Summary
There is an emblematic clinical and genetic heterogeneity associated with inherited retinal diseases (IRDs). The most common form is retinitis pigmentosa (RP), a rod-cone dystrophy caused by pathogenic variants in over 80 different genes. Further complexifying diagnosis, different variants in individual RP genes can also alter the clinical phenotype. USH2A is the most prevalent gene for autosomal-recessive RP and one of the most challenging because of its large size and, hence, large number of variants. Moreover, USH2A variants give rise to non-syndromic and syndromic RP, known as Usher syndrome (USH) type 2, which is associated with vision and hearing loss. The lack of a clear genotype-phenotype correlation or prognostic models renders diagnosis highly challenging. We report here a long-awaited differential non-syndromic RP and USH phenotype in three human disease-specific models: fibroblasts, induced pluripotent stem cells (iPSCs), and mature iPSC-derived retinal organoids. Moreover, we identified distinct retinal phenotypes in organoids from multiple RP and USH individuals, which were validated by isogenic-corrected controls. Non-syndromic RP organoids showed compromised photoreceptor differentiation, whereas USH organoids showed a striking and unexpected cone phenotype. Furthermore, complementary clinical investigations identified macular atrophy in a high proportion of USH compared with RP individuals, further validating our observations that USH2A variants differentially affect cones. Overall, identification of distinct non-syndromic RP and USH phenotypes in multiple models provides valuable and robust readouts for testing the pathogenicity of USH2A variants as well as the efficacy of therapeutic approaches in complementary cell types.
Keywords: genotype-phenotype correlations, clinical testing, molecular genetics, molecular pathophysiology, pathogenesis, USH2A, Usherin, Usher Syndrome, retinal organoids, retinitis pigmentosa
Introduction
Retinitis pigmentosa (RP; MIM: 268000; http://www.omim.org), with a prevalence of 1 in 4,000 individuals,1 is the most common inherited retinal disease (IRD) causing visual impairment in adults.2 Pathogenic RP variants have been identified in more than 80 different genes (https://web.sph.uth.edu/RetNet/), which gives rise to a wide clinical landscape.3 The most common form is autosomal-recessive RP, which is characterized by night vision difficulties in childhood or adolescence and onset of mid-peripheral vision loss in adulthood.4 Furthermore, syndromic forms exist, and Usher syndrome (USH), with a prevalence of 1 in 6,000 individuals,5 is the most common. USH individuals present with congenital sensorineural hearing loss in addition to progressive RP. Depending on the clinical signs, USH can be classified into three main subtypes. USH type 2 (MIM: 276901) is the most prevalent, and up to 85% of cases are due to variants in the gene USH2A (MIM: 608400).6 In addition, USH2A variants account for 8%–22% of non-syndromic RP cases (MIM: 276901),7,8 rendering USH2A the most prevalent gene for autosomal-recessive RP.9,10
USH2A comprises 72 exons,11 and over 2,000 different variants have been reported (www.LOVD.nl/USH2A). Although a genotype-phenotype correlation has been proposed for USH2A variants,12 it has not been definitively established. The most prominent example is two variants in exon 13, c.2299delG, p.Glu767Serfs∗21 and c.2276G>T, p.Cys759Phe, which are frequently seen in affected individuals10,13 and together represent approximately 30% of all pathogenic USH2A alleles.14 The variant c.2299delG is commonly associated with USH, whereas c.2276G>T has been described as a “retina-specific” allele that leads to RP in the absence of hearing anomalies in childhood.15 However, the reason behind this phenomenon is currently unknown.
USH2A encodes usherin, which belongs to a protein complex known as the USH interactome.16 This complex is located at the periciliary membrane that is wrapped around the connecting cilium (CC) of photoreceptors.17 To date, the exact function of the USH proteins in the retina is not well understood. Furthermore, none of the murine USH models show retinal degeneration,18 except for the Ush2a mutant mice, which show only a mild and late-onset degeneration phenotype.19 This is most likely due to anatomical differences in photoreceptor structures between humans and rodents.20 The zebrafish ush2a knockout and/or mutant lines more closely mimic the retinal degeneration of affected individuals but with variations in onset and severity.21,22,23,24,25 However, the evolutionary distance between zebrafish and humans, as well as the regenerative capacities of the zebrafish, renders pathophysiological studies of this progressive disorder highly challenging. Therefore, there is a need for development of a human retinal model to further decipher USH2A-associated disease phenotypes.
Over recent years, induced pluripotent stem cell (iPSC)-derived retinal organoids containing features of mature photoreceptors26,27,28 have proven their worth for disease modeling of diverse IRDs and for testing the efficacy of potential therapies.27,29,30,31,32,33,34 We previously reported generation of iPSC lines from two individuals carrying the most prevalent exon 13 USH2A mutations: a non-syndromic RP individual compound heterozygous for c.2299delG and c.2276G>T alleles35 and an USH individual homozygous for the c.2299delG allele.36 We further reported aberrantly increased USH2A mRNA levels in USH2A-USH iPSCs compared with control or USH2A-RP lines, which were rescued following CRISPR-Cas-mediated correction of the c.2299delG variant.37 This differential mRNA expression profile was a molecular glimpse into the putative genotype-phenotype correlation for USH2A mutations.
We hence extended our efforts to further decipher the genotype-phenotype relationship of USH2A mutations by studying fibroblasts, iPSCs, and iPSC-derived retinal organoids of affected individuals with different USH2A variants. Here, we report differential non-syndromic RP and USH phenotypes in all models investigated. Moreover, we report a mature human retinal organoid model system of USH2A-specific mutations, allowing us to detect usherin expression at the CC of human photoreceptors. Furthermore, phenotypic reversion and restored usherin expression in retinal organoids was demonstrated using CRISPR-Cas-mediated gene-corrected isogenic controls. Last, by performing additional tailored clinical investigations, the unexpected differential retinal phenotype of the human organoids was confirmed at the clinical level in RP and USH individuals. Together, these data provide long-awaited direct evidence of a genotype-phenotype correlation associated with USH2A mutations.
Material and methods
Clinical investigations
Informed consent for clinical examination and genetic analyses was obtained and signed by each individual. All methods were carried out in accordance with approved protocols of Montpellier University Hospital, Montpellier, France (ID IRB-MTP_2021_11_202100959) and the Declaration of Helsinki for medical research. Individuals underwent a comprehensive ophthalmological examination, including best corrected visual acuity and multimodal imaging. Color fundus photography was performed using an automated, non-mydriatic fundus camera (AFC 330, Nidek). Short-wave fundus autofluorescence imaging was performed with a Spectralis combined retinal angiography and spectral domain optical coherence tomography device (Heidelberg Engineering).
Fibroblast culture
Skin biopsies of the USH2A volunteers were performed under sterile conditions at the Center of Reference for Inherited Sensory Diseases (CHU Montpellier) following informed consent as part of the registered clinical trial NCT03853252. Ethics approval was provided by the local institutional review board and the French National Agency for the Safety of Medicines and Health Products (ID 2014-A00549-38). The biopsies and emerging fibroblasts were cultured in AmnioMAX C100 basal medium supplemented with GlutaMAX (Gibco), 10% decomplemented fetal calf serum (FCS; Gibco), 1% penicillin-streptomycin-amphotericin B (Gibco), and 2% AmnioMax C100 supplement (Gibco), as described previously.38
iPSC reprogramming and culture
The iPSCs were reprogrammed from USH2A fibroblasts under feeder-free conditions using the integration-free CytoTune iPSC 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific), cultured in Essential 8 (Gibco) or mTeSR Plus (STEMCELL Technologies) medium, and passaged weekly using 0.48 mM Versene solution (Gibco).39 We previously reported the generation and characterization of the USH2A-RP-1,35 USH2A-USH-1,36 USH2A-RP-2,40 and control39 iPSC lines. We also reported the CRISPR-Cas9-mediated genome-edited USH2A-RP-1 and USH2A-USH-1 lines that were corrected for the c.2276G>T and C.2299delG alleles, respectively.37 The iPSC lines USH2A-RP-3 and USH2A-USH-3 were newly generated in the framework of this study. The genome stability of the generated iPSC lines was analyzed by karyotype analysis (Chromostem, CHU Montpellier), as described previously.39 Pluripotency was determined using reverse-transcriptase PCR (RT-PCR), quantitative PCR (qPCR), immunofluorescence, and embryoid body assays as described previously without modification.40
DNA sequencing
Genomic DNA was isolated from USH2A-RP-3 and USH2A-USH-3 lines using the DNeasy Blood & Tissue Kit (QIAGEN) and PCR amplified using USH2A-specific primers (Table S1). Extra dNTPs were removed from the PCR reaction using the ExoSAP-IT PCR Clean-up Kit (GE Healthcare), and sequencing was performed using the BigDye Terminator Cycle Sequencing Ready Reaction Kit V3.1. The reaction was run on an Applied Biosystems 3130xL genetic analyzer.
iPSC-derived retinal organoid differentiation
Retinal organoids were differentiated from adherent iPSC cultures and, following excision, cultured as free-floating structures in DMEM/F12+GlutaMAX (Gibco) supplemented with taurine and retinoic acid, as described previously.28 A minimum of 5 differentiations were performed per line, and for each differentiation, retinal organoids were monitored using an Olympus CKX53 microscope. For analysis of brush border length, 5 measurements were performed and averaged per organoid.
qPCR studies
For the USH2A expression studies, RNA was extracted from iPSCs using the QIAShredder and RNeasy Mini Kits (QIAGEN) and treated with RNase-Free DNase (QIAGEN), and 500 ng was reverse transcribed using the Superscript III Reverse Transcriptase Kit (Thermo Fisher Scientific). qPCR amplification was performed using a 1/10 dilution of cDNA per reaction. Two individual clones per iPSC line were assayed in triplicate and the results averaged; three individual experiments were performed. For the iPSC-derived retinal organoids, RNA was extracted from either individual organoids or pools of 2 organoids using the NucleoSpin RNA Plus XS Kit (Macherey Nagel), and 200 ng was reverse transcribed. qPCR amplifications were performed using a 1/20 dilution of cDNA per reaction in triplicate. For all experiments, reactions were performed with the SensiFAST SYBR No-ROX (Meridian Bioscience) mix on a Roche LightCycler 480 II. Results were normalized to GAPDH expression levels.
Immunofluorescence studies
Human fibroblasts were seeded on glass coverslips at a density of 75,000 cells per 1.88 cm2 in supplemented AmnioMAX medium. The day after seeding, fetal bovine serum (FBS) was removed from the medium for 48 h, and the coverslips were fixed in ice-cold methanol at −20°C for 15 min. The fibroblasts were permeabilized using 0.1% Triton X-100 for 15 min at room temperature and blocked in 10% donkey serum (Millipore) and 1% bovine serum albumin (BSA; Sigma-Aldrich) at room temperature for 1 h. Generally, the iPSC-derived retinal organoids were fixed in 4% paraformaldehyde (PFA) at 4°C for 20 min, incubated in 30% sucrose in PBS at 4°C overnight, and embedded in Tissue-Tek O.C.T. compound (Sakura). 10-μm cryosections were blocked and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) in 10% donkey serum and 5% BSA at room temperature for 1 h. Primary antibodies (Table S2) were incubated at 4°C overnight in 1% donkey serum and 1% BSA (fibroblasts) or in 2% donkey serum, 1% BSA, and 0.1% Triton X-100 (retinal organoids). For CC antibodies, organoids were fixed in 0.5% methanol-free PFA (Pierce, Thermo Fisher Scientific) for 20 min at 4°C and incubated sequentially in 7.5%, 15%, and 30% sucrose in PBS prior to embedding. For all CC antibodies except ADGRV1, cryosections were permeabilized for 20 min with 1% Tween 20 in PBS and blocked for 1 h with blocking buffer (0.1% ovalbumin, 0.5% fish gelatine, and 1% Tween 20 in PBS). For ADGRV1, cryosections were permeabilized for 20 min with 0.01% Tween 20 in PBS and blocked for 1 h with blocking buffer (0.1% ovalbumin and 0.5% fish gelatine in PBS). Primary antibodies were diluted in blocking buffer and incubated overnight at 4°C. Secondary antibodies (Table S2) and 0.2 μg/mL Hoechst 33258 solution (Sigma-Aldrich) were incubated at room temperature for 1 h in their respective blocking buffers. All samples were mounted in fluorescent mounting medium (Dako, France). For antibodies against ciliary proteins except ADGRV1, sections were post fixed with 4% methanol-free PFA at room temperature for 10 min prior to mounting. For ADGRV1,41,42 the post-fixation step was performed in 1% methanol-free PFA at room temperature for 10 min prior to secondary antibody incubation. Samples processed without primary antibodies were used as negative controls. All samples were imaged using a Carl Zeiss ApoTome 2 upright wide-field microscope or a Carl Zeiss confocal LSM700 microscope. At least three different organoids from different differentiations were assayed for each antibody. For the quantification studies, one image was analyzed per organoid unless divergent phenotypes within an organoid were observed, in which case two images were analyzed.
