Abstract
Enterobacter cloacae is a Gram-negative rod with multidrug-resistant potential due to chromosomally-induced AmpC β-lactamase. We evaluated characteristics, antibiotic utilization, and outcomes associated with battlefield-related E. cloacae infections (2009–2014). Single initial and serial E. cloacae isolates (≥24 hours from initial isolate from any site) associated with a clinical infection were examined. Susceptibility profiles of initial isolates in the serial isolation group were contrasted against last isolate recovered. Characteristics of 112 patients with E. cloacae infections (63 [56%] with single initial isolation; 49 [44%] with serial isolation) were compared to 509 patients with bacterial infections not attributed to E. cloacae. E. cloacae patients sustained more blast trauma (78%) compared to non-E. cloacae infections patients (75%; p<0.001); however, injury severity scores were comparable (median of 34.5 and 33, respectively; p = 0.334). Patients with E. cloacae infections had greater shock indices (median 1.07 vs 0.92; p = 0.005) and required more initial blood products (15 vs. 14 units; p = 0.032) compared to patients with non-E. cloacae infections. Although E. cloacae patients had less intensive care unit admissions (80% vs. 90% with non-E. cloacae infection patients; p = 0.007), they did have more operating room visits (5 vs. 4; p = 0.001), longer duration of antibiotic therapy (43.5 vs. 34 days; p<0.001), and lengthier hospitalizations (57 vs. 44 days; p<0.001). Patients with serial E. cloacae had isolation of infecting isolates sooner than patients with single initial E. cloacae (median of 5 vs. 8 days post-injury; p = 0.046); however, outcomes were not significantly different between the groups. Statistically significant resistance to individual antibiotics did not develop between initial and last isolates in the serial isolation group. Despite current combat care and surgical prophylaxis guidelines recommending upfront provision of AmpC-inducing antibiotics, clinical outcomes did not differ nor did significant antibiotic resistance develop in patients who experienced serial isolation of E. cloacae versus single initial isolation.
Introduction
The microbiology of infections associated with combat-related injuries have transitioned to a predominance of Gram-negative bacilli since the Vietnam War, as early wound debridement and anatomic site-directed empiric antibiotics have reduced the number of infections secondary to Gram-positive organisms [1]. In the U.S. Naval Hospital in DaNang, Vietnam, 52% of severe extremity wound cultures prior to debridement grew Gram-negative organisms and 20% of the entire wounded cohort developed Enterobacter spp. bacteremia by the fifth day of hospitalization [2]. During military operations in Afghanistan, there was a similar prevalence (56%) of Gram-negative organisms from mangled lower extremities on pre-operative wound cultures [3]. Both theaters of war saw wound cultures trend toward a Gram-negative predominance as hospitalization progressed [4]. As modern combat-related injuries have led to a surge in Gram-negative hospital-related infections, multidrug-resistant Gram-negative (MDRGN) infections have become an increasing threat [5].
Microbiology of wounds and wound infections among blast casualties injured in Iraq and Afghanistan has been described, and a higher prevalence of MDR organisms was found in polymicrobial infections compared to monomicrobial infections [6]. In that analysis, Enterobacter spp. was the second most commonly isolated Gram-negative organism from polymicrobial cultures and was noted to have a shorter time from injury to first infection. As polymicrobial infections among blast wound infections were more likely to produce MDR organisms and Enterobacter spp. infections have a potentially quicker onset, earlier identification and treatment of Enterobacter spp. infections may improve patient outcomes.
As MDRGN infections were being recognized as a worsening threat to patients in military hospitals, U.S. civilian hospitals saw a similar rise in these challenging infections. In the early 2000s, an epidemic of carbapenem-resistant Enterobacterales spread across the northeastern United States [7]. A subsequent analysis by the Veterans Health Administration from 2006–2015 indicated that Enterobacter cloacae was one of the emerging pathogens with dramatically increasing resistance rates largely secondary to AmpC β-lactamase induction [8]. E. cloacae is initially phenotypically susceptible to 3rd generation cephalosporins (e.g., cefotaxime, ceftriaxone, and ceftazidime) in vitro; however, as high as 19% of isolates developed β-lactam resistance during treatment [9–12]. Making matters more difficult, clinical laboratories do not typically test for AmpC production, and molecular testing is required to differentiate between chromosomally-induced or constitutively-expressed plasmid AmpC [9]. Third-generation cephalosporins are also not the only AmpC inducers, as amoxicillin / clavulanic acid, cefoxitin, 1st generation cephalosporins, and carbapenems are similarly potent inducers [13, 14].
Guidance surrounding the treatment of E. cloacae focuses primarily on bacteremia secondary to respiratory, urinary, intravascular, or intra-abdominal sources, but largely neglects wound infections [15, 16]. Due to concerns regarding potential delays in transport/medevac of combat casualties, the Department of Defense (DoD) Tactical Combat Casualty Care (TCCC) guidelines recommend use of ertapenem for care of open traumatic wounds in the field [17]. In addition, the DoD Joint Trauma System (JTS) clinical practice guidelines, as well as non-DoD guidelines, recommend cefazolin for post-injury prophylaxis [18, 19]. As both ertapenem and cefazolin may induce AmpC production, there is concern of increasing β-lactam resistance negatively affecting patient outcomes in infections where E. cloacae is the primary pathogen. Herein, we assessed the epidemiological characteristics of patients with E. cloacae infections, characterized the antibiotic prescribing patterns used in the treatment of these infections and their effects on developing resistance, and examined clinical outcomes compared to battlefield trauma patients who developed bacterial infections associated with organisms other than E. cloacae. We described clinical characteristics and outcome differences between patients with single initial isolation of E. cloacae and patients with serial isolation to assess for antimicrobial resistance development.
Materials and methods
Study population and definitions
Data and specimens were collected through the DoD–Veterans Affairs Trauma Infectious Disease Outcomes Study (TIDOS), which is an observational, longitudinal study of infectious outcomes among military personnel who were wounded in Iraq or Afghanistan (2009–2014) [20, 21]. Criteria for inclusion in TIDOS were being ≥18 years of age, active-duty personnel or DoD beneficiaries injured during deployment, and medically evacuated to Landstuhl Regional Medical Center in Germany with subsequent transfer to a participating military hospital in the United States. The participating U.S. military hospitals were Brooke Army Medical Center (BAMC) in San Antonio, TX, and Walter Reed National Military Medical Center in the National Capital Region (NCR) (prior to September 2011 it was National Naval Medical Center and Walter Reed Army Medical Center). Patients were included if they had E. cloacae isolation associated with a clinical infection diagnosis. Patients with a clinical diagnosis of a bacterial infection attributed to an organism(s) other than E. cloacae comprised the comparator population for the analysis. The Institutional Review Board (IRB) of the Uniformed Services University of the Health Sciences (USU, Bethesda, MD) approved this study. Data and specimens were collected from individuals who provided authorization through informed consent and HIPAA authorization processes, or through an IRB-approved waiver of consent for use of de-identified data not obtained through interaction or intervention with human subjects.
