Abstract
The benefits shown by the recent introduction of PCR for the in vitro diagnosis of hepatitis C virus (HCV) infection has prompted the development of standardized, ready-to-use assays that can be implemented in routine clinical laboratories. We have evaluated the clinical performance of COBAS AMPLICOR HCV (COBAS), the first instrument system that allows the automation of HCV RNA amplification and detection, to determine its performance in the routine laboratory setting. More than 2,000 specimens collected at five centers were analyzed in parallel by the COBAS and the manual AMPLICOR HCV (AMPLICOR) tests, and the results were compared with the results for biochemical and serological markers of HCV. In this study the two PCR systems showed the same accuracy, with a concordance rate of 99.8%. As expected, the correlation between serology and PCR was not absolute because the presence of anti-HCV antibodies may be associated with a latent or past infection. On the other hand, if the presence of confirmed anti-HCV antibodies and elevated alanine aminotransferase levels are taken as the “gold standard,” indicating an active, ongoing infection, the COBAS and AMPLICOR tests show high and comparable sensitivities (100%) and specificities (98%), with positive and negative predictive values of 100 and 97%, respectively. During the study no false-positive reactions were detected. The use of an internal control allowed the identification of inhibitory substances that prevented amplification for 0.3 and 0.4% of samples tested by the COBAS and AMPLICOR tests, respectively. Compared to the manual system, the COBAS system allowed a significant reduction of hands-on time and could improve the overall laboratory work flow. In conclusion, these results support the use of the COBAS and AMPLICOR tests for the molecular diagnosis of active HCV infections.
The lack of virus isolation techniques and antigen detection assays has made nucleic acid-based amplification the method of choice for the direct identification of hepatitis C virus (HCV). Indeed, by PCR it is possible to assess the status of the infection, to detect viral replication in seropositive patients, and to diagnose the infection in immunocompromised patients and during the window that precedes seroconversion (2, 3, 15, 18, 21).
The implementation of PCR in routine diagnostic laboratories, however, requires a level of standardization that was not originally provided by in-house methods. Additionally, by in-house methods the occurrence of false-positive results due to amplified product contamination is not always preventable and false-negative results due to inhibition are not always easily detectable (5, 17, 19, 20, 24). Also, the hands-on time required by a manual in-house assay for reagent preparation as well as the testing of samples from patients limits its utility in the routine diagnostic laboratory. To address these technical concerns, a dedicated instrument, the COBAS AMPLICOR HCV (COBAS) analyzer, that automates amplification and detection has recently been introduced (6, 10). The system provides amplification and detection with standardized, ready-to-use reagents based on the reagents formerly developed for the corresponding AMPLICOR HCV manual format (AMPLICOR). The amplification reagents include the enzyme uracil-N-glycosylase (AmpErase) for the prevention of carryover contamination (12, 22) and an internal control (IC) for the identification of processed specimens containing substances that may interfere with amplification (16). The closed system provides additional protection against carryover contamination. In the present study we evaluated the performance of the COBAS test with a collection of more than 2,000 clinical specimens and compared it with that of the corresponding manual AMPLICOR test and the results for other serological and biochemical parameters.
MATERIALS AND METHODS
We analyzed 2,292 consecutive serum specimens collected and separately tested at five centers. The five centers provided results for 484, 655, 495, 498, and 160 serum specimens, respectively. Serum was separated from whole blood and was immediately stored at −80°C in several aliquots to preserve the integrity of the HCV RNA.
Serology.
All specimens were tested for the presence of anti-HCV antibodies by enzyme-linked immunosorbent assay (ELISA; Abbott Diagnostics, North Chicago, Ill.; Ortho Diagnostic Systems, Raritan, N.J.) and at most centers by RIBA (Chiron Corp., Emeryville, Calif.). This confirmatory assay was performed with 2,056 (89%) samples. In addition, biochemical markers of liver inflammation and necrosis including aspartate aminotransferase and alanine aminotransferase (ALT) levels were evaluated.
