Award from the Paul G. Allen Frontiers Group (12171), the National Institutes of Health (AR061988, GM078484, AR055993), the G. Harold and Leila Y. Mathers Charitable Foundation, the W. M. Keck Foundation (5903), the Templeton World Charity FoundationTWCF0089/AB55, and the National Science Foundation (DBI-1152279 and Emergent Behaviors of Integrated Cellular Systems subaward CBET-0939511).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.biosystems.2017.08.009.
References
- Aboobaker AA, 2011. Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol. 21, 304–311. [DOI] [PubMed] [Google Scholar]
- Adams DS, Levin M, 2012. General principles for measuring resting membrane potential and ion concentration using fluorescent bioelectricity reporters. Cold Spring Harb. Protoc 2012, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Levin M, 2013. Endogenous voltage gradients as mediators of cell–cell communication: strategies for investigating bioelectrical signals during pattern formation. Cell Tissue Res. 352, 95–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Robinson KR, Fukumoto T, Yuan S, Albertson RC, Yelick P, Kuo L, McSweeney M, Levin M, 2006. Early, H+-V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development 133, 1657–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Masi A, Levin M, 2007. H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development 134, 1323–1335. [DOI] [PubMed] [Google Scholar]
- Adams DS, Tseng AS, Levin M, 2013. Light-activation of the Archaerhodopsin H(+)-pump reverses age-dependent loss of vertebrate regeneration sparking system-level controls in vivo. Biol. Open 2, 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Lemire JM, Kramer RH, Levin M, 2014. Optogenetics in Developmental Biology: using light to control ion flux-dependent signals in Xenopus embryos. Int. J. Dev. Biol 58, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Uzel SG, Akagi J, Wlodkowic D, Andreeva V, Yelick PC, Devitt-Lee A, Pare JF, Levin M, 2016a. Bioelectric signalling via potassium channels: a mechanism for craniofacial dysmorphogenesis in KCNJ2-associated Andersen-Tawil Syndrome. J. Physiol 594, 3245–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams RA, Huys QJ, Roiser JP, 2016b. Computational Psychiatry: towards a mathematically informed understanding of mental illness. J. Neurol. Neurosurg. Psychiatry 87, 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M, 2008. Biosemiotics: a new understanding of life. Die Naturwissenschaften 95, 577–599. [DOI] [PubMed] [Google Scholar]
- Barkai N, Ben-Zvi D, 2009. ‘Big frog, small frog’?maintaining proportions in embryonic development. FEBS J. 276, 1196–1207. [DOI] [PubMed] [Google Scholar]
- Baslow MH, 2011. Biosemiosis and the cellular basis of mind how the oxidation of glucose by individual neurons in brain results in meaningful communications and in the emergence of mind. Biosemiotics-Neth 4, 39–53. [Google Scholar]
- Beane WS, Morokuma J, Adams DS, Levin M, 2011. A Chemical genetics approach reveals H,K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chem. Biol 18, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane WS, Morokuma J, Lemire JM, Levin M, 2013. Bioelectric signaling regulates head and organ size during planarian regeneration. Development 140, 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgacem YH, Borodinsky LN, 2015. Inversion of Sonic hedgehog action on its canonical pathway by electrical activity. Proc. Natl. Acad. Sci. U. S. A 112, 4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentrup FW,Jaffe LF, 1968. Analyzing the group effect: rheotropic responses of developing fucus eggs. Protoplasma 65, 25–35. [DOI] [PubMed] [Google Scholar]
- Bentrup F, Sandan T,Jaffe L, 1967. Induction of polarity in fucus eggs by potassium ion gradients. Protoplasma 64, 254. [Google Scholar]
- Benya EGF, Leal-Zanchet AM, Hauser J, 2017. Polyploidy as a chromosomal component of stochastic noise: variable scalar multiples of the diploid chromosome complement in the invertebrate species Girardia schubarti from Brazil. Braz. J. Biol 0. [DOI] [PubMed] [Google Scholar]
- Birnbaum KD, Alvarado AS, 2008. Slicing acros kingdoms: regeneration in plants and animals. Cell 132, 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackiston DJ, Silva Casey E, Weiss MR, 2008. Retention of memory through metamorphosis: can a moth remember what it learned as a caterpillar? PLoS One 3, e1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackiston DJ, McLaughlin KA, Levin M, 2009. Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle. ABBV Cell Cycle 8, 3519–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackiston D,Adams DS, Lemire JM, Lobikin M, Levin M, 2011. Transmembrane potential of GlyCl-expressing instructor cells induces a neoplastic-like conversion of melanocytes via a serotonergic pathway. Dis. Models Mech 4, 67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondos S, 2006. Variations on a theme: hox and Wnt combinatorial regulation during animal development. Sci. STKE 2006, pe38. [DOI] [PubMed] [Google Scholar]
- Borgens RB, Vanable JW Jr., Jaffe LF, 1977a. Bioelectricity and regeneration: large currents leave the stumps of regenerating newt limbs. Proc. Natl. Acad. Sci. U. S. A 74, 4528–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgens RB, Vanable JW Jr., Jaffe LF, 1977b. Bioelectricity and regeneration: i. Initiation of frog limb regeneration by minute currents. J. Exp. Zool 200, 403–416. [DOI] [PubMed] [Google Scholar]
