Abstract
Major depressive disorder (MDD) is a chronic, generally episodic and debilitating disease that affects an estimated 300 million people worldwide, but its pathogenesis is poorly understood. The heritability estimate of MDD is 30–40%, suggesting that genetics alone do not account for most of the risk of major depression. Another factor known to associate with MDD involves environmental stressors such as childhood adversity and recent life stress. Recent studies have emerged to show that the biological impact of environmental factors in MDD and other stress-related disorders is mediated by a variety of epigenetic modifications. These epigenetic modification alterations contribute to abnormal neuroendocrine responses, neuroplasticity impairment, neurotransmission and neuroglia dysfunction, which are involved in the pathophysiology of MDD. Furthermore, epigenetic marks have been associated with the diagnosis and treatment of MDD. The evaluation of epigenetic modifications holds promise for further understanding of the heterogeneous etiology and complex phenotypes of MDD, and may identify new therapeutic targets. Here, we review preclinical and clinical epigenetic findings, including DNA methylation, histone modification, noncoding RNA, RNA modification, and chromatin remodeling factor in MDD. In addition, we elaborate on the contribution of these epigenetic mechanisms to the pathological trait variability in depression and discuss how such mechanisms can be exploited for therapeutic purposes.
Subject terms: Epigenetics in the nervous system, Epigenetics
Introduction
Major depressive disorder (MDD) affects an estimated 300 million people worldwide.1 The condition is characterized by episodes of low mood, anhedonia or loss of interest, feelings of guilt or worthlessness, suicidal thoughts, psychomotor retardation or agitation, impaired cognitive function, and physical symptoms such as changes in appetite and disrupted sleep patterns.2,3 It has become one of the leading cause of disease burden worldwide,4–6 impairs human capability in terms of education, employment and relationships, and is also linked to premature mortality from suicide and other diseases.7–10 It has been reported that the prevalence and burden of depressive and anxiety disorders have increased dramatically worldwide (more than 25% during the first year of the pandemic) during the COVID-19 pandemic,11 thus posing grave challenges for mental health services during and after the epidemic.12 To date, causal mechanisms and pathogenesis of MDD are only partly understood. The heritability of MDD is estimated to be about 35%,13 which is lower than estimates of genetic contributions to other psychiatric disorders like schizophrenia and bipolar disorder (with heritability rates thought to be 65–70%).14 Genome-wide association studies (GWAS) have recently identified over 80 reproducible loci contributing to MDD, each with only a small effect.15,16 Moreover, the variance explained by major depression polygenic risk scores based on these genomic loci is still a very low fraction of the total heritable risk.15 These findings suggested that we are yet to discover most gene variants contributing to the genetic risk and that genetics alone do not account for most of the risk of major depression.17
Epidemiological studies indicate that environmental factors are strongly associated with the risk of developing MDD and other stress-related disorders.18–22 Early studies examined how stressful life experiences affected MDD, usually in the year preceding its onset.23,24 These documented stressful events occur mainly in adulthood. They include bereavement, financial crisis, loss of employment, separation, academic setbacks, life-threatening or chronic health problems, persistent physical pain, and exposure to violence.25 Adverse experiences early in life, such as maternal stress during pregnancy and poor maternal care after childbirth, childhood physical and sexual abuse, emotional neglect, bullying, or early separation from parents, are associated with subsequent onset, severity and chronicity of MDD.26,27 Epigenetics is a molecular mechanism that has attracted attention as it helps explain the biological impact of environmental factors.28,29
Epigenetics refers to short- and long-term gene expression variations that are caused by non-DNA-encoded mechanisms.30,31 These mechanisms include DNA methylation or hydroxymethylation, chemical changes occurring on histone proteins (histone modification), expression of noncoding RNAs, chromatin remodeling, and RNA modification. These interconnected mechanisms can mold how a cell responds at a molecular level.31,32 Epigenetic regulation mediates direct epigenetic effects or gene-by-environment interactions and can lead to complex diseases.33,34 The importance of epigenetic alterations and their effects on almost every biological pathway involved in the pathophysiology of MDD and other stress-related disorders, such as anxiety disorders and post-traumatic stress disorder (PTSD) is increasingly appreciated.35–37 Epigenetics can regulate neuronal plasticity and memory consolidation.38–41 Epigenetic regulation plays a mediating role for abnormal stress response systems, monoamine neurotransmitter dysfunction and neuroinflammation in MDD, and other stress-related disorders in animal models.42,43
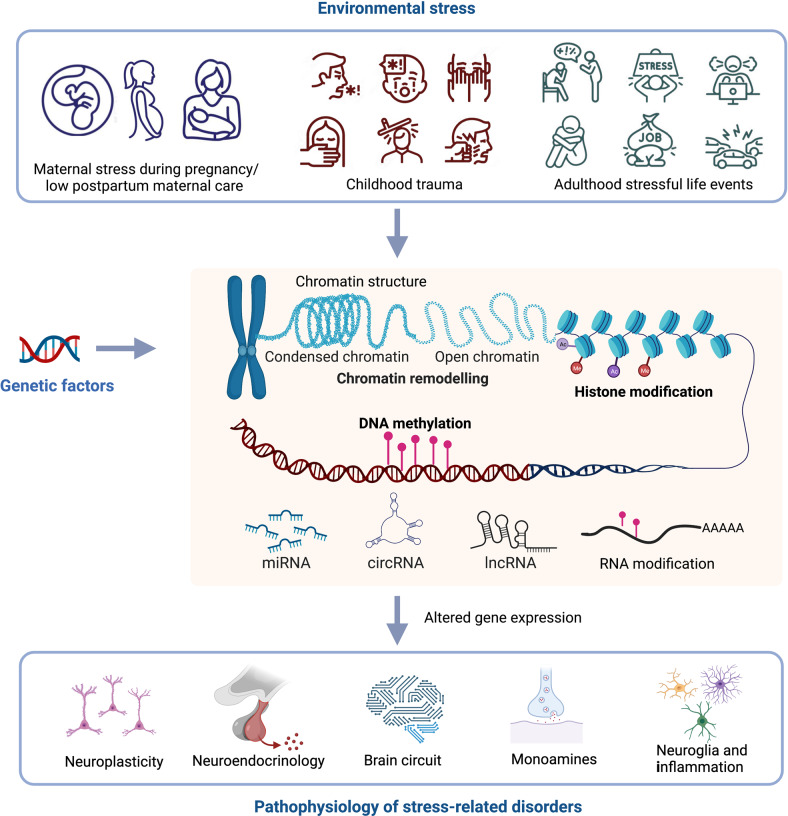
In this review, we first provide an overview of our current understanding of the functional role of different types of epigenetic regulation, including DNA methylation, histone modification, noncoding RNAs and some newly studied modifications such as RNA modification and chromatin structure remodeling factor in stress-related disorders (Fig. 1). Specifically, we discuss the roles of these epigenetic alterations in MDD pathophysiology, including neuroplasticity, neuroendocrinology, neurotransmission, and neuroinflammation. We explain how these epigenetic mechanisms might facilitate diagnosis and treatment of MDD.
Fig. 1.
Overview of the role of epigenetic processes on the pathophysiology of stress-related disorders. Environmental factors, including early life stress, childhood trauma, and stressful life events in adulthood contribute to the development of stress-related disorders through direct epigenetic regulation or gene-by-environment interactions. Epigenetic mechanisms include chromatin structure changes, histone modifications, DNA modification, noncoding RNA changes, and RNA modifications. These epigenetic processes play crucial roles in different aspects of the pathophysiology of stress-related disorders
DNA methylation regulates MDD progression
DNA methylation is the covalent addition of a methyl group to DNA’s cytosine residues, resulting in a methylcytosine (mC) base. In the human genome, mC most frequently occurs at CpG sites (cytosine followed by a guanine base in the DNA sequence).44,45 In addition, cytosines followed by a non-guanine base, such as cytosine, adenine, or thymine, might also experience DNA methylation. In brain tissues, such non-CpG methylation is a common alteration that increases in frequency during development.46 DNA methyltransferases (DNMTs) include DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMTL (Fig. 2).47,48 Methylated DNA triggers a recognition response by proteins known as methyl-CpG-binding domain (MBD) proteins, which lead to the recruitment of other proteins that either activate or repress gene expression. Methyl-CpG binding protein 2 (MeCP2) and methyl-CpG-binding domain 1 (MBD1) are two examples of MBD proteins that detect methylated DNA and are known for their association with neurodevelopment.49–51 DNA methylation is widely recognized as the most extensively researched epigenetic mechanism and is generally believed to be stable over the course of an organism’s lifetime.52
Fig. 2.
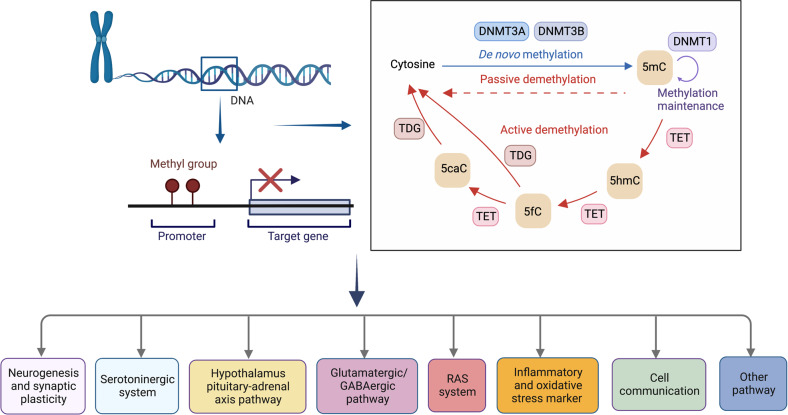
DNA methylation is involved in the progression of MDD and stress-related disorders. DNA methylation is a biological process by which methyl groups are added to the DNA at position 5′ in cytosine (5mC), which is mainly found at CpG dinucleotides. In contrast to DNA methylation, which is set up by methyltransferases (DNMT3A and DNMT3B) and maintained by DNMT1, 5mC is oxidized to 5hmC by the ten-eleven translocation (TET) family of dioxygenase proteins. In successive steps, TET enzymes further hydroxylate 5hmC to 5fC and then 5caC, which are recognized and removed by TDG, generating an unmodified cytosine. Many clinical and animal studies have examined DNA methylation of genes involved in multiple biological pathways. 5mC methylation at position 5′ in the cytosine, DNMT DNA methyltransferase, TET ten-eleven translocation, 5hmC 5-hydroxymethylcytosine, 5fC 5-formylcytosine, 5caC 5-carboxylcytosine, TDG thymine-DNA glycosylase
The location and functional effects of DNA methylation
It is often postulated that increased methylation of CpG islands in promoter regions results in the suppression of gene expression, and decreased methylation leads to increased gene expression. For example, increased CpG methylation in the promoter region of the gene encoding brain-derived neurotrophic factor (BDNF) has been found to correlate with a decreased synthesis of BDNF in neurons.53 However, this only reliably occurs in the promoter region surrounding the first exon. For other genomic locations, this is different. One study showed that the methylation of non-proximal promoters, which is dependent on DNMT3a, enhances the expression of a large cohort of neurogenic genes.54 Another study demonstrated a positive correlation between gene expression and gene body methylation.55 In clinical studies, variable associations of DNA methylation and MDD have been shown to occur in regions outside the promoter. One example showed hypomethylation of synapsins (SYN2) linked to depression.56 In contrast, a study found that patients with MDD had increased methylation in the TESC gene, negatively correlated with the right parahippocampal cingulum integrity.57 These studies suggest that the association between methylation changes and MDD is diverse, with no common effects throughout the genome or specific genomic locations.
Methylation adds to the diversity of genomic responses because DNA methylation impairs access of transcription factors to gene regulatory regions. When a recruited transcription factor is inhibitory, DNA methylation results in enhanced gene expression.58 Another possible reason for different transcriptional effects of DNA modifications may be the presence of variant forms of 5-methylcytosine, such as 5-hydroxymethylcytosine, which is produced by the addition of hydroxyl groups to 5-methylcytosine under the action of the ten-eleven translocation (TET) enzymes (Fig. 2).59,60 Currently, the majority of reports in the literature do not differentiate between DNA methylation and hydroxymethylation, and assays that rely on bisulfite conversion measure both modifications indiscriminately. Future studies must investigate these two changes in parallel to provide more information.
Non-CpG methylation is highly concentrated in neurons and glial cells61 and accumulates in neurons as they mature.62 It is a rare occurrence in the frontal cortex of human fetuses; however, it increases significantly during later stages of life. This increase in non-CpG methylation is accompanied by the development of synapses and increased synaptic density.61 As more research is conducted, it becomes increasingly evident that non-CpG methylation plays a significant role in regulating gene activity and can continue to function in the adult brain, where it may act similarly to CpG methylation in repressing transcription.63 A previous study showed that the binding of MeCP2 to non-CpG methylates DNA sequences is critical for BDNF expression. This process influenced the onset timing for Rett syndrome.64 Although the importance of non-CpG methylation in the nervous system has been demonstrated, whether it contributes to the pathophysiology of MDD and other stress-related disorders remains poorly investigated and requires further study.
Sensitive periods of DNA methylation in stress vulnerability
Exposure to environmental factors such as stress, toxins, or viruses at particularly vulnerable times of fetal development or early infancy may predispose the body to diseases in adulthood.65 Some of these effects may be mediated by epigenetic mechanisms.66,67 Early stages of life, from embryonic development through adolescence, include the ages during which the development and the later plasticity capacity of neuronal circuits are formed, along with immune, stress response, and hormone regulation pathways. These early stages are a time window when there is greater susceptibility to environmental toxins than at later periods in life.68–70
Previous studies undertaken in the rat explored potential mechanisms to explain how maternal care practice variations impact the development of individual variances in the stress response.71–74 Female Long-Evans rats differ significantly in how frequently they lick/groom (LG) their pups, which is a stable feature of the maternal phenotype. When compared to animals raised by high LG mothers, the adult offspring from low LG mothers had less hippocampal glucocorticoid receptor (GR) and protein expression, lower plasma pituitary adrenocorticotropin (ACTH) and impaired corticosterone responses to acute stress,71,74,75 and was more vulnerable to show learned helplessness to environmental stress.76 In the hippocampus of adult rats with low maternal care, the transcription factor nerve growth factor-inducible factor A (NGFI-A) binding region of the GR promoter 17 gene is hypermethylated, but in those with high maternal care, it is hypomethylated. Cross-fostering reverses these methylation discrepancies.74 However, when the timing of the stressor was shifted to adulthood, there was little effect on the GR promoter methylation levels in the brain (neither in the hippocampus or hypothalamic paraventricular nucleus).77 However, an increase in methylation levels in the peripheral hypothalamic-pituitary-adrenal (HPA) axis tissues was found to be accompanied by chronic stress.77 The findings of these animal studies illustrate how DNA methylation is affected by the timing of stress relative to sensitive periods.
Studies using human postmortem tissues also showed a link between early life adversity and epigenetic regulation of GR expression in the hippocampus. Lower GR expression along with higher levels of cytosine methylation of the GR promoter exon 1 F have been reported in suicide decedents with a history of childhood abuse than in suicide decedents without a history of abuse as well as in non-suicide controls.78 Cell type-specific alterations in the methylation of DNA in oligodendrocyte genes along with a general disruption of the transcriptional program related to myelin, were reported in depressed suicide decedents with childhood maltreatment.79 Impaired myelination in the anterior cingulate cortex in those with childhood abuse was also observed. Furthermore, recent clinical studies demonstrate that the developmental timing of childhood adversity, with sensitive periods before three years of age, explains more variability in DNA methylation than the accumulation or recency of exposure.39 This suggests that early childhood is a crucial time when exposure to life stress predicts altered DNA methylation patterns.
Tissue- and cell-type-specific changes in MDD-associated DNA methylation
Tissue specificity
DNA methylation changes in major depression and other stress-related disorders are observed in the brain and other tissues. While brain tissue is usually not available in living human studies, DNA derived from peripheral blood cells, saliva, and cheek swabs is accessible from live subjects. It may enable extensive epigenetic research using samples from various age groups, as well as repeated sampling over time.80,81 Whether findings from peripheral tissues are meaningful indicators of the pathogenesis of MDD remains to be delineated. Previous work indicates some overlap between MDD-associated differentially methylated regions (DMRs) in blood and the MDD-associated DMRs in the prefrontal cortex and other brain regions.82–85 One study, despite a small sample size, reported that three loci located in GABBR2, RUFY3, and in an intergenic region on chromosome 2, were replicated in blood and some cortical regions (Brodmann area (BA) 10 and 25).82 These genes are involved in normal brain development and function.86
However, several studies have reported that most DNA methylation markers detected in the peripheral tissues cannot accurately predict the DNA methylation status of the brain.87 The latest advancements in array techniques enabled the use of hypothesis-free paradigms to examine the association of DNA methylation changes across the entire genome to study different phenotypes.88 Unbiased, genome-wide studies using peripheral blood have reported epigenetic alterations in genes predominantly unassociated with established candidate genes selected based on known pathogenic findings, such as the serotonin transporter gene (SLC6A4) and brain-derived neurotrophic factor (BDNF),89–93 indicating alternative or additional pathogenic mechanisms.94–97 Another study comparing DNA methylation across the whole genome in live human brain tissue with that in peripheral (blood, saliva, and buccal) tissues concluded that the patterns unique to the target genomic region must be considered when selecting the best surrogate tissue to mimic brain DNA methylation.98 Nevertheless, DNA methylation detected in the brain or peripheral tissue could offer instructive insights into different biological pathways implicated in the etiopathogenesis of MDD and other stress-related psychiatric disorders (Fig. 2, Table 1).
Table 1.
Summary of DNA methylation alterations in depression and related clinical or biological outcomes
| Targeted gene (and location) | Candidate or epigenome-wide approach | Sample characteristic (human/animal) | Tissue | Clinical/biological outcome | Study |
|---|---|---|---|---|---|
| Neurogenesis and synaptic plasticity | |||||
| BDNF | |||||
| Promoter | Candidate | Human (community residents with and without late-life depression) | Whole blood | ↑BDNF DNA methylation (DNAm) → higher depression prevalence and increased depressive severity | 92 |
| Promoter | Methylome-wide association studies | Human (Monozygotic twins) | Blood (leukocytes) | BDNF, NR3C1, and SLC6A4 DNAm were positively associated with depressive symptoms; BDNF and NR3C1 DNAm mediate the association between childhood trauma and depression. | 93 |
| Promoter | Candidate | Human (MDD patients and healthy volunteers) | Whole blood | DNAm inversely correlated with the integrity (fractional anisotropy) of the anterior corona radiata in MDD patients. | 431 |
| Promoter | Candidate | Human (MDD patients and healthy volunteers) | Whole blood | BDNF DNAm mediates the association between neurocognitive performance and two BDNF single nucleotide polymorphisms (SNPs; rs908867 and rs925946). | 432 |
| Promoter | Candidate | Human (mothers with interpersonal violence-related PTSD) | Saliva-derived DNA | ↑BDNF DNAm → maternal anxiety | 433 |
| PRIMA1 | |||||
| – | Genome-wide | Human (patients with MDD and matched controls) | Postmortem frontal cortex brain tissue/ lymphoblastoid cell lines | ↑PRIMA1 DNAm in MDD → decreased PRIMA1 immunoreactivity for acetylcholinesterase and mRNA levels. | 434 |
| POU3F1 | |||||
| – | Genome-wide | Human (suicidal subjects with/without child abuse experience, sudden death controls) | Brian tissue (Anterior cingulate cortex) | Changes in DNAm of oligodendrocyte genes is associated with previous childhood abuse. | 79 |
| ID3 | |||||
| Gene body | Genome-wide | Human (children with and without maltreatment) | Saliva-derived DNA | ID3 methylation is correlated with morning cortisol levels in depressive children. | 435 |
| TPPP | |||||
| Gene body | Genome-wide | Human (children with and without maltreatment) | saliva-derived DNA | One CpG site in the gene body of TPPP predicts children’s depression. | 435 |
| PSD-95 and GJA-1 | |||||
| – | Candidate | Human (MDD patients and healthy controls) | Brain tissue (prefrontal cortex and hippocampus) | MDD patients did not show differences in PSD-95 and GJA-1 DNA methylation compared with healthy controls. | 97 |
| TESC | |||||
| – | Candidate | Human (MDD patients and healthy controls) | Whole blood | ↑TESC gene DNAm → significantly correlated with right PHC integrity in the MDD group. | 57 |
| SYN2 | |||||
| Promoter | Candidate | Human (MDD, BD patients and healthy controls) | Brain tissue (BA10) | ↓SYN2 DNAm → inversely correlated with SYN2a mRNA expression. | 56 |
| Serotoninergic system | |||||
| SLC6A4 | |||||
| Promoter | Candidate | Human (patients with MDD) | Whole blood | ↑SLC6A4 promoter methylation → higher childhood adversities, family history of depression, perceived stress, and the manifestation of more serious psychopathology. | 89 |
| Promoter, exon1, intron1 | Candidate | Human (Adolescent participants from a cohort) | Buccal cell | ↑ 5HTT DNAm who carried 5HTTLPR short-allele → more common depressive symptoms. | 90 |
| Promoter | Candidate | Human (Iowa Adoption Study) | Blood (lymphoblastoid cell lines) | ↑DNAm → lifetime history of major depression | 436 |
| Promoter | Candidate | Human (Monozygotic twins with MDD) | Blood (leukocytes) | ↑Mean DNAm → increase in the difference in depressive symptom scores. The 5-HTTLPR genotype does not modulate this association. | 437 |
| Promoter | Candidate | Human (MDD patients and healthy controls) | Whole blood | Negative emotional content significantly correlated positively with anterior insula activation and SLC6A4 methylation levels. | 438 |
| Up upstream to the transcription start site | Candidate | Human (MDD patients and healthy controls) | Whole blood | ↑SLC6A4 DNAm at CpG2 in MDD → ↓white matter integrity in the corpus callosum. | 439 |
| First extron/intron | Candidate | Human (PTSD patients and healthy controls) | Whole blood | ↓SLC6A4 DNAm and traumatic events → PTSD | 440 |
| MAO-A | |||||
| Promoter and exon1/intron1 region | Candidate | Human (MDD patients) | Whole blood | ↓MAO-A promoter DNAm hypomethylation →impaired treatment response in female patients with MDD. | 441 |
| Promoter | Candidate | Human (MDD patients) | Saliva samples | ↓MAO-A DNAm hypomethylation → higher MAO-A expression and depression in female patients. | 442 |
| First exon region | Candidate | Human (MDD patients) | Saliva samples | ↓DNAm → a history of depression; ↑DNAm in female individuals compared to males. | 443 |
| Hypothalamus pituitary-adrenal axis pathway | |||||
| NR3C1 | |||||
| Promoter | Candidate | Human (adult residents) | Whole blood | ↓DNAm associated with MDD, ↑DNAm associated with childhood maltreatment. | 444 |
| Exon 1F | Candidate | Human (MDD patients, healthy controls) | Whole blood | ↑NR3C1 exon 1F DNAm → ↑morning cortisol concentrations. | 445 |
| 1F promoter | Candidate | Human (MDD patients, healthy controls) | Whole blood | ↓DNAm at two CpG sites in MDD associated with hippocampal subfields. | 446 |
| Promoter | Candidate | Human (MDD patients, healthy controls) brain | Post-mortem tissues | MDD patients did not show differences in NR3C1 DNAm compared with healthy controls. | 447 |
| Promoter | Candidate | Animal model (adult rat) | Brain tissue | Females showed ↑NR3C1 DNAm in hippocampus | 448 |
| Promoter | Candidate | Animal model | Brain tissue | ↑Hippocampal NR3C1 DNAm in maternally separated males; ↑hippocampal BDNF IX DNAm in male and female maternally separated mice. | 449 |
| 1F promoter | Candidate | Human (combat veterans/PTSD) | Peripheral blood mononuclear cells (PBMCs) | ↓NR3C1 DNAm → glucocorticoid activity, PTSD symptoms↑ | 450 |
| 1F promoter | Candidate | Human (generalized anxiety disorder patients, healthy controls) | PBMCs | ↑ NR3C1 DNAm negatively correlated with serum basal cortisol levels and GR sensitivity in the PBMCs | 451 |
| FKBP5 | |||||
| Intron 7 GR response element region | Candidate | Human (MDD patients, healthy controls) | Whole blood | ↓FKBP5 introns DNAm → childhood adversity in MDD patients carrying the high-risk T allele rs1360780; bilaterally higher activation during valence recognition in MDD. | 452 |
| Intron 7 | Candidate | Human (MDD patients, healthy controls) | Whole blood | MDD patients did not show differences in DNAm at FKBP5 intron 7 compared with healthy controls. | 445 |
| Intron 7 | Candidate | Human (general population sample) | Whole blood | FKBP5 methylation levels were not related to FKBP5 transcription levels. | 453 |
| Intron 7 | Candidate | Human (childhood abuse/PTSD) | Whole blood | ↓FKBP5 introns DNAm → altered glucocorticoid responsiveness of FKBP5, altered hippocampal volume | 454 |
| GLU1 | |||||
| Promoter | Candidate | Human (MDD patients and healthy controls) | Whole blood | ↑GLUT1 DNAm → acute phase of MDD and mild insulin resistance | 455 |
| OXTR | |||||
| 4 bp proximal to an estrogen receptor binding region | Candidate | Human (women cohort) | Whole blood | OXTR DNAm negatively correlated with postpartum depression (PPD); a PPD specific negative correlation of DNAm with serum estradiol levels. | 456 |
| Promoter | Candidate | Human (depressed women, healthy controls) | Leukocyte cell | ↓OXTR exon 1 DNAm in depressed female patients compared to nondepressed women. Exon 1 DNAm was associated with depressed traits, whereas rs53576 genotype affected exon 2 methylation. | 457 |
| CpG site -934 | Candidate | Human (depressed women, healthy controls) | Whole blood | rs53576 interacts with DNAm in the OXTR gene among women who developed PPD. | 458 |
| Glutamatergic/GABAergic pathway | |||||
| GRIN1 | |||||
| Gene body | Genome-wide methylation | Human (maltreated and healthy children) | Saliva-derived DNA | DNAm changes in GRIN1 predicted depression independently, beyond the maltreatment history effects. | 435 |
| RAS systems | |||||
| ACE | |||||
| Promoter and exon 1 | Candidate | Human (MDD patients and healthy controls) | Blood (leukocyte)/human post mortem brain tissue | DNAm frequency at the ACE promoter inversely correlated with the serum concentrations of cardiovascular disorders risk markers (ICAM-1, E-selectin, and P-selectin) in depressed patients. | 459 |
| Inflammatory and oxidative stress marker | |||||
| YOD1 | |||||
| — | Epigenome-wide association study | Human (community-based elderly participants) | Brain tissue (dorsal lateral prefrontal cortex) | A significant relationship between DNAm at four CpG sites in YOD1 and late-life MDD, and the effects were more strongly related to late-life MDD in men than in women. | 460 |
| IL-6 | |||||
| Promoter | Candidate | Human (community-based elderly participants) | Buccal cells | ↓IL6 DNAm → current MDD or high depressive symptoms; ↑IL-6 DNAm at the same site → antidepressant use. | 461 |
| CRP | |||||
| – | Candidate | Human (a large community-based sample) | Whole blood | DNAm of CRP associated with global gray matter/cortical volume reduction and widespread white matter tract integrity impairment. | 462 |
| Cell communication | |||||
| DEPDC7 | |||||
| – | Candidate | Human (general monozygotic twins) | Whole blood | ↓ cg09090376DNAm in a cotwin → ↑ depressive symptom score. | 463 |
| Other pathway | |||||
| – | Methylome-wide association study | Human (Monozygotic twins with a lifetime history of MDD) | Blood (monocytes) | A significant relationship between 39 DMRs, 30 differentially expressed genes and lifetime history of MDD. These DMRs are involved in various processes including synaptic activity, neuropsychiatric disorders, neuronal plasticity and social behavior. | 464 |
Cell-type specificity
In addition to tissue specificity considerations, cell-type specificity in epigenetic research could provide more precise insights into the molecular pathology of MDD. In mice that were subjected to chronic stress and exhibiting severe depressive behavior, DNMT3A levels were found to be higher in the nucleus accumbens (NAc) than controls.99 Studies examining postmortem brain tissue reported lower DNMT1 levels and higher DNMT3B levels in the frontopolar cortex. The study also reported reduced expression of DNMT1 and DNMT3B in the amygdala, and increased expression of DNMT3B in the paraventricular nucleus of depressed suicide decedents.100 Therefore, changes in DNMT mRNA expression occurred in specific cells of autopsied brain tissue in depressed suicide decedents. Fluorescence-activated cell sorting detected oligodendrocyte-specific DNA methylation changes in MDD postmortem.79 Consistent with the consideration of cell-type specificity in preclinical studies, some clinical studies using peripheral blood to examine DNA methylation changes in MDD corrected for cell composition,101,102 with the authors focusing mainly on peripheral immune cells implicated in the pathophysiology of psychiatric disorders.103
Changes in DNA methylation of particular genes within specific brain cells that are linked to depression are also evident. DNA methylation at the glial cell-derived neurotrophic factor (Gdnf) promoter with distinct epigenetic modulator complexes between depression-susceptible and depression-resistant animals was increased by chronic stress.104 A methylation map specific to astrocytes throughout the genome indicated less methylation in GRIK2 (glutamate receptor, ionotropic kainate 2) and BEGAIN (brain-enriched guanylate kinase-associated protein) associated with depressive psychopathology.105 Single-cell epigenomics106 is based on sequencing single nuclei, barcoding all the transcripts from each nucleus, and using the expression pattern to sort the nuclei into specific cell subtypes. This technique may be an excellent approach to further investigate changes in specific genes in specific brain cells in MDD-associated DNA methylation; however, single-nucleus, postmortem brain studies of MDD are rare.
