Abstract
Human CD56br natural killer (NK) cells represent a small subset of CD56+ NK cells in circulation and are largely tissue-resident. The frequency and number of CD56br NK cells in blood has been shown to increase following administration of low-dose IL-2 (LD-IL2), a therapy aimed to specifically expand CD4+ regulatory T cells (Tregs). Given the potential clinical application of LD-IL-2 immunotherapy across several immune diseases, including the autoimmune disease type 1 diabetes, a better understanding of the functional consequences of this expansion is urgently needed. In this study, we developed an in vitro co-culture assay with activated CD4+ T cells to measure NK cell killing efficiency. We show that CD56br and CD56dim NK cells show similar efficiency at killing activated CD4+ conventional T (Tconv) and Treg cell subsets. However, in contrast to CD56dim cells, CD56br NK cells preferentially target highly proliferative cells. We hypothesize that CD56br NK cells have an immunoregulatory role through the elimination of proliferating autoreactive CD4+ Tconv cells that have escaped Treg suppression. These results have implications for the interpretation of current and future trials of LD-IL-2 by providing evidence for a new, possibly beneficial immunomodulatory mechanism of LD-IL-2-expanded CD56br NK cells.
Keywords: Natural killer (NK) cells, CD56br NK cells, CD4+ T cells, in vitro NK killing assay, low-dose IL-2 immunotherapy
Introduction
Natural killer (NK) cells represent a population of innate immune cells that have evolved to mediate their effector function through the acquisition of a cytotoxic profile, while avoiding tissue damage [1]. Although typically associated with a multitude of effector functions, including tumour cell recognition and anti-viral immunity, NK cells have also been more recently shown to display immunomodulatory functions through the regulation of T cell responses [2–6]. In humans, circulating CD56+ NK cells can be differentiated into two main subtypes based on the expression of CD56 and CD16: a rare CD56br CD16–/low (CD56br) subset and a more abundant CD56dim CD16+ (CD56dim) subset, usually representing > 90% of circulating NK cells. In contrast to blood, CD56br NK cells are predominantly found in tissues, where they represent the majority of tissue-resident NK cells [7]. While it is known that CD56+ NK cells develop from CD34+ haematopoietic stem cells, there is conflicting data on whether CD56br NK cells are precursors of CD56dim NK cells or if they constitute an independent NK cell lineage [8, 9]. More recently, the application of multi-parametric multiomics technologies have revealed a more complex picture of the phenotypic heterogeneity of NK cell subsets in blood and in tissues [10–15].
In addition to CD4+ regulatory T cells (Tregs), circulating CD56br NK cells expand greatly in response to low-dose interleukin-2 (LD-IL-2) treatment [16–19]. While both CD56+ NK cell subsets express high levels of the IL-2 receptor β chain (CD122), the increased sensitivity of CD56br NK cells to LD-IL-2 is mediated by the concomitant increased expression of the high-affinity α chain (CD25) [18]. The initial therapeutic success and ongoing clinical trials of LD-IL-2 immunotherapy in a variety of autoimmune and inflammatory diseases [20, 21] therefore provide an important rationale to better understand the potential contribution of CD56br NK cells in the mechanism of action of LD-IL-2 immunotherapy. However, the exact function of CD56br NK cells is still poorly understood in the context of their expansion by LD-IL-2 and the therapeutic potential of this drug. One of the main challenges is the scarcity of these cells in blood, which strongly limits the access to the cell numbers required for the development of robust cellular assays. Here, we detail the development of an in vitro NK:CD4+ T cell co-culture assay. We demonstrate that in contrast to CD56dim NK cells, CD56br NK cells preferentially kill the more highly proliferative CD4+ T cells, suggesting a regulatory function in the elimination of pathogenic proliferating T cells in inflamed tissues.
Methods
Subjects
A total of 29 healthy study participants were recruited from the Oxford BioBank. Approximately 50 ml of peripheral venous blood was collected in Ethylenediaminetetraacetic acid tubes from each donor. All samples and information were collected with written and signed informed consent. The study conformed to the principles of the Helsinki Declaration and Good Clinical Practice and was approved by the East of England—Cambridgeshire and Hertfordshire research ethics committee (05/Q0106/20).
Cell preparation
For assays using freshly isolated peripheral blood mononuclear cells (PBMCs; n = 14 donors), cells were isolated from human peripheral blood samples within 2 h of venepuncture. Whole blood samples were diluted at a 1:1 ratio with Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma) and then gently layered over density gradient medium Lymphoprep™ (STEMcell). PBMCs were isolated by centrifugation at 800 g for 20 min. PBMCs were washed twice in phosphate-buffered saline (PBS; Gibco) with 0.2% bovine serum albumin (BSA; Miltenyi Biotec) and re-suspended in 10 ml PBS + 0.2% BSA for cell counting.
For assays using cryopreserved cells (n = 15 donors), PBMCs were re-suspended at a concentration of 106 cells/ml in CryoStor. Samples were incubated overnight at −80°C in a cryogenic storage container (CoolCell™, Corning) and then stored in liquid nitrogen until the day of the assay. Samples were then thawed at 37°C, re-suspended drop-by-drop in RPMI with 1% heat-inactivated, filtered human AB serum (AB HS; Sigma), and washed twice with PBS + 0.2% BSA.
Cell staining and fluorescence-activated cell sorting (FACS)
To isolate the CD4+ T cell subsets for our assays, approximately 5% of the PBMCs (T cell fraction) were washed with PBS only and then incubated for 10 min at 37°C with cell proliferation dye eFluor™ 450 (Invitrogen) at a concentration of 10 µM. Following incubation, the cell proliferation dye was quenched with ice-cold PBS + 10% BSA and incubated on ice for a further 5 min, then washed with PBS + 0.2% BSA. The remaining 95% of PBMCs (NK cell fraction) were dedicated for the isolation of the CD56+ NK subsets and were not incubated with the cell proliferation dye prior to cell sorting.
Both cell fractions were then incubated with Brilliant Stain Buffer (BD Biosciences) and incubated for 30 min at 4°C with the following fluorochrome-conjugated antibodies: CD3-BUV737, CD4-BUV395, CD25-PE (BD Biosciences), CD127-PE Cy7 (eBioscience), CD56-BV711, and CD8-AF700 (Biolegend). PBMCs were then washed and re-suspended in PBS + 0.2% BSA for cell sorting on a BD FACSAria™ Fusion cell sorter (BD Biosciences). The following cell subsets were sorted: CD3+CD4+CD127+/lowCD25–/low [CD4+ conventional T (Tconv)] and CD3+CD4+ CD127lowCD25hi (CD4+ Tregs) from the T cell fraction (stained with proliferation dye); and CD3–CD56hiCD127+/low (CD56br NK) and CD3–CD56lowCD127– (CD56dim NK) from the NK cell fraction (proliferation dye-negative). Post-sorting cell purity rates were found to be > 94% and > 97% for the NK and T cell subsets, respectively.
Cell culture
All cell subsets were washed with PBS + 0.2% BSA and then re-suspended in master media [RPMI + 100U penicillin + 100 µg streptomycin + 2 mM l-Glutamine (Gibco) + 5% AB HS] + 100 IU/ml IL-2 (Proleukin®/aldesleukin; Novartis) at a target concentration of 5 × 105 cells/ml and 100 µl (50 000 cells) were seeded in a 96 well U-bottom plate (Greiner). The main limitation for the practical implementation of these in vitro co-culture assays is the number of CD56br NK cells that can be sorted from each donor, as their frequency is variable between donors and they represent only a small fraction of the total NK cells. This was particularly challenging for cryopreserved samples, where we achieved a median of 41 198 [95% confidence interval (CI) 24 047–73 024] sorted CD56br NK cells. By contrast, we obtained much higher numbers of sorted CD56br NK cells from freshly isolated PBMCs (median = 122 700; 95% CI 69 416–210 000). We, therefore, opted to use only cells sorted from freshly isolated PBMCs to set up the killing assays described in this study.
