Abstract
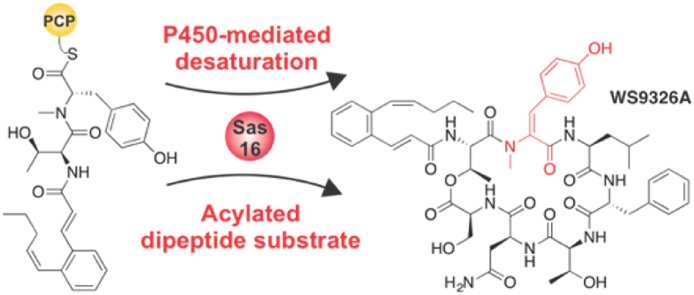
WS9326A is a peptide antibiotic containing a highly unusual N-methyl-E-2-3-dehydrotyrosine (NMet-Dht) residue that is incorporated during peptide assembly on a non-ribosomal peptide synthetase (NRPS). The cytochrome P450 encoded by sas16 (P450Sas) has been shown to be essential for the formation of the alkene moiety in NMet-Dht, but the timing and mechanism of the P450Sas-mediated α,β-dehydrogenation of Dht remained unclear. Here, we show that the substrate of P450Sas is the NRPS-associated peptidyl carrier protein (PCP)-bound dipeptide intermediate (Z)-2-pent-1′-enyl-cinnamoyl-Thr-N-Me-Tyr. We demonstrate that P450Sas-mediated incorporation of the double bond follows N-methylation of the Tyr by the N-methyl transferase domain found within the NRPS, and further that P450Sas appears to be specific for substrates containing the (Z)-2-pent-1′-enyl-cinnamoyl group. A crystal structure of P450Sas reveals differences between P450Sas and other P450s involved in the modification of NRPS-associated substrates, including the substitution of the canonical active site alcohol residue with a phenylalanine (F250), which in turn is critical to P450Sas activity and WS9326A biosynthesis. Together, our results suggest that P450Sas catalyses the direct dehydrogenation of the NRPS-bound dipeptide substrate, thus expanding the repertoire of P450 enzymes that can be used to produce biologically active peptides.
Key words: Cytochrome P450, Non-ribosomal peptide synthetase, Protein crystal structure, Enzyme mechanism, Natural products, Peptide antibiotic
Graphical abstract
WS9326A biosynthesis contains a dehydrotyrosine residue that we show is directly installed by the cytochrome P450 enzyme Sas16 during peptide assembly on the non-ribosomal peptide synthetase assembly line.
1. Introduction
Nonribosomal peptides (NRPs) are an important class of natural products that exhibit a wide range of biological activities and include diverse antimicrobial compounds (daptomycin, glycopeptide antibiotics) central to clinical treatment of bacterial infections1,2. The importance and complexity of NRPs has inspired significant efforts to understand their biosynthesis, itself motivated by a desire to be able to generate alternate, modified forms of natural compounds for various applications1,2. The chemical diversity that underpins the wide range of activities of NRPs is made possible through the nature of the machinery that produces these compounds. Known as nonribosomal peptide synthetases (NRPSs), these large peptide assembly lines typically rely on a modular architecture of catalytic domains to generate their specific peptide products3. The selection and activation of monomers for incorporation into the peptide is controlled by adenylation (A) domains, which generate activated amino acid adenylates via consumption of ATP3. Subsequent interception of the adenylate by the 4′-phosphopantetheine (PPant) group of a neighbouring peptidyl carrier protein (PCP) domain generates a protein-tethered thioester intermediate, which then serves as a substrate for all additional modifications performed during peptide biosynthesis, including peptide extension. Peptide extension is performed by condensation (C) domains4, which catalyse the attack of the amine group of a downstream (acceptor) aminoacyl-PCP on the thioester linkage of the upstream (donor) substrate. The modular-domain architecture of NRPSs is often further complemented by additional domains that can modify the peptide (or monomeric building blocks) via addition of chemical moieties (e.g., methylation, hydroxylation, halogenation)5, 6, 7 or can alter the stereochemistry of the peptide backbone. These modification domains can be found either covalently linked within the main NRPS assembly line (such as epimerisation (E) domains) or as separate trans-interacting enzymes that interact with PCP-bound substrates3. The modular nature of NRPS systems and the ability of trans-interacting enzymes to further modify the peptides produced by NRPS-assembly lines leads to huge diversity of peptide structure and makes understanding natural NRPS biosynthesis of great interest from a biotechnology perspective.
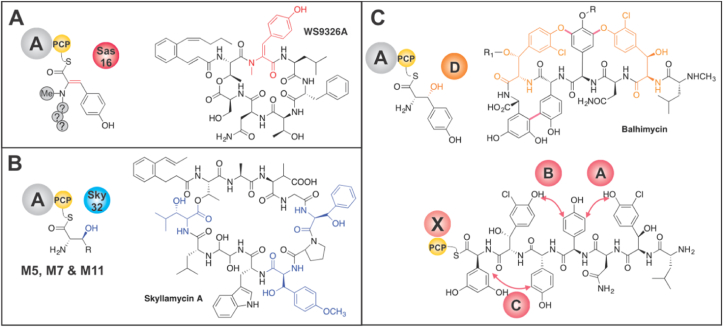
One important class of enzymes that is commonly found in NRPS pathways is cytochrome P450 (P450) enzymes, which can perform a range of oxidative transformations of substrates before, during or after NRPS biosynthesis8. Whilst the oxidative phenolic coupling performed by P450s to produce (poly)cyclic glycopeptide antibiotics (GPAs) exemplifies a highly complex P450-mediated transformation in NRPS-mediated antibiotic biosynthesis9, a more common role of P450s in NRPS pathways is the installation of β-hydroxyl groups in amino acids that are bound to the PCP domains of the NRPS machinery (Fig. 1)10,11. Such β-hydroxylating P450s are unusual in that they can be predicted from their sequence by inspection of a small number of active site residues that maintain a common structure to enable interaction with the PCP domain; they display a low specificity for the amino acid itself10,11. Whilst it has been postulated that the elimination of P450-introduced β-hydroxyl groups leads to the formation of C=N bond in during luzopeptin biosynthesis12, it is not clear if a similar route (i.e., via elimination of P450-introduced β-hydroxyl group) can account for the presence of desaturated residues (commonly Phe, Tyr, Trp) in other NRPs. Whilst a variety of routes have been postulated for the incorporation of desaturated residues, few have been investigated in detail. Given this, we chose to investigate a cryptic P450-mediated installation of α,β-dehydrotyrosine (Dht) within the peptide antibiotic WS932613. WS9326 contains several unusual features, including the presence of a (Z)-2-pent-1′-enyl-cinnamoyl moiety in addition to the Dht residue. Furthermore, the biosynthesis of WS9326 involves type II thioesterase (TE) domain mediated transfer of activated substrates during peptide assembly by the NRPS14. Previous investigations into WS9326 biosynthesis have shown that the P450 Sas16 (P450Sas) is involved in Dht formation, with deletion of this P450 leading to isolation of WS9326 derivatives bearing a Tyr instead of the Dht residue13,15. Whilst P450s are known to directly desaturate substrates (i.e., not through introduction of a transient hydroxyl group)16, this has not previously been observed for an NRPS-produced peptide8. Given that both the timing (and hence substrate) and the mechanism underlying the P450-mediated incorporation of the Dht residue has not been resolved, we commenced an investigation into the biosynthesis of this intriguing building block (Fig. 1A).
Figure 1.
Examples of the involvement of P450 enzymes in nonribosomal peptide biosynthesis. (A) Introduction of a dehydrotyrosine (Dht) residue in WS9326A by P450Sas (Sas16, red sphere; Dht residue shown in red). (B) Introduction of β-OH groups into residues of skyllamycin by P450 P450sky (Sky32, blue sphere), with this P450 targeting PCP-bound amino acid substrates during NRPS-mediated peptide assembly through specific interactions with the PCP domain (modified residues shown in blue; other examples of such P450s are have been characterised in the biosynthesis of novobiocin and nikkomycin). R = alkyl side chain. (C) Examples of P450s interacting with the NRPS machinery of the balhimycin GPA: this includes β-hydroxylation of PCP-bound Tyr residues by P450 OxyD (upper, orange sphere labelled D; residues shown in orange) and the side chain crosslinking of NRPS-bound heptapeptide intermediates by P450s OxyA–C (lower panel, red spheres labelled A‒C, location of crosslinks installed shown as red arrows). The former process is governed by P450/PCP interactions, whilst the latter is mediated by P450 interactions with the X-domain that is specific to GPA biosynthesis (shown as a pink sphere labelled X). R, R1 = sugar moiety. Domain key: PCP–peptidyl carrier protein (yellow sphere; PPant shown as black curved line); A–adenylation domain (grey sphere).
2. Materials and methods
2.1. General methods
2.1.1. Metabolic analysis
The metabolic analysis of mutants was performed by LC‒MS and UPLC–HRMS. The LC–MS conditions used were reported previously13. HRESI MS analyses were executed on an UPLC system (Ultimate 3000, Thermo-Scientific, Germany) equipped with to a Thermo QExactive HF mass spectrometer (Thermo Fisher Scientific, USA). The instrument was equipped with a ZORBAX Eclipse XDB C18 column (100 mm × 4.6 mm, 3.5 μm). A linear gradient analysis from 5% to 100% phase B was performed over 30 min with mobile phase A (H2O with 0.1% formic acid) and mobile phase B (MeCN with 0.1% formic acid). For each sample 1 μL was injected onto the column at a flow rate of 0.3 mL/min. Mass spectra were acquired in m/z in a positive ionization mode with auto MS2 fragmentations. All small-scale fermentations and analyses followed the same method as described above.
2.1.2. General NMR methods for chemical analysis
Nuclear magnetic resonance (NMR) was employed to elucidate chemical structures. The 1D NMR spectra [1H NMR (400 MHz), 13C NMR (100 MHz)] and 2D NMR spectra [1H/1H-COSY (correlation spectroscopy), HSQC (heteronuclear single quantum coherence), HMBC (heteronuclear multiple bond correlation)] of these compounds were measured on a Bruker 400-MHz spectrometer in 150 μL DMSO-d6 at T = 35 or 25 °C. Residual solvent signals were used as an internal standard (DMSO- d6: δH = 2.5 ppm, δC = 39.5 ppm).
