Abstract
The M-protein genes (emm genes) of 103 separate impetiginous Streptococcus pyogenes isolates were sequenced and the sequence types were compared to the types obtained by Vir typing. Vir typing is based on restriction fragment length polymorphism (RFLP) analysis of a 4- to 7-kb pathogenicity island encoding emm and other virulence genes. By using both HaeIII and HinfI to generate RFLP profiles, complete concordance between Vir type and emm sequence type was found. Comparison of the emm sequences with those in GenBank revealed new sequence types sharing less than 90% identity with known types. Diversity in the emm sequence was generated by corrected frameshift mutations, point mutations, and small in-frame mutations.
Infections caused by group A streptococci (GAS) lead to a spectrum of disease (4, 5) ranging from relatively benign conditions such as impetigo to more severe invasive diseases and serious nonsuppurative sequelae: acute poststreptococcal glomerulonephritis (APSGN) and acute rheumatic fever (ARF). Despite the recent reemergence of severe invasive diseases in North America and Europe, sequelae following streptococcal infection are now comparatively rare in much of the developed world. Nevertheless, both APSGN and ARF still cause significant morbidity and mortality within the Aboriginal population of northern Australia (6, 24, 36).
Because a significant proportion of isolates of GAS from the Northern Territory are nontypeable by serology, a molecular method called Vir typing (9) was developed. This method is based on the restriction fragment length polymorphisms (RFLPs) of a long PCR product of the Vir regulon of GAS encoding the M-protein family of genes and other virulence factors. More than 400 GAS isolates from skin sores of Australian Aboriginal children have been genotyped by this method, yielding 43 distinct RFLP patterns or Vir types (VTs) (11, 12). A further 193 isolates from four communities involved in a widespread outbreak of APSGN produced 17 distinct VTs (13). However, for a small minority of VTs, the HaeIII restriction profiles were found to be uninformative, yielding only a single RFLP fragment in addition to the RFLP fragments which were common to the majority of VTs. In order to determine if these VTs could be subdivided, a second restriction enzyme, HinfI, was used in this study.
Selected VTs were also subjected to sequence analysis of the hypervariable region of the emm gene in order to determine if the diversity observed in the VT patterns of these isolates was due to architectural diversity of the regulon or to variation in emm. Other studies have focused on the sequencing of reference M types (1, 2, 38, 39) which are collections of isolates predominantly from North America and Europe. Unique sequence types (STs) from M-nontypeable (MNT) isolates that are related to reference STs have also been described (18, 29–31). In this study, we examined isolates collected over the past 6 years from the skin of individuals in a small but widespread population from the Northern Territory of Australia. The nucleotide sequences of the hypervariable region of a number of emm genes divided the isolates into four categories: (i) isolates with 99 to 100% identity with reference types (group I), variants of reported reference types (group II), isolates with 100% identity with emm sequence types previously reported from among MNT isolates from this region (29, 31) (group III), and isolates with 70 to 90% identity with previously reported emm STs (group IV). For the isolates in this last group, there was sufficient divergence from previously reported emm STs for them to be considered new STs. Point mutations, corrected frameshift mutations, and short in-frame mutations accounted for the majority of the changes in the hypervariable regions of these new STs.
MATERIALS AND METHODS
Vir typing of Streptococcus pyogenes isolates.
The 103 isolates of GAS selected for VT and emm sequence analysis were collected between 1990 and 1996 mainly from pyoderma lesions of children (10, 13). They were processed as described previously by the agarose microtiter tray DNA extraction procedure and long PCR (9, 11). Cycling conditions consisted of an initial denaturation step for 30 s at 95°C, followed by 30 cycles of 94°C for 15 s, 60°C for 60 s, and 68°C for 6 min. Five microliters of the PCR mixture was electrophoresed on a 1% agarose gel to determine the quality of the DNA that was amplified. Vir typing was conducted by digesting approximately 0.5 μg of PCR product (from 8 to 25 μl) with 2 U of HaeIII or 2 U of HinfI (Pharmacia).
Vir typing with HaeIII results in very distinct RFLP profiles for the majority of isolates. Certain HaeIII RFLP patterns were thought to possess a lower information content due to the presence of only a single RFLP fragment, in addition to the RFLP fragments which were common to the majority of VTs. Because it generates a considerable number of bands, HinfI was then used to determine if the designation obtained by typing with HaeIII could be further subdivided. Previous studies have shown that HinfI digestion, in addition to HaeIII digestion, of the Vir regulon is as discriminatory as multilocus enzyme electrophoresis with 20 alloenzymes in distinguishing strains of GAS (12).
emm sequence analysis.
