Abstract
During early embryogenesis, the transcription factor SOX17 contributes to hepato-pancreato-biliary system formation and vascular-hematopoietic emergence. To better understand Sox17 function in the developing endoderm and endothelium, we developed a dual-color temporal lineage-tracing strategy in mice combined with single-cell RNA sequencing to analyze 6934 cells from Sox17-expressing lineages at embryonic days 9.0-9.5. Our analyses showed 19 distinct cellular clusters combined from all 3 germ layers. Differential gene expression, trajectory and RNA-velocity analyses of endothelial cells revealed a heterogenous population of uncommitted and specialized endothelial subtypes, including 2 hemogenic populations that arise from different origins. Similarly, analyses of posterior foregut endoderm revealed subsets of hepatic, pancreatic, and biliary progenitors with overlapping developmental potency. Calculated gene-regulatory networks predict gene regulons that are dominated by cell type-specific transcription factors unique to each lineage. Vastly different Sox17 regulons found in endoderm versus endothelial cells support the differential interactions of SOX17 with other regulatory factors thereby enabling lineage-specific regulatory actions.
Keywords: single-cell RNA sequencing, Sox17, endoderm, hepato-pancreato-biliary system, endothelium, hematopoiesis
Graphical abstract
Graphical Abstract.
Introduction
Sets of so-called “master” regulatory genes, such as those in the Sry-related SOX transcription factor (TF) family, play critical roles in a spatiotemporally modulated and hierarchically organized cascade of such networks, enabling progressively more complex specification of the many cellular lineages required for completing organogenesis. Sox17 is a crucial contributor to multiple developmental processes in early vertebrate embryogenesis.1-3 A defining feature of all SOX proteins is a high mobility group (HMG) box domain4 that binds the minor groove of the DNA double helix, resulting in DNA bending that enables the overlaying chromatin topology to be modulated.5 Interactions between SOX17 and other protein partners during development enable Sox17 to dynamically regulate target genes6-8 in multiple lineages, including extra-embryonic endoderm, definitive endoderm, vascular endothelium, hematopoietic stem cells, and oligodendrocytes.9-11
Previous analyses of bulk transcriptomic changes in Sox17-null embryos and Sox17-overexpressing cells in culture produced highly variable predictions of Sox17 downstream targets, suggesting cell type-specific effects.12-15 Single-cell RNA sequencing (scRNAseq) has enabled investigation of transcriptomic features across different cell types and developmental trajectories. While endoderm development (especially posterior foregut), and endothelium-derived hematopoiesis have been broadly subjected to scRNAseq transcriptomic analyses,16-19 a more focused decoding of Sox17 lineages is necessary to gain deeper insights into the development of subsets of endodermal and mesodermal tissues.
In this study we used a Sox17-centric strategy, utilizing a Sox17GFPCre allele alone or in combination with a R26LSL.TdTomato Cre reporter, to isolate Sox17-expressing cells from E9.0 - E9.5 embryos. scRNAseq analysis of these cells revealed multiple clusters of endoderm, endothelium, and hematopoietic cells. Trajectory analysis of the endothelial compartment identified 2 pools of hemogenic endothelial cells of different origins. By integrating the endodermal cluster with a previously published dataset,20 we observed a spectrum of overlapping transcriptomic profiles among the hepatic, pancreatic, and biliary progenitors as they segregate within the posterior foregut domain. Applying pySCENIC,21 which infers scRNAseq co-expression patterns between genes and further refines direct targets using existing TF binding-motif databases to predict active gene regulatory networks (GRNs) in various cell types, we identified prospective distinct endodermal and endothelial-specific gene targets for SOX17.
Methods
Mouse Lines
The Sox17GFPCre allele (Sox17tm1.3(CreGFP)Mgn, MMRRC_036463-UNC), containing a GFPCre fusion protein in lieu of SOX17 coding sequences,22 was bred into a CD1 background and maintained in a heterozygous state. Mice that are heterozygous for the Sox17GFPCre allele exhibit no obvious phenotype, are fertile, born at the expected Mendelian ratios, and have normal life expectancy. Sox17GFPCre heterozygous embryos for sequencing experiments were generated by crossing Sox17GFPCre heterozygous males (CD1 background) with either wild-type (CD1 background) or R26LSL.TdTomato (B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, The Jackson Laboratory, #007914) homozygous (C57BL/6J background) females. Cre-negative, Sox17 wild-type embryos harvested from the same dam established baseline fluorescence for cell sorting. Embryos at embryonic day (E) 9.0-9.5 were dissected in cold PBS and staged by Theiler’s criteria. Genotypes were from PCR analysis of yolk sac DNA and fluorescence signal (green indicating presence of Sox17GFPCre and red indicating recombination of R26LSL.TdTomato by Sox17GFPCre).
Genotyping
Tail or yolk-sac tissue digested with proteinase K was analyzed by PCR assay detecting wild-type or mutant Sox17 allele with EconoTaq PLUS 2X Master Mix (Biosearch Technologies, #30035). Primers (forward: 5ʹ CAGAGGTATGCAGATCTCTGT 3ʹ and reverse: 5ʹ CATTCTGGTCAACATGTAAGGT 3ʹ) were annealed at 57 °C.
Fluorescence-Activated Cell Sorting
Embryos were incubated for 10 min at 37 °C with Accumax (Millipore Sigma, #A7089) supplemented with DNase (Life Technologies, #AM2222; 1:1000 dilution ratio), and further disaggregated by vigorous pipetting during incubation. Passing the cell suspension through 35 µm strainer caps (Corning, #352235) led to single cells, which were washed with FACS buffer (R&D systems, #FC001) and pelleted at 4 °C, 1000 rpm for 3 min. Cells were resuspended in FACS buffer containing DNaseI (1:1000 dilution) and stained with 7AAD (15 min, room temperature—RT). Live cells (7AAD-negative) expressing either GFP or TdTomato fluorescence were then sorted into DMEM with 10% FBS at the Vanderbilt Flow Cytometry Shared Resource.
Sequencing and Computational Analysis
Cell suspensions were processed for 3ʹ-scRNAseq by the 10X Chromium system. A NovaSeq 6000 device was used to acquire 150 bp paired-end reads. Base calling was performed by RTA (version 2.4.11; Illumina), and further analysis was carried out using 10X Genomics Cell Ranger software v3.0.2. Initial quality control filtration, unsupervised clustering, and differential gene expression analysis used Seurat 4.23 Doublet prediction used DoubletFinder.24 Pseudotime analysis used Monocle 3,25 while RNA velocities were calculated with scVelo.26 pySCENIC21 was used to infer gene regulatory networks with downstream visuals by Cytoscape27 and ComplexHeatmap.28
Data Availability
Raw scRNAseq data is at ArrayExpress (E-MTAB-12719), Seurat objects are at https://zenodo.org/record/7725887#.ZA5jRR_MLkI, and scripts used to perform quality control, clustering, differential gene expression analysis, Monocle3 and RNAvelocity analyses are at https://github.com/markmagnuson/2023-Linh-scRNASeq-Mouse-Sox17-Expressing-Lineages.
Immunofluorescence Staining
Embryos were fixed with 2% PFA (1 h, RT), incubated in 30% sucrose (overnight, 4 oC), embedded in OCT (Sakura, #4583). Eight micrometre-thick embryonic sections were post-fixed with ice-cold acetone (30 min, RT), permeabilized with 0.3% Triton X-100 (10 min, RT), and blocked with 3% BSA (1 h, RT). Primary antibodies (Supplementary Table S1) were diluted in 1% BSA for staining (overnight, 4 °C). After 3 PBST (0.2% Tween in PBS) washes, secondary antibodies (Supplementary Table S1) were incubated (1 h, RT), followed by 3 PBST washes and autofluorescence quenching with Vector TrueVIEW (following manufacturer protocol; Vector Laboratories, #SP-8400-15). Stained sections were mounted with Vectashield (Vector Laboratories, #H-1700) and imaged using Zeiss LSM710 confocal microscope.