2.1.3. HRMS and MS2 methods for trace WS9326A derivatives
High-resolution electron spray ionization mass spectra (HR-ESI-MSMS) were measured on a Thermo Q Exactive HF and on an Orbitrap Fusion equipped with the UltiMate 3000 RSLCnano system (Thermo Scientific). The percentage concentration indicated in HPLC solvents/conditions are v/v. Agilent Ultivo LCMS was used for sample analysis. QBH preparative HPLC was used for compound purification. Agilent1260 Analytical/Preparative HPLC was used for sample analysis and purification.
2.2. Molecular biology
2.2.1. Generation of Sas16 point mutants by site-directed mutagenesis
To explore the active sites of Sas16, three possible active site residues were selected and mutated through site-specific mutation to those of OxyDbal (A246G, Y248N, F250T; detailed in Supporting Information Fig. S3). The sas16 gene and its promoter region was amplified from the genome of Streptomyces asterosporus DSM 41452 using primers sas16-MFrg1-F and sas16-MFrg1-R (Supporting Information Table S2), with this fragment and the XbaI/EcoRV digested plasmid of pSET152-hyg subsequently ligated to form the pSET152-sas16 plasmid through Gibson assembly. The site-directed mutagenesis of Sas16 was performed using overlap-extension PCR method according to the manufacturers protocol using the primers listed in Table S217. The three residues under investigation were mutated individually, as pairs and altogether to the residues found at comparable positions in the active site of OxyDbal, yielding the mutated plasmids pSET152-sas16-F250T, pSET152-sas16-A246G, pSET152-sas16-Y248N, pSET152-sas16-AY246-248 GN, pSET152-sas16-YF248-250NT, pSET152-sas16-AF246-250 GT and pSET152-sas16-AYF-246-248-250GNT. These plasmids were confirmed by DNA sequencing before the recombinant plasmids were individually transformed into E. coli ET12567(pUZ8002) and transferred into the Δsas16 mutant strain by conjugation. The exconjugants were selected based on phenotypes showing apramycin resistance and confirmed by PCR.
2.2.2. Generation of Sas17 module 2 NMt domain point mutants in pBAC1F16 plasmid by counterselection method
The seamless point mutations in the NRPS biosynthetic genes sas17 were created by ccdB counterselection-based recombineering as reported previously18. This approach used two rounds of recombination, with the site of interest first replaced by a counterselection cassette amp-ccdB in E. coli Gbred-gyrA462. Subsequently, a synthetic single strand oligonucleotide containing the mutation of interest was used to replace this cassette under the counterselection of the toxicity marker inferred by the counter-selectable gene ccdB. Mutants D2014A and D2014Q of sas17 (plasmid pBAC1F16-D2014A and pBAC1F16-D2014Q) were produced using this method and verified by sequencing. All primers used for point mutations are listed in the Table S2. The verified plasmids were individually transformed into Streptomyces lividans 1326 for heterologous expression and metabolic analysis. Detailed information concerning the construction of plasmids and related primers/strains are found in Supporting Information Tables S1‒S3.
2.3. Chemical synthesis
2.3.1. N-Methyl-tyrosine
Boc-N-Me-tyrosine-OH. DCHA was purchased from Mjinbiotech Chemicals. Boc-N-Me-Tyr-OH.DCHA (15 mg) was dissolved in dichloromethane/THF (8 mL, 7:1 v/v′) and trifluoracetic acid (4 mL) added with stirring at 0 °C. The solution was allowed to warm to RT over 4 h, before the solvent was removed in vacuo and the crude product purified using reverse phase C18 flash chromatography in water/MeCN with 0.1% formic acid added to afford N-methyl-tyrosine (3 mg, 49%); spectra match reported values19. 1H NMR (400 MHz, DMSO-d4) δ 9.53 (s, 1H), 8.91 (s, 1H), 7.05 (d, J = 8.2 Hz, 2H), 6.71 (d, J = 8.2 Hz, 2H), 4.10 (s, 1H), 3.13 (dd, J = 14.1 Hz, J = 4.0 Hz, 1H), 3.05 (dd, J = 14.1 Hz, J = 4.0 Hz, 1H), 2.53 (s, 3H). 13H NMR (100 MHz, DMSO-d4) δ 170.4, 157.1, 130.9 (x2), 124.7, 115.8 (x2), 61.7, 34.3, 32.3. ESI-MS m/z [M‒H]‒ 194.1, [M+H]+ 196.1 (calcd. for C10H13NO3, 195.1).
2.3.2. N-Methyl-tyrosine aminoacyl-SNAC
N-Methyltyrosine SNAC (N-acetylcysteamine) was prepared using the approach reported previously20,21, and purified by semipreparative RP-HPLC (column: YMC-Pack ODS-A C18; 5 μm, 250 mm × 10 mm, gradient elution: 0–5 min, 2% MeOH; 5–25 min, 2%–30% MeOH; 25.1–35 min, 95% MeOH; 35–40 min, 2% MeOH). 1H NMR (400 MHz, MeOD) δ 7.06 (d, J = 8.3 Hz, 2H), 6.76 (d, J = 8.3 Hz, 2H), 4.43 (t, J = 6.9 Hz, 1H), 3.37 (m, 1H), 3.27 (m, 1H), 3.12 (m, 2H), 3.08 (m, 2H), 2.67 (s, 3H), 1.92 (s, 3H). 13H NMR (100 MHz, MeOD) δ 197.2, 173.7, 158.6, 131.8 (×2), 124.9, 117.1 (×2), 69.2, 39.4, 37.3, 32.6, 30.2, 22.9. ESI-MS m/z [M+H]+ 296.8 [M+Na]+ 318.8, (calcd. for C14H20N2O3S, 296.1195).
2.3.3. Synthesis of aminoacyl and peptide-CoAs
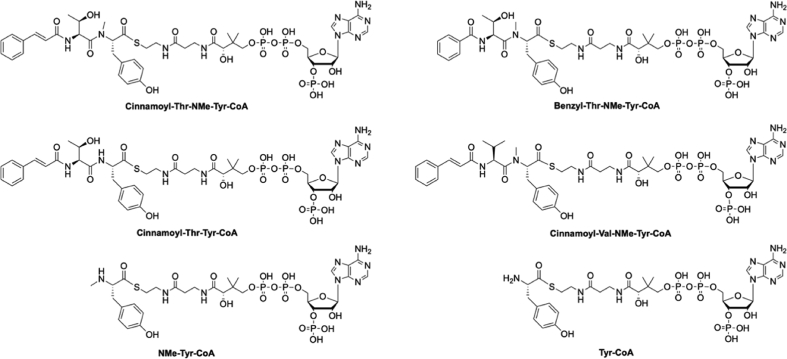
Peptidyl-CoAs bearing C-terminal tyrosine residues were obtained through a protocol involving the pre-activation of the unprotected peptide hydrazide to form the peptide acyl-azide with sodium nitrate and subsequent reaction of the acyl-azide with the coenzyme A to form the peptidyl-CoA22. This strategy was unable to generate peptidyl-CoAs containing a C-terminal N-methylated-tyrosine residue23. To overcome this, peptidyl-CoAs with a C-terminal N-methyl-tyrosine were prepared using a two-step process from the protected dipeptide using a protocol also applied to amino acyl-CoA synthesis24,25. The protected dipeptide was obtained by solid phase peptide synthesis using 2-chlorotrityl resin (0.1 mmol scale), standard peptide synthesis conditions (piperidine for Fmoc removal and DIC/Oxyma for the coupling reaction) with mild acidic cleavage AcOH/TFE/DCM (10:20:70 v/v′/v′′)26.
Briefly, the Boc-protected amino acid or dipeptide (1.0 eq) were dissolved in DMF (∼50 mmol/L) and pre-activated using DIC (1.0 eq) and Oxyma (2 eq) for 15 min at 4 °C. Next, a suspension of coenzyme A in DMF (0.6 eq) was added and the reaction left at room temperature for 18 h. The Boc-protected amino acyl-CoAs or protected peptide-CoAs were precipitated in cold diethyl ether, dried under nitrogen, and purified by RP-HPLC. Cleavage of the protecting groups (Boc, tBu) was accomplished using TFA/TIS/H2O (95:2.5:2.5 v/v′/v′′, 1 mL) with shaking at room temperature for 1 h. Subsequently, the solution was concentrated under a N2 stream to ∼0.1 mL and the aminoacyl or dipeptide-CoAs precipitated with ice cold diethyl ether (∼1.5 mL) before being collected by centrifugation in a flame-resistant centrifuge. Finally, the purified compounds were dissolved in MeCN/H2O and freeze dried in 50 mg aliquots before use in enzyme assays. For LCMS and HRMS analysis see Supporting Information Figs. S17‒S22 and S23–S26.
Structures of aminoacyl and peptidyl-CoAs synthesised in this study.
2.3.4. NRPS offloading probes
Three probes, a tyrosine probe (N-(2-(2-amino-3-(4-hydroxyphenyl)propanamido)-ethyl)heptanamide), β-alanine probe (N-(2-(3-aminopropanamido)ethyl)heptanamide) and glycine probe (N-(2-(2-aminoacetamido)ethyl)heptanamide) were prepared as previously reported27.
2.4. Strain cultivation and supplementation
2.4.1. N-Methyltyrosine and N-methyltyrosine SNAC supplementation of S. lividans 1326/1F16 NmetD2104Q and S. lividans 1326/1F16 NmetD2104A
To explore whether the supplementation of N-methyltyrosine or N-methyltyrosine SNAC was able to restore the production of WS9326A derivatives from NMt inactivation strains, N-methyltyrosine or N-methyltyrosine SNAC (200 μmol/L) was individually supplemented to growing cultures of S. lividans 1326/1F16 NmetD2104Q and S. lividans 1326/1F16 NmetD2104A at three time points (24, 48 and 72 h). After 5 days cultivation at 30 °C and 200 rpm, the culture was extracted by acetyl acetate and concentrated before the secondary metabolites were analysed by LCMS using the general method for LCMS analysis.