Transmission electron microscopy
The iPSC-derived retinal organoids were processed and embedded as described previously without modification.28 Counterstained 70-nm sections were observed using a Tecnai F20 transmission electron microscope at 200 kV in the in-house CoMET facility.
Image quantification and statistical analyses
Quantification of primary cilium length in human fibroblasts and brush border length in retinal organoids was performed using ImageJ software (https://imagej.nih.gov/ij/). Quantification of the area of fluorescence in retinal organoids was performed using Imaris software (Bitplane). Quantification of fluorescent cones was performed using QuPath software.43 All data are presented as dot plots with bars to indicate the mean, and the sample size for each condition is shown in the bar or stated in the corresponding legend. Statistical analyses were performed using GraphPad Prism 8.3 software. Comparisons of 2 groups of data were performed using an unpaired Student’s t test.
Results
USH2A variants differentially affect ciliogenesis in non-syndromic RP and USH fibroblasts
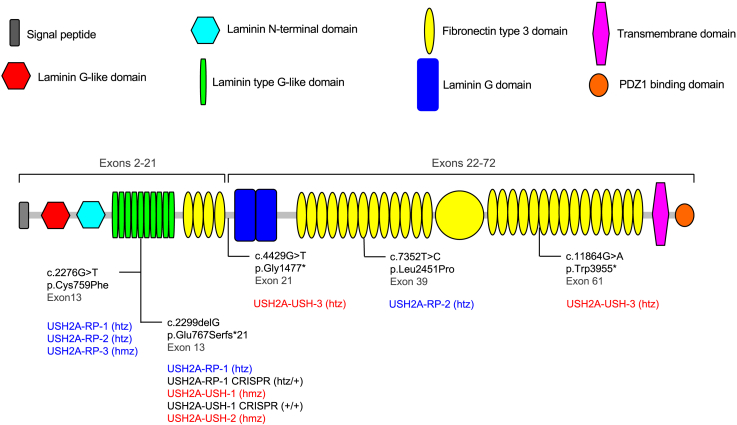
Either previously or as part of this study, we performed skin biopsies (Table S3) and isolated fibroblasts of three non-syndromic RP and three USH individuals carrying USH2A variants (Figure 1). The three RP individuals carried the recurrent exon 13 c.2276G>T missense variant in the heterozygous state with either c.2299delG (USH2A-RP-135) or the missense variant c.7352T>C in exon 39 (USH2A-RP-240), or in the homozygous state (USH2A-RP-3). By contrast, all the USH individuals carried loss-of-function variants. Two of the individuals (USH2A-USH-136 and USH2A-USH-2) carried the recurrent exon 13 c.2299delG variant in the homozygous state, whereas USH2A-USH-3 was compound heterozygous for two nonsense mutations, c.4429G>T in exon 21 and c.11864G>A in exon 61.
Figure 1.
Usherin protein structure and position of the variants in this study
Shown is a graphic representation of the usherin protein structure as determined using the SMART prediction tool (smart.embl-heidelberg.de). The signal peptide (gray), laminin G-like domain (red), laminin N-terminal domain (light blue), and the first four fibronectin type 3 domains (yellow) are encoded by USH2A exons 2–21. The laminin G domains (royal blue), the remainder of the fibronectin type 3 domains, the transmembrane domain (pink), and the C-terminal PDZ1 binding domain (orange) are encoded by exons 22–72. The USH2A variants carried by the individuals recruited for this study are indicated, along with their corresponding exonic position in black. The protein change for each mutation is also indicated. It is also stated whether the variants are carried in the compound heterozygous (htz) or homozygous (hmz) state by RP (blue) and USH (red) individuals. The genotypes of the non-syndromic RP and USH CRISPR-corrected iPSC clones are also indicated in black.
Because usherin is thought to be localized at the periciliary membrane of photoreceptors,17 we first investigated whether USH2A mutations affected ciliogenesis in serum-starved fibroblasts. Using immunofluorescence studies, we assayed the classic ciliary markers ARL13B, polyglutamylated tubulin (GT335), acetylated α-tubulin, and pericentrin (PCN). We observed a similar spatial distribution of the ciliary proteins between control and USH2A-RP and USH2A-USH cells; ARL13B (Figures 2A and 2B) extended the length of the ciliary shaft, GT335 (Figure 2B) and acetylated α-tubulin (Figure S1) labeled the axoneme, and PCN was found in the basal body (Figure 2A). We also assayed the localization of intraflagellar transport (IFT) particles using the marker IFT88 and observed a correct distribution that extended beyond acetylated α-tubulin at the apical and basal ends in control and USH2A cells (Figure S1).
Figure 2.
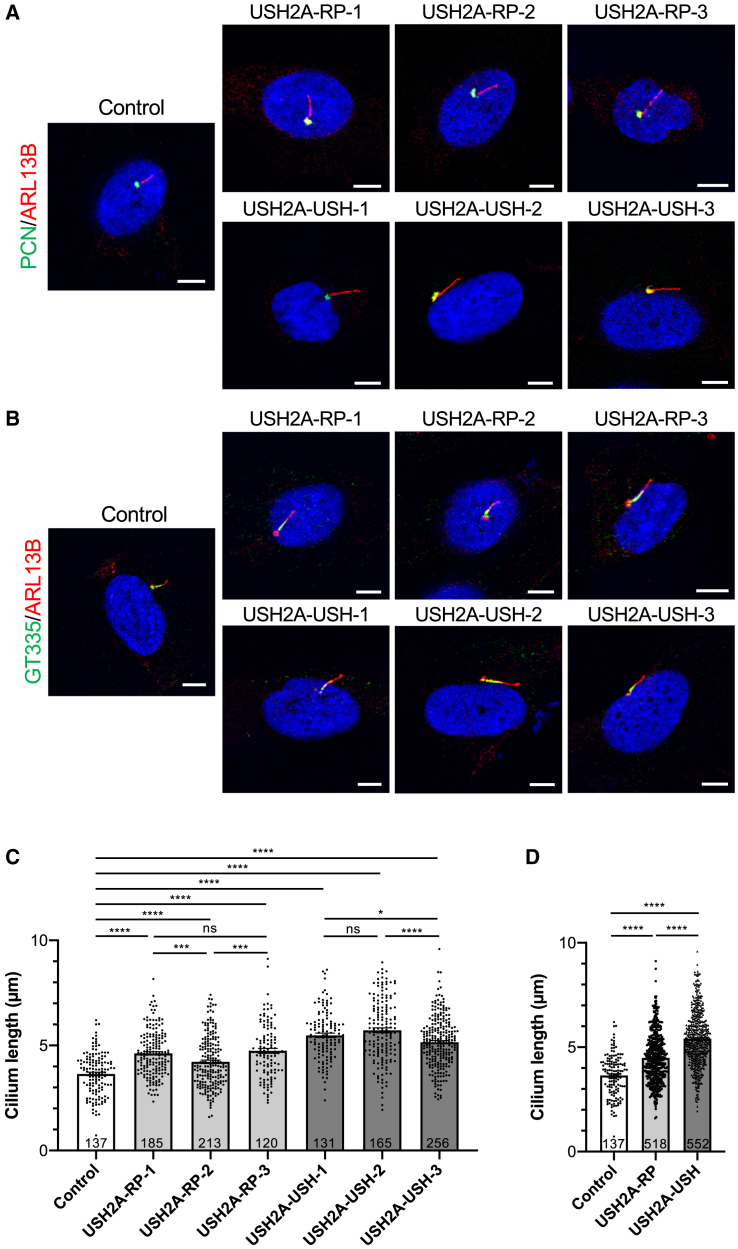
Ciliogenesis in human USH2A-RP and USH2A-USH fibroblasts
(A–C) IF staining of primary cilia and maximum-intensity plane confocal imaging of control, USH2A-RP, and USH2A-USH fibroblasts using antibodies against (A) PCN (green) and ARL13B (red) and (B) GT335 (green) and ARL13B (red). Scale bars, 5 μm.
(C) Length of the primary cilium of control (white) and individual USH2A-RP (light gray) and USH2A-USH (dark gray) fibroblasts. Data are represented as mean ± SEM, and the number of cells analyzed per group is indicated in the bars. ns, non-significant, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Student's t-test.
(D) Cilium length of combined USH2A-RP (light gray) and USH2A-USH (dark gray) values in comparison with controls (white). Data are represented as mean ± SEM, and sample sizes are indicated in the bars. ∗∗∗∗p < 0.0001, Student's t-test.
We then quantified the length of the ARL13B-labeled primary cilium in control and USH2A-RP and USH2A-USH cells. Overall, the ciliary length of the fibroblasts from each USH2A individual was significantly longer than that of the control cells (Figure 2C). On average, the length of the primary cilium of the combined USH2A-RP fibroblasts (4.5 ± 0.05 μm) was significantly (1.2-fold) longer than that of controls (3.7 ± 0.09 μm) (Figure 2D). Furthermore, the combined USH2A-USH fibroblasts displayed an even more pronounced elongated cilium (5.44 ± 0.066 μm), which was significantly (1.55-fold and 1.2-fold) longer than that of control and USH2A-RP fibroblasts, respectively.
Taken together, these results suggest defective ciliogenesis associated with USH2A mutations that is more pronounced in fibroblasts from USH than non-syndromic RP individuals.
USH2A variants differentially affect USH2A expression in non-syndromic RP and USH iPSCs
To validate our previous observations concerning differential USH2A mRNA levels in RP and USH iPSCs,37 we generated pluripotent and genetically stable iPSC lines for the additional individuals USH2A-RP-2,40 USH2A-RP-3 (Figure S2), and USH2A-USH-3 (Figure S3); we did not reprogram the fibroblasts of USH2A-USH-2 into iPSCs because they had a genotype identical to USH2A-USH-1 (Figure 1). We then assayed USH2A expression in two clones of each iPSC line and observed slightly increased levels in USH2A-RP-1 iPSC clones compared with controls (Figure 3A). The USH2A levels in USH2A-RP-2 and USH2A-RP-3 clones were not significantly different from controls. Between the RP lines, the USH2A levels in USH2A-RP-1 and USH2A-RP-3 iPSCs were significantly higher than that of USH2A-RP-2. Consistent with our previous data, USH2A expression levels in USH2A-USH-1 clones were significantly (6-fold) higher than that of controls. Similarly, USH2A expression levels in USH2A-USH-3 clones were significantly (4-fold) higher than that of controls. No significant differences were detected between the USH lines. More globally, when grouped by disease phenotype, the USH2A levels in the iPSC lines of non-syndromic RP individuals were not significantly different from controls, whereas those of USH individuals were significantly (5- and 3.5-fold) higher than those of the control and USH2A-RP lines, respectively (Figure 3B).
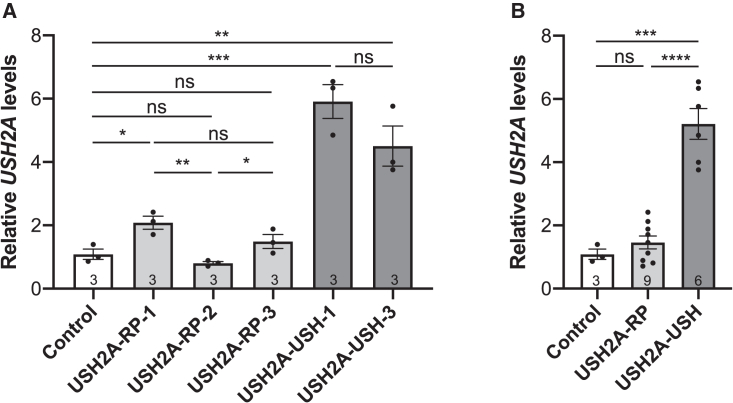
Figure 3.
USH2A levels in USH2A-RP and USH2A-USH iPSC lines
(A) qPCR analysis of USH2A expression levels in control, USH2A-RP, and USH2A-USH iPSCs using primers specific to exon 39. Data are represented as mean ± SEM; n = 3 independent experiments. ns, non-significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Student's t-test.
(B) Mean USH2A levels of combined USH2A-RP (light gray) and USH2A-USH (dark gray) values in comparison with controls (white). Data are represented as mean ± SEM, and sample sizes are indicated in the bars. ns, non-significant, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Student’s t test.
In conclusion, these data confirm our previous observations that USH2A expression levels are elevated in iPSC lines from USH but not RP individuals.