Results
Sox17 GFPCre Expression Faithfully Reflects Known Sox17-Expressing Lineages and Suggests Several New Sox17-Expressing Cell Types
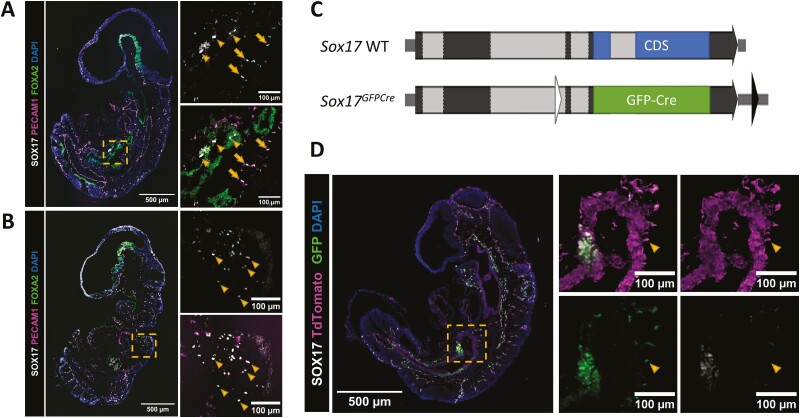
To better define Sox17 gene expression at E9.5, a dynamic time during gastrulation when multiple Sox17-associated cellular diversifications are occurring, we began with immunofluorescence labeling of SOX17, PECAM1, and FOXA2, marking previously known Sox17-expressing lineages (Fig. 1A). As expected, SOX17 was detected mainly in PECAM1+ vascular cells and FOXA2+ endoderm. Nuclear signals were also present in scattered SOX17+ cells not producing either PECAM1 or FOXA2, located mostly along the dorsal side of embryos (Fig. 1B), suggesting previously unreported Sox17-expressing lineages or sites of aberrant gene expression.
Figure 1.
Temporal lineage tracing of Sox17 expression with a dual color system in mice. (A) Representative image and inset of immunofluorescence labelling of an E9.5 mouse embryonic section showing nuclear SOX17 staining (colocalizes with DAPI staining) seen in both endodermal cells (positive for FOXA2—arrowheads) and endothelial cells (positive for PECAM1—arrows). (B) Representative image and inset of immunofluorescence labelling of an E9.5 mouse embryonic section showing SOX17 positive cells that are not positive for either FOXA2 or PECAM1 staining, arrowheads. (C) Sox17 temporal lineage tracing allele with full-length SOX17 protein coding sequence (CDS) replaced with a sequence coding for GFP-Cre fusion protein (black boxes—exon; gray boxes—intron; blue and green boxes—protein coding sequences; black and white triangles—retained loxP sites after recombinase-mediated cassette exchange strategy used to generate the allele.22 (D) Immunofluorescence labelling of SOX17, GFP, and TdTomato in Sox17GFPCre/+R26LSL.TdTomato/+ E9.5 embryonic sections. Inset shows a group TdTomato positive cells with the majority are negative for both GFP and SOX17 and a small portion positive for GFP but not SOX17, as indicated by arrowhead.
To characterize Sox17-expressing populations we developed a dual-color lineage-tracing strategy by intercrossing Sox17GFPCre/+ (Fig. 1C) and R26LSL.TdTomato Cre-reporter mice, to enable identification of short and long-term progeny of Sox17-expressing lineages based on GFP and TdTomato production. Immuno-labeling of SOX17, GFP, and TdTomato in E9.5 embryonic sections confirmed SOX17 colocalization with all GFP, while most TdTomato-positive cells did not have GFP or SOX17 (Fig. 1D), indicating short-term longevity of the GFP-Cre fusion protein and robustness of the dual-color tracing strategy. While TdTomato intensity was uniform throughout the embryos, GFP fluorescence varied within the emerging endoderm at E8.5-E9.5 reflecting temporally varying Sox17 expression throughout the foregut, midgut, and hindgut (Supplementary Fig. S1). Within mesodermal tissues, TdTomato but not GFP was present mostly in the heart at E8.5-9.0, while a mixture of both fluorescence signals was observed within the vasculature in other embryonic regions by E9.5 (Supplementary Fig. S1).
Single-Cell Transcriptomic Analysis Reveals Canonical and Non-Canonical Sox17-Expressing Cells
Two separate E9.0-9.5 single-cell collections were prepared from our dual-color strategy (Fig. 2A) using Sox17GFPCre/+ males mated with wild type or R26LSL.TdTomato/LSL.TdTomato females. The first sample reporting Sox17 short-term expression was prepared on the basis of GFP produced from a mixture of Sox17GFPCre/+; R26+/+ and Sox17+/+; R26+/+ embryos. The second sample was isolated from Sox17GFPCre/+; R26LSL.TdTomato/+ embryos were identified by red fluorescence, and comprised cells derived over a short period after Sox17 expression indicated from their green/red co-fluorescence, or over a longer period by being only red. scRNAseq produced 2 datasets, hereafter referred to as GFP and TdTomato datasets, which were analyzed separately to filter out low-quality cells and predicted doublets (Supplementary Figs. S2 and S3). The integrated (GFP and TdTomato) dataset contained 6934 cells segregated into 19 clusters (Fig. 2B) belonging to 3 main lineages: endoderm (Epcam+), endothelium (Pecam1+), and hematopoietic cells (Hba-a1+) (Supplementary Fig. S3).
Figure 2.
Single-cell RNA sequencing of murine Sox17-expressing lineages using the temporal dual color tracing system captured both expected and non-canonical Sox17-expressing cell types.
(A) Experimental layout of 2 scRNAseq experiments. At around E9.5, single cells from embryos with desired genotypes are isolated for subsequent flow cytometry sorting and RNA sequencing with 10X Chromium platform. Datasets were analyzed using Seurat package version 4.0. (B) UMAP visualization of the integrated Sox17-expressing lineage dataset. Cells were colored based on clusters identified by unsupervised clustering. Cell types were defined based on differential gene expression analysis. (C) Expression of representative marker genes for each cell cluster. Epcam, Cldn6, and Krt19 served as shared markers for 3 endodermal clusters; Pecam1 and Eng for 7 endothelial clusters; and Gata1, Itga2b, and Hba-a1 for 8 hematopoietic clusters. The rest of the genes are differentially expressed by individual clusters (P-adjusted < .05). Dot color and size indicate gene expression intensity and percentage of cell expressing genes of interest, respectively.
Cell identities were assigned via differential expression analysis (Fig. 2C). The endothelial lineage (Cd31/Pecam1+, Tie2/Tek+) consisted of hemangioblasts (Etv2+, Tal1+), 3 different clusters of endothelial cells (EC1 to EC3) (differing by expression of histone H1 genes, extracellular matrix Vwa1, and Hox genes), endocardium (Hand2+), liver sinusoidal endothelial cells (LSEC—Oit3+, Stab2+), and hemogenic endothelium (HE—Cd44+, Sox17+). The next most abundant cell group represented hematopoietic lineages, with 4 distinct erythroid clusters, one of which lies proximate to the endothelial island. Three endodermal clusters were identified with scattered expression of hepatic (Afp), pancreatic (Pdx1), anterior endoderm (Sox2), and posterior endoderm (Cdx2) markers. A small number of neuronal progenitors (SoxB+, Nkx6.1+) were Sox17−, suggesting putative emergence from Sox17-expressing progenitors. Together, the 2 datasets reflected known Sox17 expression patterns in endodermal, endothelial, and hematopoietic cells, and also suggested Sox17 expression in rare neuronal-lineage cells.
scRNAseq of Sox17-Expressing Endothelium Captures Transcriptional Signatures of Early Vasculogenesis, Angiogenesis, and Erythropoiesis
Based on Cd31/Pecam1 expression and tight clustering, the endothelial compartment was defined as EC1/2/3, hemangioblast, HE, erythroid 1, and LSEC (Fig. 3A, 3B). Since there were 3 distinct clusters of ECs that did not differentially express any endothelial subtype (arterial/venous/capillary/lymphatic) markers, we distinguished them based on their transcriptional profiles. EC3 expressed relatively high levels of Hox genes, also a feature of the HE cluster, which is known for regulating hematopoiesis29 (Supplementary Fig. S4A), suggesting hematopoietic potential. Of note, although the yolk sac was specifically omitted from our dissections, a small portion of EC3 expressed signature placental genes Plac1 and Nrk30,31 (Supplementary Fig. S4B), suggesting an extraembryonic (perhaps mobile) origin of EC3. EC1 and EC2 differed mostly by cell-cycle gene expression (Supplementary Fig. S4D). EC1 expressed significantly higher levels (P-adjusted < .05) of Cdc20, Cenpa, Ccnb2, and Tuba1c, suggesting mitotic status, whereas EC2 had higher expression of histone H1, Top2a, Cdk1, and Aurkb more fitting the G2-like state.32 However, Seurat cell-cycle scoring predicts a large portion of EC2 cells are in S phase whereas those in the EC1 cluster are in either the G1, G2/M, or S phases (Supplementary Fig. S4C).