CD4+ T cells were activated using αCD3/CD28 beads (Gibco) at a ratio of 1 bead: 3 T cells in addition to IL-2 (100 IU/ml), whereas activation of NK subsets only required 100 IU/ml IL-2. Cell subsets were cultured separately for up to 3.5 days in vitro. To assess the proliferation dynamics of both CD4+ T and CD56+ NK cell subsets, cells were processed as described above using 10 × 106 cryopreserved PBMCs from four independent healthy donors and stained with the proliferation dye eFluor™ 450 (Invitrogen), before culture. CD4+ T cells were cultured as described above and CD56+ NK cells were cultured either in the presence or absence of 100 IU/ml IL-2. All cells were harvested at days 3 and 7 to assess proliferation on BD LSRFortessa™ (BD Biosciences).
NK killing assay
At 2.5 or 3.5 days of in vitro cell culture, CD4+ T and CD56+ NK cell subsets were incubated for 20 min at room temperature with CD4-BUV395 (BD) and CD56-BV711 (BioLegend), respectively. CD4+ T cell subsets were then washed and co-cultured with the respective CD56+ NK cell subset. Before co-culture, each CD4+ T and CD56+ NK cell subsets were carefully counted using a haemocytometer to achieve the intended 5 NK: 1 T cell ratio. Due to the limiting number of NK cells, the number of cells used for co-culture in each assay were determined by the available NK cells. Sufficient activation of CD56br NK cells was achieved for most of the donors, except for one donor pair, where we were only able to use 5000 CD56br NK cells for the co-culture assays. We obtained much higher numbers of CD56br NK for all other donor pairs (median = 28 000 cells, 95% CI 13 000–67 500). Co-cultures were incubated for 5 h, at 37°C in master media with 100 IU/ml IL-2. Immediately before cells were analysed, co-cultures were incubated in the dark with fluorescent cell viability dye (DRAQ7; BioLegend) for 15 min at room temperature. Each well was rinsed with PBS + 0.2% BSA and re-suspended in a final volume of 300 µl for analysis on BD LSR Fortessa™ (BD Biosciences). To ensure no dead T cells were removed from the analysis—hence skewing the assessment of the frequency of specific killing—we analysed all acquired cells, without prior forward scatter (FSC) versus side scatter (SSC) exclusion. CD4+ T were discriminated from CD56+ NK cells based on the expression of CD4 and proliferation dye. Frequency of specific killing was then calculated by subtracting the percentage of background spontaneous cell death, assessed in both CD4+ Tconv (median = 8.4%, 95% CI 4.9–19.5%) and Tregs (median = 9.6%, 95% CI 6.8–13.0%) from each donor incubated in the absence of CD56+ NK cells.
To calculate the relative frequency of specific killing according to the proliferative stage of the responder T cells, we have defined “highly proliferative” T cells, as CD4+ Tconvs and Tregs having undergone ≥2 proliferation cycles at day 2.5 or ≥3 proliferation cycles at day 3.5. The different thresholds used at day 2.5 and day 3.5 were set to account for the lower proliferation dynamics of CD4+ T cells at day 2.5 compared to day 3.5. The frequency of specific killing in highly proliferative and lowly proliferative cells was then assessed by determining the percentage of dead responder CD4+ T cells in each respective population, after subtracting the percentage of background spontaneous cell death (assessed in responder T cells cultured in the absence of CD56+ NK cells) in both the high (Tconv: median = 12.6%, 95% CI 6.4–20.1%; Treg: median = 10.7%, 95% CI 7.7–15.0%) and low (Tconv: median = 7.6%, 95% CI 3.6–17.0%; Treg: median = 10.4%, 95% CI 6.4–13.3%) proliferation groups.
Data analysis
The software package FlowJo (BD Biosciences, V10.8.1) was used for the analysis of the FCS files obtained from the flow cytometer. FlowJo’s proliferation modelling software was used to analyse the proliferative capacity of cell subsets. Proliferative capacity was calculated using both the division and proliferation index (PI) methods implemented in FlowJo. The division index (DI) models proliferation for the whole population, while the PI, only models the cells that have initiated cell division. Differences in the proliferative capacity of cell subsets was compared using two-tailed paired t-tests.
Statistical analyses of NK killing assay data were performed by Bayesian inference on a Beta regression model [22], with cell type and sample pair as predictor variables, and killing proportion as response variable. We depict posterior distributions of killing proportion means for each group and fold changes across groups. As a summary of posterior distributions, we report the median and a 95% credible interval. As a measure of uncertainty, we refer to the false sign rate (FSR) [23]. This quantity denotes the support of the posterior distribution for an effect with a sign different than the median, i.e. the probability that killing proportions are higher on the group whose killing proportion is estimated to be lower. Posterior distributions were simulated using a No-U-Turn sampler (Stan, v2.31) [24] with default parameters and four chains that drew 2500 samples each during warmup, followed by another 2500 samples for estimation. Prior distributions were set to default weakly informative choices for regularization and stabilization.
Results
Assessing the proliferative capacity of in vitro stimulated immune cell subsets
The investigation of the functional relevance of NK cell subsets has been hampered by the lack of suitable in vitro assays. Currently, the most widely used NK functional assays include co-culture assays, aiming at quantifying NK killing of the target T cells subset. Previous studies have shown that both the NK cells and target T cells require appropriate activation for efficient autologous killing to occur in vitro [25]. One of the main limitations for the development of more robust in vitro assays is the availability of sufficient cell numbers, particularly for the rare subset of circulating CD56br NK cells. To overcome this limitation, we aimed to establish an in vitro NK cell killing assay that minimizes the required number of CD56+ NK and CD4+ T cell subsets sorted from PBMCs.
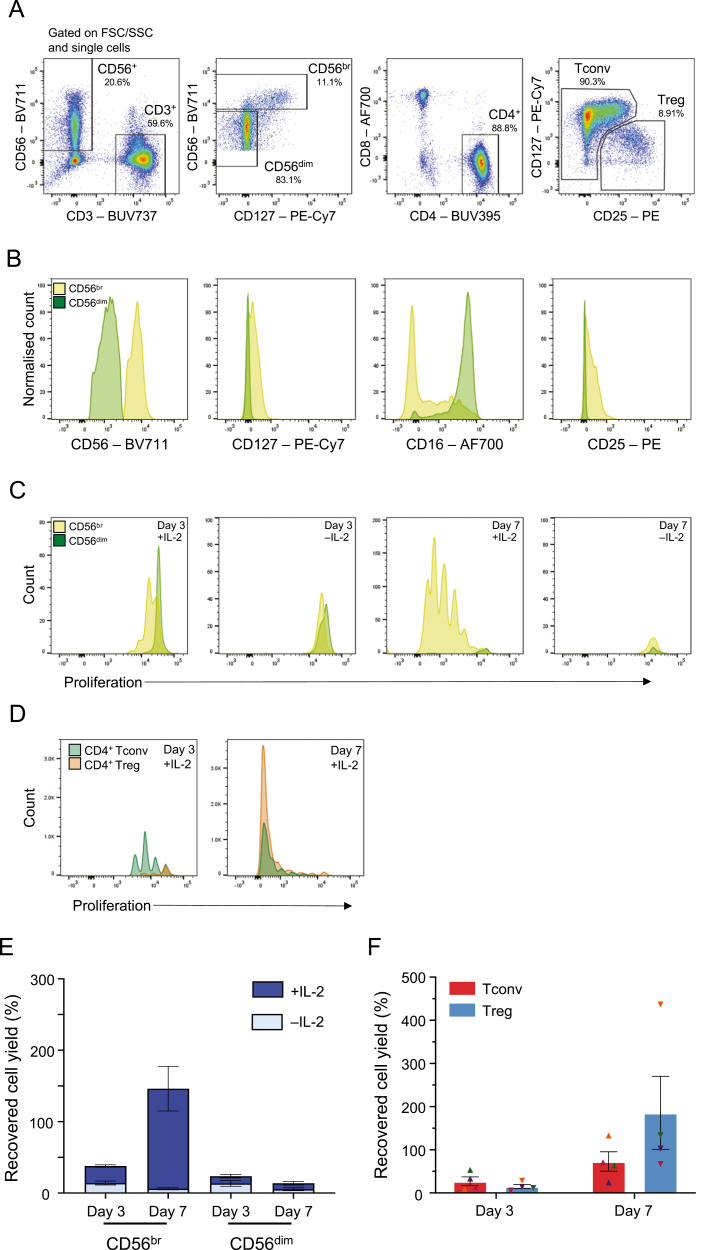
Initially, we explored the use of cryopreserved PBMCs to set up the in vitro assay. We flow-sorted CD56br NK, CD56dim NK, CD4+CD25–/low Tconv, and CD4+ Treg cell subsets (Fig. 1A) and cultured both TCR-stimulated T cell and NK cell subsets for a period of 3 or 7 days in vitro in the presence of 100 IU/ml IL-2. As expected, sorted CD56br and CD56dim NK cell subsets displayed distinct phenotypic profiles, as evidenced by the expression of key markers, such as CD56, CD127, CD16, and CD25 (Fig. 1B). A feature of our assay was the ability to assess the proliferation status of the different cell subsets at the single-cell level using an intracellular cell proliferation dye, resulting in distinct proliferation peaks. To investigate the dynamics of cell proliferation in both CD56+ NK cell and TCR-stimulated CD4+ T cell subsets, we initially assessed the proliferative capacity of both populations at days 3 and 7 of culture to determine the optimal duration of the initial stimulation period. Notably, CD56+ NK cell proliferation in vitro was mostly restricted to the CD56br NK cells and was strictly dependent on the presence of IL-2 in culture (Fig. 1C). In contrast to CD56+ NK cells, both T cell subsets demonstrated strong in vitro proliferative capacity following stimulation with αCD3/CD28 beads (1 bead: 3 T cells) and IL-2 (Fig. 1D). At day 7, the majority of CD4+ Tconv and Tregs in culture had undergone over four-division cycles suggesting excessive proliferation that may compromise the potential to assess cell subset-specific functional differences.