- Borgens RB, McGinnis ME, Vanable JW Jr., Miles ES, 1984. Stump currents in regenerating salamanders and newts. J. Exp. Zool 231, 249–256. [DOI] [PubMed] [Google Scholar]
- Borgens R, Robinson K, Vanable J, McGinnis M, 1989. Electric Fields in Vertebrate Repair. Alan R. Liss, New York. [Google Scholar]
- Borgens RB, 1984. Are limb development and limb regeneration both initiated by an integumentary wounding? A hypothesis. Differentiation 28, 87–93. [DOI] [PubMed] [Google Scholar]
- Borgens RB, 1986. The role of natural and applied electric fields in neuronal regeneration and development. Prog. Clin. Biol. Res 210, 239–250. [PubMed] [Google Scholar]
- Brandts WA, 1993. A field model of left-right asymmetries in the pattern regulation of a cell. IMA J. Math. Appl. Med. Biol 10, 31–50. [DOI] [PubMed] [Google Scholar]
- Broccoli V, Colasante G, Sessa A, Rubio A, 2015. Histone modifications controlling native and induced neural stem cell identity. Curr. Opin. Genet. Dev 34, 95–101. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Sousounis K, Farkas JE, Bryant S, Thao N, Guzikowski AR, Monaghan JR, Levin M, Whited JL, 2017. Repeated removal of developing limb buds permanently reduces appendage size in the highly-regenerative axolotl. Dev. Biol 424, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubenik AB, Pavlansky R, 1965. Trophic responses to trauma in growing antlers. J. Exp. Zool 159, 289–302. [DOI] [PubMed] [Google Scholar]
- Bugaj LJ, O’Donoghue GP, Lim WA, 2017. Interrogating cellular perception and decision making with optogenetic tools. J. Cell Biol 216, 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr HS, Bullock TH, 1941. Steady state potential differences in the early development of Amblystoma. Yale J. Biol. Med 14, 51–57. [PMC free article] [PubMed] [Google Scholar]
- Burr HS, Northrop F, 1935a. The electrodynamic theory of life. Q. Rev. Biol 10, 322–333. [Google Scholar]
- Burr HS, Northrop FSC, 1935b. The electro-dynamic theory of life. Q. Rev. Biol 10, 322–333. [Google Scholar]
- Burr HS, Sinnott EW, 1944. Electrical correlates of form in cucurbit fruits. Am. J. Bot 31, 249–253. [Google Scholar]
- Burr HS, 1944. The meaning of bio-Electric potentials *. Yale J. Biol. Med 16, 353–360. [PMC free article] [PubMed] [Google Scholar]
- Buznikov G, Shmukler Y, Lauder J, 1996. From oocyte to neuron: do neurotransmitters function in the same way throughout development? Cell Mol. Neurobiol 16, 537–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buznikov GA, Lambert HW, Lauder JM, 2001. Serotonin and serotonin-like substances as regulators of early embryogenesis and morphogenesis. Cell Tissue Res. 305, 177–186. [DOI] [PubMed] [Google Scholar]
- Cao L, Wei D, Reid B, Zhao S, Pu J, Pan T, Yamoah E, Zhao M, 2013. Endogenous electric currents might guide rostral migration of neuroblasts. EMBO Rep. 14, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, McCaig CD, Scott RH, Zhao S, Milne G, Clevers H, Zhao M, Pu J, 2014. Polarizing intestinal epithelial cells electrically through Ror2. J. Cell Sci 127, 3233–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro K, Donnet C, Rejtar T, Karger BL, Barisone GA, Diaz E, Kortagere S, Lemire JM, Levin M, 2011. Histone deacetylase activity is necessary for left-right patterning during vertebrate development. BMC Dev. Biol 11, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Minc N, 2014. Electrochemical control of cell and tissue polarity. Ann. Rev. Cell Dev. Biol 30, 317–336. [DOI] [PubMed] [Google Scholar]
- Chernet BT, Levin M, 2013. Transmembrane voltage potential is an essential cellular parameter for the detection and control of tumordevelopment in a Xenopus model. Dis. Models Mech 6, 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernet BT, Levin M, 2014. Transmembrane voltage potential of somatic cells controls oncogene-mediated tumorigenesis at long-range. Oncotarget 5, 3287–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernet BT, Fields C, Levin M, 2015. Long-range gap junctional signaling controls oncogene-mediated tumorigenesis in Xenopus laevis embryos. Front. Physiol 5, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernet BT, Adams DS, Lobikin M, Levin M, 2016. Use of genetically encoded, light-gated ion translocators to control tumorigenesis. Oncotarget 7, 19575–19588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone CD, Tongier M, 1971. Control of somatic cell mitosis by simulated changes in the transmembrane potential level. Oncology 25, 168–182. [DOI] [PubMed] [Google Scholar]
- Cone CD Jr., 1970. Variation of the transmembrane potential level as a basic mechanism of mitosis control. Oncology 24, 438–470. [DOI] [PubMed] [Google Scholar]
- Cone CD, 1971. Unified theory on the basic mechanism of normal mitotic control and oncogenesis. J. Theor. Biol 30, 151–181. [DOI] [PubMed] [Google Scholar]
- Corning WC, 1966. Retention of a position discrimination after regeneration in planarians. Psychanomic Sci 5, 17–18. [Google Scholar]
- Corning WC, 1967. Regeneration and Retention of Acquired Information. NASA. [Google Scholar]
- Cortese B, Palama IE, D’Amone S, Gigli G, 2014. Influence of electrotaxis on cell behaviour. Integr. Biol. (Camb.) 6, 817–830. [DOI] [PubMed] [Google Scholar]