Figure 3.
Heterogenous continuum of endothelial cell states during vasculogenesis and endothelium-derived hematopoiesis. (A) UMAP visualization of the Sox17-expressing endothelium subset colorized by initial unsupervised clustering of the Sox17-expressing dataset. (B) Transcriptional level of top 3 differentially expressed genes by each cluster. Dot color and size indicate gene expression intensity and percentage of cell expressing genes of interest, respectively. (C) Developmental trajectory analysis of the Sox17-expressing endothelium subset. Pseudotime assignment was performed using Monocle3. RNA velocity streamlines were generated with scVelo and then overlayed with UMAP embeddings colored by cell types in (A). (D) Proposed model of multiple developmental pathways from distinct origins to specified endothelial subtypes. Hemangioblasts can give rise to hemogenic endothelial cells (HE) via endothelial cell 1 (EC1)—pathway (a), or directly to EC2—pathway (b). EC3 can differentiate to either EC2—pathway (c) or directly to erythroid cells—pathway (d). Liver sinusoidal endothelial cells (LSEC) appear to be in pathway (c). (E) Expression of arterial marker genes in HE and EC3. (F) Expression of hemogenic marker genes in HE and EC3.
Observing progenitor-type and further-differentiated endothelial cells both near a cluster of erythroid cells suggested that Sox17-lineages include cells progressing through vasculogenesis and endothelium-derived hematopoiesis. To assess the transcriptomic alterations of such cells occurring within these individual processes, we performed Monocle 3 trajectory analysis to order cells in pseudotime33 and used scVelo RNA velocity to reveal putative transitional directions between cells26,34 (Fig. 3C). While scVelo uses the unbiased RNA spliced/unspliced ratio, Monocle 3 requires supervised input of the trajectory root, which we selected based on Etv2 defining the conventional hemangioblast population. These analyses recapitulated a well-established model whereby hemangioblasts give rise either indirectly to hemogenic endothelium through the differentiation of EC1 cells (pathway a), or directly to the non-hemogenic EC2 cells (pathway b) (Fig. 3D). In addition, they revealed a potentially bipotent progenitor group EC3, which may originate from mobile yolk-sac cells that become either EC2 (pathway c) or erythroid cells (pathway d).
Because HE and EC3 clusters both express multiple Hox genes known to drive hematopoiesis, we further resolved the transcriptional distinctions between these 2 populations. Differential expression analysis (P-adjusted < .05) revealed arterial-related signatures for HE that include gap-junction genes (Gja4, Gja5) that become upregulated by Notch signaling (marked by Hey1) under shear stress.35 Consistent with Sox17-expression itself being a marker for arterial specification,36 HE cells also express Ephrin B2, a classical arterial marker37 (Fig. 3E). On the other hand, EC3 express Hspd1 (heat-shock gene involved in yolk-sac erythropoiesis38), Npm1 (important modulator of primitive hematopoiesis39), Ncl (promoter of the hemogenic GRN40), and Lyar (necessary for hemocyte generation41) (Fig. 3F), suggesting the extraembryonic origin and strong hemogenic potential of EC3. Together, these data are consistent with contribution of extra-embryonic ECs to intra-embryonic blood-vessel formation and suggest the existence of at least 2 EC populations with hemogenic potency from different origins at E9.5.
Distinct Hematopoietic, Vasculo-Angiogenic, and Homeostatic Regulons in the Endothelial Lineage
Given that scRNAseq of Sox17-expressing lineages unbiasedly captured the dynamic transcriptional changes of vascular formation and endothelial-to-hematopoietic transition of cells from extra- and intra-embryonic sources, we attempted to unveil the key GRNs underlying these important developmental processes. By performing pySCENIC regulon analysis to infer groups of active genes that are regulated as units, with putative direct regulation defined by TF binding motifs located within likely cis-regulatory regions of target genes,21 we identified 287 regulons among the endothelial compartment clusters. Plotting the top 10% of such regulons revealed 3 distinct GRN clusters for erythropoiesis, vasculo-angiogenesis, and potential homeostasis of endothelial cells (Fig. 4A). Regulons of the well-established Klf1/Gata1/Tal1 triplet—master regulators of hematopoiesis42,43—clustered tightly together (Fig. 4A) and are active specifically in erythroid 1 cluster (Fig. 4B), validating the reliability of pySCENIC predictions.
Figure 4.
Predicted active regulons in the endothelium. (A) Network plot of top 10% predicted regulons ranked by “importance” metric (signifying strength of TF-target gene relation). TF (pink nodes) and connected to target genes (white nodes) via edges (gray). Sox17 node is highlighted in yellow. Thicker to thinner edges indicate stronger to weaker relation of TF-target respectively. The size of each node represents the “betweenness centrality” measurements (possible measure of “hub” gene due to high TF-targets connectivity) of a given TF within the network. Regulon networks are manually classified into bigger gene modules (blue circles) based on known biological functions of master regulator genes. (B) Single-cell AUC scores (area under the recovery curve across genes ranked by expression value indicating the enrichment of each regulon in each cell) measure for 6 most prominent regulons embedded on UMAP.
The 2 biggest nodes in the GRN are Etv2 and Ets1, known pioneer TFs during embryonic vasculo-angiogenesis.44,45 Although Ets1 may have redundant functions with Etv2 (from studies in zebrafish46), not many shared targets are observed by our regulon prediction, suggesting cross-species variation. Regulons of Elk3 and Sox18, which contribute to lymphatic vascular development,47,48 are clustered closely and overlap with parts of the Etv2 and Ets1 regulons, consistent with Elk3 and Sox18 regulating a more focused lymphatic subset of endothelial differentiation. Of note, Elk3 may antagonize Ets1 transcriptional activation to suppress angiogenesis,49 likely through shared targets of Elk3 and Ets1. Tagln2, a marker of smooth-muscle cells recently documented for its function during angiogenesis,50 also forms a large node in the endothelial GRN.
Somewhat distinct from the erythropoietic and vasculo-angiogenetic GRNs are the regulons for Ybx1 and Tfdp1 (Fig. 4A), general TFs that regulate proliferation, apoptosis, and differentiation in a non-cell-type-specific manner, often being linked to cancer progression.51-54 Some angiogenic-specific functions such as proliferation, apoptosis, migration, and tubulogenesis are associated with Ybx151,55,56 but its regulon is notably different from other angiogenic regulons. Its activity in both endothelial and erythroid cells (Fig. 4B) suggests a more general cellular homeostatic function.
Unsupervised clustering of endothelial PySCENIC regulons revealed a cluster specifically activated in hemogenic endothelium (Supplementary Fig. S5) that contains genes previously linked to the emergence of hematopoietic stem cells, such as Runx1, Hox genes, Bcl11a57,58 and the more recently investigated Gata3, Cdx4, Cdx1, and Gli3.59-61 Although Sox17, required for endothelial-to-hematopoietic transition,62 was not found in this cluster, 4 other Sox members were: Sox7 (interacts with Runx1 to specify HE in the yolk sac63), Sox6 (suppresses embryonic globin genes during definitive erythropoiesis64), and Sox2 and Sox21 (unknown functions in vascular and hematopoietic lineages). Foxf1, which is important for endothelial progenitors and vessel sprouting,65 and Foxn3, which is detected in human hemogenic endothelium but has unknown function,66 also appear in this HE-specific regulon cluster, suggesting previously unknown functions of these genes during endothelium-derived hematopoiesis.
