Abstract
Introduction
Critical Care is a specialty in medicine providing a service for severely ill and high-risk patients who, due to the nature of their condition, may require long periods recovering after discharge. Consequently, focus on the routine data collection carried out in Intensive Care Units (ICUs) leads to reporting that is confined to the critical care episode and is typically insensitive to variation in individual patient pathways through critical care to recovery.
A resource which facilitates efficient research into interactions with healthcare services surrounding critical admissions, capturing the complete patient’s healthcare trajectory from primary care to non-acute hospital care prior to ICU, would provide an important longer-term perspective for critical care research.
Objective
To describe and apply a reproducible methodology that demonstrates how both routine administrative and clinically rich critical care data sources can be integrated with primary and secondary healthcare data to create a single dataset that captures a broader view of patient care.
Method
To demonstrate the INTEGRATE methodology, it was applied to routine administrative and clinical healthcare data sources in the Secure Anonymised Data Linking (SAIL) Databank to create a dataset of patients’ complete healthcare trajectory prior to critical care admission. SAIL is a national, data safe haven of anonymised linkable datasets about the population of Wales.
Results
When applying the INTEGRATE methodology in SAIL, between 2010 and 2019 we observed 91,582 critical admissions for 76,019 patients. Of these, 90,632 (99%) had an associated non-acute hospital admission, 48,979 (53%) had an emergency admission, and 64,832 (71%) a primary care interaction in the week prior to the critical care admission.
Conclusion
This methodology, at population scale, integrates two critical care data sources into a single dataset together with data sources on healthcare prior to critical admission, thus providing a key research asset to study critical care pathways.
Keywords: intensive care, critical care, electronic health records, linkable research data, ICNARC
Introduction
Increasing volumes of electronic healthcare data are being collected on a routine basis by clinical and administrative health systems and represent a valuable resource for research [1–6]. Given the complexities of critical illness and of critical care monitoring and interventions, Intensive Care Units (ICUs) and High Dependency Units (HDUs) represent a particularly data-rich environment [7, 8].
Existing critical care data collection and reporting is typically insensitive to variation in individual patient pathways prior to the admission and recovery from critical illness, with an emphasis focusing overwhelmingly on the patient’s stay in critical care, and to a lesser extent the non-acute hospital or emergency episode [9, 10] that may have preceded it. Furthermore, in the UK, standard critical care data collection coordinated by the Intensive Care National Audit and Research Centre (ICNARC), has historically been relatively selective in capture of past medical history and health status and does not routinely report on longer-term outcomes [11].
The integration of clinical critical care data with other existing administrative healthcare data sources may therefore provide an important longer-term perspective and create an opportunity to define more thoroughly a patient’s clinical journey, such as the healthcare interactions prior to critical illness, or subsequent re-admission after hospital discharge. Integration of additional data on chronic illnesses and health status would provide a longitudinal platform to study healthcare trajectories before and after critical illness, and to support research into “survivorship” [12, 13].
While attempts to link multiple data sources relating to a critical care episode must address a number of common issues (e.g. data systems access, “cleaning,” data resource linkage, and patient privacy [10, 13–15]), such integration could provide a rich “research ready resource”, primed for interrogation once appropriate governance requirements have been met [16–20]. Requirements for timely data provision to enable novel research and inform quality improvement initiatives have been brought even more sharply into focus by the COVID-19 pandemic [21, 22].
Objectives
The paper has two main objectives:
Describing a transparent and reproducible methodology (INTEGRATE) which can be applied to both routine administrative and clinically rich, critical care data sources to integrate and combine them into a single, enhanced dataset for research.
Applying the INTEGRATE methodology to the data held in the SAIL Databank (https://saildatabank.com) to demonstrate the value and range of admission data made available and to describe the advantages this has for critical care research.
Method
Data sources
The INTEGRATE methodology has been designed to run within a Trusted Research Environment (TRE) and is contingent on the availability of appropriate critical care and healthcare data sources. The SAIL Databank is the TRE in which the methodology is demonstrated, details of which are found below, together with a description of the data sources upon which the methodology is applied.
SAIL databank
The Secure Anonymised Information Linkage (SAIL) Databank is a privacy-protecting, remotely accessible Trusted Research Environment, containing de-identified, continuously updated, individual-level, routinely collected health and social care data about the population of Wales; approximately 3.2 million people [24–27].
Data in the SAIL Databank are de-identified by a trusted third party and access is gained through a privacy-protecting safe haven and remote access system called the SAIL Gateway.
