Abstract
C-Glycosides are important natural products with various bioactivities. In plant biosynthetic pathways, the C-glycosylation step is usually catalyzed by C-glycosyltransferases (CGTs), and most of them prefer to accept uridine 5′-diphosphate glucose (UDP-Glc) as sugar donor. No CGTs favoring UDP-rhamnose (UDP-Rha) as sugar donor has been reported, thus far. Herein, we report the first selective C-rhamnosyltransferase VtCGTc from the medicinal plant Viola tricolor. VtCGTc could efficiently catalyze C-rhamnosylation of 2-hydroxynaringenin 3-C-glucoside, and exhibited high selectivity towards UDP-Rha. Mechanisms for the sugar donor selectivity of VtCGTc were investigated by molecular dynamics (MD) simulations and molecular mechanics with generalized Born and surface area solvation (MM/GBSA) binding free energy calculations. Val144 played a vital role in recognizing UDP-Rha, and the V144T mutant could efficiently utilize UDP-Glc. This work provides a new and efficient approach to prepare flavonoid C-rhamnosides such as violanthin and iso-violanthin.
Key words: Flavonoid C-glycoside, C-rhamnosyltransferase, Biosynthesis, Catalytic mechanisms, Sugar donor selectivity
Graphical abstract
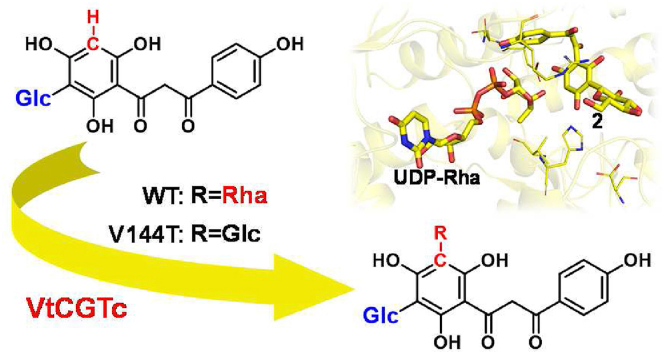
The first selective plant C-rhamnosyltransferase VtCGTc was reported and its sugar donor selectivity mechanisms were dissected.
1. Introduction
C-Glycosides are an important class of bioactive natural products in plants1, 2, 3. Due to the presence of C–C glycosidic bonds, C-glycosides are generally stable and can resist chemical and enzymatic degradation, which bring distinctive pharmacokinetic properties compared to O-glycosides4,5. The formation of C-glycosidic bonds is catalyzed by C-glycosyltransferases (CGTs). Up to now, a range of CGTs have been reported, most of which prefer uridine 5′-diphosphate glucose (UDP-Glc) as sugar donor6, including AbCGT7, MiCGT8, GgCGT9 and TcCGT110. In our previous study, a series of CGTs were discovered to catalyze the arabinosylation of mono-C-glucosides of 2-hydroxyflavanone, which led to the biosynthesis of (iso)schaftosides in higher plants11.
l-Rhamnose is a popular sugar unit of plant glycosides. Thus far, several O-rhamnosyltransferases have been identified12, 13, 14, such as a highly specific saponin 2″-O-rhamnosyltransferase GuRhaGT from Glycyrrhiza uralensis15. However, no CGTs with high selectivity towards UDP-Rha have been reported, thus far, though flavonoid C-rhamnosides have been reported in many plant species16, 17, 18.
Viola tricolor is a heat-clearing and detoxifying Chinese herbal medicine19. It contains abundant violanthin (3a) and minor iso-violanthin (3b), which are di-C-glycosides with glucosyl and rhamnosyl substitutions at C-6 and C-8 (Fig. 1A)20. (Iso)violanthin could inhibit acetylcholinesterase (AchE)21 and the SARS-CoV-2 virus main protease 3CLpro22. However, the CGTs involved in their biosynthesis are still unknown.
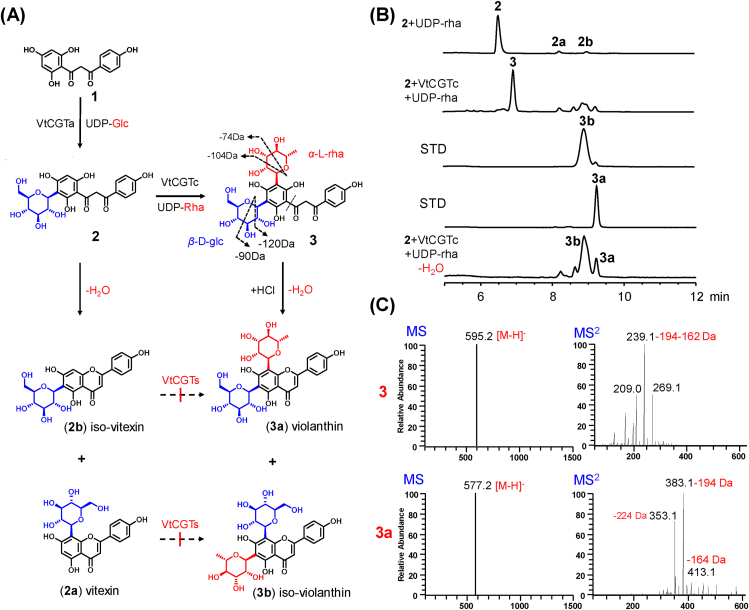
Figure 1.
Stepwise di-C-glycosylation for the biosynthesis of (iso)violanthin by VtCGTa and VtCGTb. (A) The catalytic reactions. (B) UHPLC/UV chromatograms for the enzymatic reaction mixtures and reference standards (300 nm). (C) (‒)-ESI-MS and MS/MS spectra of 3.
Here, we report the first selective C-glycosyltransferase VtCGTc from V. tricolor. It shows high sugar donor selectivity towards UDP-Rha, and could efficiently catalyze 5-C-rhamnosylation of 2-hydroxynaringenin 3-C-glucoside. The catalytic mechanisms were also investigated by theoretical calculations.
2. Materials and methods
2.1. Chemical reagents
Compounds 1 and 2 were prepared in our previous work11. The other substrate compounds were purchased from Chengdu Must Biotechnology Co., Ltd. (Chengdu, China), Chengdu Dexter Technology Co., Ltd. (Chengdu, China), and Shanghai Titan Scientific Co., Ltd. (Shanghai, China). The UDP-sugar donors were purchased from Guangzhou Angfei Biological Technology Co., Ltd. (Guangzhou, China). Methanol and acetonitrile (Fisher Scientific, USA) were of HPLC grade. All other chemicals and reagents were purchased from Sigma–Aldrich (St. Louis, MO, USA) and Beijing Chemical Corporation (Beijing, China) unless otherwise specified.