PCR analysis.
Serum specimens were analyzed by the COBAS test (Roche Diagnostic Systems, Somerville, N.J.) by following the manufacturer’s instructions. Briefly, 100 μl of serum was incubated with a lysis buffer, and the RNA was then precipitated with isopropanol by centrifugation, washed once with ethanol, and resuspended in 1 ml of specimen diluent. An IC RNA was also introduced into the specimen with the diluent and served as an amplification control for each individually processed specimen. The HCV IC consists of a synthetic RNA transcript with primer binding regions identical to those of the HCV target sequence, a randomized internal sequence with a length and a base composition similar to those of the HCV target sequence, and a unique probe binding region that differentiates the IC from the target amplicon.
Each specimen or control was amplified in the thermal cycler section of the COBAS analyzer with primers KY78 (biotinylated) and KY80, which identify a 244-bp sequence of the highly conserved 5′ untranslated region of the HCV genome.
After amplification, the analyzer automatically denatured the double-stranded, biotinylated amplified products and captured them with a suspension of magnetic particles coated with an oligonucleotide probe specific for HCV (or IC). The unbound material was washed away, and the biotinylated amplicon was detected with an avidin-horseradish peroxidase conjugate–tetramethylbenzidine-H2O2 colorometric reaction. The absorbance (660 nm) of each sample was recorded and was compared with a predefined cutoff value to determine positive or negative results for both HCV and IC.
An aliquot of each specimen was tested in parallel by the AMPLICOR assay (Roche Diagnostic Systems) as described in the manufacturer’s instructions.
The results of the PCR assay as well as all other clinical, serological, and biochemical data were collected and analyzed. Discrepancies between PCR, both automated and manual, and between PCR and serology were resolved by repeat testing of frozen aliquots. Repeat testing was also done with specimens with no IC signal or a low IC signal to ascertain the presence of substances inhibitory to the PCR.
At two of the participating centers a workload analysis was carried out by the method of the College of American Pathologists (4). To determine core timing values, data were collected for the following steps of the procedure: (i) initial handling of the specimen, (ii) specimen testing, (iii) daily and periodic activities, and (iv) recording of test data from the instrument. Individual times for each step were summed, and “hands-off” times were also calculated to give the total time from the initiation of the test to retrieval of the final test result.
RESULTS
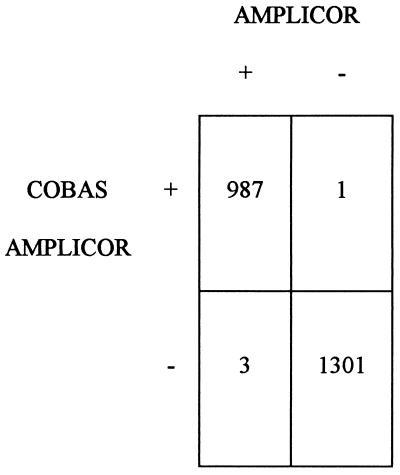
The 2,292 consecutive specimens obtained from patients at risk of HCV infection were simultaneously analyzed by the COBAS and the AMPLICOR tests. The concordance between the two systems was 98%, but it rose to 99.8% after repeat testing (Fig. 1). Discrepant results were observed for 4 (0.2%) of all samples.
FIG. 1.
Overall performance of COBAS and AMPLICOR tests. Numbers indicate the numbers of samples with the indicated test results.
Samples presenting negative IC values due to the presence of inhibitory substances that prevented amplification of the IC were resolved by retesting another aliquot of the specimen. After resolution of the results, invalid results were obtained for 0.3 and 0.4% of all specimens analyzed by the COBAS and AMPLICOR tests, respectively. The values obtained prior to repeat testing were 3 and 2% for the COBAS and AMPLICOR tests, respectively.