The SAIL Databank provides access to linkable, anonymised data from primary and secondary care health systems for residents, living in Wales, and all those receiving treatment in NHS Wales services. Within this environment, it is possible to link together different sources of health data at individual level, using an Anonymised Linkage Field (ALF).
Table 1 lists the SAIL data sources which underpin the INTEGRATE methodology and which have been used to demonstrate it.
Table 1: Data sources available in the SAIL Databank which are used to underpin the methodology.
| SAIL data source** | Data source full name | Data source description | Start date of coverage |
|---|---|---|---|
| ADDE | Annual district death extract | Monthly update – Office for national statistics (ONS) deaths register | January 2003 |
| CCDS | Critical care data set | Monthly update – Critical care admissions and details | April 2006 |
| EDDS | Emergency department data set | Monthly update – emergency department events | April 2008 |
| ICNC | Intensive care national audit & research centre (ICNARC) | Quarterly update – ICNARC admissions and details | April 2009 |
| PEDW | Patient episode database for wales | Weekly update – in-patient hospital admissions and details | January 1995 |
| WDSD | Welsh demographic service dataset | Weekly update – wales population registration and residence history, and demographic details | January 1990 |
| WLGP | Welsh longitudinal general practice | Monthly update – primary care general practice events* and details | January 1990 |
*A primary care, general practice event includes records for any interactions with primary care such as visits, prescriptions, diagnoses, test results and administrative actions.
**Further details on the abbreviations used can be found in Supplementary Appendix 2.
The date when data sources become available in the SAIL Databank varies over time.
Table 1 provides the overall coverage available in the individual data sources, a description of each data source and the frequency with which it is updated.
Critical care data sources
The SAIL Databank contains two independent data sources that record clinical and administrative details of critical care episodes for patients:
Intensive Care National Audit & Research Centre data (ICNARC) [11].
The Critical Care Minimum Dataset (CCDS) provided by Digital Health Care Wales (DHCW) [28].
Both data sources record details of a patient’s admission to critical care.
ICNARC was established in the mid 1990’s to collect detailed clinical data for patients admitted to a care critical unit. ICNARC’s goal is to assist in the development of effective practices in the management of critical care and treatments [29, 33]. The first set of ICUs joined the programme in 1995 and by 2005 there were over 500,000 patients in the database; in the subsequent period, the number of cases added to the programme has steadily increased. ICNARC provides a particularly rich clinical data source in relation to the first 24 hours in critical care.
The Critical Care Minimum Dataset (CCDS) is a summary of the organ support modalities received during the critical care admission. It is a nationally mandated collation of primarily administrative data, exported from Welsh ICUs and the Patient Episode Database for Wales (PEDW) on a monthly basis.
Ideally, the data in ICNARC should reflect the locally captured data in CCDS. However, this is not always the case and for various reasons there are sometimes admissions recorded in one but not both data sources.
By combining admissions common to both data sources, we can identify duplicate admissions for a patient and reduce them to a single record while capturing those admissions present in one data source and not in the other. This ensures that all available admissions from both sources are captured into a single dataset while still maintaining the pseudo-anonymised identity Anonymised Linkage Field (ALF) of the patient, the source(s) of the admission data, and the date of the admission.
Description of the INTEGRATE methodology
The methodology is designed to capture all available critical care admissions from ICNARC and CCDS data sources into a single research ready dataset. This captures the Anonymised Linkage Fields for individuals, the source(s) of the admission data, and the date of the critical admission. The date for a critical care admission is utilised to search through other healthcare data sources for any activity recorded within a chosen time frame which is adjacent to or near the date of the critical care admission. This enables a broader view of patient care to be created. Furthermore, the time frame for the search can be adapted to the specific requirements of the research question.
This approach has been developed to allow maximum flexibility in the way the methodology can be applied, however, certain parameters will need to be defined beforehand and are dependent on the specific research question.
The project-dependent parameters of the methodology are:
The period over which the critical care admissions are to be studied
The time frame around the critical care admission date in which to search for other healthcare interactions
The inclusion/exclusion criteria relating to characteristics of the critical care patients to be studied.
There are two stages to the methodology. These are illustrated in Figures 1 and 2 below and describe how the tables in the SAIL Databank are used to identify patients and their critical care admissions.