USH2A variants differentially affect outer segment elongation in RP and USH retinal organoids
Having observed a genotype-phenotype correlation for USH2A variants linked with non-syndromic RP or USH in fibroblasts and iPSCs, we differentiated iPSC-derived retinal organoids from the USH2A-RP-1 and USH2A-USH-1 lines. In parallel, we differentiated their isogenic control iPSC lines (Figure 1), which we generated previously.37 We obtained retinal organoids for all lines; however, efficiencies varied depending on the cell line, as has been well documented.44,45,46
We first looked at brush border formation, which represents the inner segment (IS)- and outer segment (OS)-like structures of rod and cone photoreceptors,28 in mid-stage organoids on day 150 of differentiation (Figures 4A, 4B, and 4E). We observed a shorter or sometimes absent brush border in USH2A-RP-1 organoids (Figure 4A), which was significantly (3-fold) shorter (7.2 ± 1 μm) compared with control (22.1 ± 1 μm) organoids (Figure 4E). Moreover, the brush border of USH2A-RP-1 organoids was also significantly (2.4-fold) shorter than that of USH2A-RP-1 CRISPR-corrected organoids (17 ± 1.7 μm). Although, at first glance, we did not observe a clear difference in brush border formation in USH2A-USH-1 organoids compared with control organoids (Figure 4A), the USH2A-USH-1 brush border was significantly (1.2-fold) shorter (17.8 ± 0.7 μm). By contrast, it was not significantly different from that of isogenic USH2A-USH-1 CRISPR-corrected organoids (21.5 ± 2.4 μm) (Figure 4E). To confirm these differences, we performed immunofluorescence (IF) staining with the pan-photoreceptor marker rhodopsin kinase (RK) and the outer limiting membrane (OLM) marker ZO-1 (Figure 4B). RK and ZO-1 expression was observed for all retinal organoids, but differences in the extension of RK staining beyond the OLM of the developing IS- and OS-like structures were exclusively visible for USH2A-RP-1 organoids, consistent with the brush border measurements.
Figure 4.
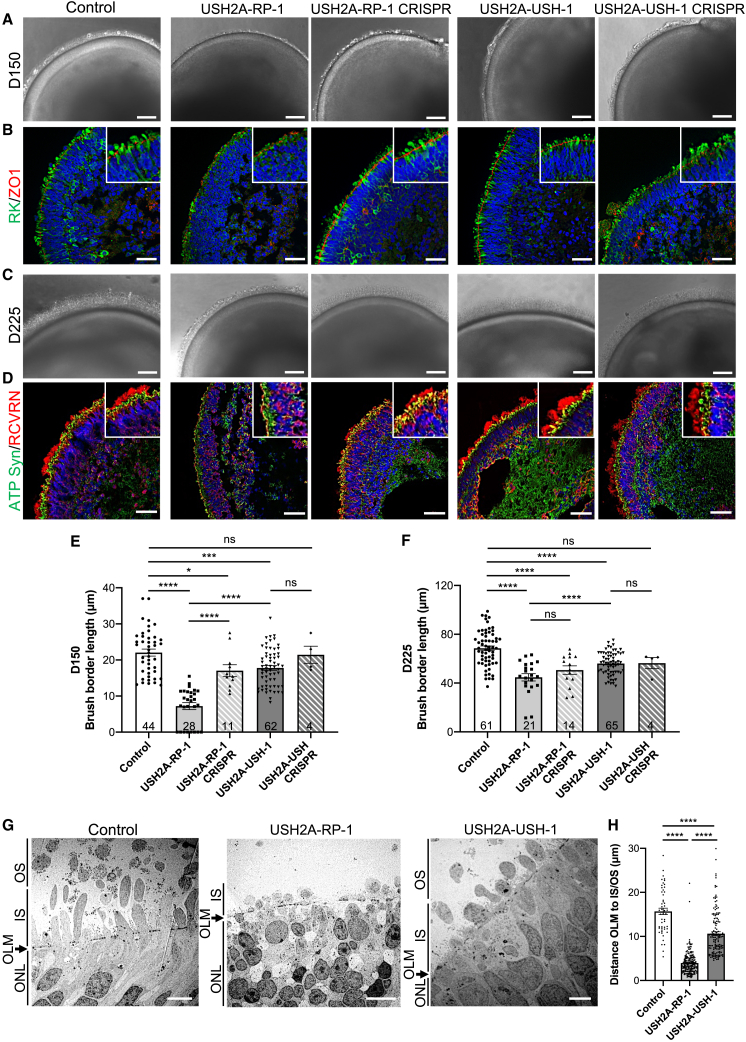
Morphological characteristics of human USH2A-RP and USH2A-USH iPSC-derived retinal organoids
(A) Bright-field microscopy of control, affected (USH2A-RP-1 and USH2A-USH-1), and isogenic CRISPR-corrected (USH2A-RP-1 CRISPR and USH2A-USH-1 CRISPR) retinal organoids on day 150 (D150) of differentiation. Scale bars, 100 μm.
(B) IF of day 150 retinal organoids using antibodies directed to rhodopsin kinase (RK; green) and ZO1 (red). Insets: higher magnifications. Scale bars, 30 μm.
(C) Bright-field microscopy of control, affected, and isogenic CRISPR-corrected retinal organoids on day 225. Scale bars 100 μm.
(D) IF of day 225 (D225) retinal organoids using antibodies directed to ATP synthase (ATP Syn; green) and recoverin (RCVRN; red). Insets: higher magnifications. Scale bars, 50 μm.
(E and F) Brush border length of control, affected (USH2A-RP-1 and USH2A-USH-1), and isogenic CRISPR-corrected (USH2A-RP-1 CRISPR and USH2A-USH-1 CRISPR) retinal organoids on day 150 (E) and day 225 (F). Data are represented as mean ± SEM, and the number of organoids analyzed is indicated in the bars. ns, non-significant, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Student’s t test.
(G) TEM images of control and affected organoids on day 225, showing the position of the ONL, OLM, IS-, and/or OS-like structures. Scale bars, 5 μm (control and USH2A-USH-1) and 10 μm (USH2A-RP-1).
(H) Distance measured from the OLM to the farthest IS- (USH2A-RP-1) or OS-like (control and USH2A-USH-1) structure.
Data are represented as mean ± SEM. n = 51 (control), 182 (USH2A-RP-1), and 146 (USHA-USH-1) measurement. ∗∗∗∗p < 0.0001, Student’s t test.
We then assayed the brush border in retinal organoids on day 225 (Figures 4C, 4D, and 4F) and observed an increase in length for all organoids (Figure 4C) compared with day 150 (Figure 4A). However, the brush border of control organoids (68.6 ± 1.9 μm) was significantly longer (on average 1.4-fold) than that of USH2A-RP-1 (44.8 ± 3 μm), USH2A-RP-1 CRISPR-corrected (50.7 ± 3.5 μm), and USH2A-USH-1 (56.1 ± 1.2 μm) organoids (Figure 4F). By contrast, no significant difference was observed with the USH2A-USH-CRISPR organoids (56.4 ± 4.5 μm). IF studies for expression of the IS marker ATP synthase and the pan-photoreceptor marker recoverin (RCVRN) again revealed a notable difference in the extension of IS- and OS-like structures in USH2A-RP-1 organoids compared with the other organoids (Figure 4D). In addition, although we did not observe a complete distinction in IS and OS staining with each marker in USH2A-RP-1 CRISPR-corrected organoids, as could be seen with controls, we did observe an improvement in OS formation compared with USH2A-RP-1 organoids. No clear differences in the staining profiles were observed between control, USH2A-USH-1, and USH2A-USH-1 corrected organoids.
To further investigate the differences in formation of IS- and OS-like structures, we analyzed the ultrastructure of photoreceptors in RP and USH organoids on day 225 using transmission electron microscopy (TEM) (Figure 4G). The control organoids had IS- and OS-like structures that extended well beyond the OLM, whereas USH2A-RP-1 organoids had small IS protrusions but no evident OS-like structures. By comparison, USH2A-USH-1 organoids presented with IS- and OS-like structures extending beyond the OLM, although not as far as those of control organoids. These observations were further confirmed quantitatively by measuring the distance from the OLM to the end of the farthest detectable IS (in the case of USH2A-RP-1) or OS-like (control and USH2A-USH-1) structures in all organoids (Figure 4H). In control organoids, this distance was, on average, 15.7 ± 0.7 μm, whereas it was significantly (4-fold) shorter in USH2A-RP-1 (4 ± 0.2 μm) and 1.5-fold shorter in USH2A-USH-1 (10.6 ± 0.4 μm) organoids. Furthermore, there was also a significant 2.7-fold difference between USH2A-RP-1 and USH2A-USH-1 organoids.
Taken together, the USH2A-mutant organoids present differences in IS/OS formation and/or elongation that are more pronounced in organoids from RP than USH individuals.
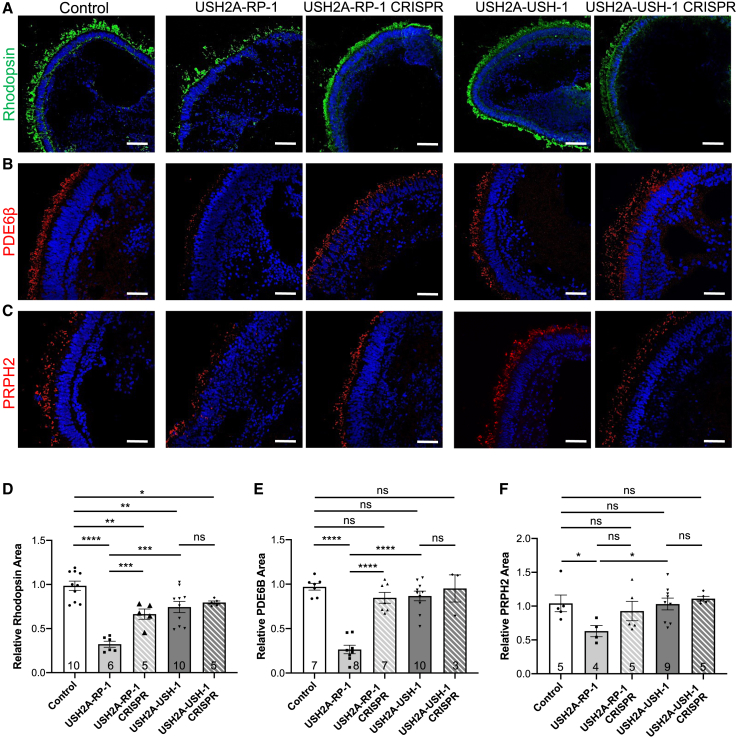
USH2A variants preferentially affect rod photoreceptors in non-syndromic RP organoids
Because we observed differences in photoreceptor IS/OS formation and/or elongation in USH2A organoids, we next specifically examined the rod photoreceptor markers rhodopsin (Figure 5A), PDE6β (Figure 5B), and PRPH2 (Figure 5C) in day 225 retinal organoids. By IF of control organoids, rhodopsin staining could be seen throughout photoreceptor cells but primarily in the surrounding brush border (Figure 5A). We observed a similarly distributed but less homogeneous rhodopsin profile in USH2A-RP-1 organoids. Importantly, in CRISPR-corrected USH2A-RP-1 organoids, this phenotype was reversed, and more regular rhodopsin staining was observed. To validate these observations, we measured the area of rhodopsin staining as a function of Hoechst staining in the outer nuclear layer (ONL) and observed a significantly smaller (3- and 2-fold) area of rhodopsin staining in USH2A-RP-1 organoids compared with control and CRISPR-corrected organoids, respectively (Figure 5D). Interestingly, the area of rhodopsin staining in isogenic USH2A-RP-1 CRISPR-corrected organoids was also significantly smaller (1.5-fold) than in control organoids. By comparison, the rhodopsin staining of USH2A-USH-1 and isogenic CRISPR-corrected organoids was more homogeneous than that of USH2A-RP-1 organoids (Figure 5A), but the area was significantly smaller (1.3- and 1.2-fold) than in control organoids, respectively (Figure 5D).
Figure 5.
Rods in human USH2A-RP and USH2A-USH iPSC-derived retinal organoids
(A–C) IF of the rod markers rhodopsin (A), PDE6β (B), and PRPH2 (C) in control, affected (USH2A-RP-1 and USH2A-USH-1), and isogenic CRISPR-corrected (USH2A-RP-1 CRISPR and USH2A-USH-1 CRISPR) retinal organoids on day 225 of differentiation. Scale bars, 100 μm (rhodopsin) and 50 μm (PDE6β and PRPH2).
(D–F) Area of rhodopsin (D), PDE6β (E), and PRPH2 (F) staining in control, affected, and isogenic CRISPR-corrected retinal organoids on day 225 and normalized to controls.
Data are represented as mean ± SEM, and the number of images analyzed is indicated in the bars. ns, non-significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Student’s t test.
We then specifically studied rod photoreceptor OS-like structures using PDE6β as a marker. We detected specific PDE6β staining of the brush border of control organoids that appeared similar to that in USH2A-RP-1 CRISPR-corrected, USH2A-USH-1, and USH2A-USH-1 CRISPR-corrected organoids. PDE6β staining, however, was visibly reduced in USH2A-RP-1 organoids (Figure 5B), similar to the observations for rhodopsin. Quantification of the area of PDE6β staining as a function of Hoechst staining further confirmed these results. Only USH2A-RP-1 organoids showed a significantly lower (3.7- and 3.2-fold) level of PDE6β staining compared with control and isogenic-corrected organoids, respectively (Figure 5E). By contrast, PDE6β staining in USH2A-USH-1 and USH2A-USH-1 CRISPR-corrected organoids was again at levels similar to that of the USH2A-RP-1 CRISPR-corrected organoids and not significantly different from controls. Although less striking, a similar profile was observed for the rod OS PRPH2 (Figure 5C), where a significant difference in the area of PRPH2 staining was only detected for USH2A-RP-1 organoids compared with control organoids (Figure 5F).