Demographics, injury characteristics, and early casualty care data were obtained from the DoD Trauma Registry (DoDTR). Infection-related data (e.g., infection syndromes, microbiology, and antibiotic management) were collected from the TIDOS Infectious Disease module of the DoDTR [22]. Data on use of ertapenem in the prehospital setting were not collected by the DoDTR. Tetracycline use was excluded from the analysis as doxycycline was prescribed to military personnel deployed to Afghanistan for antimalarial prophylaxis and continued for 28 days following departure from the country, per DoD guidelines.
Infections were identified using a combination of clinical (e.g., signs and symptoms from direct observations) and laboratory (e.g., microbiology) findings, and classified based on National Healthcare Safety Network definitions, as previously described [20, 23]. Isolates recovered during workups for clinical infection were classified as infecting. Inclusion criteria for the single initial E. cloacae isolate group required patients to have an infecting E. cloacae isolate collected from the initial culture (may be either monomicrobial or polymicrobial) with no isolation from subsequent cultures. For the serial E. cloacae isolate group, all patients with multiple non-colonizing E. cloacae isolates cultured at least one day apart were included, as prior studies have determined that E. cloacae may develop resistance to β-lactams after one day of therapy [10]. If multiple isolates were collected on the first day of E. cloacae isolation, the isolates collected from sterile body sites (more likely to be a true infection) or more proximal wound sites (wounds less likely to undergo early amputation) were given preference. In addition, all isolates must have been stored in the TIDOS specimen repository.
Laboratory analysis
Identification and susceptibility testing of the E. cloacae isolates were performed using the BD Phoenix Automated Microbiology System (NMIC/ID-308 and NMIC-311 panels, BD Diagnostics, Sparks, MD). Antimicrobial susceptibility testing results were interpreted in accordance with the Clinical Laboratory Standards Institute (CLSI M100 30th edition) breakpoints to construct an antibiogram [24]. Initial and last isolates cultured from patients in the serial isolate group were compared to assess the changing resistance patterns and we regarded any isolates with intermediate susceptibility as resistant. As molecular assays to assess for AmpC were not routinely conducted at the military hospitals during the study period, data on AmpC induction were not available.
Statistical analysis
Patients with E. cloacae infections were analyzed against the comparator population of patients with non-E. cloacae infections. Characteristics of E. cloacae patients with single initial or serial isolates and characteristics were compared. Categorical variables were assessed using Χ2 and Fisher’s Exact Tests, where appropriate. Continuous variables were analyzed using Mann-Whitney U. Statistical analysis was performed using IBM SPSS Statistics 22 (Version 22 IBM, NY, 2013.). A p value of <0.05 was considered statistically significant.
Results
Population characteristics
A total of 112 patients with infections due to E. cloacae and 509 patients with non-E. cloacae infections met inclusion criteria for the analysis. The majority of patients were young males with median age of 24 years (interquartile range [IQR] 21–28) in the U.S. Army who suffered blast injuries from improvised explosive devices while on foot patrol in Afghanistan (Table 1). Patients diagnosed with E. cloacae infections sustained more blast injuries (89% vs 75%; p = 0.001) and burns (16% vs. 9%; p = 0.027), had higher first documented shock indices (1.07 vs 0.92; p = 0.005), received more blood products within the first 24 hours of injury (15 vs. 14 units; p = 0.032), and required a greater number of visits to the operating room (5 vs. 4; p<0.001; Table 1).
Table 1. Characteristics of patients with and without Enterobacter cloacae infection.
| Characteristic, No. (%) | Patients with E. cloacae infection (N = 112) | Patients with non-E. cloacae infection (N = 509)a | All Patients (N = 621) |
p-value |
|---|---|---|---|---|
| Age at injury, median (IQR) | 23 (21–27) | 24 (22–29) | 24 (21–28) | 0.065 |
| Male | 111 (99.1) | 502 (98.6) | 613 (98.7) | 1.000 |
| Branch of Service | 0.449 | |||
| Air Force and Navy | 5 (4.5) | 36 (7.1) | 41 (6.6) | |
| Army | 68 (60.7) | 298 (58.5) | 366 (58.9) | |
| Marine | 39 (34.8) | 168 (33.0) | 207 (33.3) | |
| Other | 0 (0) | 7 (1.4) | 7 (1.1) | |
| Combat Theater | 0.069 | |||
| Afghanistan | 108 (96.4) | 460 (90.4) | 568 (91.5) | |
| Iraq | 4 (3.6) | 31 (6.1) | 35 (5.6) | |
| Non-theater | 0 (0) | 18 (3.5) | 18 (2.9) | |
| Combat Injury | 108 (96.4) | 476 (93.5) | 584 (94.0) | 0.239 |
| Mechanism of Injury | 0.001 | |||
| Blast | 100 (89.3) | 383 (75.2) | 483 (77.8) | |
| Non-blast | 12 (10.7) | 126 (24.8) | 138 (22.2) | |
| Blast Type | 0.410 | |||
| IED | 93 (83.0) | 346 (68.0) | 439 (70.7) | |
| Non-IED | 7 (6.2) | 37 (7.2) | 44 (7.1) | |
| Injured on foot patrol | 76 (67.9) | 286 (56.2) | 362 (58.3) | 0.213 |
| Burn | 18 (16.1) | 46 (9.0) | 64 (10.3) | 0.027 |
| 1st documented shock index, median (IQR) | 1.07 (0.74–1.49) | 0.92 (0.70–1.22) | 0.93 (0.70–1.27) | 0.005 |
| 1st 24 hour blood transfusion, median units (IQR) | 15 (8–31) | 14 (6–24) | 14 (6–25) | 0.032 |
| Body Region of Injury | 0.016 | |||
| Lower extremity | 21 (18.7) | 89 (17.5) | 110 (17.7) | |
| Upper extremity | 7 (6.2) | 26 (5.1) | 33 (5.3) | |
| Both lower and upper extremity | 82 (73.2) | 334 (65.6) | 416 (67.0) | |
| Non extremity | 2 (1.8) | 60 (11.8) | 62 (10.0) | |
| Injury severity score, median (IQR) | 34.5 (24–45) | 33 (24–43) | 33 (24–43) | 0.334 |
| U.S. military hospital | 0.004 | |||
| BAMC | 36 (32.1) | 94 (18.5) | 130 (20.9) | |
| NCR | 73 (65.2) | 389 (76.4) | 462 (74.4) | |
| Both BAMC and NCR | 3 (2.7) | 26 (5.1) | 29 (4.7) | |
| Mechanical ventilation | 0.030 | |||
| LRMC only | 16 (14.3) | 126 (24.7) | 142 (22.9) | |
| LRMC & U.S. hospital ≤1 week | 55 (49.1) | 242 (47.5) | 297 (47.8) | |
| LRMC & U.S. hospital ≥2 weeks | 3 (2.7) | 4 (0.8) | 7 (1.1) | |
| None | 38 (33.9) | 137 (26.9) | 175 (28.2) | |
| ICU admission | 90 (80.4) | 456 (89.6) | 546 (87.9) | 0.007 |
| Number of operating room visits, median (IQR) | 5 (4–6) | 4 (3–6) | 5 (3–6) | <0.001 |
| Hospitalization, median days (IQR) | 57 (40.5–84.5) | 44 (30–62) | 45 (33–66) | <0.001 |
| Death | 5 (4.5) | 12 (2.4) | 17 (2.7) | 0.216 |
BAMC–Brooke Army Medical Center; ICU–intensive care unit; IED–improvised explosive device; IQR–interquartile range; LRMC–Landstuhl Regional Medical Center; NCR–National Capital Region
a Predominant non-E. cloacae infections include coagulase-negative staphylococci (13%), Pseudomonas aeruginosa (12%), Escherichia coli (10.5%), Acinetobacter calcoaceticus-baumannii complex (8%), and Enterococcus faecium (8%).