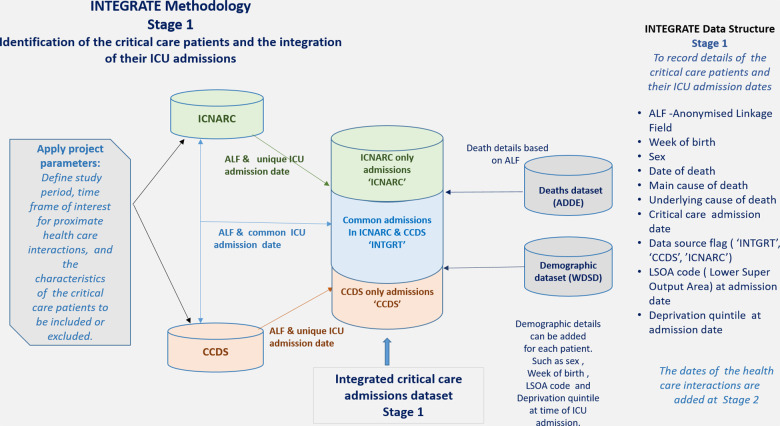
Figure 1: Stage 1 of the INTEGRATE methodology: This illustrates how critical care patients are identified and in which source their critical care admissions are captured and flagged.
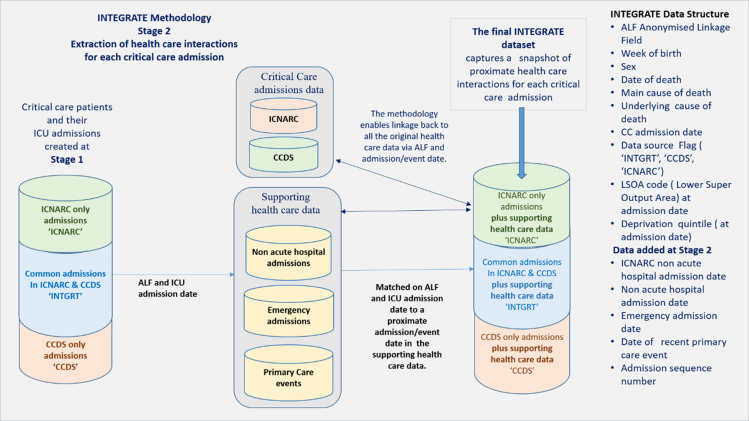
Figure 2: Stage 2 of the INTEGRATE methodology: This illustrates how the proximate or adjacent non-acute hospital and emergency admission dates and interactions with primary care services can be identified and merged into the final data set.
Stage 1: Identification of patients and integration of their critical care admissions from two critical care data sources.
During Stage 1, admissions from the two critical care data sources are extracted and the common admissions (those matched on both the ALF and critical care admission date) are merged into a single record and flagged as ‘INTGRT’ indicating they represent a common, integrated admission between the two data sources. Additional single admissions from either data source are then added and flagged with the appropriate identifier (either ‘CCDS’ or ‘ICNARC’), indicating they originate from the CCDS or the ICNARC data respectively. The two fields ‘ALF plus critical care admission date’ then form a unique identifier for each admission.
Additional demographic details are then added as required. This is dependent on the data sources available in the supporting TRE. For example, birth details, sex, date of death and main and underlying causes of death can be found in SAIL with linkage to the Office for National Statistics records. Given their importance for understanding a patient’s clinical journey through critical care, it is also possible to add further background details to include frailty and /or comorbidity scores from primary or secondary care data [32].
This completes Stage 1, where a dataset of critical care patients has been identified with their admission dates captured from both critical care data sources and where additional project defined background details have been added. As the methodology captures every critical care admission for an individual, an admission sequence number is assigned so multiple admissions can also be identified and examined.
Stage 2: Extraction of admission dates for the supporting healthcare events adjacent to each critical care admission.
Stage 2 uses the Stage 1 integrated spine of critical care admissions and the unique ‘ALF plus critical care admission date’ to capture adjacent or proximate non-acute hospital and emergency admission dates and the dates of recent primary care interactions. The time frame around which this supporting data is captured can be determined beforehand and adapted to examine events and admissions before or after the critical care admission. Table 6 in the Results section below, shows an example of how it is possible to capture the nearest non-acute hospital admission dates, emergency admission dates and the most recent primary care interactions in the week prior to the critical care admission.
Table 6: Numbers of critical care admissions captured with supporting adjacent healthcare data.
| Associated healthcare admission/events | N | % |
|---|---|---|
| Admissions with a non-acute hospital admission recorded in the 7-days prior to the critical care admission | 90,632 | 99 |
| Admissions with an emergency admission recorded in the 7-days prior to the critical care admission | 48,979 | 53 |
| **Admissions with a primary care interaction/event recorded in the 7-days prior to the critical care admission | 64,832 | 71 |
**Not all primary care general practices contribute their data to the SAIL Databank. Currently there is approximately 80% coverage of practices and people in Wales.