Taken together, organoids from a non-syndromic RP individual showed an altered rod photoreceptor phenotype, which was improved upon CRISPR correction, whereas those from an USH individual had a phenotype that was relatively similar to that of controls.
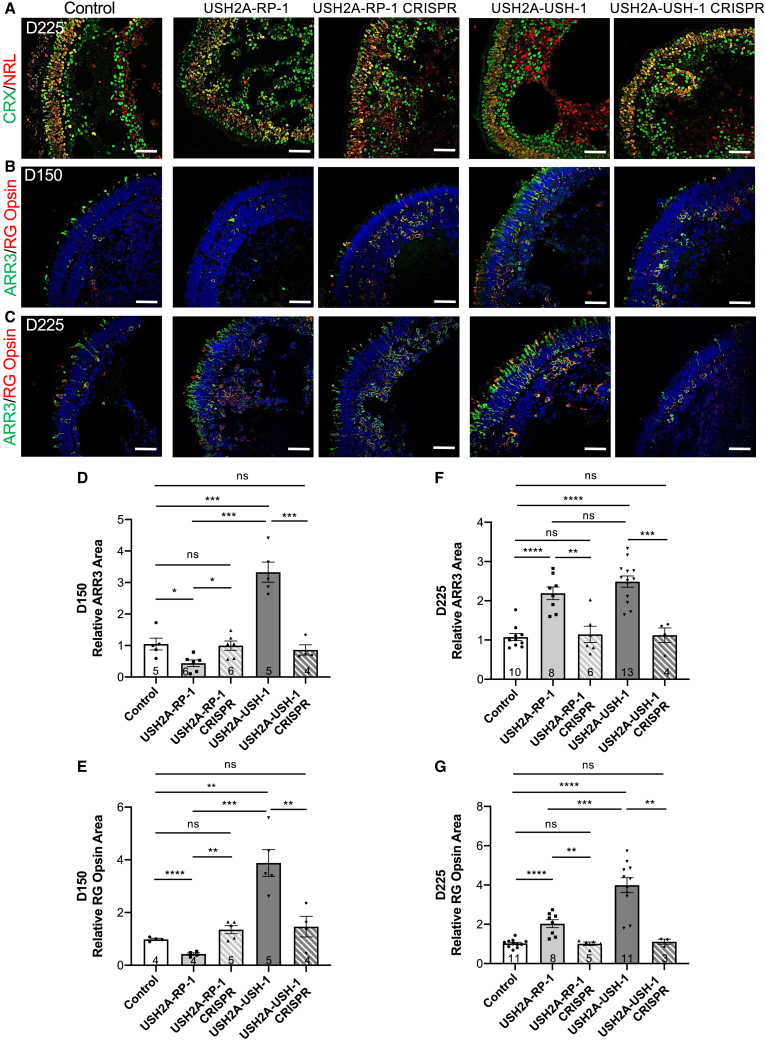
USH2A variants preferentially affect cone photoreceptors in USH organoids
Because of the differential rod phenotype in USH2A mutant organoids, we then specifically analyzed cone photoreceptors. We first tested day 225 retinal organoids by IF for expression of two transcription factors partners regulating cone/rod fate: the neural retina-specific leucine zipper protein (NRL), which is restricted to rod nuclei, and the cone-rod homeobox protein (CRX), which labels cone and rod nuclei. Analysis of control organoids demonstrates clear labeling of a structured ONL at the outer edge (Figure 6A). A similar structure was seen for all affected and isogenic control organoids except for USH2A-RP-1 organoids. Furthermore, in all organoids, we could distinguish that the majority of the nuclei were co-labeled for CRX and NRL, identifying them as rods, except for some nuclei at the outer edge that were exclusively labeled by CRX, identifying them as cones. Interestingly, a well-defined outer rim of exclusively CRX-positive nuclei was noted in USH2A-RP-1 and USH2A-USH-1 organoids, which was absent in control or the CRISPR-corrected organoids. We therefore quantified the number of CRX-positive/NRL-negative nuclei per number of Hoechst-stained nuclei in the ONL and observed a significantly increased number of cones in USH2A-RP-1 and USH2A-USH-1 organoids compared with control or CRISPR-corrected organoids (Figure S4A).
Figure 6.
Cones in human USH2A-RP and USH2A-USH iPSC-derived retinal organoids
(A) IF of the transcription factors CRX (green) and NRL (red) in control, affected (USH2A-RP-1 and USH2A-USH-1), and isogenic CRISPR-corrected (USH2A-RP-1 CRISPR and USH2A-USH-1 CRISPR) retinal organoids on day 225 (D225) of differentiation.
(B and C) IF of ARR3 (green) and RG opsin (red) in day 150 (B) and day 225 (C) retinal organoids. Scale bars, 50 μm.
(D and E) Area of ARR3 (D) and RG opsin (E) staining in control, affected, and isogenic CRISPR-corrected retinal organoids on day 150 and normalized to controls. Data are represented as mean ± SEM, and the number of images analyzed is indicated in the bars. ns, non-significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Student’s t test.
(F and G) Area of ARR3 (F) and RG opsin (G) staining in retinal organoids on day 225 and normalized to controls. Data are represented as mean ± SEM, and the number of images analyzed is indicated in the bars. ns, non-significant, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Student’s t test.
We therefore further analyzed retinal organoids for the cone-specific markers cone arrestin-3 (ARR3) and red/green (RG) opsin) on day 150 (Figure 6B) and day 225 (Figure 6C). On day 150 in control organoids, we clearly observed ARR3-positive cones with IS/OS-like structures extending beyond the ONL (Figure 6B). Only a small number expressed the more mature marker RG opsin. Cones were barely detectable in USH2A-RP-1 organoids; however, following CRISPR correction, ARR3-positive and ARR3-positive/RG opsin-positive cones were again clearly visible throughout the ONL in these isogenic controls. Unexpectedly, for USH2A-USH-1 organoids, we observed an excessive amount of cone photoreceptors in the ONL compared with control or CRISPR-corrected isogenic organoids, and many were also labeled for RG opsin. This tendency was further confirmed on day 225, where the ONL of USH2A-USH-1 organoids contained a large quantity of highly structured cones compared with control or CRISPR-corrected organoids (Figure 6C). Furthermore, the majority of these cones were also positive for RG opsin, suggesting a higher degree of maturation. Surprisingly, on day 225, there was a striking increase in the amount of cones for USH2A-RP-1 organoids compared with control and CRISPR-corrected organoids, although only a small proportion was co-labeled with RG opsin. However, neither the ONL nor the cones were as nicely structured as in the control, CRISPR-corrected, or even USH2A-USH-1 organoids.
To validate these observations, we quantified the area of ARR3 and RG opsin fluorescence relative to Hoechst staining in the ONL (Figures 6D–6G). We detected no significant differences between control or CRISPR-corrected isogenic organoids regardless of marker or time point. By contrast, on day 150, the area of ARR3 staining was significantly decreased by 2.4-fold in USH2A-RP-1 organoids and significantly increased by 3.2-fold in USH2A-USH-1 organoids compared with controls (Figure 6D). Similarly, the area of RG opsin staining was significantly decreased by 2.3-fold in USH2A-RP-1 and significantly increased by 4-fold in USH2A-USH-1 organoids compared with controls (Figure 6E). On day 225, the tendency for USH2A-RP-1 organoids had reversed, as suggested by IF images. The area of ARR3 staining was significantly increased by 2- and 2.3-fold in USH2A-RP-1 and USH2A-USH-1 organoids, respectively (Figure 6F). Similarly, the area of RG opsin staining was significantly increased by 2- and 3.9-fold (Figure 6G) in USH2A-RP-1 and USH2A-USH-1 organoids, respectively. We further confirmed these observations by counting the number of ARR3-positive or RG opsin-positive cones per number of Hoechst-stained nuclei in the ONL at both time points and observed identical profiles (Figures S4B–S4E). Last, we assayed expression of the corresponding genes ARR3 (Figure S4H) and OPN1MW (Figure S4I) on days 150 and 225 by qPCR. In day 150 organoids, a significant increase in ARR3 expression was observed in USH2A-USH-1 organoids compared with controls. Although a tendency of increased levels was also detected on day 225, high variations in expression between organoids made it difficult to confirm (Figure S4H). By contrast, a significant increase in OPN1MW expression was observed in USH2A-USH-1 organoids on day 150 and day 225 compared with control and USH2A-RP-1 organoids (Figure S4I), consistent with the area of RG opsin staining following IF analysis (Figure 6G).
To conclude, there is a clear differential cone phenotype, in terms of number and structure, in organoids from non-syndromic RP and USH individuals.
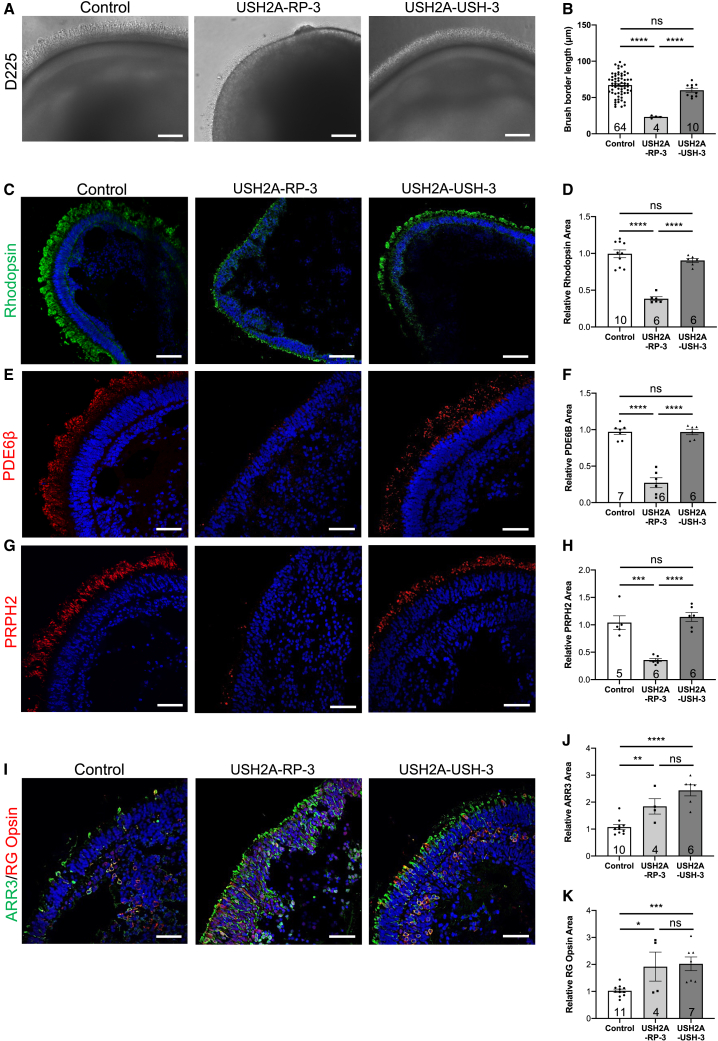
Differential RP and USH disease phenotypes in organoids of additional individuals
Because a differential RP or USH phenotype as a result of USH2A mutations in the retina had not been reported previously, we wanted to confirm these phenotypes in organoids of additional individuals carrying other variant combinations. We therefore generated retinal organoids from an additional RP (USH2A-RP-3) and an additional USH (USH2A-USH-3) individual, which were studied on day 225. Similar to our aforementioned observations, USH2A-RP-3 organoids showed a shorter and sometimes absent non-homogeneous brush border compared with controls, whereas USH2A-USH-3 organoids showed a similar brush border length (Figure 7A). Quantification of the brush border length confirmed a significant (3- and 2.6-fold) decrease in USH2A-RP-3 organoids (23.3 ± 0.9 μm) compared with control (67.2 ± 2 μm) and USH2A-USH-3 (60 ± 2.8 μm) organoids, respectively (Figure 7B). We then checked the rod-specific markers by IF and again observed a reduced area of rhodopsin (Figure 7C), PDE6β (Figure 7E), and PRPH2 (Figure 7G) staining in USH2A-RP-3 organoids compared with control and USH2A-USH-3 organoids. For rhodopsin, we detected a significant 2.6- and 2.3-fold lower area of staining in USH2A-RP-3 organoids compared with control and USH2A-USH-3 organoids, respectively (Figure 7D). Similarly, for PDE6β, we detected significantly (3.6-fold) smaller area of staining in USH2A-RP-3 organoids compared with control and USH2A-USH-3 organoids (Figure 7F). Last, for PRPH2, we observed a significantly (2.9- and 3.2-fold) smaller area of staining in USH2A-RP-3 organoids compared with control and USH2A-USH-3 organoids, respectively (Figure 7H).