- Dahal GR, Rawson J, Gassaway B, Kwok B, Tong Y, Ptacek LJ, Bates E, 2012. An inwardly rectifying K+ channel is required for patterning Development 139, 3653–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal GR, Pradhan SJ, Bates EA, 2017. Inwardly rectifying potassium channels influence Drosophila wing morphogenesis by regulating Dpp release. Development 144, 2771–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson LA,Joshi SD, Kim HY, von Dassow M, Zhang L, Zhou J, 2010. Emergent morphogenesis: elastic mechanics of a self-deforming tissue. J. Biomech 43, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JA, Cachat E, 2016. Synthetic biology meets tissue engineering. Biochem. Soc. Trans 44, 696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarse TB, Dockendorf KP, 2005. Adaptive flight control with living neuronal networks on microelectrode arrays. Ieee Ijcnn, 1548–1551. [Google Scholar]
- Deglincerti A, Etoc F, Ozair MZ, Brivanlou AH, 2016. Self-Organization of spatial patterning in human embryonic stem cells. Curr. Top. Dev. Biol 116, 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Singla S, Toda H, Monje M, Palmer TD, Malenka RC, 2004. Excitation-neurogenesis coupling in adult neural stem/progenitor cells. Neuron 42, 535–552. [DOI] [PubMed] [Google Scholar]
- Dennett D, 1987. The Intentional Stance. MIT Press, Cambridge, Mass. [Google Scholar]
- Doursat R, Sanchez C, 2014. Growing fine-grained multicellular robots. Soft Rob. 1, 110–121. [Google Scholar]
- Doursat R, Sayama H, Michel O, 2012. Morphogenetic Engineering: Reconciling Self-Organization and Architecture. Morphogenetic Engineering: Toward Programmable Complex System., pp. 1–24. [Google Scholar]
- Doursat R, Sayama H, Michel O, 2013. A review of morphogenetic engineering. Nat. Comput 12, 517–535. [Google Scholar]
- Durant F, Morokuma J, Fields C, Williams K, Adams DS, Levin M, 2017. Long-term, stochastic editing of regenerative anatomy via targeting endogenous bioelectric gradients. Biophys. J 112, 2231–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons-Bell M, Durant F, Hammelman J, Bessonov N, Volpert V, Morokuma J, Pinet K, Adams DS, Pietak A, Lobo D, Levin M, 2015. Gap junctional blockade stochastically induces different species-specific head anatomies in genetically wild-type girardia dorotocephala flatworms. Int. J. Mol. Sci 16, 27865–27896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinella-Ferruzza N, 1956. The transformation of a tail into a limb after xenoplastic transformation. Experientia 15, 304–305. [Google Scholar]
- Fernandez JD, Lobo D, Martin GM, Doursat R, Vico FJ, 2012. Emergent diversity in an open-ended evolving virtual community. Artif. Life 18, 199–222. [DOI] [PubMed] [Google Scholar]
- French V, Bryant PJ, Bryant SV, 1976. Pattern regulation in epimorphic fields. Science 193, 969–981. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Daunizeau J, Kilner J, Kiebel SJ, 2010. Action and behavior: a free-energy formulation. Biol. Cybern 102, 227–260. [DOI] [PubMed] [Google Scholar]
- Friston K, Sengupta B, Auletta G, 2014. Cognitive dynamics: from attractors to active inference. Proc. IEEE 102, 427–445. [Google Scholar]
- Friston K, 2013. Active inference and free energy. Behav. Brain Sci 36, 212–213. [DOI] [PubMed] [Google Scholar]
- Fukumoto T, Levin M, 2003. Serotonin is a novel very early signaling mechanism in left-right asymmetry. Dev. Biol 259, 490–490. [Google Scholar]
- Fukumoto T, Kema IP, Levin M, 2005. Serotonin signaling is a very early step in patterning of the left-right axis in chick and frog embryos. Curr. Biol 15, 794–803. [DOI] [PubMed] [Google Scholar]
- Gao R, Zhao S, Jiang X, Sun Y, Gao J, Borleis J, Willard S, Tang M, Cai H, Kamimura Y, Huang Y,Jiang J, Huang Z, Mogilner A, Pan T, Devreotes PN, Zhao M, 2015. A large-scale screen reveals genes that mediate electrotaxis in Dictyostelium discoideum. Sci. Signal 8, ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding A, Guay JA, Herrera-Rincon C, Levin M, Kaplan DL, 2016. A tunable silk hydrogel device for studying limb regeneration in adult xenopus laevis. PLoS One 11, e0155618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin BC, Pateromichelakis S, 1979. The role of electrical fields, ions, and the cortex in the morphogenesis of acetabularia. Planta 145, 427–435. [DOI] [PubMed] [Google Scholar]
- Goodwin BC, 1985. Developing organisms as self-organizing fields. In: Antonelli PL (Ed.), Mathematical Essays on Growth and the Emergence of Form. University of Alberta Press, Alberta. [Google Scholar]
- Gruler H, Nuccitelli R, 1991. Neural crest cell galvanotaxis – new data and a novel-approach to the analysis of both galvanotaxis and chemotaxis. Cell Motil. Cytoskeleton 19, 121–133. [DOI] [PubMed] [Google Scholar]
- Halley JD, Smith-Miles K, Winkler DA, Kalkan T, Huang S, Smith A, 2012. Self-organizing circuitry and emergent computation in mouse embryonic stem cells. Stem Cell Res. 8, 324–333. [DOI] [PubMed] [Google Scholar]
- Hammelman J, Lobo D, Levin M, 2016. Artificial neural networks as models of robustness in development and regeneration: stability of memory during morphological remodeling. Artif. Neural Netw. Model 628, 45–65. [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW, 1999. From molecular to modular cell biology. Nature 402, C47–52. [DOI] [PubMed] [Google Scholar]
- Hoffmeyer J, 2000. Code-duality and the epistemic cut. Ann. N. Y. Acad. Sci 901, 175–186. [DOI] [PubMed] [Google Scholar]
- Hopfield JJ, 1982. Neural networks and physical systems with emergent collective computational abilities. Proc. Natl. Acad. Sci. U. S. A 79, 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotary KB, Robinson KR, 1990. Endogenous electrical currents and the resultant voltage gradients in the chick embryo. Dev. Biol 140, 149–160. [DOI] [PubMed] [Google Scholar]
