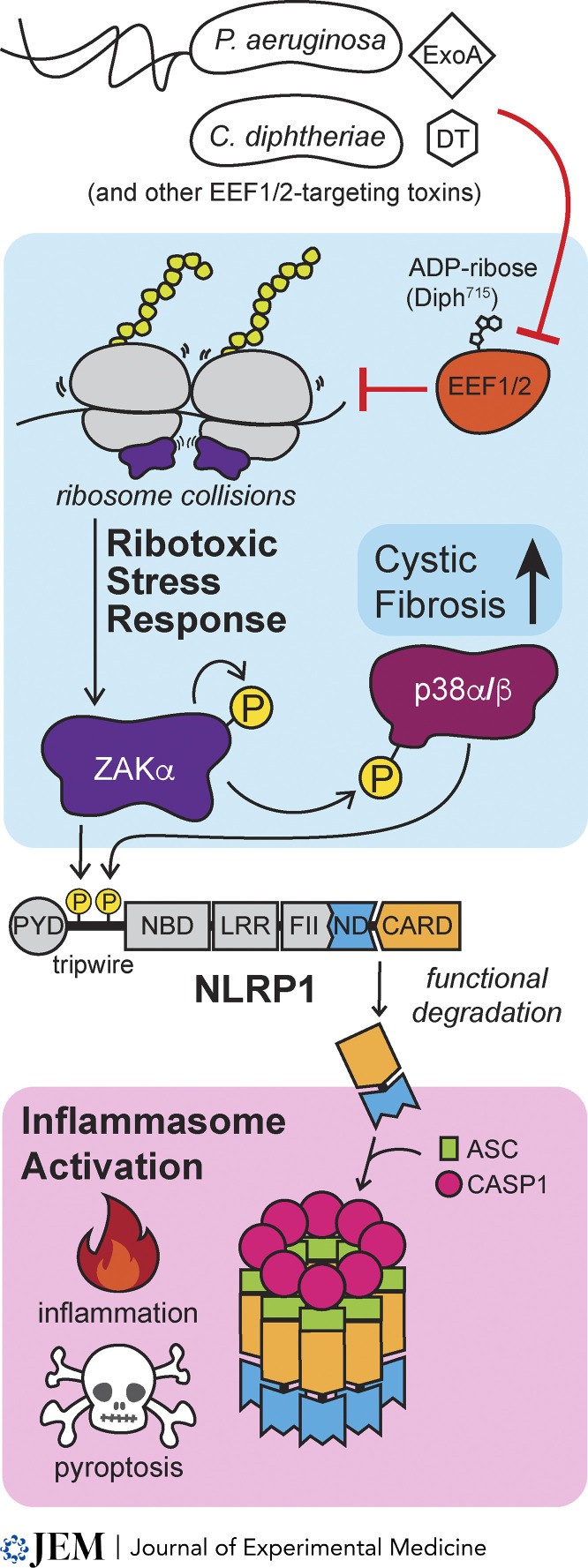
In this issue of JEM, companion articles from Pinilla et al. and Robinson et al. demonstrate that ribotoxic stress induced by P. aeruginosa and C. diphtheriae EEF2-targeting exotoxins leads to NLRP1 inflammasome activation, representing a new mechanism of effector-triggered immunity.
Abstract
In this issue of JEM, companion articles from Pinilla et al. (2023. J. Exp. Med. https://doi.org/10.1084/jem.20230104) and Robinson et al. (2023. J. Exp. Med. https://doi.org/10.1084/jem.20230105) demonstrate that ribotoxic stress induced by Pseudomonas aeruginosa and Corynebacterium diphtheriae EEF2-targeting exotoxins leads to NLRP1 inflammasome activation, representing a new mechanism of effector-triggered immunity.
The ability of our innate immune system to accurately recognize microbial threats is critical for mounting an appropriate immune response. In the complex microbial milieu of our barrier tissues, distinguishing non-harmful or commensal microbes from pathogens is particularly challenging, as both produce ligands that stimulate the innate immune system (Vance et al., 2009). However, some intracellular innate immune sensors mitigate this conundrum by recognizing the activities of pathogen virulence factors—a defense strategy known as effector-triggered immunity (ETI; Jones and Dangl, 2006; Lopes Fischer et al., 2020; Remick et al., 2023). In this issue of JEM, Pinilla et al. (2023) and Robinson et al. (2023) show that inactivation of host protein synthesis by Pseudomonas aeruginosa and Corynebacterium diphtheriae exotoxins leads to ribotoxic stress and subsequent NLRP1 inflammasome activation. These findings highlight the capacity of innate immune sensors, such as NLRP1, to detect pathogens via sensing cellular disruptions caused by virulence factor activity.