Figure 7.
Phenotype of day 225 USH2A-RP-3 and USH2A-USH-3 iPSC-derived retinal organoids
(A) Bright-field microscopy of control, USH2A-RP-3, and USH2A-USH-3 retinal organoids. Scale bars, 100 μm.
(B) Brush border length of control, USH2A-RP-3, and USH2A-USH-3 organoids. Data are represented as mean ± SEM, and the number of images analyzed is indicated in the bars. ns, non-significant, ∗∗∗∗p < 0.0001, Student’s t test.
(C, E, and G) IF of the rod markers rhodopsin (C), PDE6β (E), and PRPH2 (G) in control, USH2A-RP-3, and USH2A-USH-3 retinal organoids. Scale bars, 100 μm (rhodopsin) and 50 μm (PDE6β and PRPH2).
(D, F, and H) Area of rhodopsin (D), PDE6β (F), and PRPH2 (H) staining in retinal organoids normalized to controls. Data are represented as mean ± SEM, and the number of images analyzed is indicated in the bars. ns, non-significant, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Student’s t test.
(I) IF of the cone markers ARR3 (green) and RG opsin (red) in control, USH2A-RP-3, and USH2A-USH-3 retinal organoids. Scale bars, 50 μm.
(J and K) Area of ARR3 (J) and RG opsin (K) staining in retinal organoids and normalized to controls. Data are represented as mean ± SEM, and the number of images analyzed id indicated in the bars. ns, non-significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Student’s t test.
We then assayed the cone-specific markers by IF and again observed increased staining of ARR3 in USH2A-RP-3 and USH2A-USH-3 organoids compared with control organoids (Figure 7I). Furthermore, in USH2A-USH-3 organoids, many of the ARR3-positive cones appeared to be co-labeled with RG opsin, whereas, in USH2A-RP-3 organoids, some co-localization could be seen, but RG opsin seemed to be mostly mislocalized to the nuclei, suggestive of immature cones. Quantification demonstrated a 1.7-fold and 2.3-fold increase in the area of ARR3 and a 1.9- and 2-fold increase in the area of RG opsin staining in USH2A-RP-3 and USH2A-USH-3 organoids, respectively, compared with control organoids. These results were again confirmed by counting cones labeled for ARR3 (Figure S4F) and RG opsin (Figure S4G) per Hoechst-labeled nuclei in the ONL as similar profiles were observed.
Thus, taken together, the differential rod and cone phenotypes associated with non-syndromic RP and USH organoids are not individual or mutation dependent but disease dependent.
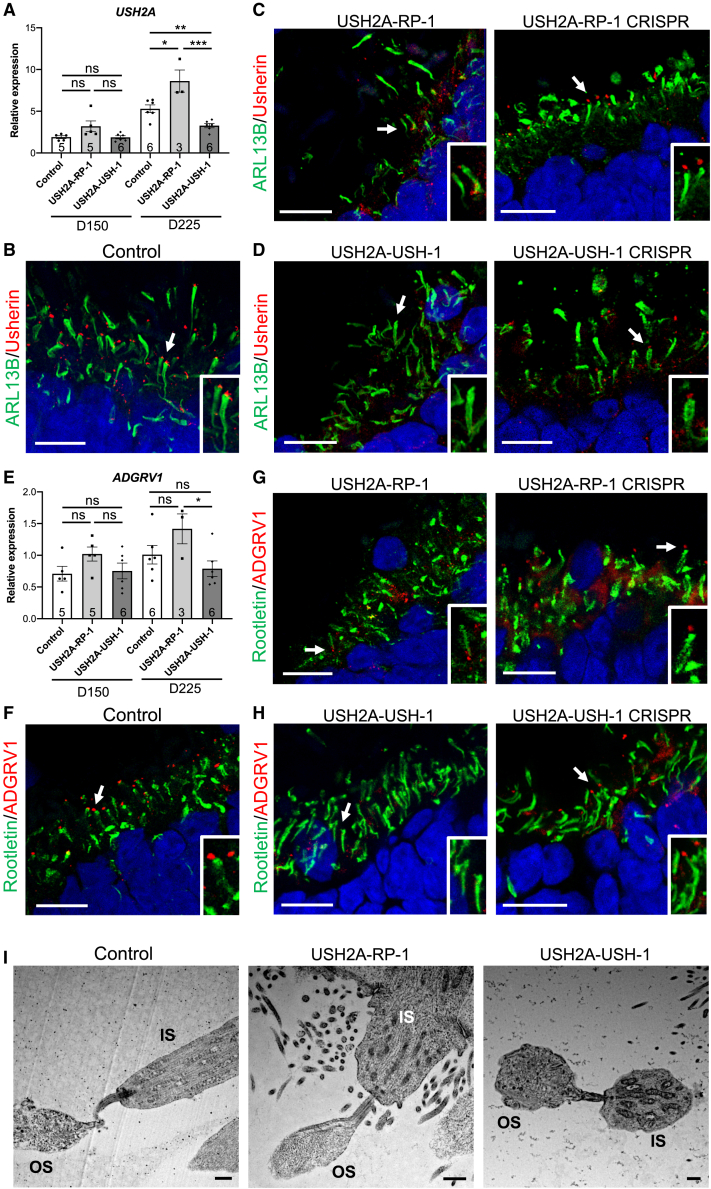
USH2A variants differentially affect human usherin expression
In addition to disease modeling, generation of these mature organoids for USH2A also provided the unique opportunity to study usherin expression and localization in control and diseased human retina. As a first screen, we analyzed USH2A expression in human retinal organoids by qPCR on day 150 and day 225 (Figure 8A). On day 150, USH2A levels in the USH2A-RP-1 and USH2A-USH-1 lines were not significantly different from controls, although USH2A-RP-1 showed a tendency of increased levels. On day 225, USH2A levels were significantly increased in the USH2A-RP-1 line compared with controls, whereas the levels in USH2A-USH-1 were significantly decreased. Surprisingly, this profile was in contrast to that observed in the iPSC lines. We therefore assayed usherin in day 225 retinal organoids using an anti-usherin antibody that was directed to a C-terminal epitope. In control organoids, we detected punctate usherin staining in the basal body region. More precisely, the signal was situated at the tip of the ciliary rootlet, which projects into the IS and was labeled by an anti-ARL13B antibody (Figure 8B). It should be noted that the staining of ARL13B co-localized with that of the well-defined ciliary rootlet marker rootletin47 (Figure S5A). In USH2A-RP-1 organoids, less usherin staining was detectable at the tip of the ciliary rootlet, whereas mislocalized usherin could be detected near the ONL (Figure 8C). By contrast, in isogenic CRISPR-corrected organoids, we observed correct usherin localization at the tip of the ciliary rootlet. Consistent with the decreased USH2A expression in the qPCR data, usherin was not detectable in USH2A-USH-1 organoids, whereas usherin was restored to the tip of the ciliary rootlet in the isogenic controls (Figure 8D). Last, we tested the localization of usherin in parallel to CEP290, which is present in the basal body region, and consistently observed adjacent usherin and CEP290 signals in wild-type and CRISPR-corrected RP and USH organoids, whereas in USH2A-RP-1 and USH2A-USH-1 organoids, CEP290 staining was isolated (Figure S5B).
Figure 8.
Usherin and ADGRV1 expression in USH2A-USH and USH2A-RP retinal organoids
(A) qPCR analysis of USH2A expression in control, USH2A-RP-1, and USH2A-USH-1 retinal organoids on day 150 and day 225. Data are represented as mean ± SEM, and the number of organoids analyzed is indicated in the bars. ns, non-signficant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Student’s t test.
(B–D) IF of ARL13B (green) and usherin (red) in control (B), USH2A-RP-1 and CRISPR-corrected isogenic control (C), and USH2A-USH-1 and CRISPR-corrected isogenic control (D) retinal organoids on day 225. Scale bars, 10 μm. Insets: magnification of the cilia, indicated by arrows, in each type of organoid.
(E) qPCR analysis of ADGRV1 expression in control, USH2A-RP-1, and USH2A-USH-1 retinal organoids on day 150 and day 225. Data are represented as mean ± SEM, and the number of organoids analyzed is indicated in the bars. ns, non-significant, ∗p < 0.05, Student's t-test.
(F–H) IF studies of Rootletin (green) and ADGRV1 (red) in control (F), USH2A-RP-1 and CRISPR-corrected isogenic control (G), and USH2A-USH-1 and CRISPR-corrected isogenic control (H) retinal organoids on day 225. Scale bars, 10 μm. Insets: magnification of the cilia, indicated by arrows, in each type of organoid.
(I) TEM images of the CC linking the IS- and OS-like structures in control, USH2A-RP-1, and USH2A-USH-1 retinal organoids. Scale bars, 1 μm.
To further validate these results, we assayed the expression of an usherin partner, ADGRV1 (USH2C; MIM: 602851), by qPCR. Interestingly, ADGRV1 levels in the USH2A-RP-1 and USH2A-USH1 lines showed the same profiles as for USH2A in day 150 and day 225 organoids, but because of higher variability in expression between organoids, the differences were not significant (Figure 8E). We thus assayed the localization of the protein ADGRV1 in day 225 organoids by IF studies. In control organoids, ADGRV1 was localized to the tip of the ciliary rootlet labeled by an anti-rootletin antibody (Figure 8F), similar to usherin. Furthermore, ADGRV1 was again detectable but mislocalized in USH2A-RP-1 organoids (Figure 8G) compared with isogenic CRISPR-corrected organoids. Interestingly, ADGRV1 was not detectable in USH2A-USH-1 organoids (Figure 8H), whereas it was correctly localized to the tip of the ciliary rootlet in the isogenic controls. We further confirmed the presence of a CC by TEM (Figure 8I), which could be detected linking an IS- to an OS-like structure in control and USH2A-USH-1 organoids.
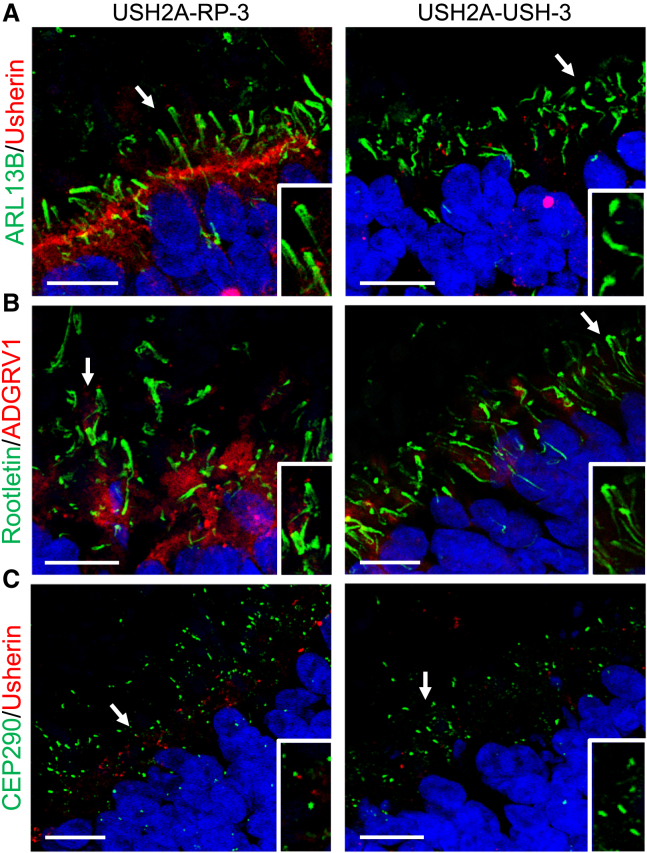
To finish, we assayed the localization profiles of ARL13B/Usherin (Figure 9A), rootletin/ADGRV1 (Figure 9B), and CEP290/usherin (Figure 9C) in the retinal organoids of USH2A-RP-3 and USH2A-USH-3. Consistently, we observed reduced and mislocalized usherin and ADGRV1 staining in USH2A-RP-3 organoids and the absence of staining in USH2A-USH-3 organoids. Interestingly, the usherin and ADGRV1 signals at the tip of the ciliary rootlet for USH2A-RP-3 organoids, which are homozygous for c.2276G>T, were more visible than for USH2A-RP-1 organoids, which are compound heterozygous c.2276G>T/c.2299delG, even though they were still less evident than in control organoids (Figures 8B and 8F).
Figure 9.
Ciliary markers in day 225 USH2A-RP-3 and USH2A-USH-3 iPSC-derived retinal organoids
(A–C) IF studies of ARL13B (green) and usherin (red) (A), rootletin (green) and ADGRV1 (red) (B), and CEP290 (green) and usherin (red) (C) in USH2A-RP-3 and USH2A-USH-3 retinal organoids. Scale bars, 10 μm. Insets: magnification of the cilia, indicated by arrows, in each type of organoid.
