This work demonstrates that diphtheria toxin is a potent trigger for the ZAKα-dependent ribotoxic stress response and the NLRP1 inflammasome in human skin cells. ZAKα inhibition is effective at limiting the skin damage caused by diphtheria toxin in vitro.
Abstract
The ZAKα-driven ribotoxic stress response (RSR) is activated by ribosome stalling and/or collisions. Recent work demonstrates that RSR also plays a role in innate immunity by activating the human NLRP1 inflammasome. Here, we report that ZAKα and NLRP1 sense bacterial exotoxins that target ribosome elongation factors. One such toxin, diphtheria toxin (DT), the causative agent for human diphtheria, triggers RSR-dependent inflammasome activation in primary human keratinocytes. This process requires iron-mediated DT production in the bacteria, as well as diphthamide synthesis and ZAKα/p38-driven NLRP1 phosphorylation in host cells. NLRP1 deletion abrogates IL-1β and IL-18 secretion by DT-intoxicated keratinocytes, while ZAKα deletion or inhibition additionally limits both pyroptotic and inflammasome-independent non-pyroptotic cell death. Consequently, pharmacologic inhibition of ZAKα is more effective than caspase-1 inhibition at protecting the epidermal barrier in a 3D skin model of cutaneous diphtheria. In summary, these findings implicate ZAKα-driven RSR and the NLRP1 inflammasome in antibacterial immunity and might explain certain aspects of diphtheria pathogenesis.
Introduction
Before the introduction of a worldwide vaccination campaign, diphtheria, caused by the Gram-positive extracellular bacterium Corynebacterium diphtheriae, was a deadly scourge throughout modern human history, with a case fatality rate of up to 20% in children under five (Sharma et al., 2019; Acosta and Tiwari, 2020). The respiratory form of diphtheria causes tissue necrosis of tonsils, throat, and larynx, leading to the formation of “pseudomembrane”—an eponymous hallmark symptom (latin, diphthera, “leather hide”). Fatality is typically associated with respiratory failure as well as myocarditis, polyneuropathy, and kidney damage caused by systemic dissemination of bacterial toxins. C. diphtheriae and related Corynebacterium ulcerans also infect skin wounds and cause cutaneous diphtheria, which is characterized by deep ulcers and extensive epidermal and dermal necrosis (Hadfield et al., 2000).
As early as 1888, Roux and Yersin and others discovered that the sterile filtrate of C. diphtheriae culture broth was sufficient in eliciting most of the diphtheria symptoms when injected into small animals. This was the first demonstration that diphtheria, in the strictest sense, is a toxin-mediated disease. Subsequent work demonstrated that a single secreted toxin, named diphtheria toxin (DT), was responsible for diphtheria pathogenesis (Holmes, 2000). These landmark discoveries paved the way for modern-day diphtheria treatment and prevention. When administered early, antisera against DT together with antibiotics are a curative treatment for diphtheria. Vaccines that elicit a humoral immune response against DT, rather than the bacterium itself, have proven highly effective and are a cornerstone component of childhood vaccinations across the world (reviewed in Murphy, 1996). As a result, worldwide incidence of diphtheria has been low in the last few decades, but localized outbreaks continue to occur even in 2023 in overcrowded settings with inadequate vaccine coverage (Blumberg et al., 2018; Wise, 2022).
DT is a typical A/B exotoxin: DTB binds to the host receptor human pro-HBEGF (heparin-binding EGF-like growth factor) and delivers DTA into the cytosol, where DTA catalyzes the conjugation of an ADP-ribose moiety to a posttranslationally acquired unusual amino acid known as diphthamide at position 715 of human Eukaryotic Translation Elongation Factor 2 (EEF2; Holmes, 2000). This reaction inactivates EEF2 and shuts down host protein synthesis. As continuous translation is a prerequisite for cellular survival, DT is well-known to cause apoptosis in human cells. As a result, DT has been adapted as a toxin payload in antibody conjugates for cancer treatment (Frankel et al., 2002). It is also widely used as a research tool to carry out cell ablation experiments in engineered mouse models (Palmiter et al., 1987). However, other cellular effects of DT have received considerably less attention.
Here, and in a companion manuscript (Pinilla et al., 2023), we demonstrate that DT and other EEF1/EEF2-targeting bacterial toxins also cause inflammasome-driven pyroptosis, in addition to apoptosis, of primary human epithelial cells. Using DT-treated human keratinocytes and 3D reconstructed skin as a model of cutaneous diphtheria, we found that DT-driven pyroptosis requires ZAKα-driven ribotoxic stress response (RSR) and the inflammasome sensor NLRP1. Pharmacologic inhibitors of ZAKα completely block IL-1 secretion and partially prevent epidermal damage by purified and toxigenic C. diphtheria. Our results define DT as a bona fide ribotoxic stress inducer and a species-specific trigger for IL-1–driven inflammation in human epithelia. These results also suggest a role of RSR and the human NLRP1 inflammasome in the pathogenesis of diphtheria and likely other bacterial infections.
Results and discussion
EEF1/2-targeting bacterial exotoxins activate the NLRP1 inflammasome
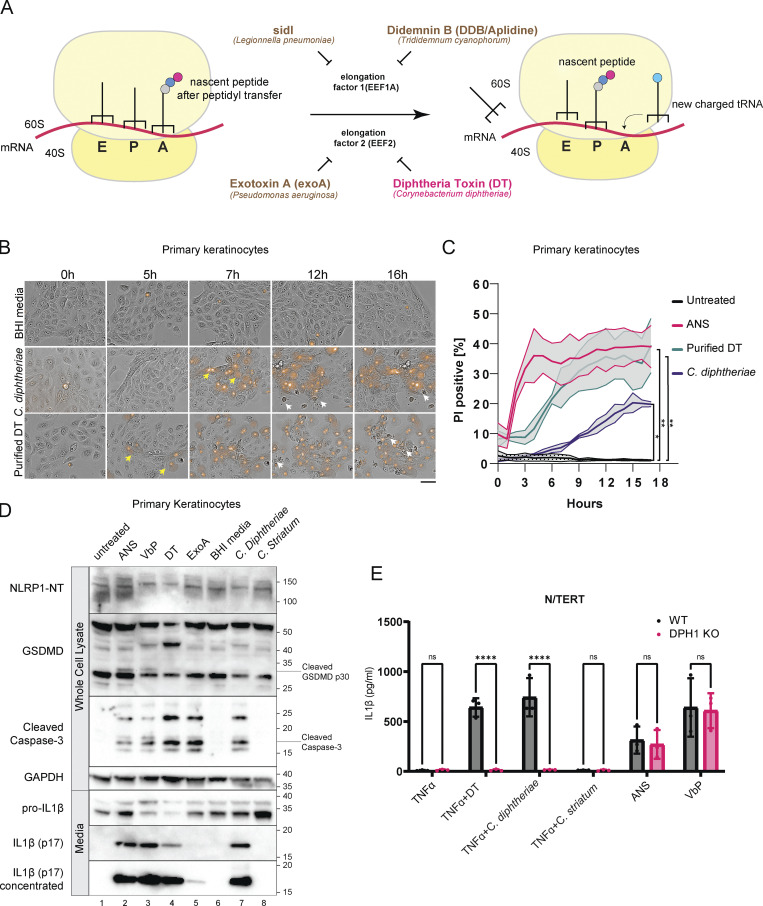
NLRP1 is a versatile sensor for the cytosolic inflammasome complex (Martinon et al., 2002; Mitchell et al., 2019; Bachovchin, 2021). Human NLRP1 was recently found to sense agents that cause ribosome stalling and/or collisions, e.g., UVB and anisomycin (ANS; Robinson et al., 2022; Jenster et al., 2023). This discovery prompted us to investigate whether bacterial virulence factors that are known to affect host translation could also activate human NLRP1. We tested three secreted bacterial exotoxins, C. diphtheriae DT, Pseudomonas aeruginosa exotoxin A (exoA), and Legionella pneumophila sidI (Fig. 1 A). DT and exoA are ADP-ribosylating enzymes that inactivate EEF2 at a conserved diphthamide residue, whereas sidI acts as a transfer RNA mimic that inactivates EEF1 and ribosome components (Subramanian et al., 2022 Preprint; Joseph et al., 2019 Preprint). A small molecule inhibitor of EEF1, didemnin B (DDB, also known as Aplidine; Crews et al., 1994; Fig. 1 A), was also tested, with ANS as a positive control. SidI and DDB have recently been linked to the RSR pathway downstream of ZAKα (Subramanian et al., 2022 Preprint).
Figure 1.
C. diphtheriae, DT, and exoA activate pyroptosis in primary keratinocytes. (A) Simplified cartoon representation of eukaryotic ribosome translocation and EEF1/EEF2-targeting toxins. E, exit; P, peptidyl; A, aminoacyl. (B) Live cell imaging of primary keratinocytes treated with BHI media, sterile filtrates of C. diphtheriae grown in iron-restricted media or recombinant DT (150 ng/ml). The concentration of DT in C. diphtheriae media was normalized to ∼150 ng/ml. Brightfield images were overlaid with PI fluorescence (red). Images were taken at 20× magnification. Images are representative of three independent experiments. Distinct cell death morphology is marked by yellow arrows (pyroptosis, PI+ve cells with membrane swelling) and white arrows (apoptosis, cell shriveling). Only select representative cells are marked. Scale bar represents 100 μm. (C) Quantification of the percent of PI+ve cells over time in B. Error bars represent three biological replicates, with each drug treatment considered as one replicate. Significance values were calculated from Student’s t test at the 7-h time point. *, P ≤ 0.05. **, P ≤ 0.01. (D) Immunoblot of inflammasome activation markers in primary keratinocytes 24 h after treatment. ANS (1 µM); VbP (1 µM); DT (150 ng/ml), exoA (1 µg/ml). Cells in lane 6 were incubated with BHI broth, while lanes 7 and 8 were incubated with sterile-filtered C. diphtheriae or C. striatum conditioned BHI media. Immunoblot is representative of three replicate experiments. (E) IL-1β ELISA from TNFα-primed wild-type or DPH1 KO N/TERT culture media. Media were harvested 18 h after treatment. Significance values were calculated from one-way ANOVA with multiple group comparisons, from three biological replicates. ns, not significant. ****, P ≤ 0.0001. Source data are available for this figure: SourceData F1.