2.2. Plant materials and transcriptome analysis
The seeds of V. tricolor were purchased from Shanghai (China) and were sown in our laboratory under natural conditions. One month after sowing, the seedlings were collected and washed, then immediately frozen in liquid nitrogen for RNA extraction. The transcriptome of fresh seedling was sequenced and assembled by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (China). The original data have been uploaded onto NCBI (BioProject ID: PRJNA946018). For BLAST analysis, the gene sequences of 12 plant C-glycosyltransferases were used as templates, and 2 CGT candidate genes (VtCGTa and VtCGTc) were obtained (Supporting Information Fig. S1)11.
2.3. Molecular cloning, heterologous expression, and protein purification
The total RNA of V. tricolor was extracted from the seedlings using the TranZol™ kit (Tiangen Biotech, Beijing, China) and was reverse-transcribed to cDNA by TransScript supermix (Transgen Biotech) following the manufacturer's instructions. TransScript KD Plus and Fast-Pfu DNA polymerases (Transgen Biotech) were used to amplify candidate genes. The primer sequences are given in Supporting Information Table S1. The candidate CGTs were cloned into pET-28a (+) vectors (Invitrogen, USA) by homologous recombination method at the BamHI site. After verification of the sequences, the recombinant plasmids pET-28a (+)-VtCGTa and pET-28a (+)-VtCGTc were transformed into Escherichia coli BL21 (DE3) (TransGen Biotech, China) for heterologous expression. The E. coli cells were grown in 500 mL Luria–Bertani (LB) medium containing 50 μg/mL kanamycin at 37 °C with shaking (200 rpm). After OD600 reached 0.6, the cells were induced with 0.1 mmol/L Isopropyl β-d-thiogalactoside (IPTG) for 18 h at 18 °C (200 rpm). The cells were then collected by centrifugation (7500 rpm at 4 °C for 3 min) and resuspended in 15 mL of lysis buffer (pH 7.4, 20 mmol/L Tris, 200 mmol/L NaCl, 2% glycerol). Then the suspensions were ruptured by sonication on ice for 15 min. The cell debris was removed by centrifugation at 12,000 rpm for 45 min at 4 °C. The supernatant including crude protein was applied to a nickel-affinity chromatographic column (GE HealthCare) which was pre-equilibrated with 30 mmol/L imidazole elution buffer (pH 7.4, 20 mmol/L Tris, 200 mmol/L NaCl, 2% glycerol, 30 mmol/L imidazole). The miscellaneous proteins were eluted by 30 mmol/L imidazole elution buffer, and the target proteins were purified by 300 mmol/L imidazole elution buffer (pH 7.4, 20 mmol/L Tris, 200 mmol/L NaCl, 2% glycerol, 300 mmol/L imidazole). The purified proteins were analyzed by SDS-PAGE (Supporting Information Fig. S2), and stored in a storage buffer (pH 7.4, 20 mmol/L Tris, 200 mmol/L NaCl, 20% glycerol) at −80 °C.
2.4. Phylogenetic analysis
Molecular phylogenetic analysis was conducted using MEGA6 software with the maximum likelihood method (Fig. S1). The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed.
2.5. Enzyme activity assay
To characterize the catalytic function of VtCGTa and VtCGTc, the reaction mixtures contained 10 μg purified protein, 0.1 mmol/L sugar acceptor, and 0.5 mmol/L sugar donor in 100 μL buffer (37 °C, 1 h). 50 mmol/L Tris-HCl buffer (pH 8.0) and 50 mmol/L NaH2PO4/Na2HPO4 buffer (pH 7.0) were used for VtCGTc and VtCGTa, respectively. To probe the sugar donor promiscuity of VtCGTa and VtCGTc, UDP-rhamnose (UDP-Rha), UDP-glucose (UDP-Glc), UDP-xylose (UDP-Xyl), UDP-galactose (UDP-Gal), UDP-arabinose (UDP-Ara), UDP-N-acetylglucosamine (UDP-GlcNAc), UDP-aminoglucose (UDP-GlcN), UDP-N-acetylgalactosamine (UDP-GalNAc) and UDP-galactosamine (UDP-GalN) were tested using 2-hydroxynaringenin 3-C-glucoside (2) and 2-hydroxynaringenin (1) as acceptor, respectively. 5 μg of purified enzyme was used (37 °C for 0.5 h). The other reaction parameters were the same as described above. All reactions were terminated with 200 μL ice cold MeOH and then centrifuged at 15,000 rpm for 20 min. The glycosylated products were analyzed by a Vanquish UHPLC system (Thermo Fisher Scientific Inc., Waltham, MA, USA). Samples were separated on an Agilent Zorbax Eclipse Plus C18 column (Rapid Resolution HT 4.6 mm × 100 mm, 1.8 μm) protected with an Agilent 1290 Infinity in-line filter. The mobile phase consisted of methanol (A) and water containing 0.1% formic acid (v/v, B). The mobile phase gradient was as follows: 0–1.5 min, 20%–30% A; 1.5–5 min, 30% A; 5–8 min, 30%–40% A; 8–14 min, 40% A; 14–16 min, 40%–80% A; 16–19 min, 80% A. The flow rate was 0.3 mL/min. The column temperature was 50 °C. The detection wavelength was 300 nm. MS analysis was performed on an UHPLC-Q-Exactive HRMS instrument. The MS parameters were as follows: negative ion mode, sheath gas pressure 45 arb, aux gas pressure 10 arb, discharge voltage 4.5 kV, capillary temperature 350 °C. MS resolution was set as 70,000 FWHM, AGC target 1∗E6, maximum injection time 50 ms, and scan range m/z 100–1000. MS2 resolution was set as 17,500 FWHM, AGC target 1∗E5, maximum injection time 100 ms, NCE 35.
2.6. Effects of reaction time, pH, temperature, and divalent metal ions on enzyme activity
To characterize the enzymatic properties of VtCGTa and VtCGTc, the optimal reaction time was determined to be 30 min for VtCGTa and 20 min for VtCGTc, respectively. To optimize the reaction pH, the enzymatic reactions were carried out in various buffers with pH 4.0–6.0 (citric acid-sodium citrate buffer), 6.0–8.0 (Na2HPO4–NaH2PO4 buffer), 7.0–9.0 (Tirs-HCl buffer), and 9.0–11.0 (Na2CO3–NaHCO3 buffer). To investigate the optimal reaction temperature, the enzymatic reactions were incubated at different temperatures (4–60 °C). To test the dependence of divalent metal ions for VtCGTa and VtCGTc activity, different divalent cations Mg2+, Ba2+, Fe2+, Co2+, Cu2+, Zn2+, Mn2+, Ca2+, Ni2+ and EDTA in a final concentration of 5 mmol/L were added individually. Enzymatic reactions of VtCGTc were conducted with UDP-Rha as sugar donor and 2-hydroxynaringenin 3-C-glucoside (2) as sugar acceptor, while enzymatic reactions of VtCGTa were conducted with UDP-Glc as sugar donor and 2-hydroxynaringenin (1) as acceptor. All experiments were performed in triplicate and the mean value was used. The reactions were terminated with pre-cooled methanol and centrifuged at 12,000 rpm for 20 min for LC analysis. The conversion rates were calculated from UHPLC peak areas of glycosylated products and substrates.