Insights from Ryan Tibble, Marisa A. Yonemitsu, and Patrick S. Mitchell.
NLRP1: A multifaceted molecular tripwire
NLRP1 is one of the predominant inflammasome-forming sensors in human epithelia and recognizes diverse pathogen-associated activities (Bauernfried and Hornung, 2022). Like other NLR inflammasome–forming sensors, NLRP1 contains an N-terminal Pyrin domain, a nucleotide-binding domain, and leucine-rich repeats. NLRP1 also has an atypical, C-terminal function-to-find domain and caspase activation and recruitment domain (CARD). The function-to-find domain undergoes limited autoproteolysis, resulting in two noncovalently associated fragments. NLRP1’s N-terminal fragment functions like a molecular tripwire to detect virulence factor activities, harboring substrate mimics of pathogen-encoded proteases and E3 ubiquitin ligases. Molecular recognition events that trip the tripwire result in the “functional degradation” of the N-terminal fragment. The liberated C terminus recruits the adaptor protein apoptosis-associated speck-like protein containing a CARD (ASC) to activate pro-inflammatory caspases, including caspase-1 (CASP1). CASP1 then processes IL-1β and IL-18 cytokines and the pore-forming protein gasdermin D (GSDMD) into their active forms, leading to inflammation and pyroptotic cell death.
In addition to directly sensing pathogen effectors, NLRP1 can also indirectly sense pathogen- and stress-induced perturbations to host proteostasis caused by UVB irradiation and small molecule toxins (Jenster et al., 2022; Robinson et al., 2022). These NLRP1-activating stimuli converge in their induction of the ribotoxic stress response (RSR). Ribosome collisions (but not other forms of translational inhibition) trigger the RSR upon autophosphorylation of the mitogen-activated protein kinase kinase kinase ZAKα (Vind et al., 2020; Wu et al., 2020). Activated ZAKα initiates a signaling cascade that includes p38α and p38β (p38α/β) kinases, and p38α/β and ZAKα phosphorylation of the NLRP1 tripwire induces inflammasome activation by an unclear mechanism. Thus, NLRP1 is positioned to detect translational blockade, a common feature of pathogenic infections.
Here, reports by Pinilla et al. (2023) and Robinson et al. (2023) extend this mode of ETI to the detection of RSR-triggering bacterial pathogen exotoxins. Pinilla et al. (2023) found that P. aeruginosa infection of primary human nasal and corneal epithelial cells caused elevated cell lysis and IL-1β/IL-18 cytokine stimulation. Similarly, Robinson et al. (2023) observed a rapid onset of pyroptotic cell death and IL-1β secretion following C. diphtheriae infection of primary human keratinocytes. Both phenotypes were dependent on NLRP1, leading the authors to next determine how NLRP1 detects these diverse bacterial pathogens.
P. aeruginosa and C. diphtheriae secrete exotoxin A (ExoA) and diphtheria toxin (DT), respectively, to inhibit host protein synthesis. ExoA and DT accomplish this by ADP-ribosylating a diphthamide modification in eukaryotic translation elongation factor 2 (EEF2). Accordingly, recombinant ExoA and DT were both sufficient to induce NLRP1 inflammasome activation. Mutation of the catalytic residues of ExoA and DT or knockout of the host gene DPTH1, which is critical for generating diphthamide in EEF2, prevented NLRP1 inflammasome activation as evidenced by the loss of ASC speck formation and propidium iodide staining in ExoA- and DT-treated cells. The authors further demonstrated that NLRP1 activation by ExoA and DT goes through the RSR because NLRP1 activation was abolished upon genetic and chemical ablation of ZAKα, and a NLRP1 variant lacking ZAKα/p38 phosphosites in its N-terminal tripwire failed to mount a response to ExoA and DT. Thus, ZAKα/p38 phosphorylation of NLRP1 integrates innate immune recognition into the RSR, permitting ETI by NLRP1 inflammasome activation upon RSR sensing of EEF2-targeting exotoxins from P. aeruginosa and C. diphtheriae (see figure).