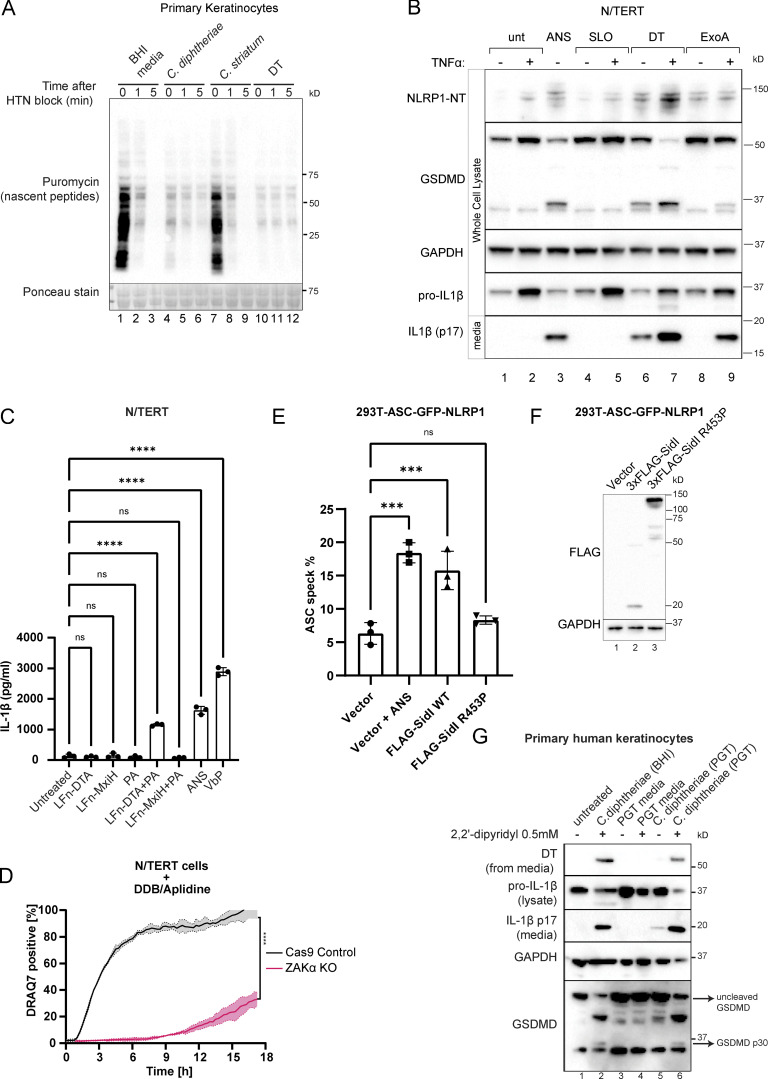
In primary keratinocytes, purified DT potently inhibits ribosome elongation and abrogates total protein synthesis (Fig. S1 A), consistent with EEF2 inactivation. Prolonged DT treatment (16–24 h) leads to caspase-3 cleavage, similar to prior observations in other human cell types (Fig. 1 D; Narayanan et al., 2005; Cheng, 2010). However, the early effects of DT intoxication (∼5 h) in human keratinocytes are more characteristic of pyroptosis instead of apoptosis, as evident by membrane swelling, propidium iodide (PI) uptake (Fig. 1 B, yellow arrows; Fig. 1 C), IL-1β p17 secretion, and the processing of full-length Gasdermin D (GSDMD) into the p30 form (Fig. 1 D). Dying cells with classical “shriveled” apoptotic morphology only became apparent much later (>10 h; Fig. 1 B, white arrows). In immortalized N/TERT-1 (N/TERT) cells, DT also triggered inflammasome activation and pyroptosis, albeit with delayed kinetics and less potency (Fig. S1 B). exoA induced minimal pyroptosis in N/TERT cells as measured by IL-1β secretion and GSDMD cleavage (Fig. S1 B). This difference is likely due to lower expression levels of the respective entry receptors pro-HBEGF and LRP1 as compared with primary cells. TNFα significantly sensitized N/TERT cells both to DT and exoA-induced pyroptosis (Fig. S1 B), in agreement with published findings that it upregulates LRP1 and pro-HBEGF (Morimoto et al., 1991; Morimoto and Bonavida, 1992). TNFα additionally increased the expression of IL-1β and NLRP1 itself, thus acting as a priming signal in N/TERT cells. Bypassing the requirement of pro-HBEGF using an engineered anthrax factor conjugated DTA (LFn-DTA) in combination with protective antigen (PA) also robustly induced IL-1β in N/TERT cells (Milne et al., 1995; Fig. S1 C).
Figure S1.
Additional data on the effect of EEF2 targeting bacterial toxins, including DT. (A) Anti-puromycin immunoblot of primary keratinocytes subjected to the harringtonine run-off assay. Cells were treated with BHI media, sterile filtrates of C. diphtheriae, C. striatum for 6 h, or purified DT (150 ng/ml) for 3 h before harringtonine addition (2 µg/ml). Nascent peptides were then labeled with 10 µg/ml puromycin for 10 min at different intervals. (B) Immunoblot of inflammasome components NLRP1, GSDMD, and IL-1β in N/TERT with and without TNFα priming (25 ng/ml, 8 h). Streptolysin O (SLO, 1 µg/ml) was included as additional negative control. (C) IL-1β ELISA of culture media from N/TERT cells treated with the indicated recombinant proteins or compounds. LFn (150 ng/ml): Lethal Factor from B. anthracis (aa 34–288). PA (300 ng/ml). MxiH: Shigella flexneri type 3 secretion needle protein. All cells were primed with TNFα. Significance values were calculated from one-way ANOVA with multiple group comparisons. Error bars derived from data from three technical replicates. The graph represents one of two biological replicates. (D) Kinetics of PI uptake in unprimed N/TERT cells treated with DDB/Aplidine. Significance values were calculated from Student’s t test at 4 h. Error bars are derived from data from three technical replicates. Data represent one of two biological replicates. (E) The percentage of cells with ASC-GFP specks among 293T-ASC-GFP-NLRP1 cells transfected with the indicated plasmids. Cells were fixed 24 h after transfection. ASC-GFP specks were visualized using GFP epifluorescence and normalized to the total number of cells per field of view using DAPI nuclear counterstain. Error bars are derived from data from three technical replicates. Data represent one of two biological replicates. (F) Immunoblot of overexpressed 3xFLAG-sidI and R453P glycolysase-defective mutant. Note that the level of wild-type sidI is much lower than the R453P mutant due to its strong inhibitory effect on translation, as reported previously by Subramanian et al. (2022) Preprint. (G) Immunoblot of inflammasome activator markers GSDMD and IL-1β in primary keratinocytes treated with the indicated bacterial filtrate. C. diphtheriae was cultured in BHI broth and PGT media (see Materials and methods). ***, P ≤ 0.001; ****, P ≤ 0.0001. Source data are available for this figure: SourceData FS1.
Aside from exoA and DT, EEF1 inhibitor DDB also triggered pyroptosis in N/TERT cells (Fig. S1 D). Transient overexpression of the proteinaceous L. pneumoniae ribotoxic toxin sidI, but not its glycosylase-defective mutant, similarly induced the formation of ASC (apoptosis-associated speck-like protein containing a CARD)-GFP specks in a 293T-based NLRP1 inflammasome reporter cell line (Fig. S1, E and F). Together, these data demonstrate that multiple bacterial EEF1 and EEF2 toxins can activate the human NLRP1 inflammasome.
C. diphtheriae induces pyroptosis along with other forms of cell death in human keratinocytes
Next, we tested the effect of DT+ve pathogenic C. diphtheriae on human keratinocytes. In all clinical C. diphtheriae strains, DT is encoded on a prophage under the control of an iron-responsive repressor known as DtxR (Holmes, 2000). In agreement with previous findings, the secretion of DT from the toxigenic C. diphtheriae strain was low in iron-containing basal media but strongly induced by the iron chelator 2-2′-dipyridyl within 4 h (Fig. S1 G). Consequently, the cytotoxic effects of C. diphtheriae on primary keratinocytes were dependent on iron depletion, as C. diphtheriae grown in iron-containing media was significantly attenuated at inducing IL-1β secretion and GSDMD cleavage (Fig. S1 G). A DT−ve Corynebacterium species, C. striatum, which also colonizes the human skin, does not elicit pyroptosis or caspase-3 cleavage in primary keratinocytes or N/TERT cells (Fig. 1, D and E).
To further confirm that C. diphtheriae–driven pyroptosis is strictly downstream of EEF2 inactivation, we knocked out DPH1, an enzyme that is required to convert EEF2 His715 into diphthamide, in N/TERT cells (Liu et al., 2004). DPH1 deficient cells were completely unable to secrete IL-1β in response to purified DT, but their ability to respond to other NLRP1 agonists is unaffected (Fig. 1 E). Taken together, these results demonstrate that DT is the predominant cytotoxic factor secreted by toxigenic C. diphtheria and the inactivation of EEF2 by DT is essential for C. diphtheriae–induced cell death in human keratinocytes.
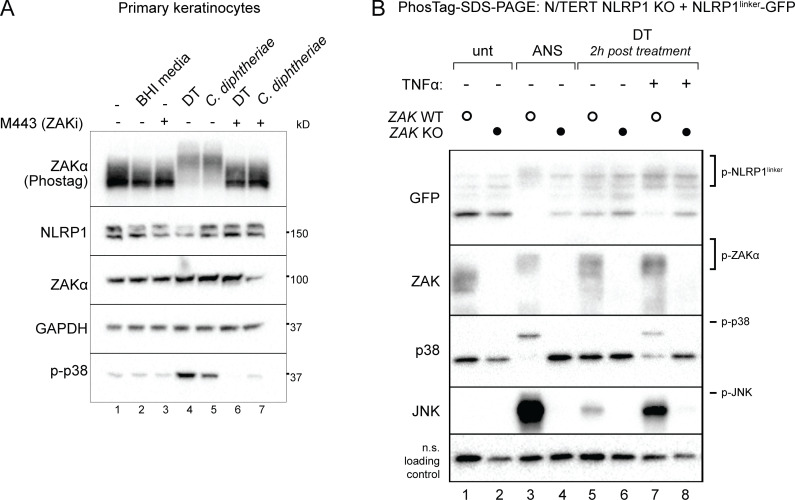
We next examined DT-induced signaling events upstream of cell death. Recently, we and others found that ribosome stalling agents cause NLRP1-dependent pyroptosis through the RSR-sensing MAP3K ZAKα and its effector p38 (Vind et al., 2020a, 2020b; Wu et al., 2020; Robinson et al., 2022; Jenster et al., 2023). Thus, we tested if DT can trigger RSR similarly to ANS. Primary keratinocytes treated with C. diphtheriae conditioned media or purified DT undergo p38 phosphorylation and ZAKα autophosphorylation within 3 h (Fig. 2 A). In addition, using a previously published N/TERT reporter cell line expressing GFP-tagged NLRP1 linker domain (NLRP1linker-GFP, which lacks the inflammasome activating domains and is not targeted for proteasomal degradation; Robinson et al., 2022), we found that DT induced similar patterns of NLRP1linker hyperphosphorylation as ANS (Fig. 2 B). Genetic deletion of ZAKα or a small molecule inhibitor of ZAKα kinase activity, M443, abrogated all hallmarks of ribotoxic stress signaling, including the phosphorylation of ZAKα itself, p38, JNK, and the hyperphosphorylation of NLRP1linker (Fig. 2, A and B). Thus, DT indeed functions as a bona fide ribotoxic stress inducer.