The dates extracted at Stage 2 are then merged into the record for each critical care admission based on ‘ALF plus critical care admission date’. The critical admissions data together with the healthcare events around that admission date build a broader picture of the patient’s trajectory through the healthcare system and create a spine of all the dates of related healthcare interactions.
In addition, a further advantage is that all of the original, underlying data in the supporting data sources are still accessible and can be linked back to and examined by using the ‘ALF and the admission/event date’ for that data source.
Details of INTEGRATE dataset produced by applying this methodology and the fields it contains can be found in Supplementary Appendix 1.
The following section describes the application of the methodology using SAIL data sources and comments on some of the practical factors to be considered.
Demonstrating the methodology in SAIL
The data sources available in SAIL to which this methodology is applied are described in Table 1. RECORD guidelines [23] have been followed.
ICNARC data in SAIL contains admissions for all patients admitted to Welsh Intensive Care Units. That includes those who are residents of Wales, as well as those who reside outside Wales. It also includes admissions to any non-Welsh Intensive Care Unit for residents living in Wales. As only individuals living in Wales, who are treated within a Welsh hospital, can be linked to the wider, individual level resources in SAIL, all persons living outside Wales were excluded, as were the residents in Wales treated in non-Welsh hospitals.
To demonstrate the methodology, the characteristics chosen for the critical care patients were individuals resident in Wales, treated in Welsh hospitals, aged 17 years and over. The study period chosen was January 2010 to December 2019. To capture adjacent healthcare interactions in Stage 2, a period of 7 days prior to the critical care admission date was chosen.
Results
Given the parameters above, Table 2 provides details on the number of critical care admissions captured by the INTEGRATE methodology. This includes the number of integrated admissions common to both ICNARC and CCDS and the number of unmatched admissions recorded in CCDS or ICNARC only.
Table 2: The number of critical care admissions and unique individuals admitted to critical care recorded in each data source.
| Data source | Admissions | Patients |
|---|---|---|
| Critical care admissions unique to the ICNARC data | 1,349 (1%) | 1,325 (1%) |
| Common, integrated admissions recorded in both ICNARC and CCDS | 84,952 (93%) | 71,214 (94%) |
| Critical care admissions unique to the CCDS administrative data | 5,281 (6%) | 4,893 (6%) |
| Total admissions 91,582 | *Total unique individuals = 76,019 |
*Total unique individuals: Some patients have multiple critical care admissions; a unique individual may have more than one admission recorded in each flagged group.
Summary of the critical care patients
A brief analysis of the 76,019 unique individuals who were admitted to critical care between 2010 and 2019 showed that 41,461 (55%) were male and 34,558 (45%) were female.
Some 36,250 (48%) died within the study period after their last admission to critical care; of those, 15,812 died within 14 days of their last critical care admission, and 10,352 died after 1 year.
The majority of patients were in the older age groups when first admitted to critical care, as shown in Table 3.
Table 3: Age groups of patients on first admission to critical care.
| Age group at first ICU admission | N | % |
|---|---|---|
| 17 to 30 | 4,690 | 6 |
| 31 to 40 | 4,316 | 6 |
| 41 to 50 | 6,999 | 9 |
| 51 to 60 | 11,125 | 15 |
| 61 to 70 | 17,720 | 23 |
| 71 to 80 | 19,762 | 26 |
| 80+ | 11,407 | 15 |
Some 64,769 (85%) of the patients have a single admission to critical care. However, a number of patients have multiple admissions recorded, as seen in Table 4.
Table 4: Numbers of patients with multiple admissions recorded in critical care.
| Multiple admissions | N | % |
|---|---|---|
| Patients with 2 admissions | 8,562 | 11 |
| Individuals with 3-5 admissions | 2,518 | 3 |
| Individuals with 6+ admissions | 170 | <1 |
Summary of the critical care admissions
The tables below provide background details on the critical care admissions that can be captured by the methodology. These include deprivation quintiles, based on residency on the date of the critical care admission and the number of critical care admissions with accompanying data on non-acute hospital, and/or emergency admissions and interactions with primary care, in the week prior to that critical care admission.
Table 5 shows that critical admissions occur more often in the more deprived quintiles than the least deprived quintiles of the population.
Table 5: Numbers of admissions occurring in each deprivation quintile.
| Deprivation quintile at admission | Admissions (91,582) | % (100) |
|---|---|---|
| 1 (Most deprived) | 21,131 | 23 |
| 2 | 19,470 | 21 |
| 3 | 19,116 | 21 |
| 4 | 15,457 | 17 |
| 5 (Least deprived) | 13,020 | 14 |
| Missing | 3388 | 4 |
* Missing: In Wales, deprivation quintile is based on residency within a lower layer super output area (LSOA) [30]. Where residential data are missing for persons on their critical care admission date the deprivation quintile is unknown.