- Hotary KB, Robinson KR, 1991. The neural tube of the Xenopus embryo maintains a potential difference across itself. Brain Res. Dev. Brain Res 59, 65–73. [DOI] [PubMed] [Google Scholar]
- Hotary KB, Robinson KR, 1992. Evidence of a role for endogenous electrical fields in chick embryo development. Development 114, 985–996. [DOI] [PubMed] [Google Scholar]
- Humphries J, Xiong L, Liu J, Prindle A, Yuan F, Arjes HA, Tsimring L, Suel GM, 2017. Species-independent attraction to biofilms through electrical signaling. Cell 168, 200–209 (e212). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimatsu K, Takamatsu A, Takeda H, 2010. Emergence of traveling waves in the zebrafish segmentation clock. Development 137, 1595–1599. [DOI] [PubMed] [Google Scholar]
- Jaffe LF, Nuccitelli R, 1977. Electrical controls of development. Ann. Rev. Biophys. Bioeng 6, 445–476. [DOI] [PubMed] [Google Scholar]
- Jaffe LF, 1979. Control of development by ionic currents. In: Cone RA, John E Dowling (Eds.), Membrane Transduction Mechanisms. Raven Press, New York. [Google Scholar]
- Jerka-Dziadosz M, Jenkins LM, Nelsen EM, Williams NE, Jaeckel-Williams R, Frankel J, 1995. Cellular polarity in ciliates: persistence of global polarity in a disorganized mutant of Tetrahymena thermophila that disrupts cytoskeletal organization. Dev. Biol 169, 644–661. [DOI] [PubMed] [Google Scholar]
- Kamm RD, Bashir R, 2014. Creating living cellular machines. Ann. Biomed. Eng 42, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzer F, van Duijn M, Lyon P, 2013. What nervous systems do: early evolution, input-output, and the skin brain thesis. Adapt. Behav 21, 67–85. [Google Scholar]
- Kiortsis V, Moraitou M, 1965. Factors of regeneration in Spirographis spallanzanii. In: Trampusch V.K.a.H.A.L. (Ed.), Regeneration in Animals and Related Problems. , pp. 250–261 (Amsterdam: ). [Google Scholar]
- Kline D, Nuccitelli R, Robinson K, 1981. Ion currents in the cleaving egg and mid-cleavage blastomeres of the amphibian embryo. J. Cell Biol 91, A181–A181. [Google Scholar]
- Kline D, Robinson KR, Nuccitelli R, 1983. Ion currents and membrane domains in the cleaving Xenopus egg.J. Cell Biol 97, 1753–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Schnapp E, Epperlein HH, Tanaka EM, 2008. Novel insights into the flexibility of cell and positional identity during urodele limb regeneration. Cold Spring Harb. Symp. Quant. Biol 73, 583–592. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM, 2009. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65. [DOI] [PubMed] [Google Scholar]
- Kralj JM, Hochbaum DR, Douglass AD, Cohen AE, 2011. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science 333, 345–348. [DOI] [PubMed] [Google Scholar]
- Kucerova R, Walczysko P, Reid B, Ou J, Leiper LJ, Rajnicek AM, McCaig CD, Zhao M, Collinson JM, 2011. The role of electrical signals in murine corneal wound re-epithelialization. J. Cell. Physiol 226, 1544–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Canaday DJ, Doong H, 1993. A review of the biophysical basis for the clinical application of electric fields in soft-tissue repair. J. Burn Care Rehabil 14, 319–335. [DOI] [PubMed] [Google Scholar]
- Levin M, Thorlin T, Robinson KR, Nogi T, Mercola M, 2002. Asymmetries in H+/K+-ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell 111, 77–89. [DOI] [PubMed] [Google Scholar]
- Levin M, Buznikov GA, Lauder JM, 2006. Of minds and embryos: left-right asymmetry and the serotonergic controls of pre-neural morphogenesis. Dev. Neurosci 28, 171–185. [DOI] [PubMed] [Google Scholar]
- Levin M, Pezzulo G, Finkelstein JM, 2017. Endogenous bioelectric signaling networks: exploiting voltage gradients for control of growth and form. Annu. Rev. Biomed. Eng 19, 353–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, 2011a. Endogenous bioelectric signals as morphogenetic controls of development, regeneration, and neoplasm. In: Pullar CE (Ed.), The Physiology of Bioelectricity in Development, Tissue Regeneration, and Cancer. CRC Press, Boca Raton, FL, pp. 39–89. [Google Scholar]
- Levin M, 2011b. The wisdom of the body: future techniques and approaches to morphogenetic fields in regenerative medicine, developmental biology and cancer. Regen. Med 6, 667–673. [DOI] [PubMed] [Google Scholar]
- Levin M, 2014. Molecular bioelectricity: how endogenous voltage potentials control cell behavior and instruct pattern regulation in vivo. Mol. Biol. Cell 25, 3835–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Levin M, Kaplan DL, 2016. Bioelectric modulation of macrophage polarization. Sci. Rep 6, 21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH, 2011. Evolution of sodium channels predates the origin of nervous systems in animals. Proc. Natl. Acad. Sci. U. S. A 108, 9154–9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH, 2015. Convergence of ion channel genome content in early animal evolution. Proc. Natl. Acad. Sci. U. S. A 112, E846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Tonegawa S, 2014. Inception of a false memory by optogenetic manipulation of a hippocampal memory engram. Philos. Trans. R Soc. Lond. B Biol. Sci 369, 20130142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobikin M, Chernet B, Lobo D, Levin M, 2012. Resting potential, oncogene-induced tumorigenesis, and metastasis: the bioelectric basis of cancer in vivo. Phys. Biol 9, 065002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobikin M, Lobo D, Blackiston DJ, Martyniuk CJ, Tkachenko E, Levin M, 2015. Serotonergic regulation of melanocyte conversion: a bioelectrically regulated network for stochastic all-or-none hyperpigmentation. Sci. Signal 8, ra99. [DOI] [PubMed] [Google Scholar]