Figure 1:
Assessing the proliferative capacity of the CD56+ NK cell subsets in vitro. (A) Sorting strategy for the isolation of the four assessed immune cell subsets: (i) CD3–CD56br NK cells (CD56br NK); (ii) CD3–CD56dim NK cells (CD56dim NK); (iii) CD3+CD4+CD25–/low (Tconv); and (iv) CD3+CD4+CD127lowCD25hi (Treg). Plots represent an illustrative example from the sorting of cryopreserved PBMCs from four healthy donors. (B) Representative histograms depicting the expression of CD56, CD127, CD16, and CD25 in sorted CD56br (light green) and CD56dim (dark green) NK cells at day 0. (C) Representative histograms depicting the proliferative capacity of CD56br (light green) and CD56dim (dark green) NK cells after 3 (left panels) or 7 (right panels) days of culture in the presence or absence of 100 IU/ml IL-2. (D) Representative histograms depicting the proliferative capacity of CD4+ Tconvs and Tregs after 3 (left panel) or 7 (right panel) days of culture in the presence of 100 IU/ml IL-2 and TCR stimulation with αCD3/CD28 beads (1 bead: 3 T cells). (E and F) Bar plots depict the distribution (mean ± SEM) of the recovered cell yields (defined as the percentage of plated cells harvested after the initial 3 or 7 days in vitro culture period) in both CD56+ NK cell (E) and CD4+ T cell (F) subsets. CD4+ Tconvs (depicted by triangles) and Tregs (depicted by inverted triangles) isolated from the same donor are depicted using the same colour.
In agreement with their lower proliferative capacity, we obtained much lower cell recovery (defined as the percentage of the initial plated cells that were harvested) for the CD56+ NK cell subsets compared to the T cell subsets after 7 days of culture (Fig. 1E and F). This was particularly pronounced in the CD56dim NK cell subset at day 7, where we obtained only a marginal 8.41% median cell recovery (Fig. 1E). These numbers reflect the lack of proliferative capacity of CD56dim NK cells in this in vitro model and demonstrate the reduced viability of CD56dim NK cells in vitro, thereby justifying the use of an initial 3-day stimulation period to set up our co-culture assay. Importantly, the low cell yields obtained for the CD56+ NK cell subsets using cryopreserved PBMCs compromised the technical integrity of the co-culture assays, leading to insufficient numbers of target T cells being plated for the assay (at an intended ratio of 5 NK cells: 1 T cell) to robustly estimate the proportion of T cell death after co-culture. To mitigate this issue, we subsequently opted to perform this assay using freshly isolated PBMCs, which yielded much higher numbers of the sorted NK cell subsets, most notably of the limiting CD56br subset.
Development of an in vitro NK cell-killing assay
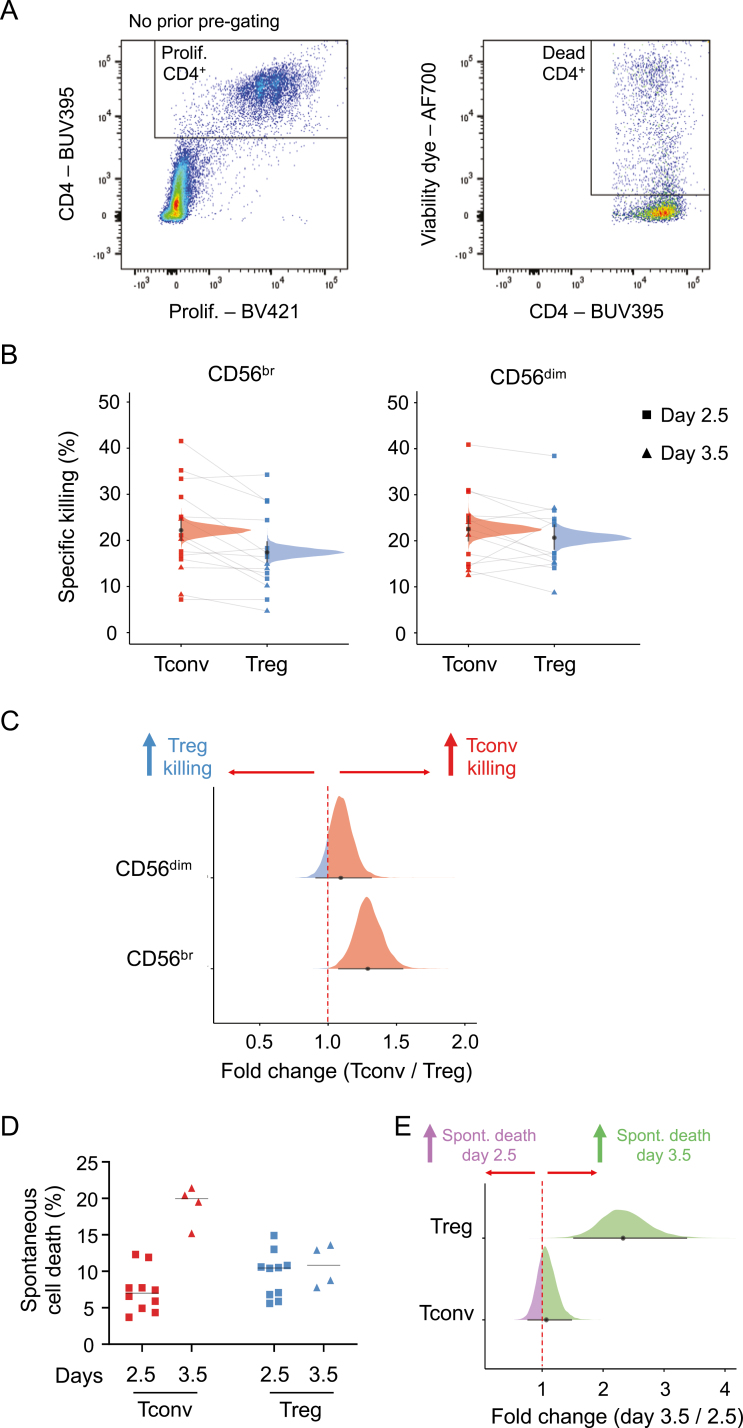
Having established the duration of the initial activation period, we then proceeded to perform the killing assays using sorted NK and T cells from freshly isolated PBMCs from seven pairs of donors. After an initial activation period of up to 3.5 days, activated NK and T cells were then co-cultured at a target ratio of five effector cells (CD56+ NK cell subset) to one target cell (CD4+ T cell subset). The proportion of dead T cells was then assessed by FACS after a 4 h co-culture period, and a frequency of specific killing was calculated by subtracting the background frequency of spontaneous cell death obtained in each CD4+ T cell subset incubated in the absence of CD56+ NK cells (Fig. 2A and Methods). Both CD56br and CD56dim NK cells displayed similar capacity to kill autologous CD4+ T cells in our in vitro assay. Of note, CD56br NK cells displayed a small increased frequency of specific killing for CD4+ Tconv (median posterior probability (PP) = 22.2%, 95% credible interval = 20.2–24.4%) compared to CD4+ Treg (median PP = 17.4%, 95% credible interval = 15.5–19.5%) cells (FSR = 0.2%; Fig. 2B and C). In contrast, we observed no difference in the frequency of specific killing of CD4+ Tconv (median PP = 22.5%, 95% credible interval = 20.3–25.0%) or Treg (median PP = 20.7%, 95% credible interval = 18.5–23.1%) cells by CD56dim NK cells (1.09 fold change, FSR = 12%; Fig. 2B and C).