In conclusion, usherin and its partner ADGRV1 are partially mislocalized in organoids from non-syndromic RP individuals but absent in organoids from USH individuals.
Differential retinal anomalies in RP and USH individuals
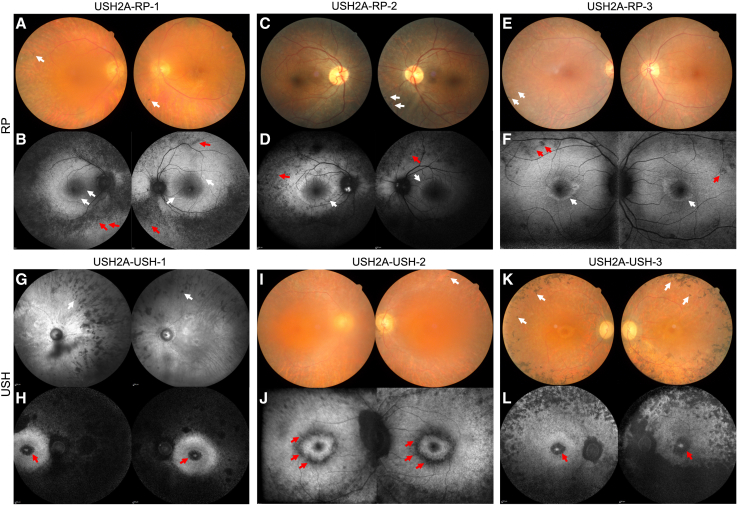
Combined, our data strongly suggest that, in retinal organoids, cones are differentially affected according to non-syndromic RP versus USH phenotypes. We thus reappraised the six affected individuals included in this study to determine whether this differential cone involvement translated into a clinical difference in the RP, either isolated or as part of USH type 2. The USH2A-RP-1 (Figures 10A and 10B), USH2A-RP-2 (Figures 10C and 10D), and USH2A-RP-3 (Figures 10E and 10F) individuals were last seen at the ages of 73, 39, and 47 years, respectively. All three showed classic signs of RP, such as alterations in peripheral pigmentation, thinning of the retinal vessels, and pallor of the optic nerve on color fundus images (Figures 10A, 10C, and 10E). Furthermore, a characteristic hyperautofluorescent ring was detected by short-wave fundus autofluorescence (Figures 10B, 10D and 10F), a technique highly sensitive for detecting small areas of atrophy. There were no macular hypoautofluorescent (dark or black) lesions at the inner or outer boundaries of the hyperautofluorescent ring (white) in any of these three individuals, indicating that they had not developed macular atrophy. Consistently, their visual acuity was relatively preserved with respect to age (Table S3).
Figure 10.
Multimodal imaging of USH2A individuals included in the study
(A, C, and E) Imaging of non-syndromic RP individuals. Shown are color fundus images of USH2A-RP-1 (A) at 73 years, USH2A-RP-2 (C) at 39 years, and (E) USH2A-RP-3 at 47 years of age. Note typical pigmentary changes of the peripheral retina (white arrows).
(B, D, and F) Short-wave fundus autofluorescence images of USH2A-RP-1 (B), USH2A-RP-2 (D), and USH2A-RP-3 (F), showing a large annular perifoveal ring (white arrows) and peripheral hypoautofluorescent dots (red arrows). Note the absence of macular atrophy.
(G–L) Imaging of USH individuals. (G) Infrared fundus images of USH2A-USH-1 at 64 years of age. (I and K) Color fundus images of USH2A-USH-2 (I) at 42 years and USH2A-USH-3 (K) at 48 years of age. Note typical pigmentary changes of the peripheral retina (white arrows).
(H, J, and L) Fundus autofluorescence images of USH2A-USH-1 (H), USH2A-USH-2 (J), and USH2A-USH-3 (L), showing macular atrophy occurring inside or outside of the perifoveal ring in both eyes (red arrows).
The USH2A-USH-1 (Figures 10G and 10H), USH2A-USH-2 (Figures 10I and 10J), and USH2A-USH-3 (Figures 10K and 10L) individuals were seen at the ages of 64, 42, and 48 years, respectively. All three showed signs of a more severe RP on color fundus images (Figures 10G, 10I, and 10K) than that observed in the non-syndromic RP individuals. However, in contrast to the RP individuals, all three USH individuals developed macular atrophy, as documented by fundus autofluorescence imaging (Figures 10H, 10J and 10L), which impacted visual acuity (Table S3). The atrophy was perifoveal as it began at the external border of the hyperautofluorescent ring (Figure 10J) and extended into the fovea in more advanced cases (Figures 10H and 10L). The appearance of the macular atrophy was well documented for USH2A-USH-2; it was not detected at the individual’s previous clinical examination at the age of 36 years (Figures S6A and S6B).
We then screened our clinical and genetic database to identify all individuals with biallelic pathogenic variants in USH2A in whom macular atrophy could be assessed on autofluorescence images of sufficient quality. Ninety people were retained, of whom 39 had non-syndromic RP (with the c.2276G>T, p.Cys759Phe pathogenic variant; mean visual acuity 20/30 and age 46 years), and 51 had USH type 2 (mean visual acuity 20/40 and age 42 years). In the case of the non-syndromic RP individuals, macular atrophy was detected in only 8 (20.5%; Figure S6C) and occurred relatively late, later than 55 years of age. By contrast, in the USH group, 20 individuals (39.2%) developed perifoveal macular atrophy. Furthermore, this atrophy appeared early, during the fourth decade, as noted for USH2A-USH-2.
Therefore, the clinical data suggest that cones are impacted more severely in USH individuals with USH2A variants than in non-syndromic RP individuals and support the differential cone phenotype observed in USH and RP human retinal organoids.
Discussion
We report here a human 3D retinal organoid-based, disease-modeling study of non-syndromic RP and USH because of variants in USH2A. Using three different human cellular models, we describe a clear genotype-phenotype correlation associated with USH2A mutations in each model, which is disease dependent. Furthermore, we identify a differential retinal disease phenotype in non-syndromic RP and USH models, which includes a previously unsuspected cone defect seen predominantly in USH lines that is consistent with the clinical data. These data provide invaluable information for elaborating a diagnosis, clarifying the pathogenicity of novel variants, and testing therapeutic efficiency.
Phenotypic variability in individuals presenting pathogenic variants in genes associated with USH has been extensively documented.17,48 In the case of USH2A, a genotype-phenotype correlation was suggested by the concept of retinal disease-specific USH2A alleles.15 Individuals presenting with at least one such allele, notably c.2276G>T, had preserved hearing, and thus these variants were proposed to be disease causing exclusively for non-syndromic RP. In the current study, we go one step further and demonstrate that the differential phenotype observed in non-syndromic RP or USH individuals with USH2A variants goes beyond differential hearing impairment because differences in ciliogenesis, mRNA expression, and molecular and structural retinal phenotypes were also observed, depending on the disease.
The first genotype-phenotype correlation from this study is the disease-dependent, and perhaps mutation-dependent, elongated primary cilium in affected fibroblasts. A similar observation has been reported in fibroblasts of two individuals with either early onset IRD or congenital blindness homozygous for a nonsense mutation in CEP290.49 The cilia of the individual with congenital blindness were significantly longer than those of the early-onset IRD counterpart, which were also longer than the control. These results are reminiscent of our findings; thus, we could speculate that there is a direct correlation between cilium length and disease severity associated to USH2A mutations. If confirmed in a larger number of control and affected samples, then this would suggest that the study of ciliogenesis in skin fibroblasts could be used as a rapid screening test by laboratories to determine the pathogenicity of newly identified USH2A variants.
The second clear genotype-phenotype correlation described in the current study is the confirmation of our previous observations concerning differential USH2A expression profiles in affected iPSCs.37 Globally, we confirm that missense RP-associated variants result in USH2A mRNA levels similar to control, whereas USH-associated variants that lead to premature termination codons result in significantly increased USH2A mRNA levels, even when they are localized outside of exon 13. These results further support our previous hypothesis that mutant USH2A mRNA transcripts housing premature stop codons are not degraded by the nonsense-mediated mRNA decay pathway in iPSCs. Importantly, these observations could be exploited to differentiate between non-syndromic RP- or USH-causing USH2A variants. It should be noted that this phenomenon is only observed in iPSCs and that the USH2A mRNA levels in retinal organoids do not follow this same pattern, suggesting activation of nonsense-mediated decay. Moreover, by using additional mutant iPSC lines, we show that not only the USH but also the RP lines carrying the c.2299delG allele have the highest USH2A expression levels of the USH and RP groups, respectively. Furthermore, within the non-syndromic RP group, we show that the iPSC lines carrying biallelic exon 13 variants have higher USH2A levels than the line carrying a mono-allelic exon 13 variant. This profile was mirrored in terms of ciliary length in disease-specific fibroblasts, making it tempting to speculate that exon 13 variants may exacerbate the corresponding phenotypes. This is consistent with a previous study showing that exon 13 missense mutations and deletions disrupt disulfide bonds and may affect local protein folding, thus having consequences for protein function.50
The third and most compelling genotype-phenotype correlation of this study is the differential retinal phenotype identified in the iPSC-derived organoids from USH and non-syndromic RP individuals. To date, there have been two reports of immature day 8651 or day 18052 retinal organoids from an RP individual presenting with a recurrent USH2A mutation in the Asian population (c.8559-2A>G). Even though the differentiation protocol used44 was slightly different than that in our study, the reported RP organoids presented a disorganized presumptive ONL, consistent with our observations. However, the lack of isogenic controls prevented validation of the phenotype, and no conclusions could be drawn concerning USH2A-associated disease phenotype in more mature photoreceptors. Our results suggest differential retinal phenotypes in organoids on day 225 that are not individual or mutation dependent but disease dependent and that, importantly, were validated by parallel differentiation of isogenic control lines. The RP organoids present a more disorganized phenotype in terms of IS/OS elongation and formation compared with USH or control organoids. In addition, aberrant formation of rod photoreceptors in RP organoids is much more pronounced than in USH organoids. By contrast, the cone phenotype is far more striking for USH organoids than RP organoids. This may be due to the differences in the structure of photoreceptor cilia in cones and rods. In cones, the axoneme of the CC extends the length of the OS, suggesting an important role in disc morphogenesis and turnover, whereas in rods, the microtubules of the axoneme end at a lower level of the OS.53 Furthermore, human cones also possess more calyceal processes than rod photoreceptor cells.20 As cellular protrusions of the IS, these calyceal processes contain USH proteins and presumably serve to stabilize the OS against mechanical forces.54 Therefore, we can speculate that absence of usherin, such as in the case of USH organoids, could have a more dramatic effect on cone morphology and function.
Several reports have underlined a difference in the severity of the RP phenotype between USH and non-syndromic RP individuals, with USH individuals being more severely affected.12,55,56 Similarly, previous observations in ush2a-mutant zebrafish lines suggested that progression and onset of retinal degeneration depend on the introduced mutation.21,22,23,24,25 However, there have not been any studies focusing specifically on cone differences. Our clinical data using short-wave fundus autofluorescence imaging demonstrate that there is perifoveal atrophy more frequently associated with USH individuals and at a younger age. This atrophy exacerbates the visual handicap by perifovealar damage and then impacts visual acuity by its progression toward the fovea. Similarly, Sengillo et al.57 previously noted that perifoveal atrophy was more frequently observed in USH as opposed to non-syndromic RP cases associated with USH2A variants. These authors further suggested that genotype-phenotype correlation of subjective hearing loss in USH2A individuals may be predictive of worse cone function and, thus, visual phenotype. Our retinal organoid data definitively confirm the cone involvement in USH individuals and furthermore suggest that this difference is not simply due to more rapid progression of the rod-cone phenotype but, rather, due to an autonomous cone defect.
The clinical differences observed between RP and USH individuals could indeed reflect the cone dissimilarities observed in the organoid models. The occurrence of perifoveal atrophy concerns a zone where cones and rods are present with an estimated ratio of 1:18.58 This zone is specifically affected by macular atrophy as a final stage of cone dystrophy and other genetic or multifactorial macular diseases. In IRDs specifically, an alteration in cone density in the area of the hyperautofluorescent perifoveal ring was demonstrated by adaptive optics on the cone mosaic of RP individuals59 and preferential perifoveal cone anomalies in early Stargardt’s disease.60 We have reported previously that our retinal organoid model presents a rod-cone ratio of 1:3, which is similar to that described for the parafoveal region of the human retina.28 The two USH individuals we modeled using retinal organoids showed the most advanced stages of perifoveal atrophy, reaching well into the fovea. This may be why we were able to detect a striking cone phenotype in the retinal organoids of these patients. It would now be interesting to generate iPSC-derived retinal organoids of younger USH individuals with less advanced or no signs of perifoveal atrophy to see whether a cone phenotype is still detectable.
