Abstract
A previously undescribed Helicobacter sp. was recovered from a cat with severe diarrhea. Based upon the absence of any other identifiable cause of diarrhea, this helicobacter may be involved in the development of the disease signs. The organism could not be cultured but was described on the basis of 16S rRNA gene sequence analysis and morphology and appeared to be a new species, with Helicobacter canis being the most genetically similar species. The presence of a diarrhea-inducing helicobacter in a companion animal may pose a risk of zoonosis.
Diarrhea is common in humans and animals, although it is frequently impossible to identify a causative agent. Although Helicobacter spp. are better known as gastric pathogens, accumulating reports describe enteric pathogenic helicobacters, including Helicobacter canis in dogs (3, 22), Helicobacter cinaedi in humans (11, 24), Helicobacter fennelliae in humans (24), Flexispira rappini (a misnamed helicobacter) in humans (1), and Helicobacter pullorum in poultry and humans (4, 23). Helicobacter pamatensis from bird feces has been described, but it is of unknown pathogenicity (7). Some Helicobacter species appear to exist primarily in animal reservoirs in nature but may be zoonotic, such as Helicobacter pullorum (23) and perhaps Helicobacter heilmannii (21). The pathogenic potential of many gastrointestinal bacteria remains poorly understood. For example, experimentally induced diarrhea apparently flushed helicobacter organisms from the intestinal crypts of healthy rats, suggesting that the helicobacter was not a primary pathogen in these animals (15). The current paper describes a newly recognized, possibly pathogenic Helicobacter isolated from a cat with severe diarrhea. Although we were unable to culture the organism, we have identified it by microscopy and 16S rRNA gene sequence analysis, and we describe its phylogenetic association with other Helicobacter spp.
MATERIALS AND METHODS
Clinical evaluation.
Antibodies against viruses were evaluated by indirect immunofluorescence with FIPV-UCD1-infected Felis catus whole fetal (fcwf-4) cells as substrate for feline enteric coronavirus (19), FIV-Petaluma-infected fcwf-4 cells for feline immunodeficiency virus (25), F11/C2-7A-infected Crandell feline kidney cells for canine distemper virus, and N/CEK3-infected Crandell feline kidney cells for feline panleukopenia virus. Feline leukemia virus was evaluated by enzyme-linked immunosorbent assay for p27 antigen (16), and canine parvovirus was evaluated by the Cite fecal test (Idexx, Westbrook, Maine). Parasites and ova were evaluated by fecal flotation on a saturated zinc sulfate solution; fecal Cryptosporidium spp. and Giardia spp. were tested by direct smear and fluorescent antibody assay (Merifluor C/G; Meridian Diagnostics, Cincinnati, Ohio).
Microbiological methods.
Feces were plated onto MacConkey agar (PML, Rancho Cordova, Calif.); campylobacter agar containing cefoperazone, vancomycin, and amphotericin B (CVA; PML); fresh brain heart infusion (BHI) agar containing 2.5 μg of trimethoprim per ml, 5 μg of vancomycin per ml, 1.25 IU of polymyxin B per ml, and 2 μg of amphotericin B per ml; and fresh brucella agar containing fetal calf serum, trimethoprim, vancomycin, polymyxin B, and amphotericin B (Anaerobe Systems, San Jose, Calif.). A portion of the sample was also placed into selenite broth. After overnight incubation, the selenite broth was subcultured onto XLT4 (xylose-lysine-tergitol 4) agar (PML). The selenite broth and the MacConkey and XLT4 agar plates were incubated in air at 37°C. The CVA plate was incubated at 42°C and the BHI and brucella agar plates were incubated at 37°C. The CVA and the BHI and brucella agar plates were held under microaerophilic conditions with a CampyPakPlus system. The presence in feces of Clostridium difficile toxin A was evaluated by the monoclonal enzyme-linked immunosorbent assay-based C. difficile Toxin A kit (Pet RPLA; Unipath, Hampshire, United Kingdom). The presence of enterotoxigenic Clostridium perfringens was evaluated by PET-reverse passive latex agglutination assay.
Experimental inoculations.