Histone modifications in the pathogenesis of MDD
Histones are essential proteins rich in lysine and arginine residues in eukaryotic somatic chromatin. The DNA molecule (approximately 150 bp) is enveloped by histone octamers, which comprise two sets of fundamental histones (H2A, H2B, H3, and H4), to form a single nucleosome, the basic repeating subunit of chromatin.107 Histones regulate gene expression both positively and negatively.108 This is mainly governed by posttranslational modifications catalyzed by enzymes that act on particular amino acid residues located on histones. The H3-H4 tetramer is stable and allows histone modifications to be heritable epigenetic marks.109 The lengthy tails extending from the nucleosome of H3 and H4 histones can be covalently changed in several locations. Methylation, acetylation, phosphorylation, ubiquitination, SUMOylation, crotonylation, citrullination, and ADP-ribosylation are a few modifications of the tail.110,111 Acetylation or methylation of lysine or arginine residues are the most common modifications observed, which alter the interactions between histones or transcription factors and DNA, regulating gene expression.112 Furthermore, studies have suggested that the patterns of DNA methyltransferase localization, DNA methylation, and actively transcribed gene bodies may be specified by histone modifications.113,114
Histone acetylation in stress response and neuroplasticity
Histone acetylation at lysine residues (mainly including K9, K14, K18, K23, and K27), is commonly linked to the activation of gene transcription. Acetylation is typically observed at the transcriptional start sites and enhancers of genes that are actively transcribed.115 The “writers” that mediate histone acetylation include histone acetyltransferases (HATs) for the addition of the acetylation mark. The “eraser” histone deacetylases (HDACs) eliminate acetyl groups from lysine residues (Fig. 3c); as a result, the ionic interaction between histones and DNA increases, DNA packs more tightly and chromatin is more highly condensed. HDACs are grouped into two families: the traditional HDACs and the NAD+-dependent, silent information regulator (SIR2) family of HDACs.116 The former includes three phylogenetic classes: I (HDAC1, 2, 3, and 8), II (HDAC4, 5, 6, 7, 9, and 10), and IV (HDAC11). The SIR2 family of HDACs, sometimes called class III HDACs or sirtuin (SIRT), deacetylates both histone and nonhistone proteins, modulating many cellular processes117,118, and influencing gene expression.119
Fig. 3.
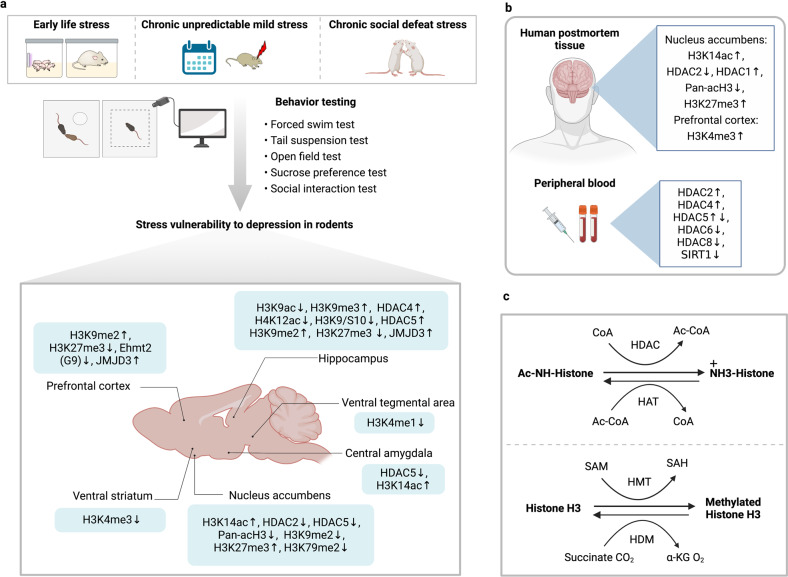
Different types of histone modification changes in different brain regions in stressed animals and depressed humans. a Animal models and behavior analyses for studying the relationship between stress vulnerability to epigenetic changes and depression. Recent studies using animal models show brain region-specific histone modification changes. The NAc104,130,158,167,168,211 and hippocampus136,137,148,170,175,513 are the most studied brain regions for histone modification, with both consistent and conflicting findings across different studies. Different types of histone modification are also observed in other brain regions, such as the prefrontal cortex175. b Histone modification changes based on human postmortem tissue130,158,165,168 and peripheral blood, collected in both cases from depressed individuals159,160. c Chemical reactions involved in histone acetylation and methylation. NAc nucleus accumbens, CoA crotonyl-coenzyme A, HDAC histone deacetylases, HAT histone acetyl transferases, SAM S-adenosyl methionine, HMT histone methyltransferases, HDM histone demethylases, SAH S-adenosyl homocysteine, α-KG α-ketoglutarate. ↑ Increased changes compared with controls; ↓ decreased changes compared with controls; ↑↓ both increased and decreased changes were reported across different studies
As a stress-related disorder, major depression is associated with an abnormal stress response system. For example, excessive cortisol release in response to stress and impaired GR-mediated feedback inhibition have been known for decades.120,121 The NR3C1 gene encodes for the GR,122,123 which is an essential component of the HPA axis. As a transcription factor, the GR interacts with and influences the histone landscape, playing a critical role in shaping it.124,125 Animal models for studying depression have been established based on exposure to different forms of stressors in rodents such as early life stress (ELS), chronic social defeat stress, and chronic unpredictable mild stress.126 These models, combined with the ability to assess anhedonia, anxiety- and depression-related behavior objectively and learned helplessness in rodents, have helped to elucidate the link between stress and vulnerability to depression as mediated by epigenetic changes (Fig. 3a). Histone acetylation changes, primarily mediated by HDAC, impact the stress response, depression-like behavior, and antidepressant effects.104,127–129
Class I HDACs involved in antidepressant effects and neuronal plasticity
Early evidence supporting the involvement of HDACs in the stress response and antidepressant action showed that mice exposed to chronic social defeat stress experienced a temporary reduction followed by a lasting escalation in histone acetylation levels (H3K14ac), which was connected to decreased histone deacetylase 2 (HDAC2) levels in the NAc.130 Infusion of HDAC inhibitors (i.e., MS-275) into the NAc reversed global gene expression patterns in the NAc and improved depression-like behaviors. The observation that MS-275 had antidepressant effects suggested that histone acetylation plays an adaptive role in the response to stress. This was supported by another study showing that animals overexpressing HDAC2 in the NAc region exhibit more depression-like behavior.104
Although primarily manifested as depression, MDD also results in cognitive impairments.131,132 Neuronal plasticity and cognitive function are associated with transcriptional changes regulated by HDAC-mediated epigenetic modification.133 The overexpression of HDAC2 specifically in neurons resulted in a decrease in dendritic spine density, synapse number, synaptic plasticity, and memory formation in mice. However, prolonged administration of HDAC inhibitors improved synapse number and learning impairment.134 Furthermore, a promoter occupancy analysis has revealed a link between HDAC2 and the promoters of genes that play a role in synaptic plasticity and memory formation.134 Consistent with these findings, viral-mediated knockdown of HDAC2 can restore both structural and synaptic plasticity, ultimately leading to an improvement in memory loss associated with neurodegeneration.135
Class II HDACs have opposite roles in different brain regions related to depression
Studies with class II HDACs (i.e., HDAC4 and HDAC5) revealed different roles for these molecules. The AChE gene, which encodes the acetylcholine-hydrolyzing enzyme acetylcholinesterase, was downregulated in the hippocampus after stress and accompanied by decreased acetylation and increased trimethylation of H3K9 at the corresponding promoter, as well as HDAC4 accumulation in hippocampal neurons. These effects were reversed drastically by administration an HDAC inhibitor, and reduced hippocampal HDAC4 levels restored the long-lasting behavioral deficit.136 In addition, overexpression of HDAC4 in the hippocampus caused adult rats to be depressive but not anxious.137
HDAC4 exhibits high expression levels in the forebrain and is enriched in neurons.138,139 It shuttles between the cell nucleus and cytoplasm.140 Cytoplasmic localization of HDAC4 is maintained by HDAC phosphorylation, suppressing the binding of the transcription factor MEF2,141 whereas HDAC4 dephosphorylation by calcineurin allows nuclear translocation.142 Nuclear HDAC4 functions as a transcriptional repressor, downregulating the expression of numerous plasticity-related genes, that may mediate effects on learning and memory impairment in mice.143,144 In contrast, the role of cytoplasmic HDACs is less understood. HDAC4 in the cytoplasm may have neuroprotective effects.145–147 Inhibiting HDAC4 delayed the formation of huntingtin protein cytoplasmic aggregates, while the levels of BDNF transcripts were restored, resulting in the restoration of neuronal and cortico-striatal synaptic function in mouse models of neurological disorders.147
Regarding HDAC5, chronic imipramine (a tricyclic antidepressant) administration to mice with chronic social defeat behavior was linked to a selective downregulation of HDAC5 in the hippocampus; viral-mediated HDAC5 overexpression blocked the antidepressant-like effect of imipramine.148 In another study of chronically stressed rats, a significant decrease in histone acetylation (H4K12Ac) and phosphor-acetylation (H3K9/S10) was observed in CA3 and dentate gyrus (DG) in stressed animals compared with control animals, along with increased HDAC5 expression. HDAC5 seems to play a role of promoting depression in the hippocampus, but in other brain regions, HDAC5 may have the opposite effect. For example, mice exposed to chronic social defeat stress showed decreased HDAC5 expression in the NAc, whereas chronic administration of imipramine increased HDAC5 expression in NAc,149 suggesting a pro-resilient role of HDAC5 in the NAc. Rats exposed to variable mild stress manifested decreased HDAC5 in the central amygdala (CeA).150 Acetylation in the amygdala could potentially be an advantageous adaptation. Transiently increased H3K14 acetylation in the amygdala of mice followed by chronic social defeat stress. Conversely, the injection of an HDAC inhibitor into the amygdala reversed social avoidance.127 Changes of HDAC5 in the amygdala resemble the observations made in the NAc of mice following exposure to stress, suggesting a homeostatic role, in contrast to the pro-depressive effect in the hippocampus. The reason why HDAC5 plays opposite roles in different brain areas may lie in its involvement in diverse complexes that target specific gene subsets associated with depression.
Class III HDACs associate with depression
In addition to classical HDACs, class III HDAC (i.e., SIRT), which deacetylates histones and nonhistone proteins, also appears to be associated with hippocampal neuroplasticity and depression-like behavior.151–153 Activation of SIRT in the hippocampus exerts antidepressant effects and blocks abnormal dendritic structures. Blocking SIRT1 function in the hippocampus increases depression-like behaviors.151 SIRT1 is also one of the first genes (SNPs; rs12413112) identified using GWAS to be associated with MDD.154,155 Furthermore, lower SIRT expression levels in peripheral blood samples of MDD patients156 align with findings from other studies.157 Taken together, these findings suggest that activating SIRT1-dependent pathways may be a potential therapeutic strategy for MDD.
Differential histone modification in human samples
Some studies examined histone modifications in the human postmortem brain or the expression of histone-modifying enzymes in the peripheral blood cells in MDD (Fig. 3b). Expression of Rac1, involved in synaptic structure regulation, was low in NAc postmortem tissues in MDD and associated with lower histone H3 pan acetylation and more histone H3K27 trimethylation.158 In addition, differential brain expression of HDACs was found in the postmortem in MDD.130,159 Expression of HDAC2 and HDAC5 was higher during a depressive state, compared with remission, in peripheral white blood cells of patients diagnosed with MDD and bipolar disorder (BPD).159 The expression of HDAC6 and HDAC8 was lower regardless of mood states compared with controls in BPD, while the HDAC4 expression was higher only in a depressive state.159 This study links altered expression of HDACs with depression and is consistent with a potential homeostatic role of histone acetylation in response to stress and depression. Antidepressant treatment caused higher peripheral HDAC5 expression to decrease to control levels after about eight weeks of treatment.160 It is impossible to separate in clinical studies whether biology that fluctuates with the severity of illness is a homeostatic response, a measure of the stress due to the illness severity, or part of the pathophysiology responsible for illness severity.
Collectively, stress-induced depression is associated with decreased histone acetylation. HDACs appear to be involved in neuroplasticity, neuronal survival, and cognition and may become potential targets of antidepressant intervention. HDAC inhibitors restore memory deficits in mice,161 but it remains to be seen whether modifying HDACs ameliorates cognitive impairments and depression in humans.
Histone methylation in stress response and depression
Histone methylation involves the transfer of methyl groups to amino acids in histone proteins,162 occurring at various locations along the histone tails, resulting in the addition of one, two, or three methyl groups to the lysine or arginine residues. In contrast to histone acetylation, histone methylation is in general linked to transcriptional repression. However, in some cases, the relationship between histone methylation and transcription depends on the level of methylation and the site of the residue. For example, methylation of H3K9 and H3K27 leads to repressed gene expression;163 while histone methylation of H3K4 in the promoter regions results in relaxed chromatin, which promotes gene expression.164 The enzymes that facilitate histone methylation include histone methyltransferases (HMTs) and histone demethylases (HDMs), which transfer or remove methyl groups from target residues using S-Adenosyl methionine as a methyl donor.
Trimethylation of histone 3 lysine 4 (H3K4me3) is one of the most characteristic histone modifications. When exposed to chronic unpredictable mild stress (CUMS), the level of H3K4me3 was decreased at the promoter region of the Gdnf gene, which led to altered Gdnf expression in the ventral striatum in mice.104 Interestingly, enhancement of H3K4me3 was found in postmortem brain tissue (BA10) in MDD165: the enriched H3K4me3 was found at transcriptional start sites of synapsin 1, relevant to increased expression of synapsin 1a and synapsin 1b. These synapsins are critical in synapse function and plasticity.166 Other forms of histone methylation include H3K9me2 and H3K27me3, are repressive in the gene promoter region in response to stress.167,168 Rats exposed to the early stress of maternal separation (ES) exhibited decreased H3K9me2 modification at the BDNF IV promoter site, along with increased BDNF levels, enhanced hippocampal neurogenesis, and better cognitive performance in both the postnatal life and young adulthood. Interestingly, middle-aged rats that experienced early maternal separation exhibited impaired cognition, reduced H3K9me2 regulation of the BDNF expression, and opposite changes in the hippocampal neurogenesis, suggesting both biphasic and distinct, age-dependent changes in the histone methylation in response to ES.169 Another study showed elevated H3K9me2 levels and reduced BDNF levels in the hippocampus and medial prefrontal cortex (mPFC) in CUMS rats showing depression-like behaviors.170 In addition, higher H3K9me2 levels at the calmodulin-dependent protein kinase II α (CaMKIIα) promoter and inhibition of CaMKIIα were found in MDD and mice following antidepressant administration.171 The activation of H3K27me3 in the NAc of mice exposed to social defeat stress inhibits the expression of RAS-related C3 botulinum toxin substrate 1 (Rac1), a Rho GTPase–related gene known for synaptic structure regulation.158
In parallel to prominent findings for HDACs, related enzymes for histone methylation and demethylation (HMTs and HDMs) also were potentially crucial in depression. Higher expression of Setdb1, an H3K9-specific HMT in the mouse forebrain, was associated with changes in the composition of NMDA receptor subunits, as well as other molecular modifications resulting from suppressive chromatin remodeling at specific target genes.172 This regulation led to anhedonia, despair, and learned helplessness in behavioral paradigms. In line with these findings, downregulation of HMT Ehmt2 (G9a) was associated with loss of repressive histone methylation (H3K9meX) after chronic stress.173 Protein arginine methyltransferase 1 (PRMT1), another HMT, when knocked out, improved depression-like behavior, along with upregulated expression of BDNF and postsynaptic density protein 95 (PSD95).174 The histone lysine demethylase jumonji domain-containing 3 was also upregulated in the prefrontal cortex and hippocampus of rats exposed to CUMS.175
Other types of histone modifications affecting depression
Several uncommon histone modifications, such as histone crotonylation, histone phosphorylation, and histone β-hydroxybutyrylation, are shown to be involved in the pathophysiology of MDD.176–178 Histone crotonylation was previously shown to occur at the promoters or enhancers of gens that are actively transcribed, and play a role in spermatogenesis.111,179 Similar to histone acetylation, histone crotonylation is catalyzed by HATs, which add a crotonyl group from crotonyl-coenzyme A (CoA) to amino acid residues of histones,111 while HDACs 1/2/3 and sirtuin 1/2/3, act as decrotonylases.180–182 In the medial prefrontal cortex of mice vulnerable to chronic social defeat stress, histone crotonylation was inhibited. In contrast, chromodomain Y-like protein (CDYL), which acts as both a crotonyl-CoA hydratase and a histone methyllysine reader, was selectively upregulated.183
Histone phosphorylation mainly occurs on serine and threonine residues, though some studies have found that tyrosine residues also can be phosphorylated.184 Histone phosphorylation in the central nervous system is involved in stress response. For example, the expression of H3S10 phosphorylation (H3S10p) was increased in the hippocampus in mice exposed to fear conditioning.185 Rats exposed to novelty stress also showed an induction of H3S10p at the c-fos promoter site in the dentate gyrus.186 To date, few studies have investigated the relationship between histone phosphorylation and MDD. Rats exposed to forced swimming stress showed an increase in H3 phosphorylation in the infralimbic (ILCx) and prelimbic area (PrLCx) of the prefrontal cortex.187
In 2006, histone β-hydroxybutyrylation was reported as a newly-histone modification. It is dynamically controlled by the concentration of hydroxybutyrate in cells.177 However, the mechanism by which a β-hydroxybutyryl group is added to histones remains unknown.177,188 It has been demonstrated that β-hydroxybutyrate exerts an antidepressant-like effect in a rodent model of chronic unpredictable stress189,190. Injection of β-hydroxybutyrate improved depression symptoms and reversed the reduction of H3K9 β-hydroxybutyrylation and increased BDNF expression. This study emphasized that metabolite changes after stress contribute to histone modification.191 However, further research is necessary to investigate the homeostasis of other types of histone modifications and the pathophysiological changes associated with MDD.
Susceptibility and adaptation to stress mediated by histone modification
When exposed to stressful events, only a subgroup of individuals develops major depression.192–195 Two genetically diverse mouse strains show how epigenetic changes underlie susceptibility and adaptation to chronic stress during adulthood.104 The interplay of genetic factors and environmental stressors can be mediated by histone modifications and DNA methylation in the ventral striatum, thus contributing to behavioral responses to stress.104 Specific cell types and epigenetic changes affect stress susceptibility in an opposing manner.196 Fosb-targeted histone acetylation or methylation in different types of medium spiny neurons (i.e., D2-type vs. D1-type) controls stress susceptibility and resilience to social stress.
Experiencing stress in the early life, commonly known as Early life stress (ELS), increases the likelihood of developing depression, suicidal behavior, and various other psychiatric disorders during adulthood.26,197–200 Depressive patients reporting childhood trauma show earlier onset, chronic course, and poorer response to antidepressant treatment.201 In animal models, long-lasting effects of ELS have been documented.202–207 ELS increased the vulnerability to adult stress in rodents208. ELS may not cause immediate behavioral abnormalities but instead causes long-lasting transcriptional alterations that prime critical brain regions to be hyper-responsive to later stress and prone to develop a depression-like state.209,210 This was illustrated by a “two-hit” stress model in mice wherein ELS (stress hit number 1) increased susceptibility to depression-like behavior in response to social defeat stress (stress hit number 2) during adulthood.209
Enduring transcriptional alterations were observed in the ventral tegmental area (VTA) in ELS, mediated by the developmental transcription factor orthodenticle homeobox 2 (OTX2). Furthermore, histone methylation (H3K4me1) changes were found in genes targeted by OTX2 binding. The same stress model211 revealed long-lasting histone modifications in the NAc after ELS. ELS-induced depression susceptibility was caused by decreased histone methylation (H3K79me2) and induction of the DOT1L and KDM2B enzymes that regulate this histone methylation in D2-type medium spiny neurons. In addition, systemic delivery of DOT1L inhibitor reversed ELS-induced behavioral deficits without detectable side effects, suggesting a potential therapeutic target.211 The regulation of epigenetic markers is complex, involving upregulation or downregulation at hundreds of loci on a genome-wide scale. Thus, more studies are needed using genome-wide histone code profiling such as chromatin immunoprecipitation followed by sequencing.212
Noncoding RNAs participate in the development and treatment of MDD
MicroRNAs (miRNAs), circular RNAs (lncRNAs), long noncoding RNAs (lncRNAs), and other as yet unidentified RNAs are referred as noncoding RNAs (ncRNAs), which do not code for peptides or proteins.213,214 Although ncRNAs cannot encode proteins, they can influence the expression of other genes through multiple mechanisms to cause a wide range of disorders.215–218 ncRNAs can regulate expression of genes via the translation or transcription of mRNAs, the methylation of DNA and RNA, or as a modular scaffold of histone modification complexes.219 Studies have demonstrated that significantly altered ncRNAs expression in MDD relative to healthy subjects. At the same time, antidepressants can also alter the abnormal expression of ncRNAs.220 ncRNAs may influence the pathophysiology of depression by regulating neuronal function, neurotransmitter release, and microglia (Fig. 4).213,221,222
Fig. 4.