2.7. Kinetic parameters of glycosyltransferases
For kinetic studies of VtCGTc and VtCGTc-V144T, enzymatic assays were performed in a final volume of 50 μL containing 50 mmol/L Tris-HCl (pH 8.0). The reactions were conducted at 37 °C for 10 min, and then quenched with 100 μL ice cold methanol. Samples were centrifuged at 12,000 rpm for 20 min and analyzed by HPLC. All experiments were performed in triplicate. Michaelis–Menten method was used (mean ± SEM) with GraphPad Prism software.
For the kinetic study of VtCGTc, 2-hydroxynaringenin 3-C-glucoside (2) and UDP-Rha, 0.08333 μg of purified VtCGTc, 2 mmol/L of saturated UDP-Rha, and varying concentrations (2–200 μmol/L) of 2 were added. For the kinetic study of VtCGTc, 2 and UDP-Glc, 1.25 μg of purified VtCGTc, 2 mmol/L of saturated UDP-Glc, and varying concentrations (2–200 μmol/L) of 2 were added. For the kinetic study of VtCGTc, 2 and UDP-Xyl, 0.8333 μg of purified VtCGTc, 2 mmol/L of saturated UDP-Xyl, and varying concentrations (2–200 μmol/L) of 2 were added. For the kinetic study of VtCGTc-V144T, 2 and UDP-Rha, 0.3125 μg of purified VtCGTc-V144T, 2 mmol/L of saturated UDP-Rha, and varying concentrations (2–600 μmol/L) of 2 were added. For the kinetic study of VtCGTc-V144T, 2 and UDP-Glc, 0.1667 μg of purified VtCGTc-V144T, 2 mmol/L of saturated UDP-Glc, and varying concentrations (2–200 μmol/L) of 2 were added.
2.8. QC calculations
The QC (quantum chemistry) calculations were performed using Gaussian 16, rev C.01. Dispersion corrected density functional with Beck-Johnson damping23, 24 and the implicit polarizable continuums model (PCM)25, 26 solvent environment were used for all calculations. Two levels of theory were applied, levels 1 and 2, corresponding to B3LYP-D3(BJ)/6-31+G (d,p)/PCM and B3LYP-D3(BJ)/6-311++G (2d,2p)/PCM, respectively. Geometry optimizations for ground state, flexible coordinate scan, transition state (TS), and intrinsic reaction coordinates (IRC)27, as well as vibrational frequency calculations, were carried out at level 1 of theory, while all the single point energies were calculated at level 2 of theory. The flexible coordinate scan was performed to locate an initial guess of TS structure, followed by a full TS optimization procedure implemented in Gaussian 16 and validated by the existence of a unique imaginary frequency corresponding to the max vibrational mode along the reaction path. IRC calculations were further carried out to verify the reactant and product of a TS structure. The bond dissociation energies of the interested O–H bonds in compound 3 were estimated by the difference of energies between the compound and corresponding radical species.
2.9. Molecular modeling
The crystal structure of GgCGT was used as a template for homology modeling, which has a high sequence identity of 46.7% from the BLAST search. The sequences of GgCGT and VtCGTc/VtCGTa were aligned using ClustalX2.028. UDP-Glc and UDP-Rha were modeled into the active site of the template structure (PDB 6L5S)9 using the positions from the structures of GgCGT (PDB 6L5P) and UGT89C1 (PDB 6IJA)29, respectively. The homology structures of VtCGTa and VtCGTc were modeled using MODELLER 9.1030 with the UDP-sugar moiety and prepared using the protein-preparation wizard module implemented in Schrödinger suite (version 2021-4)31. All the side-chain atoms of Asn and Gln residues were not flipped. The protonation state of His24 (VtCGTc)/His22 (VtCGTa) was assigned at δ-nitrogen. For the other histidine residues, the protonation states were determined by PROPKA 3.0 at pH=7 and the surrounding environment32. Compounds 2 and 3 were prepared using the Ligprep module in Schrödinger suite, followed by the induced fit docking (IFD) module33 to dock into the active sites of VtCGTa and VtCGTc, respectively. In the IFD procedure, the binding sites were defined as the geometrical center of residues 24/22, 90/91, 93/94, 390/386, and 391/387 for VtCGTc/VtCGTa, with the dimension of inner and outer boxes assigned to 10 and 32 Å for grid generation, respectively. The default values of other parameters were retained for IFD.
2.10. MD simulations
Molecular dynamics (MD) simulations were performed for the optimal complexes of VtCGTc/2/UDP-Rha, VtCGTc/2/UDP-Glc, VtCGTc-V144T/2/UDP-Rha, VtCGTc-V144T/2/UDP-Glc, VtCGTa/1/UDP-Rha, and VtCGTa-T145V/1/UDP-Rha. All the MD simulations were carried out for 100-ns production with OPLS-4 force field34 by the Desmond module. In each simulation, the protein‒ligand complex was centered into an orthorhombic box with the boundary buffer of 12 Å and added with ∼19,000 TIP3P water molecules and counter ions to neutralize the system35. Additional Na+ and Cl‒ were added to reach a 0.15 mol/L salt concentration. Before the production simulations, the system is energy minimized and equilibrated with Nose-Hoover chain thermostat (300.0 K)36 and Martyna-Bobias-Klein barostat (1.0 atm)37 using the default protocol implemented in Desmond. In our trialed simulations, the conformations and positions of compounds 1 and 2 were highly flexible and unstable. In order to seek an ensemble of conformations close to the reactant complex of enzyme and substrate, we applied 4 distance restraints to maintain the position of 1/2 in the active site of VtCGTa/VtCGTc. The harmony constraints were defined for the following atom pairs for VtCGTc:
| {CG@Asp120—ND1@His24, force_constant = 20.0 ref = 3.4 sigma = 0.2} |
| {NE2@His24—O4@2, force_constant = 20.0 ref = 2.8 sigma = 0.2} |
| {C5@2—C1′@UDP-Rha/Glc, force_constant = 20.0 ref = 3.5 sigma = 1.0} |
| {O4′@UDP-Rha/Glc—CG@Asp390, force_constant = 20.0 ref = 3.9 sigma = 0.2} |
and added into the configuration file for Desmond simulation. The equilibrium length of distance was assigned by the keyword ref for each distance, with the deviation assigned by the keyword sigma. For the VtCGTa systems, the atoms in the corresponding residues listed above were used for restraints with the same sets of force constants, reference distances and deviations. With a small force constant of 20, we were able to adequately sample the conformations of substates without leaving the active site of enzymes.