Figure 2.
DT and C. diphtheriae trigger RSR downstream of EEF2 inactivation. (A) Immunoblot of ZAKα, p38 NLRP1 in primary keratinocytes treated with the indicated triggers in the presence of DMSO or M443 (1 µM). M443 was added 10 min before DT or bacterial filtrate. Lysates were harvested 3 h after treatment. Immunoblot representative of three replicate experiments. (B) Immunoblot of RSR kinases and GFP-NLRP1linker in control and ZAKα KO N/TERT cells. ANS-treated cells were harvested 2 h after treatment. Indicated cells were primed with 25 ng/ml TNFα overnight and then treated with 150 ng/ml DT. Immunoblot representative of three replicate experiments. Source data are available for this figure: SourceData F2.
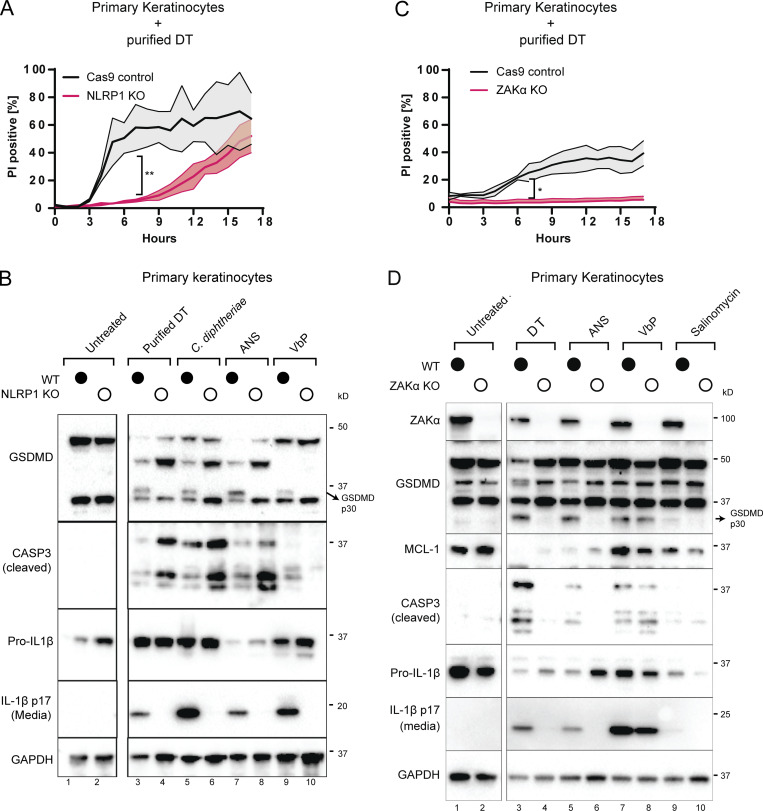
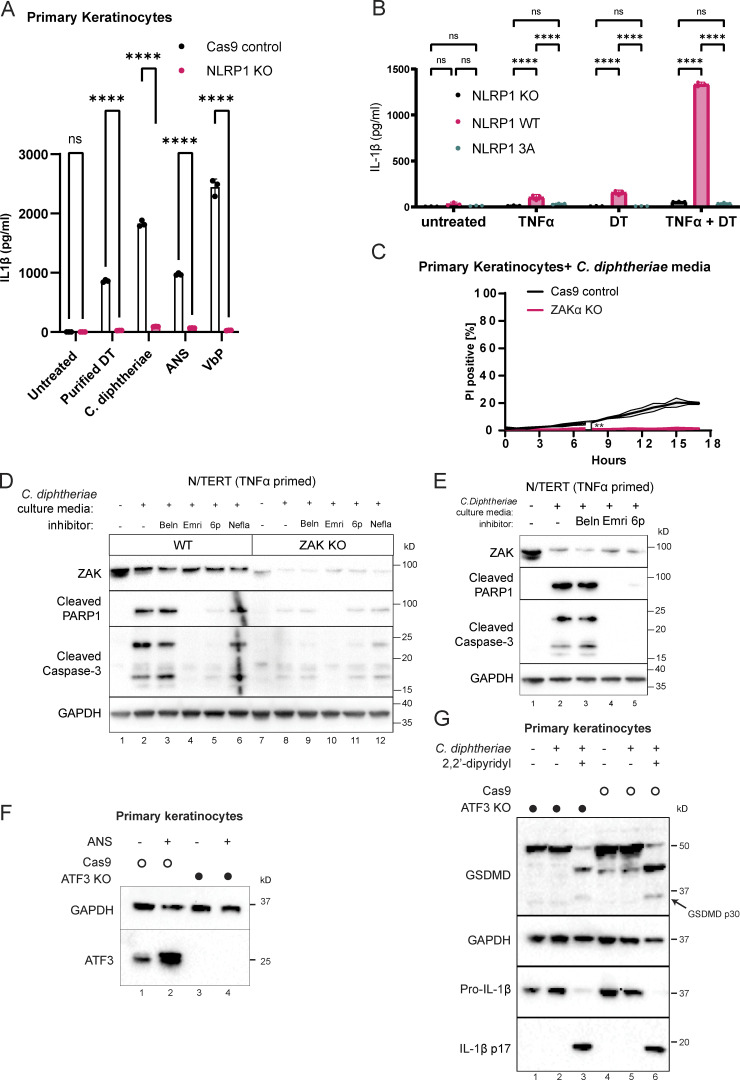
General protein synthesis inhibitors including DT are well known to induce apoptosis via a variety of mechanisms, such as the rapid depletion of unstable anti-apoptotic proteins and Fas associated via death domain–dependent caspase-8 activation (Orzalli et al., 2021; Thorburn et al., 2003; Cheng, 2010; Narayanan et al., 2005). Given our observations that DT also triggers pyroptosis in primary human keratinocytes, we sought to dissect the genetic regulation of pyroptotic vs. nonpyroptotic cell death downstream of DT. In line with the prior finding that NLRP1 is the main inflammasome sensor expressed in primary epithelial cells, NLRP1 KO abrogated the rapid PI uptake (<5 h) that is characteristic of pyroptosis (Fig. 3 A), but did not alter the extent of PI uptake at later points, which is typically associated with late-stage apoptosis (Fig. 3 A). NLRP1 KO also abrogated other hallmarks of inflammasome activation, including IL-1β secretion and GSDMD p30 cleavage (Fig. 3 B and Fig. S2 A), as did mutating the ZAKα/p38 recognition motif on the NLRP1 linker domain (Fig. S2 B). By contrast, NLRP1 KO cells showed increased levels of caspase-3 cleavage (Fig. 3 B), suggesting that apoptosis becomes the predominant mode of cell death following DT exposure when the NLRP1 inflammasome pathway is blocked. A similar effect was observed when pyroptosis was blocked using the caspase-1 inhibitor, belnacasan (Fig. S2 E).
Figure 3.
DT and C. diphtheriae cause pyroptosis in a ZAKα and NLRP1-dependent manner but also cause other forms of cell death. (A) Comparison of PI uptake kinetics between control and NLRP1 KO primary keratinocytes in response to purified DT. Error bars represent three biological replicates, with each drug treatment considered as one replicate. Significance values were calculated from Student’s t test at the 7-h time point. **, P ≤ 0.01. (B) Immunoblot of GSDMD, cleaved caspase-3, and IL-1β in WT and NLRP1 KO keratinocyte lysates or media 24 h after the indicated treatment. Immunoblot is representative of three replicate experiments. (C) Comparison of PI uptake kinetics between control and ZAKα KO primary keratinocytes in response to purified DT. Error bars represent three biological replicates, with each drug treatment considered as one replicate. Significance values were calculated from Student’s t test at the 7-h time point. *, P ≤ 0.05. (D) Immunoblot of ZAKα, GSDMD, MCL-1, cleaved caspase-3 and IL-1β in WT and ZAKα KO keratinocyte lysates or media 24 h after the indicated treatment. Salinomycin (10 µM) does not activate the NLRP1 inflammasome and was used as a negative control. Immunoblot is representative of three replicate experiments. Source data are available for this figure: SourceData F3.
Figure S2.
Additional characterization of NLRP1 phosphorylation, ZAKα, and ATF3 in DT-induced pyroptosis and apoptosis. (A) IL-1β ELISA from control and NLRP1 KO human primary keratinocytes. Media were harvested 18 h after treatment. Purified DT (150 ng/ml). ANS (1 µM). VbP (3 µM). (B) IL-1β ELISA from NLRP1 KO N/TERT cells rescued with wild-type NLRP1 or NLRP1 3A mutant primed or treated with the indicated conditions. TNFa priming 18 h, media harvested 18 h later after treatment. (C) Kinetics of PI uptake for control and ZAKα KO primary keratinocytes. Error bars are derived from data from three technical replicates. Data represent one of two biological replicates. Significance values were calculated from Student’s t test at the 7-h time point. (D) Immunoblot of apoptotic markers (cleaved caspase-3 and PARP1) from the lysates of TNFα-primed N/TERT cells treated with C. diphtheriae BHI media filtrate and the indicated inhibitors. Beln: caspase-1 inhibitor belnacasan (5 µM). Emri: pan-caspase inhibitor emricasan (5 µM). 6p (0.5 µM). (E) Immunoblot of apoptotic markers from the lysates of TNFα-primed WT and ZAKα KO N/TERT cells treated with C. diphtheriae BHI media filtrate and the indicated inhibitors. Beln: caspase-1 inhibitor belnacasan (5 µM). Emri: pan-caspase inhibitor emricasan (5 µM). 6p (0.5 µM). Nefla: p38 inhibitor neflammapimod (0.5 µM). (F) ATF3 immunoblot in control and ATF3 KO primary keratinocytes. Lysates were harvested 3 h after ANS. (G) Immunoblot of GSDMD and IL-1β comparing inflammasome activation of Cas9 control and ATF3 KO keratinocytes treated with C. diphtheriae media (normalized to ∼150 ng/ml DT). ns, not significant; **, P ≤ 0.01; ****, P ≤ 0.0001. Source data are available for this figure: SourceData FS2.