2.4.2. Capturing biosynthetic intermediates of the WS9326A assembly line via offloading
The offloading probes were supplemented into solid and liquid cultures of S. asterosporus DSM 41452, singly or portion wise over 4 days, to a final concentration of 2 mmol/L. S. asterosporus DSM 41452 strain (100 μL glycerol spore stock) was grown in TSB medium (10 mL) for 3 days (250 rpm at 30 °C) in 50 mL Erlenmeyer flasks. For liquid cultures, a starter culture (100 μL) was used to inoculate SG liquid medium (10 mL, duplicates, in 50 mL Erlenmeyer flasks with spring), which was incubated at 30 °C, 250 rpm for 1 day. After the first day of fermentation, the probes were added portion wise (50 μL of 100 mmol/L probe dissolved in MeOH, per day) over 4 days to reach the final concentration of 2 mmol/L. For solid cultures, a starter culture (50 μL) was plated on SG solid medium (5 mL, duplicates, in 40 mm diameter petri dish) already containing 2 mmol/L of chemical probe dissolved in MeOH, and incubation at 30 °C for 5 days followed. All cultures (including control liquid and solid cultures in the absence of the probes) were also prepared duplicates. After 5 days, the liquid cultures were lyophilised overnight and the lyophilised residue was extracted with MeOH (10 mL), then concentrated, whereas for solid cultures, the agar medium was cut into small pieces that were extracted with MeOH (10 mL) and then concentrated. The residues were redissolved in HPLC-grade methanol (500 μL) for LC‒HR MS/MS analysis. Putative biosynthetic intermediates intercepted by the chemical probes were identified and characterised by high-resolution mass detection and MS2 fragments as previously described10.
2.5. Construct cloning, expression and characterisation
2.5.1. Cloning sas16 into the pET28 vector
A 1224bp DNA fragment containing sas16 was amplified from S. asterosporus DSM 41452 genomic DNA using oligonucleotides P450pET-F and P450pET-R (Table S2). The PCR product was ligated into EcoRV-digested pBluescript II sk(+) plasmid to yield pBSK-SAS16. Next, a fragment containing sas16 was obtained from pBSK-Sas16 and ligated into pET28a(+) vector to yield plasmid pET28-Sas16, with the sequence confirmed by sequencing (Supporting Information Table S1).
2.5.2. Construction of mutant sas16-F250T expression vector
The gene sas16-F250T was amplified from the plasmid pSET152-sas16-F250T using primer pET28a-sas16-NdeI-GA-F and pET28a-sas16-HindIII-GA-R (Table S2). Then, this fragment was ligated into pET28a vector (linearized at the NdeI and HindIII sites) through Gibson assembly to yield plasmid pET28-Sas16-F250T (Table S1).
2.5.3. Expression and purification of Sas16
pET28-Sas16 was transformed into chemically competent E. coli BL21 star(DE3) cells. A 7 mL culture transformed was grown overnight at 37 °C in LB medium supplemented with kanamycin (50 mg/L) to provide a starter culture for Sas16 protein expression. A 700 mL culture of LB supplemented with kanamycin (50 mg/L) was inoculated with 1% (v/v) of overnight culture and grown at 37 °C to an OD absorbance (600 nm) of 0.4, at which point the culture temperature was reduced to 28 °C and expression of Sas16 induced using 0.2 mmol/L IPTG. After 6 h, the cell pellet was collected by centrifugation, the pellet resuspended in buffer A (50 mmol/L Tris-HCl (pH 8.0), 300 mmol/L NaCl), and lysed using French press. After centrifugation, the supernatant was purified in a two-step procedure commencing with Ni-NTA affinity chromatography using a 2 mL Ni-NTA column pre-equilibrated in buffer A and connected to a Äkta FPLC system. After washing with 5 CV of buffer A, bound protein was eluted with 1.5 CV of buffer B (buffer A + 250 mmol/L imidazole). Fractions with red colour were collected and concentrated by ultrafiltration (molecular mass cut-off 10 kDa), before being further purified using gel filtration (Sephadex G-25 column (200 mm × 40 mm), gel filtration buffer (Tris-HCl 50 mmol/L, pH 7.4, NaCl 150 mmol/L)). After elution, red fractions were combined, concentrated using an Amicon Ultra centrifugal filter with a 5 kDa molecular weight cut-off and the buffer was then exchanged to buffer (20 mmol/L Tris-HCl, pH 8.0, 100 mmol/L NaCl) with 15% glycerol. Subsequently, the protein solution was aliquoted and flash cooled in liquid nitrogen before being stored at −80 °C. For SDS-PAGE, see Supporting Information Fig. S7.
2.5.4. Expression and purification of Sas16-F250T
pET28-Sas16-F250T was firstly transformed into E. coli BL21 (DE3) cells. One single colony picked from LB agar plate was inoculated in 10 mL LB medium supplemented with kanamycin (50 mg/mL) and grown overnight at 37 °C. This overnight culture 1% (v/v) was transferred into 1 L LB medium supplemented with kanamycin (50 mg/mL) and grown at 37 °C until the OD absorbance (600 nm) reached to 0.6, at which point 0.1 mmol/L IPTG was added to induce protein expression. After 18 h, the cell pellet was collected by centrifugation, and then resuspended in buffer A (50 mmol/L Tris-HCl (pH 8.0), 300 mmol/L NaCl) before being lysed using French press. After centrifugation, the supernatant was purified in a two-step procedure commencing with Ni-NTA affinity chromatography using a 2 mL Ni-NTA column pre-equilibrated in buffer A. After washing with 5 CV buffer A, bound protein was eluted with 1.5 CV buffer B (buffer A + 250 mmol/L imidazole). The red coloured fractions were collected and concentrated by ultrafiltration. Next, the protein sample was exchanged into buffer C (20 mmol/L Tris-HCl, pH 8.0, 100 mmol/L NaCl) via ultrafiltration and flash cooled in liquid nitrogen before being stored at −80 °C. For SDS-PAGE, see Fig. S7.
2.5.5. Reduced CO spectrum of Sas16 and F250T mutant
Two anaerobic cuvettes were filled with buffer (20 mmol/L Tris pH 8.0, 150 mmol/L NaCl) and the baseline of buffer absorption recorded with a dual-beam spectrophotometer (Molecular Devices SpectraMax M2) in the range of 350–500 nm. Next, the sample cuvette was replaced with either Sas16 (105 μmol/L) or F259T mutant (75 μmol/L) protein solution in the same buffer, before the cuvette was degassed and the gas phase was replaced with oxygen-free CO. A small amount (a few microliters) of saturated sodium dithionite (Na2S2O4) solution was injected into the protein solution to reduce the hemoprotein, and the spectra were recorded every 2 min until the hemoprotein was totally reduced and CO-bound. See Figs. S1 and S7.
2.5.6. Soluble substrate binding to Sas16
Solutions of tyrosine, cyclic WS9326B, and linear acylated-peptide analogues of WS9326A (chemically synthesized by solid phase peptide synthesis, GL Biochem (Shanghai) Ltd.) were prepared at a concentration of 1 mmol/L in binding buffer (20 mmol/L Tris-HCl and 500 mmol/L NaCl). Substrate binding of Sas16 was then monitored by UV‒visible spectroscopy. Sas16 protein in buffer A was prepared to a final concentration of 5.6 μmol/L, with aliquots (500 μL) divided into reference and sample cuvette. After equilibration at 30 °C, the baseline was recorded between 200 and 700 nm. The possible substrates were individually titrated into the Sas16 protein solution (25 μL aliquots), gently mixed and the corresponding absorbance spectrum recorded using UV-visible spectrophotometer (Shortwave NIR) wavelengths from 350 to 600 nm (USB-ISS-UV-VIS-2, Ocean Optics). See Fig. S1.
2.5.7. Characterisation of module 2 of the Sas17 NRPS protein (Trx-A-NMt-PCP)
A thioredoxin (Trx) fusion protein encoding the A-NMt-PCP domains from module two of the Sas NRPS (Sas17, UniProt ID A0A514JVM2, residues 1553-2567) was prepared from genomic DNA following amplification with primers (For_A_NMT_PCP_NcoI, Rev_A_NMT_PCP_XhoI; Table S2) and subsequent digestion with NcoI/XhoI restriction cloning into the pET-Trx-1c vector28,29. Protein expression was performed in autoinduction media at 18 °C for 72 h, with cells harvested, lysed and protein isolated using a three-step purification protocol (sequential Ni-NTA affinity chromatography, ion-exchange chromatography, and gel filtration) as has been previously reported30.
A-domain: A-domain activity towards Tyr was tested following conversion of the PCP domain (35 μmol/L) into the holo-form using Sfp (R4-4 mutant31, 5 μmol/L) and CoA (150 μmol/L) and incubation for 1 h at 30 °C with the following conditions: 50 mmol/L HEPES (pH 7.0), 10 mmol/L MgCl2 and 50 mmol/L NaCl (35 μmol/L). Washing of the sample after CoA loading was accomplished using sequential concentration/dilution steps (4) in centrifugal concentrators (30 kDa molecular weight cut-off). Tyr activation was confirmed following incubation of the holo-NRPS construct with ATP, Mg2+ and Tyr via LCMS analysis of the products of cleavage of the PCP-bound thioesters using methylamine (analysis used a gradient of MeCN (+0.1% formic acid) in water (+0.1% formic acid) of 0–30% over 35 min); formation of Tyr-NMe-amide confirmed the activation and PCP-loading of Tyr by the NRPS construct (see Supporting Information Fig. S8)29. A-domain activity was also demonstrated through the generation of pyrophosphate, which was detected using a coupled enzyme assay previously reported (data not shown)32.
NMt-domain: NMt-domain activity was examined using Tyr-loaded Trx-A-NMt-PCP, which was prepared from Tyr-CoA using the Sfp loading protocol above with the replacement of CoA with Tyr-CoA. The solution was supplemented with SAM (5 mmol/L) and the reaction was incubated for 1 h. Analysis was then performed using LCMS as indicated above (see Fig. S8); control incubations where Tyr was not bound to the NRPS showed no methylation (data not shown).