The template DNA used to determine the hypervariable region of emm was either the PCR product from Vir typing as described above or an emm-specific amplification product (18, 28). PCR primers were removed by isopropanol precipitation before cycle sequencing with pF (28). To ensure that there was no sampling bias, the isolates of each VT used for sequencing were chosen, whenever possible, from different communities at different time points over the period of the study. Between 2 and 11 isolates of each VT were sequenced.
Sequences were examined by a suite of programs at the Australian National Genomic Information Service. Pairwise comparison of the nucleotide identities for the first 90 to 189 bases of the hypervariable region was conducted after the conserved leader sequence was excluded.
Statistical analysis.
The concordance of Vir typing and emm sequence typing was determined by the use of a contingency table. For example, two isolates had identical VTs (VT 4) and also had identical STs (NS27). What is the expected number of such identities? To illustrate the statistical analysis, note that 103 isolates had been both Vir typed and sequenced typed. Of these, the first occurrence of an observed concordance between VT 4 and emm NS27 serves to identify this pair. This reduces the observed numbers for testing the hypothesis by one, and we calculated the expected number of excess identities (over and above the first) as 1 × 1/102 = 0.01. For the 16 VTs with an apparently unique ST, there were 53 excess identities compared with 3.42 excess identities expected if the null hypothesis of no association between VT and emm ST is correct (X1 = 718.8). For some VTs (e.g., VTs 3, 7, 17, and 29), several STs were identified. By a similar argument, as outlined above, there were 23 excess identities compared to the expected excess of 0.86 (X1 = 570.0). When both of these groups were amalgamated, there were a total of 76 excess identities, compared to the expected excess of 4.38 (X1 = 1171.1) (P < 0.001).
Nucleotide sequence accession numbers.
Ten unique emm sequences (AF018176 to AF018185) were submitted to GenBank.
RESULTS
Improving the discriminatory power of Vir typing by HinfI restriction.
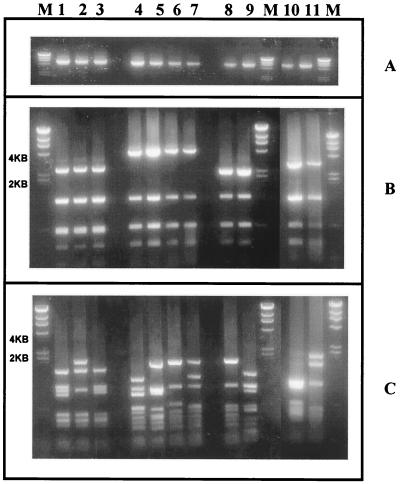
HaeIII restriction of the PCR product derived from the amplification of the Vir regulon of GAS from impetiginous isolates of GAS generally gives rise to between four and eight fragments which vary in size from 200 to 4,000 bp (11). Of these fragments, up to three (1,400, 500, and 275 bp) may correspond to fragments common to the majority of isolates tested. These common fragments do not contribute to the discrimination of RFLP patterns without the presence of other higher-molecular-mass fragments. In the case of VTs 3, 7, and 17, only one other fragment not common to the majority of isolates was present. To improve the discrimination in such cases, another restriction enzyme was used. HinfI could distinguish three subtypes of VT 3, four subtypes of VT 17, and two subtypes of VT 7 (Fig. 1). VTs which could be split by the use of HinfI were designated by their primary HaeIII VT type followed by a decimal point and their HinfI RFLP type (namely, VT 3.1 and VT 3.2).
FIG. 1.
Ethidium bromide-stained gel of RFLP profiles of VT 3 (lanes 1 to 3), VT 17 (lanes 4 to 7), VT 29 (lanes 8 and 9), and VT 7 (lanes 10 and 11). (A) Undigested PCR product. (B) PCR product digested with HaeIII. (C) PCR product digested with HinfI. Lane 1, VT 3.1; lane 2, VT 3.2; lane 3, VT 3.3; lane 4, VT 17.1; lane 5, VT 17.2; lane 6, VT 17.3; lane 7, VT 17.4; lane 8, VT 29.1; lane 9, VT 29.2; lane 10, VT 7.1; lane 11, VT 7.2; lane M, bacteriophage λ DNA digested with HindIII.
As expected, for VTs which had two or more fragments other than the common fragments, HinfI restriction did not appear to provide any further discrimination except for VT 29, in which two HinfI subtypes were found (VT 29.1 and VT 29.2). HinfI has been found to generate high numbers of fragments (9, 12; this study) and is more useful in further discriminating the RFLP patterns obtained by HaeIII digestion than as the primary restriction enzyme in Vir typing.
5′ emm sequence analysis.