New Insights Into Hepato-Pancreato-Biliary Development by scRNAseq Analysis of Sox17- and Prox1-Expressing Endodermal Lineage
Toward better understanding endodermal, and especially hepato-pancreato-biliary (HPB) development, we re-clustered 392 endodermal cells identified in both the GFP and TdTomato datasets (“Sox17 endoderm dataset”) to identify early progenitor cells of various organs (Supplementary Fig. S6A, S6B). Within this dataset, pancreatic progenitors (Pdx1+ and Rfx6+), and hepatobiliary progenitors (Sox17+, Hhex+, and Prox1+) were most abundant. Since HPB cell numbers were insufficient to resolve the transcriptional segregation of progenitor cells from the 3 independent organ anlagen we combined our Sox17 endodermal dataset with a dataset recently reported from the Spagnoli group obtained via sorting Prox1-expressing cells,20 hereafter called the “Prox1 dataset.” The Prox1 dataset contains 346 posterior-foregut cells from embryos at multiple stages during mid-gestation. Analysis of 738 cells combined from the Sox17 endoderm (392 cells) and Prox1 (346 cells) datasets, hereafter the “Sox17-Prox1 endoderm” dataset, resulted in 10 distinct clusters consisting mostly of endoderm and a few mesoderm-derived cell types such as cardiomyocytes, HPSCs, pancreatic and lung mesenchyme (Fig. 5A). There was high integration across the original Sox17 endoderm and Prox1 datasets (Supplementary Fig. S6C; Supplementary Table S2). Cell-type assignments of the Sox17-Prox1 endoderm dataset based on differentially expressed genes (Fig. 5B) were mostly consistent with the previous manual-dissection-based assignments of the Prox1 dataset (Supplementary Fig. S6D; Supplementary Table S2).
Figure 5.
Dynamic expression profiles of hepato-pancreato-biliary cells during foregut endoderm organogenesis. (A) UMAP visualization of the integrated endodermal subset. Cells were colored based on clusters identified by unsupervised clustering. Cell types were defined based on differential gene expression analysis. (B) Transcriptional level of top 3 marker genes defining cell type of each cluster. Dot color and size indicate gene expression intensity and percentage of cell expressing genes of interest, respectively. (C) Heatmap of top 20 differential expressed genes by distinct groups of progenitor cells within the developing hepato-pancreato-biliary system. Hepatic and biliary progenitor clusters are further divided into subgroups based on the expression level of organ-specific genes (lo—low; med—medium; hi—high).
After obtaining 3 distinct clusters of HBP progenitors, we performed differential transcriptomic analysis during their segregation to identify new markers for each progenitor population. We not only identified 287 significantly upregulated genes for pancreatic buds, 624 for liver bud, and 23 for biliary (P adjusted < .05) (top 20 differentially expressed genes are listed in Fig. 5C), but also observed substantially overlapping transcriptional profiles among the HBP progenitors.
Pancreatic markers included: Nepn (expressed in the dorsal pancreatic bud and duodenum by E9.567), Cldn4, Krt7, and Krt19 (epithelial markers highly expressed in pancreatic but absent from hepatic bud68), Ociad2 (unknown function in embryonic pancreatic development but consistent with previous observations69), App, S100a10, and S100a11 (involved in pancreatic cancer and diseases, patterns not yet reported during embryonic development70,71), Cdx2 (marking caudal gut tube), and Sox4 (expressed in pancreatic buds and important for endocrine-cell development72). Other genes identified as hepatic markers were previously described as highly expressed in liver buds such as Apo, Serpin, and Fibrinogen gene families, Alb, Reln,73,74 and those active in mature liver cells such as Itih2, Shbg, Lpl, and Trf.75-78Sox17, the most established marker of the biliary bud,79 was also the most highly upregulated gene in biliary progenitors. Flrt3, reported in the original manuscript of the Prox1 dataset,20 exhibited similar expression to Sox17, and may be a new marker of biliary progenitors. Itih2, Reln, Hhex, and Paqr9 shared the pattern of Sox17 expression in biliary progenitors but were also upregulated in the liver, suggesting shared characteristics between biliary and hepatic progenitor cells.
Other top biliary-specific genes exhibited high, medium, or low expression, subdividing the biliary cluster into 3 groups (Fig. 5C, P-adjusted < .05). The biliary-low group had a high level of pancreatic markers, suggesting that they were not-yet developmentally resolved but more akin to the ventral pancreatic state. The biliary-high group was likely more-mature biliary epithelial cells whereas the biliary-medium group showed medium levels of pancreatic genes, indicating putative pancreato-biliary bipotent progenitor status. A similar group of biliary-medium/pancreatic-medium genes was found in part of the hepatic cluster carrying low levels of liver markers. These findings are consistent with the existence of multi- and bi-potent pools of progenitor cells during progressive segregation of the 3 foregut anlagen that is under way at E9.0-9.5. Besides having high Sox17 expression, these putative multi- and bipotent progenitors expressed Phlda2, previously verified as expressed in ventral definitive endoderm80 yet without a known function in early endodermal organogenesis.
Active Regulons During Endodermal Development
To identify putative GRNs governing HBP segregation from the posterior foregut, specifically not yet addressed in the literature, we performed pySCENIC analysis on the Sox17-Prox1 endoderm dataset, which identified 238 regulons. The top decile was shown in the network plot (Fig. 6A) with further clustering of regulons corresponding to specific cell types based on node proximity and cell types in which regulons were imputed as most active (Fig. 6B).
Figure 6.
Predicted active regulons in the endoderm. (A) Network plot of top 10% predicted regulons ranked by “importance” metric signifying strength of TF-target relation. TF (pink nodes) and connected to target genes (white nodes) via edges (gray). Sox17 node is highlighted in yellow. Thicker to thinner edges indicate stronger to weaker relation of TF-target respectively. The size of each node represents the “betweenness centrality” measurements (possible measure of “hub” gene due to high TF-targets connectivity) of a given TF within the network. Regulon networks are manually classified into bigger gene modules (blue circles) based on known biological functions of master regulator genes. (B) Single-cell AUC scores (area under the recovery curve across genes ranked by expression value indicating the enrichment of each regulon in each cell) measure for 8 most prominent regulons embedded on UMAP.
Besides regulons specific to non-endodermal cell types such as cardiovascular-hematopoietic lineages and pancreatic mesenchyme, we classified 3 separate regulon hubs related to the development of the early endoderm and HBP system. We inferred a dense cluster of medial and caudal Hox regulons that, together with Cdx2 and Cdx4 regulons, are predicted in posterior endoderm. Besides a role for Cdx2 in establishing the caudal part of the gut-tube axis, genes in its regulon are more active in pancreatic buds than in liver, biliary, or islet cells, suggesting additional contribution to early pancreas formation.
Another distinct pancreatic endocrine-centric regulon cluster had major nodes of Neurod1 and Rfx6, connected with Neurog3, Neurog1, Isl1, Mafb, Pax6, Pou3f4, Arx, and Pax4, all encoding TFs involved in islet endocrine-cell differentiation programs.81,82Nhlh1 and Npdc1 regulons were specific to the islet cluster, but as of now, lack identified functions here.
The rest of the network was classified as anterior endoderm and HBP-focused regulon hubs. Gata4, Sox9, Foxa1, Hnf1b, Gata6, Jun, and Fos regulons are inferred as strongly active in pancreatic progenitor cells. Hnf4a, Foxa3, and Nr5a2, which shared some target genes, were upregulated in hepatic progenitors, suggesting their coordinate regulation of hepatic-bud formation and outgrowth.83,84 While the Onecut2 regulon was equally active among the 3 HPB domains, the Onecut3 regulon was restricted to hepato-biliary progenitors. Sox17 was the only biliary-major regulon in the mapped network. Sox1, Foxb1, Hoxd3, Hoxa1, and Pou3f2 regulons were identified specifically in the anterior DE population. Of note, regulons associated with the same cell type were not clustered tightly together, reflecting their few shared targets, and implying diverse molecular functions for each regulon responsible during the development of each individual organ. In contrast, the mingling together of regulons important for pancreas, liver, biliary, and anterior endoderm suggests shared target genes among these cell types, in keeping with their overlapping transcriptomic profiles.