2.5.8. In vitro NRPS-bound substrate activity assays for Sas16
The apo-Trx-A-NMt-PCP construct of the SAS (module 2) was loaded with aminoacyl-CoA (Tyr, NMe-Tyr) or acyl-dipeptidyl-CoA substrates (cinnamoyl-Val-NMe-Tyr-CoA, cinnamoyl-Thr-NMeTyr, benzyl-Thr-NMe-Tyr-CoA) using the phosphopantetheinyl transferase Sfp R4-4 mutant as described above (1 h, 30 °C, 50 mmol/L HEPES (pH 7.0), 10 mmol/L MgCl2 and 50 mmol/L NaCl) in a total volume of 100 μL; the PCP: peptidyl-CoA: Sfp-R4-4 ratio was 1: 4: 0.1. The holo-PCP constructs were then used immediately. The enzyme turnover reaction was performed with a P450: peptidyl-PCP: PuR: PuxB ratio of 10:50:1:5 respectively in 50 mmol/L HEPES (pH 7.0), 0.33% glucose, 0.033 mg/mL glucose dehydrogenase and 2 mmol/L NADH. The reactions were then allowed to incubate at 30 °C for 18 h before the PCP-bound substrates were cleaved via the addition of methylamine, purified using solid phase extraction (Strata™-X-33 μm Polymeric Reversed Phase Tubes (30 mg/mL) (Phenomenex)) and analysed using LCMS and HRMS/MS2 as has been previously described (see Supporting Information Fig. S11 for acylated dipeptide results)33.
2.5.9. NRPS-bound acyl-dipeptide binding to Sas16
Spectra were obtained using a Jasco V-750 spectrophotometer at 30 °C between 390 and 600 nm. The P450 was diluted to 2.5 μmol/L in Tris-HCl (50 mmol/L, pH 7.4) in two cuvettes and a baseline recorded. Next, substrate was added to the sample cuvette over a concentration range of 1–10 μmol/L (with the same volume of buffer added to the reference cuvette) and stirred briefly before each measurement was recorded (see Fig. S11).
2.6. Structural biology and modelling
2.6.1. Protein crystallization and data collection
For protein crystallization and structural determination, the concentrated Sas16 protein (10 mg/mL) after size exclusion chromatography (Superdex S200 26/60) in buffer (20 mmol/L Tris pH 8.0, 150 mmol/L NaCl) was used for initial screens using sitting drop vapor-diffusion at 20 °C. Initial crystallization experiments were established using an automatic crystallization drop-set system (Oryx Nano, Douglas Instrument); drops of 0.6 μL in total with different protein to reservoir ratios (33%, 50%, 67%) were set and equilibrated again reservoir solutions (50 μL). Reddish rod-like crystals initial appeared after two weeks in the buffer with 25% polyethylene glycol (PEG) 3350, 0.2 mol/L magnesium chloride and 0.1 mol/L HEPES pH 7.5. Then these crystals were crushed for seeding using an Oryx Nano. Large diamond shape single crystals were obtained in a condition comprising 4% polyethylene glycol 6000, 4% polyethylene glycol 8000, 4% polyethylene glycol 10,000, 0.1 mol/L potassium thiocyanate, 0.1 mol/L sodium bromide and 0.1 mol/L MES pH 6.5.
Crystals were mounted on cryoloops and flash frozen in liquid nitrogen prior to data collection. Multiple datasets to 2.0 Å was collected at beamline X06DA at Swiss Light Source (Villigen, Switzerland) with the PILATUS pixel detector (Dectris). The data was processed by using iMosflm34 and XDS package35, the crystal was assigned to the space group P42212 with unit cell dimensions a = b = 112.8 Å and C = 146.2 Å. The asymmetric unit contains two subunits, with a solvent content of 52.45%.
2.6.2. Structure determination and refinement
Crystal structure of Sas16 was determined by single-wavelength anomalous dispersion (SAD) with AutoSol and AutoBuild in PHENIX suite36 using dataset collected at the Fe X-ray absorption edge (K-edge, 7172 eV). The low resolution model after AutoBuild was used for molecular replacement in MOLREP37 with the higher resolution dataset. Refinement of the initial electron density was carried out using cycles of refinement in REFMAC538 in the CCP4 Suite39, and model building was performed in COOT40. The final structure was refined to Rcryst = 20% and Rfree = 24% at resolution of 2.0 Å. The quality of the structure was validated by MOLPROBITY41. Data collection and refinement statistics are shown in Supporting Information Table S4. The interface area between the C-terminal loop of Chain A and the neighbouring chain in the asymmetric unit of the crystal structure was calculated using the ‘Protein interfaces, surfaces and assemblies’ service PISA (v1.52) at the European Bioinformatics Institute (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html).
2.6.3. Structural modelling of the Sas17 NMet domain
A model of the Sas17 module 2 NMt domain was generated using the Phyre 2 server42. The resulting model was overlayed with structure of the A-NMt domain TioS from thiocoraline biosynthesis (PDB Code 5WMM)5 and the active site inspected, particular with reference to the residues interacting with SAM. From this analysis, two positions (E2081 and D2104) were selected for mutation in the Sas17 NMt domain, the former E2081 to alanine (E2081A) and the latter D2104 to both alanine (D2104A) and glutamine (D2104Q).
2.6.4. Modelling of Sas16 in absence of C11 disulfide bond
A physics-based model of Sas16 was generated using the AlphaFold Colab notebook, which runs a simplified AlphaFold v2.0 protocol43.
2.6.5. Molecular dynamics simulations of Sas16
Molecular dynamics (MD) simulations were performed using Desmond (Schrödinger Release2020-4, Desmond Molecular Dynamics System, DE Shaw Research, New York, NY, 2020). The Protein Preparation Wizard was used to prepare a model of Sas16 without the C11 disulfide bond (i.e., Chain A only): all waters and buffer molecules were removed, bond orders were assigned, hydrogens were added, hydrogen-bond optimization was performed using default parameters, and the protonation states of residues were assigned using PROPKA at pH 7.0. Restrained minimization was then performed (converging heavy atoms to RMSD 0.3 Å) using the OPLS3e forcefield44. The protein was enclosed in an orthorhombic box with a minimum buffer distance of 12 Å around the protein and solvated using SPC water molecules. The system was neutralized by the addition of 20 Na+ ions. The system was relaxed using the default “relax system” protocol of Desmond prior to production runs. Two random-seed production runs were performed (one to 750 ns and one to 1 μs), totalling 1.75 μs of simulation time.
2.7. Structural analysis of biosynthesis products
2.7.1. Isolation and identification of WS9326M, WS9326N and WS9326X
For purification of WS9326M and WS9326N, the cultures of S. asterosporus Δsas16:pSET152-sas16-F250T were maintained on the Tryptic soy broth (TSB) medium, a small loop of spores growing on a TSB solid plate was inoculated into a 250 mL Erlenmeyer flask containing 75 mL liquid productive SG medium (soy peptone 10 g, glucose 20 g, l-valine 2.34 g CaCO3 2 g, CoCl2-solution 1 mg/mL 1 mL, tap water 1000 mL, pH 7.2) and cultured at 28 °C for 3 days on rotary shaker at 180 rpm. Subsequently, 10 mL of the preculture was inoculated into a 500 mL Erlenmeyer flask containing 150 mL of the SG medium, then incubated for 4 days, 28 °C, 180 rpm. The fermentation broth (20 L) was separated by high-speed centrifugation, yielding the supernatant and cell pellet. The supernatant was extracted using twice the volume of ethyl acetate, before the organic solvent was evaporated to yield 2.661 g of crude extract. The crude extract was dissolved in 20 mL methanol and fractionated. The fractions collected were analysed by LC‒MS. Among those fractions, fractions 3 and 4 were subsequently subjected to further purification by a semi-preparative HPLC, equipped with a C18 column (100 mm × 4.6 mm), with fractions eluted using an isocratic method (60% CH3OH, 0.5% acetic acid, 3 mL/min) to yield compound WS9326M (7.5 mg) and WS9326N (1.5 mg).
For purification of WS9326X, strain S. lividans:1F16 was cultured by large-scale fermentation (10 L). After extraction, 7.4 g crude extract was obtained. The sample was firstly subject to an open silica gel column (CH2Cl2/MeOH/EtOAc system was used as mobile phase) with a gradient method used (30%, 50%, 70% and 100%). Fraction No.5 containing the desired molecular mass was further purified by semi-preparative HPLC (10 mm × 250 mm, 5 μm) and compound WS9326X eluted using an isocratic method (60% CH3CN‒40% H2O, 0.5% acetic acid, 1.5 mL/min) to yield compound WS9326X (8.5 mg).
2.7.2. Determination of absolute configuration of WS9326X by Marfey’s method
In WS9326X, the integration of the Asp residue is distinct to that of other congeners of WS9326A; thus to determine its absolute configuration of this Asp residue in WS9326X Marfey’s method was performed following the standard protocol (Marfey’s reagent was purchased from Thermo Scientific (Number 48895)). The amino acid standards involved in the assembly of WS9326X were obtained commercially from Sigma‒Aldrich. Each of these (1 mg) were dissolved in 100 μL H2O before the amino acids were derivatized by adding 1 mol/L NaHCO3 (40 μL) and 1% FDAA (in acetone, 200 μL). The reaction mixture was heated at 40 °C for 1 h, before the mixture was neutralized with 2 mol/L HCl after cooling to room temperature. The derivatives were then dried and dissolved in CH3OH, before being analyzed by HPLC–MS (Zorbax Eclipse XDB-C18 column, 100 mm × 4.6 mm) using a gradient elution method (A: CH3CN with 0.1% HAc; B: H2O with 0.1% HAc; 10%‒40%–90% CH3CN in 50 min, 0.5 mL/min at 25 °C, detection at wavelengths of 270 nm, and 340 nm, m/z range from 100 to 2000).