In contrast to NLRP1 KO, ZAK KO reduced DT- or C. diphtheriae–induced PI uptake throughout the 18-h time course, both at early and later time points (Fig. 3 C and Fig. S2 C). Thus, the effect of ZAKα on DT toxicity is more profound and extends beyond pyroptosis. ZAKα KO cells not only showed reduced GSDMD cleavage and IL-1β secretion but also had reduced caspase-3 cleavage (Fig. 3 D). Blocking the ZAKα kinase activity using compound 6p also reduced caspase-3 cleavage (Fig. S2, D and E) in DT-treated N/TERT cells. Thus, ZAKα, as the apex sensor in the RSR cascade, is not only essential for NLRP1-driven pyroptosis but is also partially involved in caspase-3–driven apoptosis. It is important to note that ZAKα KO cells still succumbed to DT over time and showed a loss of anti-apoptotic protein MCL-1 (Fig. 3 D). Thus, our results suggest that ZAKα regulates multiple cell death pathways triggered by DT, but is unlikely to be the only means by which cells die after ribosome inactivation.
Transcriptomic analysis of RSR supports a broader role of ZAKα
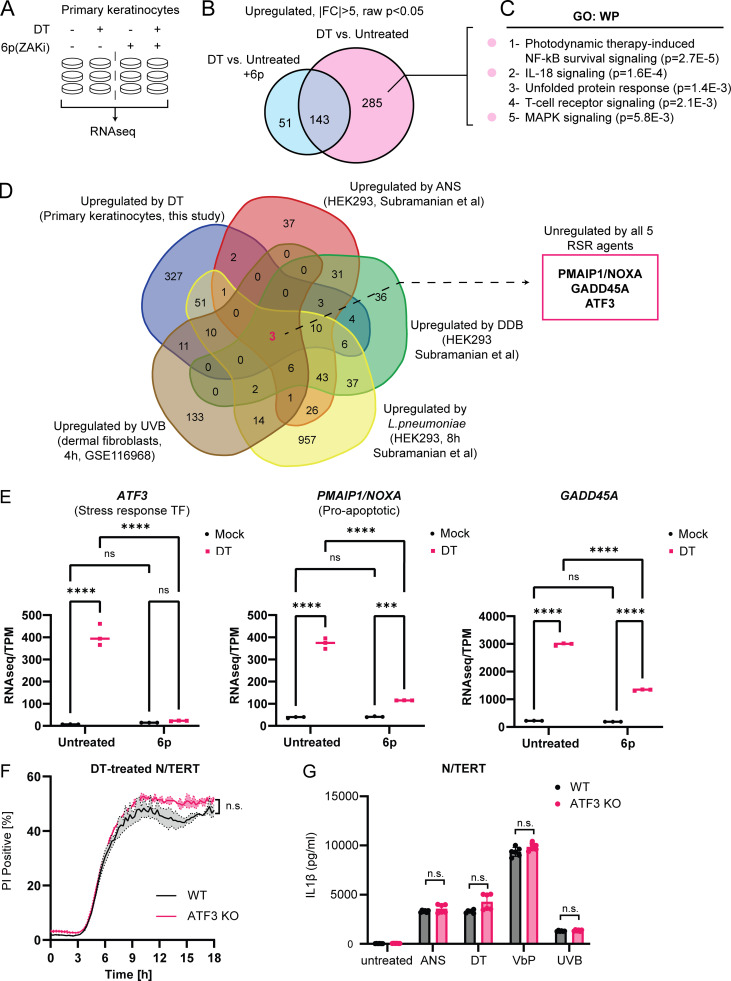
To gain more mechanistic insight into the effect of RSR signaling in DT cytotoxicity, we performed bulk RNA sequencing (RNAseq) on primary keratinocytes treated with DT for 4 h in the presence or absence of compound 6p, a specific ZAKα inhibitor (Yang et al., 2020; Fig. 4 A). Adding further support to the finding that DT activates both ZAKα-driven RSR and pyroptosis, Gene Ontology analysis of ZAKα-dependent DT-induced transcripts revealed enrichment for MAPK and IL-18 signaling pathway components (Fig. 4, B and C). Interestingly, this analysis also uncovered a significant overlap between DT and photodynamic therapy (Fig. 4 C), a treatment modality for skin hyperplasia that relies on laser-induced cytotoxicity and is mechanistically similar to UV-induced cell death (Choi et al., 2015). As UVB irradiation is known to trigger ZAKα-driven RSR, this finding suggests that different ribotoxic stress agents might induce a common transcriptional program. By intersecting our data with those from published datasets, three genes were found to be consistently induced by five distinct RSR agents: DT, UVB, ANS, DDB, and L. pneumophila in a cell type agnostic manner (Fig. 4 D; Subramanian et al., 2022 Preprint). These include the known pro-apoptotic regulator PMAIP5/NOXA (Fig. 4 D), stress-response transcriptional factor, ATF3, and a known stress-responsive gene GADD45A. 6p treatment significantly blunted the induction of all three genes by DT in primary keratinocytes (Fig. 4 E). Given the known roles of NOXA and GADD45A (Salvador et al., 2013; Oda et al., 2000) in promoting apoptosis, it is likely both contribute to ZAKα-dependent non-pyroptotic cell death induced by DT, although the detailed mechanisms require further investigation. ATF3 has a known role in the cellular response to UVB and was recently shown to mediate cell survival in L. pneumophila infection (Turchi et al., 2008; Subramanian et al., 2022 Preprint). We thus tested whether ATF3 is upstream of the NLRP1 inflammasome or represents a distinct arm of RSR in primary human keratinocytes. ATF3 KO did not alter either the kinetics of DT- or ANS-induced pyroptosis in either primary keratinocytes or N/TERT cells as measured by PI uptake and IL-1β secretion (Fig. 4, F and G; and Fig. S2, F and G). Thus, ATF3 and NLRP1 represent two distinct arms of RSR, both downstream of ZAKα.
Figure 4.
Identification of transcripts that are induced by multiple RSR agents across multiple cell types. (A) Design of RNAseq experiment using primary keratinocytes. Compound 6p is a ZAKα inhibitor. (B) Venn diagram showing the identification of 285 ZAKα-dependent, DT-induced upregulated transcripts in primary keratinocytes. (C) Gene Ontology (GO) analysis of ZAKα-dependent DT-induced upregulated transcripts. (D) Venn diagram demonstrating the overlap of transcriptional changes elicited by the indicated RSR agents. The source cell types are indicated in brackets. Fold change (FC) > 2 used a cutoff for datasets published in Subramanian et al. (2022) (Preprint; DDB, ANS, and L. pneumoniae). FC > 1.5 was used as the cutoff for UVB (GEO accession: GSE116968) in accordance with the depositing authors’ analysis. (E) Transcripts per kilobase million (TPM) transcript levels of ATF3, PMAIP1/NOXA, and GADD45A in mock and DT-treated control or 6p-treated primary keratinocytes. Significance values were calculated from -way ANOVA from three biological replicates shown. ns, not significant; ****, P ≤ 0.0001. (F) Kinetics of PI uptake of primed wild-type and ATF3 KO N/TERT cells treated with DT. Significance values were calculated from Student’s t test at the 7-h time point. n.s., not significant. (G) IL-1β ELISA of control and ATF3 KO primed N/TERT cells after the drug/toxin treatments. Significance values were calculated from one way ANOVA from two biological replicates. n.s., not significant.
ZAKα and p38 inhibitors rescue epithelial integrity in a 3D skin model of cutaneous diphtheria
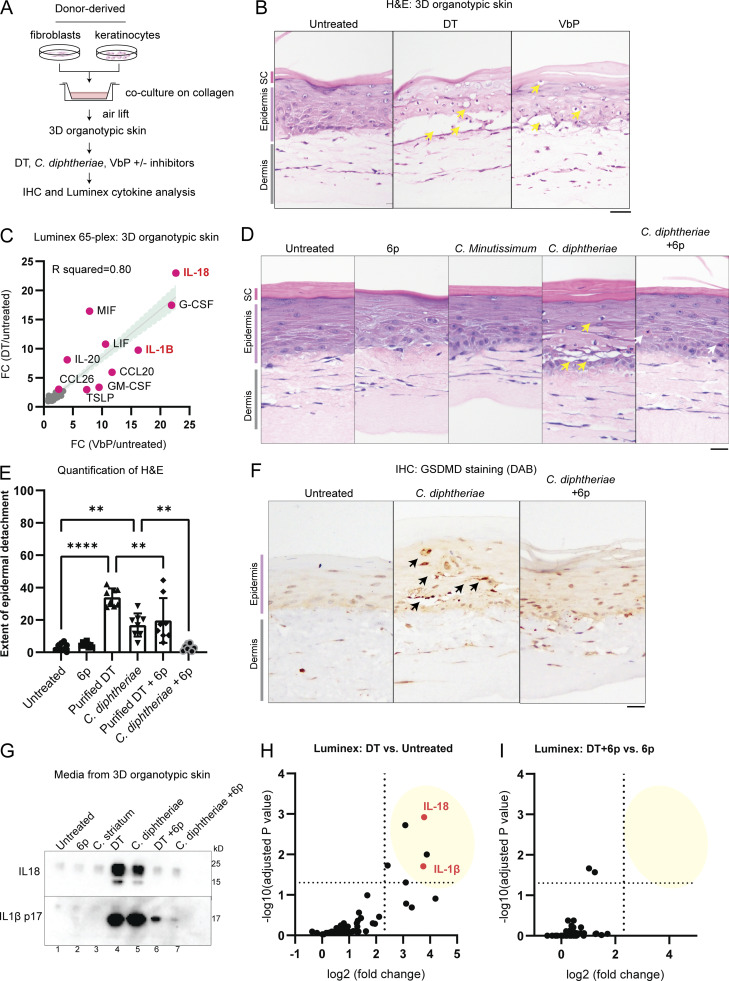
Next, we employed human 3D organotypic skin as an approximate model for cutaneous diphtheria (Fig. 5 A). This system offers distinct advantages over murine models since (1) NLRP1 is not expressed in murine skin (Sand et al., 2018), (2) muNLRP1 lacks the linker region that harbors the ZAKα and p38 phosphorylation sites (Robinson et al., 2022; Jenster et al., 2023), and (3) murine pro-HBEGF does not function as a DT receptor (Ivanova et al., 2005). Using H&E staining, we observed that recombinant DT causes significant damage to the epidermis in 3D organotypic skin culture, as evidenced by vacuolated keratinocytes with condensed nuclei in basal and spinous layers and significant detachment of the epidermis from the dermis (Fig. 5 B, yellow arrows; Fig. S3 A). Next, we measured the levels of 65 inflammatory cytokines and chemokines in DT- and Val-boro-Pro (VbP)–induced 3D organotypic skin using Luminex. Remarkably, DT and VbP led to the secretion of a nearly identical set of pro-inflammatory cytokines/chemokines, with IL-18 and IL-1β among the most induced (Fig. 5 C). This suggests that even though VbP and DT both have unrelated, non-inflammasome-dependent activities, their effects on cytokine secretion are dominated by the inflammasome response.
Figure 5.