Figure 2:
Development of a CD56br and CD56dim NK cell in vitro killing assay. (A) Gating strategy for the quantification of dead responder T cells from the in vitro killing assays. Killing assays were performed using T and NK cells sorted from freshly isolated PBMCs from 14 healthy donors. (B) Scatter plot depicts the frequency of specific killing of CD4+ Tconvs (red) and Tregs (blue). Frequency of specific killing was determined as the relative proportion of dead responder T cells after co-culture with CD56br (left panel) and CD56dim (right panel) NK cells, after subtracting the relative proportion of background spontaneous cell death, estimated in both responder T cell subsets (Tconvs and Tregs) incubated in the absence of NK cells (see Methods). Individuals where the initial stimulation period was 2.5 days (n = 10) or 3.5 days (n = 4) are depicted by squares or triangles, respectively. Densities depict the mean posterior frequency of cell death values in each group estimated by beta regression, with an annotation for the median and a 95% credible interval. CD4+ Tconv and Treg cells isolated from the same donor are identified by a connecting line. (C) Densities depict the effect size distribution (measured as fold change) for the observed relative rates of specific killing in CD4+ Tconv (red) and Treg (blue) by CD56br (bottom panel) and CD56dim (top panel) NK cells. (D) Scatter plots depicts the percentage of spontaneous cell death of CD4+ Tconv (red) and Treg (blue) cells. The frequency of spontaneous cell death was assessed in each donor by culturing each T cell subset in the absence of CD56+ NK cells. Data were stratified according to whether the CD4+ T cells subsets were stimulation for an initial period of 2.5 (depicted by squares) or 3.5 (depicted by triangles) days. (E) Densities depict the effect size distribution (measured as fold change) for the observed relative rates of spontaneous cell death at days 2.5 and 3.5 in CD4+ Tconv (bottom panel) and Treg (top panel) cells.
To assess whether the observed differences in the frequencies of specific killing were partly due to increased apoptosis in more activated cells, we next compared the frequency of spontaneous cell death between the two responder T cell subsets. CD4+ Tconv cells stimulated for 3.5 days showed a 2.33-fold increased frequency (FSR = 0.04%; Fig. 2D and E) of spontaneous cell death compared with 2.5 days of stimulation (median PP = 7.0%, 95% credible interval = 5.5–8.9% and 16.2%, 95% credible interval = 12.5–20.2% and at 2.5 and 3.5 days, respectively). In contrast, no discernible differences could be observed in CD4+ Treg cells (median PP = 8.6% and 9.7% at 2.5 and 3.5 days, respectively; Fig. 2D and E). Nevertheless, we note that the increased rate of spontaneous cell death in more activated CD4+ T conv cells did not cause noticeable differences in the frequency of spontaneous cell death between the two responder T cell subsets (Fig. 2D and E). These data, therefore, indicate that the reported differences in the frequency of specific killing of CD4+ Tconvs and Tregs by CD56br NK cells are not simply caused by differences in the rate of spontaneous cell death between the two subsets.
CD4+ Tconv and Treg cells display different proliferation dynamics in vitro
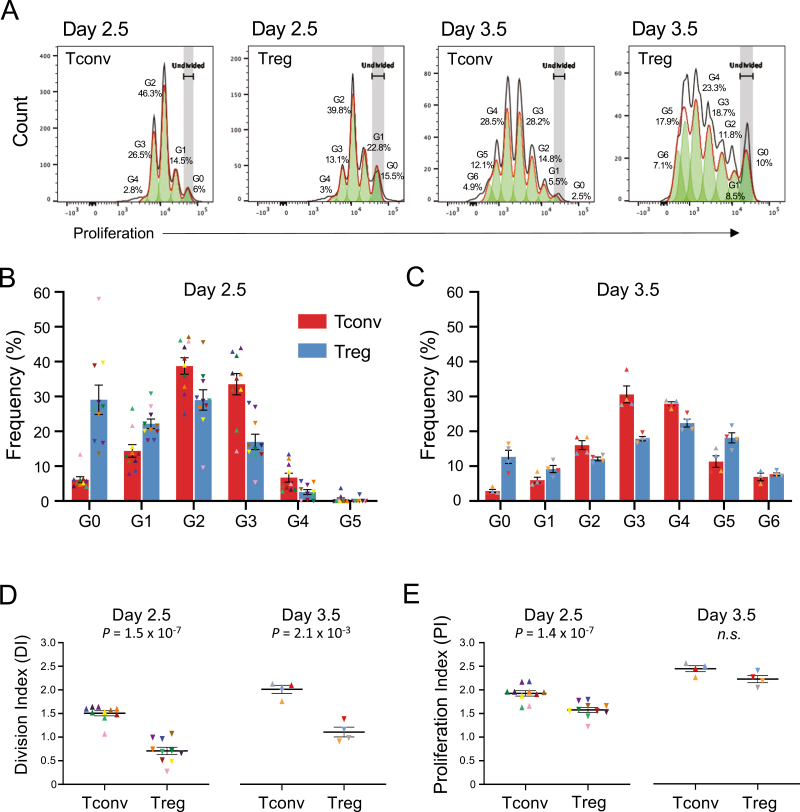
To further investigate the causes leading to the slight preferential killing of CD4+ Tconvs by CD56br NK cells, we next investigated whether intrinsic differences in the response of the two CD4+ T cell subsets to stimulation in our in vitro model could explain this result. We first assessed whether differences in the in vitro proliferation dynamics of CD4+ Tconvs and Tregs stimulated with αCD3/CD28 beads and IL-2 could contribute for this result (Fig. 3A). We note that because only the CD4+ T cell subsets were stained with the intracellular cell proliferation dye in our NK cell killing assay, the combination of the proliferation dye and CD4 antibody staining allowed a very clear distinction of the CD4+ T and the CD56+ NK cell subsets following co-culture. This stratification of the responder T cell subsets allowed us to compare the dynamics of response to αCD3/CD28 stimulation in both CD4+ Tconv and Treg subsets. One notable difference between the T cell subsets was associated with the timing of initial response to stimulation, and entry into cell cycle. CD4+ Tconvs showed a more rapid response to stimulation, as illustrated by the substantial increased proportion of cells having divided at least once by day 2.5 (94% and 71% in CD4+ Tconvs and Tregs, respectively; Fig. 3B). In contrast, one day later (day 3.5 post-stimulation), this difference was much less pronounced, with a mean of 97% and 87% of CD4+ Tconvs and Tregs, respectively, having divided at least once (Fig. 3C). Another notable difference between CD4+ Tconv and Treg proliferation dynamics was the range of proliferation cycles achieved by both subsets. At day 2.5 post-stimulation only 0.51% and 0.19% of CD4+ Tconvs and Tregs, respectively, had already achieved the fifth proliferation cycle (G5; Fig. 3B). However, by day 3.5 we observed a striking difference in the frequency of cells that had undergone five or more cell cycles, with CD4+ Tregs showing a substantial increased proportion of highly proliferative cells compared to Tconvs (25.81% and 16.52%, respectively; Fig. 3C).