One hundred three isolates of GAS of 20 VTs obtained by HaeIII digestion (not including subtypes) were analyzed for the DNA sequence corresponding to the hypervariable region of emm. Isolates from all the subtypes of VTs 3, 7, 17, and 29 obtained by HinfI digestion were also included. Twenty-seven distinct N-terminal emm STs were found among the 20 VTs. Every VT obtained by HaeIII digestion other than VTs 3, 7, 17, and 29 had a single corresponding unique ST, whereas every subtype of VTs 3, 7, 17, and 29 had a unique ST.
The concordance between VT and emm ST is presented in Table 1. There was a statistically significant concordance between VTs and STs (P < 0.0001); each VT represents at least one distinct emm ST, and in the case of VT 3, 7, 17, and 29, the VT represents more than one distinct emm ST. When VTs 3, 7, 17, and 29 are divided into their subtypes, there is an absolute concordance between the RFLP digests of the Vir regulon and the emm ST.
TABLE 1.
Concordance between VT and ST
| No. of emm STs and VT | ST | No. of Vir isolates typed | No. of isolates sequenced | No. of isolates with identical VTs and STs (no. of excess identities) | Expected no. of excess identities |
|---|---|---|---|---|---|
| One emm ST | |||||
| 4 | NS27 | 14 | 2 | 2 (1) | 0.01 |
| 5 | BSA29 | 4 | 2 | 2 (1) | 0.01 |
| 6 | BSB19 | 2 | 2 | 2 (1) | 0.01 |
| 8 | 57 | 10 | 6 | 6 (5) | 0.33 |
| 9 | 25 | 14 | 11 | 11 (10) | 1.32 |
| 10 | 2.3 | 18 | 3 | 3 (2) | 0.05 |
| 11 | DRX4 | 11 | 2 | 2 (1) | 0.01 |
| 12 | BL18 | 11 | 2 | 2 (1) | 0.01 |
| 16 | 1.4 | 15 | 6 | 6 (5) | 0.33 |
| 18 | 55 | 15 | 6 | 6 (5) | 0.33 |
| 19 | 74 | 9 | 3 | 3 (2) | 0.05 |
| 23 | NS1 | 11 | 4 | 4 (3) | 0.12 |
| 26 | PL1 | 10 | 7 | 7 (6) | 0.47 |
| 27 | 33 | 5 | 3 | 3 (2) | 0.05 |
| 39 | 80 | 9 | 5 | 5 (4) | 0.21 |
| 40 | 19.2 | 7 | 5 | 5 (4) | 0.21 |
| More than one emm ST | |||||
| 3.1 | 44/66 | 2 | 2 (1) | 0.01 | |
| 3.2 | 11 | 5 | 5 (4) | 0.22 | |
| 3.3 | 49.2 | 2 | 2 (1) | 0.01 | |
| 7.1 | 63 | 3 | 3 (2) | 0.05 | |
| 7.2 | 9 | 3 | 3 (2) | 0.05 | |
| 17.1 | NS5 | 2 | 2 (1) | 0.01 | |
| 17.2 | BL12 | 2 | 2 (1) | 0.01 | |
| 17.3 | Donald | 5 | 5 (4) | 0.22 | |
| 17.4 | DRV8 | 2 | 2 (1) | 0.01 | |
| 29.1 | 52 | 5 | 5 (4) | 0.22 | |
| 29.2 | PK14 | 3 | 3 (2) | 0.05 | |
| Total | 103 | 103 (76) | 4.38 |
Comparison with known emm STs.
Twelve of the emm sequences corresponded to known emm STs (group I, 99 to 100% identity) which have been characterized in North America and Europe (Table 2). However, in the case of the ST emm1.4 (25), the original strains used in this study came from either Australia or New Zealand and may represent a geographically local emm ST. Three other VTs (VTs 3.3, 10, and 40) had STs that had significant sequence homology to known reference STs and were considered variants of these known STs (group II). One of these variants, emm2.3 (accession no. AF018178), has been reported previously (30). In group III, five STs corresponded to previously characterized STs in northern Australia. Finally, seven STs (group IV) showed only 70 to 90% identity with any previously published emm sequences. These STs appear to be unique to this geographic location.
TABLE 2.