Table 6 shows the level of supporting data captured by the methodology from non-cute hospital and/or emergency admissions and primary care interactions in the week before critical care admission.
Comments
The INTEGRATE methodology is designed to combine critical care admission records from two independent critical care data sources into a single dataset. It enhances the data by also capturing any non-acute or emergency hospital admission or primary care interactions immediately prior to the critical care admission.
This not only provides a snapshot of the events leading up to the critical care event but also facilitates the linkage back to the respective, supporting healthcare data sources (non-acute hospital, emergency, and primary care interactions). This allows a more detailed examination into pathways of care and the specific characteristics and underlying conditions of the critical care patient recorded in other data sources.
However, there are a number of practical considerations when applying the methodology to the data sources in the SAIL Databank.
As mentioned previously, not all Welsh Intensive Care Units contributed to the ICNARC programme from the outset and there have been a small number of breaks in ICNARC data collection due to site staffing shortages. In such circumstances, admission episodes will have been more readily captured within the local routine administrative CCDS data, leading to some discrepancies between the locally recorded cases and those in ICNARC. However, with a more consistent contribution from hospitals over time, such discrepancies have become less frequent. For example, just under 93% of the critical care admissions are common to both data sources with just over 7% recorded in a single data source only.
In this latter group (admissions recorded in single data source), there are a number of admission records for individuals with very slight differences in the dates between the two data sources which could be interpreted as the same admission. An examination into those with a difference of 1 to 3 days between admission dates for the same person accounted for fewer than 0.2% of the admissions.
One interesting observation was that a higher percentage of ‘CCDS only’ admissions were found to have a recorded date of death within 5 days of the critical care admission date. Out of the 753 admissions recorded in either ICNARC or CCDS where the death occurred within 5 days of the admission, 144 (19%) were recorded in ICNARC and 609 (81%) were recorded in CCDS. This finding could help inform ICNARC data collection policies to ensure that patients who died shortly after admission are not under-represented in the dataset.
In Stage 2, when extracting the dates for the adjacent/recent admissions to a non-acute hospital episode of care a further challenge was identified. This was due to the disparity in approach that different hospitals adopt when recording admissions and discharges between the non-acute hospital episode of care and the transfers to and from critical care. Some hospitals record the critical care admission within a complete, single non-acute hospital admission/discharge period while others create two separate hospital admission/discharge records; one before transfer to critical care and another when the patient is transferred back to the non-acute ward. The methodology manages this by recording the nearest non-acute hospital admission date extracted from the hospital data and when available, the date of the admission to hospital that is recorded within the ICNARC data. In addition, when a patient is discharged from and then re-admitted to critical care on the same day, it is recorded as a single admission.
By integrating both ICNARC and the local routine administrative CCDS admissions data into a cleaned dataset, the methodology identifies and captures all possible critical care admissions. The findings have shown that this was an important step in curating the critical care cohort due to the discrepancy between the two data sources. The transparency of applying the methodology means that there is less chance of introducing bias in the cohort of interest by only considering admissions from a single data source and reduces the risk of over-counting admissions due to duplicated records and/or multiple admissions. A further advantage is that it permits linkage back to all the original healthcare data sources for the development and integration of associated clinical, demographic, and administrative research questions. Therefore, providing an efficient spine of all related healthcare interactions for each critical care admission.
Although the value of the data created through use of this methodology has been demonstrated within the SAIL Databank, the methodology provides sufficient flexibility to be adapted for use in other TREs where similar, regularly updated electronic healthcare data are available.
Future developments
Within the SAIL Databank, the methodology has the potential to form the basis of a research ready data asset that could be run periodically to provide access to up-to-date admission data for all individuals admitted to critical care.
Having access to primary care exposure and previous interactions due to chronic health conditions prior to critical admission, could help determine the relative contributions of chronic decline in health versus severity of acute illness on outcomes. Furthermore, examining characteristics associated with critical care and hospital re-admissions, could help gain a better understanding of patterns of healthcare utilisation.
Such an asset has the potential to provide a broader platform from which to evaluate the healthcare interactions surrounding critical admissions. It would also provide a maintainable and consistent approach for ongoing critical care related research.
There is also a recognition of the need, escalated by the COVID-19 pandemic, to deliver high quality critical care healthcare information that is accurate, accessible, and as up to date as possible.