Interestingly, and providing further support for our data, two animal models of USH2A exon 13 mutations were reported during preparation of this manuscript. The first was a rabbit model carrying a frameshift mutation in the exon equivalent to human exon 13.61 Similar to our data, this model showed decreased USH2A expression and absence of usherin in the retina. Furthermore, from 7 months of age, USH2A knockout rabbits showed progressive deterioration of rod and, notably, cone function by electroretinography (ERG) recordings, mimicking the reduced cone function in humans. Interestingly, these USH-related reduced cone ERGs are also consistent with observations in another large animal model for USH, the USH1C porcine model.62 The second was a humanized mouse model for the murine equivalent of the c.2299delG mutation.63 Consistent with our results, full-length usherin (as well as ADGRV1) was not detected at the base of the CC, although use of a FLAG tag suggested that the truncated protein accumulates in the IS. The knockin mice also showed decreased rod and cone ERG responses but only at late stages (>3 years of age). In contrast to our fibroblast and organoid data, this murine study suggested shortening of the CC of photoreceptors in the mutant mice. This discrepancy may stem from the well-described differences in the structural architecture between the human and murine ciliary region,20 further arguing for the pertinence of a human model. Regardless, an interesting observation in this animal model was that the CC changes occurred early, preceding photoreceptor degeneration and visual loss, which suggested a developmental defect of the CC structure. These data further validate the differences we observed on the human retinal organoids, which were studied on day 225 (32 weeks) and could thus be considered equivalent to the fetal period.64
There are some additional noteworthy aspects of this retinal organoid study. First, the phenotype of the organoids from USH2A-RP-3 homozygous for the c.2276G>T variant allows us to confirm the pathogenicity of this variant, which was recently determined in a zebrafish model.24 Second, we determined in a human retinal model that usherin is present at the tip of the ciliary rootlet. Third, we show that the mislocalization/absence of usherin in RP/USH organoids, respectively, also results in mislocalization/absence of its partner ADGRV1, thus suggesting impaired formation and function of the USH interactome in human cells. The ciliogenesis defects in the affected fibroblasts therefore raise the question of whether usherin may have roles in other ciliated tissues in addition to the retina or the ear.65 This is in line with identification of at least two protein isoforms for USH2A:11 a long transmembrane isoform that is predominant in the retina19 and a short secreted N-terminal isoform that is more widely expressed.11 Further studies concerning actin cytoskeleton assembly are needed to better understand the USH2A-related ciliogenesis defects.
Last, the effect of CRISPR-Cas-mediated correction on the phenotype of the affected organoids also yielded some interesting observations. Correction of the c.2276G>T variant in the RP organoids markedly improved OS formation and/or elongation but did not fully restore a control phenotype, particularly for rods. This is likely due to the presence of the c.2299delG variant on the non-corrected allele in the isogenic organoids, suggesting that carriers of loss-of-function USH2A variants may harbor subclinical retinal alterations. Interestingly, the cone phenotype was restored to control after gene correction in the RP and USH organoids, further validating the unexpected phenotype as being directly due to USH2A variants. Similarly, usherin and ADGRV1 localization were correctly restored in RP and USH organoids. Taken together, these data additionally provide a proof of concept that, in photoreceptors, this USH2A genome-editing strategy is a viable therapeutic option.
In conclusion, collectively, our data provide long-awaited evidence of a genotype-phenotype correlation associated with USH2A variants in a sophisticated human retinal model. Furthermore, they provide robust readouts in multiple cellular models for testing the pathogenicity of variants of uncertain significance as well as the efficacy of therapeutic approaches.
Acknowledgments
The authors thank K. Damodar, G. Dubois, D. Mamaeva, and H. Boukhaddaoui for technical support and helpful comments, B. Bocquet for medical data, C. Vaché and L. Mansard for informative discussions and/or critical reading of the manuscript, and C. Cazevieille for electron microscopy expertise (INM CoMET facility). The authors also thank the MRI (Biocampus) and the qPHD (University of Montpellier) platforms. The authors are grateful to the individuals who participated in the study. This work was funded by Aviesan-Unadev (N° AAK200003FSA; to V.K.), Vaincre Usher 2 (to V.K.), Fondation de France (N° 00112525; to V.K.; post-doctoral grant for C.S.-S.), ANR-EJP RD (GET-READY) (N° ANR-20-RAR4-0006-04; to V.K.; post-doctoral grant for L.C.), Deutsche Forschungsgemeinschaft (SPP2127, Gene and cell-based therapies to counteract neuroretinal degeneration) (N°s 399443882 (to K.N.-W.) and 399487434 (to U.W.)), Gelderse Blinden Stichting (to E.V.W.), Stichting Ushersyndroom (to E.V.W.), and Foundation Fighting Blindness (N° PPA-0517-0717-RAD; to E.V.W.).
Author contributions
C.S.-S. and V.K. designed the study. C.S.-S., C.J.-M., N.E., and L.C. performed experiments. A.L. and I.M. performed clinical investigations. K.N.-W., U.W., and E.V.W. provided reagents and discussed data. A.-F.R. performed genetic investigations. V.K. provided financial support. C.S.-S. and V.K. analyzed the data and wrote the manuscript. All authors edited the manuscript.
Declaration of interests
C.S.-S. and V.K. are the inventors of a published patent on genome editing for USH2A (WO/2020/221832).
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2023.100229.
Web resources
OMIM: http://www.omim.org
RetNet: https://web.sph.uth.edu/RetNet/
LOVD: https://databases.lovd.nl/shared/genes/USH2A
ImageJ: https://imagej.nih.gov/ij/
Supplemental information
Data and code availability
All data are available in the main text or the supplemental materials.
References
- 1.Pagon R.A. Retinitis pigmentosa. Surv. Ophthalmol. 1988;33:137–177. doi: 10.1016/0039-6257(88)90085-9. [DOI] [PubMed] [Google Scholar]
- 2.Wright A.F., Chakarova C.F., Abd El-Aziz M.M., Bhattacharya S.S. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat. Rev. Genet. 2010;11:273–284. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- 3.Verbakel S.K., van Huet R.A., Boon C.J., den Hollander A.I., Collin R.W., Klaver C.C., Hoyng C.B., Roepman R., Klevering B.J. Non-syndromic retinitis pigmentosa. Prog. Retin. Eye Res. 2018;66:157–186. doi: 10.1016/j.preteyeres.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Hamel C. Retinitis pigmentosa. Orphanet J. Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimberling W.J., Hildebrand M.S., Shearer A.E., Jensen M.L., Halder J.A., Trzupek K., Cohn E.S., Weleber R.G., Stone E.M., Smith R.J.H. Frequency of Usher syndrome in two pediatric populations: implications for genetic screening of deaf and hard of hearing children. Genet. Med. 2010;12:512–516. doi: 10.1097/GIM.0b013e3181e5afb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan D., Liu X.Z. Genetics and pathological mechanisms of Usher syndrome. J. Hum. Genet. 2010;55:327–335. doi: 10.1038/jhg.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenberger T., Neuhaus C., Khan A.O., Decker C., Preising M.N., Friedburg C., Bieg A., Gliem M., Charbel Issa P., Holz F.G., et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PLoS One. 2013;8:e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge Z., Bowles K., Goetz K., Scholl H.P.N., Wang F., Wang X., Xu S., Wang K., Wang H., Chen R. NGS-based Molecular diagnosis of 105 eyeGENE((R)) probands with Retinitis Pigmentosa. Sci. Rep. 2015;5:18287. doi: 10.1038/srep18287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baux D., Blanchet C., Hamel C., Meunier I., Larrieu L., Faugère V., Vaché C., Castorina P., Puech B., Bonneau D., et al. Enrichment of LOVD-USHbases with 152 USH2A genotypes defines an extensive mutational spectrum and highlights missense hotspots. Hum. Mutat. 2014;35:1179–1186. doi: 10.1002/humu.22608. [DOI] [PubMed] [Google Scholar]
- 10.Pennings R.J.E., Te Brinke H., Weston M.D., Claassen A., Orten D.J., Weekamp H., Van Aarem A., Huygen P.L.M., Deutman A.F., Hoefsloot L.H., et al. USH2A mutation analysis in 70 Dutch families with Usher syndrome type II. Hum. Mutat. 2004;24:185. doi: 10.1002/humu.9259. [DOI] [PubMed] [Google Scholar]
- 11.van Wijk E., Pennings R.J.E., te Brinke H., Claassen A., Yntema H.G., Hoefsloot L.H., Cremers F.P.M., Cremers C.W.R.J., Kremer H. Identification of 51 novel exons of the Usher syndrome type 2A (USH2A) gene that encode multiple conserved functional domains and that are mutated in patients with Usher syndrome type II. Am. J. Hum. Genet. 2004;74:738–744. doi: 10.1086/383096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierrache L.H.M., Hartel B.P., van Wijk E., Meester-Smoor M.A., Cremers F.P.M., de Baere E., de Zaeytijd J., van Schooneveld M.J., Cremers C.W.R.J., Dagnelie G., et al. Visual prognosis in USH2A-associated retinitis pigmentosa is worse for patients with Usher syndrome type IIa than for those with nonsyndromic retinitis pigmentosa. Ophthalmology. 2016;123:1151–1160. doi: 10.1016/j.ophtha.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Slijkerman R.W., Vaché C., Dona M., García-García G., Claustres M., Hetterschijt L., Peters T.A., Hartel B.P., Pennings R.J., Millan J.M., et al. Antisense oligonucleotide-based splice correction for USH2A-associated retinal degeneration caused by a frequent deep-intronic mutation. Mol. Ther. Nucleic Acids. 2016;5:e381. doi: 10.1038/mtna.2016.89. [DOI] [PubMed] [Google Scholar]
- 14.Mansard L., Baux D., Vaché C., Blanchet C., Meunier I., Willems M., Faugère V., Baudoin C., Moclyn M., Bianchi J., et al. The study of a 231 french patient cohort significantly extends the mutational spectrum of the two major usher genes MYO7A and USH2A. Int. J. Mol. Sci. 2021;22:13294. doi: 10.3390/ijms222413294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenassi E., Vincent A., Li Z., Saihan Z., Coffey A.J., Steele-Stallard H.B., Moore A.T., Steel K.P., Luxon L.M., Héon E., et al. A detailed clinical and molecular survey of subjects with nonsyndromic USH2A retinopathy reveals an allelic hierarchy of disease-causing variants. Eur. J. Hum. Genet. 2015;23:1318–1327. doi: 10.1038/ejhg.2014.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delmaghani S., El-Amraoui A. The genetic and phenotypic landscapes of Usher syndrome: from disease mechanisms to a new classification. Hum. Genet. 2022;141:709–735. doi: 10.1007/s00439-022-02448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathur P., Yang J. Usher syndrome: Hearing loss, retinal degeneration and associated abnormalities. Biochim. Biophys. Acta. 2015;1852:406–420. doi: 10.1016/j.bbadis.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams D.S. Usher syndrome: animal models, retinal function of Usher proteins, and prospects for gene therapy. Vis. Res. 2008;48:433–441. doi: 10.1016/j.visres.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X., Bulgakov O.V., Darrow K.N., Pawlyk B., Adamian M., Liberman M.C., Li T. Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc. Natl. Acad. Sci. USA. 2007;104:4413–4418. doi: 10.1073/pnas.0610950104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahly I., Dufour E., Schietroma C., Michel V., Bahloul A., Perfettini I., Pepermans E., Estivalet A., Carette D., Aghaie A., et al. Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice. J. Cell Biol. 2012;199:381–399. doi: 10.1083/jcb.201202012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dona M., Slijkerman R., Lerner K., Broekman S., Wegner J., Howat T., Peters T., Hetterschijt L., Boon N., de Vrieze E., et al. Usherin defects lead to early-onset retinal dysfunction in zebrafish. Exp. Eye Res. 2018;173:148–159. doi: 10.1016/j.exer.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulla K., Slijkerman R., van Diepen H.C., Albert S., Dona M., Beumer W., Turunen J.J., Chan H.L., Schulkens I.A., Vorthoren L., et al. Antisense oligonucleotide-based treatment of retinitis pigmentosa caused by USH2A exon 13 mutations. Mol. Ther. 2021;29:2441–2455. doi: 10.1016/j.ymthe.2021.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han S., Liu X., Xie S., Gao M., Liu F., Yu S., Sun P., Wang C., Archacki S., Lu Z., et al. Knockout of ush2a gene in zebrafish causes hearing impairment and late onset rod-cone dystrophy. Hum. Genet. 2018;137:779–794. doi: 10.1007/s00439-018-1936-6. [DOI] [PubMed] [Google Scholar]