Classification of 5′ emm ST found among 27 VTs and Vir subtypes
| Group and VT | Closest match (accession no.) | % Identity (no. of bases compared)a | New accession no. |
|---|---|---|---|
| I | |||
| VT 3.1 | emm44/61 (U11964) | 100 (168) | |
| VT 3.2 | emm11 (U11938) | 99 (150) | |
| VT 7.1 | emm63 (U11982) | 100 (189) | |
| VT 7.2 | emm9 (U12002) | 100 (147) | |
| VT 8 | emm57 (X60959) | 100 (138) | |
| VT 9 | emm25 (U11952) | 100 (138) | |
| VT 16 | emm1.4 (U20098) | 99 (162) | |
| VT 18 | emm55 (U11973) | 100 (135) | |
| VT 19 | emm74 (U11994) | 99 (153) | |
| VT 27 | emm33 (U11942) | 100 (144) | |
| VT 29.1 | emm52 (L27098) | 100 (132) | |
| VT 39 | emm80 (L27097) | 100 (135) | |
| II | |||
| VT 3.3 emm49.2 | emm49 (M31789) | 88 (111) | AF018176 |
| VT 10 emm2.3 | emm2.1 (X56398) | 90 (184) | AF018178 |
| VT 40 emm19.2 | emm19.1 (U39838) | 93 (90) | AF018177 |
| III | |||
| VT 4 | ST NS27 (L27094) | 100 (150) | |
| VT 17.1 | ST NS5 (L27093) | 100 (150) | |
| VT 17.3 | ST Donald (L05017) | 100 (129) | |
| VT 23 | ST NS1 (L05022) | 100 (144) | |
| VT 26 | ST PL1 (L28822) | 100 (120) | |
| IV | |||
| VT 5 (BSA29) | emm6 (M11338) | 76 (159) | AF018179 |
| VT 6 (BSB19) | emm13 (AF025950) | 84 (100) | AF018183 |
| VT 11 (DRX4) | emm52 (L27098) | 86 (153) | AF018181 |
| VT 12 (BL18) | emm70-emm33 (L27095-U11942) | 92/85 (144) | AF018184 |
| VT 17.2 (BL12) | emm71 (L46652) | 71 (150) | AF018182 |
| VT 17.4 (DRV8) | emmPT2110 (U11957) | 75 (132) | AF018180 |
| VT 29.2 (PK14) | emm70-emm33 (L27095-U11942) | 73/83 (102) | AF018185 |
Number of bases corresponding to the hypervariable region of the mature protein (i.e., conserved leader sequences were not used in the comparison).
Variants of the characterized emm types.
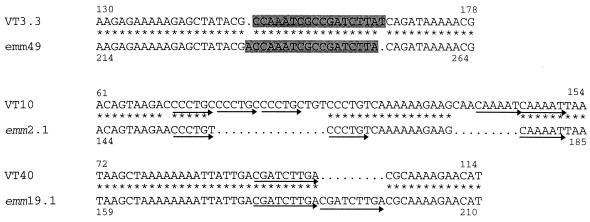
VT 3.3 represented by a variant of emm49, designated emm49.2 (accession no. AF018176), shares 88% identity with emm49 in the first 112 bases of the hypervariable region. VT 3.3 has three point mutations and one corrected frameshift region of 18 bases (Fig. 2). This is expected to change six amino acid residues in relation to the sequence of emm49 in this region. Excluding the frameshift mutation, the level of identity of these two sequences increases to 95%.
FIG. 2.
Alignment of nucleotide sequences of VT 3.3 (accession no. AF018176) and emm49 (accession no. M31789), VT 10 (accession no. AF018178) and emmL2.1 (accession no. X56398), and VT40 (accession no. AF018177) and emm19.1 (accession no. U39838) in the region of in-frame and frameshift insertions or deletions. Asterisks represent identity of the corresponding nucleotide, and dots represent missing nucleotides. Highlighted regions in VT 3.3 and emm49 represent the position of the frameshift mutation, while the arrows in VT 10, VT 40, emm2.1, and emm19.1 represent tandem repeats. The numbers attached to the sequences correspond to the position in the nucleotide sequence in GenBank.
In VT 10, which has 90% identity with emmL2.1 over 184 bases, there are two point mutations and two insertions (insertions 15 and 9 bases) within the hypervariable region of the emm gene (Fig. 2). In the first insertion, the nucleotide sequence CCCTGY has been tandemly repeated three times, while in the second insertion the nucleotide sequence CAAAAT has been repeated twice. In emm2.1, only two repeats of CCCTGY were found, and interestingly, pyrimidine transitions were observed in both of these repeats. Due to the significant similarity of the majority of the nucleotide sequence to that of emmL2.1 and to prevent confusion with the enn gene called emmL2.2 (3), this ST has been designated emm2.3 (accession no. AF018178). emm2.3 also shares 92% identity with the recently described ST 2967 (2).
VT 40 corresponds closely to emm19.1 with 93% identity in the first 100 bases, with five point mutations and one deletion of 9 bases corresponding to one tandem repeat (Fig. 2). This sequence has been designated emm19.2 (accession no. AF018177).
New STs.
VT 5, represented by STBSB29 (accession no. AF018177), resembled M6, but they showed 76% homology in the hypervariable region. Most of the changes seen were due to point mutations (45 in 150 bases), with two insertions and a corrected frameshift. Of the 45 point mutations, 26 were transversions rather than transitional changes. An excess of transversion point mutations was also noted for VT 11 (STDRX4; accession no. AF018181) and VT 17.2 (STBL12; accession no. AF018182) when the sequences of these emm STs were compared to those of their closest corresponding reference type (Table 2).