Figure 3:
Assessing the proliferative capacity of CD4+ T cells in vitro. (A) Representative histograms depicting the proliferative capacity of CD4+ Tconv and Treg cells after 2.5 or 3.5 days of in vitro culture in the presence of 100 IU/ml IL-2 and TCR stimulation with αCD3/CD28 beads (1 bead: 3 T cells). The respective cell generation (Gn) are shown in each histogram. G0 represents undivided CD4+ T cells. Data were generated from freshly isolated PBMCs from 14 healthy donors (n = 10 and n = 4 for day 2.5 and day 3.5, respectively). (B) Histograms depict the frequency (mean ± SEM) of CD4+ Tconv (red) and Treg (blue) cells at each generation of cell division after 2.5 (B) and 3.5 (C) days of stimulation with αCD3/CD28 beads and 100 IU/ml IL-2, followed by an additional 4 h of co-culture in the presence or absence of the respective CD56+ NK cell subset. The reported frequencies of proliferating CD4+ Tconvs and Tregs for each donor at each generation represent the average of the three co-culture conditions assessed in this study: (i) co-culture with CD56br NK cells; (ii) co-culture with CD56dim NK cells; and (iii) cultured alone in the absence of NK cells (background control to assess spontaneous cell death). The T cell proliferation rates were not affected by the different co-culture conditions, and were therefore averaged to increase the number of T cells assessed. CD4+ Tconvs (depicted by triangles) and Tregs (depicted by inverted triangles) isolated from the same donor are depicted using the same colour. (D) Scatter plots depict the distribution (mean ± SEM) of the observed DI for CD4+ Tconv and Treg cells after co-culture at 2.5 (left panel) or 3.5 (right panel) days of stimulation. (E) Same as for D, but depicting the PI of the cells. P values were calculated using two-tailed paired t-tests. DI denotes the average number of cell divisions undergone by a cell in the original population and PI denotes the average number of divisions among the cells that have divided at least once. n.s., P value not significant.
To further understand the differences in the proliferation dynamics of the CD4+ Tconv and Treg subsets, we then modelled their proliferation rate using two different methods, namely the DI (average number of cell divisions undergone by a cell in the original population) and the PI (average number of divisions among the cells that have divided at least once). At both days 2.5 and 3.5 post-stimulation, we observed a consistently higher DI in CD4+ Tconvs compared to Tregs (P = 1.5 × 10−7 and P = 2.1 × 10−3, respectively; Fig. 3D), indicating that on average more CD4+ Tconvs have divided at least once compared to their respective Treg counterparts. In contrast to the DI, while CD4+ Tconvs also displayed an increased PI compared to Tregs at day 2.5 (P = 1.4 × 10−7), this difference was nullified at day 3.5 (Fig. 3E), indicating that although less Treg cells are actively proliferating, on average the proliferating fraction has undergone more cell cycles than the proliferating Tconv counterparts. Taken together these results support different dynamics of response to αCD3/CD28 stimulation in vitro by CD4+ Tconvs and Tregs, with Tconvs showing increased sensitivity to initial stimulation, but displaying a more limited potential to proliferate compared to Tregs.
CD56br NK cells preferentially target highly proliferative CD4+ T cells
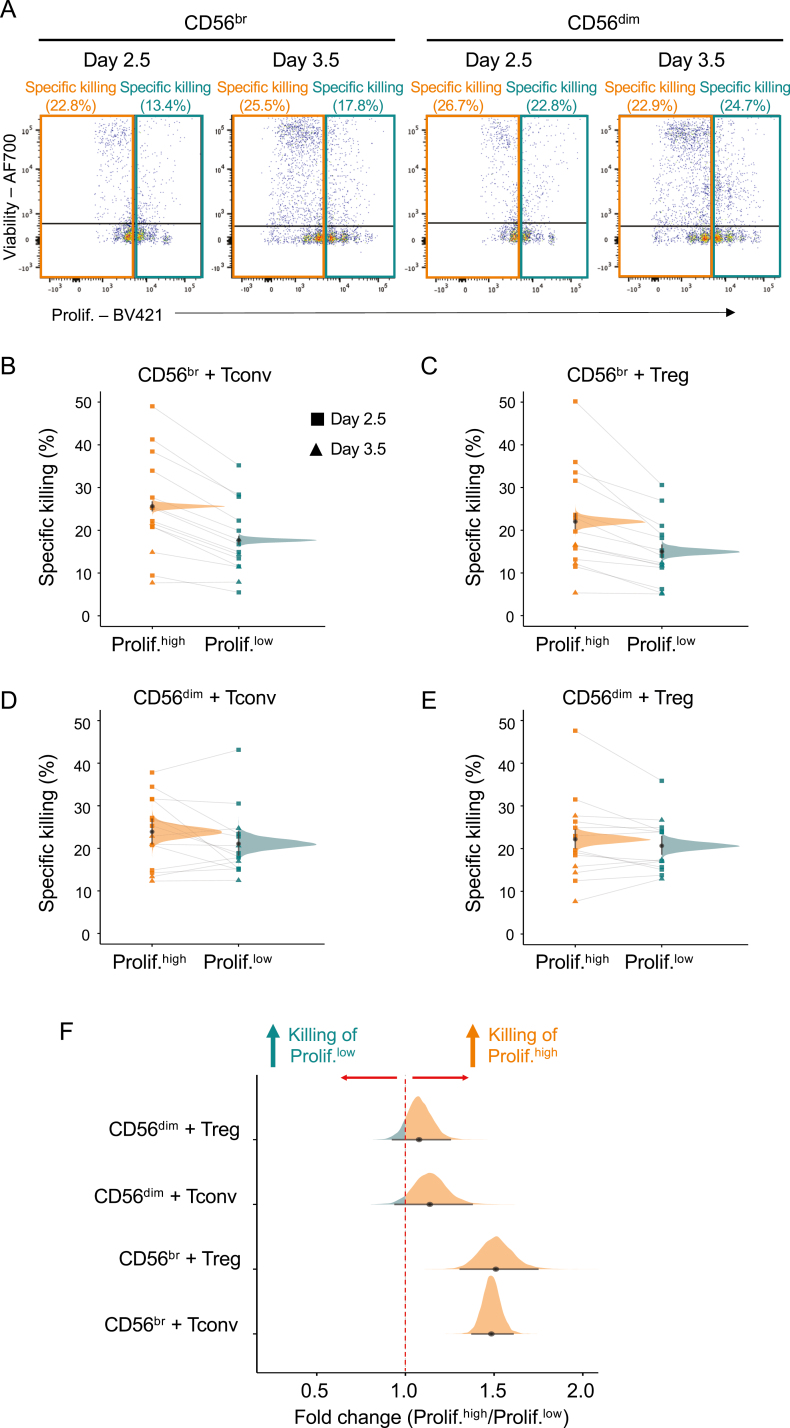
In many autoimmune diseases, CD4+ T cells have been implicated in mediating aspects of inflammation. Therefore, the body must regulate the number of CD4+ T cells to maintain tolerance. Given the high sensitivity of our assay to specifically interrogate T cell proliferation state, we next investigated whether CD56br NK cells and/or CD56dim NK cells could be differentially targeting T cells according to their proliferative state. Given the observed dynamics of T cell proliferation under our assay conditions, we have defined in this study highly proliferative CD4+ Tconvs and Tregs as cells having undergone two or more proliferation cycles at day 2.5 and more than three proliferative cycles at day 3.5 (Fig. 4A). We then assessed the relative frequency of dead responder CD4+ T cells in each respective proliferative group. We found that CD56br NK cells differentially killed target T cells according to their proliferative state.