- Lobo D, Beane WS, Levin M, 2012. Modeling planarian regeneration: a primer for reverse-engineering the worm. PLoS Comput. Biol 8, e1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo D, Solano M, Bubenik GA, Levin M, 2014. A linear-encoding model explains the variability of the target morphology in regeneration. Journal of the Royal Society. Interface/ R. Soc 11, 20130918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, 1947. Bioelectric Fields and Growth. Univ. of Texas Press, Austin. [Google Scholar]
- MacFarlane SN, Sontheimer H, 2000. Changes in ion channel expression accompany cell cycle progression of spinal cord astrocytes. Glia 30, 39–48. [DOI] [PubMed] [Google Scholar]
- Maden M, 2008. Axolotl/newt. Methods Mol. Biol 461, 467–480. [DOI] [PubMed] [Google Scholar]
- Mange D, Sipper M, Stauffer A, Tempesti G, 2000a. Toward robust integrated circuits: the embryonics approach. Proc. Ieee 88, 516–541. [Google Scholar]
- Mange D, Sipper M, Stauffer A, Tempesti G, 2000b. Toward self-repairing and self-replicating hardware: the Embryonics approach. Second NASA/DoD Workshop on Evolvable Hardware, Proceedings, 205–214. [Google Scholar]
- Marsh G, Beams HW, 1952. Electrical control of morphogenesis in regenerating dugesia tigrina: I. Relation of axial polarity to field strength. J. Cell Comp. Physiol 39, 191–213. [DOI] [PubMed] [Google Scholar]
- Martens JR, O’Connell K, Tamkun M, 2004. Targeting of ion channels to membrane microdomains: localization of KV channels to lipid rafts. Trends Pharmacol. Sci 25, 16–21. [DOI] [PubMed] [Google Scholar]
- Martin-Granados C, McCaig CD, 2014. Harnessing the electric spark of life to cure skin wounds. Adv. Wound Care (New Rochelle) 3, 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews J, Levin M, 2016. Gap junctional signaling in pattern regulation: physiological network connectivity instructs growth and form. Dev. Neurobiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews AP, 1903. Electrical polarity in the hydroids. Am. J. Physiol 8, 294–299. [Google Scholar]
- McCaig CD, Zhao M, 1997. Physiological electrical fields modify cell behaviour. Bioessays 19, 819–826. [DOI] [PubMed] [Google Scholar]
- McCaig CD, Rajnicek AM, Song B, Zhao M, 2005. Controlling cell behavior electrically: current views and future potential. Physiol. Rev 85, 943–978. [DOI] [PubMed] [Google Scholar]
- McCaig CD, 1986. Electric fields, contact guidance and the direction of nerve growth. J. Embryol. Exp. Morphol 94, 245–255. [PubMed] [Google Scholar]
- McCusker CD, Gardiner DM, 2013. Positional information is reprogrammed in blastema cells of the regenerating limb of the axolotl (Ambystoma mexicanum). PLoS One 8, e77064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker CD, Gardiner DM, 2014. Understanding positional cues in salamander limb regeneration: implications for optimizing cell-based regenerative therapies. Dis. Models Mech 7, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara HM, Zhang H, Werley CA, Cohen AE, 2016. Optically controlled oscillators in an engineered bioelectric tissue. Phys. Rev. X 6, 031001. [Google Scholar]
- Mondia JP, Levin M, Omenetto FG, Orendorff RD, Branch MR, Adams DS, 2011. Long-distance signals are required for morphogenesis of the regenerating Xenopus tadpole tail, as shown by femtosecond-laser ablation. PLoS One 6, e24953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D, Walker SI, Levin M, 2017. Cancer as a disorder of patternng information: computational and biophysical perspectives on the cancer problem. Convergent Sci. Phys. Oncol (in press). [Google Scholar]
- Moran Y, Barzilai MG, Liebeskind BJ, Zakon HH, 2015. Evolution of voltage-gated ion channels at the emergence of Metazoa.J. Exp. Biol 218, 515–525. [DOI] [PubMed] [Google Scholar]
- Morgan TH, 1904. The control of heteromorphosis in Planaria maculata. Arch f Entw Mech Bd 17, 683–694. [Google Scholar]
- Morokuma J, Blackiston D, Adams DS, Seebohm G, Trimmer B, Levin M, 2008. Modulation of potassium channel function confers a hyperproliferative invasive phenotype on embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A 105, 16608–16613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustard J, Levin M, 2014. Bioelectrical mechanisms for programming growth and form: taming physiological networks for soft body robotics. Soft Rob. 1, 169–191. [Google Scholar]
- Mycielska ME, Djamgoz MB, 2004. Cellular mechanisms of direct-current electric field effects: galvanotaxis and metastatic disease. J. Cell Sci 117, 1631–1639. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Zhu K, Sun YH, Hegyi B, Zeng Q, Murphy CJ, Small JV, Chen-Izu Y, Izumiya Y, Penninger JM, Zhao M, 2015. KCNJ15/Kir4.2 couples with polyamines to sense weak extracellular electric fields in galvanotaxis. Nat. Commun 6, 8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naselaris T, Prenger RJ, Kay KN, Oliver M, Gallant JL, 2009. Bayesian reconstruction of natural images from human brain activity. Neuron 63, 902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhof M, Levin M, Rechavi O, 2016. Vertically- and horizontally-transmitted memories – the fading boundaries between regeneration and inheritance in planaria. Biol Open 5, 1177–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto S, Vu AT, Naselaris T, Benjamini Y, Yu B, Gallant JL, 2011. Reconstructing visual experiences from brain activity evoked by natural movies. Curr. Biol.: CB 21, 1641–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura O, Hosoda K, Kawaguchi E, Yazawa S, Hayashi T, Inoue T, Umesono Y, Agata K, 2015. Unusually large number of mutations in asexually reproducing clonal planarian dugesia japonica. PLoS One 10, e0143525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi T, Levin M, 2005. Characterization of innexin gene expression and functional roles of gap-junctional communication in planarian regeneration. Dev. Biol 287, 314–335. [DOI] [PubMed] [Google Scholar]