NLRP1 recognizes P. aeruginosa and C. diphtheriae through the RSR. DT and ExoA exotoxins ADP-ribosylate eukaryotic translation elongation factor 2 (EEF2) to inhibit protein synthesis and induce ribosome collisions that activate the MAP kinase kinase kinase ZAKα, initiating a signaling cascade culminating in p38α and p38β phosphorylation. Both ZAKα and p38α/β phosphorylate NLRP1’s N-terminal “tripwire,” leading to its functional degradation. The bioactive C terminus of NLRP1 complexes with ASC and CASP1 to form an active inflammasome and drive subsequent inflammation and pyroptotic cell death.
Excitingly, several other bacterial virulence factors are known to cause ribotoxic stress, including Vibrio cholerae Cholix toxin, which also ADP-ribosylates EEF2, and Shigella dysenteriae Shiga toxins that directly inactivate the ribosome. Indeed, disruption of host translation is a common tactic employed by a wide range of pathogens to impede host immunity and enhance their replicative fitness, raising the possibility that the integration of NLRP1 into the RSR, at least in some vertebrate lineages, was an evolutionary adaptation in response to pathogen-induced translational blockade.
NLRP1, ETI, and potential links to bacterial pathogenesis
C. diphtheriae infections induce a DT-mediated disease of respiratory and cutaneous tissues known as diphtheria, which results in extensive necrosis of affected tissues stemming from inhibition of host protein synthesis and cell death. Using a 3D organotypic skin model of cutaneous diphtheria, Robinson et al. (2023) found that both DT and bacterial infection induce RSR and epidermal damage characteristic of diphtheria. Treatment with ZAKα and p38α/β inhibitors abrogated the extent of epidermal damage by DT and C. diphtheriae infection, implicating RSR and subsequent NLRP1 inflammasome activation as a driver of C. diphtheriae pathogenesis. However, CASP1 inhibition nominally impacted the extent of epidermal damage, suggesting that other caspases may contribute to NLRP1-dependent inflammation. Alternatively, ZAKα also mediates inflammasome-independent cell death pathways that contribute to tissue damage, and the relative impact of these responses versus pro-inflammatory cytokines and pyroptosis in the innate response to C. diphtheriae and diphtheria pathogenesis remains unclear.
P. aeruginosa is an opportunistic pathogen that can cause chronic, life-threatening infections, and is the primary bacterial pathogen associated with cystic fibrosis (CF). Pathogenic mutations in the CF transmembrane conductance regulator (CFTR) gene reduce CFTR expression, stability, and/or function, leading to defects in ion and fluid transport, aberrant mucus accumulation, and other dysfunctions that underlie CF pathogenesis. CFTR deficiency also induces p38α/β kinase activity (Bérubé et al., 2010). Pinilla et al. (2023) found that epithelial cells from people living with CF (PwCF) underwent a more rapid and robust cell death in response to ExoA treatment compared to healthy donor cells. In contrast, RSR-independent activation of NLRP1 did not cause heightened inflammasome responses in PwCF cells, implying that CFTR dysfunction specifically lowers the threshold for or otherwise licenses RSR-mediated NLRP1 inflammasome activation. Indeed, CFTR corrector therapy (i.e., TRIKAFTA) and chemical inhibition of ZAKα alleviated the hypersensitivity of NLRP1 to ExoA in PwCF donor epithelia. Although several aspects of the molecular basis linking CFTR, p38, and NLRP1 remain unknown, these findings support a model in which the increased sensitivity to RSR-triggered NLRP1 inflammasome activation contributes to CF pathogenesis.
Despite these exciting advances, the impact of NLRP1 inflammasome activation on bacterial burden, host immunity, and bacterial pathogenesis remains an open question at the organismal level, a particularly challenging pursuit given that the RSR does not activate murine orthologs of NLRP1.
What’s next?