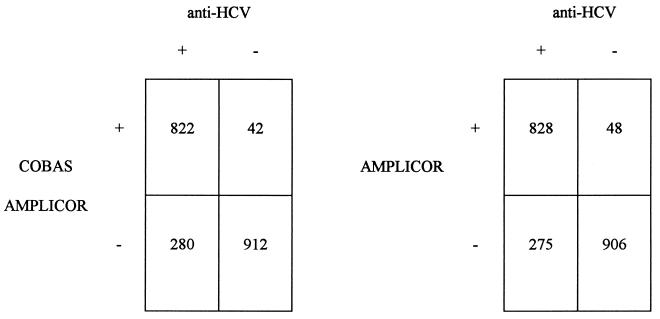
When compared to the patients’ serological status, as assessed by anti-HCV ELISA and RIBA, the results of PCR showed an overall concordance rate of 84% (Fig. 2).
FIG. 2.
Comparison between COBAS and AMPLICOR test results and the HCV serological status as defined by ELISA and RIBA. Numbers indicate the numbers of samples with the indicated test results.
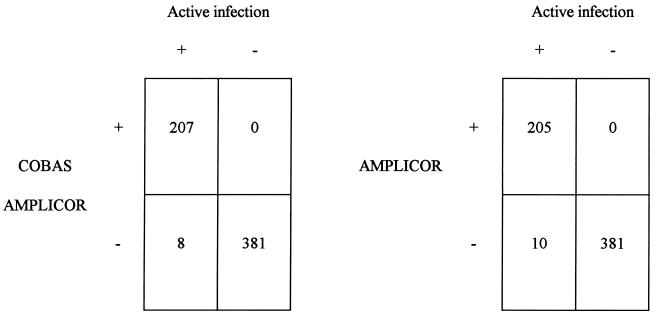
To better define the relationship between serological and molecular data for our patients, we compared the results obtained with the COBAS and AMPLICOR tests with those of ALT level and RIBA determinations. The combination of the latter two parameters served to identify active HCV infection. By this approach the concordance rate increased to 98% (Fig. 3), indicating that most anti-HCV-positive, PCR-negative discrepant results were due to past or latent infections. Moreover, if PCR results are compared to the active infection status, with use of active infection status as the “gold standard,” the COBAS and AMPLICOR tests show sensitivities and specificities of 96 and 100% and of 95 and 100%, respectively. The corresponding positive predictive and negative predictive values were 100 and 98%, respectively, for the COBAS test and 100 and 97%, respectively, for the AMPLICOR test.
FIG. 3.
Comparison between COBAS and AMPLICOR test results and the presence or absence of an active HCV infection as defined by the detection of specific antivirus antibodies and elevated ALT levels. Numbers indicate the numbers of samples with the indicated test results.
The turnaround time for the complete COBAS test including specimen preparation and the amplification and detection of HCV and IC was 5 h for a batch of 20 specimens. The operator was required to be present for less than 2 h to perform manual steps that included sample and reagent preparation and loading of the amplification tubes in the thermal cyclers and in the detection racks. The overall workload time was about 5 min/specimen, compared to 7 min/specimen for the manual test. By using the parallel-run feature of the instrument, the throughput can be increased to three to four runs per day, including an unattended overnight run.
DISCUSSION
HCV infection is a typical example of a disease in which direct detection of the virus is essential for a correct diagnosis. In contrast to the other available in vitro assays, PCR has the potential for high diagnostic value because it offers a definitive identification of HCV. However, the poor reproducibility of in-house-developed assays, shown in previous studies, has indicated the need for standardized and more accurate procedures (5, 24). The recent introduction of an automated instrument for the amplification and detection of HCV RNA has prompted us to evaluate its performance with a large collection of clinical specimens obtained from different institutions. The results of this study have shown that the COBAS test has the same performance as the established manual AMPLICOR test because only 0.2% of all results remained discordant after repeat testing. Initial discrepancies were due mostly to invalid results caused by low values for the IC.