ZAKα inhibition rescues epidermal integrity by limiting pyroptosis in a model of cutaneous diphtheria. (A) Outline of the 3D human skin model of cutaneous diphtheria. (B) H&E staining demonstrating the histological changes caused by DT and VbP. Yellow arrows indicate dyskeratotic keratinocytes with vacuolated cytoplasm and condensed nuclei. Images representative of three biological replicates. Tissues were fixed 24 h after treatment. (C) Correlation between the fold change (FC) of 65 cytokines/chemokines after 24 h of DT or VbP treatment. (D) H&E staining of 3D skin treated with the indicated bacteria filtrate or compound. Yellow arrows indicate dyskeratotic keratinocytes with vacuolated cytoplasm and condensed nuclei. Gray arrows mark putative apoptotic cells with eosin-rich cytosol and condensed nuclei. Compound 6p is a ZAKα inhibitor. Images representative of three biological replicates. Tissues were fixed 24 h after treatment. (E) Quantification of the extent of the detachment between the dermal–epidermal layer based on the H&E staining of 3D skin in D. Significance values were calculated from one-way ANOVA. **, P ≤ 0.01. ****, P ≤ 0.0001. (F) GSDMD p30 staining of 3D skin treated with C. diphtheriae filtrate with and without compound 6p. Black arrows indicate membranous staining around foci of epithelial barrier damage. (G) Western blot of IL-18 and IL-1β p17 in the cultured media of 3D skin after the indicated treatment. (H) Significantly upregulated cytokines/chemokines in DT-treated 3D skin culture relative to untreated 3D skin from D. Log10(adjusted P values) and log2(fold change) were calculated from a multiparametric t test. Fold change cutoff is set at 5. (I) Significantly upregulated cytokines/chemokines in DT + 6p 3D skin culture relative to 6p only 3D skin from D. Log10(adjusted P values) and log2(fold change) were calculated from a multiparametric t test. Fold change cutoff is set at 5. Source data are available for this figure: SourceData F5.
Figure S3.
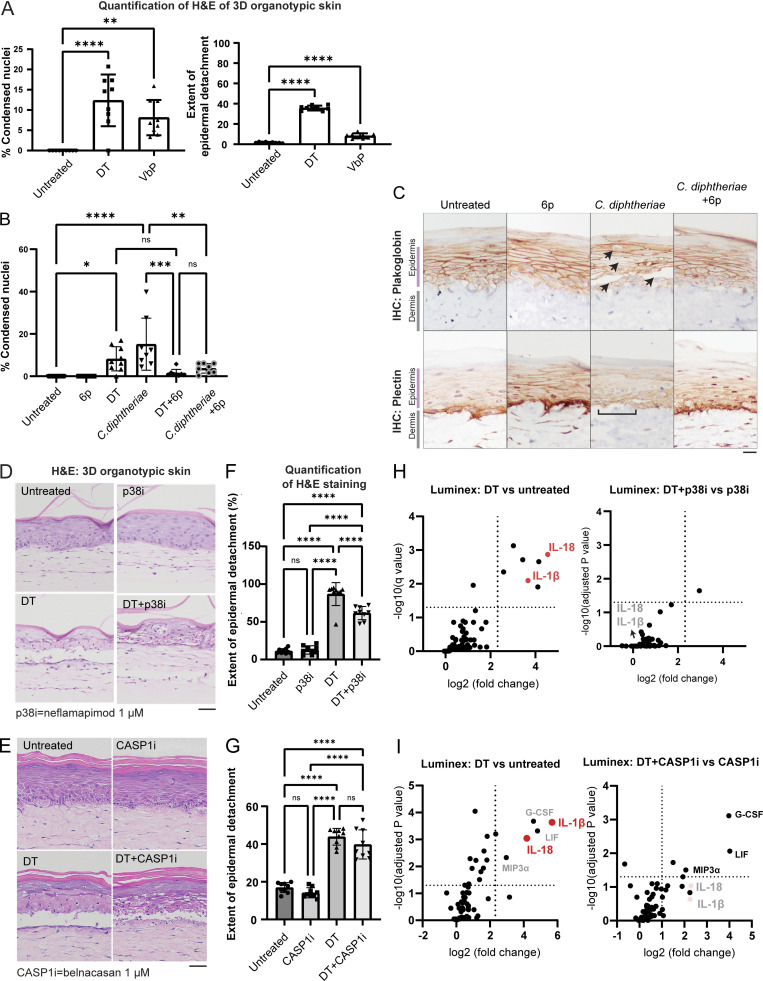
Further data on C. diphtheriae–induced skin damage in 3D skin cultures. (A) Quantification of the percentage of cells with condensed nuclei and the detachment between the dermal–epidermal layer, based on the H&E staining of 3D skin. (B) Quantification of the percentage of cells with condensed nuclei based on the H&E staining of 3D skin sections shown in Fig. 5. (C) Immunohistochemistry (IHC) staining of plakoglobin (desmosome and adherens junction marker) and plectin (hemidesmosome marker) in 3D skin sections treated with the indicated conditions. Black arrows indicate foci of epidermal damage with loss of plakoglobin staining. Brackets indicate areas of DEJs with disorganized plectin staining. Scale bar = 50 µm. (D) H&E staining of 3D skin treated with DT and neflamapimod (p38i). Tissues were fixed 24 h after treatment. Scale bar = 50 µm. (E) H&E staining of 3D skin treated with DT and belnacasan (CASP1i). Tissues were fixed 24 h after treatment. Scale bar = 50 µm. (F) Quantification of the extent of the detachment between the dermal–epidermal layer based on the H&E staining of p38i-treated organotypic skin samples. (G) Quantification of the extent of the detachment between the dermal–epidermal layer based on the H&E staining of CASP1i-treated organotypic skin samples. (H) Significantly upregulated cytokines/chemokines in the indicated organotypic skin samples. Log10(adjusted P value) and log2(fold change) were calculated from a multiparametric t test. The samples correspond to D and F. Media from organotypic skin collected 24 h after treatment with DT and the inhibitors. (I) Significantly upregulated cytokines/chemokines in the indicated organotypic skin samples. Log10(adjusted P value) and log2(fold change) were calculated from a multiparametric t test. The samples correspond to E and G. Media from organotypic skin were collected 24 h after treatment with DT and the inhibitors. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
We next compared the relative efficacy of ZAKα, p38, and caspase-1 inhibition at limiting DT- and C. diphtheriae–induced epidermal damage using chemical inhibitors: 6p (ZAKi), Neflamapimod (p38i), and belnacasan (CASP1i; Fig. 5, D–F; and Fig. S3, B–E). All three inhibitors proved effective at blocking IL-18 and IL-1β secretion in DT-treated 3D skin cultures (Fig. 5, G–I; and Fig. S3, H and I), but they differed in their abilities to limit the epidermal disruption. CASP1i did not significantly alter DT-induced epidermal detachment (Fig. S3, E–G), while p38i proved partially effective (Fig. S3, D–F). ZAKα inhibitor 6p had the most dramatic effect. 6p significantly reduced the extent of epidermal detachment and the frequency of vacuolated nuclei (Fig. 5, D and E; and Fig. S3 B), as well as other signs of epidermal damage caused by DT and C. diphtheriae media, including (1) focal loss of adherens junctions and desmosomes in the spinous layer, (2) disruption of hemidesmosomes in the basal layer (Fig. S3 C), and (3) GSDMD p30 accumulation (Fig. 5 F). Remarkably, Luminex analyses revealed that in the presence of 6p, DT treatment no longer induced any of the inflammatory cytokines profiled in the Luminex panel (Fig. 5, H and I). This stands in contrast with CASP1i (belnacasan) treatment, which selectively abrogated inflammasome-dependent cytokines IL-1β and IL-18 but had much less pronounced effects on inflammasome-independent cytokines such as G-CSF (Fig. S3 I). It is important to note that 6p is unable to rescue all aspects of DT-associated toxicity. For instance, eosin-rich, but non-vacuolated keratinocytes could be readily observed in 3D skin treated with C. diphtheria even in the presence of 6p (Fig. 5 D, right panel, gray arrows). We hypothesize that these cells represent keratinocytes undergoing non-pyroptotic cell death, likely as a result of prolonged protein synthesis block. Notwithstanding, these findings confirm the observations in 2D culture and establish that NLRP1 and ZAKα have distinct roles in DT-induced signaling and cytotoxicity. While NLRP1 specifically controls the inflammasome response, ZAKα plays a much broader role by acting as an upstream regulator of both NLRP1-driven pyroptosis and non-pyroptotic cell death.
In summary, we report that multiple bacterial exotoxins that target ribosome elongation factors EEF1 and EEF2 activate ZAKα-dependent RSR and the NLRP1 inflammasome in primary human epithelial cells. Using cutaneous diphtheria as a model, we demonstrate that one of these toxins, DT, causes keratinocyte-intrinsic ribotoxic stress and IL-1–driven inflammation via the NLRP1 inflammasome. While NLRP1 exclusively activates pyroptosis in C. diphtheria–treated primary keratinocytes and 3D skin culture, ZAKα, the apex sensor of the RSR pathway, seems to control both pyroptosis and non-pyroptotic cell death in DT-treated skin culture. As a result, pharmacologic inhibition of ZAKα alleviates both C. diphtheria–induced skin damage and inflammation.
Our results suggest that RSR constitutes a pathogen-triggered proinflammatory signaling network in which the human NLRP1 inflammasome forms a specific sub-branch. Since the activation of NLRP1 by ribotoxic stress requires a human-specific linker domain, NLRP1 likely became an RSR effector relatively recently during mammalian evolution, enabled by the acquisition of the ZAKα/p38 phosphorylation sites in higher primates. It is tempting to speculate that this evolutionary change conferred better immunity to a pathogen, such as diphtheria or certain viruses, and hence became fixed in the human genome (Tsu et al., 2021).