- Novák B, Bentrup FW, 1972. An electrophysiological study of regeneration in Acetabularia Mediterranea. Planta 108, 227–244. [DOI] [PubMed] [Google Scholar]
- Nuccitelli R, Erickson CA, 1983. Embryonic cell motility can be guided by physiological electric fields. Exp. Cell Res 147, 195–201. [DOI] [PubMed] [Google Scholar]
- Nuccitelli R, Robinson K,Jaffe L, 1986. On electrical currents in development. Bioessays 5, 292–294. [DOI] [PubMed] [Google Scholar]
- Nuccitelli R, 1983. Transcellular ion currents: signals and effectors of cell polarity. Mod. Cell Biol 2, 451–481. [Google Scholar]
- Nuccitelli R, 1986. A two-dimensional extracellular vibrating probe for the detection of trans-cellular ionic currents.J. Cell Biol 103, A519–A519. [Google Scholar]
- Nuccitelli R, 1987. A two-dimensional extracellular vibrating probe for the detection of trans-cellular ionic currents. Biophys. J 51, A447–A447. [Google Scholar]
- Nuccitelli R, 1988. Ionic currents in morphogenesis. Experientia 44, 657–666. [DOI] [PubMed] [Google Scholar]
- O’Connell KM, Tamkun MM, 2005. Targeting of voltage-gated potassium channel isoforms to distinct cell surface microdomains. J. Cell Sci 118, 2155–2166. [DOI] [PubMed] [Google Scholar]
- O’Connell KM, Rolig AS, Whitesell JD, Tamkun MM, 2006. Kv2.1 potassium channels are retained within dynamic cell surface microdomains that are defined by a perimeter fence. J. Neurosci 26, 9609–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouadid-Ahidouch H, Ahidouch A, 2008. K+ channel expression in human breast cancer cells: involvement in cell cycle regulation and carcinogenesis. J. Membr. Biol 221, 1–6. [DOI] [PubMed] [Google Scholar]
- Ouadid-Ahidouch H, Ahidouch A, 2013. K(+) channels and cell cycle progression in tumor cells. Front. Physiol 4, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouadid-Ahidouch H, Ahidouch A, Pardo LA, 2016. Kv10.1 K(+) channel: from physiology to cancer. Pflugers Arch. 468, 751–762. [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Beane WS, 2009. Regeneration: the origin of cancer or a possible cure? Semin. Cell Dev. Biol 20, 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo NJ, Nicolas CL, Adams DS, Levin M, 2008. Live imaging of planarian membrane potential using DiBAC4(3). Cold Spring Harb. Protoc 2008 (pdb.prot5055). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo NJ, Morokuma J, Walentek P, Kema IP, Gu MB, Ahn JM, Hwang JS, Gojobori T, Levin M, 2010. Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev. Biol 339, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai VP, Aw S, Shomrat T, Lemire JM, Levin M, 2012. Transmembrane voltage potential controls embryonic eye patterning in Xenopus laevis. Development 139, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai VP, Lemire JM, Chen Y, Lin G, Levin M, 2015a. Local and long-range endogenous resting potential gradients antagonistically regulate apoptosis and proliferation in the embryonic CNS. Int. J. Dev. Biol 59, 327–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai VP, Lemire JM, Pare JF, Lin G, Chen Y, Levin M, 2015b. Endogenous gradients of resting potential instructively pattern embryonic neural tissue via notch signaling and regulation of proliferation.J. Neurosci 35, 4366–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai VP, Martyniuk CJ, Echeverri K, Sundelacruz S, Kaplan DL, Levin M, 2016. Genome-wide analysis reveals conserved transcriptional responses downstream of resting potential change in Xenopus embryos, axolotl regeneration, and human mesenchymal cell differentiation. Regeneration (Oxf.) 3, 3–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios-Prado N, Bukauskas FF, 2009. Heterotypic gap junction channels as voltage-sensitive valves for intercellular signaling. Proc. Natl. Acad. Sci. U. S. A 106, 14855–14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Borgens RB, 2012. Strict perpendicular orientation of neural crest-derived neurons in vitro is dependent on an extracellular gradient of voltage. J. Neurosci. Res 90, 1335–1346. [DOI] [PubMed] [Google Scholar]
- Pattee HH, 1982. Cell psychology: an evolutionary approach to the symbol-matter problem. Cogn. Brain Theory 5, 325–341. [Google Scholar]
- Pattee HH, 2001. The physics of symbols: bridging the epistemic cut. Biosystems 60, 5–21. [DOI] [PubMed] [Google Scholar]