Figure 4:
CD56br NK cells preferentially kill highly proliferative CD4+ T cell subsets. (A) Representative dot plots depicting the gating strategy used to define the frequency of T cell death among highly (prolif.high; orange) and lowly (prolif.low; teal) proliferative CD4+ T cells co-cultured with CD56br (left panels) and CD56dim (right panels)NK cells. The relative frequency of cell death was calculated in each proliferative group and takes the background levels of spontaneous CD4+ T cell death (CD4+ T cells co-cultured without NK cells) into account (see Methods). Data were generated from freshly isolated PBMCs from 14 healthy donors for day 2.5 (n = 10; squares) and day 3.5 (n = 4; triangles), respectively. (B and C) Scatter plots depict the relative frequency of cell death in CD4+ Tconv (B) and Treg (C) cells from the prolif.high and prolif.low proliferation groups after co-culture with CD56br NK cells. (D and E) Same as B and C but for CD4+ Tconvs (D) and Tregs (E) co-cultured with CD56dim NK cells. Densities depict the mean posterior frequency of cell death values in each group estimated by Beta regression, with an annotation for the median and a 95% credible interval. (F) Densities depict the effect size distribution (measured as fold change) for the observed relative rates of specific killing in highly and lowly proliferative for each of the CD4+ responder T cell subsets by either CD56br (top panels) or CD56dim (bottom panels) NK cells.
Both CD56br and CD56dim NK cell subsets displayed similar frequencies of specific killing of the autologous CD4+ T cell subsets irrespective of their proliferative state (Fig. 4B–E). Notably, highly proliferative CD4+ Tconv and Tregs were preferentially killed by CD56br NK cells (median PP = 25.6%, 95% credible interval = 24.6–26.6% and 22.0%, 95%credible interval = 20.5–23.7%, respectively) compared to their respective less proliferative counterparts (median = 17.7%, 95%credible interval 16.8–18.6% and 15.0%, 95% credible interval 13.6–16.4%, respectively; Fig. 4B and C). These differences corresponded to a median fold-change increase of 1.49 and 1.51 (FSR < 0.001%) in the frequency of specific killing of highly proliferative CD4+ Tconv and Treg cells, respectively (Fig. 4F). We note that the increased preference of CD56br NK cells to kill proliferative autologous T cells can also explain the observed small increased relative killing of CD4+ Tconvs compared to Tregs (Fig. 2B), which could be attributed to the increased proliferative state of Tconvs compared to Tregs. In contrast to CD56br NK cells, CD56dim NK cells showed no preferential killing of either responder T cell subset according to their respective proliferative state (median 0.96 and 0.94 fold change, respectively, and FSR > 5% in both cases; Fig. 4E and F), suggesting that the observed preferential killing of highly proliferative T cells in our in vitro model is a specific feature of CD56br NK cells.
Discussion
The characterization of NK cell biology has been hampered by technical limitations with in vitro assay development, ranging from limited cell numbers in peripheral blood to the variability in assay conditions (e.g. the combination of cytokines used in each specific assay). In this study, through the development of a NK killing assay, we demonstrated the specific consequences of these limitations and the necessity to distinguish between the CD56br and CD56dim NK cell subsets. In agreement with their terminally differentiated phenotype [13], CD56dim NK cells display very limited proliferative capacity in vitro when stimulated with IL-2 only. This contrasts with the CD56br NK subset that expands in vitro in the presence of IL-2 stimulation. These differences underscore the importance of stratifying the CD56br and CD56dim NK cell subsets in blood to understand the specific contribution of each subset. The distinct proliferative capacities of these two NK cell subsets can lead to erroneous conclusions when culturing bulk NK cell populations, due to the strong preferential survival of the CD56br NK cell subset in vitro, which can easily become predominant after a few days in culture. Further to the specific changes in the NK cell subsets, our data also demonstrate the relevance of limiting the functional heterogeneity of the target cell population to reduce the variability of in vitro NK co-culture assays. In this study we show that CD4+ Tconvs and Tregs also display distinct dynamics of response to αCD3/CD28 stimulation, leading to the induction of different proliferative rates. These differences highlight the need to carefully normalize assay conditions to limit the variability associated with changes in the cell’s proliferative state. These considerations are particularly relevant when directly comparing CD4+ Tconvs and Tregs using in vitro functional assays, as they can lead to changes in the assay readout due to specific differences in proliferation rates.
Recently, we have characterized the changes in CD56br NK cells in patients treated with LD-IL-2 using a single-cell multiomics approach [19]. Notably, we observed increased proliferative capacity and activated phenotype in CD56br NK cells after a month-long treatment period with a 3-day dosing interval schedule. The finding that CD56br NK cells kill proliferating Tconv and Tregs similarly could entail the detrimental preferential killing of the expanded Treg subset in patients treated with LD-IL-2. However, in the context of LD-IL-2 immunotherapy, the expanded Tregs in blood are unlikely to be acutely activated, and therefore not susceptible to killing by CD56br NK cells. A more plausible mechanism is that CD56br NK cells activated in response to LD-IL-2 treatment rapidly migrate to tissues where they could exert an immunoregulatory function in the elimination of chronically activated T cells, thereby providing a rationale for the clinical benefit observed in patients undergoing LD-IL-2 immunotherapy [26–30]. This could be relevant not only in the context of autoimmune diseases, by targeting autoreactive T cells expanded at the sites of inflammation, but also in the context of potentially detrimental anti-tumour responses, by eliminating hyperproliferative effector T cells. We also note that although we did not observe a preferential killing of proliferating T cells within the broader CD56dim NK subset, the very poor proliferative potential and survival of these cells in our in vitro assay conditions may significantly reduce our power to identify these changes. One consistent observation stemming from LD-IL-2 immunotherapy studies [16–19], is that even very low amounts of IL-2 can significantly expand the proportion and numbers of CD56br NK cells, but not CD56dim NK cells, in blood. The original observation in the non-obese diabetic (NOD) mouse model of autoimmune diabetes that activated NK cells infiltrating the pancreatic islets were involved in the pathogenesis of the disease [31, 32] raised questions over their putative detrimental role in the human disease. These findings led pharmaceutical companies to explore the development of IL-2 mutein molecules characterized by a reduced capacity to elicit NK cell proliferation [33, 34]. However, the recent promise of LD-IL-2 trials in several autoimmune and inflammatory diseases [20, 21] demonstrates the need to better understand the function of the IL-2-expanded CD56br NK cells and their specific contribution to the drug’s mechanism of action. Given the technical limitations with functional in vitro NK killing assays, it is critical to develop better tools to investigate these cells in clinical samples from completed and ongoing trials.
Our current study presents several limitations to consider. The first limitation is that we did not define the exact molecular mechanism underlying the preferential killing of proliferating T cells by CD56br NK cells. It is likely that some of the mechanisms used by CD56br NK cells to kill activated T cells [25] are acting preferentially on the proliferating cells. The activation of T cells in vitro with αCD3/CD28 stimulation induces a vast number of changes to the transcriptional profile of T cells, and therefore there are a variety of targets that may affect the dynamics of NK:T cell interaction and mediate the preferential targeting of proliferative cells. Furthermore, it is also plausible that αCD3/CD28 stimulation induces the upregulation of pro-apoptotic responses to limit overt T cell activation and effector T cell expansion [35], thereby leading to increased susceptibility to NK cell killing. Further work is therefore warranted to investigate the mechanism of NK-cell mediated killing of proliferative T cells and how it could potentially be modulated in vivo. Another limitation of our study was the relatively small change of ~1.5-fold increased killing efficiency in proliferating T cells. It is difficult to estimate the exact physiological impact of these observed changes in our in vitro assay, and how they could translate to a much more complex system in vivo. Relatively small changes (<2-fold) have been previously shown to lead to robust and quantifiable phenotypic alterations. For example, we have previously shown that a genetic variant in the IL2 locus provided only a 2-fold increased expression of IL-2 in the NOD mouse model, which was associated with a robust 80% increased survival [36]. It is notable that in organisms with long life span, such modest effects can have significant cumulative effects over time, thereby contributing to much greater biological implications. This is particularly relevant in a setting such as life-long NK cell-mediated protection from potentially pathogenic proliferative T effector cells. Finally, a third limitation is the high cell number requirement for the in vitro NK killing assay described in this study. This requirement is particularly relevant in the context of clinical studies, which, in its current format, may preclude the application of the assay to limited cryopreserved clinical samples.