Ninety percent of the 621 patients in the population sustained extremity injuries with the patients with E. cloacae infections having a greater proportion compared to the non-E. cloacae infected patients (98% vs. 88%; p = 0.016; Table 1). Despite the higher initial shock indices and blood product requirements, there was no significant difference in the injury severity scores between E. cloacae and non-E. cloacae infected patients (35 vs. 33; p = 0.33). A higher proportion of patients admitted to BAMC developed a E. cloacae infection (32% vs 19% among patients with non-E. cloacae infections; p = 0.004). Although fewer patients with E. cloacae infections required mechanical ventilation (64% vs. 73%; p = 0.03) or intensive care unit (ICU) admission (80% vs. 90%; p = 0.007), they did have longer hospitalizations (57 vs. 44 days; p<0.001). Patients with E. cloacae infections received significantly more total days of antibiotic therapy (43.5 vs. 34 days p <0.001). These patients also were treated significantly more days with carbapenems, 1st generation cephalosporins, fluoroquinolones, and vancomycin (Table 2). There was no significant mortality difference between the groups and 97% of the total population survived (Table 1).
Table 2. Total duration of antibiotic use among patients with and without E. cloacae infectionsa.
| Duration of Antibiotic Use, median days (IQR) | ||||
|---|---|---|---|---|
| Antimicrobials | Patients with E. cloacae infection (N = 112) | Patients with non-E. cloacae infection (N = 509) | All Patients (N = 621) | p-value |
| Aminoglycoside | 1 (0–7) | 1 (0–4) | 1 (0–4) | 0.256 |
| Carbapenem | 12.5 (4–21.5) | 9 (1–18) | 9 (2–18) | 0.003 |
| Cephalosporin- 1st generation | 8.5 (4–15) | 7 (3–12) | 7 (4–13) | 0.018 |
| Fluoroquinolone | 9 (1.5–15.5) | 5 (0–11) | 5 (1–13) | 0.003 |
| Vancomycin | 10 (2–27) | 0 (0–0) | 0 (0–0) | <0.001 |
| Total antibiotic durationb | 43.5 (32.5–71.0) | 34 (24–50) | 36 (26–52) | <0.001 |
IQR–interquartile range
a Antibiotics that were used for a median of zero days in both groups are not shown and include aminopenicillin, anti-pseudomonal penicillin, 2nd generation cephalosporin, 3rd generation cephalosporin, 4th generation cephalosporin, clindamycin, linezolid, macrolide, monobactam, penicillin, penicillinase-resistant penicillin, polymyxin, trimethoprim-sulfamethoxazole, and topical antibiotic therapy.
b Total antibiotic duration was calculated as the total number of days at least one antibiotic was administered. Any days on which no antibiotics were administered are not counted in this measure.
E. cloacae isolates were linked to 49 patients with serially infecting cultures and 63 patients with single initial infecting cultures. The patients with single initial and serial E. cloacae isolation were of similar median age (23 and 24 years, respectively), with the majority sustaining blast injuries (85.7% and 93.9%, respectively) resulting in a minority of burn wounds (15.9% and 16.3%, respectively) and a similar proportion of ICU admissions (81% and 79.6%, respectively; Table 3). There was a higher proportion of polymicrobial infections among the patients who had serial isolates compared to single initial isolates (86% and 67%, respectively; p = 0.021). Single initial vs serial isolation was not associated with a difference in number of operating room visits (median of 5 for both groups), length of hospitalization (median of 57 days for both groups), or death (5% and 4%, respectively; Table 3). Although there was not a significant difference in the duration of antibiotic therapy (median of 39 and 47 days for single initial and serial isolation respectively), there was a trend toward greater 1st generation cephalosporin utilization in patients who experienced serial isolation of E. cloacae (median of 11 vs. 7 days with single initial isolation; p = 0.052; Table 4); however, this does not control for duration of hospitalization and number of visits to the operating room, which would drive use of 1st generation cephalosporins in these trauma patients.
Table 3. Clinical characteristics of infected patients with single initial isolation of E. cloacae versus infected patients with serial isolation of E. cloacae.