Two BALB/c mice were orally inoculated with 0.2 ml of previously frozen feces from the original cat; two specific-pathogen-free cats received 20 mg of methylprednisolone acetate intramuscularly and were orally inoculated with 1 ml of feces. Cats and mice were observed daily after inoculation for fever, dehydration, lethargy, inappetance, and diarrhea. Daily fecal samples were collected for Gram staining and PCR. After 10 days, the animals were killed with an intravenous (cats) or intracardiac (mice) overdose of barbiturates, and sections of the intestines were collected and placed into 10% formalin, fixed overnight, embedded in paraffin, cut into thin sections, and stained with hematoxylin and eosin (H&E) and Warthin-Starry stains.
Pathology and light microscopy.
Midcerebral, hippocampal, cerebellar, and medullary transverse sections were submitted to the Yolo County Health Department for rabies virus detection by indirect immunofluorescence. Sections of bladder, bone marrow, brain, cecum, cerebellum, colon, duodenum, eye, heart, ileum, jejunum, kidney, liver, lung, mesenteric lymph nodes, pancreas, skeletal muscle, spleen, and stomach were fixed by immersion in 10% buffered formalin, embedded in paraffin, cut into 5-μm sections, and stained with H&E stain. The Warthin-Starry and Steiner stains were applied to the sections of stomach and intestines.
Electron microscopy.
The formalin-fixed tissues were rinsed in phosphate buffer and were then transferred to Karnovsky’s medium and fixed overnight at 4°C. The samples were then dehydrated with a 50, 75, 95, and 100% ethanol series and dried to the critical point in liquid carbon dioxide. They were mounted onto electron microscopy stubs and coated with gold in a sputter coater. The images were visualized with a Philips 501 scanning electron microscope at 75 kV.
A slurry of frozen feces from the original cat was negatively stained with 2% (wt/vol) phosphotungstic acid (pH 7.2) for 20 to 30 s and applied to a Formvar-coated, carbon-backed, 200-mesh copper grid. The specimens were examined with a Zeiss 10C transmission electron microscope at 80 kV.
Fecal extraction and PCR.
Fresh feces was suspended in a 1:1 (vol/vol) ratio of sterile phosphate-buffered saline solution, and the solution was then centrifuged at 2,600 × g for 10 min. The nucleic acids were extracted from the fecal supernatant by a modified Boom method of acid silica extraction (5). A forward eubacterial primer designated 8FPL (5′-CTGCAGAGTTTGATCCTGGCTCAG-3′) from the 16S rRNA and a Helicobacter genus-specific (7) reverse primer designated 300R (5′-TCTCAGGCCGGATACCCGTCATAGCCT-3′) were used to amplify a 292-bp fragment for initial screening. The generation of PCR products for sequencing was performed with primers 8FPL and 300R and primer 300F (the reverse complement of primer 300R) and eubacterial primer 1492RPL (5′-CGGGTTACCTTGTTACGACTT-3′). A second PCR was performed with H. pullorum-specific primers 5′-ATGAATGCTAGTTGTTGTCAG-3′ and 5′-GATTGGCTCCACTTCACA-3′ as described previously (23).
Amplification mixtures consisted of 10 μl of extracted DNA and 90 μl of a PCR mixture containing 50 mM KCl, 10 mM Tris-HCl, 0.1% Triton X-100, 2 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphates, 20 pmol each of the forward and reverse primers, and 2 U of Taq DNA polymerase. Amplification was performed in a thermal cycler (MJ Research, Watertown, Mass.) as follows: 1 cycle of denaturation at 95°C for 2 min and then 35 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 2 min, followed by a 7-min elongation step at 72°C and cooling at 4°C. The H. pullorum PCR was run as described previously (23). The products were separated on a 1% agarose gel by electrophoresis and were visualized with ethidium bromide.
DNA sequencing.
PCR products were purified with a Microcon 50 column according to the manufacturer’s instructions (Amicon, Beverly, Mass.). Sequencing reactions were performed by dye terminator cycle sequencing chemistry in a Ready Reaction Kit with AmpliTaq DNA polymerase FS (ABI Prism, Foster City, Calif.). Reactions were run on a 4.25% acrylamide/bisacrylamide gel on an ABI Prism 377 DNA Sequencer. The products were analyzed with ABI Prism Sequencing (version 2.1.1) software.
Statistical methods.