Noncoding RNA in depression: generation, mechanisms of function, and effects. DNA constitutes an essential part of the human genome and contributes to the formation of noncoding RNA after transcription. For lncRNAs, multiple mechanisms are involved in the pathophysiology of depression: transcriptional activation: lncRNAs can activate the expression of target genes via (1) recruiting transcriptional factors to upstream open reading frames; (2) suppressing transcriptional factors to upstream open reading frames; (3) recruiting chromatin modifying factors to alter chromatin structure; (4) suppressing interacting proteins and RNP-complexes to target genes. For circRNAs, they are generated from back-splicing and canonical splicing of an mRNA transcript. These circRNAs are associated with homer scaffolding protein 1, regulate cell proliferation, and inhibit JAK2/STAT3 signal, leading to neuropathological changes related to depression. For miRNAs, multiple mechanisms, including mRNA degradation or inhibition by the RISC complex, activate depression. These mechanisms induce gene expression changes, which are associated with several molecular pathways of depression. RISC RNA-induced silencing complex, RNP ribonucleoprotein complexes, STAT3 signal transducer and activator of transcription 3, SERT serotonin transporter, BDNF brain-derived neurotrophic factor, GR glucocorticoid receptor, JAK2-STAT3 Janus kinase 2 and signal transducer and activator of transcription 3
miRNAs and the development of depression
MicroRNA molecules are highly conserved small (21–24 nucleotides) RNA molecules.223 Over 60% of human protein-coding genes are dynamically regulated by miRNAs, thereby influencing cell growth and differentiation.224 The mechanistic details of miRNA synthesis and maturation have been reviewed225 and are summarized here. RNA polymerase II converts an intragenic or intergenic miRNA gene into a pri-miRNA (Fig. 5).226 Following several cleavage and maturation steps, the pre-miRNA is exported to the cytoplasm and incorporated with RNA-induced silencing complexes (RISCs).227,228 miRNA sequences guide the RISC to target mRNA transcripts, resulting in translational cleavage or repression of mRNA.226 Traditionally it was thought that the miRNAs targeted the 3′ UTRs of transcripts but it has become apparent that 5′ UTRs, coding sequences, and promoters also can be targeted for repression.229 Because miRNAs have imperfect sequence recognition, they may bind to multiple target genes and modulate their expression.226 In contrast, aberrant miRNA can contribute to cancers, cardiovascular diseases, metabolic diseases, immune-mediated disorders, and neurological disorders, etc.230,231 For example, miR-106b-5p is an oncogenic miRNA that appears to be upregulated in different cancers, such as hepatic cancer,232 cervical cancer,233 gliomas,234 and gastric cancer.235 Likewise, miR-34 dysregulation is related to different psychiatric disorders, including schizophrenia236, Alzheimer’s disease,237 and MDD.238
Fig. 5.
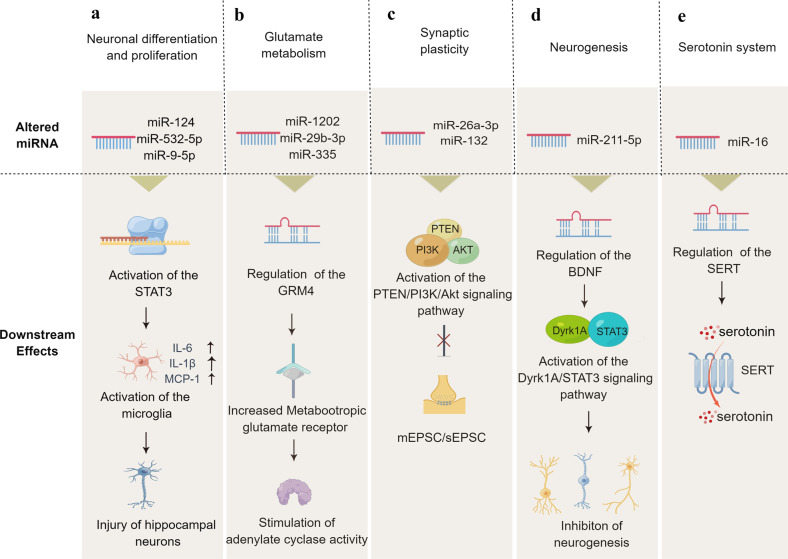
Functional mechanisms of representative miRNAs involved in depression. A variety of miRNAs can influence neurodevelopment, synaptic plasticity and neurotransmitters by regulating their target genes, thus leading to depression. PTEN phosphatase and tensin homolog deleted on chromosome ten, PI3K phosphoinositide 3-kinase, GRM metabotropic glutamate receptor, mEPSC/sEPSC miniature excitatory postsynaptic current/spontaneous excitatory postsynaptic current, SERT serotonin transporter, BDNF brain-derived neurotrophic factor
miRNAs have been evaluated in MDD by examining their level in peripheral blood, cerebrospinal fluid, and postmortem cortex of depressive patients and in animal models of depression.239–241 The depression susceptibility is highly associated with the miRNA polymorphisms.242 In peripheral blood of depressed patients, multiple miRNAs, such as miR-330-3p, miR-345-3p, miR-425-3p, and miR-24-3p, were reportedly altered.243 In the prefrontal cortex (BA9) of MDD patients, other 21 miRNAs, including miR-142-5p, miR-101, miR-137, and miR-301a, was downregulated.244 MiRNA expression levels were also found to be altered in BA10,244 BA44,245 anterior cingulate cortex (ACC),246 and other brain regions in MDD.247 The link between miRNAs and the pathophysiology of depression248–252 may be implicated by the fact that miRNAs regulate neuronal regulation, neurotransmitter regulation, and microglia activation.253,254
Neuronal regulation by miRNAs
In depressed patients, fewer pyramidal neurons are found in the hippocampus and prefrontal cortex.255,256 Cell loss, neuronal atrophy, and alterations in synapse density are also observed in depression animal models.257 MiRNAs may mediate these effects in depression by influencing neuronal production, differentiation, proliferation and synaptic plasticity.258
As shown in Fig. 5, the expression of miR-124, miR-532-5p, and miR-9-5p alters in tissues from depression patients/animal models, activating signal transducer and activator of transcription 3 (STAT3) and mediating the disruption of neuronal differentiation and proliferation in depression.259–262 Similarly, miR-26a-3p263 and miR-132264 can induce depressive symptoms by activating the phosphatase and tensin homolog (PTEN)/phosphoinositide 3-kinase (PI3K)/Akt signaling pathway, which is one of the main pathways for regulating synaptic plasticity. Low hippocampal miR-211-5p can inhibit neuronal neurogenesis and induce depression-like behaviors via the promotion of the Dyrk1A/STAT3 signaling pathway.265 Low miR-218 expression may influence synapse formation and synaptic plasticity of the mPFC by repressing the gene expression of Netrin-1 guidance cue receptor gene DCC in mouse models of social defeat stress.266 These data demonstrate the crucial role of miRNAs in neuronal differentiation, production, and maintenance.
miRNAs regulate neuroglial cells
The peripheral and central nervous systems (CNS) both contain neuroglial cells, such as microglia, astrocytes, and oligodendrocytes. The neuroglial cells serve as the first line of defense for the brain, not only by recognizing pathogens and repairing damaged tissue but also by secreting neurotrophic factors and producing chemokines and cytokines.267 Chronic unpredictable stress (CUS) model mice showed changes in neuroglial numbers and morphologies in the hippocampus. Postmortem studies of MDD cases show deficits in neuroglial numbers in the neocortex.268 Therefore, the occurrence and development of depression may be associated with neuroglial cells.269
The protein WNT2 plays pivotal role in embryonic development, the adult hippocampal neurogenesis, and development of the nervous system. In depression, more miR-199a-5p was noted in microglia suppressing WNT2 expression through the CREB/BDNF signaling regulation.270 The downregulation of miR-124 in depression also promotes the activation of BV2 microglia by activating STAT3.259 This increases inducible nitric oxide synthase and proinflammatory cytokine (MCP-1, IL-6, TNF-α, and IL-1β) secreted by BV2 cells, which induces depressive symptoms. In addition, miR-146a-5p is transferred by microglia to the hippocampus DG, where it targets Krüppel-like factor 4 (KLF4) and inhibits neurogenesis and the spontaneous discharge of excitatory neurons.254
Regulation of neurotransmitters by miRNAs
Studies have shown that miRNA-mediated disturbance of neurotransmitters, including glutamate, dopamine, serotonin and norepinephrine, can lead to neuronal damage, and loss of trophic effects and thereby potentially contribute to the pathogenesis of depression.271–274 Glutamate and its receptors are associated with many neuropsychiatric illnesses, including depression.275,276 MiR-335, miR-1202, miR-29b-3p, and miR-134-5p may regulate depression through inhibiting the expression of GRM4 gene (Fig. 5), that in turn affect the influx of Ca2+ into the prefrontal cortex neurons and the extracellular concentration of glutamate.277–283 The monoamine oxidase A (MAO-A) enzyme modulates monoamine neurotransmitter levels. miR-142, miR-34a, and miR-34c indirectly regulate MAO-A, potentially impacting monoamine neurotransmitters.284 In addition, miR-16 upregulation in depression inhibits the translation of the serotonin transporter (SERT) gene, affecting serotonin function in the hippocampus.285,286
circRNAs engage in the development of MDD
circRNAs are covalently closed circular RNA molecules that typically contain exons, formed by direct splicing 3′ and 5′ terminal ends generated from precursor mRNA.220,287 These RNA molecules function as a class of post-transcriptional regulators.288 Typically, circRNAs regulate the expression levels of downstream genes at several levels, including mRNA transcription, splicing, and translation287. In addition, it has been reported that some circRNAs can translate to peptides.289 Several studies have demonstrated that circRNAs are abundantly expressed in the brain. And also, circRNAs are involved in the pathogenesis of neuropsychiatric disorders, impacting neuron development, cognitive function, and synaptic plasticity290–292.
Neuronal regulation by circRNAs
CircRNAs involve in regulating central neural system development. They are highly expressed in the brain from the embryonic stage to adulthood and substantially upregulated during the differentiation and maturation of neurons.293 Many circRNAs are significantly upregulated at different time points of neuron maturation in vitro.293 In vitro studies have shown that overexpression of circPTK2 can induce neuronal apoptosis via miR-29b inhibition, preventing inflammatory response and protecting against neuronal apoptosis.294 Conversely, circPTK2 silencing inhibited JAK2/STAT3 and decreased IL-1β, thereby ensuring neuronal survival.294
circRNAs regulate neuroglial cells
It was reported that cirRNAs regulate neuroglial activities. For example, one study revealed that the gut microbiota-circHIPK2-astrocyte axis was involved in the development of depression in a stress mouse model.295 Fecal microbiota transplantation significantly alleviated astrocyte dysfunction and depression-like behaviors in recipient depressive mice via inhibition of circHIPK2 expression.295 Similarly, another study demonstrated that overexpression of circSTAG1 significantly ameliorated depression-like behaviors and astrocyte dysfunction in the stress mice model.296 In addition, a recent study using two different MDD mouse models demonstrated the presence of a close relationship between depression and decreased circDYM and revealed that increased circDYM expression could significantly attenuate depressive-like behavior and inhibit microglial activity.290
Regulation of neurotransmitters and synaptic plasticity by circRNAs
circRNAs were also observed to be linked to neurotransmitter function and synaptic plasticity and have a higher expression in the neuropil area compared to their mRNA isoforms.297 Studies have shown many host genes of circRNA were enriched in the pathway of synaptic activities and neurotransmitter secretion.289,297,298 For instance, circHomer1 derived from the homer scaffolding protein 1 pre-RNA can regulate some synaptic structure during the development and neuronal plasticity.297 Furthermore, a study using a murine cell line model of Huntington’s disease identified 23 differentially expressed circRNAs significantly enriched in the MAPK, dopaminergic synapse, and long-term depression pathways.299
lncRNAs participate in the process of depression
lncRNAs are more than 200 nucleotides in length and cannot encode proteins, but they share some characteristics with messenger RNAs.300 lncRNAs, for instance, are transcribed by RNA polymerase II; they are 5′ capped with a polyA tail and have multiple exons. Because of their lack of conservation across species and lower expression levels than mRNAs, lncRNAs were initially regarded as transcriptional noise or junk.301 However, some long noncoding RNAs are involved in regulating gene expression, including transcriptional regulation, RNA regulation, chromatin modification, and posttranscriptional regulation of protein activity and localization.302
Highly expression of lncRNAs in the brain is crucial for maintaining neural stem cells, neurogenesis, neural plasticity and cognitive function, etc.303 For example, brain cytoplasmic lncRNA (BC1), one of the earliest lncRNAs studied in the brain, regulates metabotropic receptor signaling.304 In addition, neurogenesis-associated lncRNAs may act as guides for proteins associated with neurogenesis, including REST, SOX2 and SUZ12. Furthermore, RNAi knockdown of some lncRNAs impairs neuronal differentiation, suggesting that lncRNAs play critical roles in neurogenesis.305 Several recent studies have indicated that lncRNAs are crucial in the development of numerous neurological disorders, including autism, schizophrenia, and depression.306–309 However, further research is needed to determine their signal pathway targets and regulation.
Levels of 60% (217 out of 364) of the 364 lncRNAs expressed in the rostral cingulate cortex of normal individuals differed from those in MDD.310 Using whole-genome sequencing, a recent study found that lncRNAs accounted for 30% of the differences between patients with depression and control populations.311 In MDD patients, four lncRNAs (PCAT1, MER11C, Y5, and PCAT29) levels were higher, and one lncRNA (RMRP) was lower in peripheral blood leukocytes.312
Regulation of neuronal cells by lncRNAs
In MDD patients, LncRNA TCONS 00019174 levels were lower, and phosphorylated-GSK3 and -catenin was higher in the hippocampus, potentially causing neuronal damage.313 Furthermore, viral-mediated overexpression of lncRNA TCONS 00019174 aggravated depression status in depressed mice model, indicating that lncRNA TCONS 00019174 may be linked to the pathogenesis of depression.313 Differentially expressed lncRNAs showed correlations with 18 synapse-related functions, which showed that lncRNA-directed regulatory machinery might mediate synaptic dysfunction in depression.314
Regulation of neuroglial cell by lncRNAs
Administration of lncRNAs can treat depression at the neuroglial cell level, which promotes the pathogenesis of depression. More M1 microglia were found in the hippocampus of depressed rats and fewer M2 microglia, as well as a negative regulation factor of the lncRNA uc.80.315 Elevated expression of lncRNA uc.80 can reduce depression-like behaviors in rats and hippocampal neuron apoptosis in vitro and in vivo.315 Consequently, lncRNA uc.80 may be a potential target of treatment for depression.
Neurotransmitters regulated by lncRNAs
Researchers have shown that upregulated lncRNA NONHSAG045500 inhibits the expression of the 5-HT transporter SERT which influences 5-HT transmission in depression.316 Meanwhile, the concentration of 5-HT was increased by interference with the levels of NONHSAG045500. NONHSAG045500 may modify the transport of monoamine neurotransmitters.316
Effective antidepressant treatment is associated with ncRNA alteration
There are approximately six major classes of antidepressants used worldwide. Classical antidepressants include the noradrenaline reuptake inhibitors (NRIs), selective serotonin reuptake inhibitors (SSRIs), combination serotonin and norepinephrine reuptake inhibitors (SNRIs), and older drugs like tricyclic antidepressants and monoamine oxidase inhibitors.317,318 New-generation antipsychotics and drugs targeting the alpha2-adrenergic autoreceptor like mirtazapine are also used.319 In 2019, the Food and Drug Administration in the United States approved Ketamine, a newer medication for patients with depression and suicidal tendencies not responding to standard antidepressants. It is reported that almost 60% of patients fail to recover after undergoing one antidepressant trial,320–322 and misleading reviews have ignored a vast body of efficacy data and even questioned whether antidepressants work at all.323,324 Accordingly, a better understanding of how antidepressants work and why they do not work in some patients is needed. Some evidence suggests that SSRIs, SNRIs, or ketamine may exert their influence by affecting miRNAs.249,325–328
Noncoding RNA modulation in response to SSRIs
Application of escitalopram in depression of patients altered the levels of 30 miRNAs,329 have triggered studies finding that miRNAs may be downstream regulators mediating the antidepressant effects of SSRIs.
Fluoxetine upregulated the expression of miR-16 in the raphe nucleus of mouse,330 and downregulated miR-598-5p and miR-451 in hippocampus of mouse brain.331 In vitro, fluoxetine upregulated miR-320a, miR-663a, miR-572, and miR-489 in both SK-N-SH and SHSY5Y cell lines.332 In addition, fluoxetine can regulate hippocampal lncRNA levels to improve CUMS-induced depression symptomatology.333 Among them, lncRNA Gm26917 was significantly upregulated. Paroxetine upregulated the expression of miRNA-451a and downregulated the expression of miRNA-221-3p and miRNA-34a-5p.334 A relationship is reported between clinical symptom changes and miRNA expression levels.334 Higher miR-1202 levels in MDD patients are reported following eight weeks of citalopram, but no differences in controls.281 Plasma miR-132 levels MDD were reduced after two months of citalopram and plasma miR-124 levels increased.248 Citalopram increased miRNA-16 in mouse brains and decreased SERT protein levels, while exerting antidepressant effects.330 30 miRNAs were expressed differently in the blood of MDD patients following three months of escitalopram therapy.329 Escitalopram administration for four weeks normalized abnormal levels of miR-326 in the NAc of depressed rats, indicating that miR-326 may be a novel target of escitalopram.335 The problem with most studies is that they cannot distinguish between miRNA normalization as part of the antidepressant action or miRNA normalization as a consequence of clinical improvement in the patient.
Noncoding RNA modulation in response to SNRIs
It has been suggested that SNRIs have better therapeutic efficacy and higher tolerability compared to commonly used antidepressants since of their impact on both norepinephrine (NE) and 5-HT.336–338 One of the SNRIs widely used in clinical settings is duloxetine.339 Downregulation of miR-3074-5p, miR-146a-5p, miR-24-3p, miR-425-3p, and miR-146b-5p expression in the peripheral blood of depression patients after duloxetine treatment was found.340 Administration of duloxetine to rats undergoing CUMS upregulated miR-18a and miR-132 in hippocampus, and downregulated levels of miR-124a and miR-134.341 Desvenlafaxine reduced miR-1202 levels in depressed individuals after two months of treatment.342 Furthermore, miR-1202 levels in peripheral blood were associated with alterations in brain activity and connectivity in several brain regions.343 Perhaps miR-1202 levels may mediate some of the desvenlafaxine’s antidepressant effect through brain circuitry function, possibly via the glutamatergic system.342
Noncoding RNA modulation in response to ketamine for depression
Ketamine inhibits the N-methyl-D-aspartate (NMDA) receptor, enhances mushroom spine growth in 1–2 days via the mTOR pathway and is known to reduce depression symptoms quickly and robustly.343 Ketamine downregulated 18 miRNAs levels and upregulated 22 miRNAs levels in rats.344 MiR-206 suppressed ketamine’s ability to up-graduate BDNF expression and is downregulated by ketamine.344 Thus, miRNA-206 may moderate ketamine’s antidepressant effect. Ketamine can also increase the expression of miR-29b-3p and result in lower levels of metabotropic glutamate receptor 4 (GRM4) in the prefrontal cortex of depressed rat model, suggesting that the lncRNA Gm26917/miR29b-3p/GRM4 pathway may plays a critical role in both fluoxetine and ketamine therapies in CUMS rats.345 Understanding how lncRNA regulation is related to antidepressant treatment effects has become increasingly important to find potential therapeutic targets to improve the treatment of MDD.
RNA modifications participate in the molecular mechanisms underpinning MDD
There are over 100 types of RNA modifications.346 Due to their ubiquitous role in cell biology, RNA modifications have been associated with the development of various illnesses, such as cancer, neurological and developmental disorders, and metabolic diseases.347 To date, among all RNA modifications, N6-methyladenosine (m6A) accounts for two-thirds of all RNA modifications that can be “written”, “read”, and “erased” via the actions of a complex network of proteins348,349 and has been widely studied since it was discovered and proposed by Desrosiers in 1975.350 M6A is a methylation modification of the N at the sixth position of adenosine, which exists not only in mRNA, but also in tRNA, rRNA, and lncRNA.351 m6A interacts with different reading proteins and related complexes to broadly influence gene expression at multiple levels. There are also many m6A modification sites on the mRNA of some proteins that regulate histone modification. Therefore, inhibition of the m6A-regulating enzymes may result in increased or decreased histone modification levels.352 Here, we review prospective evidence concerning the involvement of RNA modifications, especially m6A, in the pathogenesis of depression (Fig. 6), drawing on studies from different species and multiple experimental designs. We discuss the biological effects of RNA modifications in pharmacological and nonpharmacological antidepressant therapies.
Fig. 6.