In the meantime, WS9326X (0.5 mg) was hydrolyzed with 2 mol/L HCl (2.0 mL) at 45 °C for 2 h. The solution was evaporated to dryness, and derivatized with Marfey’s reagent. The retention times of amino acid standards derivatized with Marfey’s reagent (l-Asp FDAA derivative and d-Asp FDAA derivative) are shown in Supporting Information Fig. S47). The derivatives of WS9326X and its acid hydrolysates were analysed by LC‒MS (see Supporting Information Figs. S48‒49). The resulting amino acid FDAA derivatives from WS9326X hydrolysates show HPLC profiles that are in agreement with the FDAA derivatives of standard amino acids.
3. Results and discussion
Our investigations into P450Sas commenced with heterologous expression of this P450, which afforded heme-bound enzyme with the anticipated Soret absorption maximum at 418 nm. Reduction of P450Sas and complexation of the resultant ferrous heme with CO led to a shift in this absorption to 450 nm, indicating that the enzyme was correctly folded and potentially catalytically competent (Fig. S1A). Next, we assayed substrate binding to P450Sas by monitoring the spin state shift of the heme iron upon titration with the cyclic peptide WS9326B (with a Tyr-2 residue) and soluble analogues of various early (Tyr) and late-stage linear peptide (formylated penta- and hexapeptides) intermediates of WS9326 biosynthesis (Fig. S1B). However, none of the molecules tested generated a spectral shift upon titration (Fig. S1). This led us to postulate that P450Sas interacts primarily with a substrate bound to a PCP domain in the NRPS assembly line during WS9326 biosynthesis (Fig. S1C). Indeed, several P450s–including those involved with side chain crosslinking of peptides during GPA biosynthesis (OxyA-C, which is mediated by a modified C-type recruitment domain, the X-domain, Fig. 1C)9,45 or the β-hydroxylation of aminoacyl-PCPs10,11–have shown to only interact with PCP-bound substrates as opposed to soluble analogues. Given that elimination of a β-hydroxyl group was one possible pathway to form Dht, we next performed sequence alignments between P450Sas and P450s known to catalyse the β-hydroxylation of PCP-bound substrates (Supporting Information Figs. S2‒3). This included P450s that perform such β-hydroxylation to produce hydroxylated amino acid substrates for incorporation via the main NRPS assembly line (OxyD in type I GPAs, Fig. 1C)11 as well as those that perform β-hydroxylation during peptide assembly (e.g., skyllamycin, Fig. 1B)10. This revealed only low levels of sequence similarity between P450Sas and P450s known to catalyse β-hydroxylation and importantly showed that the characteristic amino acids conserved in these P450s were missing, suggesting a different substrate for P450Sas (Figs. S2‒3).
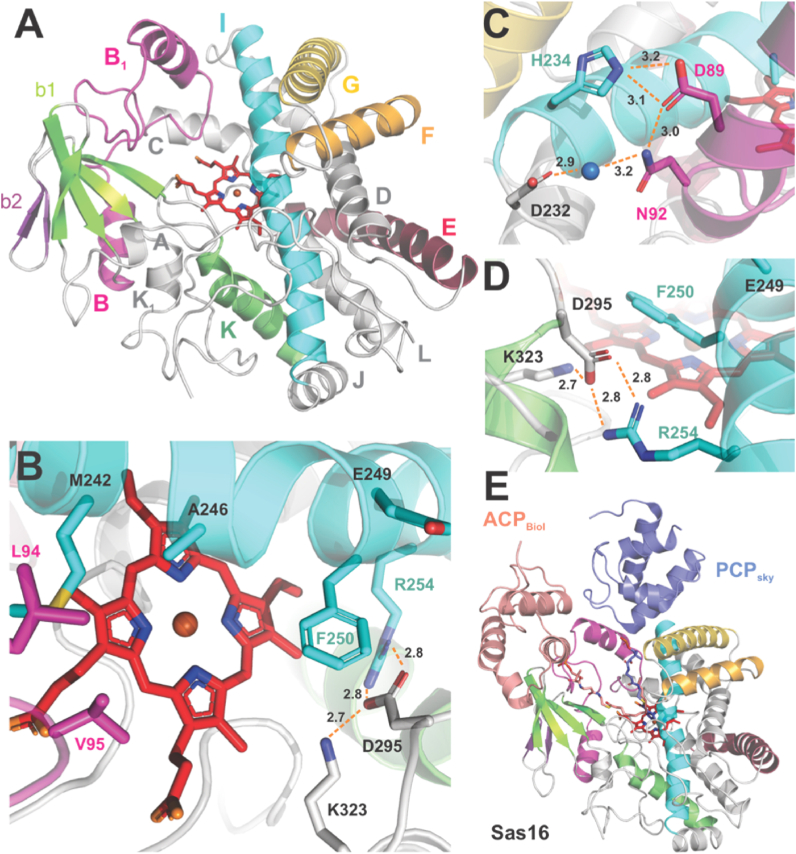
To further investigate whether Dht formation would proceed via β-hydroxylation, we structurally characterised P450Sas and compared its structure with those of P450s that only accept PCP-bound substrates. An X-ray crystal structure of P450Sas was solved to a resolution of 2.0 Å (2 molecules/ASU; subsequent analyses performed on chain A) and phased using single-wavelength anomalous diffraction (Fig. 2A). The structure of this enzyme conforms to the general structure known for P450s, with the structure dominated by α-helices (A‒L, including B1 and K2) and a small region comprised of two β-sheets8. The heme moiety lies beneath the long I-helix, which includes residues typically involved in oxygen activation, with the heme iron ligated from below by an axial thiolate ligand donated from a cysteine residue (C358) immediately prior to the L-helix. The heme propionate groups are coordinated by H-bonds to R300/H102 and H356, whilst the remainder of the active site around the heme is almost exclusively populated by residues with hydrophobic side chains (Fig. 2B). The B–C loop region of P450Sas, which is commonly found disordered in substrate free P450 structures (although not in the case of P450s that β-hydroxylate aminoacyl-PCP substrates such as OxyD and P450sky, see Supporting Information Fig. S4)7,10,11, contains a B1 helix, the C-terminus of which is stabilised by H-bonding interactions from D89 and N92 to I-helix residues H234 and D232 (the latter mediated by a water molecule (Fig. 2C). The B1 helix runs essentially parallel to the G-helix that caps the active site (together with the F-helix), which would allow the binding of a PCP domain in a similar manner to aminoacyl-PCP β-hydroxylating P450s albeit with the position of the PCP further away from the P450 active site (Fig. 2E)7,46.
Figure 2.
Structure of P450Sas. (A) The overall structure of P450sas (Sas16), which conforms to the anticipated P450 fold and is dominated by alpha helices surrounding the central heme moiety (red sticks). The secondary structure elements are labelled for clarity and adopt the following conserved colour scheme: B-B1, magenta; E, red; F, orange; G, gold; I, cyan; K, green; β1 sheet, apple green; β2 sheet, purple. Colour scheme is retained for all panels. (B) P450Sas active site, showing the putative substrate binding site above the heme. (C) Interactions between the B1 and I helices; water molecule shown as a blue sphere. (D) Interactions between the I-, K- and K2-helices that alter the environment of the F250 residue in the central active site. (E) Comparison of the position of carrier protein substrates taken from the P450BioI/ACP complex (pink, PDB code 3EJB) and the P450sky/PCP complex (slate, PDB code 4PXH). Whilst the ACP positioning is not favourable with the structure of P450Sas, the PCP position would be possible, albeit with the PCP moved away from the P450Sas active site.
Turning to residues close to the heme moiety of P450Sas, the catalytically important and typically highly conserved acid/alcohol pair found in the I-helix above the heme is not present in this P4508. Whilst the acid residue (E249) is found in P450Sas, F250 occupies the position normally occupied by an alcohol residue (S or T) in other P450s (Fig. 2D). The acid/alcohol pair are believed to act in concert to facilitate the activation of molecular oxygen and produce the iron-oxo species (so-called Compound I) that is believed to be responsible for most of the chemistry catalysed by P450s. Given that the alcohol residue is central to oxygen activation by most P450s, this is clearly an unusual substitution and one that might speak to differences in the mechanism of this P450. The phenyl ring of F250 sits at a 45-degree angle to the heme plane and would be predicted to interact with the substrate. Furthermore, there is an alteration to the typical trajectory of the end of the K-helix and the start of the third strand of the β-1 sheet, which sees an extension of the K2 helix to provide the space for the side chain of F250. This divergence in structure from typical bacterial P450s is mediated through several ionic/H-bonding interactions between R254 in the I-helix and K323 in the K2-helix with D295 found in the K-helix (2 × 2.8 Å, 2.7 Å respectively); residues in the position of D295 in other structurally characterised P450s typically are Pro or hydrophobic in nature (Fig. 2C). Notable differences also occur at the C-terminal loop; while the apex of this loop is typically found close to the heme and contributes to substrate binding in other P450s, the C-terminal loop residues do not protrude as far towards the active site in P450Sas (Supporting Information Fig. S5). However, the presence of a C11 disulfide bond–an artefact of crystallisation–between the two molecules in the asymmetric unit (and the nature of the resulting crystal packing) likely contributes to the reorganisation of this loop; the disulfide bond stabilises a non-canonical conformation of the neighbouring N-terminal region and the C-terminal loop conformation is further stabilised by crystal packing interactions (the interface area between one C-terminal loop and the neighbouring chain is 455.5 Å2) (Fig. S5A). It is difficult to predict what conformations these regions would adopt in solution; two independent molecular dynamics simulations (totalling 1.75 μs) initiated from chain A of the crystal structure (i.e., no C11 disulfide bond or packing interactions present) showed slight rearrangements of these regions trending towards conformations observed in other P450s (Fig. S5) but may remain trapped in a local energy minimum during these simulations. On the other hand, a physics-based structural model of the monomer (generated using AlphaFold2)43 shows these regions adopting conformations more like that found in other P450s (Fig. S5).
The highly conserved alcohol residue plays a key role in efficient activation of molecular oxygen for catalysis by the majority of vast P450s that employ it as the stoichiometric oxidant8. Mutation of this residue generally results in a P450 characterised by significant loss of catalytic oxidative activity and the production of large amounts of hydrogen peroxide through incomplete oxygen activation. Sometimes the effect of the mutation is ameliorated by the correct substrate/residue pairing where the lack of an alcohol side chain is overcome through the adventitious generation of a highly ordered water network47, 48, 49. In addition, the lack of an active site alcohol residue has previously been observed in a number of wild-type P450s. Some of these function by substitution of the protein alcohol moiety through the involvement of OH groups from within the substrate50,51. Other P450s naturally replace the alcohol with another H-bonding residue; CYP176A1 (P450cin) possesses a asparagine instead of the alcohol, although mutagenesis experiments have indicated that a H-bonding residue at this position is not essential for activity52. Replacement with phenylalanine is unusual however, and the deviations seen in P450Sas near this residue do suggest its importance to the formation of Dht.