Distinct Sox17-Regulons Between Endoderm and Endothelium
Perhaps the most important part of this study is our identification of highly independent regulons for Sox17 in endoderm versus endothelium. While pySCENIC predicts 81 endothelial targets for Sox17, only 11 were endoderm-selective (Fig. 7A). To partially validate these targets, we surveyed 3 recently published SOX17 ChIP-seq and CUT&RUN datasets from cell-line-derived human definitive endoderm and mouse primitive endoderm and found the majority of pySCENIC-predicted endodermal targets also exhibit SOX17 binding within promoter-proximal regions (Supplementary Fig. S7). Among these genes regulated by Sox17, only Efnb2 was shared between the endodermal and endothelial Sox17 regulons. The marked diversity of Sox17 regulons as assessed by gene ontology (GO) terms is consistent with SOX17 driving largely different molecular functions in these 2 populations. Indeed, GO-term enrichment patterns suggest that Sox17 works in endothelial cells to regulate blood-vessel morphogenesis, cell migration, and sprouting during vasculo-angiogenesis, whereas in endoderm it is more geared toward regulating chemotaxis and signaling pathways such as growth-factor stimulus, serine/threonine kinase and Wnt signaling (Fig. 7B).
Figure 7.
Predicted Sox17-regulons in endoderm and endothelial cells. (A) pySCENIC predictions of Sox17 (pink) downstream targets (blue) in the endothelium and endoderm. (B) Gene ontology enrichment analysis of Sox17 target genes in the endothelium and endoderm.
Discussion
Our robust 2-color lineage-tracing strategy, scRNASeq, and use of several computational methods have further characterized Sox17-expressing cell populations and expanded our understanding of putative Sox17 functions during early stages of lineage diversification and tissue specification. Our results not only integrate previous knowledge of Sox17-expressing cells but also putatively reveal distinct regulons operating during endothelial and endodermal differentiation, which suggest multiple future lines of investigation.
Tracing Sox17-Expressing Lineages Effectively Captures Early-Stage Vasculo-Angiogenesis and Endothelium-Derived Hematopoiesis
Previous attempts to profile single-cell transcriptomes of the endothelium and hematopoietic system used an antibody panel against multiple cell-surface markers.19,85 Using just 2 endogenously expressed fluorescent alleles that report short- or long-term expression of Sox17, we effectively recorded transcriptomic changes during vasculo-angiogenesis and endothelial-to-hematopoietic transition.
Notably, our findings further delineate some controversial and complex concepts in endothelial and hematopoietic development. We captured a classical hemangioblast population (Etv2 and Tal1 positive) that, through our trajectory analysis, gives rise to 2 distinct subtypes of ECs, one of which can advance to become the canonical hemogenic endothelium, supporting the model of in vivo unipotency of hemangioblasts.86 Furthermore, we identified a hemangioblast-like endothelial population (EC3—negative for Etv2 and Tal1), that expresses many genes suggestive of an extra-embryonic origin and appears to differentiate directly to both erythroids and ECs. Based on recent live imaging87 and the fact that yolk sacs were omitted from our scRNAseq, we propose that these bipotent progenitors arise in the yolk sac, migrate to the embryo proper, and contribute to both intra-embryonic vascular (potentially at the dorsal aorta, head vasculature, and endocardium—known secondary hematopoiesis sites in the developing embryo) and hematopoietic system likely through Hox-dependent mechanisms.
Our analyses of the endothelium raise several areas for further investigation. First, 2 types of ECs seem to arise from hemangioblasts, which differ mostly by expression of Histone H1 and other cell-cycle genes, but only one can become hemogenic. Based on the known importance of linker histone H1 in cell-fate determination,88 and of cell-cycle regulation association with endothelial-to-hematopoietic transition,89 we hypothesize an active participation of histone H1 during HE specification. Second, whether Ybx1 and Tfdp1 regulons, emerging on top of the important endothelial GRN, are critical for endothelial homeostasis or other molecular functions such as migration and morphology, is not known. Lastly, functions of several regulons identified specifically for HE emergence remain to be explored such as Sox2, Sox21, Foxn3, and Foxf1.
Although Sox17 is early and widely expressed within the emerging vasculature system (reflected in our usage of Sox17GFPCre allele), its regulon did not appear among the top 10% most “important” ones in our GRN analysis, indicating moderate, redundant (to other SoxF members), or ambiguous functions of Sox17 in primitive endothelial cells. These results are consistent with Sox17 having limited functions in the adult endothelial system despite being widely expressed. Indeed, mutations of Sox17 in the adult vasculature are reported to affect vascular integrity only under challenging conditions such as hypertensive stress.90-92
Transcriptomic Insights Into Hepato-Pancreato-Biliary Development
Unsupervised clustering of the Sox17-Prox1 endoderm dataset showed 3 distinct clusters of the HPB system, allowing identification of new markers and understudied genes as plausible determinants for each progenitor type. Ociad2, encoding a mitochondria-associated protein and top pancreatic marker was recently reported as specifically upregulated in β- rather than α-cell differentiation from endocrine progenitors,69 suggesting links between mitochondrial metabolism and insulin-secreting fate acquisition. Similarly, App (amyloid precursor protein), expressed in adult pancreatic islets,93 could influence pancreatic specification from the posterior foregut through autocrine/paracrine signaling. S100 (Ca2+-binding protein with EF-hand motif) family members are also prominent in pancreatic compared to liver and biliary buds, consistent with our previous work showing high S100a11 in endocrine progenitors and S100a10 in early Sox17+ endoderm.94S100 functions in cell proliferation, migration, and other signaling pathways are implicated in many cell types,70 but knowledge of their function during pancreatic development is poor. Flrt3 (fibronectin leucine-rich transmembrane protein), which contributes to cell adhesion and receptor signaling in other contexts, is present in biliary progenitors,20 yielding a promising candidate not-yet-determined cellular interactions in biliary development.
Previous attempts to profile single-cell transcriptomes of the developing HPB, perhaps due to technical challenges in obtaining adequate cell numbers, could not resolve all 3 cell types of the system, such as liver-biliary using FoxA2eGFP, dorsal and ventral pancreas using Pdx1GFP, and liver-pancreatobiliary using Prox1eGFP mice. Our Sox17-centric strategy complements other datasets (especially Prox1eGFP as described above), enabling all 3 populations to be distinguished and, further, identification of a subset of liver progenitors with low-hepatic but medium-pancreatobiliary profile, and 3 subsets of biliary progenitors with high-pancreatic, high-biliary, or medium-pancreatobiliary features. These findings indicate the presence of multi- and bipotent progenitors during the gradual segregation of progenitors of the 3 anlagens. These early-stage progenitor cells, besides sharing a high expression of Sox17 and Flrt3 with the biliary-committed cells, also express Phlda2, a cell-surface Pleckstrin homology-like domain family A member, with potential utility in future cell-sorting analyses. Notably, Phlda2 is expressed in the ventral gut tube, suggesting a functional association with HPB development, perhaps by regulating epithelial-to-mesenchymal transition via PI3K/AKT during diversification from the posterior foregut.
We also identified several yet-to-be characterized endoderm-active GRNs linked to HPB development including regulons for Nhlh1 (Nescient Helix-Loop-Helix 1) and Npdc1 (Neural Proliferation, Differentiation And Control 1), 2 neurogenesis TFs expressed in pancreatic islets and that parallel studies of the human endocrine lineage.95,96 Our results also unveiled previously unknown but plausibly stochastic functions of the highly redundant Foxa TF members,97 with the Foxa1 regulon operating in early pancreatic progenitors, the Foxa2 regulon biased toward mature pancreatic endocrine cells, and the Foxa3 regulon predominantly active in the hepatic bud.
Different Sox17 Regulons in Endoderm and Endothelium
To potentially distinguish Sox17 functions in the endoderm and endothelium, we compared the pySCENIC-predicted Sox17-regulons from the endothelial subset with those derived from the Sox17-Prox1 endoderm dataset. Of 81 target genes in endothelium compared to 11 predicted in endoderm, Efnb2 is the only common imputed downstream Sox17 target. Efnb2 (an ephrin family member) together with related receptor tyrosine kinases modulates cell-cell communication have well-established roles in arterial/venous specification of endothelial cells, consistent with Sox17 being indispensable for arterial fate acquisition.36Efnb2 was recently proposed as important in defining the dorsal-ventral axis of foregut,98 suggesting an unappreciated role of Sox17 in the endodermal gut tube.