The COBAS and AMPLICOR tests also showed the same sensitivities and specificities compared to those of serology and clinical diagnosis, which were taken as the gold standards. We used clinical diagnosis because serology alone does not necessarily reflect the presence of an active infection but reflects only a previous exposure to the virus (2, 21). On the other hand, the presence of anti-HCV antibodies in conjunction with elevated ALT levels and histological evidence of chronic hepatitis better represent the features of an ongoing, clinically relevant infection (1, 3, 13, 21). In this regard, the samples with discrepant results that were anti-HCV positive and HCV RNA negative were from patients who had normal ALT levels and hence a latent, inactive infection. The few patients (2%) who had a clinical diagnosis of type C hepatitis but who were PCR negative had borderline ALT level elevations and probably had HCV RNA concentrations below the analytical sensitivity of the AMPLICOR system (7).
We did not compare the COBAS test with nested PCR techniques because the manual AMPLICOR test, which served as the reference for the automated system in this study, had been previously extensively analyzed (8, 9, 14, 23).
An interesting feature of the COBAS test is the optional detection of an IC that is coamplified with the target and that serves to identify the possible presence of interfering substances that inhibit DNA polymerization, causing false-negative reactions (16).
Inhibition occurred with 3% of all samples and was readily solved by repeat testing of frozen aliquots. This may indicate that modification or degradation of inhibitory substances occurred during storage or that inhibitors are not equally distributed throughout a sample (16).
On the other hand, the presence of uracil-N-glycosylase (AmpErase), which destroys a dUTP-containing amplicon from previous reactions, and the use of proper precautions taken at the time of specimen preparation prevented false-positive reactions due to carryover contamination or cross contamination from positive to negative samples (11, 12).
In this respect, an additional benefit of the COBAS system is the closed containment in which all amplification and detection steps take place. Amplification products are kept in sealed tubes that are never opened but that are pierced by the transfer tip during the detection part of the procedure. Throughout the duration of the study we assessed the robustness of the instruments and never had failures requiring external assistance. Taken together, these data suggest that the COBAS test may be a suitable system for the routine analysis of samples obtained from anti-HCV-positive individuals.
REFERENCES
- 1.Alberti A, Morsica G, Chemello L, Cavalletto D, Noventa F, Pontisso P, Ruol A. Hepatitis C viraemia and liver disease in symptom-free individuals with anti-HCV. Lancet. 1992;340:697–698. doi: 10.1016/0140-6736(92)92234-7. [DOI] [PubMed] [Google Scholar]
- 2.Alter H J. To C or not to C: these are the questions. Blood. 1995;85:1681–1695. [PubMed] [Google Scholar]
- 3.Bonino, F., M. R. Brunetto, F. Negro, M. Baldi, G. Saracco, M. L. Abate, A. Fabiano, G. Verme, et al. 1993. Hepatitis C virus infection and disease. Diagnostic problems. J. Hepatol. 17(Suppl. 3):78–82. [DOI] [PubMed]
- 4.College of American Pathologists. Workload recording method and personnel management manual. Northfield, Ill: College of American Pathologists; 1992. [Google Scholar]
- 5.Damen M, Cuypers H T M, Zaaijer H W, Reesink H W, Schaasberg W P, Gerlich W H, Niesters H G M, Lelie P N. International collaborative study on the second EUROHEP HCV-RNA reference panel. J Virol Methods. 1996;58:175–185. doi: 10.1016/0166-0934(96)02011-3. [DOI] [PubMed] [Google Scholar]
- 6.DiDomenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy Z G, Rosenstraus M. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine PCR. Clin Chem. 1996;42:1915–1923. [PubMed] [Google Scholar]