Viewed in a different light, our discovery represents an example of “effector triggered immunity” in humans (Stuart et al., 2013; Fontana and Vance, 2011; Lopes Fischer et al., 2020), where a sensor protein (ZAKα, upstream of NLRP1) becomes activated in response to perturbations of an essential cellular process (ribosome elongation) caused by a pathogen virulence factor (DT, exoA, and sidI). It is noteworthy that a similar mechanism has been demonstrated for exoA in nematodes (McEwan et al., 2012; Dunbar et al., 2012). While RSR- and inflammasome-dependent cytokines could promote bacterial clearance in early stages of diphtheria infection, e.g., by recruiting neutrophils to the infected sites, unchecked NLRP1-driven inflammation could also be maladaptive and host-detrimental by hindering the wound healing response at later stages of infection. We speculate that the loss of epithelial barrier integrity caused by ZAKα-driven RSR promotes the dissemination of DT to internal organs such as the heart at later stages of infection—a frequent fatal complication of diphtheria (Murphy, 1996; Sharma et al., 2019; Holmes, 2000). In an alternative context, ZAKα-driven RSR and NLRP1-driven pyroptosis might constitute a host-protective response for diseases caused by intracellular pathogens, such as Legionnaires’ disease. These hypotheses await characterization. It would also be interesting to test in future work if pyroptotic epithelial cells can directly facilitate bacterial growth in their native niche, e.g., by providing nutrients that are sequestered by intact cells. Notwithstanding, the data shown here should inform future studies that seek to further understand the arms race between RSR, inflammasome, and microbial ribotoxins, which are very abundant in nature (Dmitriev et al., 2020). In proof-of-concept experiments, we showed that inhibition of ZAKα limits DT-induced skin damage in 3D cultures. Although ZAKα inhibition is unlikely to supplant current treatment and prevention options for diphtheria, ZAKα and/or NLRP1 inhibitors might find utility for other bacterial infections for which effective vaccines and antibiotics are lacking.
Materials and methods
Bacterial strains and culture conditions
Two non-toxigenic commensal Corynebacterium species, Corynebacterium striatum (6940; ATCC) and Corynebacterium minutissimum (23347; ATCC), and one toxigenic strain, C. diphtheriae (13812; ATCC) were cultured in Brain Heart Infusion (BHI; 53286; Sigma-Aldrich) broth at 37°C overnight in a shaking incubator at 230 rpm. Homemade 611 PGT media with the recipe described in Table S2 was used for quantification of bacterial growth.
Bacterial growth kinetics and correlation of DT induction
To track bacterial growth under iron starving conditions and correlate the amount of DT, C. striatum and C. diphtheriae were incubated at 37°C in a shaking incubator in 611 PGT media containing 0.5 mM 2,2′-bipyridyl (D2163050; Sigma-Aldrich). Growth was closely monitored by measuring OD600 every hour for 4 h: the culture was serially diluted 10-fold by sequential transfer of 20 μl into 180 μl of phosphate-buffered saline in triplicate wells by a factor of 10−7 every hour, for 4 h. To enumerate bacterial colony forming units, 5 μl of each dilution was spotted in triplicates on prewarmed blood agar (PP1325P90; Thermo Fisher Scientific) and incubated overnight at 37°C. Subsequently, colonies were counted and bacterial concentrations in the original sample were estimated. 200 μl of bacteria culture was also harvested every hour for 4 h, whereby it was centrifuged and supernatant kept to perform immunoblotting to correlate the levels of DT induction. 5 ng of purified DT was used as a loading control. The levels of band intensity were used to compare and estimate the amount of DT produced in a given volume of bacteria culture.
DT induction and harvesting of bacterial sterile filtrates
C. diphtheriae strain was incubated at 37°C in a shaking incubator in BHI broth (53286; Merck) and was grown to an OD600 of 0.4–0.6. 2,2′-bipyridyl was added to a final concentration of 0.5 mM and incubated for 4 h to induce DT production under iron-starving conditions. The bacteria were then removed by centrifugation at 3,000 × g for 10 min and the supernatant containing DT was harvested and filtered through a 0.2-µm syringe filter. The presence of DT in sterile filtrates is validated through immunoblotting assays with 5 ng of purified DT used as loading control to estimate DT concentration. For cellular and 3D skin culture experiments, unless otherwise indicated, the amount of DT in bacterial filtrate was normalized to be the same as concomitant DT treatment (150 ng/ml).
Cell culture and chemicals
293Ts (#CRL-3216; ATCC) and A549-ASC-GFP-NLRP1 (a549-ascg-nlrp1; Invivogen) were cultured according to manufacturer's protocols. Immortalized human keratinocytes (N/TERT herein) were provided by J. Rheinwald (Material Transfer Agreement) and cultured in Keratinocyte Serum Free Media (17005042; Gibco) supplemented with final concentration of 25 mg/liter bovine pituitary extract (13028-014; Gibco), 294.4 ng/liter human recombinant EGF (10450-013; Gibco), and 300 µM of CaCl2 (07058-00; Kanto Chemicals). Cell culture experiments involving immortalized cells or cell lines are conducted under Biological Project Number (BPN-72-2021) and approved by the Institutional Biosafety Committee (Nanyang Technological University, Singapore). Primary human keratinocytes were derived from the skin of healthy donors and obtained with informed consent from the Asian Skin Biobank (https://www.a-star.edu.sg/sris/technology-platforms/asian-skin-biobank). All primary keratinocyte and 3D organotypic skin experiments were carried out with approval from the Agency for Science, Technology and Research Human Biomedical Research Office (A*STAR Full IRB-2020-209). All cell lines underwent routine mycoplasma testing with MycoGuard (#LT07-118; Genecopoeia). For the 3D organoid cultures, primary human keratinocytes and fibroblasts were obtained from a single donor aged 39, male of Chinese ethnicity. For 2D experiments, primary human keratinocytes were obtained from two donors both of which were derived from foreskin of Malay ethnicity.
The following drugs and chemicals were used as part of this study: talabostat (HY-13233; VbP, MCE), ANS (HY-18982; ANS, MCE), neflamapimod (HY-10328; MCE), M443 (HY-112274; MCE), harringtonine (HYN0862; HTN, MCE), puromycin (P9620; Sigma-Aldrich), emricasan (HY-10396; MCE), belnacasan (HY-13205; MCE), aplidine (HY-16050; MCE), vemurafenib (HY-12057; MCE), and salinomycin (HY-15597; MCE). Compound 6p is a kind gift from X. Lu (Jinan University, Guangzhou, China). The following recombinant protein were used in this study: TNFα (210-TA; R&D Systems), DT (D0564; Sigma-Aldrich), exoA (P0184; Sigma-Aldrich), and Streptolysin O (SAE0086; Sigma-Aldrich). Recombinant LFn-DT and PA were a gift from F. Schmidt (University of Bonn, Bonn, Germany).
Organotypic 3D skin culture
Organotypic cultures were generated by adapting a previously described protocol (Arnette et al., 2016). Briefly, 2 ml of collagen I (4 mg/ml; #354249; Corning) mixed with 7.5 × 105 human fibroblasts were allowed to polymerize over 1 ml of acellular collagen I in 6-well culture inserts (#353102; Falcon) placed in 6-well deep well plates (#355467; Falcon). After 24 h, 1 × 106 primary human keratinocytes were seeded into the inserts and kept submerged in a 3:1 DMEM (#SH30243.01; Hyclone) and F12 (#31765035; Gibco) mixture with 10% FBS (#SV30160.03; Hyclone), 100 U/ml of penicillin–streptomycin (#15140122; Gibco), 10 µM Y-27632 (#1254; Tocris), 10 ng/ml of EGF (#E9644; Sigma-Aldrich), 100 pM cholera toxin (#BML-G117-001; Enzo), 0.4 µg/ml of hydrocortisone (#H0888; Sigma-Aldrich), 0.0243 mg/ml adenine (#A2786; Sigma-Aldrich), 5 µg/ml of insulin (#I2643; Sigma-Aldrich), 5 µg/ml of transferrin (#T2036; Sigma-Aldrich), and 2 nM 3,3′,5′-triiodo-L-thyronine (#T6397; Sigma-Aldrich). After another 24 h, the organotypic cultures were then raised at the air–liquid interface and fed with the submerged media (without Y-27632 and EGF) below the insert to induce epidermal differentiation. The air-lifting medium was replaced every 2 d and treatments began 10–14 d after airlifting. Organotypic treatments were performed as three technical replicates. Organotypic cultures were then harvested 24 h after treatment and formalin-fixed for 24 h. Fixed tissues were then embedded into wax for histological purposes before being cut and stained using a standard H&E protocol, the previously described method for GSDMD staining (Robinson et al., 2020) plectin (#611348; BD Transduction Labs) or plakoglobin (#61005S; Progen) using standard 3, 3'-diaminobenzidine staining method.
Quantification of epithelial damage in 3D organotypics
H&E images from three individual sections were captured for each 3D organotypic technical replicate within the same experiment. To quantify the splitting at the dermal-epidermal junction (DEJ), ImageJ was used to create a binary, black-and-white image; the polygon selection tool was used to select the area starting from the DEJ to the top of the epidermis not including the stratum corneum. Image J was then used to calculate the number of white pixels and black pixels and then the ratio was calculated.
Cytokine analysis
Human IL-1β enzyme-linked immunosorbent assay (ELISA) kit (#557953; BD) was used according to the manufacturer's protocol. Culture supernatants of 3D skin were also collected and sent for Luminex analysis using the ProcartaPlex, Human Customized 65-plex Panel (Thermo Fisher Scientific) to measure the following targets: APRIL; BAFF; BLC; CD30; CD40L; ENA-78; Eotaxin; Eotaxin-2; Eotaxin-3; FGF-2; Fractalkine; G-CSF; GM-CSF; Gro α; HGF; IFN-α; IFN-γ; IL-10; IL-12p70; IL-13; IL-15; IL-16; IL-17α; IL-18; IL-1α; IL-1β; IL-2; IL-20; IL-21; IL-22; IL-23; IL-27; IL-2 R; IL-3; IL-31; IL-4; IL-5; IL-6; IL-7; IL-8; IL-9; IP-10; I-TAC; LIF; MCP-1; MCP-2; MCP-3; M-CSF; MDC; MIF; MIG; MIP1α; MIP-1β; MIP-3 A; MMP-1; NGF β; SCF; SDF-1 α; TNF-β; TNF-α; TNF-R2; TRAIL; TSLP; TWEAK; and VEGF-α.
RNAseq sample preparation and analysis
Primary human keratinocytes or N/TERT cells were grown to 80% confluence in a 6-well plate before performing treatments. Cells were treated with stated treatments and harvested after 5 h. Total RNA was isolated from each treatment using RNAeasy mini kit (cat. # 74004; Qiagen). The quantity and quality of each RNA sample were then assayed using the NanoDrop (Thermo Fisher Scientific) but also by running the RNA on an agarose gel to check for RNA degradation. RNAseq was performed at Macrogen Asia using the Novaseq 6000 platform. Library construction and sequencing followed the standard sequencing protocols and were performed by Macrogen Asia. Preprocessing and analysis were also performed by Macrogen Asia, in brief reads were mapped to reference genome (Homo sapiens, GRCh38) with HISAT2, and transcript was assembled by StringTie with aligned reads. Expression profiles were represented as read count and normalization values, which were calculated as transcripts per kilobase million for each sample, based on transcript length and depth of coverage. Using pairwise comparisons, differentially expressed transcripts were calculated using an R package (DEseq2). The DEseq2 analysis was performed on read counts of expressed genes, and for each gene, the P value and fold change were calculated per comparison pair. The raw and processed data are available in the GEO repository with the accession number GSE236652.