Our observation that CD56br NK cells kill proliferating T cells more efficiently lends support to a putative regulatory function of this subset in vivo by eliminating proliferative T cells in the context of an inflammatory response in the tissues. This may be a key regulatory mechanism to control overt T effector cell proliferation and activation in response to local IL-2 production at the sites of inflammation and is consistent with the higher proportion of CD56br NK cells in tissues compared to circulation [37]. A regulatory role for CD56br NK cells has been previously suggested in the context of multiple sclerosis patients treated with anti-CD25 (daclizumab) [38, 39]. Response to treatment in these patients was associated with a significant expansion of CD56br NK cells and concomitant decline in the frequency of both CD4+ and CD8+ T cells in blood, thereby supporting a regulatory role of CD56br NK cells via NK cell-mediated negative immunoregulation of activated T cells. Furthermore, CD56br NK cells have previously been shown to be able to recognize and kill early activated T cells [6], which together with the observations that CD56br NK cells kill tumour cells [40] and increased proportions of CD56br NK cells in blood are associated with better prognosis in several types of tumours [40–42], warrants further work to understand how they may contribute to the regulation of anti-tumour responses. Taken together, these findings support a physiological regulatory role of CD56br NK cells in an inflammatory context. Our results raise the possibility that CD56br NK cell expansion by low-dose IL-2 is a positive feature and could contribute to the previous reports of clinical efficacy in diverse inflammatory conditions [26–30].
Acknowledgements
We thank the volunteers from the Oxford Biobank (www.oxfordbiobank.org.uk) for their participation in this study. We also thank Heather McMurray, Shannah Donhou and Sarune Kacinskaite, University of Oxford, for sample collection and preparation and members of the Diabetes and Inflammation Laboratory for critical discussion. The Editor-in-Chief, Simon Milling and handling editor, Meera Ramanuiam, would like to thank the reviewers for their contribution to the publication of this article.‘
Glossary
Abbreviations:
- BV
Brilliant Violet™
- BUV
Brilliant™ Ultraviolet
- CD56br
CD56bright natural killer cell
- CI
confidence interval
- CV
coefficient of variation
- FACS
fluorescence-activated cell sorting
- IL-2
interleukin-2
- IU
international units
- LD-IL2
low-dose IL-2
- NK
natural killer
- Tconv
CD4+ conventional T cell
- Treg
CD4+ regulatory T cell
Contributor Information
Mercede Lee, JDRF/Wellcome Diabetes and Inflammation Laboratory, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford, UK.
Charles J M Bell, JDRF/Wellcome Diabetes and Inflammation Laboratory, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford, UK.
Arcadio Rubio Garcia, JDRF/Wellcome Diabetes and Inflammation Laboratory, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford, UK.
Leila Godfrey, JDRF/Wellcome Diabetes and Inflammation Laboratory, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford, UK.
Marcin Pekalski, JDRF/Wellcome Diabetes and Inflammation Laboratory, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford, UK.
Linda S Wicker, JDRF/Wellcome Diabetes and Inflammation Laboratory, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford, UK.
John A Todd, JDRF/Wellcome Diabetes and Inflammation Laboratory, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford, UK.
Ricardo C Ferreira, JDRF/Wellcome Diabetes and Inflammation Laboratory, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford, UK.
Ethical approval
All samples and information were collected with written and signed informed consent. The study conformed to the principles of the Helsinki Declaration and Good Clinical Practice and was approved by the East of England—Cambridgeshire and Hertfordshire research ethics committee (05/Q0106/20).
Conflict of Interests
J.A.T. is a member of the science advisory boards of GSK, Precion, Qlife, and Vesalius Therapeutics M.L. was supported by a grant from Bristol Myers Squibb (BMS 2338). The remaining authors declare no competing interests.
Funding
The Diabetes and Inflammation Laboratory was supported by a strategic award from the Wellcome (107212/A/15/Z) and the JDRF (4-SRA-2017-473-A-A). The research was supported by a Wellcome Trust Core Award Grant Number 203141/Z/16/Z with additional support from the NIHR Oxford BRC and a grant from Bristol Myers Squibb (BMS 2338). The Oxford BioBank and Oxford Bioresource are funded by the NIHR Oxford Biomedical Research Centre (BRC).
Data availability
The code and data used for the statistical analyses are availabe at https://github.com/arcadio/nk-models.
Author contributions
C.J.M.B., A.R.G., L.S.W., J.A.T., and R.C.F. conceptualized the work. L.S.W., J.A.T., and R.C.F. designed the experiments. M.L. performed the experiments with help from L.G., M.L.P., and R.C.F. M.L. performed the statistical analysis of the data under the supervision of A.R.G. M.L., L.S.W., J.A.T., and R.C.F. interpreted the results and wrote the manuscript with input from all authors.
References
- 1.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S.. Functions of natural killer cells. Nat Immunol 2008, 9, 503–10. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 2.Martín-Fontecha A, Thomsen Lindy L, Brett S, Gerard C, Lipp M, Lanzavecchia A.et al. Induced recruitment of NK cells to lymph nodes provides IFN-γ for TH1 priming. Nat Immunol 2004, 5, 1260–5. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 3.Mailliard RB, Alber SM, Shen H, Watkins Simon C, Kirkwood JM, Herberman RB.et al. IL-18–induced CD83+CCR7+ NK helper cells. J Exp Med 2005, 202, 941–53. doi: 10.1084/jem.20050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivier E E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM.et al. Innate or adaptive immunity? The example of natural killer cells. Science 2011, 331, 44–9. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong JL, Berk E, Edwards RP, Kalinski P.. IL-18–primed helper NK cells collaborate with dendritic cells to promote recruitment of effector CD8+ T cells to the tumor microenvironment. Cancer Res 2013, 73, 4653–62. doi: 10.1158/0008-5472.CAN-12-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pallmer K, Oxenius A.. Recognition and regulation of T cells by NK cells. Front Immunol 2016, 7, 251. doi: 10.3389/fimmu.2016.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melsen JE, Lugthart G, Lankester AC, Schilham MW.. Human circulating and tissue-resident CD56bright natural killer cell populations. Front Immunol 2016, 7, 262. doi: 10.3389/fimmu.2016.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michel T, Poli A, Cuapio A, Briquemont B, Iserentant G, Ollert M.et al. Human CD56bright NK cells: an update. J Immunol 2016, 196, 2923–31. doi: 10.4049/jimmunol.1502570. [DOI] [PubMed] [Google Scholar]