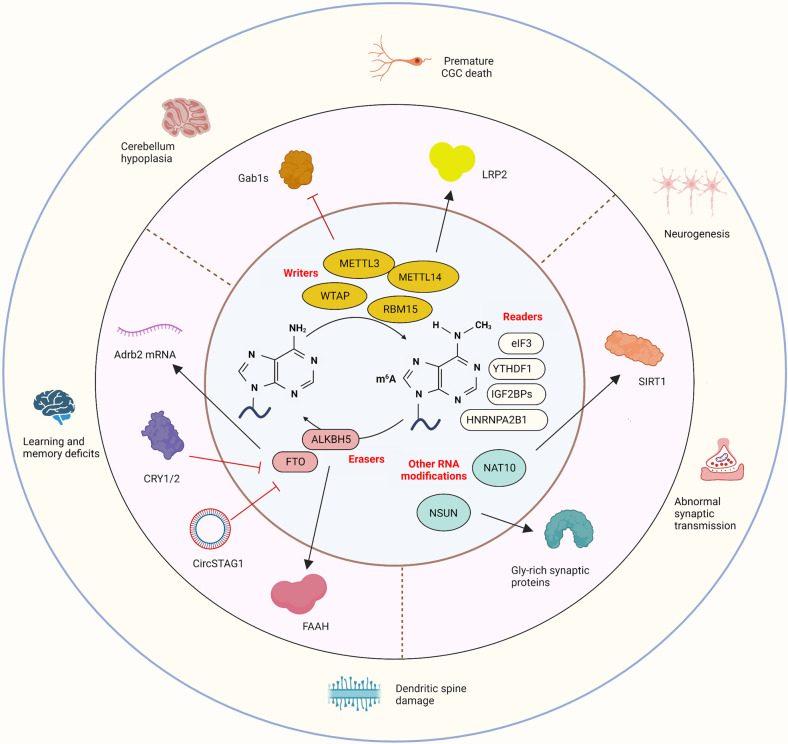
Schematic diagram illustrating how the molecular basis of m6A and other reported RNA modifications relates to depression. The modification of m6A in MDD is regulated by the action of a complex network of proteins: “writers”, which include METTL3, METTL14, WTAP and RBM15–methylate RNA; “erasers”, which include ALKBH5 and FTO–demethylate RNA; and “readers”, which include eIF3, YTHDF1, IGF2BPs and HNRNPA2B1-recognize m6A. Other RNA modifications associated with MDD include N4-acetylcytidine (ac4C) and 5-methylcytosine (m5C). NAT10 can regulate mRNA translation efficiency by catalyzing ac4C while NSUN catalyzes m5C. The molecular consequences of these enzymes involve a variety of pathophysiological mechanisms related to MDD. METTL3 methyltransferase-like 3, METTL14 methyltransferase-like 14, WTAP Wilms tumor 1-associated protein, RBM15 RNA-binding motif protein 15, FTO obesity-associated protein, ALKBH5 Alkb homolog 5, eIF3 eukaryotic initiation factor 3, YTHDF1 YT521-B homology N6-Methyladenosine RNA Binding Protein 1, IGF2BPs insulin-like growth factor 2 mRNA binding proteins, CGC cerebellar granule cells, SIRT sirtuin, Adrb2 Adrenoceptor 2, CRY1/2 circadian regulator cryptochrome 1 and 2, FAAH fatty acid amide hydrolase, LRP2 lipoprotein receptor-related protein 2, Gab1s growth factor receptor-bound protein 2 associated binding protein 1
Preclinical and clinical research on m6A modification in depression
Writers
Scientists have previously shown that the consensus recognition sites for methyltransferase complexes are highly conserved in most eukaryotes.353 Core m6A methyltransferase complex components include methyltransferase-like 14 (METTL14), Wilms tumor 1-associated protein (WTAP), methyltransferase-like 3 (METTL3), and RNA-binding motif protein 15 (RBM15).354 The complex employs S-adenosine methionine (SAM) as a methyl donor to catalyze the formation of methyl groups on the sixth nitrogen element of adenine present in RNA. As the earliest identified component of m6A methyltransferase-possessing methylation catalysts, METTL3 plays a role in neural development, and its absence causes hypoplasia of the cerebellum in mice, resulting in ataxia.355 METTL3 catalyzes the m6A modification of the low-density lipoprotein receptor-related protein 2 (LRP2) mRNA to improve its stability and efficiency in translation. This process relies on reader protein YTH domain containing 2 (Ythdc2), thereby facilitating neurogenesis.356 A deficit of METTL3 in mice lessens hippocampal neurogenesis, which induces spatial memory decline, and depression-like behaviors. Targeting the METTL3-Ythdc2-LRP2 axis to regulate neurogenesis may be a promising antidepressant strategy.356 Evidence from other studies suggests that upregulated METTL3 can aggravate cognitive impairment in rats exposed to CUMS by mediating m6A modification to promote the processing and maturation of pri-miR-221. In addition, this upregulation of METTL3 could increase miR221-3p levels, leading to growth factor receptor-bound protein 2 associated binding protein 1 (Gab1s) inhibition.357
Erasers
RNA modifications are removed by demethylases, allowing dynamic adjustment. The m6A erasers associated with mental stress disorders primarily consist of fat mass and obesity-associated protein (FTO) and Alkb homolog 5 (ALKBH5). The FTO variant rs9939609 in human trials was associated with a lower risk of developing MDD.358 A correlation is reported between FTO variants and the severity of MDD.359 Lack of the FTO gene impacts anxiety-depression-like behavior in mice.360 Adrenoceptor 2 (Adrb2) mRNA appears to be a target of FTO. FTO expression is also related to cognition. Activation of circadian regulator cryptochrome 1 and 2 to inhibit FTO transcription, which in turn inhibits the expression of TrκB in the hippocampus through m6A-dependent posttranscriptional regulation, resulted in impaired cognition in mice.361 Anxiety-like and depression-like behavior induced by peripheral nerve injury could be reversed in mice by blocking FTO downregulation in the anterior cingulate cortex.362 The m6A demethylase gene ALKBH5 variant rs12936694 showed an allelic association and genotypic association with MDD.360,363 Huang et al. reported that circular RNA STAG (circSTAG1) overexpression hindered the translocation of ALKBH5 into the nucleus, leading to elevated m6A methylation of fatty acid amide hydrolase (FAAH) mRNA and degradation of FAAH in astrocytes followed by reduced depression-like behavior and reduced astrocyte loss.296
Readers
Readers can identify modification signals that react to RNA output, splicing, translation, and degradation by preventing the writer or eraser from adding the modified base, or by recruiting other RNA-binding proteins to facilitate the chemical modification of the RNA. The heterogeneous nuclear ribonucleoprotein (HNRNP) protein family, YT521-B homology domain family (YTHDF), insulin-like growth factor 2 mRNA binding proteins (IGF2BPs) and eukaryotic initiation factor 3 (eIF3) are all members of the “reader” subfamily of m6A modification enzymes. According to a previous study, the HNRNP proteins HNRNPA2B1 and HNRNPC exhibit selective binding to mRNAs containing m6A.364,365 HNRNPA2B1 may be one of eight genes associated with postpartum depression and may have diagnostic value.366 By accelerating the translation of specific transcripts in the adult mouse hippocampus, YTHDF1 enhances learning and memory of neuronal stimuli, whereas its absence affects synaptic transmission and long-term enhancement in the hippocampus, leading to deficiencies in learning and memory.367
RNA modifications other than m6A are also involved in depression
RNA modifications other than m6A have been poorly studied in the field of depression with scant coverage of modifications such as N4-acetylcytidine (ac4C) and 5-methylcytosine (m5C). N-acetyltransferase 10 (NAT10) is a member of the general control non-repressible 5 (GCN5)-related N-acetyltransferase (GNAT) family involved in epigenetic events.368 It can regulate mRNA translation efficiency by catalyzing the addition of ac4C to the N4 position on thymidines. In a study conducted on mice, chronic mild stress (CMS) was used to trigger anxiety and depression-like behavior, and the results indicated an increase in NAT10 expression in the hippocampus following the administration of CMS. Pharmacological methods prevented NAT10’s anti-anxiety and antidepressant-like effects.369 As one of the most abundant modifications on tRNA and rRNA, m5C is enzymatically catalyzed by members of the NOL1/NOP2/SUN domain (NSUN) family and DNA methyltransferase homolog DNMT2.370 By altering the translational dynamics of Glycine (Gly)-rich synaptic proteins, the impaired cortical NSUN2, which is a tRNA methyltransferase, exerts bidirectional impacts on tRNA methylation and induces depression-like behavior in the mouse brain.371
Antidepressant treatments are associated with RNA modification
Altered epigenetic modification of m6A mRNA may contribute to therapeutic responses in psychiatric disorders. The complex protein network that maintains the m6A homeostasis can also be served as therapeutic target for depression potentially. Altered levels of RNA demethylase, the “erasers”, are the most frequently reported m6A-regulating enzymes associated with antidepressant effects. Tricyclic antidepressants increase FTO expression and activate its epigenetic function in the VTA; eliminating m6A modification in the VTA is postulated to cause an antidepressant effect.372 Moreover, activation of FTO in the hippocampus relieves depression-like behaviors and reduces the density of dendritic spines and the number of branches in mice induced by chronic restraint stress, improving the synaptic plasticity.373 In contrast, only a few studies reported the association between the writer and the eraser protein and effects of antidepressants. One study found that physical exercise alleviates anxiety-like behaviors and restores m6A levels in mouse medial prefrontal cortex; the restoration of m6A is related to increased expression of RNA Methylase (i.e., METTL3, METTL14, METTL16) and downregulation of RNA demethylase (i.e., ALKBH5), as well as upregulated expression of the RNA-binding factor YTHDF3.374 Another study showed that NAc-deep brain stimulation (DBS) could reverse the impacts of CUMS on gene expression and m6A-mRNA modification for certain genes, but the m6A-regulating enzymes were not detected.375
Chromatin remodeling factor alteration is associated with MDD
Modifying chromatin structure by ATP-dependent protein complexes is also recognized as an essential epigenetic process. These protein complexes depend on ATP hydrolysis to control the histone arrangement, steering a dynamic process of DNA accessibility.376 ATP-dependent complexes can be classified into four groups (referring to the specificity of distinct ATPase subunits): switch/sucrose nonfermentable (SWI/SNF), imitation switch (ISWI), inositol (INO80) and chromodomain helicase DNA-binding (CHD).376,377 Among these, ATP-utilizing chromatin assembly and remodeling factors (ACF) are the most extensively studied protein complexes of the ISWI family. It consists of various subunits, including the bromodomain adjacent to zinc finger domain 1A (BAZ1A). Mice with ACF overexpression in NAc exhibit depression-like behaviors after social defeat stress.378 This study also showed that ACF overexpression regulates nucleosome architecture at transcriptional start sites, inhibiting the expression of a subset of genes, and increases susceptibility to stress-induced depression in mice and humans.378 The inactivation of ATPase subunit of the SWI/SNF complexes, Brahma Related Gene 1, reduces the expression of stress- and cocaine-induced immediate early genes, increases heterochromatin levels, and generally reduces chromatin accessibility in striatal medium spiny neurons.379 Other studies reported altered levels of selective ATP-dependent chromatin remodeling factors in mice with high anxiety-related behavior, including SNF2H in the amygdala and CHD3 and CHD5 in the ventral hippocampus.380 However, the impact of ATP-dependent nucleosome remodeling complexes on depression is not well-understood, and more research is needed to explore this area.
A translational perspective on epigenetic modifications in MDD
The molecular machinery and epigenetic mechanisms related to MDD have become more understood; it is possible to test whether these epigenetic alterations have the translational potential to diagnose and aid in the treatment of MDD. Therapeutic strategies targeting writers, erasers and readers of different epigenetic components hold promise in the context of major depression and related cognitive impairments (Fig. 7).
Fig. 7.
Targeting epigenetic modifications by potential antidepressants and relevant behavioral changes observed in rodents. a Different HDAC inhibitors, including MS-275, sodium, and butyrate can block histone deacetylation and promote the expression of target genes. DNMT inhibitors, including cannabidiol, s-adenosyl methionine and quetiapine activate DNA demethylation patterns to promote the expression of target genes. b These epigenetic variations lead to important recovery in the neural circuits, synaptic plasticity, astrocyte morphological changes, etc. c Improved depression-like behaviors in rodents after implementing drugs interfering with epigenetic mechanisms
Biomarkers for the diagnosis of depression
Large numbers of clinical trials are currently in progress to determine whether DNA methylation, miRNAs and lncRNAs are biomarkers for different types of cancer,381–383 perhaps profiles of such molecules could also be used for diagnosing depression.220,384 It is imperative that a biomarker for the diagnosis of depression be specific for MDD, determined by whether it is a state or trait marker and whether it is affected by antidepressant treatments.106 Since blood samples can be collected easily, they are attractive options for diagnostic biomarkers.385,386 Because miRNA and lncRNA levels in blood distinguish depressed patients from healthy controls,243,310,387 for a diagnostic tool, it is not essential if blood and brain exhibit significant similarities.388,389 Results are summarized in Table 2.
Table 2.
Summary of promising epigenetic biomarkers for diagnosis in depressed patients or depression-like animals
| ncRNA | Species | Tissue | Mechanism/effects | References |
|---|---|---|---|---|
| miR-124↑ | Human | Serum | ↑miR-124 → SAT1, SMOX genes → altered synaptic plasticity | 465–469 |
| Rodents | Peripheral blood mononuclear cells | ↑miR-124 → BDNF-TrkB signaling pathway → inhibit neurogenesis | ||
| Plasma | ↑miR-124 → neuronal differentiation | |||
| Prefrontal cortex (BA46) | ↑miR-124 → synaptogenesis and neuronal proliferation; | |||
| Hippocampus | ↑miR-124 → microglial activation | |||
| miR-139-5p↑ | Human | Prefrontal cortex (BA44) | ↑miR-139-5p → inhibit neural stem cell proliferation and neuronal differentiation | 245,470 |
| Rodents | Blood-derived exosome | ↑miR-139-5p → target SAT1 and SMOX genes → influence neurotransmitter transmission | ||
| Hippocampus | ||||
| Blood-derived exosome | ||||
| miR-221↑ | Human | Prefrontal cortex (BA10) | ↑miR-221 → activate the Wnt2/CREB/BDNF axis → depression symptom | 334,471–474 |
| Rodents | Cerebrospinal fluid | ↑ miR-221 → activate the IRF2/IFN-a pathway → depression symptom | ||
| Cerebrospinal fluid | ||||
| Serum | ||||
| Serum | ||||
| Hippocampus | ||||
| miR-218↓ | Human | Prefrontal cortex (BA44) | ↓miR-218 → target Netrin-1 guidance cue receptor DCC | 266,475 |
| Rodents | Plasma | ↓miR-218 → regulating density of thin dendritic spines | ||
| Prefrontal cortex (BA44) | ||||
| Medial prefrontal cortex | ||||
| miR-17-5p↓ | Human | Locus coeruleus | ↓miR-218 → target CREB1, CHRM2, NTRK3, and SLC17A7 genes | 247,476 |
| Plasma | ||||
| miR-335↓ | Human | Prefrontal cortex (BA9) | ↓miR-335 → target GRM4, SOX4, PTPRN2, and MERTK genes | 244,477 |
| Whole blood | ||||
| miR-1202↓ | Human | Prefrontal cortex (BA44) | ↓miR-1202 → target GRM4 gene | 340,478 |
| Serum | ↓miR-1202 → regulating the metabolism of glutamate | |||
| miR-135-a↓ | Human | Dorsal raphe/raphe magnus | ↓miR-135-a → Serotonin transporter and serotonin receptor-1a transcripts | 479 |
| Whole blood | ||||
| miR-184↓ | Human | Anterior cingulate cortex | ↓miR-184 → target NCOR2 and PDE4B genes | 478,480 |
| Plasma | ||||
| Serum | ||||
| miR-34c-5p↑ | Humans | Peripheral blood leukocytes | ↑miR-34c-5p → target SAT1, SMOX, and NOTCH1 | 238,245 |
| Prefrontal cortex (BA44) | ||||
| miR-24-3p↑ | Human | Prefrontal cortex (BA44) | ↑miR-24-3p → active MAPK/Wnt signaling pathway | 243,340 |
| Whole blood | ||||
| miR-146a↓ | Human | Prefrontal cortex (BA9) | ↓miR-146a → TLR4 signaling pathway | 244,481 |
| Peripheral blood mononuclear cells | ||||
| miR-425-3p↑ | Human | Prefrontal cortex (BA44) | ↑miR-425-3p → active MAPK/Wnt signaling pathway | 243,340,482 |
| Whole blood | ||||
| Peripheral blood mononuclear cells | ||||
| RP1-269M15.3↑ | Human | Frontal cortex | Not reported | 483 |
| Nucleus accumbens gangli | ||||
| RMRP↓ | Human | Peripheral blood leukocytes | Not reported | 312 |
| Mouse | Blood | |||
| Y5↓ | Human | Peripheral blood leukocytes | Not reported | 312 |
| MER11C↓ | ||||
| PCAT1↓ | ||||
| PCAT29↓ | ||||
| LINC01108↑ | Human | Peripheral blood cells | Not reported | 484 |
| LINC00998↓ | Human | Peripheral blood cells | Not reported | 484 |
| LINC00473↓ | Mouse | Prefrontal cortex (PFC) neurons | Not reported | 311 |
| TCONS_00019174↑ | Human | PBMCs | ↑ TCONS_00019174 → Regulate the phosphorylated GSK3β protein and β-catenin in the hippocampus → depression-like behaviors | 485 |
| ENST00000566208↑ | ||||
| NONHSAG045500↑ | ||||
| ENST00000517573↑ | ||||
| NONHSAT034045↑ | ||||
| NONHSAT142707↑ | ||||
| ENST00000505825↑ | Human | PBMCs | Not reported | 485 |
| NONHSAG017299↑ | ||||
| NONHSAT078768↑ | ||||
| lncRNAGm26917↑ | Mouse | Hippocampus | Not reported | 345 |
Biomarkers for predicting the response to antidepressant treatment
The ideal epigenetic molecular marker for antidepressant treatment should satisfy three conditions: (1) the expression level in the blood changes significantly after taking antidepressant drugs; (2) the expression level in the brain tissue changes significantly after taking antidepressant drugs; and (3) the molecular mechanism(s) of DNAm/ncRNA-mediated antidepressant effects have been clarified. However, the key elements for a convenient blood-based biomarker, are that it shows a robust relationship to clinical response. We summarize the DNAm/ncRNAs findings in blood or brain tissue related to antidepressant response in Table 3. The findings suggest that some epigenetic marks could be helpful in predicting responses to psychotherapeutic intervention.390–392
Table 3.
Promising epigenetic biomarkers for predicting antidepressant treatment
| DNAm/ncRNA | Antidepressants | Species | Sample type | Antidepressant mechanism | References |
|---|---|---|---|---|---|
| BDNF Exon IV promoter | SSRI, SNRI Mirtazapine, TCA, Monoamine oxidase inhibitor | Human | Peripheral blood | DNAm at CpG-87↓ → higher nonresponse probability | 486 |
| BDNF Exon IV promoter | Escitalopram | Human | Peripheral blood | DNAm percentage at BDNF exon VI ↓ → higher rates of remission | 487 |
| SLC6A4 exon 1A | Selective serotonin reuptake inhibitor | Human | Peripheral blood | DNAm at SLC6A4 exon 1A ↓ → impaired response to antidepressant treatment | 488 |
| SLC6A4 promoter | Not mentioned | Human | Peripheral blood | CpG-2 methylation ↑ → higher clinical improvement with therapy | 489 |
| HTR1A promoter regions | Escitalopram | Human | PBMCs | DNAm at four CpG sites within the HTR1A/1B promoters ↑ → remission, DNAm at CpG-668 and CpG-1401 ↓ → poor treatment response | 490 |
| HTR1B promoter regions | Fluoxetine | Human | Peripheral blood | Negative association between the clinical response as assessed by GAF3/CGAS4 and the average DNAm at the HTR1B promoter | 491 |
| HTR1B promoter regions | Escitalopram | Human | PBMCs | DNAm at HTR1B_2 amplicon CpG-100 and HTR1B_4 amplicon CpG-1401 ↑ → remission | 490 |
| Interleukin-11 (IL11) | Escitalopram or nortriptyline | Human | Peripheral blood | DNAm at CpG-5 ↓ → better antidepressant response, DNAm at CpG-4 ↑ → better antidepressant response, but with worse response in those taking nortriptyline. | 492 |
| MAOA exon promoter | Escitalopram | Human | Peripheral blood | DNAm at CpG-1 CpG-5 ↓ → worse treatment response in females. | 493 |
| PPFIA4 exon I | Paroxetine | Human | Peripheral blood | DNAm within PPFIA4 ↑ → worst responders | 494 |
| CHN2 | Escitalopram | Human | Peripheral blood | Responders showed relative decreases in both DNA methylation and mRNA expression at two CpG probes (cg23687322; cg06926818) compared to non-responders | 495 |
| JAK2 | Escitalopram | Human | Peripheral blood | In comparison to non-responders, responders showed relative reductions in DNA methylation and mRNA expression at one CpG probe (cg08339825). | 495 |
| RHOJ 5ʹ-UTR and first exon | Fluoxetine | Human | Peripheral blood | RHOJ displayed four CpGs in non-responders that were noticeably hypermethylated. | 496 |
| OR2L13 | Fluoxetine | Human | Peripheral blood | OR2L13 presented three CpGs that were significantly hypomethylated in non-responders | 496 |
| SORBS2 | Escitalopram | Human | Peripheral blood | Seven CpG-sites in an enhancer region of SORBS2, differentially methylated between responders and non-responders, and hypermethylated in the responder group | 497 |
| miR-335↑ | Citalopram | Human | Blood samples | miR-335↑ → inhibit GRM4 expression | 477 |
| miR-135↑ | SSRI | Human | Blood and brain samples | miR-135↑ → regulate the serotonin transporter and serotonin receptor | 479 |
| miR-124↑ | Citalopram | Human | Blood samples | miR-124↑ → decrease the expression of GR | 498 |
| Gypenosides | Mice | ||||
| Blood samples | |||||
| miR-155↓ | Citalopram | Human | Neural progenitor cells Hippocampal neurons | miR-335↓ → increase SIRT1 expression | 499,500 |
| Saikosaponin d | Rats | miR-335↓ → increase the expression of FGF2 | |||
| miR-16↑ | citalopram | Rats | brain samples | miR-16↑ → decrease SERT protein levels | 330 |
| miR-29b-3p↑ | Ketamine | Rats | Prefrontal cortex and primary neurons | miR-29b-3p↑ → decrease the expression of GRM4 | 345 |
| miR-18↓ | Kampo medicine Yokukansan | Mice | hypothalamus | miR-18↓ → regulate the expression of GR | 501 |
| miR-16↑ | Dingzhi Xiaowan | Rats | Hippocampus | miR-16↓ → inhibit the reuptake of 5-HT | 502 |
| hippocampal neurons | |||||
| miR-144-3p↓ | Ketamine | Mice and human | Peripheral blood | miR-144-3p ↓ → prior to stress and following ketamine treatment in ketamine responsive mice only | 503 |
| hsa_circRNA_103636 | Citalopram | Human | Peripheral blood | Not reported | 504 |
| rno_circRNA_014900 | Ketamine | Rats | Hippocampus | rno_circRNA_014900 ↑, rno_circRNA_005442 ↓ →Wnt signaling, long-term depression, PI3K-Akt signaling, etc. | 505 |
| rno_circRNA_005442 | |||||
| circDYM | Repetitive transcranial magnetic stimulation | Human | Peripheral blood | Baseline plasma circDYM levels positively correlated with the scores of depression and retardation. | 506 |
Drugs interfering with epigenetic mechanisms as potential antidepressants
Several antidepressants targeting DNA methylation are in clinical development, including quetiapine (drug repurposing), S-adenosyl methionine, and cannabidiol.
Quetiapine
An atypical antipsychotic, quetiapine, exerts an antidepressant effect and has the protective effect of reducing epigenetic changes induced by stress in early life. Rats with maternal deprivation showed depression-like behavior in the forced swimming test (FST), and increased activity of HDACs and DNMTs in the hippocampus and NAc. Quetiapine reversed depression-like behavior in mice and reduced DNMT activity in the hippocampal region.
S-adenosyl methionine
S-adenosylmethionine (SAM) serves as a methyl donor with a broad range of applicability and is occasionally administered to individuals who exhibit unresponsiveness to primary antidepressant treatments. The anti-inflammatory actions of SAM might be beneficial in treating depression. SAM controls the activation of inflammatory genes by inducing changes in DNA methylation at specific gene promoters. SAM reduces the expression of the proinflammatory cytokine TNFa and the chemoattractant CCL2 and its receptor CCR2, in association with DNA methylation changes in specific gene promotor.393 In contrast, DNA methylation by SAM increases IL-10 expression, an anti-inflammatory cytokine.393
Cannabidiol
Cannabidiol (CBD) is cannabis’ most abundant non-psychoactive component.394 CBD’s therapeutic potential for reducing depression-like behavior has been shown, since it interacts with a wide range of brain molecules395,396. In one study,397 stress induced by FST increased levels of global DNA methylation levels and DNMT activity in the hippocampus while decreasing DNA methylation and DNMT activity in the prefrontal cortex. Interestingly, CBD administration prevented stress-induced epigenetic changes in both the hippocampus and PFC.397 A preliminary study reported that several histone modifications were changed in some brain regions of rats after CBD administration.398 But the molecular mechanism of CBD action remains to be investigated.
DNMT inhibitor
Different DNMT inhibitors (DNMTi) inhibit DNMTs through different mechanisms, including nucleosides and nonnucleosides. Nucleosidic DNMTi, including 5-AzaC, 5-AzaD, and zebularine, are chemical analogs of cytidine, which are integrated into DNA molecules during replication (S-phase). Their covalent binding to the DNMTs prevents DNA methylation and irreversibly blocks these enzymes.399 It has been demonstrated that 5-AzaD and 5-AzaC lead to a decrease in DNA methylation and increased BDNF expression in the hippocampus, displaying effects similar to those of antidepressants in several preclinical settings.400 In addition, injecting zebularine into the cerebral ventricles of adult rats for seven days reduced DNA methylation of the BDNF promoter.401 The reduced methylation of the promoter enabled this drug regimen to restore BDNF expression, suggesting the potential of zebularine as a therapeutic agent in clinical use. Some nonnucleoside inhibitors, such as RG108 and procaine, have different mechanisms of action, including noncovalent inhibition within the DNA catalytic sites, to impede enzymatic activity. RG108 alters stress-induced DNA methylation bidirectionally in the hippocampus and PFC and triggers antidepressant-like effects as demonstrated in the forced swimming test.402
The therapeutic potential of HDAC inhibitors (HDACis) is suggested because HDACis may augment the efficacy of antipsychotic medications on coadministration.403,404 Animal models and postmortem brain results show some HDACis exert antidepressant-like effects (Table 4, Fig. 7). HDACis, such as sodium butyrate and SAHA, promote cognitive function, and may provide therapeutic options for depressed patients with cognitive impairment.405–407 Although medications targeting these epigenetic molecules are proving effective in preclinical animal studies, there is a lack of successful clinical trials of HDACi in neuropsychiatric disorders, perhaps because of the complexity of different HDAC isoforms (i.e., HDAC2 and HDAC5) that are associated with pro-depressant and antidepressant functions.408 The complexity is further heightened because HDAC and DNMT are widely distributed in the CNS and peripheral tissues. Interference in nonneuronal or inappropriate neuronal locations may induce side effects and off-target effects.409 Drugs that target brain tissue-specific epigenetic modulators may ultimately prove to be helpful new therapies for depression, but the studies that show this remain to be done.
Table 4.