VT 6, represented by STBSB19 (accession no. AF018183), shows 84% homology to emm13, excluding a corrected frameshift region spanning 45 nucleotides. STBSB19 also has a 9-base deletion and two 3-base insertions. Another large frameshift region of 33 nucleotides was present in VT 17.4 (STDRV8; accession no. AF018180).
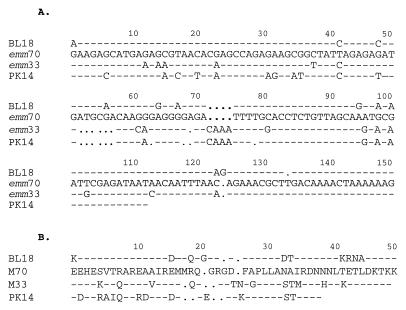
Two new types, STBL18 and STPK14, are part of the emm33 and emm70 family. The N-terminal emm gene sequence for M70 was first described as a local ST, type STBSB75, by Relf et al. (31) and was later identified by Beall and colleagues (1). The emm70 ST shares 83% identity with emm33 (Fig. 3A), an ST also found among our local isolates (VT 27). STBL18 (accession no. AF018184) from VT 12 has 92% identity with emm70 and 85% identity with emm33 (Fig. 3A). The similarity, which includes a contiguous stretch of 14 amino acids at the N-proximal region, between the translated sequence of STBL18 and emm70 is striking (Fig. 3B). A corrected frameshift spanning 9 bases is responsible for changes in 5 amino acid residues in the sub-N-terminal region.
FIG. 3.
(A) Aligned nucleotide sequences of STBL18, emm70 (STBSB75), emm33, and STPK14. The emm33 sequence was published by Whatmore et al. (38), while the emm70 sequence was first described from the Northern Territory but was identified more recently (1, 31). Hyphens indicate identical nucleotide sequence, and dots represent missing nucleotides. (B) Aligned translated protein sequences of STBL18, emm70 (STBL25), emm33, and STPK14. Hyphens indicate identical amino acid sequence, and dots represent missing amino acids.
STPK14 (accession no. AF018185) represented by VT 29.2 (Fig. 3A) also shares identity with emm70 (73%), emm33 (83%), and STBL18 (77%); however, the translated sequence of STPK14 differs from those of both M33 and M70 (Fig. 3B).
DISCUSSION
The discriminatory power of Vir typing with HaeIII has been enhanced by the use of an additional restriction enzyme, HinfI. The complete concordance between VTs and STs among these geographically related isolates indicates that each VT profile may represent at least one unique emm or emm ST among the strains tested.
These findings highlight the significant diversity of strains of GAS within the small, widespread Aboriginal communities of northern Australia because we have found 43 VTs circulating in the community (11, 13; this study) and a further 40 VTs in isolates from hospitalized patients (10). The observation that nearly one-third of Northern Territory isolates sequenced represent new STs differs considerably from a recent reports from the United States where a significant majority of the emm STs matched known STs (1, 2). Given the considerable diversity of emm STs found within the Aboriginal community, which has the highest prevalence of rheumatic heart disease reported in the world to date (6), ARF vaccines targeted to a few selected M types are unlikely to provide more than limited protection in these communities unless the same epitopes are shared by many members of an emm family.
This study supports recent conclusions by Beall and colleagues (2) that epidemiological typing is most meaningful when it is based upon a system reflecting M specificity. A study described in a recent report used an emm PCR-RFLP analysis, in which only the 1- to 2-kb emm gene is restricted with HaeIII (35). Outbreak-related strains that were defined by serological M type were compared with the emm PCR-RFLP patterns of coexistent strains of the same serotype, and it was found that M5 could be split into five HaeIII RFLP patterns, M76 could be split into six profiles, and R28 could be split into four distinct profiles. Unfortunately, sequencing of the emm gene was not done in that study, and in our experience, serotyping may group together isolates that are genetically distinct (12, 34). Nevertheless, emm RFLP analysis may be useful when examining small numbers of outbreak-related isolates in areas where strains of GAS are not endemic. In the endemic situation in which hundreds of isolates are examined, fragments in the range of 1 to 4 kb are helpful for discriminating between the different RFLP patterns obtained with HaeIII and HinfI. For these purposes, it is more convenient to analyze the RFLP profile of a 4- to 7-kb VT PCR product than a smaller emm PCR product.