- 7.Dusheiko G M, Khakoo S, Soni P, Grellier L. A rational approach to the management of hepatitis C infection. Br Med J. 1996;312:357–364. doi: 10.1136/bmj.312.7027.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerken G, Pontisso P, Roggendorf M, Rumi M G, Simmonds P, Trepo C, Zeuzem S, Colucci G. Clinical evaluation of a single reaction, diagnostic PCR assay for the detection of hepatitis C virus (HCV) RNA. J Hepatol. 1996;24:33–37. doi: 10.1016/s0168-8278(96)80183-8. [DOI] [PubMed] [Google Scholar]
- 9.Goergen B, Meyer Zum Buschenfelde K H, Gerken G. Assessment of viral replication for routine diagnosis of HCV infection and for monitoring antiviral treatment using Amplicor HCV detection system. Klin Lab. 1994;40:1–7. [Google Scholar]
- 10.Jungkind D, DiRienzo S, Beavis K G, Silverman N S. Evaluation of automated COBAS AMPLICOR PCR system for detection of several infectious agents and its impact on laboratory management. J Clin Microbiol. 1996;34:2778–2783. doi: 10.1128/jcm.34.11.2778-2783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwok S, Higuchi R. Avoiding false positive with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 12.Longo M C, Berninger M S, Hartley H L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 13.Marin M G, Bresciani S, Puoti M, Rodella A, Gussago A, Ravaggi A, Pizzocolo G, Albertini A, Cariani E. Clinical significance of serum hepatitis C virus (HCV) RNA as marker of HCV infection. J Clin Microbiol. 1996;32:3008–3012. doi: 10.1128/jcm.32.12.3008-3012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prati D, Capelli C, Bosoni P, Mozzi F, Zanella A, Sirchia G. Determination of hepatitis C virus RNA in the serum by the Amplicor HCV PCR kit. Vox Sang. 1994;67:112–114. doi: 10.1111/j.1423-0410.1994.tb05054.x. [DOI] [PubMed] [Google Scholar]
- 15.Roggendorf M, Lu M, Meisel H, Riffelmann M, Schreier E, Viazov S. Rational use of diagnostic tools in hepatitis C. J Hepatol. 1996;24:26–34. [PubMed] [Google Scholar]
- 16.Rosenstraus, M., Z. Wang, S.-Y. Chang, D. DeBoneville, and J. P. Spadoro. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J. Clin. Microbiol. 36:191–197. [DOI] [PMC free article] [PubMed]
- 17.Rys P N, Persing D H. Preventing false positives: quantitative evaluation of three protocols for inactivation of polymerase chain reaction amplification products. J Clin Microbiol. 1993;31:2356–2360. doi: 10.1128/jcm.31.9.2356-2360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tedeschi V, Seeff L B. Diagnostic tests for hepatitis C: where are we now? Ann Intern Med. 1995;123:383–385. doi: 10.7326/0003-4819-123-5-199509010-00009. [DOI] [PubMed] [Google Scholar]
- 19.Victor T, Jordaan A, du Toit R, Van Helden P D. Laboratory experience and guidelines for avoiding false positive polymerase chain reaction results. J Clin Chem Clin Biochem. 1993;31:531–535. [PubMed] [Google Scholar]
- 20.White T J, Madej R, Persing D H. The polymerase chain reaction: clinical applications. Adv Clin Chem. 1992;29:161–196. doi: 10.1016/s0065-2423(08)60224-3. [DOI] [PubMed] [Google Scholar]
- 21.Wilber J C, Polito A. Serological and virological diagnostic tests for hepatitis C virus infection. Semin Gastrointestinal Dis. 1995;6:13–19. [PubMed] [Google Scholar]
- 22.Young K K Y, Resnick R M, Myers T W. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J Clin Microbiol. 1993;31:882–886. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young K Y, Archer J J, Yokosuka O, Omata M, Resnick R. Detection of hepatitis C virus RNA by a combined reverse transcription PCR assay: comparison with nested amplification and antibody testing. J Clin Microbiol. 1995;33:654–657. doi: 10.1128/jcm.33.3.654-657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaaijer H J, Cuypers H T, Reesink H W. Reliability of HCV PCR results. Lancet. 1993;341:722–724. doi: 10.1016/0140-6736(93)90488-3. [DOI] [PubMed] [Google Scholar]