Most other endothelial Sox17 targets are unsurprisingly related to vascular formation, including tube morphogenesis, tip and stalk cell migration, and motility. In contrast to promoting “proactive” endothelial-cell behavior in response to the surrounding environmental cues, in endodermal cells, Sox17 may cause more “passive” responses related to sensing chemotactic, growth factor, and particularly Wnt signals. These predicted functions of Sox17 target genes in endoderm are in agreement with previous studies99,100 and reinforce the importance of coordinated movements of endodermal cells in response to mesodermal chemotactic cues during gastrulation. Furthermore, the marked difference between endoderm and endothelial Sox17-regulons emphasizes the importance of cellular context in unmasking SOX17 target-gene binding sites, suggesting that SOX17 employs the same strategy seen for other Sox family members—to engage in differential protein partnerships so as to economically diversify differentiation programs.101
Identification of Sox17 in Rare Neural Progenitor Cells
In addition to capturing cells from endodermal, hematopoietic, and endothelial lineages, all are well-established Sox17-associated lineages, several neural progenitor cells expressing the canonical markers Sox1/2/3 and Nkx6.1 were identified possibly indicating Sox17 function in these lineages. While Sox17 function is documented in oligodendrocyte development,10 little is known about its expression or function in neural progenitors. SoxF is associated with neurogenesis in fruit flies102 and zebrafish,103 perhaps supporting Sox17 having a similar role in mammals. In this regard, it is noteworthy that among the low-RNA-content cells excluded from our GFP dataset (Supplementary Methods), a collection of mesenchymal and neural crest-like genes such as Vim, Mest, Nes, Twist1, Prrx2 are expressed. In any case, further analyses and validations are necessary to determine the function of Sox17 in neural lineages.
Conclusion
Our analyses of Sox17-expressing lineages using scRNAseq consolidate prior knowledge of Sox17 and provide new insights into the transcriptomic features, GRNs, and developmental trajectories of both hepato-pancreato-biliary and hemato-endothelial systems.
Supplementary Material
Acknowledgments
This research was supported by funding provided by Vanderbilt University. We thank the staff of the Vanderbilt Flow Cytometry Shared Resource (FCSR) and the Vanderbilt Cell Imaging Shared Resource (CISR) for their expert assistance. FCSR is supported by CA068485 and DK058404, and CISR by CA68485, DK20593, DK58404, DK59637, and EY08126.
Contributor Information
Linh T Trinh, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN, USA; Department of Cell and Developmental Biology, Vanderbilt University, Nashville, TN, USA; Center for Stem Cell Biology, Vanderbilt University, Nashville, TN, USA; Program in Developmental Biology, Vanderbilt University, Nashville, TN, USA.
Anna B Osipovich, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN, USA; Center for Stem Cell Biology, Vanderbilt University, Nashville, TN, USA.
Bryan Liu, College of Arts and Sciences, Vanderbilt University, Nashville, TN, USA.
Shristi Shrestha, Center for Stem Cell Biology, Vanderbilt University, Nashville, TN, USA.
Jean-Philippe Cartailler, Center for Stem Cell Biology, Vanderbilt University, Nashville, TN, USA.
Christopher V E Wright, Department of Cell and Developmental Biology, Vanderbilt University, Nashville, TN, USA; Center for Stem Cell Biology, Vanderbilt University, Nashville, TN, USA; Program in Developmental Biology, Vanderbilt University, Nashville, TN, USA.
Mark A Magnuson, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN, USA; Department of Cell and Developmental Biology, Vanderbilt University, Nashville, TN, USA; Center for Stem Cell Biology, Vanderbilt University, Nashville, TN, USA; Program in Developmental Biology, Vanderbilt University, Nashville, TN, USA.
Conflict of Interest
The authors declared no potential conflicts of interest.
Author Contributions
L.T.T.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing. A.B.O.: conception and design, data analysis, and interpretation. B.L.: collection of data. S.S.: data analysis and interpretation. J-P.C.: data analysis and interpretation. C.V.E.W.: manuscript writing. M.A.M.: conception and design, data analysis and interpretation, financial support, manuscript writing.
Data Availability
Raw scRNAseq data is at ArrayExpress (E-MTAB-12719), Seurat objects are at https://zenodo.org/record/7725887#.ZA5jRR_MLkI. Scripts used to perform quality control, clustering, differential gene expression analysis, Monocle3 and RNAvelocity analyses are at https://github.com/markmagnuson/2023-Linh-scRNASeq-Mouse-Sox17-Expressing-Lineages.
References
- 1. Sarkar A, Hochedlinger K.. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12(1):15-30. 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kamachi Y, Kondoh H.. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140(20):4129-4144. 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 3. She ZY, Yang WX.. SOX family transcription factors involved in diverse cellular events during development. Eur J Cell Biol. 2015;94(12):547-563. 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 4. Bowles J, Schepers G, Koopman P.. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227(2):239-255. 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 5. Palasingam P, Jauch R, Ng CK, Kolatkar PR.. The structure of Sox17 bound to DNA reveals a conserved bending topology but selective protein interaction platforms. J Mol Biol. 2009;388(3):619-630. 10.1016/j.jmb.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 6. Aksoy I, Jauch R, Chen J, et al. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 2013;32(7):938-953. 10.1038/emboj.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaves-Moreira D, Mitchell MA, Arruza C, et al. The transcription factor PAX8 promotes angiogenesis in ovarian cancer through interaction with SOX17. Sci Signal. 2022;15(728):eabm2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sinner D, Rankin S, Lee M, Zorn AM.. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development. 2004;131(13):3069-3080. 10.1242/dev.01176. [DOI] [PubMed] [Google Scholar]
- 9. Kanai-Azuma M, Kanai Y, Gad JM, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129(10):2367-2379. 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 10. Sohn J, Natale J, Chew LJ, et al. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci. 2006;26(38):9722-9735. 10.1523/JNEUROSCI.1716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim I, Saunders TL, Morrison SJ.. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130(3):470-483. 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pfister S, Jones VJ, Power M, et al. Sox17-dependent gene expression and early heart and gut development in Sox17-deficient mouse embryos. Int J Dev Biol. 2011;55(1):45-58. 10.1387/ijdb.103158sp. [DOI] [PubMed] [Google Scholar]
- 13. Seguin CA, Draper JS, Nagy A, Rossant J.. Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell. 2008;3(2):182-195. 10.1016/j.stem.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 14. Niakan KK, Ji H, Maehr R, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24(3):312-326. 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qu XB, Pan J, Zhang C, Huang SY.. Sox17 facilitates the differentiation of mouse embryonic stem cells into primitive and definitive endoderm in vitro. Dev Growth Differ. 2008;50(7):585-593. 10.1111/j.1440-169x.2008.01056.x. [DOI] [PubMed] [Google Scholar]
- 16. Han L, Chaturvedi P, Kishimoto K, et al. Single cell transcriptomics identifies a signaling network coordinating endoderm and mesoderm diversification during foregut organogenesis. Nat Commun. 2020;11(1):4158. 10.1038/s41467-020-17968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nowotschin S, Setty M, Kuo YY, et al. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature. 2019;569(7756):361-367. 10.1038/s41586-019-1127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeng Y, He J, Bai Z, et al. Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Res. 2019;29(11):881-894. 10.1038/s41422-019-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hou S, Li Z, Zheng X, et al. Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res. 2020;30(5):376-392. 10.1038/s41422-020-0300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Willnow D, Benary U, Margineanu A, et al. Quantitative lineage analysis identifies a hepato-pancreato-biliary progenitor niche. Nature. 2021;597(7874):87-91. 10.1038/s41586-021-03844-1. [DOI] [PubMed] [Google Scholar]