Plasmids and preparation of lentiviral stocks
293T-ASC-GFP-NLRP1, N/TERT NLRP1 KO + NLRP1DR-GFP, N/TERT ZAKα KO, and N/TERT p38 dKO cells were previously described (Robinson et al., 2022). All expression plasmids for transient expression were cloned into the pCS2+ vector backbone and cloned using InFusion HD (Clontech). Constitutive lentiviral expression was performed using pCDH vector constructs (System Biosciences) and packaged using third-generation packaging plasmids. SidI plasmids for expression in 293Ts were gifts from S. Mukherjee (University of California, San Francisco, San Francisco, CA, USA).
CRISPR-Cas9 KO
NLRP1 KO, MAP3K20 (ZAKα) KO, and MAPK14/MAPK11 (p38α/β) dKO N/TERT keratinocytes were made and described in detail previously (Robinson et al., 2022). MAP3K20 (ZAKα) KO human primary keratinocytes were generated using lentiviral Cas9 and guide RNA plasmid (LentiCRISPR-V2, plasmid #52961; Addgene) using the following guides: sg1 (5′-TGTATGGTTATGGAACCGAG-3′), sg4 (5′-TGCATGGACGGAAGACGATG-3′). NLRP1 KO human primary keratinocytes were generated using lentiviral transduction with the following guide: sg1 (5′-GATAGCCCGAGTGCATCGG-3′). ATF3 KO N/TERT and human primary keratinocytes were generated using RNP nucleofection of both guides: 5′-CTTTTGTGATGGACACCCCG-3′, 5′-TAACCTGACGCCCTTTGTCA-3′. DPH1 KO N/TERTs were generated using RNP nucleofection of both guides: 5′-GATGGGTGACGTGACCTACG-3′, 5′-GCTGACTTCTTGGTGCACTA-3′.
KO efficiency was tested by immunoblot. Alternatively, Sanger sequencing of genomic DNA and overall editing efficiency were determined using the Synthego ICE tool (Synthego Performance Analysis, ICE Analysis. 2019. v2.0. Synthego, https://ice.synthego.com/#/).
Harringtonine ribosome runoff assay
Human primary keratinocytes were seeded at a cell density of 80,000 cells/well in a 24-well plate. The next day, cells were pretreated with 10 µM of emricasan (HY-10396; MCE) for 30 min followed by treatment with either BHI only, C. diphtheriae, or C. striatum for 6.5 h. Recombinant DT was added to the cells during the final 3 h. The respective samples were treated with 2 µg/ml of harringtonine (HY-10396; MCE) at staggered timepoints of 5, 1, and 0 min (equivalent to mock untreated). Cells were then pulsed with a final concentration of 10 µg/ml of puromycin at the same time in all wells for 10 min. Following puromycin treatment, the supernatant was discarded and cells were lysed directly in 1× Laemmli buffer. Immunoblotting of samples using an anti-puromycin antibody was done to measure the amount of nascent peptides with puromycin incorporated, which reflects the rate of elongation after DT treatment.
PI inclusion assay
N/TERT cells or human primary keratinocytes of various genotypes were seeded at a cell density of 10,000 cells/well in a black 96-well plate (CellCarrier-96 Ultra, #6055300; PerkinElmer) or a cell density of 80,000 cells/well in black 24-well plates (P24-1.5P; Cellvis). The next day, cells were treated with chemicals and stained with 0.5 µg/ml of PI (#ab14083; Abcam) before observing the cells on a high-content screening microscope (Perkin Elmer Operetta CLS imaging system, NTU Optical Bio-Imaging Centre in Nanyang Technological University, Singapore) over 18 h, capturing brightfield and fluorescent images every 15 min. For five fields of view per well in 96-well format with three wells per treatment or 14 fields of view per well in 24-well format, the ratio of PI-positive cells over normal cells was calculated. The number of live cells per field was counted using digital phase contrast images, which can identify cell borders, whereas the number of PI-stained regions identified through the PI channel (536/617 nm) was counted as PI+ cells.
ASC-GFP speck quantification
Images of ASC-GFP specks were acquired in 20 random fields in DAPI (358/461 nm) and GFP (469/525 nm) channels under 20× magnification using the Operetta CLS imaging microscope. The number of DAPI-stained nuclei and ASC specks in the GFP channel were counted using the Operetta CLS analysis system.
Immunoblotting
For SDS-PAGE using whole cell lysates, cells were resuspended in tris-buffered saline 1% NP-40 with protease inhibitors (#78430; Thermo Fisher Scientific). Protein concentration was determined using the Bradford assay (#23200; Thermo Fisher Scientific) and 20 µg of protein was loaded. To visualize cleaved GSDMD, cell debris floaters were separated after collection of media supernatants after treatment and lysed directly with 1× Laemmli buffer (30 μl of 1× Laemmli buffer for 200,000 cells). Attached cells on the wells were lysed directly with 1× Laemmli buffer (100 μl of 1× Laemmli buffer for 200,000 cells), sonicated, and boiled. Lysates from the floater fraction and the attached cell fraction were loaded into each gel lane in a 1:1 ratio (10 μl floaters, 10 μl attached cells’ lysate). For analysis of IL-1β in the media by immunoblotting, samples were concentrated using filtered centrifugation (#UFC500396; Merck, Amicon Ultra). Protein samples were run using immunoblotting and then visualized using a ChemiDoc Imaging system (Bio-Rad).
PhosTag SDS-PAGE was carried out using homemade 10% SDS-PAGE gel with addition of Phos-tag Acrylamide (AAL-107; Wako Chemicals) to a final concentration of 30 µM and manganese chloride(II) (#63535; Sigma-Aldrich) to 60 µM. Cells were directly harvested using 1× Laemmli buffer, lysed with an ultrasonicator, and loaded into the Phos-tag gel to run. Once the run was completed, the polyacrylamide gel was washed in transfer buffer with 10 mM EDTA twice, subsequently washed once without EDTA, blotted onto 0.45 µm poly(vinylidene fluoride) membranes (Bio-Rad), blocked with 3% milk, and incubated with primary and corresponding secondary antibodies. All primary and secondary antibodies used in this study are mentioned in Table S1.
Online supplemental material
Fig. S1 describes additional characterization of EEF2 targeting bacterial toxins. Fig. S2 contains supporting data for the effects of NLRP1 phosphorylation, ZAKα, and ATF3 in DT-induced pyroptosis and apoptosis. Fig. S3 shows further data on the effect of various inhibitors on C. diphtheriae–induced damage in 3D skin cultures. Table S1 lists the antibodies used in this study. Table S2 lists the composition of the 611 PGT Medium used for C. diphtheriae culture.
Supplementary Material
shows antibodies used in this study.
shows the composition of the 611 PGT Medium (adapted from ATCC).
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 5.
is the source file for Fig. S1.
is the source file for Fig. S2.
Acknowledgments
The authors are grateful for useful discussion and scientific advice from all members of the Zhong lab. We are especially grateful for Dr. Vijaya Chandra Shree Harsha (A*STAR Skin Research Labs) for his help with Corynebacterium culture. F.L. Zhong would like to thank Prof. Veit Hornung (Gene Center, Ludwig-Maximilians-University Munich), Prof. Simon Bekker-Jensen (University of Copenhagen), Prof. Lena Ho (Duke-NUS), Prof. Wu Bin (Nanyang Technological University), and Prof. Seth Masters (Walter and Eliza Hall Institute of Medical Research, Australia) for scientific advice and guidance.
F.L. Zhong’s lab is funded by the National Research Foundation Fellowship, Singapore (NRF-NRFF11-2019-0006), Nanyang Assistant Professorship, Ministry of Education Tier 2 grant (T2EP30222-0033), and The Singapore Therapeutics Development Review Grant Call (H22G0a0002). Work from J.E.A. Common’s lab is supported by funding from Agency for Science, Technology, and Research and A*STAR-EDB-NRF IAF-PP grants—H17/01/a0/004 “Skin Research Institute of Singapore” and H22J1a0040 “Asian Skin Microbiome Program 2.0.” K.C. Tham is supported by H17/01/a0/004 “Skin Research Institute of Singapore.” K.S. Robinson is supported by the A*STAR Career Development Fund (CDA-C210812053).
Author contributions: K.S. Robinson, G.A. Toh, M.J. Firdaus, K.C. Tham, P. Rozario, and Z.H. Lau carried out the investigation and analyzed, visualized, and validated the data. Y.X. Toh performed cell sorting. C.K. Lim performed Luminex experiments. S.C. Binder and J. Mayer prepared reagents for experiments on LFn-DTA. C. Bonnard, F.I. Schidmt, and J.E.A. Common jointly supervised the investigation. F.L. Zhong acquired the funding and conceptualized the initial phase of the study. F.L. Zhong, K.S. Robinson, and G.A. Toh wrote the manuscript with contributions from all co-authors.
Data availability
All data supporting the findings of this study are available from the corresponding author upon request. Cell lines and constructs are available from the corresponding author with Materials Transfer Agreements.