Drugs targeting epigenetic processes as a therapeutic strategy for depression
| Target | Drug | Species | Molecular Mechanisms of Action | Reference |
|---|---|---|---|---|
| Class I HDAC inhibitor | MS-275 | Mice | ↑H3 acetylation in the mPFC, exerting antidepressant-like effects | 507 |
| ↑Rac1 in the NAc, synapse structural plasticity normalization | 158 | |||
| ↑H3 acetylation in the hippocampus and the NAc, exerting antidepressant-like effects | 127,130 | |||
| Class I and II HDAC inhibitor | Sodium butyrate | Mice | HDAC5 downregulation, ↑H3 acetylation in BDNF gene promoter, ↓depression-like behavior | 148 |
| ↓Depression-like behavior, ↑HDAC2, ↑pCREB, ↑H3 acetylation, ↑BDNF in the hippocampus | 508 | |||
| Rats | ↓Depression-like behavior, ↑transthyretin (Ttr) expression, ↓serotonin 2A receptor, ↑H4 acetylation at Ttr gene promoter | 129 | ||
| SAHA | Mice | ↑H3 acetylation in the hippocampus and the NAc, exerting antidepressant-like effects, | 130 | |
| ↑Gdnf in the NAc, HDAC2 inhibition | 104 | |||
| ↓Depression-like behavior, ↑BDNF in the PFC | 509 | |||
| Valproic Acid | Rats | ↓Depression-like behavior, ↓corticosterone plasma level | 510 | |
| HDAC1/2 inhibitor | Cpd60 | Mice | ↑Histone acetylation at the promoter regions of upregulated transcripts | 511 |
| HDAC4/5 inhibitor | LMK-235 | Mice | ↓Depression-like behavior | 469 |
| SIRT2 inhibitor | 33i | Mice | ↑Serotonin levels, ↑glutamate receptor subunit expression, ↓depression-like behavior | 512 |
| SIRT1 activator | SRT2104 | Mice | Exerting antidepressant-like effects, ↓spine loss, ↑phosphorylation level of protein kinases 1 and 2 in the stressed condition | 151 |
| DNMT inhibitor | Zebularine, RG108 | Mice | Exerting antidepressant-like effects, ↓DMNT at Gdnf promoter | 104 |
| RG108 | Exerting antidepressant-like effects | 99 |
Concluding remarks and future directions
Environmental stressors and genetic factors shape the complex phenotype of MDD. Specifically, epigenetic processes shape and store a cell’s molecular response to its environment. The prevailing hypothesis suggests that epigenetic processes leave imprints on the genome, which then interact with an individual’s genetic predisposition to determine their susceptibility to depression over the course of their lifetime. Therefore, exploring epigenetic mechanisms provides fresh perspectives on the pathophysiology of MDD and has the potential to generate innovative biomarkers for diagnosis and treatment purposes. Established animal models for depression and video-recorded behavior analyses have greatly advanced the understanding of various epigenetic mechanisms contributing to stress responses, neuroplasticity, neurotransmission, and neuroglial function. Clinical studies have focused on DNA methylation and ncRNA using human peripheral blood samples, with few focusing on histone modifications.410 As MDD is a stress-related neuropsychiatric disorder, validating the employment of peripheral epigenetic molecular alterations as surrogates for what occurs in the brain does not advance our knowledge of pathogenesis. Still, it may be useful in developing biomarkers of disease and treatment response.98
Innovative approaches are necessary to enhance our understanding of the pathogenesis in the brain. Fluorescence-activated cell sorting techniques and single-cell epigenomics can facilitate the examination of DNA methylation various cell types.106,411 Likewise, although the patterns of histone modification usually differ between cell types, most studies conducted have examined these modifications in cell populations that are heterogeneous. A high priority for future studies is to probe the epigenetic landscape by integrating modern low and single-cell technologies in combination with next-generation sequencing, to simultaneously measure multiple molecules such as proteins and RNAs, DNA methylation, or chromatin accessibility.412,413
Some types of epigenetic modifications, such as changes in chromosome conformation are still largely unexplored in MDD.414,415 The chromosome conformation capture (Hi-C) technique can provide information on the 3D chromosome conformation and uncover the interactions between distant genomic regions that affect gene expression. The transposase-accessible chromatin sequencing (ATAC-Seq) assay is also a powerful tool for investigating genome-wide chromatin accessibility.416,417 Recent MeRIP-Seq methods map the m6A-methylated RNA418, offering a new way to explore the transcriptome-wide location of m6A in MDD. Combined with the widely used next-generation RNA sequencing techniques, these techniques for exploring the epigenome may facilitate a more comprehensive understanding of the possible mechanisms that contribute to MDD.
Epigenetic processes require a pool of metabolites, such as acetyl-CoA for histone acetylation and S-adenosyl-methionine for methylation. Recent studies provide evidence for metabolic signaling to chromatin in the brain.419 Lactate-dependent histone lactylation (H4K12a) in microglia promotes the expression of glycolytic genes, leading to microglial dysfunction and cognitive decline.420 Additional research is required to investigate epigenetic pathways that are dependent on metabolism, in the pathogenesis of depression and its potential to mimic the effects of antidepressants. In addition, several studies have established an association between the gut microbiome, the metabolites of the gut flora and epigenetics, revealing the impact of the gut microbiota on ncRNA production.421,422 Extensive evidence demonstrates that the brain-gut-microbiota axis plays a crucial role in the pathophysiology of MDD and other psychiatric disorders.423,424 Hence, investigating the effects of the combination of the gut microbiome and epigenetic modification could potentially shed light on the underlying mechanisms of depression.
Finally, manipulating targeted epigenetic states is crucial to gain a comprehensive understanding of the effects of stress-induced histone mark alterations. As mentioned above, the clinical adaptation of HDAC/DNMT inhibitors is severely constrained by their off-target effects. For example, the true causality of epigenetic signatures at DNA sites that are known to be involved in stress-related disorders can only be examined with the advent of novel mutagenesis tools such as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR-Cas9) and Transcription Activator-Like Effector Nucleases (TALENs).425,426 These technologies facilitate the targeting of epigenetic modifiers to specific genes, brain regions, and time frames to ascertain whether the regulation of epigenetic modifications at a particular locus is accountable for a certain psychiatric disorders.427 The use of CRISPR/Cas9 system enables regulation of gene expression and make epigenetic alterations without introducing DNA double-strand breaks.428 Furthermore, CRISPR-cas9 system can trans-epigenetic remodel the endogenous gene.429 Recently, a new set of epigenetic editor CRISPRoff has been developed, enabling persistent, heritable, and reversible DNA methylation modification and regulation of gene transcription.430 These tools specifically allow the targeting, cutting, and replacing a specific epigenetic-related gene to explore the gene functions in psychiatric disorders. It is believed leveraging these tools can help elucidate more potential epigenetic mechanisms in MDD.
Acknowledgements
Figures in this review were created with Biorender (https://www.biorender.com/). This work was supported by grants from the Ministry of Science and Technology of the People’s Republic of China (2021ZD0201900), 1·3·5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYJC21004, ZYGD22007), the National Natural Science Foundation of China (82120108002, 81821002, 82171527, 82200695), Distinguished Young Scholar of Zhejiang (LR20H090001 to C.W.) and Municipal Key R&D Program of Ningbo (2022Z127).
Author contributions
W.Z., C.W., and C.H. conceived this paper. M.Y. and B.Y. drafted and edited the paper. G.R., J.J.M., L.D.S., and X.D. gave valuable and constructive suggestions for the content of this paper and revised the paper. All authors have read and approved this paper.
Competing interests
Canhua Huang is the editorial board member of Signal Transduction and Targeted Therapy, but he has not been involved in the process of the manuscript handling. The other authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Minlan Yuan, Biao Yang
Contributor Information
Chuang Wang, Email: wangchuang@nbu.edu.cn.
Wei Zhang, Email: weizhang27@scu.edu.cn.
References
- 1.Herrman H, et al. Reducing the global burden of depression: a Lancet–World Psychiatric Association Commission. Lancet. 2019;393:e42–e43. doi: 10.1016/S0140-6736(18)32408-5. [DOI] [PubMed] [Google Scholar]
- 2.Association, A. P. Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-5), Vol. 21 (American Psychiatric Association, 2013).
- 3.Center C, et al. Confronting depression and suicide in physicians: a consensus statement. JAMA. 2003;289:3161–3166. doi: 10.1001/jama.289.23.3161. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, et al. Prevalence of depressive disorders and treatment in China: a cross-sectional epidemiological study. Lancet Psychiatry. 2021;8:981–990. doi: 10.1016/S2215-0366(21)00251-0. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, et al. Prevalence of mental disorders in China: a cross-sectional epidemiological study. Lancet Psychiatry. 2019;6:211–224. doi: 10.1016/S2215-0366(18)30511-X. [DOI] [PubMed] [Google Scholar]
- 6.Organization, W. H. The Global Burden of Disease: 2004 update (World Health Organization, 2008).
- 7.Patel V, et al. Addressing the burden of mental, neurological, and substance use disorders: key messages from Disease Control Priorities, 3rd edition. Lancet. 2016;387:1672–1685. doi: 10.1016/S0140-6736(15)00390-6. [DOI] [PubMed] [Google Scholar]
- 8.Han X, et al. Disease trajectories and mortality among individuals diagnosed with depression: a community-based cohort study in UK Biobank. Mol. Psychiatry. 2021;26:6736–6746. doi: 10.1038/s41380-021-01170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penninx BW, et al. Two-year course of depressive and anxiety disorders: results from the Netherlands Study of Depression and Anxiety (NESDA) J. Affect Disord. 2011;133:76–85. doi: 10.1016/j.jad.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Malone KM, et al. Protective factors against suicidal acts in major depression: reasons for living. Am. J. Psychiatry. 2000;157:1084–1088. doi: 10.1176/appi.ajp.157.7.1084. [DOI] [PubMed] [Google Scholar]
- 11.Covid- Mental Disorders Collaborators. Global prevalence and burden of depressive and anxiety disorders in 204 countries and territories in 2020 due to the COVID-19 pandemic. Lancet. 2021;398:1700–1712. doi: 10.1016/S0140-6736(21)02143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Öngür D, Perlis R, Goff D. Psychiatry and COVID-19. JAMA. 2020;324:1149–1150. doi: 10.1001/jama.2020.14294. [DOI] [PubMed] [Google Scholar]
- 13.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81:484–503. doi: 10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baselmans BML, Yengo L, van Rheenen W, Wray NR. Risk in relatives, heritability, SNP-based heritability, and genetic correlations in psychiatric disorders: a review. Biol. Psychiatry. 2021;89:11–19. doi: 10.1016/j.biopsych.2020.05.034. [DOI] [PubMed] [Google Scholar]
- 15.Howard DM, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 2019;22:343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levey DF, et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat. Neurosci. 2021;24:954–963. doi: 10.1038/s41593-021-00860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monroe SM, Anderson SF, Harkness KL. Life stress and major depression: the mysteries of recurrences. Psychol. Rev. 2019;126:791–816. doi: 10.1037/rev0000157. [DOI] [PubMed] [Google Scholar]
- 19.Schiele MA, Gottschalk MG, Domschke K. The applied implications of epigenetics in anxiety, affective and stress-related disorders - A review and synthesis on psychosocial stress, psychotherapy and prevention. Clin. Psychol. Rev. 2020;77:101830. doi: 10.1016/j.cpr.2020.101830. [DOI] [PubMed] [Google Scholar]
- 20.Danese A, Widom CS. Objective and subjective experiences of child maltreatment and their relationships with psychopathology. Nat. Human Behav. 2020;4:811–818. doi: 10.1038/s41562-020-0880-3. [DOI] [PubMed] [Google Scholar]
- 21.Fonzo GA, Huemer J, Etkin A. History of childhood maltreatment augments dorsolateral prefrontal processing of emotional valence in PTSD. J. Psychiatr. Res. 2016;74:45–54. doi: 10.1016/j.jpsychires.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monroe, S. M., Slavich, G. M. & Georgiades, K. in Handbook of Depression 3rd ed. 296–314 (The Guilford Press, 2014).
- 23.Phillips AC, Carroll D, Der G. Negative life events and symptoms of depression and anxiety: stress causation and/or stress generation. Anxiety Stress Coping. 2015;28:357–371. doi: 10.1080/10615806.2015.1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kendler KS, Karkowski LM, Prescott CAJAJOP. Causal relationship between stressful life events and the onset of major depression. Am. J. Psychiatry. 1999;156:837–841. doi: 10.1176/ajp.156.6.837. [DOI] [PubMed] [Google Scholar]
- 25.Kessler RC. The effects of stressful life events on depression. Annu. Rev. Psychol. 1997;48:191–214. doi: 10.1146/annurev.psych.48.1.191. [DOI] [PubMed] [Google Scholar]
- 26.Nelson J, Klumparendt A, Doebler P, Ehring T. Childhood maltreatment and characteristics of adult depression: meta-analysis. Br. J. Psychiatry. 2017;210:96–104. doi: 10.1192/bjp.bp.115.180752. [DOI] [PubMed] [Google Scholar]
- 27.McKay MT, et al. Childhood trauma and adult mental disorder: a systematic review and meta-analysis of longitudinal cohort studies. Acta Psychiatr. Scand. 2021;143:189–205. doi: 10.1111/acps.13268. [DOI] [PubMed] [Google Scholar]
- 28.Peedicayil J, Grayson DR. An epigenetic basis for an omnigenic model of psychiatric disorders. J. Theor. Biol. 2018;443:52–55. doi: 10.1016/j.jtbi.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klengel T, Binder EB. Epigenetics of stress-related psychiatric disorders and gene x environment interactions. Neuron. 2015;86:1343–1357. doi: 10.1016/j.neuron.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 30.Greally JM. A user’s guide to the ambiguous word ‘epigenetics’. Nat Rev. Mol Cell Biol. 2018;19:207–208. doi: 10.1038/nrm.2017.135. [DOI] [PubMed] [Google Scholar]
- 31.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 35.Yang R, et al. Epigenetic biotypes of post-traumatic stress disorder in war-zone exposed veteran and active duty males. Mol. Psychiatry. 2020;26:4300–4314. doi: 10.1038/s41380-020-00966-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian H, Hu Z, Xu J, Wang C. The molecular pathophysiology of depression and the new therapeutics. MedComm. 2022;3:e156. doi: 10.1002/mco2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiele M, Domschke K. Epigenetics at the crossroads between genes, environment and resilience in anxiety disorders. Genes Brain Behav. 2018;17:e12423. doi: 10.1111/gbb.12423. [DOI] [PubMed] [Google Scholar]
- 38.McGowan PO, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS ONE. 2011;6:e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn EC, et al. Sensitive periods for the effect of childhood adversity on DNA methylation: results from a prospective, longitudinal study. Biol. Psychiatry. 2019;85:838–849. doi: 10.1016/j.biopsych.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alberini CM, Kandel ER. The regulation of transcription in memory consolidation. Cold Spring Harb. Perspect. Biol. 2015;7:a021741. doi: 10.1101/cshperspect.a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan Z, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/S0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 42.Otte C, et al. Major depressive disorder. Nat. Rev. Dis. Primers. 2016;2:16065. doi: 10.1038/nrdp.2016.65. [DOI] [PubMed] [Google Scholar]
- 43.Malhi GS, Mann JJ. Depression. Lancet. 2018;392:2299–2312. doi: 10.1016/S0140-6736(18)31948-2. [DOI] [PubMed] [Google Scholar]
- 44.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz MD, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pradhan S, Bacolla A, Wells RD, Roberts R. Recombinant human DNA (cytosine-5) methyltransferase: I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 48.Fatemi M, Hermann A, Gowher H, Jeltsch A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur. J. Biochem. 2002;269:4981–4984. doi: 10.1046/j.1432-1033.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- 49.Du Q, Luu P-L, Stirzaker C, Clark SJJE. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015;7:1051–1073. doi: 10.2217/epi.15.39. [DOI] [PubMed] [Google Scholar]
- 50.Neul JL. The relationship of Rett syndrome and MECP2 disorders to autism. Dialogues Clin. Neurosci. 2022;14:253–262. doi: 10.31887/DCNS.2012.14.3/jneul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C, et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eckersley-Maslin MA, Alda-Catalinas C, Reik W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 2018;19:436–450. doi: 10.1038/s41580-018-0008-z. [DOI] [PubMed] [Google Scholar]
- 53.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 54.Wu H, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cruceanu C, et al. DNA hypomethylation of Synapsin II CpG islands associates with increased gene expression in bipolar disorder and major depression. BMC Psychiatry. 2016;16:286. doi: 10.1186/s12888-016-0989-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han KM, et al. TESC gene-regulating genetic variant (rs7294919) affects hippocampal subfield volumes and parahippocampal cingulum white matter integrity in major depressive disorder. J. Psychiatr. Res. 2017;93:20–29. doi: 10.1016/j.jpsychires.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 58.Heberle E, Bardet AF. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019;63:727–741. doi: 10.1042/EBC20190033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo JU, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jang HS, Shin WJ, Lee JE, Do JT. CpG and non-CpG methylation in epigenetic gene regulation and brain function. Genes. 2017;8:148. doi: 10.3390/genes8060148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen L, et al. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc. Natl Acad. Sci. USA. 2015;112:5509–5514. doi: 10.1073/pnas.1505909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barker DJ. The fetal and infant origins of adult disease. BMJ. 1990;301:1111. doi: 10.1136/bmj.301.6761.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calkins K, Devaskar SU. Fetal origins of adult disease. Curr. Probl. Pediatr. Adolesc. Health Care. 2011;41:158–176. doi: 10.1016/j.cppeds.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heindel JJ, Vandenberg LN. Developmental origins of health and disease: a paradigm for understanding disease etiology and prevention. Curr. Opin. Pediatr. 2015;27:248. doi: 10.1097/MOP.0000000000000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fagiolini M, Jensen CL, Champagne FA. Epigenetic influences on brain development and plasticity. Curr. Opin. Neurobiol. 2009;19:207–212. doi: 10.1016/j.conb.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fox SE, Levitt P, Nelson CA., III How the timing and quality of early experiences influence the development of brain architecture. Child Dev. 2010;81:28–40. doi: 10.1111/j.1467-8624.2009.01380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanherkar RR, Bhatia-Dey N, Csoka AB. Epigenetics across the human lifespan. Front. Cell Dev. Biol. 2014;2:49. doi: 10.3389/fcell.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu D, et al. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- 72.Zhang T-Y, et al. Maternal programming of defensive responses through sustained effects on gene expression. Biol. Psychol. 2006;73:72–89. doi: 10.1016/j.biopsycho.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 73.Bagot RC, et al. Maternal care influences hippocampal N-methyl-D-aspartate receptor function and dynamic regulation by corticosterone in adulthood. Biol. Psychiatry. 2012;72:491–498. doi: 10.1016/j.biopsych.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 74.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 75.Meaney MJ, Szyf M. Maternal care as a model for experience-dependent chromatin plasticity? Trends Neurosci. 2005;28:456–463. doi: 10.1016/j.tins.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 76.Kurata A, et al. Maternal postpartum learned helplessness (LH) affects maternal care by dams and responses to the LH test in adolescent offspring. Horm. Behav. 2009;56:112–120. doi: 10.1016/j.yhbeh.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 77.Witzmann SR, et al. Epigenetic regulation of the glucocorticoid receptor promoter 1(7) in adult rats. Epigenetics. 2012;7:1290–1301. doi: 10.4161/epi.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lutz PE, et al. Association of a history of child abuse with impaired myelination in the anterior cingulate cortex: convergent epigenetic, transcriptional, and morphological evidence. Am. J. Psychiatry. 2017;174:1185–1194. doi: 10.1176/appi.ajp.2017.16111286. [DOI] [PubMed] [Google Scholar]
- 80.Smith AK, et al. DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2015;168:36–44. doi: 10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hong SR, et al. DNA methylation-based age prediction from saliva: high age predictability by combination of 7 CpG markers. Forensic. Sci. Int. Genet. 2017;29:118–125. doi: 10.1016/j.fsigen.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 82.Aberg KA, et al. Methylome-wide association findings for major depressive disorder overlap in blood and brain and replicate in independent brain samples. Mol. Psychiatry. 2020;25:1344–1354. doi: 10.1038/s41380-018-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ewald ER, et al. Alterations in DNA methylation of Fkbp5 as a determinant of blood-brain correlation of glucocorticoid exposure. Psychoneuroendocrinology. 2014;44:112–122. doi: 10.1016/j.psyneuen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turecki G, Meaney MJ. Effects of the social environment and stress on glucocorticoid receptor gene methylation: a systematic review. Biol. Psychiatry. 2016;79:87–96. doi: 10.1016/j.biopsych.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang TY, et al. Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharmacology. 2013;38:111–123. doi: 10.1038/npp.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei Z, et al. Rufy3, a protein specifically expressed in neurons, interacts with actinpal glucocorticoid receptor gene expression in rodents. J. Neurochem. 2014;130:678–692. doi: 10.1111/jnc.12740. [DOI] [PubMed] [Google Scholar]
- 87.Walton E, et al. Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophrenia Bulletin. 2015;42:406–414. doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mill J, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kang HJ, et al. Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog Neuropsychopharmacol. Biol. Psychiatry. 2013;44:23–28. doi: 10.1016/j.pnpbp.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 90.Olsson CA, et al. Prospects for epigenetic research within cohort studies of psychological disorder: a pilot investigation of a peripheral cell marker of epigenetic risk for depression. Biol. Psychol. 2010;83:159–165. doi: 10.1016/j.biopsycho.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 91.Perez-Cornago A, Mansego ML, Zulet MA, Martinez JA. DNA hypermethylation of the serotonin receptor type-2A gene is associated with a worse response to a weight loss intervention in subjects with metabolic syndrome. Nutrients. 2014;6:2387–2403. doi: 10.3390/nu6062387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kang HJ, et al. Longitudinal associations between BDNF promoter methylation and late-life depression. Neurobiol. Aging. 2015;36:1764.e1761–1764.e1767. doi: 10.1016/j.neurobiolaging.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 93.Peng H, et al. Childhood trauma, DNA methylation of stress-related genes, and depression: findings from two monozygotic twin studies. Psychosom. Med. 2018;80:599–608. doi: 10.1097/PSY.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boström AE, et al. A MIR4646 associated methylation locus is hypomethylated in adolescent depression. J. Affect Disord. 2017;220:117–128. doi: 10.1016/j.jad.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 95.Oh G, et al. DNA modification study of major depressive disorder: beyond locus-by-locus comparisons. Biol. Psychiatry. 2015;77:246–255. doi: 10.1016/j.biopsych.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cordova-Palomera A, et al. Genome-wide methylation study on depression: differential methylation and variable methylation in monozygotic twins. Transl. Psychiatry. 2015;5:e557–e557. doi: 10.1038/tp.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kaut O, et al. Aberrant NMDA receptor DNA methylation detected by epigenome-wide analysis of hippocampus and prefrontal cortex in major depression. Eur. Arch. Psychiatry Clin. Neurosci. 2015;265:331–341. doi: 10.1007/s00406-014-0572-y. [DOI] [PubMed] [Google Scholar]
- 98.Braun PR, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl. Psychiatry. 2019;9:47. doi: 10.1038/s41398-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Poulter MO, et al. GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol. Psychiatry. 2008;64:645–652. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 101.Houseman EA, Molitor J, Marsit C. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30:1431–1439. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Höhne N, et al. FKBP5 genotype-dependent DNA methylation and mRNA regulation after psychosocial stress in remitted depression and healthy controls. Int. J. Neuropsychopharmacol. 2015;18:pyu087. doi: 10.1093/ijnp/pyu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Beurel E, Toups M, Nemeroff CB. The bidirectional relationship of depression and inflammation: double trouble. Neuron. 2020;107:234–256. doi: 10.1016/j.neuron.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Uchida S, et al. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 105.Nagy C, et al. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol. Psychiatry. 2015;20:320–328. doi: 10.1038/mp.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clark SJ, et al. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 2016;17:72. doi: 10.1186/s13059-016-0944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Luger K, Dechassa ML, Tremethick D. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kimura H. Histone dynamics in living cells revealed by photobleaching. DNA Repair. 2005;4:939–950. doi: 10.1016/j.dnarep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 110.Peterson CL, Laniel M-A. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 111.Sabari BR, et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell. 2015;58:203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith BC, Denu JMJ. Be. B. A.-G. R. M. Chemical mechanisms of histone lysine and arginine modifications. Biochim. Biophys. Acta. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weinberg DN, et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019;573:281–286. doi: 10.1038/s41586-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morselli M, et al. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. eLife. 2015;4:e06205. doi: 10.7554/eLife.06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ruijter AJD, et al. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/bj20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Whittle JR, et al. Sirtuins, nuclear hormone receptor acetylation and transcriptional regulation. Trend. Endocrinol. Metab. 2007;18:356–364. doi: 10.1016/j.tem.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 118.Bao J, Sack MN. Protein deacetylation by sirtuins: delineating a post-translational regulatory program responsive to nutrient and redox stressors. Cell. Mol. Life Sci. 2010;67:3073–3087. doi: 10.1007/s00018-010-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sassone-Corsi P. Minireview: NAD+, a circadian metabolite with an epigenetic twist. Endocrinology. 2012;153:1–5. doi: 10.1210/en.2011-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goodyer IM, Herbert J, Tamplin A, Altham P. Recent life events, cortisol, dehydroepiandrosterone and the onset of major depression in high-risk adolescents. Br. J. Psychiatry. 2000;177:499–504. doi: 10.1192/bjp.177.6.499. [DOI] [PubMed] [Google Scholar]
- 121.Wittenborn A, Rahmandad H, Rick J, Hosseinichimeh N. Depression as a systemic syndrome: mapping the feedback loops of major depressive disorder. Psychol. Med. 2016;46:551–562. doi: 10.1017/S0033291715002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Watkeys OJ, et al. Glucocorticoid receptor gene (NR3C1) DNA methylation in association with trauma, psychopathology, transcript expression, or genotypic variation: a systematic review. Neurosci. Biobehav. Rev. 2018;95:85–122. doi: 10.1016/j.neubiorev.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 123.Alexander N, et al. Glucocorticoid receptor gene methylation moderates the association of childhood trauma and cortisol stress reactivity. Psychoneuroendocrinology. 2018;90:68–75. doi: 10.1016/j.psyneuen.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 124.Muratcioglu S, et al. Structural modeling of GR interactions with the SWI/SNF chromatin remodeling complex and C/EBP. Biophys. J. 2015;109:1227–1239. doi: 10.1016/j.bpj.2015.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Q, Timberlake MA, 2nd, Prall K, Dwivedi Y. The recent progress in animal models of depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2017;77:99–109. doi: 10.1016/j.pnpbp.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Covington HE, III, et al. Hippocampal-dependent antidepressant-like activity of histone deacetylase inhibition. Neurosci. Lett. 2011;493:122–126. doi: 10.1016/j.neulet.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schroeder FA, Lin CL, Crusio WE, Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol. Psychiatry. 2007;62:55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 129.Yamawaki Y, et al. Antidepressant-like effect of sodium butyrate (HDAC inhibitor) and its molecular mechanism of action in the rat hippocampus. World J. Biol. Psychiatry. 2012;13:458–467. doi: 10.3109/15622975.2011.585663. [DOI] [PubMed] [Google Scholar]
- 130.Covington HE, 3rd, et al. Antidepressant actions of histone deacetylase inhibitors. J. Neurosci. 2009;29:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Talarowska M, Berk M, Maes M, Gałecki P. Autobiographical memory dysfunctions in depressive disorders. Psychiatry Clin. Neurosci. 2016;70:100–108. doi: 10.1111/pcn.12370. [DOI] [PubMed] [Google Scholar]
