Graphical abstract
Keywords: Degenerate zinc finger, Nuclear transcription factor, Single-domain zinc finger, Protein structure evolution, Psoriasis, PRR35, CCHC zinc finger
Highlights
-
•
ZNF750 is a transcription factor with functions in skin differentiation and cancers.
-
•
ZNF750 has a single zinc-finger classified as ‘degenerate’, or unlikely to bind zinc.
-
•
NMR and CD show the domain has a genuine and stable zinc finger structure.
-
•
Ligating site for zinc is CCHC rather than CCHH, Kd is pM.
-
•
Structure contextualizes function and cancer-linked mutations.
Abstract
ZNF750 is a nuclear transcription factor that activates skin differentiation and has tumor suppressor roles in several cancers. Unusually, ZNF750 has only a single zinc-finger (ZNF) domain, Z*, with an amino acid sequence that differs markedly from the CCHH family consensus. Because of its sequence differences Z* is classified as degenerate, presumed to have lost the ability to bind the zinc ion required for folding. AlphaFold predicts an irregular structure for Z* with low confidence. Low confidence predictions are often inferred to be intrinsically disordered regions of proteins, which would be the case if Z* did not bind Zn2+. We use NMR and CD spectroscopy to show that a 25–51 segment of ZNF750 corresponding to the Z* domain folds into a well-defined antiparallel ββα tertiary structure with a pM dissociation constant for Zn2+ and a thermal stability >80 °C. Of three alternative Zn2+ ligand sets, Z* uses a CCHC rather than the expected CCHH ligating motif. The switch in the last ligand maintains the folding topology and hydrophobic core of the classical ZNF motif. CCHC ZNFs are typically associated with protein–protein interactions, raising the possibility that ZNF750 interacts with DNA through other proteins rather than directly. The structure of Z* provides context for understanding the function of the domain and its cancer-associated mutations. We expect other ZNFs currently classified as degenerate could be CCHC-type structures like Z*.
Introduction
Transcription factors are sequence-specific DNA-binding proteins that control the transformation of genetic information from DNA to RNA. They account for ∼8% of human genes (Lambert et al., 2018) and are extremely important in biology and health since they determine what, when, where, how much, and for how long genes are expressed. Nearly half of human transcription factors have small zinc-finger (ZNF) domains, typically in multiple copies (Emerson et al., 2009). The ZNF domains usually adopt an antiparallel ββα folding motif upon binding divalent zinc (Zn2+). In transcription factors, ZNFs are typically used as nucleotide recognition modules that enable site-specific binding to double-stranded DNA (dsDNA).
ZNF750 is a 723 a.a. nuclear transcription factor that mediates skin differentiation (Boxer et al., 2014, Sen et al., 2012, Cohen et al., 2012, Zarnegar et al., 2012). Altered epidermal differentiation is a feature of >100 skin diseases, although the etiology is poorly understood at a mechanistic and molecular level (Cohen et al., 2012, Lopez-Pajares et al., 2013, Wikramanayake et al., 2014). The ZNF750 gene was first discovered due to an autosomal dominant frameshift mutation within its only putative CCHH ZNF domain (Birnbaum et al., 2006), the subject of this work that we will henceforth call Z*. The mutation occurred in a family that presented with seborrhea-like dermatosis with non-arthritic psoriasiform elements (SLDP), suggesting ZNF750 might play a role in genetic skin disease (Birnbaum et al., 2006). This was subsequently confirmed by mutations in the promoter for ZNF750 that cause familial psoriasis (Birnbaum et al., 2011, Yang et al., 2008). ZNF750 is expressed in keratinocytes but not fibroblasts (Cohen et al., 2012, Birnbaum et al., 2006). Based on mass spectrometry and ChIP-seq data, ZNF750 is thought to have dual roles in epithelial homeostasis and skin differentiation (Boxer et al., 2014, Sen et al., 2012, Cohen et al., 2012). In concert with the chromatin regulator KDM1A, ZNF750 inhibits progenitor genes that regulate the proliferation of self-renewing keratinocytes. With the pluripotency transcription factor KLF4 (Krüppel-like factor 4), ZNF750 activates genes that control skin differentiation. The keratinocyte-differentiation function of ZNF750 has recently been shown to be involved in the epidermal inflammatory response, through its interaction with the S/T-kinase IRAK2 in psoriatic epidermis but not in healthy skin. Thus, ZNF750 could affect the severity of skin diseases (Shao et al., 2021).
Over the last few years it has become evident that in addition to skin differentiation ZNF750 has roles in oral (Xu et al., 2022) and esophageal (Takahashi et al., 2022) squamous cell carcinoma, melanoma (Du et al., 2020), ocular sebaceous carcinoma (North, 2021), prostate (Montanaro et al., 2023), and breast cancers (Butera et al., 2020, An et al., 2022, Cassandri et al., 2020). A common mechanistic theme linking the involvement of ZNF750 in pathologies as varied as skin disease and cancers is that the transcription factor regulates cell differentiation. ZNF750 acts as a tumor suppressor by inhibiting cancer stem cells, with additional roles in regulating tumor growth, cell migration, and adhesion (Xu et al., 2022). When ZNF750 is mutated or expressed at low levels, it no longer acts as a tumor suppressor and cancer cells proliferate (Cassandri et al., 2017).
Classical biological approaches have shed considerable light on the function of ZNF750. However important open questions remain about the transcription factor that lend themselves to structural biology. ZNF750 has a single putative CCHH zinc-finger domain Z* (Fig. 1A), from which the transcription factor derives its name (Cohen et al., 2012). The CCHH designation (sometimes also called C2H2) gives the Cys, Cys, His, His order of Zn2+-ligands in the amino acid sequence. The vast majority of transcription factors have at least two or more CCHH ZNF domains, since a single copy is typically insufficient for sequence-specific DNA binding (Lee et al., 1991, Omichinski et al., 1997, Klug, 2010). Another unusual feature of the Z* domain is an amino acid sequence that differs considerably from the ZNF consensus.
Fig. 1.
Sequence properties of ZNF750 and its Z* domain.(A) Domain diagram for ZNF750 adapted from (Montanaro et al., 2023). Z*, the ZNF domain that is the subject of this study; NLS, nuclear localization signal; PLNLS, domain containing the given amino acid sequence thought to be important in mediating protein–protein interactions for ZNF750 (Boxer et al., 2014). (B) The consensus sequence of a CCHH ZNF determines its (C) Zn2+ coordination, (D) secondary structure, and (E) ββα folding motif. In B and C, Ω represents the aromatic residues Y or F, ψ hydrophobic residues, and X is any residue. (F) Sequence of the N-terminus of ZNF750. The first 30 residues have a high proportion of basic amino acids (light blue) some of which correspond to an NLS sequence (underlined). The sequence of the putative ZNF domain, Z*, is shown in bold type. The UniProt database gives the Z* domain boundary as 25–46. This leads to a highly unfavorable 8-residue spacing between the second and third Zn2+-ligands. Extending the domain boundary by five amino acids to residue 51 introduces an additional Cys. With the extended domain boundary, there are three alternative Zn2+-ligand sets (G) The second alternative CCHC, gives a more favorable 12-residue spacing between the second and third ligands. The Z*{C34A, H39A) mutant peptide was used to confirm the Zn2+-ligands identified from the NMR structure: C27, C30, H43, and C49.
The presence of Cys and His residues that can chelate Zn2+ is not sufficient to specify a ZNF domain. ZNF750 has 15 Cys and 22 His residues. Clearly, the protein would not function properly if all the potential ligands bound Zn2+ non-selectively. Rather, ZNF domains have amino acid preferences beyond the requirement of Cys/His ligands to bind Zn2+ (Miller et al., 1985, Berg and Shi, 1996). These preferences vary for different ZNF families described by the order of metal-chelating ligands, such as CCCC, CCHC, and CCCH. The families are associated with distinct functions such as DNA, RNA, protein, or lipid binding (Berg and Shi, 1996, Michalek et al., 2011, Fu and Blackshear, 2017, Wang et al., 2021, Ravasi et al., 2003). For the classical DNA-binding CCHH ZNFs of transcription factors, the consensus sequence is (Ω)-X-C-X2-5-C-X3-(Ω)-X5-ψ-X2-H-X3-5-H (Fig. 1B). Here, Ω is the aromatic residues Y or F, ψ is a hydrophobic residue, X is any residue, and the subscripts indicate the spacing between residues (Berg and Shi, 1996, Michalek et al., 2011). The conserved hydrophobic and aromatic residues (shown in pink and purple in Fig. 1B-C) are required to form a small hydrophobic core (Miller et al., 1985). The spacing between metal ligands is important in accommodating the antiparallel ββα secondary structure motif, as well as for satisfying functional requirements (Fig. 1D-E). The “finger region” refers to the 12-residue segment between the second Cys and the first His ligand for Zn2+ (Fig. 1C), that determines the dsDNA-binding specificity of CCHH-type ZNFs (Branden and Tooze, 1998).