| Characteristic, No. (%) | Patients with single initial E. cloacae isolation (N = 63) | Patients with serial E. cloacae isolation (N = 49) | Total Patients with E. cloacae isolation (N = 112) | p-value |
|---|---|---|---|---|
| Age at injury, median (IQR) | 22 (21–27) | 24 (21–27) | 23 (21–27) | 0.343 |
| Male | 62 (98.4) | 49 (100) | 111 (99.1) | 1.000 |
| Branch of Service | 0.403 | |||
| Air Force and Navy | 2 (3.2) | 3 (6.1) | 5 (5.5) | |
| Army | 36 (57.1) | 32 (65.3) | 68 (60.7) | |
| Marine | 25 (39.7) | 14 (28.6) | 39 (34.8) | |
| Combat Theater | 0.441 | |||
| Afghanistan | 60 (95.2) | 48 (98.0) | 108 (96.4) | |
| Iraq | 3 (4.8) | 1 (2.0) | 4 (3.6) | |
| Combat Injury | 59 (93.6) | 49 (100) | 108 (96.4) | 0.073 |
| Mechanism of Injury | 0.166 | |||
| Blast | 54 (85.7) | 46 (93.9) | 100 (89.3) | |
| Non-blast | 9 (14.3) | 3 (6.1) | 12 (10.7) | |
| Blast Type | 1.00 | |||
| IED | 50 (79.4) | 43 (87.7) | 93 (83.0) | |
| Non-IED | 4 (6.3) | 3 (6.1) | 7 (6.2) | |
| Injured on foot patrol | 40 (63.5) | 36 (73.5) | 76 (67.9) | 0.322 |
| Burn | 10 (15.9) | 8 (16.3) | 18 (16.1) | 0.948 |
| 1st documented shock index, median (IQR) | 1.05 (0.70–1.48) | 1.16 (0.83–1.51) | 1.07 (0.74–1.49) | 0.246 |
| 1st 24 hour blood transfusion, median (IQR) | 15 (7–31) | 14 (10–31) | 15 (8–31) | 0.930 |
| Body Region of Injury | 0.109 | |||
| Lower extremity | 8 (12.7) | 13 (26.5) | 21 (18.7) | |
| Upper extremity | 6 (9.5) | 1 (2.0) | 7 (6.2) | |
| Both lower and upper extremity | 48 (76.2) | 34 (69.4) | 82 (73.2) | |
| Non extremity | 1 (1.6) | 1 (2.0) | 2 (1.8) | |
| Injury severity score, median (IQR) | 34 (22–45) | 36 (27–45) | 34.5 (24–45) | 0.516 |
| U.S. military hospital | 0.361 | |||
| BAMC | 19 (30.1) | 17 (34.7) | 36 (32.1) | |
| NCR | 41 (65.1) | 32 (65.3) | 73 (65.2) | |
| Both BAMC and NCR | 3 (4.8) | 0 (0) | 3 (2.7) | |
| Mechanical ventilation | 0.872 | |||
| LRMC only | 9 (14.3) | 7 (14.3) | 16 (14.3) | |
| LRMC & U.S. hospital ≤1 week | 32 (50.8) | 23 (46.9) | 55 (49.1) | |
| LRMC & U.S. hospital ≥ 2 weeks | 1 (1.6) | 2 (4.1) | 3 (2.7) | |
| None | 21 (33.3) | 17 (34.7) | 38 (33.9) | |
| ICU admission | 51 (81.0) | 39 (79.6) | 90 (80.4) | 1.000 |
| Number of operating room visits, median (IQR) | 5 (4–7) | 5 (4–6) | 5 (4–6) | 0.216 |
| Polymicrobial infectiona | 42 (66.7) | 42 (85.7) | 84 (75.0) | 0.021 |
| Hospitalization, median days (IQR) | 57 (39–88) | 57 (43–84) | 57 (40.5–84.5) | 0.904 |
| Death | 3 (4.8) | 2 (4.0) | 5 (4.5) | 1.000 |
BAMC–Brooke Army Medical Center; ICU–intensive care unit; IED–improvised explosive device; IQR–interquartile range; LRMC–Landstuhl Regional Medical Center; NCR–National Capital Region
a Polymicrobial infection defined as a positive culture collected within ±3 days of the E. cloacae culture from the same anatomical site. Organisms predominantly isolated from polymicrobial infections were P. aeruginosa, E. faecium, E. coli, Acinetobacter calcoaceticus baumannii complex, Enterococcus faecalis, Aspergillus spp. and coagulase-negative staphylococci.
Table 4. Total duration of antibiotic use among patients with single initial isolation of E. cloacae versus patients with serial isolation of E. cloacaea.
| Duration of Antibiotic Use, median days (IQR) | ||||
|---|---|---|---|---|
| Antimicrobials | Patients with single initial E. cloacae infection (N = 63) | Patients with serial E. cloacae infection (N = 49) | All Patients with E. cloacae isolation (N = 112) | p-value |
| Aminoglycoside | 1 (0–8) | 1 (0–4) | 1 (0–7) | 0.153 |
| Carbapenem | 12 (5–20) | 13 (4–22) | 12.5 (4–21.5) | 0.796 |
| Cephalosporin- 1st generation | 7 (3–15) | 11 (6–15) | 8.5 (4–15) | 0.052 |
| Fluoroquinolone | 7 (0–15) | 10 (3–16) | 9 (1.5–15.5) | 0.299 |
| Vancomycin | 12 (1–27) | 9 (3–28) | 10 (2–27) | 0.911 |
| Total antibiotic durationb | 39 (30–63) | 47 (35–72) | 43.5 (32.5–71.0) | 0.236 |
IQR–interquartile range
a Antibiotics that were used for a median of zero days in both groups are not shown and include aminopenicillin, anti-pseudomonal penicillin, 2nd generation cephalosporin, 3rd generation cephalosporin, 4th generation cephalosporin, clindamycin, linezolid, macrolide, monobactam, penicillin, penicillinase-resistant penicillin, polymyxin, trimethoprim-sulfamethoxazole, and topical antibiotic therapy.
b Total antibiotic duration was calculated as the total number of days at least one antibiotic was administered. Any days on which no antibiotics were administered are not counted in this measure.
Among the 84 patients with polymicrobial infections, Pseudomonas aeruginosa was the most frequently isolated (34.5%), followed by Enterococcus faecium (30%), Escherichia coli (26%), Acinetobacter calcoaceticus baumannii complex (17%), Enterococcus faecalis (15.5%), Aspergillus spp. (14%), coagulase-negative staphylococci (14%), Enterococcus spp. (11%), Klebsiella pneumoniae (9.5%), and Staphylococcus aureus (9.5%). When E. cloacae infections were examined based on whether the infections were polymicrobial (N = 84) or monomicrobial (N = 28), there was no difference in use of mechanical ventilation (68% and 61%, respectively; p = 0.583), ICU admission (83% and 71%; p = 0.170), number of operating room visits (median of 5 for both; p = 0.125), length of hospitalization (median of 56 and 58.5 days; p = 0.898), and death (5% and 4%; p = 1.00). Polymicrobial E. cloacae infections were further examined for 29 patients who had the combination of E. cloacae plus P. aeruginosa (with/without other pathogens). To evaluate a wider group of bacteria of high virulence, 49 patients with E. cloacae plus at least one bacterium of high virulence (i.e., P. aeruginosa, E. coli, K. pneumoniae, and/or S. aureus), with/without other pathogens, were assessed. All 29 patients from the E. cloacae plus P. aeruginosa combination group were also included in the 49 patients with E. cloacae plus bacteria of high virulence group. Use of mechanical ventilation (72% for patients with polymicrobial combination of E. cloacae plus P. aeruginosa, p = 0.183; and 73.5% for patients with polymicrobial combination of E. cloacae plus bacteria of high virulence, p = 0.327), ICU admission (90%, p = 0.081; and 86%, p = 0.128), length of hospitalization (median 71 days, p = 0.131; and median 67 days, p = 0.130), and death (10%, p = 0.612; and 8%, p = 0.612) were not significantly different compared to patients with monomicrobial E. cloacae infections. There was also no significant difference in the number of operating room visits between patients with monomicrobial E. cloacae infections and polymicrobial infections with E. cloacae plus P. aeruginosa (median of 5 and 6, respectively, p = 0.078); however, patients with the combination of E. cloacae plus bacteria of high virulence had a significantly higher number of operating room visits (median 5, IQR: 5–7) compared to those with monomicrobial infections (median of 5; IQR: 4–6; p = 0.034).