- 132.Liu J, et al. Functional connectivity evidence for state-independent executive function deficits in patients with major depressive disorder. J. Affect Disord. 2021;291:76–82. doi: 10.1016/j.jad.2021.04.080. [DOI] [PubMed] [Google Scholar]
- 133.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 134.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gräff J, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sailaja BS, et al. Stress-induced epigenetic transcriptional memory of acetylcholinesterase by HDAC4. Proc. Natl Acad. Sci. USA. 2012;109:E3687–E3695. doi: 10.1073/pnas.1209990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sarkar A, et al. Hippocampal HDAC4 contributes to postnatal fluoxetine-evoked depression-like behavior. Neuropsychopharmacology. 2014;39:2221–2232. doi: 10.1038/npp.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Takase K, Oda S, Kuroda M, Funato H. Monoaminergic and neuropeptidergic neurons have distinct expression profiles of histone deacetylases. PLoS ONE. 2013;8:e58473. doi: 10.1371/journal.pone.0058473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Darcy MJ, Calvin K, Cavnar K, Ouimet CC. Regional and subcellular distribution of HDAC4 in mouse brain. J. Comp. Neurol. 2010;518:722–740. doi: 10.1002/cne.22241. [DOI] [PubMed] [Google Scholar]
- 140.Mathias RA, Guise AJ, Cristea IM. Post-translational modifications regulate Class IIa histone deacetylase (HDAC) function in health and disease. Mol. Cell Proteomics. 2015;14:456–470. doi: 10.1074/mcp.O114.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schlumm F, Mauceri D, Freitag HE, Bading H. Nuclear calcium signaling regulates nuclear export of a subset of class IIa histone deacetylases following synaptic activity. J. Biol. Chem. 2013;288:8074–8084. doi: 10.1074/jbc.M112.432773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Paroni G, et al. PP2A regulates HDAC4 nuclear import. Mol. Biol. Cell. 2008;19:655–667. doi: 10.1091/mbc.e07-06-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sando R, 3rd, et al. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell. 2012;151:821–834. doi: 10.1016/j.cell.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li J, et al. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat. Med. 2012;18:783–790. doi: 10.1038/nm.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fitzsimons HL. The Class IIa histone deacetylase HDAC4 and neuronal function: Nuclear nuisance and cytoplasmic stalwart? Neurobiol. Learn Mem. 2015;123:149–158. doi: 10.1016/j.nlm.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 146.Zhang P, et al. HDAC4 protects cells from ER stress induced apoptosis through interaction with ATF4. Cell Signal. 2014;26:556–563. doi: 10.1016/j.cellsig.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 147.Mielcarek M, et al. HDAC4 reduction: a novel therapeutic strategy to target cytoplasmic huntingtin and ameliorate neurodegeneration. PLoS Biol. 2013;11:e1001717. doi: 10.1371/journal.pbio.1001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tsankova NM, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 149.Renthal W, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 150.Sterrenburg L, et al. Chronic stress induces sex-specific alterations in methylation and expression of corticotropin-releasing factor gene in the rat. PLoS ONE. 2011;6:e28128. doi: 10.1371/journal.pone.0028128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Abe-Higuchi N, et al. Hippocampal sirtuin 1 signaling mediates depression-like behavior. Biol. Psychiatry. 2016;80:815–826. doi: 10.1016/j.biopsych.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 152.Gao J, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Michán S, et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30:9695–9707. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ledford H. First robust genetic links to depression emerge. Nature. 2015;523:268–269. doi: 10.1038/523268a. [DOI] [PubMed] [Google Scholar]
- 155.Converge consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Luo XJ, Zhang C. Down-regulation of SIRT1 gene expression in major depressive disorder. Am. J. Psychiatry. 2016;173:1046. doi: 10.1176/appi.ajp.2016.16040394. [DOI] [PubMed] [Google Scholar]
- 157.Abe N, et al. Altered sirtuin deacetylase gene expression in patients with a mood disorder. J. Psychiatr. Res. 2011;45:1106–1112. doi: 10.1016/j.jpsychires.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 158.Golden SA, et al. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat. Med. 2013;19:337–344. doi: 10.1038/nm.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hobara T, et al. Altered gene expression of histone deacetylases in mood disorder patients. J. Psych. Res. 2010;44:263–270. doi: 10.1016/j.jpsychires.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 160.Iga J, et al. Altered HDAC5 and CREB mRNA expressions in the peripheral leukocytes of major depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2007;31:628–632. doi: 10.1016/j.pnpbp.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 161.Kilgore M, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Murray K. The occurrence of epsilon-n-methyl lysine in histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 163.Li Y, Chen X, Lu C. The interplay between DNA and histone methylation: molecular mechanisms and disease implications. EMBO Rep. 2021;22:e51803. doi: 10.15252/embr.202051803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang H, et al. H3K4me3 regulates RNA polymerase II promoter-proximal pause-release. Nature. 2023;615:339–348. doi: 10.1038/s41586-023-05780-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cruceanu C, et al. H3K4 tri-methylation in synapsin genes leads to different expression patterns in bipolar disorder and major depression. Int. J. Neuropsychopharmacol. 2013;16:289–299. doi: 10.1017/S1461145712000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cesca F, Baldelli P, Valtorta F, Benfenati F. The synapsins: key actors of synapse function and plasticity. Prog Neurobiol. 2010;91:313–348. doi: 10.1016/j.pneurobio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 167.Wilkinson MB, et al. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J. Neurosci. 2009;29:7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dudek KA, et al. Molecular adaptations of the blood-brain barrier promote stress resilience vs. depression. Proc. Natl Acad. Sci. USA. 2020;117:3326–3336. doi: 10.1073/pnas.1914655117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Suri D, et al. Early stress evokes age-dependent biphasic changes in hippocampal neurogenesis, BDNF expression, and cognition. Biol. Psychiatry. 2013;73:658–666. doi: 10.1016/j.biopsych.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jiang Z, et al. H3K9me2 regulation of BDNF expression in the hippocampus and medial prefrontal cortex is involved in the depressive-like phenotype induced by maternal separation in male rats. Psychopharmacology. 2021;238:2801–2813. doi: 10.1007/s00213-021-05896-7. [DOI] [PubMed] [Google Scholar]
- 171.Robison AJ, et al. Fluoxetine epigenetically alters the CaMKIIalpha promoter in nucleus accumbens to regulate DeltaFosB binding and antidepressant effects. Neuropsychopharmacology. 2014;39:1178–1186. doi: 10.1038/npp.2013.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jiang Y, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J. Neurosci. 2010;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wei J, et al. Histone modification of Nedd4 ubiquitin ligase controls the loss of AMPA receptors and cognitive impairment induced by repeated stress. J. Neurosci. 2016;36:2119–2130. doi: 10.1523/JNEUROSCI.3056-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu H, Jiang J, Zhao L. Protein arginine methyltransferase-1 deficiency restrains depression-like behavior of mice by inhibiting inflammation and oxidative stress via Nrf-2. Biochem. Biophys. Res. Commun. 2019;518:430–437. doi: 10.1016/j.bbrc.2019.08.032. [DOI] [PubMed] [Google Scholar]
- 175.Wang R, et al. Dynamic effects of early adolescent stress on depressive-like behaviors and expression of cytokines and JMJD3 in the prefrontal cortex and hippocampus of rats. Front. Psychiatry. 2018;9:471. doi: 10.3389/fpsyt.2018.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chan JC, Maze I. Histone crotonylation makes its mark in depression research. Biol. Psychiatry. 2019;85:616–618. doi: 10.1016/j.biopsych.2019.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xie Z, et al. Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Mol. Cell. 2016;62:194–206. doi: 10.1016/j.molcel.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Morello N, et al. Effects of forced swimming stress on ERK and histone H3 phosphorylation in limbic areas of roman high- and low-avoidance rats. PLoS ONE. 2017;12:e0170093. doi: 10.1371/journal.pone.0170093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fellows R, et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018;9:1–15. doi: 10.1038/s41467-017-02651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xu W, et al. Global profiling of crotonylation on non-histone proteins. Cell Res. 2017;27:946–949. doi: 10.1038/cr.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wei W, et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27:898–915. doi: 10.1038/cr.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liu Y, et al. Chromodomain Y-like protein-mediated histone crotonylation regulates stress-induced depressive behaviors. Biol. Psychiatry. 2019;85:635–649. doi: 10.1016/j.biopsych.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 184.Brehove M, et al. Histone core phosphorylation regulates DNA accessibility. J. Biolo. Chem. 2015;290:22612–22621. doi: 10.1074/jbc.M115.661363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chwang WB, O’Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn. Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chandramohan Y, Droste SK, Reul JM. Novelty stress induces phospho-acetylation of histone H3 in rat dentate gyrus granule neurons through coincident signalling via the N-methyl-D-aspartate receptor and the glucocorticoid receptor: relevance for c-fos induction. J. Neurochem. 2007;101:815–828. doi: 10.1111/j.1471-4159.2006.04396.x. [DOI] [PubMed] [Google Scholar]
- 187.Morello N, et al. Effects of forced swimming stress on ERK and histone H3 phosphorylation in limbic areas of Roman high-and low-avoidance rats. PLoS ONE. 2017;12:e0170093. doi: 10.1371/journal.pone.0170093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Marosi K, et al. 3‐Hydroxybutyrate regulates energy metabolism and induces BDNF expression in cerebral cortical neurons. J Neurochem. 2016;139:769–781. doi: 10.1111/jnc.13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yamanashi T, et al. Beta-hydroxybutyrate, an endogenic NLRP3 inflammasome inhibitor, attenuates stress-induced behavioral and inflammatory responses. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-08055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pan S, et al. Evaluation of the antidepressive property of β-hydroxybutyrate in mice. Behav. Pharmacol. 2020;31:322–332. doi: 10.1097/FBP.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 191.Chen L, Miao Z, Xu X. β-hydroxybutyrate alleviates depressive behaviors in mice possibly by increasing the histone3-lysine9-β-hydroxybutyrylation. Biochem. Biophys. Res. Commun. 2017;490:117–122. doi: 10.1016/j.bbrc.2017.05.184. [DOI] [PubMed] [Google Scholar]
- 192.Charney DS. Psychobiological mechanisms of resilience and vulnerability: implications for successful adaptation to extreme stress. Am. J. Psychiatry. 2004;161:195–216. doi: 10.1176/appi.ajp.161.2.195. [DOI] [PubMed] [Google Scholar]
- 193.Preis H, Mahaffey B, Heiselman C, Lobel M. Vulnerability and resilience to pandemic-related stress among US women pregnant at the start of the COVID-19 pandemic. Soc. Sci. Med. 2020;266:113348. doi: 10.1016/j.socscimed.2020.113348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ménard C, Pfau ML, Hodes GE, Russo SJ. Immune and neuroendocrine mechanisms of stress vulnerability and resilience. Neuropsychopharmacology. 2017;42:62–80. doi: 10.1038/npp.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Greene G, Paranjothy S, Palmer SR. Resilience and vulnerability to the psychological harm from flooding: the role of social cohesion. Am. J. Public Health. 2015;105:1792–1795. doi: 10.2105/AJPH.2015.302709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hamilton PJ, et al. Cell-type-specific epigenetic editing at the fosb gene controls susceptibility to social defeat stress. Neuropsychopharmacology. 2018;43:272–284. doi: 10.1038/npp.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mandelli L, Petrelli C, Serretti A. The role of specific early trauma in adult depression: a meta-analysis of published literature. Childhood trauma and adult depression. Eur. Psychiatry. 2015;30:665–680. doi: 10.1016/j.eurpsy.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 198.Kalin NH. Early-life environmental factors impacting the development of psychopathology. Am. J. Psychiatry. 2020;177:1–3. doi: 10.1176/appi.ajp.2019.19111181. [DOI] [PubMed] [Google Scholar]
- 199.McKay MT, et al. Childhood trauma and adult mental disorder: a systematic review and metaoanalysis of longitudinal cohort studies. Acta Psychiatr. Scand. 2021;143:189–205. doi: 10.1111/acps.13268. [DOI] [PubMed] [Google Scholar]
- 200.Hjelseng IV, et al. Childhood trauma is associated with poorer social functioning in severe mental disorders both during an active illness phase and in remission. Schizophr. Res. 2022;243:241–246. doi: 10.1016/j.schres.2020.03.015. [DOI] [PubMed] [Google Scholar]
- 201.Williams LM, et al. Childhood trauma predicts antidepressant response in adults with major depression: data from the randomized international study to predict optimized treatment for depression. Transl. Psychiatry. 2016;6:e799. doi: 10.1038/tp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Köhler JC, et al. Early-life adversity induces epigenetically regulated changes in hippocampal dopaminergic molecular pathways. Mol. Neurobiol. 2019;56:3616–3625. doi: 10.1007/s12035-018-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yang Y, et al. Neonatal maternal separation impairs prefrontal cortical myelination and cognitive functions in rats through activation of Wnt signaling. Cereb. Cortex. 2017;27:2871–2884. doi: 10.1093/cercor/bhw121. [DOI] [PubMed] [Google Scholar]
- 204.Bagot RC, et al. Variations in postnatal maternal care and the epigenetic regulation of metabotropic glutamate receptor 1 expression and hippocampal function in the rat. Proc. Natl Acad. Sci. USA. 2012;109:17200–17207. doi: 10.1073/pnas.1204599109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Delpech JC, et al. Early life stress perturbs the maturation of microglia in the developing hippocampus. Brain Behav. Immun. 2016;57:79–93. doi: 10.1016/j.bbi.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hoeijmakers L, et al. Early-life stress lastingly alters the neuroinflammatory response to amyloid pathology in an Alzheimer’s disease mouse model. Brain Behav. Immun. 2017;63:160–175. doi: 10.1016/j.bbi.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 207.Yajima H, et al. Early-life stress induces cognitive disorder in middle-aged mice. Neurobiol. Aging. 2018;64:139–146. doi: 10.1016/j.neurobiolaging.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 208.Shi D-D, et al. Predictable maternal separation confers adult stress resilience via the medial prefrontal cortex oxytocin signaling pathway in rats. Mol. Psychiatry. 2021;26:7296–7307. doi: 10.1038/s41380-021-01293-w. [DOI] [PubMed] [Google Scholar]
- 209.Peña CJ, et al. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science. 2017;356:1185–1188. doi: 10.1126/science.aan4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pena CJ, et al. Early life stress alters transcriptomic patterning across reward circuitry in male and female mice. Nat. Commun. 2019;10:5098. doi: 10.1038/s41467-019-13085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kronman H, et al. Long-term behavioral and cell-type-specific molecular effects of early life stress are mediated by H3K79me2 dynamics in medium spiny neurons. Nat. Neurosci. 2021;24:667–676. doi: 10.1038/s41593-021-00814-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kaufmann K, et al. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP) Nat. Protoc. 2010;5:457–472. doi: 10.1038/nprot.2009.244. [DOI] [PubMed] [Google Scholar]
- 213.Lekka E, Hall J. Non-coding RNAs in disease. FEBS Lett. 2018;592:2884–2900. doi: 10.1002/1873-3468.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wei, L.-H. & Guo, J. U. Coding functions of “noncoding” RNAs. Science (2020). [DOI] [PubMed]
- 215.Balatti, V. & Croce, C. M. Small Non-Coding RNAs in Leukemia. Cancers (2022). [DOI] [PMC free article] [PubMed]
- 216.Chen, Z. et al. Long non-coding RNA in lung cancer. Clin. Chim. Acta (2019). [DOI] [PubMed]
- 217.Esfandi, F., Taheri, M., Kholghi Oskooei, V. & Ghafouri-Fard, S. Long noncoding RNAs expression in gastric cancer. J. Cell. Biochem. (2019). [DOI] [PubMed]
- 218.Salta, E. & De Strooper, B. Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. (2017). [DOI] [PubMed]
- 219.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Shi Y, et al. Non-coding RNAs in depression: promising diagnostic and therapeutic biomarkers. EBioMedicine. 2021;71:103569. doi: 10.1016/j.ebiom.2021.103569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Panni S, Lovering RC, Porras P, Orchard S. Non-coding RNA regulatory networks. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1863:194417. doi: 10.1016/j.bbagrm.2019.194417. [DOI] [PubMed] [Google Scholar]
- 222.Wu, Y.-Y. & Kuo, H.-C. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. (2020). [DOI] [PMC free article] [PubMed]
- 223.Rani, V. & Sengar, R. S. Biogenesis and mechanisms of microRNA-mediated gene regulation. Biotechnol. Bioeng. (2022). [DOI] [PubMed]
- 224.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods Mol. Biol. 2017;1509:1–10. doi: 10.1007/978-1-4939-6524-3_1. [DOI] [PubMed] [Google Scholar]
- 226.Iwakawa H-O, Tomari Y. Life of RISC: formation, action, and degradation of RNA-induced silencing complex. Mol. Cell. 2021;82:30–43. doi: 10.1016/j.molcel.2021.11.026. [DOI] [PubMed] [Google Scholar]
- 227.Kawamata T, Tomari Y. Making RISC. Trend. Biochem. Sci. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 228.Yoo BK, et al. Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology. 2011;53:1538–1548. doi: 10.1002/hep.24216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pu M, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019;76:441–451. doi: 10.1007/s00018-018-2940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Li X, et al. Applications of acupuncture therapy in modulating the plasticity of neurodegenerative disease and depression: do microRNA and neurotrophin BDNF shed light on the underlying mechanism? Neural Plast. 2020;2020:8850653. doi: 10.1155/2020/8850653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Salemi M, et al. Examples of inverse comorbidity between cancer and neurodegenerative diseases: a possible role for noncoding RNA. Cells. 2022;11:1930. doi: 10.3390/cells11121930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yu LX, et al. MicroRNA-106b-5p promotes hepatocellular carcinoma development via modulating FOG2. Onco. Targets Ther. 2019;12:5639–5647. doi: 10.2147/OTT.S203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yi Y, et al. The role of miR-106p-5p in cervical cancer: from expression to molecular mechanism. Cell Death Discov. 2018;4:36. doi: 10.1038/s41420-018-0096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Liu F, et al. MicroRNA-106b-5p boosts glioma tumorigensis by targeting multiple tumor suppressor genes. Oncogene. 2014;33:4813–4822. doi: 10.1038/onc.2013.428. [DOI] [PubMed] [Google Scholar]
- 235.Dong X, et al. BRD4 regulates cellular senescence in gastric cancer cells via E2F/miR-106b/p21 axis. Cell Death Dis. 2018;9:203. doi: 10.1038/s41419-017-0181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lai CY, et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE. 2011;6:e21635. doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kiko T, et al. MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J. Alzheimers Dis. 2014;39:253–259. doi: 10.3233/JAD-130932. [DOI] [PubMed] [Google Scholar]
- 238.Sun N, et al. Preliminary comparison of plasma notch-associated microRNA-34b and -34c levels in drug naive, first episode depressed patients and healthy controls. J. Affect. Disord. 2016;194:109–114. doi: 10.1016/j.jad.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 239.Carless M, et al. Utility of peripheral mirna expression profiles as biomarkers and pathological indicators of neuropsychiatric disease. Eur. Neuropsychopharmacol. 2019;29:S915–S916. doi: 10.1016/j.euroneuro.2017.08.241. [DOI] [Google Scholar]
- 240.Roy B, et al. Exploiting circulating MicroRNAs as biomarkers in psychiatric disorders. Mol. Diagn. Ther. 2020;24:279–298. doi: 10.1007/s40291-020-00464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yoshino Y, Dwivedi Y. Non-coding RNAs in psychiatric disorders and suicidal behavior. Front. Psychiatry. 2020;11:543893. doi: 10.3389/fpsyt.2020.543893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Xu Y, et al. A polymorphism in the microRNA-30e precursor associated with major depressive disorder risk and P300 waveform. J. Affect. Disord. 2010;127:332–336. doi: 10.1016/j.jad.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 243.Maffioletti E, et al. Peripheral whole blood microRNA alterations in major depression and bipolar disorder. J. Affect. Disord. 2016;200:250–258. doi: 10.1016/j.jad.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 244.Smalheiser NR, et al. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS ONE. 2012;7:e33201. doi: 10.1371/journal.pone.0033201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lopez JP, et al. Regulatory role of miRNAs in polyamine gene expression in the prefrontal cortex of depressed suicide completers. Int. J. Neuropsychopharmacol. 2014;17:23–32. doi: 10.1017/S1461145713000941. [DOI] [PubMed] [Google Scholar]
- 246.Fiori LM, et al. miR-323a regulates ERBB4 and is involved in depression. Mol. Psychiatry. 2021;26:4191–4204. doi: 10.1038/s41380-020-00953-7. [DOI] [PubMed] [Google Scholar]
- 247.Roy B, et al. Altered miRNA expression network in locus coeruleus of depressed suicide subjects. Sci. Rep. 2017;7:4387. doi: 10.1038/s41598-017-04300-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Fang Y, et al. Changes in miRNA-132 and miR-124 levels in non-treated and citalopram-treated patients with depression. J. Affect. Disord. 2018;227:745–751. doi: 10.1016/j.jad.2017.11.090. [DOI] [PubMed] [Google Scholar]
- 249.Kato M, et al. Multiple pre-treatment miRNAs levels in untreated major depressive disorder patients predict early response to antidepressants and interact with key pathways. Int. J. Mol. Sci. 2022;23:3873. doi: 10.3390/ijms23073873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Segaran RC, et al. Neuronal development-related miRNAs as biomarkers for Alzheimer’s disease, depression, schizophrenia and ionizing radiation exposure. Curr. Med. Chem. 2020;28:19–52. doi: 10.2174/0929867327666200121122910. [DOI] [PubMed] [Google Scholar]
- 251.Solich J, et al. Restraint stress in mice alters set of 25 miRNAs which regulate stress- and depression-related mRNAs. Int. J. Mol. Sci. 2020;21:9469. doi: 10.3390/ijms21249469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wu Z, et al. Effective biomarkers and therapeutic targets of nerve-immunity interaction in the treatment of depression: an integrated investigation of the miRNA-mRNA regulatory networks. Aging. 2022;14:3569–3596. doi: 10.18632/aging.204030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.De Pietri Tonelli D, et al. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Fan C, et al. Microglia secrete miR-146a-5p-containing exosomes to regulate neurogenesis in depression. Mol. Ther. 2021;30:1300–1314. doi: 10.1016/j.ymthe.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rajkowska G, et al. Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol. Psychiatry. 1999;45:1085–1098. doi: 10.1016/S0006-3223(99)00041-4. [DOI] [PubMed] [Google Scholar]
- 256.Stockmeier CA, et al. Cellular changes in the postmortem hippocampus in major depression. Biol. Psychiatry. 2004;56:640–650. doi: 10.1016/j.biopsych.2004.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Aleksandrova LR, Wang YT, Phillips AG. Evaluation of the Wistar-Kyoto rat model of depression and the role of synaptic plasticity in depression and antidepressant response. Neurosci. Biobehav. Rev. 2019;105:1–23. doi: 10.1016/j.neubiorev.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 258.Lopizzo N, et al. miRNAs in depression vulnerability and resilience: novel targets for preventive strategies. J. Neural Transm. 2019;126:1241–1258. doi: 10.1007/s00702-019-02048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Lou D, Wang J, Wang X. miR-124 ameliorates depressive-like behavior by targeting STAT3 to regulate microglial activation. Mol. Cell. Probes. 2019;48:101470. doi: 10.1016/j.mcp.2019.101470. [DOI] [PubMed] [Google Scholar]
- 260.Yan X, et al. MiRNA-532-5p Regulates CUMS-induced depression-like behaviors and modulates LPS-induced proinflammatory cytokine signaling by targeting STAT3. Neuropsychiatr. Dis. Treat. 2020;16:2753–2764. doi: 10.2147/NDT.S251152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Sadok SH, et al. News and views on STAT3 psychopathology. Front. Genet. 2021;12:021. doi: 10.3389/fgene.2021.791201. [DOI] [Google Scholar]
- 262.Xian X, et al. Neuron secrete exosomes containing miR-9-5p to promote polarization of M1 microglia in depression. J. Nanobiotechnol. 2022;20:122. doi: 10.1186/s12951-022-01332-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Li Y, et al. MicroRNA-26a-3p rescues depression-like behaviors in male rats via preventing hippocampal neuronal anomalies. J. Clin. Invest. 2021;131:e148853. doi: 10.1172/JCI148853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ronovsky M, et al. A role for miR-132 in learned safety. Sci. Rep. 2019;9:528. doi: 10.1038/s41598-018-37054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Li Y, et al. Hippocampal miR-211-5p regulates neurogenesis and depression-like behaviors in the rat. Neuropharmacology. 2021;194:108618. doi: 10.1016/j.neuropharm.2021.108618. [DOI] [PubMed] [Google Scholar]
- 266.Torres-Berrío A, et al. MiR-218: a molecular switch and potential biomarker of susceptibility to stress. Mol. Psychiatry. 2020;25:951–964. doi: 10.1038/s41380-019-0421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Arcuri C, et al. The pathophysiological role of microglia in dynamic surveillance, phagocytosis and structural remodeling of the developing CNS. Front. Mol. Neurosci. 2017;10:191. doi: 10.3389/fnmol.2017.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Rajkowska G. Postmortem studies in mood disorders indicate altered numbers of neurons and glial cells. Biol. Psychiatry. 2000;48:766–777. doi: 10.1016/S0006-3223(00)00950-1. [DOI] [PubMed] [Google Scholar]
- 269.Deng S-L, Chen J-G, Wang F. Microglia: a central player in depression. Curr. Med. Sci. 2020;40:391–400. doi: 10.1007/s11596-020-2193-1. [DOI] [PubMed] [Google Scholar]
- 270.Liu Z, et al. MiRNA-199a-5p targets WNT2 to regulate depression through the CREB/BDNF signaling in hippocampal neuron. Brain Behav. 2021;8:e02107. doi: 10.1002/brb3.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Fan X, et al. In situ real-time monitoring of glutamate and electrophysiology from cortex to hippocampus in mice based on a microelectrode array. Sensors. 2017;17:61. doi: 10.3390/s17010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Hao W-Z, et al. Emerging roles of long non-coding RNA in depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2022;115:110515. doi: 10.1016/j.pnpbp.2022.110515. [DOI] [PubMed] [Google Scholar]
- 273.Higuchi Y, Soga T, Parhar IS. Potential roles of microRNAs in the regulation of monoamine oxidase A in the brain. Front. Mol. Neurosci. 2018;11:339. doi: 10.3389/fnmol.2018.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Martinetz S. MicroRNA’s impact on neurotransmitter and neuropeptide systems: small but mighty mediators of anxiety. Pflügers Archiv. Eur. J. Physiology. 2016;468:1061–1069. doi: 10.1007/s00424-016-1814-9. [DOI] [PubMed] [Google Scholar]
- 275.Guan J, et al. Early life stress increases brain glutamate and induces neurobehavioral manifestations in rats. ACS Chem. Neurosci. 2020;11:4169–4178. doi: 10.1021/acschemneuro.0c00454. [DOI] [PubMed] [Google Scholar]
- 276.Ho TC, et al. Higher levels of pro-inflammatory cytokines are associated with higher levels of glutamate in the anterior cingulate cortex in depressed adolescents. Front. Psychiatry. 2021;12:642976. doi: 10.3389/fpsyt.2021.642976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Dogra S, Conn PJ. Targeting metabotropic glutamate receptors for the treatment of depression and other stress-related disorders. Neuropharmacology. 2021;196:108687. doi: 10.1016/j.neuropharm.2021.108687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Spampinato SF, et al. Metabotropic glutamate receptors in glial cells: a new potential target for neuroprotection? Front. Mol. Neurosci. 2018;11:414. doi: 10.3389/fnmol.2018.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Li J, Meng H, Cao W, Qiu T. MiR-335 is involved in major depression disorder and antidepressant treatment through targeting GRM4. Neurosci. Lett. 2015;606:167–172. doi: 10.1016/j.neulet.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 280.Dadkhah T, et al. A genetic variant in miRNA binding site of glutamate receptor 4, metabotropic (GRM4) is associated with increased risk of major depressive disorder. J. Affect. Disord. 2016;208:218–222. doi: 10.1016/j.jad.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 281.Lopez JP, et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat. Med. 2014;20:764–768. doi: 10.1038/nm.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wan Y-Q, et al. Prefrontal cortex miR-29b-3p plays a key role in the antidepressant-like effect of ketamine in rats. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Jiang T, et al. Programming changes of hippocampal miR-134-5p/SOX2 signal mediate the susceptibility to depression in prenatal dexamethasone-exposed female offspring. Cell Biol. Toxicol. 2021;38:69–86. doi: 10.1007/s10565-021-09590-4. [DOI] [PubMed] [Google Scholar]
- 284.Higuchi Y, Soga T, Parhar IS. Potential roles of microRNAs in the regulation of monoamine oxidase A in the brain. Front. Mol. Neurosci. 2018;11:339. doi: 10.3389/fnmol.2018.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Joanna, M. et al. The serotonin theory of depression: a systematic umbrella review of the evidence. Mol. Psychiatry Online ahead of print, (2022).