Mutations altering the spacing between the Zn2+-chelating residues in CCHH-type ZNFs, or replacing conserved aromatic/hydrophobic residues, can destabilize the structure or abrogate Zn2+-binding (Lachenmann et al., 2004, Párraga et al., 1990). The UniProt database (The_UniProt_Consortium, 2023) designates the domain boundaries for Z*, the putative CCHH-type ZNF in ZNF750, as residues 25–46 (Fig. 1F). With this domain assignment, the ‘finger region’ of Z* would have an 8 rather than the consensus 12-residues spacing, and none of the aromatic or hydrophobic residues that typically stabilize the ββα-fold. Due to the differences from consensus the UniProt database classifies Z* as ‘degenerate’, signifying the domain has probably lost its ability to bind Zn2+ (The_UniProt_Consortium, 2023, Aceituno-Valenzuela et al., 2020). The premiere method for protein structure prediction, AlphaFold (Jumper et al., 2021, Lewis and The, 2023), only models about 12 α-helical residues of the 723 a.a. protein with high confidence and predicts an irregular structure for the Z* domain with low confidence (PDB code AF_AFQ32MQ0F1). Clearly the folding state of Z*, whether the domain is a genuine ZNF, or an intrinsically disordered region without the ability to bind Zn2+, is critical for understanding the structure and function of ZNF750 – a transcription factor that derives its name from its sole ZNF domain. We therefore investigated the structure of Z* by nuclear magnetic resonance (NMR).
Results
The Z* domain folds in the presence of Zn2+
Anticipating the Z* domain may not be a CCHH-type ZNF because of the resulting unfavorable 8-residue spacing between the second and third Zn2+-ligands, we synthesized a slightly longer fragment of ZNF750 running from residues 25–51 (Fig. 1F). The longer fragment included C49, which could give a conventional 12-residue finger spacing if the second and third Zn2+ ligands were C30 and H43 (Fig. 1F). However, this would result in a CCHC rather than a CCHH ZNF, with C49 as the fourth ligand. A third possibility with the inclusion of C49 was a CCCC-type ZNF, since these are generally the least constrained in terms of sequence and ligand-spacing requirements. The three alternative types of Zn2+-ligand sets for the 25–51 fragment of ZNF750 are shown in Fig. 1G.
Circular dichroism (CD) and NMR spectra of the synthetic 25–51 fragment of ZNF750 showed clear evidence for protein folding in the presence of a 1.2 M excess of Zn2+ (Fig. 2). The CD spectrum is typical of an unfolded polypeptide in the absence of the metal but when Zn2+ is added matches that of folded ZNF domains (Tremblay et al., 2016, Simpson et al., 2003), with a mixture of α-helix and β-sheet secondary structure (Fig. 2A). Consistently, addition of Zn2+ leads to increased chemical shift dispersion in the NMR spectrum, and enhanced amide proton signals due to reduced solvent exchange, indicating the domain adopts a stable tertiary structure (Fig. 2C). The free and Zn2+-bound peptide are in slow exchange on the NMR timescale, with an increase in the intensity of NMR signals from the folded peptide at the expense of those from the unfolded peptide when Zn+2 is added (Matousek and Alexandrescu, 2004).
Fig. 2.
The Z* domain binds Zn2+ to form a stable folded structure. (A) CD spectra in the absence of Zn2+ (red) and in the presence of the metal at different temperatures. Note that the CD spectrum does not revert to that of the unfolded domain even at 82 °C. (B) Zn2+ binding curve for Z* in the presence of 22 mM EGTA. The data were fit to a sigmoidal curve, giving a Kd of 4.0 ± 0.4 × 10-12 M, and a Hill coefficient of 1.2 ± 0.1. (C) Downfield region of the 1H NMR spectrum of Z* in the absence (black) and presence of Zn2+ (purple). Assignments are given for some of the dispersed NMR resonances characteristic of folded structure. (D) Same as C except with the C34A, H39A mutant of Z* used to verify the Zn2+-chelating residues determined from the NMR structure.
To assess the stability of the Zn2+-bound Z* domain at pH 7, we varied the temperature from 21 to 82 °C in 4-degree increments. For clarity, data are only shown at three representative temperatures in Fig. 2A. With increasing temperature, the CD spectra show a linear increase in ellipticity at 208 nm and a decrease at 195 nm, which likely represents a pre-denaturation transition rather than the sigmoidal transition typical of protein unfolding. The overall CD spectrum remains characteristic of a folded polypeptide up to 82 °C and very different from that of the unfolded domain in the absence of Zn2+. We repeated the temperature experiment at pH 5.8, reasoning more acidic conditions could destabilize metal binding. However, we obtained similar results with Z* remaining folded up to 90 °C. We conclude that the Zn2+-bound Z* domain is highly stable to thermal unfolding.
To assess metal binding affinity we carried out a competition experiment with the metal chelator EGTA (Ivanova et al., 2008). The use of the competing chelator was necessary because Z* binds Zn2+ too strongly to measure affinity directly. The binding data gives a Kd of 4 × 10-12 M for Zn+2 at pH 7.0 (Fig. 2B). This value is within the range for ZNFs, that typically have Kd < 10-9 M but in some cases as small as 10-18 M (Kluska et al., 2018, Neuhaus, 2022, Padjasek et al., 2020).
In summary, the present results show that Z* is a genuine rather than degenerate ZNF. Upon binding Zn2+ with a pM affinity, the domain folds into a well-defined tertiary structure, with a thermal stability>80–90 °C.
Z* has a typical ZNF ββα fold
The Z* domain in the presence of Zn2+ gives excellent NMR data (Fig. 3). Each amino acid gives a single set of NMR resonances unlike other ZNF domains that exhibit spectral heterogeneity near histidines, due to interchange between aromatic ring Nδ2 and Nε1 atom coordination of Zn2+ (Matousek and Alexandrescu, 2004, Ramboarina et al., 1999). We were able to obtain virtually complete NMR assignments of Z* using standard 2D TOCSY, NOESY (Fig. 3A), DQF-COSY, and E-COSY experiments, together with natural abundance 13C-HSQC (Fig. 3B) and 15N-sofast HMQC (Fig. 3C) spectra. The NMR chemical shifts, together with short-range backbone NOE patterns, enabled us to calculate (Cheung et al., 2010, Shen and Bax, 2013) the secondary structure of Z* shown in Fig. 4. The calculated ββα secondary structure is typical for a ZNF domain (Berg, 1988). We used the Talos-N program to predict S2 order parameters from the chemical shifts of Z*. The S2 order parameter ranges from 1 for rigid sites to 0 for sites with unrestricted flexibility. Except for the last two residues in the sequence, all S2 values for the Z* domain were above 0.7. The muted flexibility of Z* is consistent with its high thermal stability.
Fig. 3.
Representative 2D-NMR data for the Z* domain. (A) Superposition of TOCSY (red) and NOESY (black) spectra, illustrating sequential walks used to obtain NMR assignments between residues M44 to F32 (purple) and C30 to Y25 (green), interrupted by P31. Blue labels indicate TOCSY HN-Hα crosspeaks, pink labels are sidechain crosspeaks. The group of peaks connected by a dashed gray line marked H2O at 4.93 ppm, are due rapid magnetization exchange between amide protons and water during the 70 ms TOCSY mixing time. (B) Fingerprint region of a natural-abundance 1H–13C HSQC spectrum. Blue labels indicate 1Hα-13Cα crosspeaks. (C) Natural-abundance 1H-15N sofast-HMQC spectrum. Blue labels indicate backbone 1H-15N correlations. Pink labels indicate correlations from sidechain –NH2 groups, and from the backbone C-terminal amide blocking group (CO-NH2).
Fig. 4.
Secondary structure of Z*. The consensus secondary structure is based on NMR short-range NOE distance contacts and chemical shift differences from random coil values, calculated with the program DANGLE (Cheung et al., 2010). The thickness of the horizontal bars denotes the intensity of backbone NOEs.