The Vir regulon of GAS shows structural as well as sequence heterogeneity among isolates (15, 27). In this study, isolates were collected from impetiginous lesions, and their Vir regulons did not show significant structural heterogeneity, as evidenced by the remarkable similarities in the sizes of the initial PCR products when compared to the 4- to 7-kb PCR product found when isolates of GAS from the skin and throats of subjects in the northern hemisphere were used (9). Thus, the heterogeneity demonstrated by Vir typing of impetiginous isolates of GAS is due mainly to variations in the emm gene rather than diversity in the architecture of the regulon. Since the same ST has not yet been found in different VTs (this study), it is reasonable to hypothesize that emm may be evolving faster than other emm-like genes in our region. This may lead to the restricted diversity of enn in comparison to that of emm, as was observed previously (22, 40).
By RFLP analysis, each fragment does not represent an independent locus, because the creation or elimination of a single restriction site will alter two restriction fragments (37). Thus, single base changes could theoretically significantly alter the RFLP profile of any individual VT without altering the 5′ emm sequence. However, the 5′ emm sequence from every VT sequenced differed significantly. The lack of transitional forms of any ST, i.e., different VTs, with either a single or a few base pair changes, even among STs with the same serological profiles, can be explained if a recent report by Gupta et al. (14) is correct. The host immune response will structure the populations of infectious pathogens into stable collections of independently transmitted strains with nonoverlapping repertoires of dominant polymorphic determinants, despite the effects of recombination. Since the M protein is the principal immunodominant protein of GAS (7, 23), the structures of strains of GAS will be based on the M protein. Previous reports (25) have ascribed a clonal population structure to GAS on the basis of the observation that specific M types are almost exclusively associated with specific multilocus enzyme electrophoretic types. However, other multilocus enzyme electrophoresis data (16) indicate that while there is strong linkage disequilibrium between M types and electrophoretic types, there is no significant linkage disequilibrium between any of the alleles of the housekeeping genes used to produce the electrophoretic profiles, indicating that recombinations are not uncommon and that the apparently “clonal” population structure (25) may be directly related to the host immune response and is not a function of the organism per se.
Antigenic variation due to corrected frameshift mutations (29–31), point mutations (17, 33), and small insertions and deletions (17) have frequently been observed within the hypervariable region of emm. Antigenic variation in the streptococcal M protein may also be due to the deletion of repeat blocks (8, 19). Corrected frameshift mutations can result in drastic changes in translated sequences, without significant changes in DNA homology. Thus, single frameshifts may not be reflected by changes in VT. Consequently, very closely related sequence types which show frameshifts may have the same VT but may have different serological M types. Corrected frameshift mutations were first described in the M52/M53/M80 family of isolates from the Northern Territory of Australia (29–31) and in an M5 family isolate, STPL1 (18). In this study we have extended those original observations and found corrected frameshift mutations in emm49, emm6, emm13, emm33-emm70, and PT2110-family STs. Generally, these STs had only a few point mutations. In contrast, STs that had numerous point mutations compared to the number of point mutations in their closest reference M type rarely exhibited frameshift mutations. Small insertions and/or deletions were noted in STBL18, STPK14, STBSB29, STBL12, and STBSB19, as were previously noted for M1 (17). The short deletions were associated with short repeat sequences within the hypervariable region.
Concordance between VT and emm ST is a significant observation because it gives insight into the overall diversity of the virulence locus and allows discrimination of different emm STs. Vir typing has clear-cut advantages over randomly amplified polymorphic DNA analysis (9, 12) and M typing with oligonucleotide probes (20, 21, 26, 32). It has recently been proposed that routine emm sequencing may be used to designate the M type (1, 2). This is not feasible for outbreaks or for laboratories that do not have the resources to run an automated sequencer. Vir typing has been shown to be a rapid method of sorting large numbers of diverse GAS into distinct genotypes (9–12) and is a method applicable to areas where GAS are endemic and where the majority of isolates are MNT by serotyping.
ACKNOWLEDGMENTS
We thank the district medical officers and nursing staff of the Rural Health Service and the Aboriginal health workers of the six communities for help in obtaining the streptococcal isolates. We thank John Mathews and Wendy Relf for review of our paper, Jenny Powers for critical comments on the statistical methods used, and Elizabeth Wilson for laboratory assistance.
This work was supported by grants from the National Health and Medical Research Council of Australia and the National Heart Foundation.