References
- Acosta, A.M., and Tiwari T.S.P.. 2020. 41: Diphtheria. In Hunter’s Tropical Medicine and Emerging Infectious Diseases. Tenth edition. Ryan E.T., Hill D.R., Solomon T., Aronson N.E., and Endy T.P., editors. Elsevier, London. 439–445. 10.1016/B978-0-323-55512-8.00041-7 [DOI] [Google Scholar]
- Arnette, C., Koetsier J.L., Hoover P., Getsios S., and Green K.J.. 2016. In vitro model of the epidermis: Connecting protein function to 3D structure. Methods Enzymol. 569:287–308. 10.1016/bs.mie.2015.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin, D.A. 2021. NLRP1: A jack of all trades, or a master of one? Mol. Cell. 81:423–425. 10.1016/j.molcel.2021.01.001 [DOI] [PubMed] [Google Scholar]
- Blumberg, L.H., Prieto M.A., Diaz J.V., Blanco M.J., Valle B., Pla C., and Durrheim D.N.. 2018. The preventable tragedy of diphtheria in the 21st century. Int. J. Infect. Dis. 71:122–123. 10.1016/j.ijid.2018.05.002 [DOI] [PubMed] [Google Scholar]
- Cheng, J.-S. 2010. Molecular Mechanisms of Cytotoxicity Induced by Ribosome-inactivating Proteins in Mammalian Cells. Rutgers University Press, New Brunswick. 77 pp. [Google Scholar]
- Choi, Y.M., Adelzadeh L., and Wu J.J.. 2015. Photodynamic therapy for psoriasis. J. Dermatolog. Treat. 26:202–207. 10.3109/09546634.2014.927816 [DOI] [PubMed] [Google Scholar]
- Crews, C.M., Collins J.L., Lane W.S., Snapper M.L., and Schreiber S.L.. 1994. GTP-dependent binding of the antiproliferative agent didemnin to elongation factor 1 alpha. J. Biol. Chem. 269:15411–15414. 10.1016/S0021-9258(17)40692-2 [DOI] [PubMed] [Google Scholar]
- Dmitriev, S.E., Vladimirov D.O., and Lashkevich K.A.. 2020. A quick guide to small-molecule inhibitors of eukaryotic protein synthesis. Biochemistry. 85:1389–1421. 10.1134/S0006297920110097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar, T.L., Yan Z., Balla K.M., Smelkinson M.G., and Troemel E.R.. 2012. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe. 11:375–386. 10.1016/j.chom.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana, M.F., and Vance R.E.. 2011. Two signal models in innate immunity. Immunol. Rev. 243:26–39. 10.1111/j.1600-065X.2011.01037.x [DOI] [PubMed] [Google Scholar]
- Frankel, A.E., Powell B.L., and Lilly M.B.. 2002. Diphtheria toxin conjugate therapy of cancer. Cancer Chemother. Biol. Response Modif. 20:301–313. [PubMed] [Google Scholar]
- Hadfield, T.L., McEvoy P., Polotsky Y., Tzinserling V.A., and Yakovlev A.A.. 2000. The pathology of diphtheria. J. Infect. Dis. 181:S116–S120. 10.1086/315551 [DOI] [PubMed] [Google Scholar]
- Holmes, R.K. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 181:S156–S167. 10.1086/315554 [DOI] [PubMed] [Google Scholar]
- Ivanova, A., Signore M., Caro N., Greene N.D.E., Copp A.J., and Martinez-Barbera J.P.. 2005. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis. 43:129–135. 10.1002/gene.20162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenster, L.-M., Lange K.-E., Normann S., vom Hemdt A., Wuerth J.D., Schiffelers L.D.J., Tesfamariam Y.M., Gohr F.N., Klein L., Kaltheuner I.H., et al. 2023. P38 kinases mediate NLRP1 inflammasome activation after ribotoxic stress response and virus infection. J. Exp. Med. 220:e20220837. 10.1084/jem.20220837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, A.M., Pohl A.E., Ball T.J., Abram T.G., Johnson D.K., Geisbrecht B.V., and Shames S.R.. 2019. The Legionella pneumophila metaeffector Lpg2505 (SusF) regulates SidI-mediated translation inhibition and GDP-dependent glycosyltransferase activity. bioRxiv. 10.1101/845313 (Preprint posted November 16, 2019). [DOI]
- Liu, S., Milne G.T., Kuremsky J.G., Fink G.R., and Leppla S.H.. 2004. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol. Cell. Biol. 24:9487–9497. 10.1128/MCB.24.21.9487-9497.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes Fischer, N., Naseer N., Shin S., and Brodsky I.E.. 2020. Effector-triggered immunity and pathogen sensing in metazoans. Nat. Microbiol. 5:14–26. 10.1038/s41564-019-0623-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon, F., Burns K., and Tschopp J.. 2002. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 10:417–426. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- McEwan, D.L., Kirienko N.V., and Ausubel F.M.. 2012. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe. 11:364–374. 10.1016/j.chom.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne, J.C., Blanke S.R., Hanna P.C., and Collier R.J.. 1995. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol. Microbiol. 15:661–666. 10.1111/j.1365-2958.1995.tb02375.x [DOI] [PubMed] [Google Scholar]
- Mitchell, P.S., Sandstrom A., and Vance R.E.. 2019. The NLRP1 inflammasome: New mechanistic insights and unresolved mysteries. Curr. Opin. Immunol. 60:37–45. 10.1016/j.coi.2019.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, H., and Bonavida B.. 1992. Diphtheria toxin- and Pseudomonas A toxin-mediated apoptosis. ADP ribosylation of elongation factor-2 is required for DNA fragmentation and cell lysis and synergy with tumor necrosis factor-alpha. J. Immunol. 149:2089–2094. 10.4049/jimmunol.149.6.2089 [DOI] [PubMed] [Google Scholar]
- Morimoto, H., Safrit J.T., and Bonavida B.. 1991. Synergistic effect of tumor necrosis factor-alpha- and diphtheria toxin-mediated cytotoxicity in sensitive and resistant human ovarian tumor cell lines. J. Immunol. 147:2609–2616. 10.4049/jimmunol.147.8.2609 [DOI] [PubMed] [Google Scholar]
- Murphy, J.R. 1996. Corynebacterium diphtheriae. In Medical Microbiology. Fourth edition. Baron S., editor. University of Texas Medical Branch at Galveston, Galveston, TX. [PubMed] [Google Scholar]
- Narayanan, S., Surendranath K., Bora N., Surolia A., and Karande A.A.. 2005. Ribosome inactivating proteins and apoptosis. FEBS Lett. 579:1324–1331. 10.1016/j.febslet.2005.01.038 [DOI] [PubMed] [Google Scholar]
- Oda, E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., Tokino T., Taniguchi T., and Tanaka N.. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 288:1053–1058. 10.1126/science.288.5468.1053 [DOI] [PubMed] [Google Scholar]
- Orzalli, M.H., Prochera A., Payne L., Smith A., Garlick J.A., and Kagan J.C.. 2021. Virus-mediated inactivation of anti-apoptotic Bcl-2 family members promotes Gasdermin-E-dependent pyroptosis in barrier epithelial cells. Immunity. 54:1447–1462.e5. 10.1016/j.immuni.2021.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter, R.D., Behringer R.R., Quaife C.J., Maxwell F., Maxwell I.H., and Brinster R.L.. 1987. Cell lineage ablation in transgenic mice by cell-specific expression of a toxin gene. Cell. 50:435–443. 10.1016/0092-8674(87)90497-1 [DOI] [PubMed] [Google Scholar]
- Pinilla M., Mazars R., Vergé R., Gorse L., Paradis M., Suire B., Santoni K., Robinson K.S., Toh G.A., Prouvensier L., et al. 2023. EEF2-inactivating toxins engage the NLRP1 inflammasome and promote epithelial barrier disruption. J. Exp. Med. 220. 10.1084/jem.20230104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, K.S., Teo D.E.T., Tan K.S., Toh G.A., Ong H.H., Lim C.K., Lay K., Au B.V., Lew T.S., Chu J.J.H., et al. 2020. Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science. 370:eaay2002. 10.1126/science.aay2002 [DOI] [PubMed] [Google Scholar]
- Robinson, K.S., Toh G.A., Rozario P., Chua R., Bauernfried S., Sun Z., Firdaus M.J., Bayat S., Nadkarni R., Poh Z.S., et al. 2022. ZAKα-driven ribotoxic stress response activates the human NLRP1 inflammasome. Science. 377:328–335. 10.1126/science.abl6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador, J.M., Brown-Clay J.D., and Fornace A.J. Jr. 2013. Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv. Exp. Med. Biol. 793:1–19. 10.1007/978-1-4614-8289-5_1 [DOI] [PubMed] [Google Scholar]
- Sand, J., Haertel E., Biedermann T., Contassot E., Reichmann E., French L.E., Werner S., and Beer H.-D.. 2018. Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death Dis. 9:24. 10.1038/s41419-017-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, N.C., Efstratiou A., Mokrousov I., Mutreja A., Das B., and Ramamurthy T.. 2019. Diphtheria. Nat. Rev. Dis. Primers. 5:81. 10.1038/s41572-019-0131-y [DOI] [PubMed] [Google Scholar]
- Stuart, L.M., Paquette N., and Boyer L.. 2013. Effector-triggered versus pattern-triggered immunity: How animals sense pathogens. Nat. Rev. Immunol. 13:199–206. 10.1038/nri3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Wang L., Moss T., Voorhies M., Sangwan S., Stevenson E., Pulido E.H., Kwok S., Krogan N.J., Swaney D.L., et al. 2022. A Legionella toxin mimics tRNA and glycosylates the translation machinery to trigger a ribotoxic stress response. bioRxiv. 10.1101/2022.06.10.495705 (Preprint posted June 10, 2022). [DOI]
- Thorburn, J., Frankel A.E., and Thorburn A.. 2003. Apoptosis by leukemia cell-targeted diphtheria toxin occurs via receptor-independent activation of Fas-associated death domain protein. Clin. Cancer Res. 9:861–865. [PubMed] [Google Scholar]
- Tsu, B.V., Beierschmitt C., Ryan A.P., Agarwal R., Mitchell P.S., and Daugherty M.D.. 2021. Diverse viral proteases activate the NLRP1 inflammasome. Elife. 10:e60609. 10.7554/eLife.60609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchi, L., Aberdam E., Mazure N., Pouysségur J., Deckert M., Kitajima S., Aberdam D., and Virolle T.. 2008. Hif-2alpha mediates UV-induced apoptosis through a novel ATF3-dependent death pathway. Cell Death Differ. 15:1472–1480. 10.1038/cdd.2008.74 [DOI] [PubMed] [Google Scholar]
- Vind, A.C., Genzor A.V., and Bekker-Jensen S.. 2020a. Ribosomal stress-surveillance: Three pathways is a magic number. Nucleic Acids Res. 48:10648–10661. 10.1093/nar/gkaa757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vind, A.C., Snieckute G., Blasius M., Tiedje C., Krogh N., Bekker-Jensen D.B., Andersen K.L., Nordgaard C., Tollenaere M.A.X., Lund A.H., et al. 2020b. ZAKα recognizes stalled ribosomes through partially redundant sensor domains. Mol. Cell. 78:700–713.e7. 10.1016/j.molcel.2020.03.021 [DOI] [PubMed] [Google Scholar]
- Wise, J. 2022. Home office is told to “get a grip” on overcrowded and unsafe immigration detention centre. BMJ. 379:o2615. 10.1136/bmj.o2615 [DOI] [PubMed] [Google Scholar]
- Wu, C.C.-C., Peterson A., Zinshteyn B., Regot S., and Green R.. 2020. Ribosome collisions trigger general stress responses to regulate cell fate. Cell. 182:404–416.e14. 10.1016/j.cell.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Shibu M.A., Kong L., Luo J., BadrealamKhan F., Huang Y., Tu Z.C., Yun C.H., Huang C.Y., Ding K., and Lu X.. 2020. Design, synthesis, and structure-activity relationships of 1,2,3-triazole benzenesulfonamides as new selective leucine-zipper and sterile-α motif kinase (ZAK) inhibitors. J. Med. Chem. 63:2114–2130. 10.1021/acs.jmedchem.9b00664 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows antibodies used in this study.
shows the composition of the 611 PGT Medium (adapted from ATCC).
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 5.
is the source file for Fig. S1.
is the source file for Fig. S2.
Data Availability Statement
All data supporting the findings of this study are available from the corresponding author upon request. Cell lines and constructs are available from the corresponding author with Materials Transfer Agreements.