- 21. Van de Sande B, Flerin C, Davie K, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15(7):2247-2276. 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 22. Choi E, Kraus MR, Lemaire LA, et al. Dual lineage-specific expression of Sox17 during mouse embryogenesis. Stem Cells. 2012;30(10):2297-2308. 10.1002/stem.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573-3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McGinnis CS, Murrow LM, Gartner ZJ.. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329-337.e4. 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao J, Spielmann M, Qiu X, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496-502. 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ.. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38(12):1408-1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 27. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gu Z, Eils R, Schlesner M.. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847-2849. 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 29. Argiropoulos B, Humphries RK.. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766-6776. 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 30. Cocchia M, Huber R, Pantano S, et al. PLAC1, an Xq26 gene with placenta-specific expression. Genomics. 2000;68(3):305-312. 10.1006/geno.2000.6302. [DOI] [PubMed] [Google Scholar]
- 31. Lestari B, Naito S, Endo A, et al. Placental mammals acquired functional sequences in NRK for regulating the CK2-PTEN-AKT pathway and placental cell proliferation. Mol Biol Evol. 2022;39(2):msab371. 10.1093/molbev/masab371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whitfield ML, Sherlock G, Saldanha AJ, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13(6):1977-2000. 10.1091/mbc.02-02-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381-386. 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature. 2018;560(7719):494-498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fang JS, Coon BG, Gillis N, et al. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat Commun. 2017;8(1):2149. 10.1038/s41467-017-01742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Corada M, Orsenigo F, Morini MF, et al. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat Commun. 2013;4:2609. 10.1038/ncomms3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shin D, Garcia-Cardena G, Hayashi S, et al. Expression of ephrinB2 identifies a stable genetic difference between arterial and venous vascular smooth muscle as well as endothelial cells, and marks subsets of microvessels at sites of adult neovascularization. Dev Biol. 2001;230(2):139-150. 10.1006/dbio.2000.9957. [DOI] [PubMed] [Google Scholar]
- 38. Duan Y, Wang H, Mitchell-Silbaugh K, et al. Heat shock protein 60 regulates yolk sac erythropoiesis in mice. Cell Death Dis. 2019;10(10):766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grisendi S, Bernardi R, Rossi M, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437(7055):147-153. 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 40. Mahotka C, Bhatia S, Kollet J, Grinstein E.. Nucleolin promotes execution of the hematopoietic stem cell gene expression program. Leukemia. 2018;32(8):1865-1868. 10.1038/s41375-018-0090-4. [DOI] [PubMed] [Google Scholar]
- 41. Jariyapong P, Pudgerd A, Cheloh N, et al. Hematopoietic tissue of Macrobrachium rosenbergii plays dual roles as a source of hemocyte hematopoiesis and as a defensive mechanism against Macrobrachium rosenbergii nodavirus infection. Fish Shellfish Immunol. 2019;86:756-763. 10.1016/j.fsi.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 42. Kang Y, Kim YW, Yun J, Shin J, Kim A.. KLF1 stabilizes GATA-1 and TAL1 occupancy in the human beta-globin locus. Biochim Biophys Acta. 2015;1849(3):282-289. [DOI] [PubMed] [Google Scholar]
- 43. Tallack MR, Whitington T, Yuen WS, et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 2010;20(8):1052-1063. 10.1101/gr.106575.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu F, Li D, Yu YY, et al. Induction of hematopoietic and endothelial cell program orchestrated by ETS transcription factor ER71/ETV2. EMBO Rep. 2015;16(5):654-669. 10.15252/embr.201439939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koyano-Nakagawa N, Garry DJ.. Etv2 as an essential regulator of mesodermal lineage development. Cardiovasc Res. 2017;113(11):1294-1306. 10.1093/cvr/cvx133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Casie Chetty S, Sumanas S.. Ets1 functions partially redundantly with Etv2 to promote embryonic vasculogenesis and angiogenesis in zebrafish. Dev Biol. 2020;465(1):11-22. 10.1016/j.ydbio.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park JI, Kim KS, Kong SY, Park KS.. Novel function of E26 transformation-specific domain-containing protein ELK3 in lymphatic endothelial cells. Oncol Lett. 2018;15(1):55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Francois M, Caprini A, Hosking B, et al. Sox18 induces development of the lymphatic vasculature in mice. Nature. 2008;456(7222):643-647. 10.1038/nature07391. [DOI] [PubMed] [Google Scholar]
- 49. Heo SH, Cho JY.. ELK3 suppresses angiogenesis by inhibiting the transcriptional activity of ETS-1 on MT1-MMP. Int J Biol Sci. 2014;10(4):438-447. 10.7150/ijbs.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsuji-Tamura K, Morino-Koga S, Suzuki S, Ogawa M.. The canonical smooth muscle cell marker TAGLN is present in endothelial cells and is involved in angiogenesis. J Cell Sci. 2021;134(15):jcs254920. 10.1232/jcs.254920 [DOI] [PubMed] [Google Scholar]
- 51. Gopal SK, Greening DW, Mathias RA, et al. YBX1/YB-1 induces partial EMT and tumourigenicity through secretion of angiogenic factors into the extracellular microenvironment. Oncotarget. 2015;6(15):13718-13730. 10.18632/oncotarget.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN.. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc). 2011;76(13):1402-1433. 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 53. Zhan W, Wang W, Han T, et al. COMMD9 promotes TFDP1/E2F1 transcriptional activity via interaction with TFDP1 in non-small cell lung cancer. Cell Signal. 2017;30:59-66. 10.1016/j.cellsig.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 54. Morimoto Y, Mizushima T, Wu X, et al. miR-4711-5p regulates cancer stemness and cell cycle progression via KLF5, MDM2 and TFDP1 in colon cancer cells. Br J Cancer. 2020;122(7):1037-1049. 10.1038/s41416-020-0758-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang W, Wang HJ, Wang B, et al. The role of the Y box binding protein 1 C-terminal domain in vascular endothelial cell proliferation, apoptosis, and angiogenesis. DNA Cell Biol. 2016;35(1):24-32. 10.1089/dna.2015.2908. [DOI] [PubMed] [Google Scholar]
- 56. Pham TP, Bink DI, Stanicek L, et al. Long non-coding RNA Aerrie controls DNA damage repair via YBX1 to maintain endothelial cell function. Front Cell Dev Biol. 2020;8:619079. 10.3389/fcell.2020.619079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gao L, Tober J, Gao P, et al. RUNX1 and the endothelial origin of blood. Exp Hematol. 2018;68:2-9. 10.1016/j.exphem.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dou DR, Calvanese V, Sierra MI, et al. Medial HOXA genes demarcate haematopoietic stem cell fate during human development. Nat Cell Biol. 2016;18(6):595-606. 10.1038/ncb3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zaidan N, Nitsche L, Diamanti E, et al. Endothelial-specific Gata3 expression is required for hematopoietic stem cell generation. Stem Cell Rep. 2022;17(8):1788-1798. 10.1016/j.stemcr.2022.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Philip Creamer J, Luff SA, Yu H, Sturgeon CM.. CD1d expression demarcates CDX4+ hemogenic mesoderm with definitive hematopoietic potential. Stem Cell Res. 2022;62:102808. 10.1016/j.scr.2022.102808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Azzoni E, Frontera V, Anselmi G, et al. The onset of circulation triggers a metabolic switch required for endothelial to hematopoietic transition. Cell Rep. 2021;37(11):110103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jung HS, Uenishi G, Park MA, et al. SOX17 integrates HOXA and arterial programs in hemogenic endothelium to drive definitive lympho-myeloid hematopoiesis. Cell Rep. 2021;34(7):108758. 10.1016/j.celrep.2021.108758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lilly AJ, Costa G, Largeot A, et al. Interplay between SOX7 and RUNX1 regulates hemogenic endothelial fate in the yolk sac. Development. 2016;143(23):4341-4351. [DOI] [PubMed] [Google Scholar]
- 64. Yi Z, Cohen-Barak O, Hagiwara N, et al. Sox6 directly silences epsilon globin expression in definitive erythropoiesis. PLoS Genet. 2006;2(2):e14. 10.1371/journal.pgen.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sturtzel C, Lipnik K, Hofer-Warbinek R, et al. FOXF1 mediates endothelial progenitor functions and regulates vascular sprouting. Front Bioeng Biotechnol. 2018;6:76. 10.3389/fbioe.2018.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schnerch A, Lee JB, Graham M, Guezguez B, Bhatia M.. Human embryonic stem cell-derived hematopoietic cells maintain core epigenetic machinery of the polycomb group/trithorax group complexes distinctly from functional adult hematopoietic stem cells. Stem Cells Dev. 2013;22(1):73-89. 10.1089/scd.2012.0204. [DOI] [PubMed] [Google Scholar]