Three replicates of the 16S rRNA gene amplicon were independently sequenced in both directions, and the sequences were then compared for similarity to known Helicobacter sequences with the program Blast (Genetics Computer Group, Madison, Wis.). GenBank accession numbers of the organisms used in the comparison were L36147 (H. pullorum), L14628 (Campylobacter upsaliensis), L39122 (Helicobacter hepaticus), M88150 (H. cinaedi), U46129 (Helicobacter cholecystus), M35048 (Helicobacter mustelae), U65103 (Helicobacter trogontum), AF013464 (Helicobacter muridarum), U08906 (Helicobacter pylori), M57398 (Helicobacter felis), L14634 (H. canis), and M88154 (H. fennelliae). Uncorrected genetic distances were calculated by using the Genetics Computer Group program Distances.
RESULTS
Clinical presentation and history.
The patient was an 8-week-old domestic short-haired stray female kitten from within the Oakland, Calif., city limits. The pertinent past history was that she was smaller than her littermates and had profound diarrhea, vomiting, and inappetance for at least 1 week. She had bitten the owner on the toe 1 week prior to presentation. The cat was approximately 8% dehydrated and emaciated, with a body condition score of 2 of 9 (14), a dry unkempt coat, and numerous fleas. She had mild mesenteric lymphadenopathy and severe liquid diarrhea. The cat had a microcytic anemia with a hematocrit of 21.6% (normal hematocrit, 25.8 to 48.1%) and mean corpuscular volume of 39.9 fl (normal mean corpuscular volume, 43.4 to 52.8 fl). The total leukocyte count was 8,500 cells/μl with 10% bands and 79% neutrophils with toxic granulation. Serum chemistry was within normal limits. The cat was serologically negative for feline leukemia virus antigen and antibodies against feline enteric coronavirus, feline immunodeficiency virus, canine distemper virus, feline panleukopenia virus, and canine parvovirus. No parasites, ova, Cryptosporidium spp., or Giardia spp. were detected in her feces.
Microbiology.
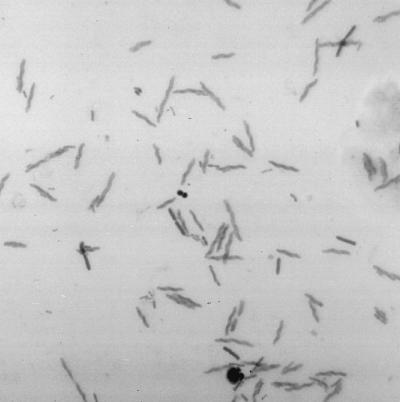
A Gram stain of the fecal smear showed large numbers of Helicobacter-like curved, gram-negative rods (Fig. 1). There was no growth of bacteria on MacConkey, campylobacter, fresh brain heart infusion, fresh brucella, or XLT4 agar plates. No clostridial toxins were detected in toxin assays.
FIG. 1.
Gram-stained feces from patient (stray female kitten).
Pathology.
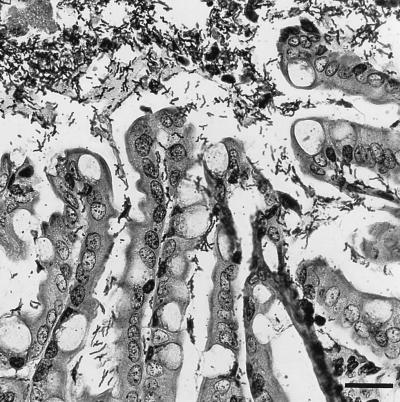
The kitten was killed with an intravenous overdose of barbiturates and was immediately necropsied. The entire length of the intestinal tract contained moderate amounts of yellow mucoid material. Microscopically, sections of the cecum and colon had a thick layer of densely packed bacteria that covered the mucosal surface and large particles of digesta (Fig. 2). The majority of the bacteria were slender and spiral shaped, with occasional large rods and rare cocci throughout. These bacteria were also frequently present throughout the lumina of the crypts, often spread over the apical surfaces of the cells. They stained strongly with Warthin-Starry stain but not with Steiner’s stain. Few histologic changes were observed in the intestinal mucosa itself. Rare small foci of neutrophil and eosinophil accumulation were found within the lamina propria of the cecum. Rare crypts were mildly dilated, had a flattened epithelium, and contained luminal cellular debris mixed with neutrophils. The cecal Peyer’s patches were well developed with well-formed germinal centers. The appearances of the duodenum, jejunum, and ileum were within normal limits. The gastric mucosa was significantly thinner than normal, with thinning of the gland layer and normal gastric pits. No significant lesions apart from a moderate histiocytic-neutrophilic bronchiolitis were observed in other tissue sections. The brain tissue was negative for rabies virus.