2.11. MM/GBSA binding free energy calculations
The MM/GBSA (molecular mechanics, the generalized Born model and solvent accessibility) binding free energies of substrates and UDP-Glc/Rha in the active sites of VtCGTa and VtCGTc were calculated from the above sampled conformational ensemble (400 snapshots for each system) by the Prime module in Schrödinger Suite. The calculations were performed in the implicit VSGB 2.0 solvent model38.
2.12. Site-directed mutagenesis
Site-directed mutagenesis of VtCGTc, VtCGTa, and 4 CGTa enzymes reported in our previous work11 (Supporting Information Table S2) were constructed using Fast Mutagenesis System kit (Transgen, China) according to the manufacturer's instructions. The primer pairs designed to construct the site-directed mutants are showed in Supporting Information Table S1. The enzyme activity assays of the mutants were performed and analyzed as described above.
3. Results
3.1. Molecular cloning and functional characterization of VtCGTa and VtCGTc
To analyze the transcriptome data of V. tricolor, 12 CGT sequences were used as templates for BLAST search (Fig. S1). Two putative glycosyltransferases VtCGTa (UGT708T5, accession number: OQ787068) and VtCGTc (UGT708T6, accession number: OQ787069) were screened out. They contain 1404 and 1431 base pairs, encoding 467 and 476 amino acids, respectively (Supporting Information Table S3). The candidate genes were cloned from cDNA using specific primers, and were then cloned into the pET-28a (+) vector. The enzymes were expressed and purified in E. coli BL21 (DE3) and were analyzed by SDS-PAGE (Fig. S2).
To characterize the catalytic function of VtCGTa and VtCGTc, in vitro catalytic reactions were carried out. Considering that the precursors of (iso)violanthin could be 2-hydroxyflavavone or flavone11,39, we selected 2-hydroxynaringenin (1), apigenin (1a), 2-hydroxynaringenin 3-C-glucoside (2), vitexin (2a), and iso-vitexin (2b) as sugar acceptors. UDP-Rha and UDP-Glc were used as sugar donor. The reaction products were then analyzed by UHPLC and LC/MS.
The results revealed that VtCGTa and VtCGTc could not catalyze the C-glycosylation of 1a, 2a or 2b (Fig. 1A, Supporting Information Fig. S3). However, VtCGTc could efficiently convert 2 into 3, with a conversion rate of up to 100% by UHPLC analysis (Fig. 1B). Its [M‒H]‒ ion appeared at m/z 595, which was 146 amu greater than 2. The MS/MS spectrum showed fragment ions at m/z 209 [M‒H‒224‒162]‒, m/z 239 [M‒H‒194‒162]‒, and m/z 269 [M‒H‒164‒162]‒, indicating that 3 was a glucose/rhamnose di-C-glycoside (Fig. 1C). With the addition of hydrochloric acid (150 mmol/L), 3 could be converted into violanthin (3a) and iso-violanthin (3b)11,39. These results indicated VtCGTc as a C-rhamnosyltransferase. To our best knowledge, VtCGTc is the first specific plant C-rhamnosyltransferase.
Likewise, VtCGTa was identified as a C-glucosyltransferase catalyzing 3-C-glucosylation of 2-hydroxynaringenin (1) (Fig. S3). Then we incubated 2-hydroxynaringenin (1), UDP-Glc, UDP-Rha, VtCGTa and VtCGTc together, and obtained violanthin (3a) and iso-violanthin (3b) after acidic dehydration (Supporting Information Fig. S4). Thus, VtCGTa and VtCGTc may be involved in the biosynthesis of (iso)violanthin in V. tricolor (Fig. 1A).
The biochemical properties of VtCGTc were investigated using 2 as acceptor and UDP-Rha as sugar donor. VtCGTc showed a maximal activity at pH 8.0 (50 mmol/L Tris-HCl) and 37 °C. The activity was independent of divalent metal ions (Supporting Information Fig. S5). Kinetic analysis demonstrated that VtCGTc exhibited Km and kcat/Km values of 18.05 μmol/L and 0.06100 μL/(mol·s), respectively (Fig. 4B). VtCGTa showed its maximal activity at pH 7.0 (NaH2PO4/Na2HPO4) and 37 °C (Fig. S5).
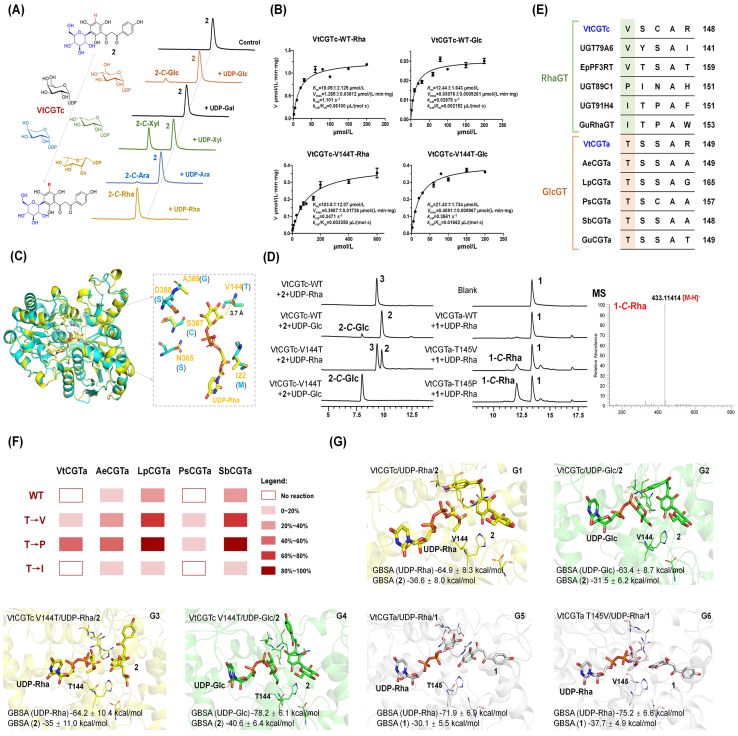
Figure 4.
Mechanisms for sugar donor selectivity of VtCGTa and VtCGTc. (A) HPLC chromatograms for sugar donor selectivity of VtCGTc using 2 as acceptor. (B) Kinetic parameters for VtCGTc and VtCGTc-V144T. 2 was used as acceptor, and UDP-Rha and UDP-Glc were used as sugar donor. (C) The superimposition of VtCGTa and VtCGTc. Amino acids around the active pocket in VtCGTc and VtCGTa are shown in orange and blue, respectively. (D) UHPLC analysis of the catalytic products of VtCGTc wild-type (WT) and V144T mutant, using 2 as acceptor. The reaction mixtures were incubated at 37 °C for 20 min, containing 2 μg purified protein. LC/MS analysis of the catalytic products of VtCGTa wild-type (WT) and mutants, using 1 as acceptor. The reaction mixtures were incubated at 37 °C for 2 h, containing 10 μg purified protein. (E) Alignment of sequences of VtCGTa/c and other UGTs, with the key residues highlighted. (F) The conversion rates of CGTa mutants. 1 was used as acceptor and UDP-Rha as sugar donor. (G) The complex structure models and MM/GBSA binding free energy of VtCGTc/UDP-Rha/2, VtCGTc/UDP-Glc/2, VtCGTc V144T/UDP-Rha/2, VtCGTc V144T/UDP-Glc/2, VtCGTa/UDP-Rha/1, and VtCGTa T145V/UDP-Rha/1. Representative snapshots were extracted from each simulation.