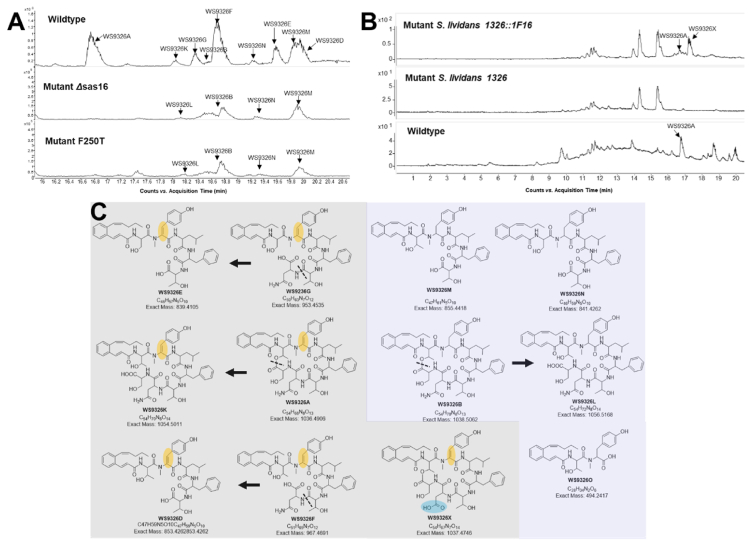
To investigate the role of the F250 in P450Sas we constructed an altered producer strain that lacked sas16 and complemented it in trans with a variety of sas16 mutants focussed around the F250 residue. Indeed, mutation of this unusual F250 residue (together with mutating combinations of neighbouring I-helix residues, see Supporting Information Fig. S6A) into the canonical threonine residue (taken from the sequence of OxyD) in S. asterosporus showed a complete loss of biosynthetic production of WS9326A (Fig. S6B). Low levels of WS9326L (the linear form of WS9326A containing Tyr-2 residue instead of Dht) biosynthetic intermediates and WS9326B (WS9326A containing a Tyr-2 residue instead of Dht) were detected as products in these mutant strains. This matches the results from the strain in which P450Sas (Sas16) has been deleted (Fig. 3A and Supporting Information Tables S5‒S7). In addition, a possible intermediate WS9326O (the linear dipeptide intermediate containing Tyr-2 residue instead of Dht) was detected from the mutant Δsas16 (Table S5 and Supporting Information Fig. S16). These results suggest that the F250T mutant of P450Sas and related mutants are no longer catalytically competent for Dht formation. Expression and purification of the P450Sas F250T mutant under the same conditions as the WT enzyme showed that this mutant is soluble and is apparently correctly folded (as evidence by P450 formation upon reduction and CO complexation), supporting this hypothesis (Fig. S7). Overall, the structural analysis of P450Sas confirmed the general P450 fold as anticipated, with comparisons to other P450 structures showing strongest similarities to P450s involved in late-stage macrolide oxidation as opposed to PCP-interacting P450s (Supporting Information Table S8). This, combined with the unusual F250 residue, suggested a hydrophobic substrate and one that was unlikely to be an aminoacyl-PCP. To further explore this hypothesis, we next expressed and purified a module tridomain (A-Mt-PCP) construct of the Sas NRPS, first confirming that A-domain was able to activate Tyr and load this onto the PCP domain (Fig. S8)32. Reconstitution of the Mt domain using SAM then led to the methylation of the PCP-bound Tyr residue, with no appreciable double methylation despite the stalled nature of the assembly line in this case. Both Tyr- and N-Me-Tyr loaded NRPS constructs were then used to explore binding and possible oxidation of tyrosyl-PCP by Sas16. Neither substrate showed any binding response or activity with P450Sas, which supports the analysis of the structure indicating an alternate substrate to an aminoacyl-PCP.
Figure 3.
HPLC‒MS analysis of the natural products produced by the WS9326 assembly line for both wild-type and mutant strains. (A) Region of HPLC-MS traces from S. asterosporus DSM41452 (wildtype), S. asterosporus DSM41452 Δsas16 (Mutant Δsas16) and S. asterosporus Δsas16:pSET152-sas16-F250T (Mutant F250T). (B) Comparative HPLC‒MS traces from mutant S. lividans 1326:1F16, S. lividans 1326 and S. asterosporus DSM41452 (wildtype). (C) The structures of the WS9326 derivatives identified from these strains.
With the substrate of P450Sas still unclear, we turned again to in vivo experiments to reveal the substrate of this P450. First, we investigated the importance of the Tyr-2 N-Me group on WS9326 biosynthesis (Fig. 4A). Previous deletion of the Mt domain from module 2 of the NRPS assembly line had led to abolition of WS9326 biosynthesis, which we hypothesised could at least in part be due to the importance of the Me group for P450 activity along with stalling of peptide extension due to C-domain selectivity13. To prevent Mt activity without completely removing the domain, we mutated two key residues predicted to be important for methyltransferase activity as selected from a homology model based on known structures of a related methyltransferase (Supporting Information Fig. S9)5,42. These mutations in the N-methyltransferase domain were performed using the BAC plasmid pBAC1F16 containing the intact sas gene cluster (Table S1 and Fig. S9). As was the case for the complete Mt domain deletion, neither mutant was able to produce WS9326A or related structures, highlighting the importance of this methyl group in the biosynthetic assembly line (Supporting Information Fig. S10). Interestingly, WS9326X, a new analogue of WS9326A, containing an N to D mutation at position 6 of the peptide was discovered from the heterologous expression host S. lividans 1326:1F16 (Fig. 3B and C, Tables S5 and S8 and Supporting Information Figs. S42‒S46). The identification of WS9326X implicates the promiscuity of the corresponding A domain (module 6, Sas23), which in accordance with the discovery of other related derivatives WS9326I and WS9326J14.
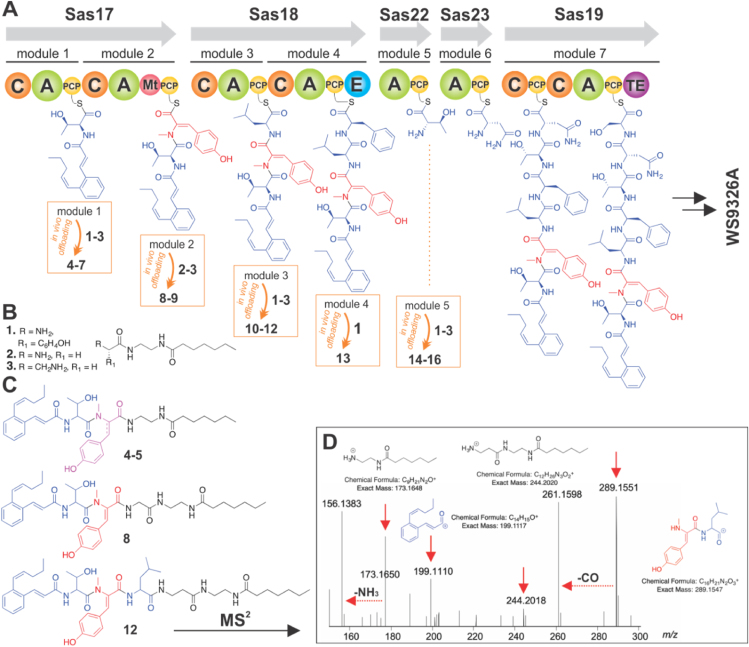
Figure 4.
The biosynthesis of WS9326A and application of in vivo offloading probes to understand the timing of Dht formation. (A) Overview of the proposed nonribosomal biosynthesis pathway producing WS9326A together with offloaded biosynthetic intermediates 4–16 obtained via the use of chemical probes 1–3 (for structures 4–16 refer to Supporting Information). (B) Structures of chemical probes 1–3. (C) Examples of putative structures obtained by offloading experiments: 4 and 5–offloaded from module 1 by 1 (Tyr probe) with concomitant N-methylation of 1 (SI); 8 and 12–offloaded from modules 2 and 3 by 2 (Gly probe, supporting Information) and 3 (β-Ala probe, supporting Information) respectively. (D) HR-MS2 characterisation of the putative offloaded intermediate species 12, showing both probe- and peptide-related fragments. Colours of the chemical structures denote synthetic probe origin (black), biosynthesis (blue), Dht intermediate (red) and desaturated Tyr probe 1 (magenta). Domain key: PCP–peptidyl carrier protein (yellow sphere; PPant shown as black curved line); A–adenylation domain (green sphere); C–condensation domain (orange sphere); E–epimerisation domain (blue sphere); Mt–N-methyl transferase domain (red sphere); TE–thioesterase domain (purple sphere).
Having seen such a dramatic effect for the alteration of methylation state, we next probed these mutants using synthetic substrates to ascertain whether the NRPS machinery was still functional. To this end, N-Me-Tyr and N-Me-Tyr N-acetylcysteamine (SNAc) thioesters were each added to Mt domain deletion strains and the cultures analysed for WS9326 production. These experiments showed that whilst N-Me-Tyr was unable to overcome the inactivation of the Mt domain (presumably due to lack of activity of the M2 A-domain towards this substrate), N-Me-Tyr SNAc supplementation led to the recovery of production of WS9326A (Fig. S10). This shows that the NRPS machinery in these mutants remains functional and reinforces the importance of the Me group for Dht formation, and consequently WS9326 biosynthesis.