FIG. 2.
Colonic epithelium of the kitten. Note the large numbers of thick, long spiral-shaped bacteria in the lumen and extending into the crypts. Warthin-Starry staining was used. Bar, 25 μm.
PCR and DNA sequencing.
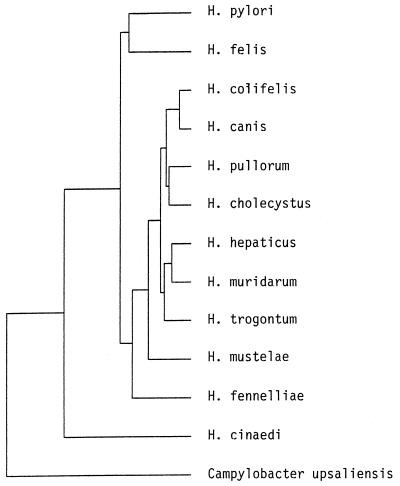
Primer pairs 8FPL-300R and 300F-1492RPL amplified a 292-bp and a 1,192-bp product, respectively. The species with the most closely related DNA base sequences were H. canis (98.3% similar), H. pullorum (from 96.9 to 96.4% similar, depending upon the strain), H. hepaticus (96.7% similar), and H. cinaedi (96.5% similar) (Table 1; Fig. 3). Specific primers for H. pullorum did not produce any detectable product, and no apparent binding sites in the sequenced DNA would correspond to these primers.
TABLE 1.
Matrix of uncorrected percent similarity on the basis of 16S rRNA gene sequence comparisons
| Species | % Similaritya
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hco | Hca | Hpu | Hhe | Hci | Hch | Hms | Htr | Hmu | Hpy | Hfl | Hfe | |
| H. colifelis | 100 | 98.3 | 96.6 | 96.7 | 96.5 | 96.3 | 95.0 | 95.6 | 94.9 | 93.4 | 93.2 | 92.5 |
| H. canis | 96.2 | 97.4 | 98.1 | 96.7 | 95.9 | 96.3 | 96.7 | 94.1 | 93.3 | 96.0 | ||
| H. pullorum | 96.1 | 95.3 | 96.8 | 94.7 | 96.2 | 95.4 | 95.2 | 94.6 | 96.3 | |||
| H. hepaticus | 96.5 | 96.4 | 95.7 | 97.0 | 97.9 | 93.9 | 93.5 | 95.8 | ||||
| H. cinaedi | 95.4 | 94.2 | 96.0 | 96.2 | 93.2 | 92.8 | 96.0 | |||||
| H. cholecystus | 96.6 | 95.8 | 96.3 | 94.5 | 94.2 | 95.7 | ||||||
| H. mustelae | 95.4 | 95.2 | 93.5 | 93.2 | 95.2 | |||||||
| H. trogontum | 96.5 | 93.0 | 92.7 | 96.3 | ||||||||
| H. muridarum | 93.1 | 92.8 | 95.5 | |||||||||
| H. pylori | 95.7 | 93.9 | ||||||||||
| H. felis | 93.7 | |||||||||||
Abbreviations: Hco, H. colifelis; Hca, H. canis; Hpu, H. pullorum; Hhe, H. hepaticus; Hci, H. cinaedi; Hch, H. cholecystus; Hms, H. mustelae; Htr, H. trogontum; Hmu, H. muridarum; Hpy, H. pylori; Hfl, H. felis; Hfe, H. fennelliae.
FIG. 3.
Genetic tree generated by the Genetics Computer Group Distances program illustrating the relationship of the 16S rRNA gene sequence of the new helicobacter isolate to other published helicobacter sequences. See text for the accession numbers of the published sequences.
Electron microscopy.