Enterobacter cloacae culture characteristics
All Enterobacter isolates were identified as E. cloacae (not E. cloacae complex). The majority of E. cloacae isolates were cultured from wounds (70%), followed by respiratory specimens (22%) and blood (6%) (Table 5). Seventy-five percent of the wound cultures were recovered from the lower extremities. Patients in the serial isolate group had a shorter duration from injury to E. cloacae isolation (median 5 days; IQR 3–13) than patients in the single initial isolate group (median 8 days; IQR 4–7; p = 0.046). For serial E. cloacae patients, the median number of days between the initial isolate and last isolate was 5 days (IQR: 2–20 days).
Table 5. Distribution of sources of initial E. cloacae isolates.
| Sites of initial E. cloacae culture | Initial Isolates (N = 112) |
|---|---|
| Wound a | 78 (70%) |
| Thigh | 26 (33%) |
| Lower leg | 19 (24%) |
| Pelvic, gluteal muscles, and genitalia | 8 (10%) |
| Knee | 7 (9%) |
| Foot and ankle | 7 (9%) |
| Upper arm and elbow | 5 (6%) |
| Forearm and hand | 3 (4%) |
| Head and neck | 2 (3%) |
| Abdomen | 1 (1%) |
| Respiratory | 25 (22%) |
| Blood | 7 (6%) |
| Urine | 1 (1%) |
| Intravascular Catheter Tip | 1 (1%) |
a The percentage for the specific wound sites is calculated using 78 as the denominator.
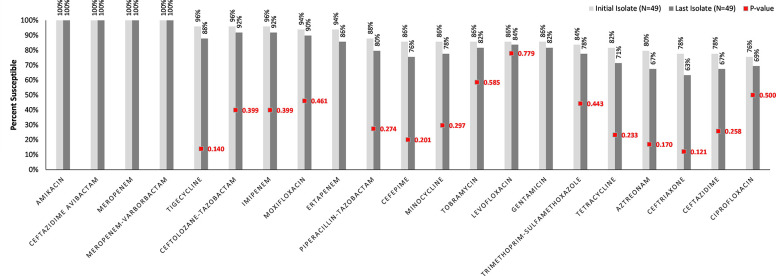
The comparative antibiogram between the initial and last E. cloacae isolates in the serial isolation group is shown in Fig 1. Amikacin, ceftazidime-avibactam, meropenem, and meropenem-vaborbactam retained 100% susceptibility between initial and last isolates. The last E. cloacae isolates were more resistant to almost all other antibiotics. The most notable decrease in susceptibility was noted for ceftriaxone, albeit not statistically significant (78% to 63% p = 0.121). All isolates were resistant to aminopenicillins, 1st generation cephalosporins, and cephamycins.
Fig 1. Comparative antibiogram of the E. cloacae isolates from the 49 patients in the serial isolation group.
Ordered by decreasing susceptibilities of the initial isolate.
Discussion
To the best of our knowledge, this study is the first to specifically characterize the significance that E. cloacae plays in battlefield trauma-related infections and broadly compare it against other bacterial infections. Battlefield trauma patients with E. cloacae infections more frequently presented with complex polytrauma resulting from blast injuries than patients with non-E. cloacae infection. Despite their critically-ill presentations, these patients did not require greater utilization of critical care, but they experienced lengthier hospitalizations, underwent a greater number of surgical interventions, and received longer durations of antimicrobial therapy. Secondarily, while patients who had serial E. cloacae isolates had a shorter duration from injury to 1st infecting isolate collection than those who had single initial isolates, there were no differences in characteristics or outcomes and no significant antibiotic resistance developed in the patients from whom E. cloacae was recovered multiple times. Although a large proportion of the E. cloacae infections were polymicrobial (75% of patients), there was no difference in outcomes (e.g., ICU admission, length of hospitalization, or death) when patients with polymicrobial and monomicrobial E. cloacae infections were compared, including when focused on specific polymicrobial combinations of clinical relevance (i.e., E. cloacae plus P. aeruginosa and E. cloacae plus bacteria of high virulence). The only significant difference between the patients with monomicrobial and polymicrobial infections was an increased number of operating visits among patients with the combination of E. cloacae plus bacteria of high virulence (i.e., P. aeruginosa, E. coli, K. pneumoniae, or S. aureus).
E. cloacae is the 4th most common Gram-negative organism causing bloodstream infections in over 200 medical centers in 45 different nations [25]. Notably, E. cloacae, as well as Klebsiella aerogenes and Citrobacter freundii have been found to be the most clinically relevant AmpC producers and AmpC’s conference of resistance to broad-spectrum β-lactams has been shown to produce significant adverse effects on clinical outcomes [26]. A 2002 study by Cosgrove et al. [27] evaluated health and economic outcomes for patients with a mean age of 63 years who had Enterobacter spp. infections cultured from several different anatomic sites that developed resistance to 3rd generation cephalosporins. Similar to other published reports [11, 12], they found that resistance developed in 10% of their population, producing an attributable longer hospital stay of 9 days, increased mortality relative risk of 5.02, and additional hospital cost of almost $30,000 [27].
In our study, compared to patients with non-E. cloacae infections, patients with E. cloacae had longer hospital stays (57 vs. 44 days) and required a significantly greater duration of antibiotic therapy (43.5 vs. 34 days); however, it should be noted that the duration of antibiotic therapy was the overall duration and not adjusted per length of hospitalization and number of operating room visits, which would impact antibiotic use (e.g., perioperative antibiotics). The comparison between patients with E. cloacae and non-E. cloacae infections did adjust for the occurrence of polymicrobial infections, including assessing clinically relevant combinations. A previous study using the TIDOS population identified that patients with P. aeruginosa infections had higher crude mortality compared to patients with infections attributed to other pathogens [28]. Therefore, we compared patients with monomicrobial E. cloacae infections to those with polymicrobial E. cloacae plus P. aeruginosa infections and there were no statistical differences in critical care or mortality between the groups. Among our total population, 17 (3% of 621) patients died and, without controlling for temporal relationship to infection or other potential factors that would potentially contribute to mortality, there was no significant mortality difference between those with and without E. cloacae infection (5% vs. 2%). In contrast to the 17% mortality reported by Cosgrove et al. [27], the low mortality in our patients is attributable to youth and overall better health prior to their battlefield wounds and E. cloacae infections.