For the demonstration of the methodology, we did not include any data from 2020 onwards as this was outside the initial scope but there is an awareness that COVID-19 has drastically impacted the recording of electronic healthcare data across the UK and the world, especially early in the pandemic [31].
The SAIL Databank regularly receives ICNARC data on a quarterly basis, but as part of the national response to COVID-19 it has also been receiving a weekly flow of ICNARC COVID-19 only related critical care admission data, which contains more recent coverage than the regular quarterly data flow. The methodology can be easily modified to include this additional third data source specifically for COVID-19.
Conclusion
The paper describes a methodology to create a single, cleaned, anonymised, individual level, population scale dataset from two independent critical care data sources, which also captures the interactions of different healthcare services adjacent to or near to each critical admission. To demonstrate the methodology, it was applied to the data in the SAIL Databank. The results showed that it can provide a means to easily capture all critical admissions with onwards linkage to the unfiltered, original data sources using the patient identifier Anonymised Linkage Field and the relevant admission/event date, ensuring no loss of data. It facilitates easy access to a data-rich environment for events surrounding a critical care admission, permitting a broad range of research questions to be explored and enhancing research that seeks a better understanding of the variation in patient pathways and outcomes. The ability to create this broader view of critical care should be beneficial for the design of appropriate follow-up care and resource planning for a group of patients requiring highly specialist care.
Ethics statement
The data used in this study are available in the SAIL Databank at Swansea University, Swansea, UK. All proposals to use SAIL data are subject to review by an independent Information Governance Review Panel (IGRP) which includes members of the public. Before any data can be accessed, approval must be given by the IGRP. The IGRP gives careful consideration to each project to ensure proper and appropriate use of SAIL data and includes informed consent of participants where applicable. When access has been approved, it is gained through a privacy-protecting safe haven and remote access system referred to as the SAIL Gateway.
Supplementary Files
Acknowledgements
This study makes use of anonymised data held in the Secure Anonymised Information Linkage (SAIL) Databank. This work uses data provided by patients and collected by the NHS as part of their care and support. We would also like to acknowledge all data providers who make anonymised data available for research. We wish to acknowledge the collaborative partnership that enabled acquisition and access to the de-identified data, which led to this output. The collaboration was led by the Swansea University Health Data Research UK team under the direction of the Welsh Government Technical Advisory Cell (TAC) and includes the following groups and organisations: the SAIL Databank, Administrative Data Research (ADR) Wales, Digital Health and Care Wales (DHCW), Public Health Wales, NHS Shared Services Partnership (NWSSP) and the Welsh Ambulance Service Trust (WAST). All research conducted has been completed under the permission and approval of the SAIL independent Information Governance Review Panel (IGRP) project number 0911.
Funding Statement
This work was supported by the Con-COV team funded by the Medical Research Council (grant number: MR/V028367/1). This work was supported by Health Data Research UK, which receives its funding from HDR UK Ltd (HDR-9006) funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation (BHF) and the Wellcome Trust. This work was supported by the ADR Wales programme of work. The ADR Wales programme of work is aligned to the priority themes as identified in the Welsh Government’s national strategy: Prosperity for All. ADR Wales brings together data science experts at Swansea University Medical School, staff from the Wales Institute of Social and Economic Research, Data and Methods (WISERD) at Cardiff University and specialist teams within the Welsh Government to develop new evidence which supports Prosperity for All by using the SAIL Databank at Swansea University, to link and analyse anonymised data. ADR Wales is part of the Economic and Social Research Council (part of UK Research and Innovation) funded ADR UK (grant ES/S007393/1). This work was supported by the Wales COVID-19 Evidence Centre, funded by Health and Care Research Wales.
Availability of data and materials
The linkable data sources used in this study are available in the SAIL Databank at Swansea University, Swansea, UK, but as restrictions apply, they are not publicly available. SAIL has established an application process to be followed by anyone who would like to access data for approved research purposes at https://www.saildatabank.com/application-process. When access has been granted, it is gained through a privacy protecting safe haven and remote access system referred to as the SAIL Gateway.