- 286.Zhao J, et al. Effect of electroacupuncture on reuptake of serotonin via miRNA-16 expression in a rat model of depression. Evid. Based Complementary Altern. Med. 2019;2019:7124318. doi: 10.1155/2019/7124318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Gan H, et al. Circular RNAs in depression: biogenesis, function, expression, and therapeutic potential. Biomed. Pharmacother. 2021;137:111244. doi: 10.1016/j.biopha.2021.111244. [DOI] [PubMed] [Google Scholar]
- 288.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 289.Mehta SL, Dempsey RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 2020;186:101746. doi: 10.1016/j.pneurobio.2020.101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zhang Y, et al. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol. Psychiatry. 2020;25:1175–1190. doi: 10.1038/s41380-018-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Shi Y, et al. Potential clinical value of circular RNAs as peripheral biomarkers for the diagnosis and treatment of major depressive disorder. EBioMedicine. 2021;66:103337. doi: 10.1016/j.ebiom.2021.103337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Singh M, et al. Circular RNA: a novel and potential regulator in pathophysiology of schizophrenia. Metab. Brain Dis. 2022;37:1309–1316. doi: 10.1007/s11011-022-00978-7. [DOI] [PubMed] [Google Scholar]
- 293.Rybak-Wolf A, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 294.Wang H, Li Z, Gao J, Liao Q. Circular RNA circPTK2 regulates oxygen-glucose deprivation-activated microglia-induced hippocampal neuronal apoptosis via miR-29b-SOCS-1-JAK2/STAT3-IL-1β signaling. Int. J. Biol. Macromol. 2019;129:488–496. doi: 10.1016/j.ijbiomac.2019.02.041. [DOI] [PubMed] [Google Scholar]
- 295.Zhang Y, et al. Gut microbiota from NLRP3-deficient mice ameliorates depressive-like behaviors by regulating astrocyte dysfunction via circHIPK2. Microbiome. 2019;7:116. doi: 10.1186/s40168-019-0733-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Huang R, et al. N(6)-methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol. Psychiatry. 2020;88:392–404. doi: 10.1016/j.biopsych.2020.02.018. [DOI] [PubMed] [Google Scholar]
- 297.You X, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Mahmoudi E, Cairns MJ. Circular RNAs are temporospatially regulated throughout development and ageing in the rat. Sci. Rep. 2019;9:2564. doi: 10.1038/s41598-019-38860-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Marfil-Marin E, et al. circRNA regulates dopaminergic synapse, MAPK, and long-term depression pathways in huntington disease. Mol. Neurobiol. 2021;58:6222–6231. doi: 10.1007/s12035-021-02536-1. [DOI] [PubMed] [Google Scholar]
- 300.Wang, W. et al. Biological function of long non-coding RNA (lncRNA) Xist. Front. Cell Dev. Biol. (2021). [DOI] [PMC free article] [PubMed]
- 301.Cloutier SC, et al. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol. Cell. 2016;62:148. doi: 10.1016/j.molcel.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 302.Mary Catherine B, Amanda CD, Antonis K. LNCcation: lncRNA localization and function. J. Cell Biol. 2021;220:e202009045. doi: 10.1083/jcb.202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Fenoglio C, Ridolfi E, Galimberti D, Scarpini E. An emerging role for long non-coding RNA dysregulation in neurological disorders. Int. J. Mol. Sci. 2013;14:20427–20442. doi: 10.3390/ijms141020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Centonze D, et al. The brain cytoplasmic RNA BC1 regulates dopamine D2 receptor-mediated transmission in the striatum. J. Neurosci. 2007;27:8885–8892. doi: 10.1523/JNEUROSCI.0548-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Aliperti V, Skonieczna J, Cerase A. Long non-coding RNA (lncRNA) roles in cell biology, neurodevelopment and neurological disorders. Non-Coding RNA. 2021;7:36. doi: 10.3390/ncrna7020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ayana R, Singh S, Pati S. Decoding crucial LncRNAs implicated in neurogenesis and neurological disorders. Stem Cell. Develop. 2017;26:541–553. doi: 10.1089/scd.2016.0290. [DOI] [PubMed] [Google Scholar]
- 308.Chen Y, Zhou J. LncRNAs: macromolecules with big roles in neurobiology and neurological diseases. Metab. Brain Dis. 2017;32:281–291. doi: 10.1007/s11011-017-9965-8. [DOI] [PubMed] [Google Scholar]
- 309.Lo Piccolo L. Drosophila as a model to gain insight into the role of lncRNAs in neurological disorders. Adv. Exp. Med. Biol. 2018;1076:119–146. doi: 10.1007/978-981-13-0529-0_8. [DOI] [PubMed] [Google Scholar]
- 310.Zhou Y, et al. Global long non-coding RNA expression in the rostral anterior cingulate cortex of depressed suicides. Transl. Psychiatry. 2018;8:224. doi: 10.1038/s41398-018-0267-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Issler O, et al. Sex-specific role for the long non-coding RNA LINC00473 in depression. Neuron. 2020;106:912–926 e915. doi: 10.1016/j.neuron.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Seki T, et al. Altered expression of long noncoding RNAs in patients with major depressive disorder. J. Psychiatr. Res. 2019;117:92–99. doi: 10.1016/j.jpsychires.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 313.Ni X, et al. Therapeutic role of long non-coding RNA TCONS_00019174 in depressive disorders is dependent on Wnt/β-catenin signaling pathway. J. Integr. Neurosci. 2018;17:125–132. doi: 10.3233/JIN-170052. [DOI] [PubMed] [Google Scholar]
- 314.Li C, et al. Profiling and co-expression network analysis of learned helplessness regulated mRNAs and lncRNAs in the mouse hippocampus. Front. Mol. Neurosci. 2017;10:454. doi: 10.3389/fnmol.2017.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Gu XH, et al. Long non-coding RNA uc.80- overexpression promotes M2 polarization of microglias to ameliorate depression in rats. IUBMB Life. 2020;72:2194–2203. doi: 10.1002/iub.2353. [DOI] [PubMed] [Google Scholar]
- 316.Liu S, et al. Therapeutic antidepressant potential of NONHSAG045500 in regulating serotonin transporter in major depressive disorder. Med. Sci. Monit. 2018;24:4465–4473. doi: 10.12659/MSM.908543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Bayes A, Parker G. How to choose an antidepressant medication. Acta Psychiatr. Scand. 2019;139:280–291. doi: 10.1111/acps.13001. [DOI] [PubMed] [Google Scholar]
- 318.Hesselink JMK. Clinical evaluation of antidepressant drugs; guidelines from authorities. Acta Neuropsychiatr. 2016;4:57–62. doi: 10.1017/S0924270800034803. [DOI] [PubMed] [Google Scholar]
- 319.Croom KF, Perry CM, Plosker GL. Mirtazapine: a review of its use in major depression and other psychiatric disorders. CNS Drugs. 2009;23:427–452. doi: 10.2165/00023210-200923050-00006. [DOI] [PubMed] [Google Scholar]
- 320.Berlim MT, Turecki G. Definition, assessment, and staging of treatment—resistant refractory major depression: a review of current concepts and methods. Can. J. Psychiatry. 2007;52:46–54. doi: 10.1177/070674370705200108. [DOI] [PubMed] [Google Scholar]
- 321.Sinyor M, Schaffer A, Levitt A. The sequenced treatment alternatives to relieve depression (STAR*D) trial: a review. Can. J. Psychiatry. 2010;55:126–135. doi: 10.1177/070674371005500303. [DOI] [PubMed] [Google Scholar]
- 322.Trivedi MH, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR* D: implications for clinical practice. Am. J. Psychiatry. 2006;163:28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 323.Broich K. Committee for Medicinal Products for Human Use (CHMP) assessment on efficacy of antidepressants. Eur. Neuropsychopharmacol. 2009;19:305–308. doi: 10.1016/j.euroneuro.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 324.Bschor T, Kilarski LL. Are antidepressants effective? A debate on their efficacy for the treatment of major depression in adults. Expert Rev. Neurother. 2016;16:367–374. doi: 10.1586/14737175.2016.1155985. [DOI] [PubMed] [Google Scholar]
- 325.Calarge C, Mills J, Coryell W. Skeletal effects of depression and SSRIs. Biol. Psychiatry. 2017;81:S82. doi: 10.1016/j.biopsych.2017.02.211. [DOI] [Google Scholar]
- 326.Cousins L, et al. Clinical characteristics associated with the prescribing of SSRI medication in adolescents with major unipolar depression. Eur. Child Adolesc. Psychiatry. 2016;25:1287–1295. doi: 10.1007/s00787-016-0849-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Oved K, et al. Genome-wide miRNA expression profiling of human lymphoblastoid cell lines identifies tentative SSRI antidepressant response biomarkers. Pharmacogenomics. 2012;13:1129–1139. doi: 10.2217/pgs.12.93. [DOI] [PubMed] [Google Scholar]
- 328.Oved K, et al. Genome-wide expression profiling of human lymphoblastoid cell lines implicates integrin beta-3 in the mode of action of antidepressants. Transl. Psychiatry. 2022;3:e313. doi: 10.1038/tp.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Bocchio-Chiavetto L, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur. Neuropsychopharmacol. 2013;23:602–611. doi: 10.1016/j.euroneuro.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 330.Baudry A, et al. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329:1537–1541. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- 331.Miao N, Jin J, Kim SN, Sun T. Hippocampal microRNAs respond to administration of antidepressant fluoxetine in adult mice. Int. J. Mol. Sci. 2018;19:671. doi: 10.3390/ijms19030671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Mundalil Vasu M, et al. Fluoxetine increases the expression of miR-572 and miR-663a in human neuroblastoma cell lines. PLoS ONE. 2016;11:e0164425. doi: 10.1371/journal.pone.0164425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Zhang CL, et al. Fluoxetine ameliorates depressive symptoms by regulating lncRNA expression in the mouse hippocampus. Zool Res. 2021;42:28–42. doi: 10.24272/j.issn.2095-8137.2020.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Kuang WH, Dong ZQ, Tian LT, Li J. MicroRNA-451a, microRNA-34a-5p, and microRNA-221-3p as predictors of response to antidepressant treatment. Braz. J. Med. Biol. Res. 2018;51:e7212. doi: 10.1590/1414-431x20187212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Zhang Y, et al. Dopamine receptor D2 and associated microRNAs are involved in stress susceptibility and resistance to escitalopram treatment. Int. J. Neuropsychopharmacol. 2015;18:pyv025. doi: 10.1093/ijnp/pyv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Goldstein DJ, et al. Duloxetine in the treatment of depression: a double-blind placebo-controlled comparison with paroxetine. J. Clin. Psychopharmacol. 2004;24:389–399. doi: 10.1097/01.jcp.0000132448.65972.d9. [DOI] [PubMed] [Google Scholar]
- 337.Schueler YB, et al. A systematic review of duloxetine and venlafaxine in major depression, including unpublished data. Acta Psychiatr. Scand. 2011;123:247–265. doi: 10.1111/j.1600-0447.2010.01599.x. [DOI] [PubMed] [Google Scholar]
- 338.Montgomery SA. Tolerability of serotonin norepinephrine reuptake inhibitor antidepressants. CNS Spectr. 2008;13:27–33. doi: 10.1017/S1092852900028297. [DOI] [PubMed] [Google Scholar]
- 339.Monteleone F, Caputo M, Tecce MF, Capasso A. Duloxetine in the treatment of depression: an overview. Cent. Nerv. Syst. Agents Med. Chem. 2011;11:174–183. doi: 10.2174/187152411798047807. [DOI] [PubMed] [Google Scholar]
- 340.Lopez JP, et al. MicroRNAs 146a/b-5 and 425-3p and 24-3p are markers of antidepressant response and regulate MAPK/Wnt-system genes. Nat. Commun. 2017;8:15497. doi: 10.1038/ncomms15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Pan B, Liu Y. Effects of duloxetine on microRNA expression profile in frontal lobe and hippocampus in a mouse model of depression. Int. J. Clin. Exp. Pathol. 2015;8:15454–15461. [PMC free article] [PubMed] [Google Scholar]
- 342.Lopez JP, et al. Co-variation of peripheral levels of miR-1202 and brain activity and connectivity during antidepressant treatment. Neuropsychopharmacology. 2017;42:2043–2051. doi: 10.1038/npp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Berman RM, et al. Antidepressant effects of ketamine in depressed patients. Biol. Psychiatry. 2000;47:351–354. doi: 10.1016/S0006-3223(99)00230-9. [DOI] [PubMed] [Google Scholar]
- 344.Yang X, et al. MicroRNA expression profile and functional analysis reveal that miR-206 is a critical novel gene for the expression of BDNF induced by ketamine. Neuromolecular Med. 2014;16:594–605. doi: 10.1007/s12017-014-8312-z. [DOI] [PubMed] [Google Scholar]
- 345.Wan YQ, et al. Prefrontal cortex miR-29b-3p plays a key role in the antidepressant-like effect of ketamine in rats. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wilkinson E, Cui YH, He YY. Context-dependent roles of RNA modifications in stress responses and diseases. Int. J. Mol. Sci. 2021;22:1949. doi: 10.3390/ijms22041949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Jung Y, Goldman D. Role of RNA modifications in brain and behavior. Genes Brain Behav. 2018;17:e12444. doi: 10.1111/gbb.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Livneh I, et al. The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020;21:36–51. doi: 10.1038/s41583-019-0244-z. [DOI] [PubMed] [Google Scholar]
- 350.Desrosiers RC, Friderici KH, Rottman FM. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5′ terminus. Biochemistry. 1975;14:4367–4374. doi: 10.1021/bi00691a004. [DOI] [PubMed] [Google Scholar]
- 351.Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods. 2016;14:23–31. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
- 352.Kan RL, Chen J, Sallam T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2022;38:182–193. doi: 10.1016/j.tig.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Bokar JA, et al. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994;269:17697–17704. doi: 10.1016/S0021-9258(17)32497-3. [DOI] [PubMed] [Google Scholar]
- 354.Roignant JY, Soller M. m(6)A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 355.Wang CX, et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16:e2004880. doi: 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Xu B, et al. Mettl3-mediated m(6) A modification of Lrp2 facilitates neurogenesis through Ythdc2 and elicits antidepressant-like effects. FASEB J. 2022;36:e22392. doi: 10.1096/fj.202200133RR. [DOI] [PubMed] [Google Scholar]
- 357.Niu J, Wang B, Wang T, Zhou T. Mechanism of METTL3-mediated m6A modification in depression-induced cognitive deficits. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2022;189:86–99. doi: 10.1002/ajmg.b.32892. [DOI] [PubMed] [Google Scholar]
- 358.Samaan Z, et al. The protective effect of the obesity-associated rs9939609 A variant in fat mass- and obesity-associated gene on depression. Mol. Psychiatry. 2013;18:1281–1286. doi: 10.1038/mp.2012.160. [DOI] [PubMed] [Google Scholar]
- 359.Milaneschi Y, et al. The effect of FTO rs9939609 on major depression differs across MDD subtypes. Mol. Psychiatry. 2014;19:960–962. doi: 10.1038/mp.2014.4. [DOI] [PubMed] [Google Scholar]
- 360.Liu S, et al. Fat mass and obesity-associated protein regulates RNA methylation associated with depression-like behavior in mice. Nat. Commun. 2021;12:6937. doi: 10.1038/s41467-021-27044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Yang C, et al. The role of m(6)A modification in physiology and disease. Cell Death Dis. 2020;11:960. doi: 10.1038/s41419-020-03143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Wang XL, et al. Downregulation of fat mass and obesity-related protein in the anterior cingulate cortex participates in anxiety- and depression-like behaviors induced by neuropathic pain. Front. Cell Neurosci. 2022;16:884296. doi: 10.3389/fncel.2022.884296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Du T, et al. An association study of the m6A genes with major depressive disorder in Chinese Han population. J. Affect. Disord. 2015;183:279–286. doi: 10.1016/j.jad.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 364.Alarcon CR, et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Deng Z, et al. Co-expression modules construction by WGCNA and identify potential hub genes and regulation pathways of postpartum depression. FBL. 2021;26:1019–1030. doi: 10.52586/5006. [DOI] [PubMed] [Google Scholar]
- 367.Shi H, et al. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–253. doi: 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Arango D, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175:1872–1886.e1824. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Guo XF, et al. Elevation of N-acetyltransferase 10 in hippocampal neurons mediates depression- and anxiety-like behaviors. Brain Res. Bull. 2022;185:91–98. doi: 10.1016/j.brainresbull.2022.05.004. [DOI] [PubMed] [Google Scholar]
- 370.Bohnsack KE, Hobartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m(5)C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes. 2019;10:102. doi: 10.3390/genes10020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Blaze J, et al. Epitranscriptomic and metabolomic signatures involved in translational mechanisms of suicide and depression. Biol. Psychiatry. 2022;91:S73–S74. doi: 10.1016/j.biopsych.2022.02.203. [DOI] [Google Scholar]
- 372.Wu PF, et al. Erasing m(6)A-dependent transcription signature of stress-sensitive genes triggers antidepressant actions. Neurobiol. Stress. 2021;15:100390. doi: 10.1016/j.ynstr.2021.100390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Shen J, Yang L, Wei W. Role of Fto on CaMKII/CREB signaling pathway of hippocampus in depressive-like behaviors induced by chronic restraint stress mice. Behav. Brain Res. 2021;406:113227. doi: 10.1016/j.bbr.2021.113227. [DOI] [PubMed] [Google Scholar]
- 374.Yan L, et al. Physical exercise prevented stress‐induced anxiety via improving brain RNA methylation. Adv. Sci. 2022;9:2105731. doi: 10.1002/advs.202105731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Song N, Du J, Gao Y, Yang S. Epitranscriptome of the ventral tegmental area in a deep brain-stimulated chronic unpredictable mild stress mouse model. Transl. Neurosci. 2020;11:402–418. doi: 10.1515/tnsci-2020-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Vignali M, et al. ATP-dependent chromatin-remodeling complexes. Mol. Cell Biol. 2000;20:1899–1910. doi: 10.1128/MCB.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Becker, P. B. & Hörz, W. J. A. R. O. B. ATP-dependent nucleosome remodeling. 71, 247–273 (2002). [DOI] [PubMed]
- 378.Sun H, et al. ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior. Nat. Med. 2015;21:1146–1153. doi: 10.1038/nm.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Zayed A, et al. SWI/SNF chromatin remodeler complex within the reward pathway is required for behavioral adaptations to stress. Nat. Commun. 2022;13:1807. doi: 10.1038/s41467-022-29380-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Wille A, et al. Dysregulation of select ATP-dependent chromatin remodeling factors in high trait anxiety. Behav. Brain Res. 2016;311:141–146. doi: 10.1016/j.bbr.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 381.Sohel MMH. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci. 2020;248:117473. doi: 10.1016/j.lfs.2020.117473. [DOI] [PubMed] [Google Scholar]
- 382.Swellam M, et al. Role of some circulating MiRNAs on breast cancer diagnosis. Archiv. Physiol. Biochem. 2018;125:456–464. doi: 10.1080/13813455.2018.1482355. [DOI] [PubMed] [Google Scholar]
- 383.Zhou S, et al. Accurate cancer diagnosis and stage monitoring enabled by comprehensive profiling of different types of exosomal biomarkers: surface proteins and miRNAs. Small. 2020;16:e2004492. doi: 10.1002/smll.202004492. [DOI] [PubMed] [Google Scholar]
- 384.Zhou L, Zhu Y, Chen W, Tang Y. Emerging role of microRNAs in major depressive disorder and its implication on diagnosis and therapeutic response. J. Affect. Disord. 2021;286:80–86. doi: 10.1016/j.jad.2021.02.063. [DOI] [PubMed] [Google Scholar]
- 385.Galvão ACDM, et al. Potential biomarkers of major depression diagnosis and chronicity. PLoS ONE. 2021;16:e0257251. doi: 10.1371/journal.pone.0257251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Xu Y-Y, et al. MicroRNA-based biomarkers in the diagnosis and monitoring of therapeutic response in patients with depression. Neuropsychiatr. Dis. Treat. 2019;15:3583–3597. doi: 10.2147/NDT.S237116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Schneider I, et al. Association of serotonin transporter gene AluJb methylation with major depression, amygdala responsiveness, 5-HTTLPR/rs25531 polymorphism, and stress. Neuropsychopharmacology. 2018;43:1308–1316. doi: 10.1038/npp.2017.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Davies MN, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:1–14. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain “‐omes”. Am. J. Med. Genet. B: Neuropsychiatr. Genet. 2013;162:595–603. doi: 10.1002/ajmg.b.32150. [DOI] [PubMed] [Google Scholar]
- 390.Yehuda R, et al. Epigenetic biomarkers as predictors and correlates of symptom improvement following psychotherapy in combat veterans with PTSD. Front. Psychiatry. 2013;4:118. doi: 10.3389/fpsyt.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Roberts S, et al. DNA methylation of FKBP5 and response to exposures of Sypsychological therapy. Am. J. Med. Genet. B: Neuropsychiatr. Genet. 2019;180:150–158. doi: 10.1002/ajmg.b.32650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Romana PG, et al. Epigenetic correlates of the psychological intervention outcomes: a systematic review and meta-analysis: epigenetic effects of psychological interventions. J. Affect. Disord. Rep. 2022;7:100310. doi: 10.1016/j.jadr.2022.100310. [DOI] [Google Scholar]
- 393.Pfalzer AC, et al. S-adenosylmethionine mediates inhibition of inflammatory response and changes in DNA methylation in human macrophages. Physiol. Genomics. 2014;46:617–623. doi: 10.1152/physiolgenomics.00056.2014. [DOI] [PubMed] [Google Scholar]
- 394.Bergamaschi MM, Queiroz RH, Zuardi AW, Crippa JA. Safety and side effects of cannabidiol, a Cannabis sativa constituent. Curr. Drug Safety. 2011;6:237–249. doi: 10.2174/157488611798280924. [DOI] [PubMed] [Google Scholar]
- 395.Campos AC, Fogaça MV, Sonego AB, Guimarães FS. Cannabidiol, neuroprotection and neuropsychiatric disorders. Pharmacol. Res. 2016;112:119–127. doi: 10.1016/j.phrs.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 396.García-Gutiérrez MS, et al. Cannabidiol: a potential new alternative for the treatment of anxiety, depression, and psychotic disorders. Biomolecules. 2020;10:1575. doi: 10.3390/biom10111575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Sales AJ, Guimarães FS, Joca SRL. CBD modulates DNA methylation in the prefrontal cortex and hippocampus of mice exposed to forced swim. Behav. Brain Res. 2020;388:112627. doi: 10.1016/j.bbr.2020.112627. [DOI] [PubMed] [Google Scholar]
- 398.Pastrana-Trejo JC, et al. Effects on the post-translational modification of H3K4Me3, H3K9ac, H3K9Me2, H3K27Me3, and H3K36Me2 levels in cerebral cortex, hypothalamus and pons of rats after a systemic administration of cannabidiol: a preliminary study. Cent. Nerv. Syst. Agents Med. Chem. 2021;21:142–147. doi: 10.2174/1871524920666200924114524. [DOI] [PubMed] [Google Scholar]
- 399.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 400.Sales AJ, et al. Antidepressant-like effect induced by systemic and intra-hippocampal administration of DNA methylation inhibitors. Br. J. Pharmacol. 2011;164:1711–1721. doi: 10.1111/j.1476-5381.2011.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Yoo CB, Cheng JC, Jones PA. Zebularine: a new drug for epigenetic therapy. Biochem. Soc. Trans. 2004;32:910–912. doi: 10.1042/BST0320910. [DOI] [PubMed] [Google Scholar]
- 402.Sales AJ, Joca SR. Effects of DNA methylation inhibitors and conventional antidepressants on mice behaviour and brain DNA methylation levels. Acta Neuropsychiatr. 2016;28:11–22. doi: 10.1017/neu.2015.40. [DOI] [PubMed] [Google Scholar]
- 403.Guidotti A, et al. Characterization of the action of antipsychotic subtypes on valproate-induced chromatin remodeling. Trends Pharmacol Sci. 2009;30:55–60. doi: 10.1016/j.tips.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 404.Calabrese F, et al. Modulation of neuronal plasticity following chronic concomitant administration of the novel antipsychotic lurasidone with the mood stabilizer valproic acid. Psychopharmacology. 2013;226:101–112. doi: 10.1007/s00213-012-2900-0. [DOI] [PubMed] [Google Scholar]