3.2. Mechanisms for the dehydration of 3
The dehydration and cyclization of 2-hydroxyflavanone basic skeleton is popular in the biosynthesis of flavonoid C-glycosides. In this work, 2-hydroxynaringenin 3-C-glucoside (2) could be converted into 2a and 2b in a ratio of around 1:1 after the addition of HCl (Supporting Information Fig. S6). This result was consistent with previous reports11,39. However, the ratio of 3a:3b was around 1:4 after dehydration of 3 (2-hydroxynaringenin 3,5-di-C-glucoside-rhamnoside) (Fig. 1B). In order to interpret this special observation, we performed quantum chemistry (QC) calculations.
Through QC calculations, we obtained the hexatomic ring transition state (TS1). The activation barrier for 3a and 3b is 20.5 and 18.4 kcal/mol, respectively (Fig. 2, Supporting Information Fig. S7). The relatively low barrier for transition state 1 (TS1) of 3b explained why 3b was the major dehydration product. On the other hand, dissociation of an O–H bonds on the phenoic ring of 3 is required to form the dehydration products. The bond dissociation energy (BDE) for 2′-OH is lower than 6′-OH (92.0 vs. 95.1 kcal/mol), and thus 3b was relatively easy to be formed (Supporting Information Fig. S8).
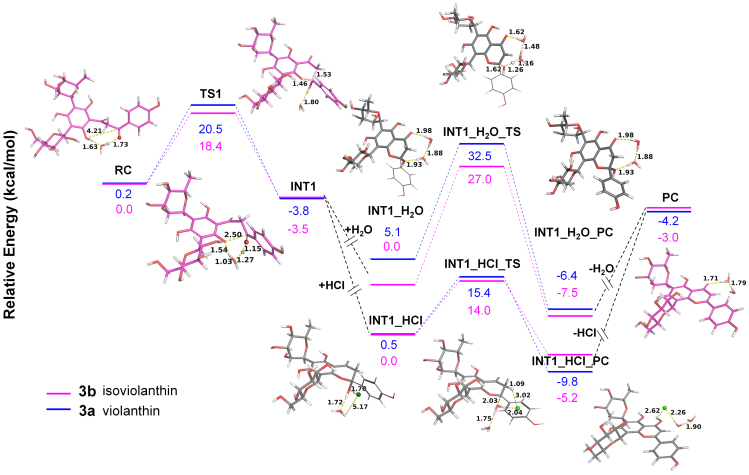
Figure 2.
Mechanisms for the dehydration of 3 based on QC calculations. The complete pathway of 3b is shown. The process of INT1_H2O, INT1_H2O_TS and INT1_H2O_PC is under natural environment, and the process of INT1_HCl, INT1_HCl_TS and INT1_HCl_PC is upon HCl treatment. RC, reactant complex; TS, transition state; INT, intermediate complex; PC, product complex.
Interestingly, the addition of HCl could speed up the dehydration process. The process includes INT1_H2O, INT1_H2O_TS and INT1_H2O_PC under natural environment, but includes INT1_HCl, INT1_HCl_TS and INT1_HCl_PC upon the addition of HCl (Fig. 2). These two conditions exhibited remarkably different energy barriers. The barrier for INT1_HCl_TS of 3b is 13.0 kcal/mol lower than INT1_H2O_TS, and explains why the dehydration reaction could be speeded up with the addition of HCl.
3.3. Substrate promiscuity and sugar donor selectivity of VtCGTc
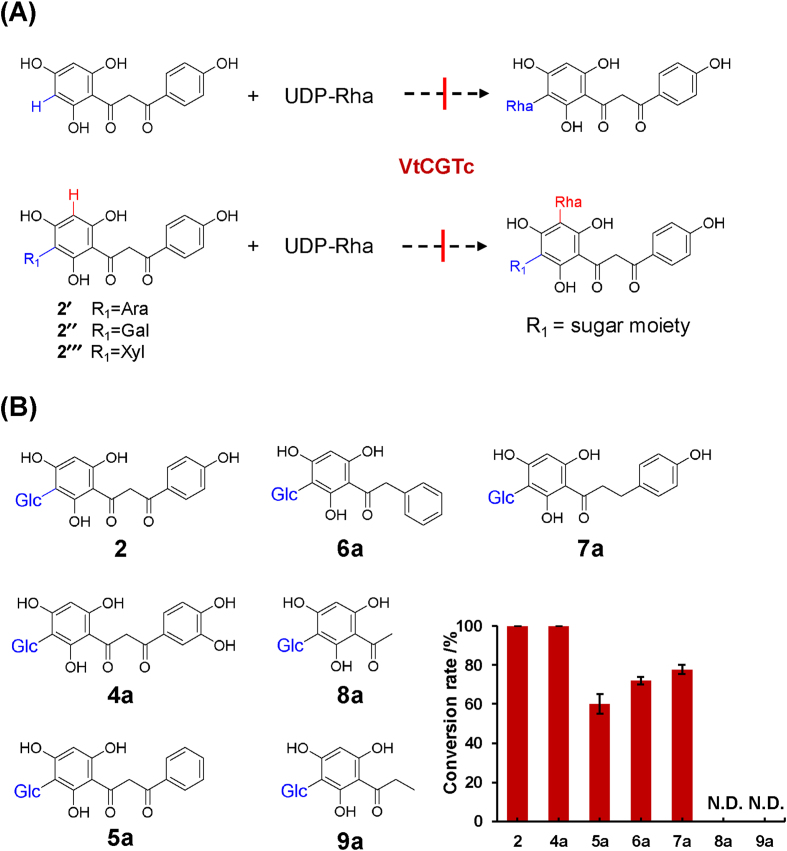
To explore the substrate promiscuity of VtCGTc, 17 substrates were screened by enzymatic assay, including phenolics, flavones, dihydrochalcones, 2-hydroxyflavanones, and their C-glycosides. UDP-Rha was used as sugar donor. Neither compounds 1a, 2a and 2b (flavones and flavone C-glycosides) nor 1 and 4–9 (2-hydroxyflavanones and phloroglucin derivatives) could be rhamnosylated by VtCGTc. VtCGTc could not accept flavonoid C-arabinoside, C-galactoside, or C-xyloside, either (Fig. 3A). The C-rhamnosylated products can only be observed for the C-glucosides of 2-hydroxyflavanones (2, 4a, 5a) or phloroglucin derivatives (6a and 7a) (Fig. 3B, Supporting Information Figs. S9‒S13). Compounds 8a and 9a could not be converted by VtCGTc.