Together, the reports from Pinilla et al. (2023) and Robinson et al. (2023) demonstrate that EEF1/2-targeting exotoxins are indirectly sensed by the NLRP1 inflammasome via the RSR. This work follows the prior observation that RSR-mediated NLRP1 inflammasome activation is triggered by environmental stimuli (i.e., UVB), raising a “chicken or egg” conundrum regarding the evolutionary origins of such responses. It remains unclear how conserved the integration of NLRP1 into the RSR is across vertebrates, with our current knowledge restricted to humans and mice. However, recurrent loss of NLRP1 across vertebrates suggests that it is not essential for the maintenance of protein homeostasis (Tsu et al., 2021). Moreover, RSR responses proceed in the absence of NLRP1 (Robinson et al., 2023). Instead, we favor a model in which the RSR-p38-NLRP1 ETI circuit evolved as an adaption to pathogen-driven evolution, consistent with NLRP1’s history of positive selection in primates (Chavarria-Smith et al., 2016).
Many outstanding questions remain regarding the molecular mechanism by which the RSR activates the NLRP1 inflammasome. RSR-induced activation of NLRP1 requires ZAKα and p38α/β kinase activity (Jenster et al., 2022; Robinson et al., 2022). However, while elevated p38α/β activation sensitizes NLRP1 to RSR agonists, p38α/β activation alone is insufficient to promote downstream NLRP1 signaling. How then does elevated p38α/β activation sensitize NLRP1 to ribotoxic stress?
It is possible that ZAKα and p38α/β phosphorylation of NLRP1 are entirely independent, and that cell type or other context-specific determinants are required for NLRP1 to respond to the RSR and/or other p38-dependent activating signals (Jenster et al., 2022; Robinson et al., 2022). Another possibility is that ZAKα and p38α/β phosphorylation of NLRP1 must occur in a stepwise manner, or that ZAKα or a downstream RSR component is required to license NLRP1 for activation by p38α/β phosphorylation of NLRP1. Given the involvement of p38α/β in several RSR-independent signaling pathways and the negative consequences of aberrant NLRP1 inflammasome activation, a ZAKα licensing model would ensure that multiple regulatory thresholds are surpassed prior to NLRP1 inflammasome activation. How such modifications initiate NLRP1 functional degradation and subsequent inflammasome activation also remains a mystery. Does phosphorylation displace a chaperone or other negative regulator and/or recruit, either directly or indirectly, an E3 ubiquitin ligase complex? A unifying model for p38-dependent NLRP1 inflammasome activation is also complicated by recent reports that double-stranded RNA and double-stranded DNA can activate NLRP1 in a p38-dependent manner (Jenster et al., 2022; Zhou et al., 2023). Nevertheless, untangling how RSR activates NLRP1 will illuminate principles of ETI that are likely to impact host immunity and pathogenesis.
References
- Bauernfried, S., and Hornung V.. 2022. J. Exp. Med. 10.1084/jem.20211405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérubé, J., et al. 2010. J. Biol. Chem. 10.1074/jbc.M109.098566 [DOI] [Google Scholar]
- Chavarria-Smith, J., et al. 2016. PLoS path. 10.1371/journal.ppat.1006052 [DOI] [Google Scholar]
- Jenster, L.-M., et al. 2023. J. Exp. Med. 10.1084/jem.20220837 [DOI] [Google Scholar]
- Jones, J.D.G., and Dangl J.L.. 2006. Nature. 10.1038/nature05286 [DOI] [Google Scholar]
- Lopes Fischer, N., et al. 2020. Nat. Microbiol. 10.1038/s41564-019-0623-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla, M., et al. 2023. J. Exp. Med. 10.1084/jem.20230104 [DOI] [Google Scholar]
- Remick, B.C., et al. 2023. Annu. Rev. Immunol. 10.1146/annurev-immunol-101721-031732 [DOI] [PubMed] [Google Scholar]
- Robinson, K.S., et al. 2022. Science. 10.1126/science.abl6324 [DOI] [Google Scholar]
- Robinson, K.S., et al. 2023. J. Exp. Med. 10.1084/jem.20230105 [DOI] [Google Scholar]
- Tsu, B.V., et al. 2021. Elife. 10.7554/eLife.60609 [DOI] [Google Scholar]
- Vance, R.E., et al. 2009. Cell Host Microbe. 10.1016/j.chom.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vind, A.C., et al. 2020. Mol. Cell. 10.1016/j.molcel.2020.03.021 [DOI] [Google Scholar]
- Wu, C.C.C., et al. 2020. Cell. 10.1016/j.cell.2020.06.006 [DOI] [Google Scholar]
- Zhou, J.Y., et al. 2023. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.2213777120 [DOI] [Google Scholar]