- 405.K VA, et al. Cognitive Improvement by Vorinostat through Modulation of Endoplasmic Reticulum Stress in a Corticosterone-Induced Chronic Stress Model in Mice. ACS Chem. Neurosci. 2020;11:2649–2657. doi: 10.1021/acschemneuro.0c00315. [DOI] [PubMed] [Google Scholar]
- 406.Ershadi ASB, Amini-Khoei H, Hosseini MJ, Dehpour AR. SAHA improves depressive symptoms, cognitive impairment and oxidative stress: rise of a new antidepressant class. Neurochem. Res. 2021;46:1252–1263. doi: 10.1007/s11064-021-03263-8. [DOI] [PubMed] [Google Scholar]
- 407.Vinarskaya AK, Balaban PM, Roshchin MV, Zuzina AB. Sodium butyrate as a selective cognitive enhancer for weak or impaired memory. Neurobiol. Learn Mem. 2021;180:107414. doi: 10.1016/j.nlm.2021.107414. [DOI] [PubMed] [Google Scholar]
- 408.Peña CJ, Bagot RC, Labonté B, Nestler EJ. Epigenetic signaling in psychiatric disorders. J. Mol. Biol. 2014;426:3389–3412. doi: 10.1016/j.jmb.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Volmar C-H, Wahlestedt CJN. Histone deacetylases (HDACs) and brain function. Neuroepigenetics. 2015;1:20–27. doi: 10.1016/j.nepig.2014.10.002. [DOI] [Google Scholar]
- 410.Lopez JP, et al. Epigenetic regulation of BDNF expression according to antidepressant response. Mol Psychiatry. 2013;18:398–399. doi: 10.1038/mp.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Ernst C, et al. Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Arch. Gen. Psychiatry. 2009;66:22–32. doi: 10.1001/archpsyc.66.1.22. [DOI] [PubMed] [Google Scholar]
- 412.Clark SJ, et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 2018;9:1–9. doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Peterson VM, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 414.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Flyamer IM, et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Xie W, et al. Single-cell RNA sequencing and assay for transposase-accessible chromatin using sequencing reveals cellular and molecular dynamics of aortic aging in mice. Arterioscler. Thromb. Vasc. Biol. 2022;42:156–171. doi: 10.1161/ATVBAHA.121.316883. [DOI] [PubMed] [Google Scholar]
- 417.Deng Y, et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature. 2022;609:375–383. doi: 10.1038/s41586-022-05094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Meng J, et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Mews P, et al. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature. 2017;546:381–386. doi: 10.1038/nature22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Pan RY, et al. Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer’s disease. Cell Metab. 2022;34:634–648.e636. doi: 10.1016/j.cmet.2022.02.013. [DOI] [PubMed] [Google Scholar]
- 421.Zhou C, et al. Hippocampus-specific regulation of long non-coding RNA and mRNA expression in germ-free mice. Funct. Integr. Genomics. 2020;20:355–365. doi: 10.1007/s10142-019-00716-w. [DOI] [PubMed] [Google Scholar]
- 422.Gao Y, et al. LncRNA lncLy6C induced by microbiota metabolite butyrate promotes differentiation of Ly6C(high) to Ly6C(int/neg) macrophages through lncLy6C/C/EBPβ/Nr4A1 axis. Cell Discov. 2020;6:87. doi: 10.1038/s41421-020-00211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Bastiaanssen TF, et al. Gutted! Unraveling the role of the microbiome in major depressive disorder. Harvard Rev. Psychiatry. 2020;28:26–39. doi: 10.1097/HRP.0000000000000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Cryan JF, et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 425.Heller EA, et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat. Neurosci. 2014;17:1720–1727. doi: 10.1038/nn.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Heller EA, et al. Targeted epigenetic remodeling of the Cdk5 gene in nucleus accumbens regulates cocaine- and stress-evoked behavior. J. Neurosci. 2016;36:4690–4697. doi: 10.1523/JNEUROSCI.0013-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Pulecio J, et al. CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell. 2017;21:431–447. doi: 10.1016/j.stem.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Liao HK, et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell. 2017;171:1495–1507.e1415. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Nuñez JK, et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 2021;184:2503–2519.e2517. doi: 10.1016/j.cell.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Choi S, et al. Association of brain-derived neurotrophic factor DNA methylation and reduced white matter integrity in the anterior corona radiata in major depression. J. Affect. Disord. 2015;172:74–80. doi: 10.1016/j.jad.2014.09.042. [DOI] [PubMed] [Google Scholar]
- 432.Ferrer A, et al. BDNF genetic variants and methylation: effects on cognition in major depressive disorder. Transl. Psychiatry. 2019;9:265. doi: 10.1038/s41398-019-0601-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Moser DA, et al. BDNF methylation and maternal brain activity in a violence-related sample. PLoS One. 2015;10:e0143427. doi: 10.1371/journal.pone.0143427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Sabunciyan S, et al. Genome-wide DNA methylation scan in major depressive disorder. PLoS ONE. 2012;7:e34451. doi: 10.1371/journal.pone.0034451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Weder N, et al. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J. Am. Acad. Child Adolesc. Psychiatry. 2014;53:417–424.e415. doi: 10.1016/j.jaac.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Philibert RA, et al. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147b:543–549. doi: 10.1002/ajmg.b.30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Zhao J, Goldberg J, Bremner JD, Vaccarino V. Association between promoter methylation of serotonin transporter gene and depressive symptoms: a monozygotic twin study. Psychosom. Med. 2013;75:523–529. doi: 10.1097/PSY.0b013e3182924cf4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Frodl T, et al. DNA methylation of the serotonin transporter gene (SLC6A4) is associated with brain function involved in processing emotional stimuli. J. Psychiatry Neurosci. 2015;40:296–305. doi: 10.1503/jpn.140180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Won E, et al. Association between reduced white matter integrity in the corpus callosum and serotonin transporter gene DNA methylation in medication-naive patients with major depressive disorder. Transl. Psychiatry. 2016;6:e866. doi: 10.1038/tp.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Koenen KC, et al. SLC6A4 methylation modifies the effect of the number of traumatic events on risk for posttraumatic stress disorder. Depress Anxiety. 2011;28:639–647. doi: 10.1002/da.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Domschke K, et al. Pharmacoepigenetics of depression: no major influence of MAO-A DNA methylation on treatment response. J. Neural Transm. 2015;122:99–108. doi: 10.1007/s00702-014-1227-x. [DOI] [PubMed] [Google Scholar]
- 442.Melas PA, et al. Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. Int. J. Neuropsychopharmacol. 2013;16:1513–1528. doi: 10.1017/S1461145713000102. [DOI] [PubMed] [Google Scholar]
- 443.Melas PA, Forsell Y. Hypomethylation of MAOA’s first exon region in depression: a replication study. Psychiatry Res. 2015;226:389–391. doi: 10.1016/j.psychres.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 444.Bustamante AC, et al. Glucocorticoid receptor DNA methylation, childhood maltreatment and major depression. J. Affect. Disord. 2016;206:181–188. doi: 10.1016/j.jad.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Farrell C, et al. DNA methylation differences at the glucocorticoid receptor gene in depression are related to functional alterations in hypothalamic-pituitary-adrenal axis activity and to early life emotional abuse. Psychiatry Res. 2018;265:341–348. doi: 10.1016/j.psychres.2018.04.064. [DOI] [PubMed] [Google Scholar]
- 446.Na KS, et al. Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS ONE. 2014;9:e85425. doi: 10.1371/journal.pone.0085425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Alt SR, et al. Differential expression of glucocorticoid receptor transcripts in major depressive disorder is not epigenetically programmed. Psychoneuroendocrinology. 2010;35:544–556. doi: 10.1016/j.psyneuen.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 448.Kosten TA, Huang W, Nielsen DA. Sex and litter effects on anxiety and DNA methylation levels of stress and neurotrophin genes in adolescent rats. Dev. Psychobiol. 2014;56:392–406. doi: 10.1002/dev.21106. [DOI] [PubMed] [Google Scholar]
- 449.Kundakovic M, Lim S, Gudsnuk K, Champagne F. Sex-specific and strain-dependent effects of early life adversity on behavioral and epigenetic outcomes. Front Psychiatry. 2013;4:78. doi: 10.3389/fpsyt.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Yehuda R, et al. Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans with posttraumatic stress disorder. Biol. Psychiatry. 2015;77:356–364. doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 451.Wang W, et al. Increased methylation of glucocorticoid receptor gene promoter 1(F) in peripheral blood of patients with generalized anxiety disorder. J. Psychiatr. Res. 2017;91:18–25. doi: 10.1016/j.jpsychires.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 452.Tozzi L, et al. Epigenetic changes of FKBP5 as a link connecting genetic and environmental risk factors with structural and functional brain changes in major depression. Neuropsychopharmacology. 2018;43:1138–1145. doi: 10.1038/npp.2017.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Klinger-König J, et al. Methylation of the FKBP5 gene in association with FKBP5 genotypes, childhood maltreatment and depression. Neuropsychopharmacology. 2019;44:930–938. doi: 10.1038/s41386-019-0319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Klengel T, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Kahl KG, et al. Altered DNA methylation of glucose transporter 1 and glucose transporter 4 in patients with major depressive disorder. J. Psychiatr. Res. 2016;76:66–73. doi: 10.1016/j.jpsychires.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 456.Kimmel M, et al. Oxytocin receptor DNA methylation in postpartum depression. Psychoneuroendocrinology. 2016;69:150–160. doi: 10.1016/j.psyneuen.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Reiner I, et al. Methylation of the oxytocin receptor gene in clinically depressed patients compared to controls: The role of OXTR rs53576 genotype. J. Psychiatr. Res. 2015;65:9–15. doi: 10.1016/j.jpsychires.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 458.Bell AF, et al. Interaction between oxytocin receptor DNA methylation and genotype is associated with risk of postpartum depression in women without depression in pregnancy. Front. Genet. 2015;6:243. doi: 10.3389/fgene.2015.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Zill P, et al. DNA methylation analysis of the angiotensin converting enzyme (ACE) gene in major depression. PLoS ONE. 2012;7:e40479. doi: 10.1371/journal.pone.0040479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Hüls A, et al. Association between DNA methylation levels in brain tissue and late-life depression in community-based participants. Transl. Psychiatry. 2020;10:262. doi: 10.1038/s41398-020-00948-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Ryan J, et al. Investigating the epigenetic profile of the inflammatory gene IL-6 in late-life depression. BMC Psychiatry. 2017;17:354. doi: 10.1186/s12888-017-1515-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Green C, et al. Structural brain correlates of serum and epigenetic markers of inflammation in major depressive disorder. Brain Behav. Immun. 2021;92:39–48. doi: 10.1016/j.bbi.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Córdova-Palomera A, et al. Further evidence of DEPDC7 DNA hypomethylation in depression: a study in adult twins. Eur. Psychiatry. 2015;30:715–718. doi: 10.1016/j.eurpsy.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 464.Zhu Y, et al. Genome-wide profiling of DNA methylome and transcriptome in peripheral blood monocytes for major depression: A Monozygotic Discordant Twin Study. Transl. Psychiatry. 2019;9:215. doi: 10.1038/s41398-019-0550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Roy B, Dunbar M, Shelton RC, Dwivedi Y. Identification of MicroRNA-124-3p as a Putative Epigenetic Signature of Major Depressive Disorder. Neuropsychopharmacology. 2017;42:864–875. doi: 10.1038/npp.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.He S, et al. Alterations of microRNA-124 expression in peripheral blood mononuclear cells in pre- and post-treatment patients with major depressive disorder. J. Psychiatr. Res. 2016;78:65–71. doi: 10.1016/j.jpsychires.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 467.Dwivedi Y, et al. Chronic corticosterone-mediated dysregulation of microRNA network in prefrontal cortex of rats: relevance to depression pathophysiology. Transl. Psychiatry. 2015;5:e682–e682. doi: 10.1038/tp.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Xu J, et al. FKBP5 and specific microRNAs via glucocorticoid receptor in the basolateral amygdala involved in the susceptibility to depressive disorder in early adolescent stressed rats. J. Psychiatr. Res. 2017;95:102–113. doi: 10.1016/j.jpsychires.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 469.Higuchi F, et al. Hippocampal microRNA-124 enhances chronic stress resilience in mice. J. Neurosci. 2016;36:7253–7267. doi: 10.1523/JNEUROSCI.0319-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Wei ZX, et al. Exosomes from patients with major depression cause depressive-like behaviors in mice with involvement of miR-139-5p-regulated neurogenesis. Neuropsychopharmacology. 2020;45:1050–1058. doi: 10.1038/s41386-020-0622-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Smalheiser NR, et al. Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS ONE. 2014;9:e86469. doi: 10.1371/journal.pone.0086469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Lian N, et al. MiR-221 is involved in depression by regulating Wnt2/CREB/BDNF axis in hippocampal neurons. Cell Cycle. 2018;17:2745–2755. doi: 10.1080/15384101.2018.1556060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Wan Y, et al. Identification of differential microRNAs in cerebrospinal fluid and serum of patients with major depressive disorder. PLoS ONE. 2015;10:e0121975. doi: 10.1371/journal.pone.0121975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Feng J, et al. Serum miR-221-3p as a new potential biomarker for depressed mood in perioperative patients. Brain Res. 2019;1720:146296. doi: 10.1016/j.brainres.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 475.Torres-Berrío A, et al. DCC confers susceptibility to depression-like behaviors in humans and mice and is regulated by miR-218. Biol. Psychiatry. 2017;81:306–315. doi: 10.1016/j.biopsych.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Camkurt MA, et al. Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. J. Psychiatr. Res. 2015;69:67–71. doi: 10.1016/j.jpsychires.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 477.Li J, Meng H, Cao W, Qiu T. MiR-335 is involved in major depression disorder and antidepressant treatment through targeting GRM4. Neurosci. Lett. 2015;606:167–172. doi: 10.1016/j.neulet.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 478.Gheysarzadeh A, et al. Serum-based microRNA biomarkers for major depression: MiR-16, miR-135a, and miR-1202. J. Res. Med. Sci. 2018;23:69. doi: 10.4103/jrms.JRMS_879_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Issler O, et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83:344–360. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]
- 480.Azevedo JA, et al. The microRNA network is altered in anterior cingulate cortex of patients with unipolar and bipolar depression. J. Psychiatr. Res. 2016;82:58–67. doi: 10.1016/j.jpsychires.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Hung YY, et al. Aberrant expression of intracellular let-7e, miR-146a, and miR-155 correlates with severity of depression in patients with major depressive disorder and is ameliorated after antidepressant treatment. Cells. 2019;8:647. doi: 10.3390/cells8070647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Belzeaux R, et al. Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl. Psychiatry. 2012;2:e185. doi: 10.1038/tp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Zeng Y, et al. Genome-wide regional heritability mapping identifies a locus within the TOX2 gene associated with major depressive disorder. Biol. Psychiatry. 2017;82:312–321. doi: 10.1016/j.biopsych.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Ye N, et al. Intergenic variants may predispose to major depression disorder through regulation of long non-coding RNA expression. Gene. 2017;601:21–26. doi: 10.1016/j.gene.2016.11.041. [DOI] [PubMed] [Google Scholar]
- 485.Cui X, et al. Can lncRNAs be indicators for the diagnosis of early onset or acute schizophrenia and distinguish major depressive disorder and generalized anxiety disorder?—A cross validation analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2017;174:335–341. doi: 10.1002/ajmg.b.32521. [DOI] [PubMed] [Google Scholar]
- 486.Tadić A, et al. Methylation of the promoter of brain-derived neurotrophic factor exon IV and antidepressant response in major depression. Mol. Psychiatry. 2014;19:281–283. doi: 10.1038/mp.2013.58. [DOI] [PubMed] [Google Scholar]
- 487.Kim JM, et al. BDNF methylation and depressive disorder in acute coronary syndrome: The K-DEPACS and EsDEPACS studies. Psychoneuroendocrinology. 2015;62:159–165. doi: 10.1016/j.psyneuen.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 488.Domschke K, et al. Serotonin transporter gene hypomethylation predicts impaired antidepressant treatment response. Int. J. Neuropsychopharmacol. 2014;17:1167–1176. doi: 10.1017/S146114571400039X. [DOI] [PubMed] [Google Scholar]
- 489.Iga J-I, et al. Association study of polymorphism in the serotonin transporter gene promoter, methylation profiles, and expression in patients with major depressive disorder. Hum. Psychopharmacol. 2016;31:193–199. doi: 10.1002/hup.2527. [DOI] [PubMed] [Google Scholar]
- 490.Wang P, et al. HTR1A/1B DNA methylation may predict escitalopram treatment response in depressed Chinese Han patients. J. Affect. Disord. 2018;228:222–228. doi: 10.1016/j.jad.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 491.Gassó P, et al. Epigenetic and genetic variants in the HTR1B gene and clinical improvement in children and adolescents treated with fluoxetine. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2017;75:28–34. doi: 10.1016/j.pnpbp.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 492.Powell TR, et al. DNA methylation in interleukin-11 predicts clinical response to antidepressants in GENDEP. Transl. Psychiatry. 2013;3:e300. doi: 10.1038/tp.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Domschke K, et al. Pharmacoepigenetics of depression: no major influence of MAO-A DNA methylation on treatment response. J. Neural Transm. 2015;122:99–108. doi: 10.1007/s00702-014-1227-x. [DOI] [PubMed] [Google Scholar]
- 494.Takeuchi N, et al. Therapeutic response to paroxetine in major depressive disorder predicted by DNA methylation. Neuropsychobiology. 2017;75:81–88. doi: 10.1159/000480512. [DOI] [PubMed] [Google Scholar]
- 495.Ju C, et al. Integrated genome-wide methylation and expression analyses reveal functional predictors of response to antidepressants. Transl. Psychiatry. 2019;9:1–12. doi: 10.1038/s41398-019-0589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Martinez-Pinteño A, et al. DNA methylation of fluoxetine response in child and adolescence: preliminary results. Pharmgenomics Pers. Med. 2021;14:459–467. doi: 10.2147/PGPM.S289480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Engelmann J, et al. Epigenetic signatures in antidepressant treatment response: a methylome-wide association study in the EMC trial. Transl. Psychiatry. 2022;12:268. doi: 10.1038/s41398-022-02032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Yi L-T, et al. miR-124 antagonizes the antidepressant-like effects of standardized gypenosides in mice. J. Psychopharmacol. 2018;32:458–468. doi: 10.1177/0269881118758304. [DOI] [PubMed] [Google Scholar]
- 499.Wang X, et al. MiR-155 is involved in major depression disorder and antidepressant treatment via targeting SIRT1. Biosci. Rep. 2018;38:BSR20181139. doi: 10.1042/BSR20181139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Chao B, et al. Saikosaponin d downregulates microRNA-155 and upregulates FGF2 to improve depression-like behaviors in rats induced by unpredictable chronic mild stress by negatively regulating NF-κB. Brain Res. Bull. 2020;157:69–76. doi: 10.1016/j.brainresbull.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 501.Shimizu S, et al. The kampo medicine yokukansan decreases microRNA-18 expression and recovers glucocorticoid receptors protein expression in the hypothalamus of stressed mice. Biomed. Res. Int. 2015;2015:797280. doi: 10.1155/2015/797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Zhu WY, et al. Effect of Dingzhi Xiaowan on miR-16 expression and 5-HT reuptake. Zhongguo Zhong Yao Za Zhi. 2018;43:3513–3518. doi: 10.19540/j.cnki.cjcmm.20180614.001. [DOI] [PubMed] [Google Scholar]
- 503.van der Zee, Y. Y. et al. Blood miR-144-3p: a novel diagnostic and therapeutic tool for depression. Mol. Psychiatry (2022). [DOI] [PMC free article] [PubMed]
- 504.Cui X, et al. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomark. Med. 2016;10:943–952. doi: 10.2217/bmm-2016-0130. [DOI] [PubMed] [Google Scholar]
- 505.Mao J, et al. Abnormal expression of rno_circRNA_014900 and rno_circRNA_005442 induced by ketamine in the rat hippocampus. BMC Psychiatry. 2020;20:1. doi: 10.1186/s12888-019-2374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Song R, et al. Plasma circular RNA DYM related to major depressive disorder and rapid antidepressant effect treated by visual cortical repetitive transcranial magnetic stimulation. J. Affect. Disord. 2020;274:486–493. doi: 10.1016/j.jad.2020.05.109. [DOI] [PubMed] [Google Scholar]
- 507.Covington HE, 3rd, Maze I, Vialou V, Nestler EJ. Antidepressant action of HDAC inhibition in the prefrontal cortex. Neuroscience. 2015;298:329–335. doi: 10.1016/j.neuroscience.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Han A, Sung YB, Chung SY, Kwon MS. Possible additional antidepressant-like mechanism of sodium butyrate: targeting the hippocampus. Neuropharmacology. 2014;81:292–302. doi: 10.1016/j.neuropharm.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 509.Meylan EM, Halfon O, Magistretti PJ, Cardinaux JR. The HDAC inhibitor SAHA improves depressive-like behavior of CRTC1-deficient mice: possible relevance for treatment-resistant depression. Neuropharmacology. 2016;107:111–121. doi: 10.1016/j.neuropharm.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Goudarzi M, et al. Valproic acid administration exerts protective effects against stress-related anhedonia in rats. J. Chem. Neuroanat. 2020;105:101768. doi: 10.1016/j.jchemneu.2020.101768. [DOI] [PubMed] [Google Scholar]
- 511.Schroeder FA, et al. A selective HDAC 1/2 inhibitor modulates chromatin and gene expression in brain and alters mouse behavior in two mood-related tests. PLoS ONE. 2013;8:e71323. doi: 10.1371/journal.pone.0071323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Erburu M, et al. SIRT2 inhibition modulate glutamate and serotonin systems in the prefrontal cortex and induces antidepressant-like action. Neuropharmacology. 2017;117:195–208. doi: 10.1016/j.neuropharm.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 513.Ferland CL, Schrader LA. Regulation of histone acetylation in the hippocampus of chronically stressed rats: a potential role of sirtuins. Neuroscience. 2011;174:104–114. doi: 10.1016/j.neuroscience.2010.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]