- Peiris TH, Ramirez D, Barghouth PG, Ofoha U, Davidian D, Weckerle F, Oviedo NJ, 2016. Regional signals in the planarian body guide stem cell fate in the presence of genomic instability. Development 143, 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perathoner S, Daane JM, Henrion U, Seebohm G, Higdon CW, Johnson SL, Nusslein-Volhard C, Harris MP, 2014. Bioelectric signaling regulates size in zebrafish fins. PLoS Genet. 10, e1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo G, Levin M, 2015. Re-membering the body: applications of computational neuroscience to the top-down control of regeneration of limbs and other complex organs. Integr. Biol. (Camb.) 7, 1487–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo G, Levin M, 2016. Top-down models in biology: explanation and control of complex living systems above the molecular level. J. R. Soc. Interface 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietak A, Levin M, 2016. Exploring instructive physiological signaling with the bioelectric tissue simulation engine (BETSE). Front. Bioeng. Biotechnol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prindle A, Liu J, Asally M, Ly S, Garcia-Ojalvo J, Suel GM, 2015. Ion channels enable electrical communication in bacterial communities. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney LK, Barber DL, 2003. Na-H exchange-dependent increase in intracellular pH times G2/M entry and transition. J. Biol. Chem 278, 44645–44649. [DOI] [PubMed] [Google Scholar]
- Qiu C, Shivacharan RS, Zhang M, Durand DM, 2015. Can neural activity propagate by endogenous electrical field? J. Neurosci 35, 15800–15811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quian Quiroga R, Panzeri S, 2009. Extracting information from neuronal populations: information theory and decoding approaches. Nature reviews. Neuroscience 10, 173–185. [DOI] [PubMed] [Google Scholar]
- Ramirez S, Liu X, Lin PA, Suh J, Pignatelli M, Redondo RL, Ryan TJ, Tonegawa S, 2013. Creating a false memory in the hippocampus. Science 341, 387–391. [DOI] [PubMed] [Google Scholar]
- Raspopovic J, Marcon L, Russo L, Sharpe J, 2014. Modeling digits: digit patterning is controlled by a Bmp-Sox9-Wnt turing network modulated by morphogen gradients. Science 345, 566–570. [DOI] [PubMed] [Google Scholar]
- Reid B, Zhao M, 2014. The electrical response to injury: molecular mechanisms and wound healing. Adv. Wound Care (New Rochelle) 3, 184–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY, 2006. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2, e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roos AD, Willems PH, Peters PH, van Zoelen EJ, Theuvenet AP, 1997. Synchronized calcium spiking resulting from spontaneous calcium action potentials in monolayers of NRK fibroblasts. Cell Calcium 22, 195–207. [DOI] [PubMed] [Google Scholar]
- Rosen R, 1985. Anticipatory Systems: Philosophical, Mathematical, and Methodological Foundations, 1 st ed. Pergamon Press Oxford, England; New York. [Google Scholar]
- Roshchina VV, 2016. New trends and perspectives in the evolution of neurotransmitters in microbial, plant, and animal cells. Adv. Exp. Med. Biol 874, 25–77. [DOI] [PubMed] [Google Scholar]
- Salo E, Abril JF, Adell T, Cebria F, Eckelt K, Fernandez-Taboada E, Handberg-Thorsager M, Iglesias M, Molina MD, Rodriguez-Esteban G, 2009. Planarian regeneration: achievements and future directions after 20 years of research. Int.J. Dev. Biol 53, 1317–1327. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Gonzalez-Zulueta M, Huang H, Herring WJ, Ahn SY, Ginty DD, Dawson VL, Dawson TM, 2000. Dynamic regulation of neuronal NO synthase transcription by calcium influx through a CREB family transcription factor-dependent mechanism. Proc. Natl. Acad. Sci. U. S. A 97, 8617–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmann Y, 2005. Induction and the Turing-Child field in development. Prog. Biophys. Mol. Biol 89, 36–92. [DOI] [PubMed] [Google Scholar]
- Shi R, Borgens RB, 1995. Three-dimensional gradients of voltage during development of the nervous system as invisible coordinates for the establishment of embryonic pattern. Dev. Dyn 202, 101–114. [DOI] [PubMed] [Google Scholar]
- Shibata N, Rouhana L, Agata K, 2010. Cellular and molecular dissection of pluripotent adult somatic stem cells in planarians. Dev. Growth Differ 52, 27–41. [DOI] [PubMed] [Google Scholar]
- Shomrat T, Levin M, 2013. An automated training paradigm reveals long-term memory in planarians and its persistence through head regeneration. J. Exp. Biol 216, 3799–3810. [DOI] [PubMed] [Google Scholar]
- Stone JR, 1997. The spirit of D’arcy Thompson dwells in empirical morphospace. Math. Biosci 142, 13–30. [DOI] [PubMed] [Google Scholar]
- Sullivan KG, Levin M, 2016. Neurotransmitter signaling pathways required for normal development in Xenopus laevis embryos: a pharmacological survey screen. J. Anat 229, 483–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KG, Emmons-Bell M, Levin M, 2016. Physiological inputs regulate species-specific anatomy during embryogenesis and regeneration. Commun. Integr. Biol 9, e1192733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundelacruz S, Levin M, Kaplan DL, 2008. Membrane potential controls adipogenic and osteogenic differentiation of mesenchymal stem cells. PLoS One 3, e3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundelacruz S, Levin M, Kaplan DL, 2009. Role of membrane potential in the regulation of cell proliferation and differentiation. Stem Cell Rev. Rep 5, 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundelacruz S, Levin M, Kaplan DL, 2015. Comparison of the depolarization response of human mesenchymal stem cells from different donors. Sci. Rep 5, 18279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swapna I, Borodinsky LN, 2012. Interplay between electrical activity and bone morphogenetic protein signaling regulates spinal neuron differentiation. Proc. Natl. Acad. Sci. U. S. A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai G, Reid B, Cao L, Zhao M, 2009. Electrotaxis and wound healing: experimental methods to study electric fields as a directional signal for cell migration. Methods Mol. Biol 571, 77–97. [DOI] [PubMed] [Google Scholar]