It is noteworthy that 22% of our initial E. cloacae isolates were resistant to 3rd generation cephalosporins. Although this is lower than the prevalence of 36.4% that was seen in a national surveillance study that measured 3rd generation cephalosporin resistance amongst E. cloacae isolates cultured from American ICU patients [29], our study population’s baseline E. cloacae resistance to 3rd generation cephalosporins may have contributed to the comparatively adverse outcomes seen in the E. cloacae infection patients compared to those with a non-E. cloacae infection; although, analysis of that relationship was outside the scope of this study. Despite fewer admissions to the ICU and less need for mechanical ventilation (80% vs 90% and 66% vs. 73% between the E. cloacae and non-E. cloacae infection patients, respectively), patients with E. cloacae infections had higher first documented shock indices (1.07 vs. 0.92), received more blood products within the first 24 hours of care (15 vs. 14 units) and had more operating room visits (5 vs. 4), which may have been secondary to the fact that patients with E. cloacae infection suffered more blast injuries (89% vs. 75%) and burns (16% vs. 9%) resulting in greater fluid loss [30]. Also, the greater number of burns in the E. cloacae infection population contributed to their significantly higher admission rate to BAMC, as BAMC is the DoD’s only specialized burn center. As burns require frequent debridement, the greater number of burn injuries also potentially led to the higher number of operating room visits in the E. cloacae infection group.
Regarding specific antibiotic utilization, patients with E. cloacae infection received significantly more carbapenems, 1st generation cephalosporins, fluoroquinolones, and vancomycin (Table 2). Burn injury commonly results in infection with S. aureus, which has led to the empiric use of vancomycin [31]. Thus, the greater number of burn injuries in the E. cloacae infection group, as well as the high proportion of polymicrobial infections (75% of patients) likely contributed to their greater receipt of vancomycin.
As previously mentioned, E. cloacae has a chromosomal AmpC β-lactamase, which is strongly induced by β-lactams, such as carbapenems and 1st generation cephalosporins. Resistance development secondary to AmpC induction is of special interest to the U.S. Armed Forces as ertapenem is recommended to be carried on the battlefield by medics in the TCCC guidelines and cefazolin is recommended as post-trauma antibiotic prophylaxis in the JTS CPG [17, 19]. These recommendations provide the possibility for the development of harmful resistance, leading to poor patient outcomes when initial wound infections are due to E. cloacae. As a result of the resistance-inducing pressure of carbapenems, there has been significant interest in seeking out carbapenem-sparing therapies, such as piperacillin-tazobactam or cefepime [32]. Our population did not receive a significant amount of therapy with piperacillin-tazobactam nor cefepime and the most prescribed antibiotic therapy for patients who had an E. cloacae infection was a carbapenem. Cefepime has also garnered significant attention as a carbapenem-sparing agent when treating AmpC-producing organisms and was only recently recommended by the IDSA as first-line therapy against E. cloacae, as well as K. aerogenes and C. freundii when the minimum inhibitory concentration (MIC) is known to be ≤2 μg/mL [26]. In our study, approximately 85% of isolates were susceptible to cefepime (MIC ≤2 μg/mL) with the majority having a MIC ≤0.5 μg/mL, while the resistant isolates largely had a MIC >16 μg/mL. As infection site inevitably affects clinical outcome data, it is important to note that our isolates differed from those assessed in other studies, such as the MERINO trials which focused on bacteremia [33, 34], in that our isolates were largely collected from infected traumatic wounds and only 6% of initial isolates were blood cultures. A significant proportion of the patients with E. cloacae infection in our study sustained burn injuries (16%), and although Enterobacter spp. cause a minority of burn wound infections, the bacteriology of burn wound infection remains largely Gram-negative [35].
Despite the presumed high risk of AmpC induction and de-repression given our study population’s frequent receipt of carbapenems, no statistically significant resistance developed to any individual antibiotic in our study when examining serial isolates. Nevertheless, there was a non-significant trend toward resistance development against almost every β-lactam antibiotic tested except for ceftazidime-avibactam and meropenem (i.e., ceftolozane-tazobactam, imipenem, ertapenem, piperacillin-tazobactam, cefepime, aztreonam, ceftriaxone, and ceftazidime). The development of β-lactam resistance in our study is similar to prior burn wound infection literature that examined the incidence of general resistance phenotypes of Enterobacterales over time [36]. However, in the 2016 study by van Duin et al. [36], significant resistance developed over the course of weeks and in our study, the median interval between initial and last isolates was 5 days (IQR 2–20).
The lack of difference in clinical outcomes between the single initial and serial isolate groups in our study may be attributed to fast source control in both groups, as evidenced by the high number of visits to the operating room for surgical debridement. Patients in our serial E. cloacae group had a shorter duration to isolation after their injuries (5 vs. 8 days p = 0.046), and similar to the findings of a prior analysis of the TIDOS population [6], 75% of the E. cloacae infections in our analysis were associated with polymicrobial infections. Nevertheless, there was no difference in outcomes between patients in the E. cloacae single initial and serial isolate groups, as well as between the E. cloacae infection patients with polymicrobial and monomicrobial infections. Given that E. cloacae carries the greatest risk for AmpC derepression, our findings regarding resistance development and clinical outcomes may be generalizable to combat trauma infections with K. aerogenes, Serratia marcescens, C. freundii, Providencia stuartii, Morganella morganii [37]. The lack of difference in clinical outcomes between the single initial and serial isolate groups bolsters both the DoD’s current combat critical care and surgical prophylaxis guidelines with regard to AmpC induction and also supports prior literature that argued against the use of expanded Gram-negative antibiotic prophylaxis after combat trauma [17, 19, 38, 39].
Our study includes limitations inherent to retrospective studies. As our analysis was not a case-control study, the non-E. cloacae patients served as a comparator group rather than a control population, so matching was not applied. A potential confounder of clinical outcome differences is that a significant proportion of the patients infected with E. cloacae were hospitalized at BAMC, of whom, 16% were admitted for burn wound care, which likely led to prolonged hospitalizations [40]. Similar to other retrospective reports of emergence of resistance while on treatment [8, 27], we did not perform a molecular assessment to ascertain the likely mechanism for resistance observed. Even if using ceftriaxone resistance as a marker for AmpC or ESBL production, the difference in resistance between first and last isolates from the serial isolate population was not statistically significant (p = 0.121). As isolates did not undergo bacterial strain typing, we cannot directly state whether the initial and serial isolates were the same. Molecular or enzymatic characterization of β-lactamase production would likely have been useful if a significant difference in 3rd generation cephalosporin resistance between the initial and last isolates was detected. Lastly, the DoDTR did not capture antibiotics that were provided in the prehospital setting (e.g., ertapenem) at the time of injury, which may have limited the evaluation for β–lactam resistance development.
To the best of our knowledge, our study is the first to specifically evaluate E. cloacae’s role as a pathogen in infection secondary to modern combat trauma. Despite DoD combat care and surgical prophylaxis guidelines recommending upfront provision of AmpC-inducing antibiotics [41], we did not see worsened clinical outcomes or significant antibiotic resistance develop in patients who experienced serial isolation of E. cloacae versus single initial isolation. Carbapenems were the most frequently prescribed antibiotics for our combat trauma population with E. cloacae infections. As IDSA and DoD guidance changes regarding antibiotic utilization, future studies on clinical outcome surveillance coupled with molecular characterization of resistance mechanisms amongst combat trauma patients are needed to ensure optimal care and support antimicrobial stewardship efforts in the Military Health System.