The NMR structure of Z* demonstrates a CCHC metal binding site
The ββα secondary structure of Z* (Fig. 4) would seem to preclude a CCHH-type ZNF structure, since this would place the putative ligands C30 in the β-hairpin turn and H39 at the beginning of the α-helix, on opposite ends of the molecule. The preeminent structure prediction algorithm AlphaFold (Lewis and The, 2023, Callaway, 2022, Service, 2021) sheds little light on the matter, as it predicts irregular structure for the Z* domain with a low confidence pLDDT score (Fig. 5A). Sequence conservation within Z* is high (Birnbaum et al., 2006), and did not allow us to distinguish between the three alternative sets of chelating residues. To determine the correct metal ligands and fold for Z*, we calculated its solution NMR structure using the experimental data in Table 1.
Fig. 5.
Structure of the Z* domain.(A) AlphaFold model for the human ZNF750 protein (PDB CSM accession code AFQ32MQ0F1). The position of the Z* domain from Y25 to N51 is indicated. The model is color-coded according to prediction confidence levels: pLDDT > 90 blue, 90 > pLDDT > 70 cyan, 70 > pLDDT > 50 yellow, 50 < pLDDT orange. (B) Folding topology and Zn2+-binding site of the Z* domain based on experimental NMR data (Table 1). The sidechains of Zn2+-chelating residues are shown, as well as C34 and H49 which do not participate in metal binding. (C) Ensemble of the top NMR structures of Z*. The backbone is shown in dark blue, sidechains of Zn2+ ligands in gray, conserved aromatic and hydrophobic residues in green, and additional nonpolar residues that are not part of the conserved consensus in orange. (D) Superposition of the NMR structure of Z* from this work with the first structure of a ZNF (Lee et al., 1989), the classical CCHH domain from Xfin-31 (PDB code 1ZNF). Note the similarity in overall structures, and in the positioning of the last ligand (H19 in Xfin-31, C49 in Z*) relative to the metal.
Table 1.
Statistics for the 20 best NMR structures of the Z* domain.
| NMR restraints (total) | 390 | |
|---|---|---|
| Distance (total) | 330 | |
| Intraresidue NOE (|i-j| = 0) | 119 | |
| Sequential NOE (|i-j| = 1) | 82 | |
| Short-range NOE (1<|i-j|<5) | 72 | |
| Long-range NOE (5≤|i-j|) | 41 | |
| Hydrogen bond (2x) | 12 | |
| Zinc restraints | 4 | |
| Dihedral (ϕ 23, ψ 21, χ1 16) | 60 | |
| Restraint violations | ||
| > 0.3 Å | 0 | |
| > 5o | 0 | |
| Residual restraint violations | ||
| Distance (Å) | 0.0685 ± 0.0007 | |
| Dihedral (o) | 0.57 ± 0.03 | |
| RMS deviations from ideal geometry | ||
| Bonds (Å) | 0.00474 ± 0.00006 | |
| Angles (o) | 0.655 ± 0.004 | |
| Improper torsions (o) | 0.379 ± 0.004 | |
| Ramachandran plot PROCHECK statistics | ||
| Residues in most favored regions | 90 % | |
| Residues in allowed regions | 10 % | |
| Residues in generously allowed regions | 0 % | |
| Residues in disallowed regions | 0 % | |
| Coordinate rms deviations (Å) | Backbone (Cα,N,C’) | All heavy |
| NMR ensemble to average | 0.25 ± 0.11 | 0.77 ± 0.12 |
| Z* vs PDB 1ZNF | 1.08 ± 0.03 | |
| Z* vs PDB 1A1H: 135–156 | 1.49 ± 0.14 | |
In the NMR structure calculations, we took great care not to bias the structures by introducing incorrect restraints between the potential ligating residues and Zn2+. We calculated an initial set of NMR structures using only dihedral and intra-protein distance restraints. The initial structures showed residues C27, C30, H43 and C49 were in proximity and poised to ligate Zn2+. By contrast residues C34 and H39 were too far to participate in metal binding. Once correct ligands were identified from the initial structures, distance restraints to the Zn2+ atom were included in the final set of NMR structure calculations (Fig. 5 B-D). As an additional check of correct ligand assignments, we used 1D NMR to demonstrate that the variant Z*(C34A, H39A), missing the cysteine and histidine not involved in ligation, retains the ability to fold with Zn2+ (Fig. 1D).
A ribbon diagram for the NMR structure of Z* closest to the ensemble average is shown in Fig. 5B. The set of 20 lowest energy structures with no violations is displayed in Fig. 5C. The Z* structure is superposed with the Xfin-31 classical CCHH ZNF in Fig. 5D. The two structures are remarkably close with a backbone RMSD of 1.1 Å. Comparison to other ZNF structures (Fig. 6A) gave similar results with a range of RMSDs between 0.8 and 1.5 Å.
Fig. 6.
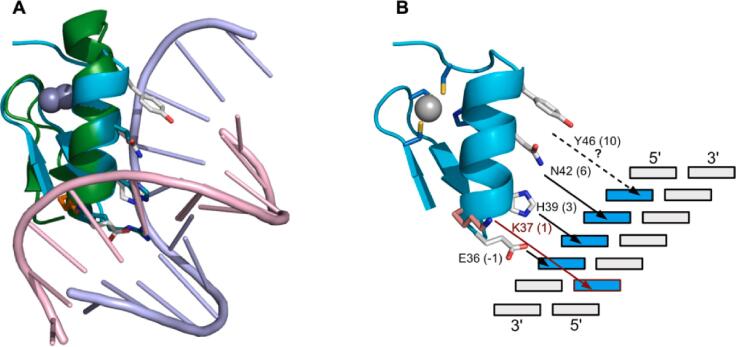
Putative DNA binding site for Z*.(A) Superposition of Z* (cyan ribbon, white sidechains) onto the middle ZNF of Egr-1 (green) in complex with its target DNA (PDB 4X9J). The 1.4 Å X-ray structure of Egr-1 (Zandarashvili et al., 2015) has two flanking ZNFs in the DNA complex that are omitted for clarity. The superposition illustrates the similarity of the ZNF structures (RMSD 1.2 Å) and the putative DNA-binding sidechains of Z* (white) relative to the Erg-1 ZNF. (B) Schematic of a possible DNA-binding residues of Z* based on the ZNF consensus. In addition to the consensus residues, Y46 may bind to the coding strand, and K37 to the noncoding strand. The DNA-binding site prediction server for ZNFs (http://zf.princeton.edu) (Persikov and Singh, 2014), suggests the Z* domain has a preference for the DNA sequence pattern (G/C)G(C/G), with only the central G of the triplet exhibiting a strong propensity. The prediction is in agreement with our NMR structure, since H39 in the 3 position of the Z* binding site would have a strong preference for guanine in the central position of the DNA triplet (Wolfe et al., 2000). Note, however, that the function of Z* may be protein-binding rather than DNA-binding, as described in the Discussion section.
Compared to Xfin-31, which is a CCHH-type ZNF, the final half-turn of α-helix in Z* is disrupted to allow C49 to become the last ligand in a tetrahedral CCHC coordination site for Zn2+ (Fig. 1D). The first ligand, C27, comes from the β1 strand; the second, C30, from the β-hairpin reverse turn, and the third, H43, from the middle of the α-helix (Fig. 5A). These are all features of the classical CCHH ZNF motif (Fig. 1C-E). To a large extent the backbone structure of the CCHC domain Z*, is very much like that of a CCHH ZNF except that the last ligand is a cysteine rather than a histidine. Structural similarity extends to the residue following the second cysteine ligand, P31, having unusual backbone dihedral angles (Simpson et al., 2003) that account for most of the deviations of Z* from Ramachandran ideality (Table 1). The structural similarity is even more striking for the sidechains around the Zn2+ ion (Fig. 5D). The first three conserved ligands give an RMSD of 0.36 Å over 15 equivalent heavy atoms, compared to 1.08 Å for the backbone atoms between the Z* and Xfin-31 structures. For the non-conserved fourth ligand, the atoms Cα, Cβ, and Sγ of C49 in Z* essentially trace the Nδ1, Cε1, and Nε2 aromatic ring atoms of H23 in Xfin-31 to result in structurally if not chemically nearly equivalent coordination sites (arrow in Fig. 5D). Of the Cys and His residues not involved in metal coordination (Fig. 5D), H39 could participate in DNA binding (see below). The cysteine that does not participate in metal coordination, C34, points to the interior of the structure with its sulfhydryl group possibly forming a hydrogen bond with the backbone carbonyl of the last residue before the start of the α-helix, E36.