References
- 1.Thayer D, Rees A, Kennedy J, Collins H, Harris D, Halcox J, et al. Measuring follow-up time in routinely-collected health datasets: Challenges and solutions. PloS ONE. 2020. Feb 11; 15(2). 10.1371/journal.pone.0228545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pugh RJ, Bailey R, Szakmany T, Al Sallakh M, Hollinghurst J, Akbari A, et al. Long-term trends in critical care admissions in Wales. Anaesthesia. 2021. May 2;76: 1291–95. 10.1111/anae.15466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollinghurst J, Akbari A, Fry R, Watkins A, Berridge D, Clegg A, et al. Study protocol for investigating the impact of community home modification services on hospital utilisation for fall injuries: a controlled longitudinal study using data linkage. BMJ Open [Internet]. 2018;8:e26290. 10.1136/bmjopen-2018-026290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rose L, Scales DC, Atzema C, Burns KE, Gray S, Doing C, Kiss A, Rubenfeld G, Lee JS. Emergency Department Length of Stay for Critical Care Admissions. A Population-based Study. Ann Am Thorac Soc. 2016. Aug;13(8):1324–32. https://www.atsjournals.org/doi/10.1513/AnnalsATS.201511-773OC [DOI] [PubMed] [Google Scholar]
- 5.Lyons RA, Turner S, Lyons J, Walters A, Snooks HA, Greenacre J, et al. All Wales Injury Surveillance System revised: Development of a population-based system to evaluate single-level and multilevel interventions. Injury Prevention. 2016;22:i50–i55. 10.1136/injuryprev-2015-041814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szakmany T, Walters AM, Pugh R, Battle C, Berridge DM, Lyons RA. Risk Factors for 1-Year Mortality and Hospital Utilization Patterns in Critical Care Survivors: A Retrospective, Observational, Population-Based Data Linkage Study. Crit Care Med. 2019;47(1):15–22. 10.1097/CCM.0000000000003424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celi LA, Mark RG, Stone DJ, Montgomery RA. “Big data” in the intensive care unit. Closing the data loop. American journal of respiratory and critical care medicine. 2013;187(11):1157–60. 10.1164/rccm.201212-2311ed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schenck EJ, Hoffman KL, Cusick M, Kabariti J, Sholle ET, Campion TR. Critical carE Database for Advanced Research (CEDAR): An Automated Method to Support Intensive Care Units with Electronic Health Record Data. Journal of Biomedical Informatics. 2021. June;118:103789. 10.1016/j.jbi.2021.103789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson Chornenki N, Liaw P, Bagshaw S, Burns K, Dodek P, English S, et al. Data initiatives supporting critical care research and quality improvement in Canada: an environmental scan and narrative review. Can J Anaesth. 2020. Jan 22;67(4):475–84. 10.1007/s12630-020-01571-1 [DOI] [PubMed] [Google Scholar]
- 10.Harris S, Shi S, Brealey D, MacCallum NS, Denaxas S, Perez-Suarez D, et al. Critical Care Health Informatics Collaborative (CCHIC): Data, tools and methods for reproducible research: A multi-centre UK intensive care database. International Journal of Medical Informatics. 2018. Apr;112:82–9. 10.17863/CAM.22169 [DOI] [PubMed] [Google Scholar]
- 11.Intensive Care National Audit & Research Centre (ICNARC) [Internet]. [cited 2021. June 21] Available from: https://www.icnarc.org/
- 12.Jouan Y, Grammatico-Guillon L, Teixera N, Hassen-Khodja C, Gaborit C, Salmon-Gandonnière C, et al. Healthcare trajectories before and after critical illness: population-based insight on diverse patients clusters. Annals of Intensive Care. 2019. Nov 9;9(1):126. 10.1186/s13613-019-0599-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McWilliams C, Inoue J, Wadey P, Palmer G, Santos-Rodriguez R, Bourdeaux C. Curation of an intensive care research dataset from routinely collected patient data in an NHS trust. F1000Res. 2019. Aug 19;8:1460. 10.12688/f1000research.20193.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson AEW, Pollard TJ, Shen L, Lehman LH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016. May 24;3(1):160035. 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data. 2018. Sep 11;5(1):180178. 10.1038/sdata.2018.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnier C, Wilkinson T, Akbari A, et al. The secure Anonymised information linkage databank dementia e- cohort (SAIL- DeC). Int J Popul Data Sci. 2020. Feb 25;5:1121. 10.23889/ijpds.v5i1.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carra G, Salluh JIF, José da Silva Ramos F, Meyfroidt G. Data-driven ICU management: Using Big Data and algorithms to improve outcomes. Journal of Critical Care. 2020. Dec, 60:300–304. 10.1016/j.jcrc.2020.09.002 [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Pinto LN, Luo Y, Churpek MM. Big Data and Data Science in Critical Care. Chest. 2018. Nov;154(5):1239–1248. 10.1016/j.chest.2018.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saeed M, Villarroel M., Reisner AT, Clifford G, Lehman LW, Moody G, et al. Multiparameter Intelligent Monitoring in Intensive Care II: a public-access intensive care unit database. Critical care medicine. 2011. May;39(5):952–960. 10.1097/CCM.0b013e31820a92c6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaulieu-Jones BK, Orzechowski P, Moore JH. Mapping Patient Trajectories using Longitudinal Extraction and Deep Learning in the MIMIC-III Critical Care Database. Pac Symp Biocomput. 2018;23: 123–132. [PubMed] [Google Scholar]