Acknowledgments
We are indebted to the Infectious Disease Clinical Research Program Trauma Infectious Disease Outcomes Study team of clinical coordinators, microbiology technicians, data managers, clinical site managers, and administrative support personnel for their tireless hours to ensure the success of this project. We also wish to thank MAJ Jack Kiley for his support and guidance.
Data Availability
All relevant data are contained in the paper. Data for this study are available from the Infectious Disease Clinical Research Program (IDCRP), headquartered at the USU, Department of Preventive Medicine and Biostatistics. Review by the USU Institutional Review Board and approval of data sharing agreements are required for use of the data collected under this protocol. Data requests may be sent to: Address: 6270A Rockledge Drive, Suite 250, Bethesda, MD 20817. Email: contactus@idcrp.org.
Funding Statement
Support for this work (IDCRP-024) was provided by the Infectious Disease Clinical Research Program (IDCRP), a Department of Defense program executed through the Uniformed Services University of the Health Sciences, Department of Preventive Medicine and Biostatistics through a cooperative agreement with The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF). This project has been funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, https://www.niaid.nih.gov/, under Inter-Agency Agreement Y1-AI-5072 to DRT, the Defense Health Program, U.S. DoD, under award HU0001190002 to DRT, the Department of the Navy under the Wounded, Ill, and Injured Program (HU0001-10-1-0014) to DRT, and the Military Infectious Diseases Research Program, https://midrp.amedd.army.mil/ (HU0001-15-2-0045) to KM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Support in the form of salaries was provided by HJF for authors KM, LS, FS, AR; HJF did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Aronson NE, Sanders JW, Moran KA. In harm’s way: infections in deployed American military forces. Clin Infect Dis. 2006;43(8):1045–51. doi: 10.1086/507539 . [DOI] [PubMed] [Google Scholar]
- 2.Tong MJ. Septic complications of war wounds. JAMA. 1972;219(8):1044–7. . [PubMed] [Google Scholar]
- 3.Wallum TE, Yun HC, Rini EA, Carter K, Guymon CH, Akers KS, et al. Pathogens present in acute mangled extremities from Afghanistan and subsequent pathogen recovery. Mil Med. 2015;180(1):97–103. doi: 10.7205/MILMED-D-14-00301 . [DOI] [PubMed] [Google Scholar]
- 4.Blyth DM, Yun HC, Tribble DR, Murray CK. Lessons of war: Combat-related injury infections during the Vietnam War and Operation Iraqi and Enduring Freedom. J Trauma Acute Care Surg. 2015;79(4 Suppl 2):S227–35. doi: 10.1097/TA.0000000000000768 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell WR, Li P, Whitman TJ, Blyth DM, Schnaubelt ER, Mende K, et al. Multi-drug-resistant Gram-negative infections in deployment-related trauma patients. Surg Infect (Larchmt). 2017;18(3):357–67. doi: 10.1089/sur.2017.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mende K, Stewart L, Shaikh F, Bradley W, Lu D, Krauss MR, et al. Microbiology of combat-related extremity wounds: Trauma Infectious Disease Outcomes Study. Diagn Microbiol Infect Dis. 2019;94(2):173–9. doi: 10.1016/j.diagmicrobio.2018.12.008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evoluation of a global menance. J Infect Dis. 2017;215(suppl_1):S28–36. doi: 10.1093/infdis/jiw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson BM, El Chakhtoura NG, Patel S, Saade E, Donskey CJ, Bonomo RA, et al. Carbapenem-resistant Enterobacter cloacae in patients from the US Veterans Health Administration, 2006–2015. Emerg Infect Dis. 2017;23(5):878–80. doi: 10.3201/eid2305.162034 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meini S, Tascini C, Cei M, Sozio E, Rossolini GM. AmpC beta-lactamase-producing Enterobacterales: what a clinician should know. Infection. 2019;47(3):363–75. doi: 10.1007/s15010-019-01291-9 . [DOI] [PubMed] [Google Scholar]
- 10.Chow JW, Fine MJ, Shlaes DM, Quinn JP, Hooper DC, Johnson MP, et al. Enterobacter bacteremia: clinical features and emergence of antibiotic resistance during therapy. Ann Intern Med. 1991;115(8):585–90. doi: 10.7326/0003-4819-115-8-585 . [DOI] [PubMed] [Google Scholar]
- 11.Choi SH, Lee JE, Park SJ, Choi SH, Lee SO, Jeong JY, et al. Emergence of antibiotic resistance during therapy for infections caused by Enterobacteriaceae producing AmpC beta-lactamase: implications for antibiotic use. Antimicrob Agents Chemother. 2008;52(3):995–1000. doi: 10.1128/AAC.01083-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang CI, Kim SH, Park WB, Lee KD, Kim HB, Oh MD, et al. Bloodstream infections caused by Enterobacter species: predictors of 30-day mortality rate and impact of broad-spectrum cephalosporin resistance on outcome. Clin Infect Dis. 2004;39(6):812–8. doi: 10.1086/423382 . [DOI] [PubMed] [Google Scholar]
- 13.Sanders CC, Bradford PA, Ehrhardt AF, Bush K, Young KD, Henderson TA, et al. Penicillin-binding proteins and induction of AmpC beta-lactamase. Antimicrob Agents Chemother. 1997;41(9):2013–5. doi: 10.1128/AAC.41.9.2013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber DA, Sanders CC. Diverse potential of beta-lactamase inhibitors to induce class I enzymes. Antimicrob Agents Chemother. 1990;34(1):156–8. doi: 10.1128/AAC.34.1.156 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris PN, Wei JY, Shen AW, Abdile AA, Paynter S, Huxley RR, et al. Carbapenems versus alternative antibiotics for the treatment of bloodstream infections caused by Enterobacter, Citrobacter or Serratia species: a systematic review with meta-analysis. J Antimicrob Chemother. 2016;71(2):296–306. doi: 10.1093/jac/dkv346 . [DOI] [PubMed] [Google Scholar]
- 16.Sanders WE Jr, Sanders CC. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev. 1997;10(2):220–41. doi: 10.1128/CMR.10.2.220 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joint Trauma System, Committee on Tactical Combat Casualty Care. TCCC Guidelines 2021. Available from: https://deployedmedicine.com/market/31/content/40. [Google Scholar]
- 18.Bratzler DW, Dellinger EP, Olsen KM, Perl TM, Auwaerter PG, Bolon MK, et al. Clinical practice guidelines for antimicrobial prophylaxis in surgery. Am J Health Syst Pharm. 2013;70(3):195–283. doi: 10.2146/ajhp120568 . [DOI] [PubMed] [Google Scholar]