The three residues Y25, F32, and L40 (green in Fig. 5C) correspond to the conserved hydrophobic core at the base of the V-like wedge between the β-hairpin and α-helix in classical ZNFs. Although, F32 (light green in Fig. 5C) is strictly not-conserved (Fig. 1B, G), it serves the consensus structural role (Fig. 1B). In addition to the classic hydrophobic core, Z* appears to have a second hydrophobic ridge that stabilizes the structure comprised of residues F28, F41, M44, Y46 and L48 (orange in Fig. 5C).
The structure of Z* provides a context for understanding sidechains that could be involved in function and cancer-associated mutations
Given the structure of Z* is so close to that of classical ZNFs, we superposed it onto the best-available 1.4 Å-resolution X-ray structure of a ZNF-DNA complex (Zandarashvili et al., 2015) to check if the domain could be compatible with DNA-binding. Indeed, we found that the Z* structure could be consistent with a DNA-binding site, however, as described in the Discussion section the presence of only a single ZNF domain with a CCHC ligand set argue the domain has a protein-binding rather than DNA-binding function.
In classical CCHH ZNFs, the DNA binding site is formed by residues −1, 3, and 6, where the numbering corresponds to the start of the α-helix (Klug, 2010, Wolfe et al., 2000). Note that the X-ray structure of Egr-1 (PDB code 4X9J) has a tandem of three ZNFs bound to DNA. For clarity only the middle finger is shown in Fig. 6A. The superposition and sequence rules for ZNFs (Klug, 2010, Wolfe et al., 2000) allow us to identify putative DNA-ligating residues in Z* as E36 (-1), H39 (3) and N42 (6) (Fig. 6B). The sidechain of Y46 at the 10 position of Z* also seems poised for interactions with the base above that recognized by the 6 position (Fig. 6B), a pattern found in some ZNFs with a canonical DNA binding mode (Wolfe et al., 2000). A common feature of the DNA recognition motif in canonical ZNFs, is that the residue in position 2 recognizes a base on the noncoding strand (Fig. 6B) to create a 4 base pair overlapping subsite in transcription factors with multiple tandem ZNFs (Klug, 2010). In Z*, the 2 position is occupied by S38 the sidechain of which seems too small to reach the opposite strand. Rather, K37 in the 1 position appears ideally positioned to interact with the noncoding strand.
Genome-wide association studies (GWAS) of cancer patients have identified several somatic missense mutations in the ZNF750 transcription factor, with some of these specifically in the Z* domain (Table 2). The mutations were compiled from the literature (Takahashi et al., 2022) and from the CBioPortal (Gao et al., 2013) database (https://www.cbioportal.org). We caution that the significance of these somatic mutations is unknown, since a higher mutation rate due to impaired DNA repair is a hallmark of cancers in general (Jackson and Loeb, 1998, Bailey et al., 2021). Of the currently catalogued cancer-associated mutations in Z*, all involve structural residues rather than amino acids that could participate in the putative DNA binding site (Table 2). This includes the C49R mutation that replaces the cysteine we established in this work as the fourth ligand for Zn2+. A large-scale analysis of CCHH ZNFs found single nucleotide polymorphisms are less abundant in DNA-contacting residues, in general (Lockwood et al., 20142014). In addition to missense mutations, there are frameshift and nonsense mutations that affect the function of the ZNF750 transcription factor. In the familial SLDP disease that led to the discovery of the ZNF750 gene (Birnbaum et al., 2006), an autosomal dominant frameshift mutation deletes everything past residue 19 of the 723-residue protein, including the Z* domain.
Table 2.
Cancer-associated somatic missense mutations in the Z* domain of ZNF750.
| Mutation | IDaor reference | Cancer type | Site role in structure |
|---|---|---|---|
| Y25D | TCGA-Z6-A8JE | Esophageal squamous cell | conserved aromatic |
| K26E | TCGA-D3-A3C7 | Cutaneous melanoma | α-helix C-cap |
| F28L | TCGA-TN-A7HL | Head and neck squamous | β-hairpin turn aromatic |
| F28Y | TCGA-TN-A7HL | Head and neck squamous | β-hairpin turn aromatic |
| P31S | TCGA-E7-A677 | Bladder urothelial | β-hairpin turn proline |
| F32L | (Takahashi et al., 2022) | Esophageal squamous cell | conserved aromatic |
| L40I | TCGA-BS-A0UV | Uterine endometrioid | conserved hydrophobic |
| L40V | TCGA-DK-A3WW | Bladder urothelial | conserved hydrophobic |
| M44R | (Takahashi et al., 2022) | Esophageal squamous cell | hydrophobic in core |
| C49R | TCGA-BB-7870 | Head and neck squamous | Loss of 4th Zn2+-ligand |
Sample IDs are from the CBioPortal (Gao et al., 2013) (https://www.cbioportal.org) for the ZNF750 entry.
Discussion
The Z* domain is classified as degenerate by the UniProt database because its sequence does not conform to the classical CCHH ZNF consensus (The_UniProt_Consortium, 2023, Aceituno-Valenzuela et al., 2020). Besides Z*, we have recently determined that the seventh ZNF domain of ZNF711, also classified as degenerate (UniProt accession code Q9Y462), folds in the presence of Zn2+ (A.J.R. and A.T.A., in preparation). We are aware of another two ZNF domains classified as degenerate by UniProt (accession codes Q9P243 domain 14, P17041 domain 7) that have NMR structures in the Protein Databank (PDB codes 2RV1 and 2EPC). The AlphaFold method that performed the best at the last few biennial CASP tournaments (Callaway, 2022, Sippl, 1999), predicts at low confidence a structure for Z* that differs from our experimental NMR structure (Fig. 5A). About 42% of residues in human proteins are predicted by AlphaFold at low confidence, defined by a pLDDT score lower than 70 (Tunyasuvunakool et al., 2021). The inference for these regions is that they are dynamic, either intrinsically disordered or becoming structured as part of a complex (Tunyasuvunakool et al., 2021). If true, AlphaFold would not only predict protein structure but flexibility. The structure of Z* is extremely stable with a melting point above 80 °C. S2 order parameters calculated from NMR chemical shifts indicate the structure is rigid except for the last two residues in the sequence, in agreement with the tight clustering of conformers in the NMR ensemble. We suspect that the low confidence of the AlphaFold prediction for Z* has less to do with dynamics than with the unfamiliarity of its sequence, making the domain underrepresented in sequence and structure databases.
The first clues that the Z* domain is a functional ZNF came from experiments demonstrating the double-mutants ZNF750(C27A, C30A), ZNF750(H39A, H43A), and the quadruple mutant ZNF750(C27A, C30A, H39A, H43A) abrogated the ability of the transcription factor to induce late terminal epidermal differentiation through its induction of the expression of the KLF4 transcription factor (Boxer et al., 2014, Sen et al., 2012). Mutagenesis can lead to ambiguous results since a missense mutation can affect structural or functional parts of a protein. Because of the assumption of a CCHH ZNF, and because the putative ligating residues were mutated in pairs, the mutagenesis experiments missed that the Z* domain has a CCHC (C27, C30, H43, C49) rather than the expected CCHH coordination site. The CCHH arrangement (C27, C30, H39, H43) is disfavored by the prohibitively short 8-residue spacing between the second and third ligands (Fig. 1F, 5B).
There are two conceivable ways a ZNF domain could fold despite sequence differences from consensus. The first is a rearrangement of the structure to preserve the character of the metal binding site. The second is to allow differences in the Zn2+ ligands, while preserving the folding motif. The second alternative is used by the Z* domain. Similarly, two previously described examples of degenerate ZNFs with structures in the PDB (codes 2RV1 and 2EPC), both have the prototypical ββα fold. This suggests selective pressure to maintain the ββα structural motif, determined by the spacing between the Zn2+ ligands and the conserved positions forming the hydrophobic core. By contrast constraints on the identity of the Zn2+ binding ligands appear laxer, as long as the ββα-fold foundation for the coordination site is preserved. From a structural point of view, the Z* domain is just like a classical CCHH-type ZNF, except that the last Zn2+ ligand histidine ligand is replaced by a cysteine (Fig. 5D). The Zn2+-ligating residues are on the opposite side of the α-helix that would determine the DNA binding-site of a CCHH-type ZNFs (Fig. 6B). In Z*, neither Zn2+-binding affinity (Fig. 2B) nor stability (Fig. 2A) are perceptibly affected by the switch of the last ligand from a histidine to a cysteine. This suggests that the association of CCHH-type ZNFs with DNA-binding functions is due to gene duplication and divergent evolution. The structure and stability properties of the ZNF folding motif appear equally well supported whether a histidine or cysteine is the last ligand for Zn2+.