With the success with these feeding experiments, we next investigated the timing of Tyr desaturation using chemical ‘offloading’ probes in vivo (Fig. 4A‒D). These chemical probes can compete at the acceptor sites of C-domains during NRPS biosynthesis in vivo, thus providing snapshots of intermediates during biosynthesis without altering the biosynthetic machinery27,53. Three probes, based around tyrosine, glycine, β-alanine (1–3) were utilised for these investigations (Fig. 4B). The tyrosine probe 1 was chosen as mimic of cognate aromatic aminoacyl substrates, in particular tyrosyl-PCP in module 2, whereas the glycine and β-alanine probes (2–3) have proven capable of acting as ‘universal’ tools for intermediate capture27,53. Following incubation, isolation and HRMSn analysis, a variety of putative offloaded intermediates from WS9326A biosynthesis (4–16) were detected and characterised, ranging from (Z)-2-pent-1′-enyl-cinnamoyl acylated Thr through to (Z)-2-pent-1′-enyl-cinnamoyl acylated pentapeptides (see Fig. 4C‒D and Supporting Information Figs. S27‒30). For the Gly and β-Ala probes, the products detected–crucially including offloaded dipeptides–contained a desaturated Tyr2 residue, indicating that this modification likely occurs during NRPS-mediated peptide biosynthesis within module 2 (Fig. 4). The Tyr-probe provided results generally consistent with those deriving from the Gly and β-Ala probes, and, crucially, it was further able to delineate on the timing of N-methylation and subsequent desaturation. The Tyr probe was found to be partially N-methylated (Fig. S27), and the offloaded (Z)-2-pent-1′-enyl-cinnamoyl acylated-Thr species from module one was also found to be methylated on the α-nitrogen of the Tyr residue (species 4) as well as partially desaturated (species 5, Fig. 4 and Supporting Information). This strongly indicates that the N-methyltransferase acts before the P450, and further that P450Sas can process substrates without the entire PCP domain, although the relative ratios of offloaded intermediates suggest that desaturation would be much more efficient if the intermediate was bound to the PCP.
Control experiments were carried out feeding the Δsas16 and complementation strains with the same chemical probes. The overall results, summarised in Supporting Information Table S10, indicate that the P450 performs desaturation at the dipeptide stage of NRPS-mediated peptide assembly. We then attempted to reconstitute this activity in vitro: without access to Z-pentenyl-cinnamic acid we instead synthesised simplified acylated dipeptide CoAs that we used to load our module 2 tridomain NRPS construct, which again showed no activity (Fig. S11). However, taken together this is unsurprising, given the hydrophobic nature of the P450 active site, the likely importance of an ordered water network for P450 activity and the structure of the (Z)-2-pent-1′-enyl-cinnamoyl group found in WS9326. The ability of the (Z)-2-pent-1′-enyl-cinnamoyl acylated-Thr-N-Me-Tyr dipeptide probe to be modified shows that the interaction with the substrate in the case of P450Sas is driven by the acylated dipeptide and not by the PCP domain (as had been seen previously with P450s performing β-hydroxylation of aminoacyl PCP substrates)7,10,11 or a recruitment domain (as is seen with Oxys in GPA crosslinking)9,30. The activity of P450Sas towards the offloaded acylated dipeptide resembles the activity seen for P450BioI for fatty acid substrates even in the absence of the ACP binding partner54. However, from a biosynthetic point of view, P450Sas must be interacting with its acylated dipeptide substrate whilst it is bound to the PCP-domain of module 2 of the Sas NRPS as this is the only point in the biosynthesis where this intermediate is present. Furthermore, no β-hydroxylated Tyr-intermediates or products were detected in any in vivo studies (and have been detected with other systems)27, which suggests that the reaction performed by this P450 is a direct desaturation. Given that many peptides produced by NRPS machineries contain β-OH amino acids8, the elimination of a transient β-OH group seems unlikely and suggests that P450Sas is a direct desaturase.
4. Conclusions
In summary, we have investigated the role of the P450Sas (Sas16) that is involved in the formation of the dehydrotyrosine (Dht) residue found in the peptide antibiotic WS9326. Our results strongly suggest that the substrate for this P450 is the N-acylated, PCP-bound dipeptide intermediate (Z)-2-pent-1′-enyl-cinnamoyl-Thr-N-Me-Tyr. The presence of the N-methyl group appears to be required for the activity of this P450, which itself is highly unusual in possessing a phenylalanine residue in place of the alcohol residue that is typically central to catalysis. This suggests that P450Sas is highly selective for the structure of the substrate, rather than domains within the NRPS, which contrasts with what has been shown for all other P450/NRPS systems studied to date. Indeed, a bioinformatic analysis showed that all close homologues of P450Sas (82% identity or higher) are found in a biosynthetic gene cluster that shares homology with the first two modules of the WS9326 NRPS assembly line and is predicted to commence with a (Z)-2-pent-1′-enyl-cinnamoyl-Thr-N-Me-Tyr dipeptide (Supporting Information Fig. S31). Thus, P450Sas is the first reported example of a P450 enzyme that targets a specific dipeptide during NRPS biosynthesis. Whilst the mechanism that P450Sas uses to generate the Dht residue of this substrate remains to be elucidated, there are strong clues that dehydrogenation of the substrate is performed directly by this P450. Furthermore, the apparent reliance of the P450 on the structure of the acylated dipeptide makes the integration of such a pathway into other NRPS pathways an intriguing challenge for biosynthetic engineering, and one that offers an alternate route to those ascribed to C-domains4 for the incorporation of alkene moieties into the side chains of NRPS peptides.
Acknowledgments
Yongwei Zhao (Monash) for assistance with binding assays; Dr Cleidiane Zampronio (School of Life Sciences, Warwick) for assistance with LC‒HRMSn Orbitrap Fusion analyses; Dr Lijiang Song (Warwick Chemistry) for preliminary MS data acquired on a BrukerMaXis Impact instrument; Prof. James de Voss (University of Queenland) for helpful discussions. This work was supported by the BBSRC (MIBTP studentship to Daniel J. Leng); the Monash Warwick Alliance (Seed Fund Award to Manuela Tosin and Max J. Cryle); the University of Warwick (Career Support Award to Manuela Tosin); Monash University, EMBL Australia, the Australian Research Council (Discovery Project DP210101752 to Max J. Cryle) and the National Health and Medical Research Council (APP1140619 to Max J. Cryle). This research was conducted by the Australian Research Council Centre of Excellence for Innovations in Peptide and Protein Science (CE200100012) and funded by the Australian Government. This research was funded by the National Natural Science Foundation of China (82104044 to Songya Zhang), the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-PTJS-003-07); We appreciate the assistance from Prof. Fu Yan (Shandong University) and Prof. Wei Zhang (Shandong University) for the P450 protein measurement. We thank the staff at beamline X06SA of the Swiss Light Source (Villigen, CH) for excellent assistance with diffraction data collection.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.03.021.
Contributor Information
Manuela Tosin, Email: M.Tosin@warwick.ac.uk.
Tong Si, Email: tong.si@siat.ac.cn.
Max J. Cryle, Email: max.cryle@monash.edu.
Author contributions
Max J. Cryle, Songya Zhang, Manuela Tosin and Tong Si designed the projects and analysed the results. Songya Zhang and Max J. Cryle wrote the manuscript. Songya Zhang performed the genetic and chemical experiments. Lin Zhang solved the crystal structure. Anja Greule, Edward Marschall and Jing Zhu performed the biochemical experiments. HRMS analysis of biochemical assays was performed by Ralf B. Schittenhelm. Julien Tailhades performed the chemical synthesis of in vitro probes. Daniel J. Leng, Panward Prasongpholchai, Jingfan Zhang, Fabrizio Alberti and Manuela Tosin synthesised the in vivo offloading tools, performed the feeding experiments and analysed the HRMS results. Joe A. Kaczmarski and Colin J. Jackson performed and analysed computational experiments. Oliver Einsle, Andreas Bechthold and Youming Zhang provided resources and critical insights during this project.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Baltz R.H. Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways. ACS Synth Biol. 2014;3:748–758. doi: 10.1021/sb3000673. [DOI] [PubMed] [Google Scholar]
- 2.Yim G., Thaker M.N., Koteva K., Wright G. Glycopeptide antibiotic biosynthesis. J Antibiot (Tokyo) 2014;67:31–41. doi: 10.1038/ja.2013.117. [DOI] [PubMed] [Google Scholar]
- 3.Süssmuth R.D., Mainz A. Nonribosomal peptide synthesis—principles and prospects. Angew Chem Int Ed Engl. 2017;56:3770–3821. doi: 10.1002/anie.201609079. [DOI] [PubMed] [Google Scholar]
- 4.Dekimpe S., Masschelein J. Beyond peptide bond formation: the versatile role of condensation domains in natural product biosynthesis. Nat Prod Rep. 2021;38:1910–1937. doi: 10.1039/d0np00098a. [DOI] [PubMed] [Google Scholar]
- 5.Mori S., Pang A.H., Lundy T.A., Garzan A., Tsodikov O.V., Garneau-Tsodikova S. Structural basis for backbone N-methylation by an interrupted adenylation domain. Nat Chem Biol. 2018;14:428–430. doi: 10.1038/s41589-018-0014-7. [DOI] [PubMed] [Google Scholar]