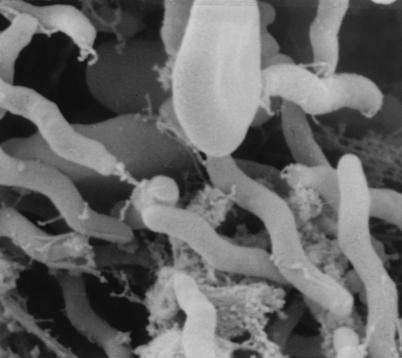
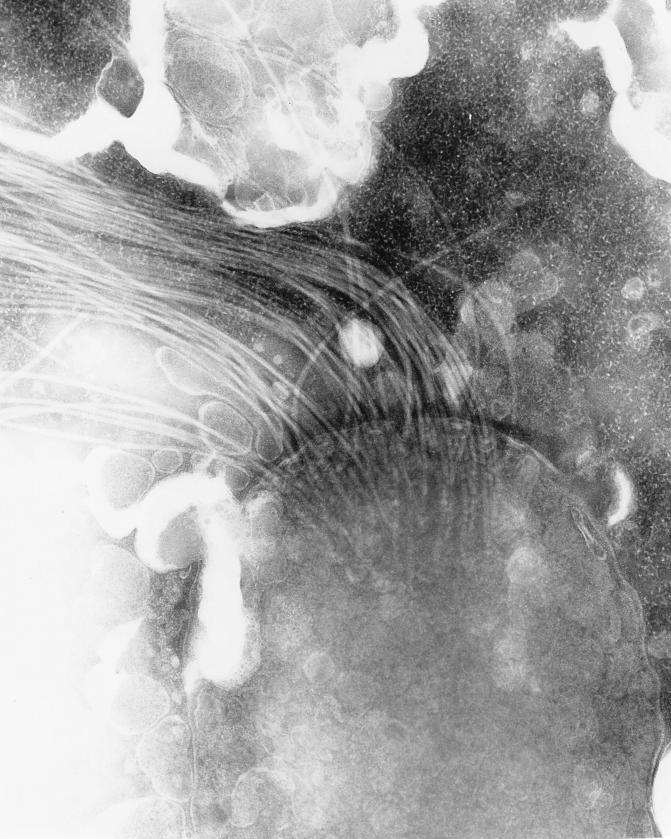
On the basis of ultrastructural analysis, bacteria were spiral shaped with two coils and measured from 4.0 to 5.9 μm in length and 0.67 μm in width (Fig. 4). Flagella were bipolar in central tufts which appeared to be connected to basal plates at the point of insertion (Fig. 5). No periplasmic fibers were observed. Extensive examination of sections revealed that all bacteria were associated with intestinal mucus but were not attached to enterocytes.
FIG. 4.
Scanning electron micrograph of helicobacter in colon section of the kitten. 7.5 mm = 1 μm.
FIG. 5.
Transmission electron micrograph of helicobacter from feces of the kitten. 10 mm = 0.1 μm.
Experimental inoculation into mice and cats.
No clinical signs of diarrhea, lethargy, dehydration, or inappetance were observed following inoculation of feces from the original cat into cats and mice. Gram stains of fecal smears from the inoculated animals revealed normal flora, and PCR of stool samples with primer pair 8FPL-300R was negative for mice on all days tested. However, cats had PCR-positive feces starting on day 2 postinoculation and continuing to day 11 postinoculation. Nevertheless, no helicobacter organism could be cultured from the fresh feces. Formalin-fixed, H&E-stained sections of intestines showed no inflammation or abnormal mucosal morphology. Few spiral bacteria were visible in the small intestinal sections of the inoculated cats.
DISCUSSION
The helicobacter from the cat described here is morphologically, ecologically, and genetically unique, and we propose it as a candidate species (17) with the specific epithet Helicobacter colifelis. It is unfortunate that the isolate could not be cultured, although it was not unexpected because it is difficult to culture many Helicobacter spp. (20), and the original feces had already been frozen before the pathologic diagnosis, reducing the organism’s viability for future passage. The natural host species was not known, and both cats and mice may have been subadequate hosts. With its close genetic relationship to H. pullorum, it is possible that H. colifelis exists in an avian reservoir in the wild. Without a cultured isolate, however, it was impossible to perform further phenotypic characterization of the organism.
Morphologically, H. colifelis was distinct from gastric helicobacters of felids, including Helicobacter acinonyx of cheetahs (8), H. pylori (12), and H. felis of cats (18). It was also morphologically dissimilar from enteric species of more disparate host species, including H. canis of dogs (3, 22), H. cinaedi of hamsters and humans (11, 24), H. fennelliae of humans (24), H. pamatensis of birds (7), and H. pullorum of poultry and humans (4, 23). The present feline isolate was large and had bipolar tufts of flagella. In contrast to H. felis, which is also relatively large with bipolar flagellar tufts, H. colifelis had only two coils and had centered flagellar tufts. Genetically, H. colifelis was distinct from H. felis and was most similar to H. canis and H. pullorum.