REFERENCES
- 1.Beall B, Facklam R R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall B, Facklam R, Hoenes T, Schwartz B. Survey of emm gene sequences and T antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J Clin Microbiol. 1997;35:1231–1235. doi: 10.1128/jcm.35.5.1231-1235.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessen D E, Fischetti V A. Nucleotide sequences of two adjacent M or M-like protein genes of group A streptococci: different RNA transcript levels and identification of a unique immunoglobulin A-binding protein. Infect Immun. 1992;60:124–135. doi: 10.1128/iai.60.1.124-135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisno A L. Group A streptococcal infections and acute rheumatic fever. N Engl J Med. 1991;325:783–793. doi: 10.1056/NEJM199109123251106. [DOI] [PubMed] [Google Scholar]
- 5.Breiman R F, Davies J P, Facklam R R, et al. Defining the group A streptococcal toxic shock syndrome: rationale and consensus definition. The Working Group on Severe Streptococcal Disease. JAMA. 1993;269:390–391. [PubMed] [Google Scholar]
- 6.Carapetis J R, Wolff D R, Currie B J. Acute rheumatic fever and rheumatic heart disease in the Top End of Australia’s Northern Territory. Med J Aust. 1996;164:146–149. doi: 10.5694/j.1326-5377.1996.tb122012.x. [DOI] [PubMed] [Google Scholar]
- 7.Fischetti V A. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischetti V A, Jarymowycz M, Jones K F, Scott J R. Streptococcal M protein size mutants occur at high frequency within a single strain. J Exp Med. 1986;164:971–980. doi: 10.1084/jem.164.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardiner D, Hartas J, Currie B, Mathews J D, Kemp D J, Sriprakash K S. Vir typing: a long-PCR typing method for group A streptococci. PCR Methods Appl. 1995;4:288–293. doi: 10.1101/gr.4.5.288. [DOI] [PubMed] [Google Scholar]
- 10.Gardiner D, Hartas J, Hibble M, Goodfellow A, Currie B, Sriprakash K S. Molecular epidemiology of group A streptococcal infection in the Northern Territory of Australia. Adv Exp Med Biol. 1997;418:317–321. doi: 10.1007/978-1-4899-1825-3_76. [DOI] [PubMed] [Google Scholar]
- 11.Gardiner D L, Sriprakash K S. Molecular epidemiology of impetiginous group A streptococci in aboriginal communities of Northern Australia. J Clin Microbiol. 1996;34:1448–1452. doi: 10.1128/jcm.34.6.1448-1452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardiner D L. Ph.D. thesis. Molecular epidemiology of Streptococcus pyogenes. Sydney, Australia: University of Sydney; 1996. [Google Scholar]
- 13.Goodfellow A M, Gardiner D L. Searching for APSGN-associated Streptococcus pyogenes in communities with endemic streptococcal skin infections. Adv Exp Med Biol. 1997;418:103–108. doi: 10.1007/978-1-4899-1825-3_26. [DOI] [PubMed] [Google Scholar]
- 14.Gupta S, Maiden M C, Feavers I M, Nee S, May R M, Anderson R M. The maintainence of strain structure in populations of recombining infectious agents. Nature Med. 1996;2:437–442. doi: 10.1038/nm0496-437. [DOI] [PubMed] [Google Scholar]
- 15.Haanes E J, Heath D G, Cleary P P. Architecture of the vir regulons of group A streptococci parallels opacity factor phenotype and M protein class. J Bacteriol. 1992;174:4967–4976. doi: 10.1128/jb.174.15.4967-4976.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haase A M, Melder A, Mathews J D, Kemp D J, Adams M. Clonal diversity of Streptococcus pyogenes within some M types reveled by multilocus enzyme electrophoresis. Epidemiol Infect. 1994;113:455–462. doi: 10.1017/s0950268800068461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbaugh M P, Podbielski A, Hugl S, Cleary P P. Nucleotide substitution and small scale insertion produce size and antigenic variation in group A streptococcal M1 protein. Mol Microbiol. 1993;8:981–991. doi: 10.1111/j.1365-2958.1993.tb01642.x. [DOI] [PubMed] [Google Scholar]
- 18.Hartas J, Goodfellow A M, Currie B, Sriprakash K S. Characteristics of group A streptococcal isolates from tropical Australia with high prevalence of rheumatic fever: probing for signature sequences to identify members of the family of serotype 5. Microb Pathog. 1995;18:345–354. doi: 10.1006/mpat.1995.0031. [DOI] [PubMed] [Google Scholar]
- 19.Hollingshead S K, Fischetti V A, Scott J R. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol Gen Genet. 1987;207:196–203. doi: 10.1007/BF00331578. [DOI] [PubMed] [Google Scholar]