As Z* is a CCHC-type ZNF it is useful to distinguish between two types of families that have this set of metal-binding ligands. One CCHC family has the sequence spacing C-X2-C-X4-H-X4-C. This family typically functions as an RNA-binding module in RNA metabolism processes (Michalek et al., 2011, Wang et al., 2021, Aceituno-Valenzuela et al., 2020). Member of the family usually have a ‘zinc knuckle’ structure, where the short four-residue segment between the second cysteine and histidine ligands, truncates the ‘finger’ to a knuckle. We think it is useful to distinguish this knuckle-CCHC family from the finger-CCHC family exemplified by the Z* domain, since the latter have structures very similar to the classical ZNFs. Besides Z*, other examples of finger-CCHC domains exist in the proteins NEMO (Cordier et al., 2008), Fog, MOZ, U-shaped (Matthews et al., 2000), and BLC11A (Grabarczyk et al., 2018, Shen et al., 2021).
All examples of the CCHC-finger motif currently described in the literature function in protein–protein rather than protein-DNA interactions (Cordier et al., 2008, Matthews et al., 2000, Grabarczyk et al., 2018, Shen et al., 2021). A possible exception is the seventh domain of ZNF32, currently classified as degenerate but with an unpublished CCHC structure from a Structural Genomics group (PDB 2EPC). Unfortunately, there is no information on the function of this domain and whether it binds proteins or DNA. In some cases such as BLC11A, CCHC domains occur together with CCHH domains in transcription factors, with the latter used for DNA-binding and the former for protein interactions (Grabarczyk et al., 2018, Shen et al., 2021). We anticipate substantially more CCHC-fingers remain to be discovered and compiled a list of examples in Table 3. These include domains currently classified as degenerate CCHH ZNFs where a cysteine could replace the missing last histidine, as well as domains where a potential fourth ligand is present in the sequence downstream of the assigned domain boundary. Ideally, the folding status and DNA or protein-binding functions of these domains would be established experimentally since there are relatively few characterized examples of CCHC fingers to date, and some of the putative examples in Table 3 have alternative sets of potential ligands.
Table 3.
Examples of potential CCHC ZNFs currently annotated as degenerate CCHH domains.
| Proteina | UniProt ID | Domain Limits | Potential Zinc Ligandsb |
|---|---|---|---|
| PRDM6-1 | Q9NQX0 | 473–495 | C475, C478, H491, C493 |
| PRDM6-4 | Q9NQX0 | 557–579 | C559, C562, H575, C582c |
| ZFAT-2 | Q9P243 | 116–141 | C118, C121, H134, C136 |
| ZFHX4-15 | Q86UP3 | 2267–2291 | C2269, C2272, H2285, C2290 |
| ZFHX4-19 | Q86UP3 | 3354–3378 | C3356, C3359, H3372, C3377 |
| ZFP57-7 | Q9NU63 | 356–378 | C358, C361, H374, C391c, d |
| ZN131-4 | P52739 | 356–381 | C358, C361, H374, C378 |
| ZN292-8 | O60281 | 1375–1397 | C1377, C1380, H1393, C1400c |
| ZN423-1 | Q2M1K9 | 67–93 | C69, C72, H85, C90e |
| ZN675-3 | Q8TD23 | 200–222 | C202, C205, H218, C224c |
| ZN746-1 | Q6NUN9 | 453–478 | C455, C458, H471, C475 |
The number following the protein is the degenerate ZNF domain number assigned by UniProt.
The underlined residue is a putative fourth Zn2+ ligand that we identified in the sequence.
The putative fourth ligand is downstream of the domain boundary specified by UniProt.
Alternatively, H388 or H389 could be the fourth ligand for this domain, giving a CCHH-type ZNF.
Alternatively, H88 could be the fourth ligand for this domain, giving a CCHH-type ZNF.
ZNF750 is an extremely rare and unusual example of a transcription factor with a single ZNF domain. We are aware of only one other such example, the single ZNF domain from the GAGA transcription factor that is a CCHH rather than a CCHC domain (Omichinski et al., 1997). With the GAGA transcription factor, NMR structural studies showed that two basic regions from the N-terminus of the protein adopt α-helical structures that augment the site-specific DNA binding modulated by the single ZNF domain (Omichinski et al., 1997). Like the GAGA transcription factor, ZNF750 has basic regions upstream of the Z* domain (light blue in Fig. 1F). In ZNF750 the N-terminal basic region includes the sequence RKPKK (underlined in Fig. 1F) that is a signature nuclear localization signal (NLS). However, there are additional NLS motifs at the C-terminus of ZNF750 (Fig. 1A), and it was shown that the N-terminal NLS was dispensable for nuclear localization (Cohen et al., 2012). The N-terminal basic region could thus be part of an extended DNA-binding site that includes the Z* domain, like in the GAGA transcription factor. Alternatively, immunoprecipitation and mutagenesis studies showed that ZNF750 interacts with the chromatin regulatory proteins KDM1A, RCOR1, CTPB1 and CTPB1, through two PLNLS amino acid sequence motifs in the 504–563 region (Fig. 1A) of the protein (Boxer et al., 2014). Moreover, mutagenesis of the PLNLS motifs affected DNA-binding (Boxer et al., 2014). The chromatin regulatory proteins that interact with ZNF750 are transcriptional regulators with inherent DNA-binding functions. Thus, it is possible ZNF750 does not bind DNA on its own but only as part of heterooligomeric complexes that extend the binding site beyond that provided by Z* alone.
Conversely, ZNF750 may not contact DNA at all but form protein interactions with other transcription regulators that bind DNA. The primary evidence that supports ZNF750 binding to DNA is ChIP-Seq data demonstrating that a segment of the ZNF750 transcription factor near the Z* domain interacts with the progenitor genes it represses and the differentiation genes it activates through a 5′-CCNNAGGC-3′ DNA consensus sequence (Boxer et al., 2014). Significantly, the quadruple ZNF750 (C27A, C30A, H39A, H43A) mutant described previously decreased DNA binding in the ChIP-Seq assay as well as binding to the proteins KLF4 and RCO1 (Boxer et al., 2014). The ChIP-Seq method uses formaldehyde crosslinking and immunoprecipitation of the target protein, together with DNA sequencing, for genome-wide profiling of protein DNA binding-sites (Park, 2009). Since formaldehyde crosslinking is nonspecific, there is a possibility the assay could identify a protein that does not interact with given DNA sequences directly but through other proteins or complexes (Narlikar and Jothi, 2012, Hoffman et al., 2015).
Factors that suggest a role in protein rather than DNA-binding for Z* include (i) that it occurs as a single ZNF in the ZNF750 transcription factor, and (ii) is a CCHC-type domain, a family so far associated with protein–protein interactions. A domain similar to Z* with only three sequence differences (N35L, F41Y, G47S) and hence probably highly similar structural properties, is present in the ZNF750 paralog PRR35 (proline-rich protein 35, UniProt accession code P0CG20). Alignment gives 77% sequence identity between residues 5–56 of ZNF750 and residues 17–68 of PRR35. Beyond the N-terminal basic region and the Z* domain (Fig. 1F), the sequences of the two proteins are dissimilar. The function of PRR35 is unknown. A family of small proline-rich proteins is involved in keratinocyte differentiation (Zabini et al., 2023) but it is unclear if PRR35 is related to these. Since the first ∼60 residues of ZNF750 and PRR35 are highly homologous, these segments either both bind similar DNA sequences through the Z*-ZNF and the conserved N-terminal basic regions, or alternatively these segments both bind similar client proteins rather than DNA.