It is unclear how pathogenic H. colifelis is and whether it is likely to be zoonotic. Even with extensive clinical, microbiological, serological, and pathological examinations, no other cause for the profound diarrhea in the kitten could be determined. The inoculated cats did not develop diarrhea and had few visible organisms after inoculation, but they did become PCR positive. If the helicobacter were a secondary infection, possible primary conditions could have included stress or food intolerance. If the helicobacter was the cause of the diarrhea, it was interesting that there was so little accompanying inflammation, suggesting a possible enterosecretory mechanism of diarrhea induction. H. colifelis was present primarily in the intestinal mucus, which is the same niche occupied by many diarrhea-inducing Campylobacter spp. In contrast, gastric helicobacters such as H. felis are more commonly intimately associated with tissue and infection with gastric helicobacters may result in inflammation. Since helicobacters appear to be a subgroup within the campylobacters, the niche within the enteric mucus may be the primitive condition for the campylobacter-helicobacter group, with inflammatory gastric niches being a secondarily evolved character.
The occupation of the intestine, with diarrhea as an efficient method of bacterial dissemination into the environment, increases the probability of zoonosis, as described for H. canis in children (3) and H. pullorum in refugees, children, and people with AIDS (4). In contrast, gastric helicobacters are only occasionally detected in feces (10) and would be less likely to spread to humans. If the diarrhea occurs in a cat, exposure of humans becomes likely. Other pathogens in feline diarrhea which may infect people include Campylobacter jejuni (2, 13), Giardia lamblia, Cryptosporidium parvum (6), and Clostridium perfringens (9).
In summary, we describe a new enteric Helicobacter sp. infecting a cat. The recent rapid discoveries of new helicobacter and campylobacter species have opened up the prospects for future evaluation of diarrhea in human and animal patients, which have often remained refractory to diagnosis in the past. However, better data on the ecologies and pathogenicities of these species will be necessary before we are able to determine accurately the relative contributions of the numerous campylobacters and helicobacters to naturally acquired diarrhea.
ACKNOWLEDGMENTS
This work was supported by grants to J. E. Foley from the Krade and Maddox Endowments to the Center for Companion Animal Health, the San Francisco Foundation, and the Center for Companion Animal Health, School of Veterinary Medicine, University of California, Davis.
We thank Amy Poland, Bob Munn, Rich Walker, and Bob Nordhausen for technical assistance and Dwight Hirsh, Carol Glaser, and Patrick Foley for suggestions and interpretations.
REFERENCES
- 1.Archer J R, Romero S, Ritchie A E, Hamacher M E, Steiner B M, Bryner J H, Schell R F. Characterization of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:101–105. doi: 10.1128/jcm.26.1.101-105.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser M, Weiss S, Barrett T. Campylobacter enteritis associated with a healthy cat. JAMA. 1982;247:816. [PubMed] [Google Scholar]
- 3.Burnens A, Stanley J, Schaad U, Nicolet J. Novel Campylobacter-like organism resembling Helicobacter fennelliae isolated from a boy with gastroenteritis and from dogs. J Clin Microbiol. 1993;31:1916–1917. doi: 10.1128/jcm.31.7.1916-1917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnens A P, Stanley J, Morgenstern R, Nicolet J. Gastroenteritis associated with Helicobacter pullorum. Lancet. 1994;344:1569–1570. doi: 10.1016/s0140-6736(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 5.Cheung R C, Matsui S M, Greenberg H B. Rapid and sensitive method for detection of hepatitis C virus RNA by using silica particles. J Clin Microbiol. 1994;32:2593–2597. doi: 10.1128/jcm.32.10.2593-2597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Current W, Garcia L. Cryptosporidiosis. Clin Microbiol Rev. 1991;4:325–358. doi: 10.1128/cmr.4.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewhirst F, Seymour C, Fraser G, Paster B, Fox J. Phylogeny of Helicobacter isolates from bird and swine feces and description of Helicobacter pamatensis sp. nov. Int J Syst Bacteriol. 1994;44:553–560. doi: 10.1099/00207713-44-3-553. [DOI] [PubMed] [Google Scholar]