- Tempesti G, Mange D, Stauffer A, 1999. The embryonics project: a machine made of artificial cells. Riv. Biol.-Biol. Forum 92, 143–188. [PubMed] [Google Scholar]
- Tempesti G, Mange D, Stauffer A, 2005. Bio-inspired computing architectures: the embryonics approach, CAMP 2005: seventh international workshop on computer architecture for machine perception. Proceedings, 3–10. [Google Scholar]
- Tempesti G, Mange D, Mudry PA, Rossier J, Stauffer A, 2007. Self-replicating hardware for reliability: the embryonics project. Acm J. Emerg. Technol. Comput 3. [Google Scholar]
- Toko K, Souda M, Matsuno T, Yamafuji K, 1990. Oscillations of electrical potential along a root of a higher plant. Biophys. J 57, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Sporns O, 2003. Measuring information integration. BMC Neurosci. 4, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Sporns O, Edelman GM, 1994. A measure for brain complexity: relating functional segregation and integration in the nervous system. Proc. Natl. Acad. Sci. U. S. A 91, 5033–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Edelman GM, Sporns O, 1998. Complexity and coherency: integrating information in the brain. Trends Cogn. Sci 2, 474–484. [DOI] [PubMed] [Google Scholar]
- Tseng A, Levin M, 2013. Cracking the bioelectric code: probing endogenous ionic controls of pattern formation. Commun. Integr. Biol 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AS, Beane WS, Lemire JM, Masi A, Levin M, 2010. Induction of vertebrate regeneration by a transient sodium current. J. Neurosci 30, 13192–13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Warton K, Musgrove EA, Campbell TJ, Breit SN, 2000. The nuclear chloride ion channel NCC27 is involved in regulation of the cell cycle. J. Physiol 529 (Pt 3), 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Morrie RD, Adams DS, 2011. V-ATPase-dependent ectodermal voltage and pH regionalization are required for craniofacial morphogenesis. Dev. Dyn 240, 1889–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Adams DS, Levin M, 2012. Normalized shape and location of perturbed craniofacial structures in the Xenopus tadpole reveal an innate ability to achieve correct morphology. Dev. Dyn 241, 863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, di Porzio U, Robinson DA, Shreeve SM, Lai J, Kerlavage AR, Fracek SP Jr., Lentes KU, Fraser CM, 1988. Evolution of neurotransmitter receptor systems. Prog. Neurobiol 30, 105–169. [DOI] [PubMed] [Google Scholar]
- Vogg MC, Owlarn S, Perez Rico YA, Xie J, Suzuki Y, Gentile L, Wu W, Bartscherer K, 2014. Stem cell-dependent formation of a functional anterior regeneration pole in planarians requires Zic and Forkhead transcription factors. Dev. Biol 390, 136–148. [DOI] [PubMed] [Google Scholar]
- Wahlgren J, De LKT, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H, 2012. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 40, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R, 2007. Neural membrane microdomains as computational systems: toward molecular modeling in the study of neural disease. Biosystems 87, 20–30. [DOI] [PubMed] [Google Scholar]
- Wang X, Veruki ML, Bukoreshtliev NV, Hartveit E, Gerdes HH, 2010. Animal cells connected by nanotubes can be electrically coupled through interposed gap-junction channels. Proc. Natl. Acad. Sci. U. S. A 107, 17194–17199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kondo S, 2015. Is pigment patterning in fish skin determined by the Turing mechanism? Trends Genet.: TIG 31, 88–96. [DOI] [PubMed] [Google Scholar]
- Witchley JN, Mayer M,Wagner DE, Owen JH, Reddien PW, 2013. Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, 1977. Studies on handedness of fiddler crab, Uca Lactea. Biol. Bull 152, 424–436. [Google Scholar]
- Yamashita T, Pala A, Pedrido L, Kremer Y, Welker E, Petersen CC, 2013. Membrane potential dynamics of neocortical projection neurons driving target-specific signals. Neuron 80, 1477–1490. [DOI] [PubMed] [Google Scholar]
- Yamashita M, 2013. Electric axon guidance in embryonic retina: galvanotropism revisited. Biochem. Biophys. Res. Commun 431, 280–283. [DOI] [PubMed] [Google Scholar]
- Yao L, Pandit A, Yao S, McCaig CD, 2011. Electric field-guided neuron migration: a novel approach in neurogenesis. Tissue Eng. Part B Rev 17, 143–153. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kaneko K, 2009. Unified description of regeneration by coupled dynamical systems theory: intercalary/segmented regeneration in insect legs. Dev. Dyn 238, 1974–1983. [DOI] [PubMed] [Google Scholar]
- Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, Wang F, Guo A, Walczysko P, Gu Y, Sasaki T, Suzuki A, Forrester JV, Bourne HR, Devreotes PN, McCaig CD, Penninger JM, 2006. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature 442, 457–460. [DOI] [PubMed] [Google Scholar]
- Zhao M, Rotgans B, Wang T, Cummins SF, 2016. REGene: a literature-based knowledgebase of animal regeneration that bridge tissue regeneration and cancer. Sci. Rep 6, 23167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jia R, Qian Y, Ye Y, Liu C, 2015. Correlation between electric potential and peristaltic behavior in Physarum polycephalum. Biol. Syst 132–133, 13–19. [DOI] [PubMed] [Google Scholar]
- Zimmermann MR, Maischak H, Mithofer A, Boland W, Felle HH, 2009. System potentials, a novel electrical long distance apoplastic signal in plants, induced by wounding. Plant Physiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.