- 21.Garland A, Yogendran M, Olafson K, Scales DC, McGowan KL, Fransoo R. The accuracy of administrative data for identifying the presence and timing of admission to intensive care units in a Canadian province. Med Care. 2012. Mar;50(3):e1–6. 10.1097/mlr.0b013e318245a754 [DOI] [PubMed] [Google Scholar]
- 22.Lyons J, Akbari A, Torabi F, et al. Understanding and responding to COVID-19 in Wales: protocol for a privacy- protecting data platform for enhanced epidemiology and evaluation of interventions. BMJ Open. 2020. Oct 2;10:e043010. https://bmjopen.bmj.com/content/10/10/e043010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benchimol EI, Smeeth L, Guttmann A, Harron K, Moher D, Petersen I, et al. The REporting of studies Conducted using Observational Routinely-collected health Data (RECORD) Statement. PLoS Med. 2015. Oct;12(10):e1001885. 10.1371/journal.pmed.1001885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones KH, Ford DV, Jones C, Dsilva R, Thompson S, Brooks CJ, et al. A case study of the Secure Anonymous Information Linkage (SAIL) Gateway: A privacy-protecting remote access system for health-related research and evaluation. J Biomed Inform. 2014. Aug 1;50(100):196–204. 10.1016/j.jbi.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyons RA, Ford DV, Moore L, Rodgers SE. Use of data linkage to measure the population health effect of non-health-care interventions. The Lancet. 2014. Apr 26;383(9927);1517–1519. 10.1016/s0140-6736(13)61750-x [DOI] [PubMed] [Google Scholar]
- 26.Ford DV, Jones KH, Verplancke J-P, Lyons RA, John G, Brown G, et al. The SAIL Databank: building a national architecture for e-health research and evaluation. BMC Health Serv Res. 2009; 9(1):157. 10.1186/1472-6963-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons RA, Jones KH, John G, Brooks CJ, Verplancke J-P, Ford DV, et al. The SAIL databank: linking multiple health and social care datasets. BMC Med Inform Decis Mak. 2009. Jan 16;9:3. 10.1186/1472-6947-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Critical Care Minimum Dataset (CCDS): NHS Data Model and Dictionary [Internet]. [cited 2021. June 21]. Available from: https://datadictionary.nhs.uk/data_sets/supporting_data_sets/critical_care_minimum_data_set.html
- 29.Intensive Care National Audit & Research Centre: Our History [Internet]. [cited 2021. Sep 16]. Available from: https://www.icnarc.org/About/Overview/Our-History
- 30.Welsh Government, Welsh Index of Multiple Deprivation (WIMD) 2014 Revised. Cardiff, UK: 2017. https://gov.wales/sites/default/files/statistics-and-research/2019-04/welsh-index-of-multiple-deprivation-2014-revised.pdf [Google Scholar]
- 31.Shioda K, Weinberger DM, Mori M. Navigating Through Health Care Data Disrupted by the COVID-19 Pandemic. JAMA Intern Med. 2020. Oct 12;180(12):1569–1570. 10.1001/jamainternmed.2020.5542. https://jamanetwork.com/journals/jamainternalmedicine/fullarticle/2771671 [DOI] [PubMed] [Google Scholar]
- 32.Szakmany T, Hollinghurst J, Pugh R, Akbari A, Griffiths R, Bailey R, Lyons RA. Frailty assessed by administrative tools and mortality in patients with pneumonia admitted to hospital and ICU: a population-based data-linkage study. Scientific Reports 2021. https://www.nature.com/articles/s41598-021-92874-w [DOI] [PMC free article] [PubMed]
- 33.Harrison David A, Brady Anthony R, Kathy Rowan. Case mix, outcome and length of stay for admissions to adult, general critical care units in England, Wales and Northern Ireland. The Intensive Care National Audit & Research Centre Case Mix Programme Database. Critical Care 2004;9:S1–S13. 10.1186/cc3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The linkable data sources used in this study are available in the SAIL Databank at Swansea University, Swansea, UK, but as restrictions apply, they are not publicly available. SAIL has established an application process to be followed by anyone who would like to access data for approved research purposes at https://www.saildatabank.com/application-process. When access has been granted, it is gained through a privacy protecting safe haven and remote access system referred to as the SAIL Gateway.