Figure 3.
Substrate promiscuity of VtCGTc. (A) Substrates that cannot be recognized by VtCGTc. (B) Screening of C-glucosides.
On the other hand, VtCGTa could efficiently catalyze the C-glucosylation of 2-hydroxyflavanones and phloroglucin derivatives, including 1–2 and 4–9 to form mono-C-glucosides or di-C-glucosides (Supporting Information Fig. S14). We also tested a number of flavanones, including pinocembrin (10), naringenin (11), eriodictyol (12), pinocembrin-6-C-glucoside (10a), pinocembrin-8-C-glucoside (10b), and naringenin-8-C-glucoside (11a). None of these substrates could be catalyzed by VtCGTa or VtCGTc (Supporting Information Fig. S15).
Furthermore, we evaluated the sugar donor selectivity of VtCGTc. UDP-Rha, UDP-Glc, UDP-xylose (UDP-Xyl), UDP-galactose (UDP-Gal), UDP-Ara, UDP-N-acetylglucosamine (UDP-GlcNAc), UDP-aminoglucose (UDP-GlcN), UDP-N-acetylgalactosamine (UDP-GalNAc), and UDP-galactosamine (UDP-GalN) were tested. VtCGTc showed potent catalytic activity towards UDP-Rha, with a conversion rate of around 100%. VtCGTc showed weak activities towards UDP-Xyl (conversion rate 45%), UDP-Glc (10%), and UDP-Ara and UDP-GlcN (<5%). No catalytic activities were observed for the other sugar donors (Fig. 4A, Supporting Information Fig. S16). Thus, VtCGTc showed a strong preference to UDP-Rha. Given that it could also accept UDP-Xyl, VtCGTc may also be involved in the biosynthesis of apigenin C-glucoside-C-xyloside in V. tricolor (Supporting Information Fig. S18).
The apparent kinetic parameters for VtCGTc with 2 and different sugar donors were measured. VtCGTc showed similar Km values (10–20 μmol/L) for 2 with saturated UDP-Rha, UDP-Glc and UDP-Xyl, respectively. Consistent with the functional characterization results, the kcat/Km value (0.06100 μL/(mol·s)) for VtCGTc with 2 and UDP-Rha was 28 times higher than that with 2 and UDP-Glc, and 19 times higher than 2 and UDP-Xyl (Fig. 4B, Supporting Information Fig. S17).
In contrast, VtCGTa showed high preference to UDP-Glc (conversion rate 100%), and could also accept UDP-Xyl (60%) and UDP-Gal (25%). However, it could not accept UDP-Rha (Fig. S16).
3.4. Mechanisms for sugar donor selectivity of VtCGTa and VtCGTc
To understand mechanisms for the remarkably different sugar donor selectivity of VtCGTa and VtCGTc, we conducted molecular modelling and site-directed mutagenesis. The structural models for VtCGTa and VtCGTc were constructed by homology modeling using MODELLER 9.1030. The models of VtCGTa/UDP-Glc and VtCGTc/UDP-Rha were established according to the crystal structures of GgCGT/UDP-Glc and UGT89C1/UDP-Rha, respectively9,29. The two CGTs showed significant difference in 6 amino acids around the active pocket, including Ile22 (Met20), Val144 (Thr145), Asn365 (Ser361), Ser367 (Cys363), Asp388 (Ser384), Ala389 (Gly385) in VtCGTc (VtCGTa) (Fig. 4C). V144 of VtCGTc and T145 of VtCGTa could form hydrophobic interaction and hydrogen bond with the sugar moieties of UDP-Rha and UDP-Glc, respectively (Fig. 4C, Supporting Information Fig. S19). Thus, we performed complementary mutagenesis for VtCGTa and VtCGTc. Interestingly, VtCGTc-V144T mutant could completely convert 2 to its 3,5-di-C-glucoside. It only showed a conversion rate of 60% with UDP-Rha (Fig. 4D). Consistently, the kcat/Km value (catalytic efficiency) of the V144T mutant for UDP-Glc was even higher than that for UDP-Rha (0.0166 vs 0.0033 μL/(mol·s)) (Fig. 4B). On the other hand, VtCGTa T145V mutant could accept UDP-Rha as sugar donor, though the conversion rate was low (Fig. 4D, Supporting Information Fig. S20). The above data indicated that Val played an important role in recognizing UDP-Rha.
Then we analyzed the same loci for the sequences of a series of rhamnosyltransferases (RhaGTs) and 2-hydroxyflavanone C-glucosyltransferases (GlcGTs) (Table S2). The threonine of GlcGTs was mapped to Pro, Val or Ile of RhaGTs (Fig. 4E). Then we selected VtCGTa, AeCGTa, LpCGTa, PsCGTa, and SbCGTa, and mutated the Thr residue to Val, Pro, or Ile, respectively. When Thr was mutated to Val or Pro, all the mutants gained C-rhamnosylation activity and showed medium to high conversion rates with 1 and UDP-Rha as substrates (Fig. 4F, Supporting Information Fig. S21). The Pro mutants even showed higher conversion rates than the Val mutants. The Ile mutants only showed weak activities.
We also found that H22, T145, S146, D386, Q387 in VtCGTa and H24, V144, S145, R287, D390 in VtCGTc were important for the C-glucosylation and rhamnosylation of VtCGTa and VtCGTc, respectively. The S146A mutant of VtCGTa even showed improved C-glucosylation activity (Supporting Information Fig. S22).
We further conducted molecular docking, followed by MD (molecular dynamics) simulations and MM/GBSA (molecular mechanics with generalized Born and surface area solvation) binding free energy calculations. Firstly, we established the complex structure models of VtCGTc/UDP-Rha/2, VtCGTc/UDP-Glc/2, VtCGTc V144T/UDP-Rha/2, VtCGTc V144T/UDP-Glc/2, VtCGTa/UDP-Rha/1, and VtCGTa T145V/UDP-Rha/1. During the 100-ns MD simulations, we found the overall structures of homology modeling derived VtCGTa and VtCGTc were well maintained, while the side chain atoms of the substrate binding domain as well as the substrates varied remarkably (Supporting Information Fig. S23). By applying knowledge-based distance restraints (see Section 2.10 for details) to keep the sugar donor and receptor in their binding domains, we obtained reasonable binding modes favoring 5-C-glycosylation.