- 9.Cichocki F, Grzywacz B, Miller JS.. Human NK cell development: one road or many?. Front Immunol 2019, 10, 2078. doi: 10.3389/fimmu.2019.02078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crinier A, Milpied P, Escalière B, Piperoglou C, Galluso J, Balsamo A.et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity 2018, 49, 971–986.e5. doi: 10.1016/j.immuni.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins PL, Cella M, Porter Sofia I, Li S, Gurewitz Greer L, Hong HS.et al. Gene regulatory programs conferring phenotypic identities to human NK cells. Cell 2019, 176, 348–360.e12. doi: 10.1016/j.cell.2018.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yudanin NA, Schmitz F, Flamar A-L, Thome JJC, Tait Wojno E, Moeller JB.et al. Spatial and temporal mapping of human innate lymphoid cells reveals elements of tissue specificity. Immunity 2019, 50, 505–519.e4. doi: 10.1016/j.immuni.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C, Siebert JR, Burns R, Gerbec ZJ, Bonacci B, Rymaszewski A.et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun 2019, 10, 3931. doi: 10.1038/s41467-019-11947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dogra P, Rancan C, Ma W, Toth M, Senda T, Carpenter DJ.et al. Tissue determinants of human NK cell development, function, and residence. Cell 2020, 180, 749–763.e13. doi: 10.1016/j.cell.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brownlie D D, Scharenberg M, Mold JE, Hard J, Kekalainen E, Buggert M, Nguyen Set al. Expansions of adaptive-like NK cells with a tissue-resident phenotype in human lung and blood. Proc Natl Acad Sci USA 2021, 118, e2016580118. doi: 10.1073/pnas.2016580118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, Bollard CM, Carlsten M, Melenhorst JJ, Biancotto A, Wang E.et al. Ultra-low dose interleukin-2 promotes immune-modulating function of regulatory T cells and natural killer cells in healthy volunteers. Mol Ther 2014, 22, 1388–95. doi: 10.1038/mt.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenzwajg M, Churlaud G, Mallone R, Six A, Dérian N, Chaara W.et al. Low-dose interleukin-2 fosters a dose-dependent regulatory T cell tuned milieu in T1D patients. J Autoimmun 2015, 58, 48–58. doi: 10.1016/j.jaut.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todd JAJ., et al. Regulatory T cell responses in participants with type 1 diabetes after a single dose of interleukin-2: a non-randomised, open label, adaptive dose-finding trial. PLoS Med 2016, 13, e1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J-Y, Hamey F, Trzupek D, Mickunas M, Lee M, Godfrey L.et al. Low-dose IL-2 reduces IL-21+ T cell frequency and induces anti-inflammatory gene expression in type 1 diabetes. Nat Commun 2022, 13, 7324. doi: 10.1038/s41467-022-34162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klatzmann D, Abbas AK.. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat Rev Immunol 2015, 15, 283–94. doi: 10.1038/nri3823. [DOI] [PubMed] [Google Scholar]
- 21.Graßhoff H, Comdühr S, Monne LR, Müller A, Lamprecht P, Riemekasten G.et al. Low-dose IL-2 therapy in autoimmune and rheumatic diseases. Front Immunol 2021, 12, 648408. doi: 10.3389/fimmu.2021.648408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrari S, Cribari-Neto F.. Beta regression for modelling rates and proportions. J Appl Stat 2004, 31, 799–815. [Google Scholar]
- 23.Gelman A, Tuerlinckx F.. Type S error rates for classical and Bayesian single and multiple comparison procedures. Comput Stat 2000, 15, 373–90. [Google Scholar]
- 24.Carpenter B, Gelman A, Hoffman MD, Lee D, Goodrich B, Betancourt M.et al. Stan: a probabilistic programming language. J Stat Softw 2017, 76, 1-32. doi: 10.18637/jss.v076.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen N, Ødum N, Ursø B, Lanier LL, Spee P.. Cytotoxicity of CD56bright NK cells towards autologous activated CD4+ T cells is mediated through NKG2D, LFA-1 and TRAIL and dampened via CD94/NKG2A. PLoS One 2012, 7, e31959. doi: 10.1371/journal.pone.0031959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koreth J, Matsuoka K, Kim HT, McDonough SM, Bindra B, Alyea EP.et al. Interleukin-2 and regulatory T cells in graft-versus-host disease. N Engl J Med 2011, 365, 2055–66. doi: 10.1056/NEJMoa1108188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saadoun D, Rosenzwajg M, Joly F, Six A, Carrat F, Thibault V.et al. Regulatory T-cell responses to low-dose interleukin-2 in HCV-induced vasculitis. N Engl J Med 2011, 365, 2067–77. doi: 10.1056/NEJMoa1105143. [DOI] [PubMed] [Google Scholar]
- 28.Castela E, Le Duff F, Butori C, Ticchioni M, Hofman P, Bahadoran P.et al. Effects of low-dose recombinant interleukin 2 to promote t-regulatory cells in alopecia areata. JAMA Dermatol 2014, 150, 748–51. doi: 10.1001/jamadermatol.2014.504. [DOI] [PubMed] [Google Scholar]
- 29.Humrich JY, von Spee-Mayer C, Siegert E, Bertolo M, Rose A, Abdirama D.et al. Low-dose interleukin-2 therapy in refractory systemic lupus erythematosus: an investigator-initiated, single-centre phase 1 and 2a clinical trial. Lancet Rheumatol 2019, 1, e44–54. doi: 10.1016/s2665-9913(19)30018-9. [DOI] [PubMed] [Google Scholar]
- 30.He J, Zhang R, Shao M, Zhao X, Miao M, Chen J.et al. Efficacy and safety of low-dose IL-2 in the treatment of systemic lupus erythematosus: a randomised, double-blind, placebo-controlled trial. Ann Rheum Dis 2020, 79, 141–9. doi: 10.1136/annrheumdis-2019-215396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurent P, Christophe B, Diane M.. Natural killer cells distinguish innocuous and destructive forms of pancreatic islet autoimmunity. Proc Natl Acad Sci USA 2004, 101, 8102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sitrin J, Ring A, Garcia KC, Benoist C, Mathis D.. Regulatory T cells control NK cells in an insulitic lesion by depriving them of IL-2. J Exp Med 2013, 210, 1153–65. doi: 10.1084/jem.20122248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson LB, Bell CJM, Howlett SK, Pekalski ML, Brady K, Hinton H.et al. A long-lived IL-2 mutein that selectively activates and expands regulatory T cells as a therapy for autoimmune disease. J Autoimmun 2018, 95, 1–14. doi: 10.1016/j.jaut.2018.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghelani A, Bates D, Conner K, Wu M-Z, Lu J, Hu Y-L.et al. Defining the threshold IL-2 signal required for induction of selective treg cell responses using engineered IL-2 muteins. Front Immunol 2020, 11, 1106. doi: 10.3389/fimmu.2020.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhan Y, Carrington EM, Zhang Y, Heinzel S, Lew AM.. Life and death of activated t cells: how are they different from Naïve T cells?. Front Immunol 2017, 8, 1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VES.et al. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet 2007, 39, 329–37. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fehniger TA, Cooper MA, Nuovo GJ, Cella M, Facchetti F, Colonna M.et al. CD56bright natural killer cells are present in human lymph nodes and are activated by Tcell–derived IL-2: a potential new link between adaptive and innate immunity. Blood 2003, 101, 3052–7. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 38.Bielekova B B, Catalfamo M, Reichert-Scrivner S, Packer A, Cerna M, Waldmann TA, McFarland H.et al. Regulatory CD56bright natural killer cells mediate immunomodulatory effects of IL-2Rα-targeted therapy (daclizumab) in multiple sclerosis. Proc Natl Acad Sci USA 2006, 103, 5941–6. doi: 10.1073/pnas.0601335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranganath T, Simpson LJ, Ferreira A-M, Seiler C, Vendrame E, Zhao N.et al. Characterization of the impact of daclizumab beta on circulating natural killer cells by mass cytometry. Front Immunol 2020, 11, 714. doi: 10.3389/fimmu.2020.00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner JA, Rosario M, Romee R, Berrien-Elliott MM, Schneider SE, Leong JW.et al. CD56bright NK cells exhibit potent antitumor responses following IL-15 priming. J Clin Invest 2017, 127, 4042–58. doi: 10.1172/JCI90387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrega P, Bonaccorsi I, Di Carlo E, Morandi B, Paul P, Rizzello V.et al. CD56 (bright) perforin (low) noncytotoxic human NK cells are abundant in both healthy and neoplastic solid tissues and recirculate to secondary lymphoid organs via afferent lymph. J Immunol 2014, 192, 3805–15. doi: 10.4049/jimmunol.1301889. [DOI] [PubMed] [Google Scholar]
- 42.Habif G, Crinier A, André P, Vivier E, Narni-Mancinelli E.. Targeting natural killer cells in solid tumors. Cell Mol Immunol 2019, 16, 415–22. doi: 10.1038/s41423-019-0224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The code and data used for the statistical analyses are availabe at https://github.com/arcadio/nk-models.