- 19.Barsoumian A, Solberg S, Maves R, Markelz E, Crouch H, Yun H, et al. Infection Prevention in Combat-related Injuries (CPG ID:24) Provides rationale and guidance for the prevention of infection after combat-related injuries. Joint Trauma System Clinical Practice Guideline. 2021. Available from: https://jts.health.mil/assets/docs/cpgs/Infection_Prevention_in_Combat-related_Injuries_27_Jan_2021_ID24.pdf. [Google Scholar]
- 20.Tribble DR, Conger NG, Fraser S, Gleeson TD, Wilkins K, Antonille T, et al. Infection-associated clinical outcomes in hospitalized medical evacuees after traumatic injury: Trauma Infectious Disease Outcome Study. J Trauma. 2011;71(1 Suppl):S33–42. doi: 10.1097/TA.0b013e318221162e . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tribble DR, Murray CK, Lloyd BA, Ganesan A, Mende K, Blyth DM, et al. After the battlefield: infectious complications among wounded warriors in the Trauma Infectious Disease Outcomes Study. Mil Med. 2019;184(Suppl 2):18–25. doi: 10.1093/milmed/usz027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tribble DR, Spott MA, Shackelford S, Gurney JM, Murray CK. Department of Defense Trauma Registry infectious disease module impact on clinical practice. Mil Med. 2022;187(Suppl 2):7–16. doi: 10.1093/milmed/usac050 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Control Centers for Disease and Prevention. CDC/NHSN Surveillance Definitions for Specific Types of Infections. 2023. Available from: http://www.cdc.gov/nhsn/pdfs/pscmanual/17pscnosinfdef_current.pdf. [Google Scholar]
- 24.Weinstein M, Lewis J, Bobenchik A, Campeau S, Cullen S, Galas M, et al. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100, 30th edition: Clinical and Laboratory Standards Institute; 2020. [Google Scholar]
- 25.Diekema DJ, Hsueh PR, Mendes RE, Pfaller MA, Rolston KV, Sader HS, et al. The microbiology of bloodstream infection: 20-year trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother. 2019;63(7):e00355–19. doi: 10.1128/AAC.00355-19 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.IDSA Guidance on the Treatment of Antimicrobial-Resistant Gram-Negative Infections: Version 2.0, (2023). Available from: https://www.idsociety.org/practice-guideline/amr-guidance-2.0/. [Google Scholar]
- 27.Cosgrove SE, Kaye KS, Eliopoulous GM, Carmeli Y. Health and economic outcomes of the emergence of third-generation cephalosporin resistance in Enterobacter species. Arch Intern Med. 2002;162(2):185–90. doi: 10.1001/archinte.162.2.185 . [DOI] [PubMed] [Google Scholar]
- 28.Ford MB, Mende K, Kaiser SJ, Beckius ML, Lu D, Stam J, et al. Clinical characteristics and resistance patterns of Pseudomonas aeruginosa isolated from combat casualties. Mil Med. 2022;187(3/4):426–34. doi: 10.1093/milmed/usab259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.System NNIS. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1990-May 1999, issued June 1999. A report from the NNIS System. Am J Infect Control. 1999;27(6):520–32. doi: 10.1016/s0196-6553(99)70031-3 . [DOI] [PubMed] [Google Scholar]
- 30.Marenco CW, Lammers DT, Morte KR, Bingham JR, Martin MJ, Eckert MJ. Shock index as a predictor of massive transfusion and emergency surgery on the modern battlefield. J Surg Res. 2020;256:112–8. doi: 10.1016/j.jss.2020.06.024 . [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Li D, Xu L, Hu Y, Sang Y, Zhang G, et al. Epidemiology and outcomes of bloodstream infections in severe burn patients: a six-year retrospective study. Antimicrob Resist Infect Control. 2021;10(1):98. doi: 10.1186/s13756-021-00969-w . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The White House. National Action Plan For Combating Antibiotic-Resistant Bacteria. 2015. Available from: https://obamawhitehouse.archives.gov/sites/default/files/docs/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf. [Google Scholar]
- 33.Harris PNA, Tambyah PA, Lye DC, Mo Y, Lee TH, Yilmaz M, et al. Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: a randomized clinical trial. JAMA. 2018;320(10):984–94. doi: 10.1001/jama.2018.12163 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart AG, Paterson DL, Young B, Lye DC, Davis JS, Schneider K, et al. Meropenem versus piperacillin-tazobactam for definitive treatment of bloodstream infections caused by AmpC beta-lactamase-producing Enterobacter spp, Citrobacter freundii, Morganella morganii, Providencia spp, or Serratia marcescens: a pilot multicenter randomized controlled trial (MERINO-2). Open Forum Infect Dis. 2021;8(8):ofab387. doi: 10.1093/ofid/ofab387 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Azzopardi EA, Azzopardi E, Camilleri L, Villapalos J, Boyce DE, Dziewulski P, et al. Gram negative wound infection in hospitalised adult burn patients—systematic review and metanalysis. PLoS One. 2014;9(4):e95042. doi: 10.1371/journal.pone.0095042 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Duin D, Strassle PD, DiBiase LM, Lachiewicz AM, Rutala WA, Eitas T, et al. Timeline of health care-associated infections and pathogens after burn injuries. Am J Infect Control. 2016;44(12):1511–6. doi: 10.1016/j.ajic.2016.07.027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohlmann R, Bahr T, Gatermann SG. Species-specific mutation rates for ampC derepression in Enterobacterales with chromosomally encoded inducible AmpC beta-lactamase. J Antimicrob Chemother. 2018;73(6):1530–6. doi: 10.1093/jac/dky084 . [DOI] [PubMed] [Google Scholar]
- 38.Lloyd BA, Murray CK, Shaikh F, Carson ML, Blyth DM, Schnaubelt ER, et al. Antimicrobial prophylaxis with combat-related open soft-tissue injuries. Mil Med. 2018;183(9–10):e260–5. doi: 10.1093/milmed/usx125 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lloyd BA, Murray CK, Shaikh F, Carson ML, Blyth DM, Schnaubelt ER, et al. Early infectious outcomes after addition of fluoroquinolone or aminoglycoside to posttrauma antibiotic prophylaxis in combat-related open fracture injuries. J Trauma Acute Care Surg. 2017;83(5):854–61. doi: 10.1097/TA.0000000000001609 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor SL, Sen S, Greenhalgh DG, Lawless M, Curri T, Palmieri TL. Real-time prediction for burn length of stay via median residual hospital length of stay methodology. J Burn Care Res. 2016;37(5):e476–82. doi: 10.1097/BCR.0000000000000332 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hospenthal DR, Murray CK, Andersen RC, Bell RB, Calhoun JH, Cancio LC, et al. Guidelines for the prevention of infections associated with combat-related injuries: 2011 update: endorsed by the Infectious Diseases Society of America and the Surgical Infection Society. J Trauma. 2011;71(2 Suppl 2):S210–34. doi: 10.1097/TA.0b013e318227ac4b . [DOI] [PubMed] [Google Scholar]