As shown in Supporting Information Table S4 and Fig. 4G1‒G2, the binding free energies for UDP-Glc and UDP-Rha in wild-type VtCGTc are similar. However, the binding free energy for 2 in VtCGTc/UDP-Rha/2 is 5.1 kcal/mol lower than in VtCGTc/UDP-Glc/2. This result indicates the 5-C-glucosylation favoring binding mode is not stable in VtCGTc. Structural analysis indicates the CH2OH moiety of Glc could form a stable hydrogen bond with Thr144 of the VtCGTc V144T mutant. Binding free energy calculations showed the single site mutagenesis could change binding stability for different substrates (Fig. 4G3‒G4). Moreover, the conformation of 2 in VtCGTc with UDP-Glc is bended, but stretchy in VtCGTc V144T mutant. The latter conformation exhibits better binding with the protein by providing more contacts. Thus, the VtCGTc V144T mutant showed improved C-glucosylation activity. On the other hand, both the sugar donor and receptor exhibit lower binding free energies (more stable) in the VtCGTa T145V mutant than in the wild type (Fig. 4G5‒G6, Table S4). The above results interpret why Val/Thr is the key residue in recognizing UDP-Rha/Glc, respectively, and preliminarily unravel mechanisms for the selective C-rhamnosylation and C-glucosylation of VtCGTc and VtCGTa.
4. Discussion
VtCGTc is the first C-rhamnosyltransferase that has been reported. It exhibits high selectivity towards UDP-Rha, and could efficiently catalyze C-rhamnosylation of 2-hydroxyflavanone C-glucosides. We also discovered VtCGTa, which could catalyze C-glucosylation of 2-hydroxyflavanones. VtCGTa and VtCGTc may be involved in the biosynthesis of violanthin and iso-violanthin in V. tricolor. In our previous study, we have discovered a series of CGTa and CGTb enzymes from plants, which sequentially catalyze C-glucosylation and C-arabinosylation of 2-hydroxyflavanones to produce schaftoside and isoschaftoside. VtCGTa shows the same function as CGTa enzymes. VtCGTc shows high selectivity towards UDP-Rha, while the CGTb enzymes prefer to accept UDP-Ara. Thus, this enzyme was named as VtCGTc instead of VtCGTb.
Structural analysis and site-directed mutagenesis indicated that V144 was a key amino acid of VtCGTc in recognizing UDP-Rha, and was interpreted by theoretical calculations. While VtCGTa shares 61.44% sequence similarity with VtCGTc, it showed selectivity towards UDP-Glc. By mutating T145 into valine, the obtained T145V mutants of VtCGTa and congenerous UDP-Glc-preferring CGTs gained C-rhamnosylation activity.
The dehydration of 2-hydroxyflavanone C-glycosides is a common step in the biosynthesis of many flavonoid C-glycosides. In previous reports, this reaction usually produces a pair of isomers at a ratio of around 1:1. In the present work, the dehydration products violanthin (3a) and iso-violanthin (3b) were produced at a ratio of 1:4. QC calculations indicated the energy barriers for the transition state of 3a was higher than that of 3b, and interpreted why 3b was the major product. However, violanthin instead of iso-violanthin is the major compound in V. tricolor20. Therefore, this dehydration reaction may not happen spontaneously in nature. A specific dehydration enzyme may participate in this final step of violanthin biosynthesis in the plant. This dehydratase deserves to be investigated in the future.
Notes
The other data needed to evaluate the conclusions are presented in the paper and/or Supporting Information. Any additional data related to this paper may be requested from Min Ye (yemin@bjmu.edu.cn).
5. Conclusions
In this work, we identified the first C-rhamnosyltransferase VtCGTc from the medicinal plant V. tricolor. VtCGTc showed high selectivity towards UDP-Rha. Through homology modeling, structural analysis, and site-directed mutagenesis, we found that Val144 was a key amino acid residue in recognizing UDP-Rha. Mechanisms for the sugar donor selectivity was interpreted by molecular docking, MD simulations, and MM/GBSA calculations. This work provides a novel and efficient approach to prepare bioactive flavonoid C-rhamnosides.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grants No. 81725023 to Min Ye and 82003614 to Yang Yi) and China National Postdoctoral Program for Innovation Talents (No. BX20220022 to Zi-Long Wang). The computations were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at the National Supercomputer Center (SNIC2022-3-34) at Linköping University (Sweden). We also thank Dr. Xiang Sheng (Tianjin Institute of Industrial Biotechnology, Chinese Academy of Science) for helpful discussions in theoretical calculations.
Author contributions
Min Ye and Zi-Long Wang for conceptualization; Zi-Long Wang and Bo-Yun Han for major investigation; Jun-Hao Li and Ågren Hans contributed to theoretical calculation; Qing Jin, Hao-Tian Wang, Kuan Chen and Yang Yi assisted with data acquisition; Min Ye, Zi-Long Wang and Yang Yi for funding acquisition; Zi-Long Wang and Bo-Yun Han for writing-original draft; Min Ye and Xue Qiao for writing-review & editing.
Conflicts of interest
There are no conflicts to declare.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.05.011.
Contributor Information
Xue Qiao, Email: qiaoxue@bjmu.edu.cn.
Min Ye, Email: yemin@bjmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Zhang Y.Q., Zhang M., Wang Z.L., Qiao X., Ye M. Advances in plant-derived C-glycosides: phytochemistry, bioactivities, and biotechnological production. Biotechnol Adv. 2022;60 doi: 10.1016/j.biotechadv.2022.108030. [DOI] [PubMed] [Google Scholar]
- 2.Xiao J.B., Capanoglu E., Jassbi A.R., Miron A. Advance on the flavonoid C-glycosides and health benefits. Crit Rev Food Sci. 2016;56:S29–S45. doi: 10.1080/10408398.2015.1067595. [DOI] [PubMed] [Google Scholar]
- 3.Kitamura K., Ando Y., Matsumoto T., Suzuki K. Total synthesis of aryl C-glycoside natural products: strategies and tactics. Chem Rev. 2018;118:1495–1598. doi: 10.1021/acs.chemrev.7b00380. [DOI] [PubMed] [Google Scholar]
- 4.Zeng P., Zhang Y., Pan C., Jia Q., Guo F.J., Li Y.M., et al. Advances in studying of the pharmacological activities and structure activity relationships of natural C-glycosylflavonoids. Acta Pharm Sin B. 2013;3:154–162. [Google Scholar]
- 5.Xiao J.B. Dietary flavonoid aglycones and their glycosides: which show better biological significance?. Crit Rev Food Sci. 2017;57:1874–1905. doi: 10.1080/10408398.2015.1032400. [DOI] [PubMed] [Google Scholar]
- 6.Putkaradze N., Teze D., Fredslund F., Welner D.H. Natural product C-glycosyltransferases‒a scarcely characterised enzymatic activity with biotechnological potential. Nat Prod Rep. 2021;38:432–443. doi: 10.1039/d0np00040j. [DOI] [PubMed] [Google Scholar]