- 20.Kaufhold A, Podbielski A, Johnson D R, Kaplan E L, Lutticken R. M protein gene typing of Streptococcus pyogenes by nonradioactively labeled oligonucleotide probes. J Clin Microbiol. 1992;30:2391–2397. doi: 10.1128/jcm.30.9.2391-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufhold A, Podbielski A, Baumgarten G, Blokpoel M, Top J, Schouls L. Rapid typing of group A streptococci by the use of DNA amplification and non-radioactive allele-specific oligonucleotide probes. FEMS Microbiol Lett. 1994;119:19–25. doi: 10.1111/j.1574-6968.1994.tb06861.x. [DOI] [PubMed] [Google Scholar]
- 22.Kehoe M A, Kapur V, Whatmore A M, Musser J M. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 1996;4:436–443. doi: 10.1016/0966-842x(96)10058-5. [DOI] [PubMed] [Google Scholar]
- 23.Lancefield R C. Current knowledge of type-specific M antigens of group A streptococci. J Immunol. 1962;89:301–313. [PubMed] [Google Scholar]
- 24.Martin D R, Sriprakash K S. Epidemiology of group A streptococcal disease in Australia and New Zealand. Recent Adv Microbiol. 1996;4:1–40. [Google Scholar]
- 25.Musser J M, Kapur V, Szeto J, Pan X, Swanson D S, Martin D R. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Podbielski A, Kuhnemund O, Lutticken R. Identification of group A type 1 streptococcal M protein gene by non-radioactive oligonucleotide detection method. Med Microbiol Immunol. 1990;179:255–262. doi: 10.1007/BF00192463. [DOI] [PubMed] [Google Scholar]
- 27.Podbielski A. Three different types of organisation of the vir regulon in group A streptococci. Mol Gen Genet. 1993;237:287–300. doi: 10.1007/BF00282810. [DOI] [PubMed] [Google Scholar]
- 28.Relf W A, Sriprakash K S. Limited repertoire of the C-terminal region of the M protein in Streptococcus pyogenes. FEMS Microbiol Lett. 1990;71:345–350. doi: 10.1016/0378-1097(90)90245-l. [DOI] [PubMed] [Google Scholar]
- 29.Relf W A, Martin D R, Sriprakash K S. Identification of sequence types among M-nontypeable group A streptococci. J Clin Microbiol. 1992;30:3190–3194. doi: 10.1128/jcm.30.12.3190-3194.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Relf W A. Ph.D. thesis. Molecular diversity of the M protein gene of Streptococcus pyogenes. Sydney, Australia: University of Sydney; 1993. [Google Scholar]
- 31.Relf W A, Martin D R, Sriprakash K S. Antigenic diversity within a family of M proteins from group A streptococci: evidence for the role of frameshift and compensatory frameshift mutations. Gene. 1994;144:25–30. doi: 10.1016/0378-1119(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 32.Saunders N A, Hallas G, Gaworzewska E T, Metherell L, Efstratiou A, Hookey J V, George R C. PCR–enzyme-linked immunosorbent assay and sequencing as an alternative to serology for M-antigen typing of Streptococcus pyogenes. J Clin Microbiol. 1997;35:2689–2691. doi: 10.1128/jcm.35.10.2689-2691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott J R. The M protein of group A streptococcus: evolution and regulation. In: Iglewski B, editor. The bacteria. XI. New York, N.Y: Academic Press, Inc.; 1990. pp. 117–203. [Google Scholar]
- 34.Single L A, Martin D R. Clonal differences within M-types of the group A streptococcus revealed by pulsed field gel electrophoresis. FEMS Microbiol Lett. 1992;91:85–90. doi: 10.1016/0378-1097(92)90567-8. [DOI] [PubMed] [Google Scholar]
- 35.Stanley J, Desai M, Xerry J, Tanna A, Efstratiou A, George R. High resolution genotyping elucidates the epidemiology of group A streptococcal outbreaks. J Infect Dis. 1996;174:500–506. doi: 10.1093/infdis/174.3.500. [DOI] [PubMed] [Google Scholar]
- 36.Streeton C L, Hanna J N, Messer R D, Merianos A. An epidemic of acute poststreptococcal glomerulonephritis among aboriginal children. Paediatr Child Health. 1995;31:245–248. doi: 10.1111/j.1440-1754.1995.tb00795.x. [DOI] [PubMed] [Google Scholar]
- 37.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whatmore A M, Kapur V, Sullivan D J, Musser J M, Kehoe M A. Noncongruent relationships between variations in emm gene sequences and population genetic structure of group A streptococci. Mol Microbiol. 1994;14:613–619. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 39.Whatmore A M, Kehoe M A. Horizontal gene transfer in the evolution of group A streptococcal emm-like genes: gene mosaics and variation in Vir regulons. Mol Microbiol. 1994;11:363–374. doi: 10.1111/j.1365-2958.1994.tb00316.x. [DOI] [PubMed] [Google Scholar]
- 40.Whatmore A M, Kapur V, Musser J M, Kehoe M A. Molecular population genetic analysis of the enn subdivision of group A streptococcal emm-like genes: horizontal transfer and restricted variation among enn genes. Mol Microbiol. 1995;15:1039–1048. doi: 10.1111/j.1365-2958.1995.tb02279.x. [DOI] [PubMed] [Google Scholar]