- 24.Reurink J., de Vrieze E., Li C.H.Z., van Berkel E., Broekman S., Aben M., Peters T., Oostrik J., Neveling K., Venselaar H., et al. Scrutinizing pathogenicity of the USH2A c.2276 G > T; p.(Cys759Phe) variant. NPJ Genom. Med. 2022;7:37. doi: 10.1038/s41525-022-00306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toms M., Dubis A.M., de Vrieze E., Tracey-White D., Mitsios A., Hayes M., Broekman S., Baxendale S., Utoomprurkporn N., Bamiou D., et al. Clinical and preclinical therapeutic outcome metrics for USH2A-related disease. Hum. Mol. Genet. 2020;29:1882–1899. doi: 10.1093/hmg/ddaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afanasyeva T.A.V., Corral-Serrano J.C., Garanto A., Roepman R., Cheetham M.E., Collin R.W.J. A look into retinal organoids: methods, analytical techniques, and applications. Cell. Mol. Life Sci. 2021;78:6505–6532. doi: 10.1007/s00018-021-03917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruczek K., Swaroop A. Pluripotent stem cell-derived retinal organoids for disease modeling and development of therapies. Stem Cell. 2020;38:1206–1215. doi: 10.1002/stem.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjurjo-Soriano C., Erkilic N., Damodar K., Boukhaddaoui H., Diakatou M., Garita-Hernandez M., Mamaeva D., Dubois G., Jazouli Z., Jimenez-Medina C., et al. Retinoic acid delays initial photoreceptor differentiation and results in a highly structured mature retinal organoid. Stem Cell Res. Ther. 2022;13:478. doi: 10.1186/s13287-022-03146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chirco K.R., Chew S., Moore A.T., Duncan J.L., Lamba D.A. Allele-specific gene editing to rescue dominant CRX-associated LCA7 phenotypes in a retinal organoid model. Stem Cell Rep. 2021;16:2690–2702. doi: 10.1016/j.stemcr.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng W.L., Gao M.L., Lei X.L., Lv J.N., Zhao H., He K.W., Xia X.X., Li L.Y., Chen Y.C., Li Y.P., et al. Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Rep. 2018;10:1267–1281. doi: 10.1016/j.stemcr.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruczek K., Qu Z., Gentry J., Fadl B.R., Gieser L., Hiriyanna S., Batz Z., Samant M., Samanta A., Chu C.J., et al. Gene therapy of dominant CRX-Leber congenital amaurosis using patient stem cell-derived retinal organoids. Stem Cell Rep. 2021;16:252–263. doi: 10.1016/j.stemcr.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane A., Jovanovic K., Shortall C., Ottaviani D., Panes A.B., Schwarz N., Guarascio R., Hayes M.J., Palfi A., Chadderton N., et al. Modeling and rescue of RP2 retinitis pigmentosa using iPSC-derived retinal organoids. Stem Cell Rep. 2020;15:67–79. doi: 10.1016/j.stemcr.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung A., Sacristan-Reviriego A., Perdigão P.R.L., Sai H., Georgiou M., Kalitzeos A., Carr A.J.F., Coffey P.J., Michaelides M., Bainbridge J., et al. Investigation of PTC124-mediated translational readthrough in a retinal organoid model of AIPL1-associated Leber congenital amaurosis. Stem Cell Rep. 2022;17:2187–2202. doi: 10.1016/j.stemcr.2022.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parfitt D.A., Lane A., Ramsden C.M., Carr A.J.F., Munro P.M., Jovanovic K., Schwarz N., Kanuga N., Muthiah M.N., Hull S., et al. Identification and correction of mechanisms underlying inherited blindness in human iPSC-derived optic cups. Cell Stem Cell. 2016;18:769–781. doi: 10.1016/j.stem.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjurjo-Soriano C., Erkilic N., Manes G., Dubois G., Hamel C.P., Meunier I., Kalatzis V. Generation of an iPSC line, INMi001-A, carrying the two most common USH2A mutations from a compound heterozygote with non-syndromic retinitis pigmentosa. Stem Cell Res. 2018;33:228–232. doi: 10.1016/j.scr.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Sanjurjo-Soriano C., Erkilic N., Manes G., Dubois G., Hamel C.P., Meunier I., Kalatzis V. Generation of a human iPSC line, INMi002-A, carrying the most prevalent USH2A variant associated with Usher syndrome type 2. Stem Cell Res. 2018;33:247–250. doi: 10.1016/j.scr.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Sanjurjo-Soriano C., Erkilic N., Baux D., Mamaeva D., Hamel C.P., Meunier I., Roux A.F., Kalatzis V. Genome editing in patient iPSCs corrects the most prevalent USH2A mutations and reveals intriguing mutant mRNA expression profiles. Mol. Ther. Methods Clin. Dev. 2020;17:156–173. doi: 10.1016/j.omtm.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cereso N., Pequignot M.O., Robert L., Becker F., De Luca V., Nabholz N., Rigau V., De Vos J., Hamel C.P., Kalatzis V. Proof of concept for AAV2/5-mediated gene therapy in iPSc-derived retinal pigment epithelium of choroideremia patients. Mol. Ther. Methods Clin. Dev. 2014;1:14011. doi: 10.1038/mtm.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torriano S., Erkilic N., Faugère V., Damodar K., Hamel C.P., Roux A.F., Kalatzis V. Pathogenicity of a novel missense variant associated with choroideremia and its impact on gene replacement therapy. Hum. Mol. Genet. 2017;26:3573–3584. doi: 10.1093/hmg/ddx244. [DOI] [PubMed] [Google Scholar]
- 40.Sanjurjo-Soriano C., Erkilic N., Vache C., Dubois G., Roux A.F., Meunier I., Kalatzis V. Generation of a human iPSC line, INMi005-A, from a patient with non-syndromic USH2A-associated retinitis pigmentosa. Stem Cell Res. 2022;60:102738. doi: 10.1016/j.scr.2022.102738. [DOI] [PubMed] [Google Scholar]
- 41.Kusuluri D.K., Güler B.E., Knapp B., Horn N., Boldt K., Ueffing M., Aust G., Wolfrum U. Adhesion G protein-coupled receptor VLGR1/ADGRV1 regulates cell spreading and migration by mechanosensing at focal adhesions. iScience. 2021;24:102283. doi: 10.1016/j.isci.2021.102283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiners J., Nagel-Wolfrum K., Jürgens K., Märker T., Wolfrum U. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp. Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G., et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capowski E.E., Samimi K., Mayerl S.J., Phillips M.J., Pinilla I., Howden S.E., Saha J., Jansen A.D., Edwards K.L., Jager L.D., et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development. 2019;146:dev171686. doi: 10.1242/dev.171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowan C.S., Renner M., De Gennaro M., Gross-Scherf B., Goldblum D., Hou Y., Munz M., Rodrigues T.M., Krol J., Szikra T., et al. Cell types of the human retina and its organoids at single-cell resolution. Cell. 2020;182:1623–1640.e34. doi: 10.1016/j.cell.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cooke J.A., Voigt A.P., Collingwood M.A., Stone N.E., Whitmore S.S., DeLuca A.P., Burnight E.R., Anfinson K.R., Vakulskas C.A., Reutzel A.J., et al. Propensity of patient-derived iPSCs for retinal differentiation: implications for autologous cell replacement. Stem Cells Transl. Med. 2023;12:365–378. doi: 10.1093/stcltm/szad028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J., Liu X., Yue G., Adamian M., Bulgakov O., Li T. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J. Cell Biol. 2002;159:431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riazuddin S., Nazli S., Ahmed Z.M., Yang Y., Zulfiqar F., Shaikh R.S., Zafar A.U., Khan S.N., Sabar F., Javid F.T., et al. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum. Mutat. 2008;29:502–511. doi: 10.1002/humu.20677. [DOI] [PubMed] [Google Scholar]
- 49.Barny I., Perrault I., Michel C., Goudin N., Defoort-Dhellemmes S., Ghazi I., Kaplan J., Rozet J.M., Gerard X. AON-mediated exon skipping to bypass protein truncation in retinal dystrophies due to the recurrent CEP290 c.4723A > T mutation. Fact or Fiction? Genes. 2019;10:368. doi: 10.3390/genes10050368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu D., Zou J., Chen Q., Zhu T., Sui R., Yang J. Structural modeling, mutation analysis, and in vitro expression of usherin, a major protein in inherited retinal degeneration and hearing loss. Comput. Struct. Biotechnol. J. 2020;18:1363–1382. doi: 10.1016/j.csbj.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo Y., Wang P., Ma J.H., Cui Z., Yu Q., Liu S., Xue Y., Zhu D., Cao J., Li Z., et al. Modeling retinitis pigmentosa: retinal organoids generated from the iPSCs of a patient With the USH2A mutation show early developmental abnormalities. Front. Cell. Neurosci. 2019;13:361. doi: 10.3389/fncel.2019.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su T., Liang L., Zhang L., Wang J., Chen L., Su C., Cao J., Yu Q., Deng S., Chan H.F., et al. Retinal organoids and microfluidic chip-based approaches to explore the retinitis pigmentosa with USH2A mutations. Front. Bioeng. Biotechnol. 2022;10:939774. doi: 10.3389/fbioe.2022.939774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mustafi D., Engel A.H., Palczewski K. Structure of cone photoreceptors. Prog. Retin. Eye Res. 2009;28:289–302. doi: 10.1016/j.preteyeres.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.May-Simera H., Nagel-Wolfrum K., Wolfrum U. Cilia - The sensory antennae in the eye. Prog. Retin. Eye Res. 2017;60:144–180. doi: 10.1016/j.preteyeres.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Colombo L., Maltese P.E., Romano D., Fogagnolo P., Castori M., Marceddu G., Cristofoli F., Percio M., Piteková B., Modarelli A.M., et al. Spectral-domain optical coherence tomography analysis in syndromic and nonsyndromic forms of retinitis pigmentosa due to USH2A genetic variants. Ophthalmic Res. 2022;65:180–195. doi: 10.1159/000520329. [DOI] [PubMed] [Google Scholar]
- 56.Hufnagel R.B., Liang W., Duncan J.L., Brewer C.C., Audo I., Ayala A.R., Branham K., Cheetham J.K., Daiger S.P., Durham T.A., et al. Tissue-specific genotype-phenotype correlations among USH2A-related disorders in the RUSH2A study. Hum. Mutat. 2022;43:613–624. doi: 10.1002/humu.24365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sengillo J.D., Cabral T., Schuerch K., Duong J., Lee W., Boudreault K., Xu Y., Justus S., Sparrow J.R., Mahajan V.B., et al. Electroretinography Reveals Difference in Cone Function between Syndromic and Nonsyndromic USH2A Patients. Sci. Rep. 2017;7:11170. doi: 10.1038/s41598-017-11679-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wells-Gray E.M., Choi S.S., Bries A., Doble N. Variation in rod and cone density from the fovea to the mid-periphery in healthy human retinas using adaptive optics scanning laser ophthalmoscopy. Eye (Lond) 2016;30:1135–1143. doi: 10.1038/eye.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tojo N., Nakamura T., Fuchizawa C., Oiwake T., Hayashi A. Adaptive optics fundus images of cone photoreceptors in the macula of patients with retinitis pigmentosa. Clin. Ophthalmol. 2013;7:203–210. doi: 10.2147/OPTH.S39879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khan K.N., Kasilian M., Mahroo O.A.R., Tanna P., Kalitzeos A., Robson A.G., Tsunoda K., Iwata T., Moore A.T., Fujinami K., et al. Early patterns of macular degeneration in ABCA4-associated retinopathy. Ophthalmology. 2018;125:735–746. doi: 10.1016/j.ophtha.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen V.P., Song J., Prieskorn D., Zou J., Li Y., Dolan D., Xu J., Zhang J., Jayasundera K.T., Yang J., et al. USH2A gene mutations in rabbits lead to progressive retinal degeneration and hearing loss. Transl. Vis. Sci. Technol. 2023;12:26. doi: 10.1167/tvst.12.2.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grotz S., Schäfer J., Wunderlich K.A., Ellederova Z., Auch H., Bähr A., Runa-Vochozkova P., Fadl J., Arnold V., Ardan T., et al. Early disruption of photoreceptor cell architecture and loss of vision in a humanized pig model of usher syndromes. EMBO Mol. Med. 2022;14:e14817. doi: 10.15252/emmm.202114817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tebbe L., Mwoyosvi M.L., Crane R., Makia M.S., Kakakhel M., Cosgrove D., Al-Ubaidi M.R., Naash M.I. The usherin mutation c.2299delG leads to its mislocalization and disrupts interactions with whirlin and VLGR1. Nat. Commun. 2023;14:972. doi: 10.1038/s41467-023-36431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leong Y.C., Di Foggia V., Pramod H., Bitner-Glindzicz M., Patel A., Sowden J.C. Molecular pathology of Usher 1B patient-derived retinal organoids at single cell resolution. Stem Cell Rep. 2022;17:2421–2437. doi: 10.1016/j.stemcr.2022.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pearsall N., Bhattacharya G., Wisecarver J., Adams J., Cosgrove D., Kimberling W. Usherin expression is highly conserved in mouse and human tissues. Hear. Res. 2002;174:55–63. doi: 10.1016/s0378-5955(02)00635-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplemental materials.