Taken together, the structure of Z* sheds new light on the role of this domain in the function of the ZNF750 transcription factor. We established in this work that ZNF750 contains a stable, folded, genuine ZNF domain, Z*. At the same time, the CCHC ligand set for Z* suggests the domain may bind proteins rather than DNA. The functional role of ZNF750 may be more akin to general transcription factors like TFIIB (Buratowski and Zhou, 1993). Clearly more information is needed on the structural properties of the full-length ZNF750 protein and its binding partners. Given the many unusual features of this transcription factor we expect these mechanistic aspects will need more experimental data to establish functions unequivocally.
Experimental procedures
Materials
The 27-residue peptides Z* and Z*(C34A, H39A), corresponding to the fragment 25–51 of human ZNF750 (UniProt Q32MQ0), were synthesized by AAPPTec (Louisville, KY). Both peptides had their termini blocked by N-acetylation and C-amidation, were ≥90% pure by HPLC, and had molecular weights by mass spectrometry within 3 Daltons of those theoretically expected. ZnSO4·H2O (purity of ≥99.9%) was from Sigma (St. Louis, MO).
Optical spectroscopy and Zn2+ affinity
Z* concentrations were determined via a BCA assay (Walker and Walker, 2002) according to the manufacturer’s instructions (Thermo Fisher Scientific; Waltham, MA) on an Ultrospec 8000 UV–Vis spectrophotometer (Thermo Fisher). Concentrations from the BCA assay were used to determine molar extinction coefficients at 280 nm of 2,954 M−1 cm−1 for unfolded Z* in the absence of Zn2+, and 2,978 M−1 cm−1 for folded Z* in the presence of a 1:1.25 ratio of Zn2+.
CD data were collected using an Applied Photophysics Chirascan V100 Spectrometer (Surrey, UK) using a 1 mm cuvette. The Z* concentration was 124 μM in 10 mM NaPO4 buffer, pH 7. CD spectra were collected between 190 and 250 nm, using a 1 nm bandwidth, a 1 nm step size, and 5 s/point data averaging for an approximate total scan time of 5 min. Thermal melt experiments were collected from 20 °C to 96 °C in 4-degree increments, with the same parameters except using 0.75 s/point data averaging, for a total scan time of 72 s.
For Zn2+-binding experiments 22 mM EGTA together with ZnSO4 concentrations of 0.1––200 μM were added to 133 µM Z*. The fraction of Zn2+-bound protein, Φ, was calculated from
| (1) |
where [Θ]i is the measured ellipticity at 200 nm for a given Zn2+ concentration, and [Θ]int and [Θ]fin are the plateau values for free and fully-bound Z*, respectively (Imanishi et al., 2010). Free Zn2+ concentrations were calculated with the WEBMACX program (Ivanova et al., 2008, MaxChelator). The binding data were modeled according to a sigmoidal function:
Φ = Φfree + (Φbound – Φfree)/(1+([Zn2+]free/Kd)n) (2)
where the fitted parameters were the dissociation constant Kd, the Hill coefficient n, and the plateau fraction bound values (Φ) at low and high [Zn2+]free concentrations (Fig. 2B).
NMR spectroscopy
To establish folding in the presence of Zn2+ (Fig. 2C, D), 1D 1H NMR experiments with 0.5 mM samples of Z* and Z*(C34A, H39A) were done at 27 °C on a Bruker Avance 500 MHz spectrometer. PFG-diffusion experiments established that the Zn2+-bound Z* domain had an Rh of 8.1 ± 1.2 Å, consistent with a monomer (Whitehead et al., 2022).
All 2D NMR experiments used for assignments and structure calculations were done at 15 °C on a Bruker Avance 600 MHz instrument equipped with a TCI cryogenic probe, that is part of the Francis Bitter Magnet Lab of MIT. A sample for 2D NMR experiments in H2O had 1.5 mM Z* and 2.4 mM ZnSO4, in 10 mM NaPO4 buffer. For experiments in D2O the sample had 3.0 mM Z*, 4.8 mM ZnSO4, and 20 mM NaPO4 buffer. Both samples had a pH of 5.9. The experiments recorded on the H2O sample included 70 ms mixing time TOCSY (mlevgpphw5), 200 ms mixing time NOESY (noesygpphw5), and a natural abundance 15N-sofast-HMQC (sfhmqcf3gpph). The names in parentheses are the Bruker pulse programs for the experiments. Experiments on the D2O sample included 50 and 200 ms mixing time NOESY, DQF-COSY (cosydfph), ECOSY (ecos3nph), and natural abundance 13C-HSQC (hsqcetgpsi2). Most of the assignments were obtained from a sequential walk using the NOESY and TOCSY spectra recorded in H2O (Fig. 3A). The natural abundance 1H–13C HSQC spectrum (Fig. 3B) proved particularly useful to confirm 1H aliphatic assignments and to extend these to the bonded carbon resonances. The 15N-sofast-HMQC experiment (Fig. 3C) was used to extend amide proton assignments to their bonded nitrogens. Experiments in D2O were used for aromatic assignments, and to observe Hα resonances without interference from solvent. The extent of assigned resonances was 1H (93%), 13C (83%, excluding C’), and 15N (77%). Stereospecific assignments were obtained from ECOSY and short mixing time (50 ms) NOESY experiments (Matousek and Alexandrescu, 2004). Cβ shifts for all four cysteine residues were in a range between 30.1 and 30.7 ppm, indicating they were all in a reduced state (Schulte et al., 2020).
NMR structure calculations
The restraints used for NMR structure calculations are summarized in Table I. Distance restraints were obtained from NOESY crosspeak intensities calculated with the program CcpNmr Analysis 2.5 (Vranken et al., 2005). Torsional restraints were calculated from 1HN; 1Hα 13Cα, 13Cβ, and 15N chemical shifts using the program TALOS-N (Shen and Bax, 2015). To identify the Zn2+ ligands an initial set of 20 lowest energy structures without violations was calculated from a starting set of 100 random conformation with the program XPLOR-NIH (Schwieters et al., 2003). These initial structures showed an anti-parallel ββα structure where C27, C30, H43, and C49 were poised to bind Zn2+ but C34 and H39 were too far to participate in the coordination site. Based on these initial structures, distance constraints were included between Zn2+ and the aforementioned ligands, and hydrogen bond (H-bond) restraints were included for amide protons protected from solvent exchange.
Two distance restraints were used per H-bond (1.5–2.5 Å for NH-O and 2.5–3.5 Å for N-O) to enforce linearity. An additional seven distance restraints were included between the protein and the Zn2+ atom: 2.33–2.37 Å for Zn2+-Sγ and rZn-Cβ = 3.25–3.51 Å for Zn2+-Sγ, for each of the three cysteines. An ambiguous restraint of 1.0–3.1 Å was set between Zn2+ and either the Nδ1 or Nε2 atom of H43. In all the final structures the Zn2+ was bonded to the Nε2 atom of H43. With all metal restraints removed, the structures retained the ββα fold, and gave an all-atom RMSD to the final structures of 0.37 ± 0.05 Å. Thus, the Z* structure is not constrained by the Zn2+ contacts alone but by the totality of dihedral and distance restraints.
Database accession numbers
NMR assignments and chemical shift values for the Z* domain were deposited in the BMRB under accession code 51951. Coordinates for the 20 lowest energy structures with no violations, together with the NOE distance and dihedral restraints used for NMR structure calculations, were deposited in the PDB under accession code 8SXM.
CRediT authorship contribution statement
Antonio J. Rua: Formal analysis, Investigation, Writing – review & editing. Richard D. Whitehead: Investigation, Methodology, Writing – review & editing. Andrei T. Alexandrescu: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Prof. Charles Giardina for useful discussion, Prof. Carolyn Teschke for use of her CD spectrophotometer, Prof. Nathan Alder for use of his UV-Vis spectrophotometer, and Prof Mei Hong for use of her group’s solution 600 MHz instrument during the sabbatical of A.T.A. The NMR experiments used equipment at the MIT-Harvard Center for Magnetic Resonance, which is supported by the P41 grant GM132079. M.H. is partially supported by NIH grant AG059661.
Data availability
Data will be made available on request.