- 67. Hou J, Wei W, Saund RS, et al. A regulatory network controls nephrocan expression and midgut patterning. Development. 2014;141(19):3772-3781. 10.1242/dev.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rodriguez-Seguel E, Mah N, Naumann H, et al. Mutually exclusive signaling signatures define the hepatic and pancreatic progenitor cell lineage divergence. Genes Dev. 2013;27(17):1932-1946. 10.1101/gad.220244.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Scavuzzo MA, Hill MC, Chmielowiec J, et al. Endocrine lineage biases arise in temporally distinct endocrine progenitors during pancreatic morphogenesis. Nat Commun. 2018;9(1):3356. 10.1038/s41467-018-05740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu Y, Zhou Q, Guo F, et al. S100 proteins in pancreatic cancer: current knowledge and future perspectives. Front Oncol. 2021;11:711180. 10.3389/fonc.2021.711180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hansel DE, Rahman A, Wehner S, et al. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003;63(21):7032-7037. [PubMed] [Google Scholar]
- 72. Wilson ME, Yang KY, Kalousova A, et al. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54(12):3402-3409. 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- 73. Lotto J, Drissler S, Cullum R, et al. Single-cell transcriptomics reveals early emergence of liver parenchymal and non-parenchymal cell lineages. Cell. 2020;183(3):702-716.e14. 10.1016/j.cell.2020.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mu T, Xu L, Zhong Y, et al. Embryonic liver developmental trajectory revealed by single-cell RNA sequencing in the Foxa2(eGFP) mouse. Commun Biol. 2020;3(1):642. 10.1038/s42003-020-01364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Winters SJ, Scoggins CR, Appiah D, Ghooray DT.. The hepatic lipidome and HNF4alpha and SHBG expression in human liver. Endocr Connect. 2020;9(10):1009-1018. 10.1530/EC-20-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yu Y, Jiang L, Wang H, et al. Hepatic transferrin plays a role in systemic iron homeostasis and liver ferroptosis. Blood. 2020;136(6):726-739. 10.1182/blood.2019002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Merkel M, Weinstock PH, Chajek-Shaul T, et al. Lipoprotein lipase expression exclusively in liver. a mouse model for metabolism in the neonatal period and during cachexia. J Clin Invest. 1998;102(5):893-901. 10.1172/JCI2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Han S, Tan C, Ding J, et al. Endothelial cells instruct liver specification of embryonic stem cell-derived endoderm through endothelial VEGFR2 signaling and endoderm epigenetic modifications. Stem Cell Res. 2018;30:163-170. 10.1016/j.scr.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 79. Spence JR, Lange AW, Lin SC, et al. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell. 2009;17(1):62-74. 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hou J, Charters AM, Lee SC, et al. A systematic screen for genes expressed in definitive endoderm by Serial Analysis of Gene Expression (SAGE). BMC Dev Biol. 2007;7:92. 10.1186/1471-213X-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bohuslavova R, Smolik O, Malfatti J, et al. NEUROD1 is required for the early alpha and beta endocrine differentiation in the pancreas. Int J Mol Sci. 2021;22(13):6713. 10.3390/ijms22136713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Duvall E, Benitez CM, Tellez K, et al. Single-cell transcriptome and accessible chromatin dynamics during endocrine pancreas development. Proc Natl Acad Sci USA. 2022;119(26):e2201267119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nissim S, Weeks O, Talbot JC, et al. Iterative use of nuclear receptor Nr5a2 regulates multiple stages of liver and pancreas development. Dev Biol. 2016;418(1):108-123. 10.1016/j.ydbio.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tachmatzidi EC, Galanopoulou O, Talianidis I.. Transcription control of liver development. Cells. 2021;10(8):2026. 10.3390/cells10082026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhu Q, Gao P, Tober J, et al. Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood. 2020;136(7):845-856. 10.1182/blood.2020004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lancrin C, Sroczynska P, Stephenson C, et al. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457(7231):892-895. 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Collart C, Ciccarelli A, Ivanovitch K, et al. The migratory pathways of the cells that form the endocardium, dorsal aortae, and head vasculature in the mouse embryo. BMC Dev Biol. 2021;21(1):8. 10.1186/s12861-021-00239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sollberger G, Streeck R, Apel F, et al. Linker histone H1.2 and H1.4 affect the neutrophil lineage determination. Elife. 2020;9:e52563. 10.7554/elife.52563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Canu G, Athanasiadis E, Grandy RA, et al. Analysis of endothelial-to-haematopoietic transition at the single cell level identifies cell cycle regulation as a driver of differentiation. Genome Biol. 2020;21(1):157. 10.1186/s13059-020-02058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lee S, Kim IK, Ahn JS, et al. Deficiency of endothelium-specific transcription factor Sox17 induces intracranial aneurysm. Circulation. 2015;131(11):995-1005. 10.1161/CIRCULATIONAHA.114.012568. [DOI] [PubMed] [Google Scholar]
- 91. Wang TM, Wang SS, Xu YJ, et al. SOX17 loss-of-function mutation underlying familial pulmonary arterial hypertension. Int Heart J. 2021;62(3):566-574. 10.1536/ihj.20-711. [DOI] [PubMed] [Google Scholar]
- 92. Park CS, Kim SH, Yang HY, et al. Sox17 deficiency promotes pulmonary arterial hypertension via HGF/c-Met signaling. Circ Res. 2022;131(10):792-806. 10.1161/CIRCRESAHA.122.320845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kulas JA, Puig KL, Combs CK.. Amyloid precursor protein in pancreatic islets. J Endocrinol. 2017;235(1):49-67. 10.1530/JOE-17-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Osipovich AB, Dudek KD, Greenfest-Allen E, et al. A developmental lineage-based gene co-expression network for mouse pancreatic beta-cells reveals a role for Zfp800 in pancreas development. Development. 2021;148(6):dev196964. 10.12422/dev.196964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Alvarez-Dominguez JR, Donaghey J, Rasouli N, et al. Circadian entrainment triggers maturation of human in vitro islets. Cell Stem Cell. 2020;26(1):108-122.e10. 10.1016/j.stem.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 96. Shrestha S, Erikson G, Lyon J, et al. Aging compromises human islet beta cell function and identity by decreasing transcription factor activity and inducing ER stress. Sci Adv. 2022;8(40):eabo3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kaestner KH. The making of the liver: developmental competence in foregut endoderm and induction of the hepatogenic program. Cell Cycle. 2005;4(9):1146-1148. 10.4161/cc.4.9.2033. [DOI] [PubMed] [Google Scholar]
- 98. Lewis AE, Kuwahara A, Franzosi J, Bush JO.. Tracheal separation is driven by NKX2-1-mediated repression of Efnb2 and regulation of endodermal cell sorting. Cell Rep. 2022;38(11):110510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nair S, Schilling TF.. Chemokine signaling controls endodermal migration during zebrafish gastrulation. Science. 2008;322(5898):89-92. 10.1126/science.1160038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chuai M, Hughes D, Weijer CJ.. Collective epithelial and mesenchymal cell migration during gastrulation. Curr Genomics. 2012;13(4):267-277. 10.2174/138920212800793357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kamachi Y, Uchikawa M, Kondoh H.. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16(4):182-187. 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 102. Cremazy F, Berta P, Girard F.. Genome-wide analysis of Sox genes in Drosophila melanogaster. Mech Dev. 2001;109(2):371-375. 10.1016/s0925-4773(01)00529-9. [DOI] [PubMed] [Google Scholar]
- 103. Hu Y, Wang B, Du H.. A review on sox genes in fish. Rev Aquacult. 2021;13(4):1986-2003. 10.1111/raq.12554. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw scRNAseq data is at ArrayExpress (E-MTAB-12719), Seurat objects are at https://zenodo.org/record/7725887#.ZA5jRR_MLkI, and scripts used to perform quality control, clustering, differential gene expression analysis, Monocle3 and RNAvelocity analyses are at https://github.com/markmagnuson/2023-Linh-scRNASeq-Mouse-Sox17-Expressing-Lineages.
Raw scRNAseq data is at ArrayExpress (E-MTAB-12719), Seurat objects are at https://zenodo.org/record/7725887#.ZA5jRR_MLkI. Scripts used to perform quality control, clustering, differential gene expression analysis, Monocle3 and RNAvelocity analyses are at https://github.com/markmagnuson/2023-Linh-scRNASeq-Mouse-Sox17-Expressing-Lineages.