- 8.Eaton K, Dewhirst F, Radin M, Fox J, Paster B, Krakowka S, Morgan D. Helicobacter acinonyx sp. nov., isolated from cheetahs with gastritis. Int J Syst Bacteriol. 1993;43:99–106. doi: 10.1099/00207713-43-1-99. [DOI] [PubMed] [Google Scholar]
- 9.Foley J E, Hirsh D C, Pedersen N C. An outbreak of Clostridium perfringens enteritis in a cattery of Bengal cats and inadvertent experimental transmission to specific pathogen free cats. Feline Pract. 1996;24(6):31–35. [Google Scholar]
- 10.Fox J G, Paster B J, Dewhirst F E, Taylor N S, Yan L-L, Macuch P J, Chmura L M. Helicobacter mustelae isolation from feces of ferrets: evidence to support fecal-oral transmission of a gastric helicobacter. Infect Immun. 1992;60:606–611. doi: 10.1128/iai.60.2.606-611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gebhart C, Fennell C, Murtaugh M, Stamm W. Campylobacter cinaedi is normal flora in hamsters. J Clin Microbiol. 1989;27:1692–1694. doi: 10.1128/jcm.27.7.1692-1694.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handt L, Fox J, Dewhirst F, Fraser G, Paster B, Yan L, Rozmiarek H, Rufo R, Stalis I. Helicobacter pylori isolated from the domestic cat: public health implications. Infect Immun. 1994;62:2367–2374. doi: 10.1128/iai.62.6.2367-2374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopkins R, Olmsted R, Istre G. Endemic Campylobacter jejuni infection in Colorado: identified risk factors. Am J Public Health. 1984;74:249–250. doi: 10.2105/ajph.74.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laflamme D, Kealy R, Schmidt D. Estimation of body fat by body condition score. J Vet Intern Med. 1994;8:154. [Google Scholar]
- 15.Leach W, Lee A, Stubbs R. Localization of bacteria in the gastrointestinal tract: a possible explanation of intestinal spirochaetosis. Infect Immun. 1973;7:961–972. doi: 10.1128/iai.7.6.961-972.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutz H, Pedersen N C, Durbin R, Theilen G H. Monoclonal antibodies to three epitopic regions of feline leukemia virus p27 and their use in enzyme-linked immunosorbent assay of p27. J Immunol Methods. 1983;56:209–220. doi: 10.1016/0022-1759(83)90413-1. [DOI] [PubMed] [Google Scholar]
- 17.Murray R, Stackebrandt E. Taxonomic note: implementation of the provisional status Candidatus for incompletely described pathogens. Int J Syst Bacteriol. 1995;45:186–187. doi: 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- 18.Paster B, Lee A, Fox J, Dewhirst F, Tordoff L, Fraser G, O’Rourke J, Taylor N, Ferrero R. Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int J Syst Bacteriol. 1991;41:31–38. doi: 10.1099/00207713-41-1-31. [DOI] [PubMed] [Google Scholar]
- 19.Pedersen N C. Serologic studies of naturally occurring feline infectious peritonitis. Am J Vet Res. 1976;37:1449–1453. [PubMed] [Google Scholar]
- 20.Seymour C, Lewis R, Kim M, Gagnon D, Fox J, Dewhirst F, Paster B. Isolation of Helicobacter strains from wild bird and swine feces. Appl Environ Microbiol. 1994;60:1025–1028. doi: 10.1128/aem.60.3.1025-1028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solnick J, O’Rourke J, Lee A, Paster B, Dewhirst F, Tompkins L. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993;168:379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 22.Stanley J, Linton D, Burnens A, Dewhirst F, Owen R, Porter A, On S, Costa M. Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J Gen Microbiol. 1993;139:2495–2504. doi: 10.1099/00221287-139-10-2495. [DOI] [PubMed] [Google Scholar]
- 23.Stanley J, Linton D, Burnens A P, Dewhirst F E, On S L W, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 24.Totten P, Fennell C, Tenover F, Wezenberg J, Perine P, Stamm W, Holmes K. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J Infect Dis. 1985;151:131–139. doi: 10.1093/infdis/151.1.131. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto J K, Hansen H, Ho E W, Morishita T Y, Okuda T, Sawa T R, Nakamura R M, Pedersen N C. Epidemiologic and clinical aspects of feline immunodeficiency virus infection in cats from the continental United States and Canada and possible mode of transmission. J Am Vet Med Assoc. 1989;194:213–220. [PubMed] [Google Scholar]