References
- Aceituno-Valenzuela U., Micol-Ponce R., Ponce M.R. Genome-wide analysis of CCHC-type zinc finger (ZCCHC) proteins in yeast, Arabidopsis, and humans. Cell. Mol. Life Sci. 2020;77(20):3991–4014. doi: 10.1007/s00018-020-03518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G., Feng L., Hou L., Li X., Bai J., He L., Gu S., Zhao X. A bioinformatics analysis of zinc finger protein family reveals potential oncogenic biomarkers in breast cancer. Gene. 2022;828:146471. doi: 10.1016/j.gene.2022.146471. [DOI] [PubMed] [Google Scholar]
- Bailey C.G., Gupta S., Metierre C., Amarasekera P.M.S., O’Young P., Kyaw W., Laletin T., Francis H., Semaan C., Sharifi Tabar M., Singh K.P., Mullighan C.G., Wolkenhauer O., Schmitz U., Rasko J.E.J. Structure-function relationships explain CTCF zinc finger mutation phenotypes in cancer. Cell. Mol. Life Sci. 2021;78(23):7519–7536. doi: 10.1007/s00018-021-03946-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J.M. Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. PNAS. 1988;85(1):99–102. doi: 10.1073/pnas.85.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J.M., Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271(5252):1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- Birnbaum R.Y., Zvulunov A., Hallel-Halevy D., Cagnano E., Finer G., Ofir R., Geiger D., Silberstein E., Feferman Y., Birk O.S. Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nat. Genet. 2006;38(7):749–751. doi: 10.1038/ng1813. [DOI] [PubMed] [Google Scholar]
- Birnbaum R.Y., Hayashi G., Cohen I., Poon A., Chen H., Lam E.T., Kwok P.-Y., Birk O.S., Liao W. Association analysis identifies ZNF750 regulatory variants in psoriasis. BMC Med. Genet. 2011;12(1) doi: 10.1186/1471-2350-12-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer L.D., Barajas B., Tao S., Zhang J., Khavari P.A. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014;28(18):2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branden C., Tooze J. Garland Science; New York: 1998. Introduction to Protein Structure. [Google Scholar]
- Buratowski S., Zhou H. Functional domains of transcription factor TFIIB. PNAS. 1993;90(12):5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butera A., Cassandri M., Rugolo F., Agostini M., Melino G. The ZNF750-RAC1 axis as potential prognostic factor for breast cancer. Cell Death Discov. 2020;6:135. doi: 10.1038/s41420-020-00371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. 'The entire protein universe': AI predicts shape of nearly every known protein. Nature. 2022;608(7921):15–16. doi: 10.1038/d41586-022-02083-2. [DOI] [PubMed] [Google Scholar]
- Cassandri M., Smirnov A., Novelli F., Pitolli C., Agostini M., Malewicz M., Melino G., Raschellà G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017;3(1) doi: 10.1038/cddiscovery.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassandri M., Butera A., Amelio I., Lena A.M., Montanaro M., Mauriello A., Anemona L., Candi E., Knight R.A., Agostini M., Melino G. ZNF750 represses breast cancer invasion via epigenetic control of prometastatic genes. Oncogene. 2020;39(22):4331–4343. doi: 10.1038/s41388-020-1277-5. [DOI] [PubMed] [Google Scholar]
- Cheung M.-S., Maguire M.L., Stevens T.J., Broadhurst R.W. DANGLE: A Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J. Magn. Reson. 2010;202(2):223–233. doi: 10.1016/j.jmr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Cohen I., Birnbaum R.Y., Leibson K., Taube R., Sivan S., Birk O.S., Brandner J.M. ZNF750 is expressed in differentiated keratinocytes and regulates epidermal late differentiation genes. PLoS One. 2012;7(8):e42628. doi: 10.1371/journal.pone.0042628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordier F., Vinolo E., Véron M., Delepierre M., Agou F. Solution structure of NEMO zinc finger and impact of an anhidrotic ectodermal dysplasia with immunodeficiency-related point mutation. J. Mol. Biol. 2008;377(5):1419–1432. doi: 10.1016/j.jmb.2008.01.048. [DOI] [PubMed] [Google Scholar]
- Du Y., Lv G., Jing C., Liu J., Liu J. ZNF750 inhibits the proliferation and invasion of melanoma cells through modulating the Wnt/b-catenin signaling pathway. Folia Histochem. Cytobiol. 2020;58:255–263. doi: 10.5603/FHC.a2020.0026. [DOI] [PubMed] [Google Scholar]
- Emerson R.O., Thomas J.H., Myers S. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5(1):e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M., Blackshear P.J. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 2017;17(2):130–143. doi: 10.1038/nri.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269) doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarczyk P., Winkler P., Delin M., Sappa P.K., Bekeschus S., Hildebrandt P., Przybylski G.K., Völker U., Hammer E., Schmidt C.A. The N-terminal CCHC zinc finger motif mediates homodimerization of transcription factor BCL11B. Mol. Cell Biol. 2018;38(5) doi: 10.1128/MCB.00368-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman E.A., Frey B.L., Smith L.M., Auble D.T. Formaldehyde crosslinking: a tool for the study of chromatin complexes. J. Biol. Chem. 2015;290(44):26404–26411. doi: 10.1074/jbc.R115.651679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi M, Nakaya T, Morisaki T, Noshiro D, Futaki S, Sugiura Y. Metal-stimulated regulation of transcription by an artificial zinc-finger protein. 2010;11:1653-5. [DOI] [PubMed]
- Ivanova, E., Ball, M., Lu, H., Zinc binding of Tim10: Evidence for existence of an unstructured binding intermediate for a zinc finger protein. 2008;71:467–75. [DOI] [PubMed]
- Jackson A.L., Loeb L.A. The mutation rate and cancer. Genetics. 1998;148:1483–1490. doi: 10.1093/genetics/148.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79(1):213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Kluska K., Adamczyk J., Krezel A. Metal binding properties, stability and reactivity of zinc fingers. Coord. Chem. Rev. 2018;367:18–64. [Google Scholar]
- Lachenmann M.J., Ladbury J.E., Qian X., Huang K., Singh R., Weiss M.A. Solvation and the hidden thermodynamics of a zinc finger probed by nonstandard repair of a protein crevice. Protein Sci. 2004;13(12):3115–3126. doi: 10.1110/ps.04866404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. The human transcription factors. Cell. 2018;172(4):650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Lee M.S., Gippert G.P., Soman K.V., Case D.A., Wright P.E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989;245(4918):635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Lee M.S., Gottesfeld J.M., Wright P.E. Zinc is required for folding and binding of a single zinc finger to DNA. FEBS Lett. 1991;279:289–294. doi: 10.1016/0014-5793(91)80170-8. [DOI] [PubMed] [Google Scholar]
- Lewis T., The A.I. Biologist: DeepMind's Demis Hassabis explains how artificial intelligence solved one of the biggest problems in biology. Sci. Am. 2023;328:28–30. doi: 10.1038/scientificamerican0223-28. [DOI] [PubMed] [Google Scholar]
- Lockwood, S.H., Guan, A., Yu, A.S., Zhang, C., Zykovich, A., Korf, I., et al. The functional significance of common polymorphisms in zinc finger transcription factors. G3 (Bethesda). 2014;4:1647-55. [DOI] [PMC free article] [PubMed]
- Lopez-Pajares V., Yan K., Zarnegar B.J., Jameson K.L., Khavari P.A. Genetic pathways in disorders of epidermal differentiation. Trends Genet. 2013;29(1):31–40. doi: 10.1016/j.tig.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matousek W.M., Alexandrescu A.T. NMR structure of the C-terminal domain of SecA in the free state. Biochim. Biophys. Acta. 2004;1702(2):163–171. doi: 10.1016/j.bbapap.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Matthews J.M., Kowalski K., Liew C.K., Sharpe B.K., Fox A.H., Crossley M., Mackay J.P. A class of zinc fingers involved in protein-protein interactions biophysical characterization of CCHC fingers from fog and U-shaped. Eur. J. Biochem. 2000;267(4):1030–1038. doi: 10.1046/j.1432-1327.2000.01095.x. [DOI] [PubMed] [Google Scholar]
- MaxChelator.