- 6.Kittilä T., Kittel C., Tailhades J., Butz D., Schoppet M., Büttner A., et al. Halogenation of glycopeptide antibiotics occurs at the amino acid level during non-ribosomal peptide synthesis. Chem Sci. 2017;8:5992–6004. doi: 10.1039/c7sc00460e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haslinger K., Brieke C., Uhlmann S., Sieverling L., Süssmuth R.D., Cryle M.J. The Structure of a transient complex of a nonribosomal peptide synthetase and a cytochrome P450 monooxygenase. Angew Chem Int Ed Engl. 2014;53:1–6. doi: 10.1002/anie.201404977. [DOI] [PubMed] [Google Scholar]
- 8.Greule A., Stok J.E., De Voss J.J., Cryle M.J. Unrivalled diversity: the many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat Prod Rep. 2018;35:757–791. doi: 10.1039/c7np00063d. [DOI] [PubMed] [Google Scholar]
- 9.Haslinger K., Peschke M., Brieke C., Maximowitsch E., Cryle M.J. X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis. Nature. 2015;521:105–109. doi: 10.1038/nature14141. [DOI] [PubMed] [Google Scholar]
- 10.Uhlmann S., Süssmuth R.D., Cryle M.J. Cytochrome P450sky interacts directly with the nonribosomal peptide synthetase to generate three amino acid precursors in skyllamycin biosynthesis. ACS Chem Biol. 2013;8:2586–2596. doi: 10.1021/cb400555e. [DOI] [PubMed] [Google Scholar]
- 11.Cryle M.J., Meinhart A., Schlichting I. Structural characterization of OxyD, a cytochrome P450 involved in b-hydroxytyrosine formation in vancomycin biosynthesis. J Biol Chem. 2010;285:24562–24574. doi: 10.1074/jbc.M110.131904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi X., Huang L., Song K., Zhao G., Liu Y., Lv L., et al. Enzymatic tailoring in luzopeptin biosynthesis involves cytochrome P450-mediated carbon‒nitrogen bond desaturation for hydrazone formation. Angew Chem Int Ed Engl. 2021;36:19821–19828. doi: 10.1002/anie.202105312. [DOI] [PubMed] [Google Scholar]
- 13.Zhang S., Zhu J., Zechel D.L., Jessen-Trefzer C., Eastman R.T., Paululat T., et al. New WS9326A derivatives and one new annimycin derivative with antimalarial activity are produced by Streptomyces asterosporus DSM 41452 and its mutant. ChemBioChem. 2018;19:272–279. doi: 10.1002/cbic.201700428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M.S., Bae M., Jung Y.E., Kim J.M., Hwang S., Song M.C., et al. Unprecedented noncanonical features of the nonlinear nonribosomal peptide synthetase assembly line for WS9326A biosynthesis. Angew Chem Int Ed Engl. 2021;36:19766–19773. doi: 10.1002/anie.202103872. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J., Zhang S., Zechel D.L., Paululat T., Bechthold A. Rational design of hybrid natural products by utilizing the promiscuity of an amide synthetase. ACS Chem Biol. 2019;14:1793–1801. doi: 10.1021/acschembio.9b00351. [DOI] [PubMed] [Google Scholar]
- 16.Wong S.H., Bell S.G., De Voss J.J. P450 catalysed dehydrogenation. Pure Appl Chem. 2017;89:841–852. [Google Scholar]
- 17.An Y., Ji J., Wu W., Lv A., Huang R., Wei Y. A rapid and efficient method for multiple-site mutagenesis with a modified overlap extension PCR. Appl Microbiol Biotechnol. 2005;68:774–778. doi: 10.1007/s00253-005-1948-8. [DOI] [PubMed] [Google Scholar]
- 18.Wang H., Bian X., Xia L., Ding X., Müller R., Zhang Y., et al. Improved seamless mutagenesis by recombineering using ccdB for counterselection. Nucleic Acids Res. 2013;42:e37. doi: 10.1093/nar/gkt1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baccile J.A., Spraker J.E., Le H.H., Brandenburger E., Gomez C., Bok J.W., et al. Plant-like biosynthesis of isoquinoline alkaloids in Aspergillus fumigatus. Nat Chem Biol. 2016;12:419–424. doi: 10.1038/nchembio.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He H.Y., Yuan H., Tang M.C., Tang G.L. An unusual dehydratase acting on glycerate and a ketoreducatse stereoselectively reducing α-ketone in polyketide starter unit biosynthesis. Angew Chem Int Ed Engl. 2014;53:11315–11319. doi: 10.1002/anie.201406602. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W., Zhou L., Li C., Deng Z., Qu X. Rational engineering acyltransferase domain of modular polyketide synthase for expanding substrate specificity. Methods Enzymol. 2019;622:271–292. doi: 10.1016/bs.mie.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 22.Tailhades J., Schoppet M., Greule A., Peschke M., Brieke C., Cryle M.J. A route to diastereomerically pure phenylglycine thioester peptides: crucial intermediates for investigating glycopeptide antibiotic biosynthesis. Chem Commun. 2018;54:2146–2149. doi: 10.1039/c7cc09409d. [DOI] [PubMed] [Google Scholar]
- 23.Teixidó M., Albericio F., Giralt E. Solid-phase synthesis and characterization of N-methyl-rich peptides. J Pept Res. 2005;65:153–166. doi: 10.1111/j.1399-3011.2004.00213.x. [DOI] [PubMed] [Google Scholar]
- 24.Izoré T., Tailhades J., Hansen M.H., Kaczmarski J.A., Jackson C.J., Cryle M.J. Drosophila melanogaster nonribosomal peptide synthetase Ebony encodes an atypical condensation domain. Proc Natl Acad Sci. 2019;116:2913–2918. doi: 10.1073/pnas.1811194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izoré T., Candace Ho Y.T., Kaczmarski J.A., Gavriilidou A., Chow K.H., Steer D.L., et al. Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity. Nat Commun. 2021;12:2511. doi: 10.1038/s41467-021-22623-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barlos K., Chatzi O., Gatos D., Stavropoulos G. 2-Chlorotrityl chloride resin. Studies on anchoring of Fmoc-amino acids and peptide cleavage. Int J Pept Protein Res. 1991;37:513–520. [PubMed] [Google Scholar]
- 27.Leng D.J., Greule A., Cryle M.J., Tosin M. Chemical probes reveal the timing of early chlorination in vancomycin biosynthesis. Chem Commun. 2021;57:2293–2296. doi: 10.1039/d0cc07421g. [DOI] [PubMed] [Google Scholar]
- 28.Bogomolovas J., Simon B., Sattler M., Stier G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr Purif. 2009;64:16–23. doi: 10.1016/j.pep.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Kaniusaite M., Tailhades J., Marschall E.A., Goode R.J.A., Schittenhelm R.B., Cryle M.J. A proof-reading mechanism for non-proteinogenic amino acid incorporation into glycopeptide antibiotics. Chem Sci. 2019;10:9466–9482. doi: 10.1039/c9sc03678d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greule A., Izoré T., Iftime D., Tailhades J., Schoppet M., Zhao Y., et al. Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics. Nat Commun. 2019;10:2613. doi: 10.1038/s41467-019-10384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunbul M., Marshall N.J., Zou Y., Zhang K., Yin J. Catalytic turnover-based phage selection for engineering the substrate specificity of Sfp phosphopantetheinyl transferase. J Mol Biol. 2009;387:883–898. doi: 10.1016/j.jmb.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Kittilä T., Schoppet M., Cryle M.J. Online pyrophosphate assay for analyzing adenylation domains of nonribosomal peptide synthetases. ChemBioChem. 2016;17:576–584. doi: 10.1002/cbic.201500555. [DOI] [PubMed] [Google Scholar]
- 33.Tailhades J., Zhao Y., Schoppet M., Greule A., Goode R.J.A., Schittenhelm R.B., et al. Enzymatic cascade to evaluate the tricyclization of glycopeptide antibiotic precursor peptides as a prequel to biosynthetic redesign. Org Lett. 2019;21:8635–8640. doi: 10.1021/acs.orglett.9b03245. [DOI] [PubMed] [Google Scholar]
- 34.Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;7873:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roos K., Wu C., Damm W., Reboul M., Stevenson J.M., Lu C., et al. OPLS3e: extending force field coverage for drug-like small molecules. J Chem Theory Comput. 2019;15:1863–1874. doi: 10.1021/acs.jctc.8b01026. [DOI] [PubMed] [Google Scholar]
- 45.Tailhades J., Zhao Y., Ho Y.T.C., Greule A., Ahmed I., Schoppet M., et al. A chemoenzymatic approach to the synthesis of glycopeptide antibiotic analogues. Angew Chem Int Ed Engl. 2020;59:10899–10903. doi: 10.1002/anie.202003726. [DOI] [PubMed] [Google Scholar]
- 46.Cryle M.J., Schlichting I. Structural insights from a P450 carrier protein complex reveal how specificity is achieved in the P450BioI ACP complex. Proc Nat Acad Sci. 2008;105:15696–15701. doi: 10.1073/pnas.0805983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimata Y., Shimada H., Hirose T., Ishimura Y. Role of THR-252 in cytochrome P450CAM: a study with unnatural amino acid mutagenesis. Biochem Biophys Res Commun. 1995;208:96–102. doi: 10.1006/bbrc.1995.1310. [DOI] [PubMed] [Google Scholar]
- 48.Cryle M.J., De Voss J.J. The role of the conserved threonine in P450BM3 oxygen activation: substrate-determined hydroxylation activity of the Thr268Ala mutant. ChemBioChem. 2008;9:261–266. doi: 10.1002/cbic.200700537. [DOI] [PubMed] [Google Scholar]
- 49.Hishik T., Shimada H., Nagano S., Egawa T., Kanamori Y., Makino R., et al. X-Ray crystal structure and catalytic properties of Thr252Ile mutant of cytochrome P450cam: roles of Thr252 and water in the active center1. J Biochem. 2000;128:965–974. doi: 10.1093/oxfordjournals.jbchem.a022848. [DOI] [PubMed] [Google Scholar]
- 50.Cupp-Vickery J.R., Poulos T.L. Structure of cytochrome P450eryF involved in erythromycin biosynthesis. Nat Struct Biol. 1995;2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 51.Cryle M.J., Bell S.G., Schlichting I. Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-l-leucyl-l-leucyl dipeptide oxidase. Biochemistry. 2010;49:7282–7296. doi: 10.1021/bi100910y. [DOI] [PubMed] [Google Scholar]
- 52.Meharenna Y.T., Slessor K.E., Cavaignac S.M., Poulos T.L., De Voss J.J. The critical role of substrate‒protein hydrogen bonding in the control of regioselective hydroxylation in P450cin. J Biol Chem. 2008;283:10804–10812. doi: 10.1074/jbc.M709722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho Y.T.C., Leng D.J., Ghiringhelli F., Wilkening I., Bushell D.P., Kostner O., et al. Novel chemical probes for the investigation of nonribosomal peptide assembly. Chem Commun. 2017;53:7088–7091. doi: 10.1039/c7cc02427d. [DOI] [PubMed] [Google Scholar]
- 54.Cryle M.J., De Voss J.J. Carbon–carbon bond cleavage by cytochrome P450BioI (CYP107H1) Chem Commun. 2004:86–87. doi: 10.1039/b311652b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.