- 7.Xie K.B., Zhang X.L., Sui S.Y., Ye F., Dai J.G. Exploring and applying the substrate promiscuity of a C-glycosyltransferase in the chemo-enzymatic synthesis of bioactive C-glycosides. Nat Commun. 2020;11:5162. doi: 10.1038/s41467-020-18990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D.W., Chen R.D., Wang R.S., Li J.H., Xie K.B., Bian C.C., et al. Probing the catalytic promiscuity of a regio- and stereospecific C-glycosyltransferase from Mangifera indica. Angew Chem Int Ed. 2015;54:12678–12682. doi: 10.1002/anie.201506505. [DOI] [PubMed] [Google Scholar]
- 9.Zhang M., Li F.D., Li K., Wang Z.L., Wang Y.X., He J.B., et al. Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra. J Am Chem Soc. 2020;142:3506–3512. doi: 10.1021/jacs.9b12211. [DOI] [PubMed] [Google Scholar]
- 10.He J.B., Zhao P., Hu Z.M., Liu S., Kuang Y., Zhang M., et al. Molecular and structural characterization of a promiscuous C-glycosyltransferase from Trollius chinensis. Angew Chem Int Ed. 2019;58:11513–11520. doi: 10.1002/anie.201905505. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z.L., Gao H.M., Wang S., Zhang M., Chen K., Zhang Y.Q., et al. Dissection of the general two-step di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants. Proc Natl Acad Sci U S A. 2020;117:30816–30823. doi: 10.1073/pnas.2012745117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones P., Messner B., Nakajima J.I., Schaffner A.R., Saito K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem. 2003;278:43910–43918. doi: 10.1074/jbc.M303523200. [DOI] [PubMed] [Google Scholar]
- 13.Casas M.I., Falcone-Ferreyra M.L., Jiang N., Mejia-Guerra M.K., Rodriguez E., Wilson T., et al. Identification and characterization of maize salmon silks genes involved in insecticidal maysin biosynthesis. Plant Cell. 2016;28:1297–1309. doi: 10.1105/tpc.16.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yano R., Takagi K., Tochigi S., Fujisawa Y., Nomura Y., Tsuchinaga H., et al. Isolation and characterization of the soybean Sg-3 gene that is involved in genetic variation in sugar chain composition at the C-3 position in soyasaponins. Plant Cell Physiol. 2018;59:797–810. doi: 10.1093/pcp/pcy019. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z.L., Zhou J.J., Han B.Y., Hasan A., Zhang Y.Q., Zhang J.H., et al. GuRhaGT, a highly specific saponin 2″-O-rhamnosyltransferase from Glycyrrhiza uralensis. Chem Commun. 2022;58:5277–5280. doi: 10.1039/d1cc07021e. [DOI] [PubMed] [Google Scholar]
- 16.Mohammed H.S., et al. Antibacterial and potential antidiabetic activities of flavone C-glycosides isolated from Beta vulgaris subspecies cicla L. var. flavescens (Amaran-thaceae) cultivated in Egypt. Curr Pharmaceut Biotechnol. 2019;20:595–604. doi: 10.2174/1389201020666190613161212. [DOI] [PubMed] [Google Scholar]
- 17.David J.P., dos Santos I.D., David J.M. A new sesquiterpene from the fruits of Allophylus laevigatus. Fitoterapia. 2004;75:795–798. doi: 10.1016/j.fitote.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Pagliuso D., et al. Flavonoids from duckweeds potential applications in the human diet. RSC Adv. 2021;10:44981–44988. doi: 10.1039/d0ra06741e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vukics V., Kery A., Bonn G.K., Guttman A. Major flavonoid components of heartsease (Viola tricolor L.) and their antioxidant activities. Anal Bioanal Chem. 2008;390:1917–1925. doi: 10.1007/s00216-008-1885-3. [DOI] [PubMed] [Google Scholar]
- 20.Vukics V., Toth B.H., Ringer T., Ludanyi K., Kery A., Bonn G.K., et al. Quantitative and qualitative investigation of the main flavonoids in heartsease (Viola tricolor L.) J Chromatogr Sci. 2008;46:97–101. doi: 10.1093/chromsci/46.2.97. [DOI] [PubMed] [Google Scholar]
- 21.Dung H.V., Cuong T.D., Chinh N.M., Quyen D., Kim J.A., Byeo J.S., et al. Compounds from the aerial parts of Piper bavinum and their anti-cholinesterase activity. Arch Pharm Res. 2015;38:677–682. doi: 10.1007/s12272-014-0432-3. [DOI] [PubMed] [Google Scholar]
- 22.Vincen S., Arokiyaraj S., Saravanan M., Dhanraj M. Molecular docking studies on the anti-viral effects of compounds from kabasura kudineer on SARS-CoV-2 3CL(pro) Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.613401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimme S., Antony J., Ehrlich S., Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys. 2010;132 doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 24.Grimme S., Ehrlich S., Goerigk L. Effect of the damping function in dispersion corrected density functional theory. J Comput Chem. 2011;32:1456–1465. doi: 10.1002/jcc.21759. [DOI] [PubMed] [Google Scholar]
- 25.Miertus S., Scrocco E., Tomasi J. Electrostatic interaction of a solute with a continuum. A direct utilizaion of AB initio molecular potentials for the prevision of solvent effects. Chem Phys. 1981;55:117–129. [Google Scholar]
- 26.Cances E., Mennucci B., Tomasi J. A new integral equation formalism for the polarizable continuum model: theoretical background and applications to isotropic and anisotropic dielectrics. J Chem Phys. 1997;107:3032–3041. [Google Scholar]
- 27.Gonzalez C., Schlegel H.B. Reaction path following in mass-weighted internal coordinates. J Chem Phys. 1990;94:5523–5527. [Google Scholar]
- 28.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Zong G.N., Fei S., Liu X., Li J., Gao Y.R., Yang X., et al. Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-β-l-rhamnose and rhamnosylation mechanism. Plant J. 2019;99:257–269. doi: 10.1111/tpj.14321. [DOI] [PubMed] [Google Scholar]
- 30.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 31.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 32.Olsson M.H.M., Sondergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theor Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 33.Sherman W., Day T., Jacobson M.P., Friesner R.A., Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 34.Lu C., Wu C.J., Ghoreishi D., Chen W., Wang L.L., Damm W., et al. OPLS4: improving force field accuracy on challenging regimes of chemical space. J Chem Theor Comput. 2021;17:4291–4300. doi: 10.1021/acs.jctc.1c00302. [DOI] [PubMed] [Google Scholar]
- 35.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 36.Martyna G.J., Klein M.L., Tuckerman M. Nosé–Hoover chains: the canonical ensemble via continuous dynamics. J Chem Phys. 1992;97:2635–2643. [Google Scholar]
- 37.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 38.Li J.N., et al. The VSGB 2.0 Model: a next generation energy model for high resolution protein structure modeling. Proteins. 2011;79:2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y.Q., Wang Z.L., Chen Z., Jin Z.T., Hasan A., Wang H.D., et al. A highly selective 2″-O-glycosyltransferase from Ziziphus jujuba and de novo biosynthesis of isovitexin 2″-O-glucoside. Chem Commun. 2022;58:2472–2475. doi: 10.1039/d1cc06949g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.