- Michalek J.L., Besold A.N., Michel S.L. Cysteine and histidine shuffling: mixing and matching cysteine and histidine residues in zinc finger proteins to afford different folds and function. Dalton Trans. 2011;40:12619–12632. doi: 10.1039/c1dt11071c. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A.D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro M, Agostini M, Anemona L, Bonanno E, Servadei F, Finazzi Agro E, et al. ZNF750: A novel prognostic biomarker in metastatic prostate cancer. Int J Mol Sci. 2023;24. [DOI] [PMC free article] [PubMed]
- Narlikar L., Jothi R. ChIP-Seq data analysis: identification of protein-DNA binding sites with SISSRs peak-finder. Methods Mol. Biol. 2012;802:305–322. doi: 10.1007/978-1-61779-400-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus D. Zinc finger structure determination by NMR: Why zinc fingers can be a handful. Prog. Nucl. Magn. Reson. Spectrosc. 2022;130–131:62–105. doi: 10.1016/j.pnmrs.2022.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North J.P. Molecular genetics of sebaceous neoplasia. Surg. Pathol. Clin. 2021;14(2):273–284. doi: 10.1016/j.path.2021.03.005. [DOI] [PubMed] [Google Scholar]
- Omichinski J.G., Pedone P.V., Felsenfeld G., Gronenborn A.M., Clore G.M. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nat. Struct. Biol. 1997;4(2):122–132. doi: 10.1038/nsb0297-122. [DOI] [PubMed] [Google Scholar]
- Padjasek M., Kocyła A., Kluska K., Kerber O., Tran J.B., Krężel A. Structural zinc binding sites shaped for greater works: Structure-function relations in classical zinc finger, hook and clasp domains. J. Inorg. Biochem. 2020;204:110955. doi: 10.1016/j.jinorgbio.2019.110955. [DOI] [PubMed] [Google Scholar]
- Park P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Párraga G., Horvath S., Hood L., Young E.T., Klevit R.E. Spectroscopic studies of wild-type and mutant “zinc finger” peptides: determinants of domain folding and structure. PNAS. 1990;87(1):137–141. doi: 10.1073/pnas.87.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov A.V., Singh M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014;42:97–108. doi: 10.1093/nar/gkt890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramboarina S., Moreller N., Fournié-Zaluski M.-C., Roques B.P. Structural investigation on the requirement of CCHH zinc finger type in nucleocapsid protein of human immunodeficiency virus 1. Biochemistry. 1999;38(30):9600–9607. doi: 10.1021/bi9905258. [DOI] [PubMed] [Google Scholar]
- Ravasi T., Huber T., Zavolan M., Forrest A., Gaasterland T., Grimmond S., Hume D.A. Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome. Genome Res. 2003;13(6b):1430–1442. doi: 10.1101/gr.949803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte L., Mao J., Reitz J., Sreeramulu S., Kudlinzki D., Hodirnau V.-V., Meier-Credo J., Saxena K., Buhr F., Langer J.D., Blackledge M., Frangakis A.S., Glaubitz C., Schwalbe H. Cysteine oxidation and disulfide formation in the ribosomal exit tunnel. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-19372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters C.D., Kuszewski J.J., Tjandra N., Marius Clore G. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Sen G., Boxer L., Webster D., Bussat R., Qu K., Zarnegar B., Johnston D., Siprashvili Z., Khavari P. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell. 2012;22(3):669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service R.F. Protein structures for all. Science. 2021;374(6574):1426–1427. doi: 10.1126/science.acz9822. [DOI] [PubMed] [Google Scholar]
- Shao S., Tsoi L.C., Swindell W.R., Chen J., Uppala R., Billi A.C., Xing X., Zeng C., Sarkar M.K., Wasikowski R., Jiang Y., Kirma J., Sun J., Plazyo O., Wang G., Harms P.W., Voorhees J.J., Ward N.L., Ma F., Pellegrini M., Merleev A., Perez White B.E., Modlin R.L., Andersen B., Maverakis E., Weidinger S., Kahlenberg J.M., Gudjonsson J.E. IRAK2 has a critical role in promoting feed-forward amplification of epidermal inflammatory responses. J, Invest. Dermatol. 2021;141(10):2436–2448. doi: 10.1016/j.jid.2021.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Bax A.d. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56(3):227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Bax A. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol. Biol. 2015;1260:17–32. doi: 10.1007/978-1-4939-2239-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Li R., Teichert K., Montbleau K.E., Verboon J.M., Voit R.A., Sankaran V.G., Girirajan S. Pathogenic BCL11A variants provide insights into the mechanisms of human fetal hemoglobin silencing. PLoS Genet. 2021;17(10):e1009835. doi: 10.1371/journal.pgen.1009835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R.J.Y., Cram E.D., Czolij R., Matthews J.M., Crossley M., Mackay J.P. CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions. J. Biol. Chem. 2003;278(30):28011–28018. doi: 10.1074/jbc.M211146200. [DOI] [PubMed] [Google Scholar]
- Sippl M.J. Who solved the protein folding problem? Structure. 1999;7(4):R81–R83. doi: 10.1016/s0969-2126(99)80048-3. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Hosomichi K., Nakaoka H., Sakata H., Uesato N., Murakami K., Kano M., Toyozumi T., Matsumoto Y., Isozaki T., Sekino N., Otsuka R., Inoue I., Matsubara H. Biased expression of mutant alleles in cancer-related genes in esophageal squamous cell carcinoma. Esophagus. 2022;19(2):294–302. doi: 10.1007/s10388-021-00900-7. [DOI] [PubMed] [Google Scholar]
- The_UniProt_Consortium UniProt: the Universal Protein Knoledgebase in 2023. Nucleic Acids Res. 2023;51:D523–D531. doi: 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay C., Bédard M., Bonin M.-A., Lavigne P. Solution structure of the 13th C2H2 Zinc Finger of Miz-1. Biochem. Biophys. Res. Commun. 2016;473(2):471–475. doi: 10.1016/j.bbrc.2016.03.034. [DOI] [PubMed] [Google Scholar]
- Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., Bridgland A., Cowie A., Meyer C., Laydon A., Velankar S., Kleywegt G.J., Bateman A., Evans R., Pritzel A., Figurnov M., Ronneberger O., Bates R., Kohl S.A.A., Potapenko A., Ballard A.J., Romera-Paredes B., Nikolov S., Jain R., Clancy E., Reiman D., Petersen S., Senior A.W., Kavukcuoglu K., Birney E., Kohli P., Jumper J., Hassabis D. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596(7873):590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Walker J.M. In: The Protein Protocols Handbook. 2nd ed. Walker J.M., editor. Humana Press; Totowa, NJ: 2002. The Bicinchoninic Acid (BCA) assay for protein quantitation; pp. 11–15. [Google Scholar]
- Wang Y., Yu Y.u., Pang Y., Yu H., Zhang W., Zhao X., Yu J. The distinct roles of zinc finger CCHC-type (ZCCHC) superfamily proteins in the regulation of RNA metabolism. RNA Biol. 2021;18(12):2107–2126. doi: 10.1080/15476286.2021.1909320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead R.D., 3rd, Teschke C.M., Alexandrescu A.T. Pulse-field gradient nuclear magnetic resonance of protein translational diffusion from native to non-native states. Protein Sci. 2022;31:e4321. doi: 10.1002/pro.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake T.C., Stojadinovic O., Tomic-Canic M. Epidermal differentiation in barrier maintenance and wound healing. Adv. Wound Care (New Rochelle). 2014;3(3):272–280. doi: 10.1089/wound.2013.0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe S.A., Nekludova L., Pabo C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29(1):183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Xu C., Yang H.L., Yang Y.K., Pan L., Chen H.Y. Zinc-finger protein 750 mitigates malignant biological behavior of oral CSC-like cells enriched from parental CAL-27 cells. Oncol. Lett. 2022;23:28. doi: 10.3892/ol.2021.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-F., Hwu W.-L., Yang L.-C., Chung W.-H., Chien Y.-H., Hung C.-F., Chen H.-C., Tsai P.-J., Fann C.S.J., Liao F., Chen Y.-T. A promoter sequence variant of ZNF750 is linked with familial psoriasis. J. Invest. Dermatol. 2008;128(7):1662–1668. doi: 10.1038/jid.2008.1. [DOI] [PubMed] [Google Scholar]
- Zabini A., Zimmer Y., Medová M. Beyond keratinocyte differentiation: emerging new biology of small proline-rich proteins. Trends Cell Biol. 2023;33(1):5–8. doi: 10.1016/j.tcb.2022.08.002. [DOI] [PubMed] [Google Scholar]
- Zandarashvili L., White M.A., Esadze A., Iwahara J. Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1. FEBS Lett. 2015;589:1748–1753. doi: 10.1016/j.febslet.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar B., Webster D., Lopez-Pajares V., Vander Stoep Hunt B., Qu K., Yan K., Berk D., Sen G., Khavari P. Genomic profiling of a human organotypic model of AEC syndrome reveals ZNF750 as an essential downstream target of mutant TP63. Am. J. Hum. Genet. 2012;91(3):435–443. doi: 10.1016/j.ajhg.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.